Abstract
A 5′ nuclease TaqMan PCR assay was developed for the quantitative detection of the major cariogenic bacteria Streptococcus mutans and Streptococcus sobrinus. The absolute and relative numbers of bacteria were measured by this method. This assay will be useful for quantifying these organisms in oral specimens and for analyzing biofilm formation.
Dental caries is one of the most common infectious diseases afflicting humans (10). Although 200 to 300 bacterial species have been found associated with dental plaque, only Streptococcus mutans (serotype c, e, and f mutans streptococci) and Streptococcus sobrinus (serotype d and g mutans streptococci) have been consistently linked with the formation of human dental caries (10-12). Additionally, S. mutans and S. sobrinus are occasionally associated with nonoral infections, principally subacute bacterial endocarditis (7, 14, 19). Consequently, various methods have been developed to identify S. mutans and/or S. sobrinus (2, 3, 9). Several of these methods are PCR-based bacterial detection systems (15, 16). Most of the PCR-based diagnosis systems reported are qualitative analyses and are therefore unsuitable for accurate evaluation of caries susceptibility or caries activity. Quantitative analysis is essential for monitoring the cell number and/or ratio of cariogenic bacteria in oral specimens, such as dental plaque and saliva. Furthermore, monitoring the number of cariogenic bacteria in oral biofilm is required from the perspective of biofilm research.
A real-time PCR assay with the TaqMan system based on the 5′-3′ exonuclease activity of Taq polymerase has been developed for the quantitative detection of DNA copy number (8). Briefly, an oligonucleotide probe with a reporter fluorescent dye attached to its 5′ end and a quencher dye attached to its 3′ end is designed to hybridize to the target gene. During PCR amplification, the quencher dye of the probe is cleaved by the 5′ nuclease activity of Taq polymerase, resulting in the accumulation of reporter fluorescence. The release of the fluorescent dye during amplification allows for the rapid detection and quantification of DNA (6).
This report describes a method for the absolute and relative quantification of human cariogenic bacteria, including S. mutans and S. sobrinus, from oral specimens by using a TaqMan PCR assay. In spite of the importance of these organisms as cariogenic dental pathogens, quantitative detection of S. mutans and S. sobrinus by TaqMan PCR has not been reported. This is the first investigation of quantitative detection of S. mutans and S. sobrinus by using a TaqMan assay.
The bacterial strains used in this study are listed in Table 1. The S. mutans and S. sobrinus strains were cultured as described previously (15). Genomic DNA was isolated and purified using a Puregene DNA isolation kit (Gentra Systems, Minneapolis, Minn.) in accordance with the manufacturer's instructions for gram-positive bacteria. Human saliva was prepared as described previously (15). Briefly, 500 μl of stimulated whole saliva and the same amount of phosphate-buffered saline (0.12 M NaCl, 0.01 M Na2HPO4, 5 mM KH2PO4 [pH 7.5]) were mixed and centrifuged at 12,000 × g for 10 min; 500 μl of cell lysis buffer (1.0% Triton X-100, 20 mM Tris-HCl, 2 mM EDTA [pH 8.0]) (18) was added to the precipitate, which was then incubated with 20 U of mutanolysin/ml and 0.2 mg of lysozyme/ml at 37°C for 2 h. The precipitate was vortexed, and the chromosomal DNA from the bacteria was extracted by boiling the precipitate at 100°C for 10 min. Plaque samples were collected from the buccal side of the upper first molar. One milligram (wet weight) of plaque was washed with phosphate-buffered saline two times. The precipitate was suspended in 100 μl of cell lysis solution and incubated with 20 U of mutanolysin/ml and 0.2 mg of lysozyme/ml at 37°C for 2 h. The lysate was boiled at 100°C for 10 min, and the chromosomal DNA was extracted.
TABLE 1.
Strains and amplification results
| Strain | Source or distribution | Amplification with primer:
|
||
|---|---|---|---|---|
| S. mutans | S. sobrinus | Universal | ||
| Oral streptococci | ||||
| Mutans group (serotype) | ||||
| Streptococcus mutans | Human | |||
| GS-5 (c) | + | − | + | |
| MT8148 (c) | + | − | + | |
| Xc (c) | + | − | + | |
| MT703R (e) | + | − | + | |
| OMZ175 (f) | + | − | + | |
| Streptococcus sobrinus | Human | |||
| MT8145 (d) | − | + | + | |
| OMZ176 (d) | − | + | + | |
| 6715 (g) | − | + | + | |
| OU8 (g) | − | + | + | |
| Streptococcus downei | Monkey | |||
| Mfe28 (h) | − | − | + | |
| S28 (h) | − | − | + | |
| Streptococcus ratti | Rat | |||
| BHT (b) | − | − | + | |
| FA1 (b) | − | − | + | |
| Streptococcus cricetus | Hamster | |||
| E49 (a) | − | − | + | |
| HS1 (a) | − | − | + | |
| Mitis-sanguinis group | ||||
| Streptococcus mitis 903 | Human | − | − | + |
| Streptococcus sanguinis ATCC 10556 | Human | − | − | + |
| Streptococcus gordonii DL1 | Human | − | − | + |
| Streptococcus oralis ATCC 10557 | Human | − | − | + |
| Salivarius group | ||||
| Streptococcus salivarius HT9R | Human | − | − | + |
| Anginosus group | ||||
| Streptococcus anginosus FW73 | Human | − | − | + |
| Other bacteria | ||||
| Porphyromonas gingivalis ATCC 33277 | Human | − | − | + |
| Actinibacillus actinomyce- temcomitans Y4 | Human | − | − | + |
| Treponema denticola | Human | |||
| ATCC 35404 | − | − | + | |
| ATCC 35405 | − | − | + | |
| Bacteroides forsythus ATCC 43037 | Human | − | − | + |
| Fusobacterium nucleatum ATCC 10953 | Human | − | − | + |
| Prevotella intermedia ATCC 25611 | Human | − | − | + |
| Haemophilus aphrophilus NCTC 5908 | Human | − | − | + |
| Eikenella corrodens 1085 | Human | − | − | + |
| Escherichia coli DH5α | GIBCO BRL | − | − | + |
Oligonucleotide primers and probes, designed using Primer Express 1.5 software (Applied Biosystems, Foster City, Calif.), are listed in Table 2. The universal primers and a probe for a broad range of bacteria were designed as previously described (4, 21). The S. mutans- and S. sobrinus-specific primers and probes were designed from the gtfB (17) and gtfT (5) genes, respectively. The specificities of the primers and probes were initially confirmed by BLAST with the National Center for Biotechnology Information server (http://www.ncbi.nlm.nih.gov/) and then confirmed by conventional PCR (Table 1) and dot blot analysis with digoxigenin-labeled probes (data not shown), respectively. Several strains of S. mutans and S. sobrinus of all serotypes served as positive controls, and the other bacteria served as negative controls for the bacterium-specific primers and probes (Table 1). Conventional PCR assays used to confirm the specificity and universality of the primers were performed as follows: 94°C for 5 min, followed by 25 cycles of 94°C for 15 s, 55°C for 30 s, and 72°C for 1 min.
TABLE 2.
Oligonucleotide primers and probes
| Designation | Sequencea | Amplicon size (bp) | Target | Source or reference |
|---|---|---|---|---|
| Primers | ||||
| Smut3368-F | 5′-GCCTACAGCTCAGAGATGCTATTCT-3′ | 114 | gtfB | UA159 |
| Smut3481-R | 5′-GCCATACACCACTCATGAATTGA-3′ | |||
| Ssob287-F | 5′-TTCAAAGCCAAGACCAAGCTAGT-3′ | 88 | gtfT | OMZ176 |
| Ssob374-R | 5′-CCAGCCTGAGATTCAGCTTGT-3′ | |||
| Uni152-F | 5′-CGCTAGTAATCGTGGATCAGAATG-3′ | 69 | 16S rRNA | 21 |
| Uni220-R | 5′-TGTGACGGGCGGTGTGTA-3′ | |||
| Fluorescent probes | ||||
| Smut3423T | 5′-FAM-TGGAAATGACGGTCGCCGTTATGAA-TAMRA-3′ | gtfB | UA159 | |
| Ssob298T | 5′-FAM-CCTGCTCCAGCGACAAAGGCAGC-TAMRA-3′ | gtfT | OMZ176 | |
| Uni177T | 5′-FAM-CACGGTGAATACGTTCCCGGGC-TAMRA-3′ | 16S rRNA | 21 |
For each real-time PCR, 20 μl of a mixture containing 1 μl of lysed cells, 1× TaqMan Universal PCR Master Mix (Applied Biosystems), 200 nM (each) sense and antisense primer, and 250 nM TaqMan probe was placed in each well of a 96-well plate. Amplification and detection were performed using the ABI PRISM 7700 sequence detection system (Applied Biosystems) with the following cycle profile: 50°C for 2 min, 95°C for 10 min, and 60 cycles of 95°C for 15 s and 58°C for 1 min. The critical threshold cycle (Ct) is defined as the cycle at which the fluorescence becomes detectable above background and is inversely proportional to the logarithm of the initial number of template molecules. The standard curves for each organism were plotted for each primer-probe set by using Ct values obtained from the amplification of genomic DNA extracted from samples containing 1.7 × 100 to 1.7 × 109 CFU of S. mutans and 1.1 × 100 to 1.1 × 109 CFU of S. sobrinus. The numbers of CFU were determined by plating culture dilutions on tryptic soy agar (Difco Laboratories, Grand Island, N.Y.) plates.
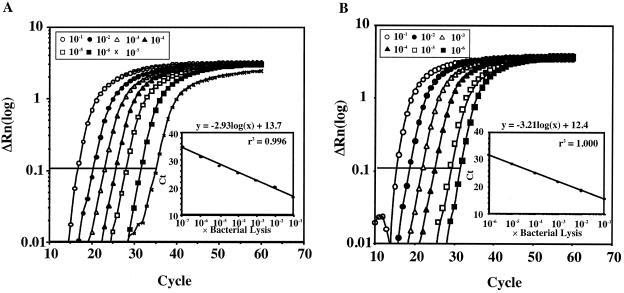
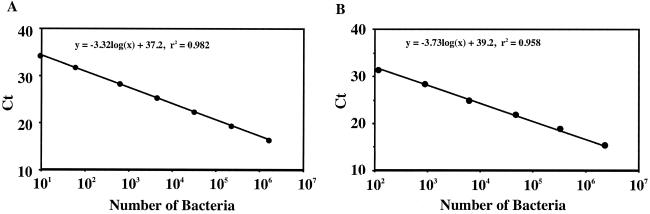
To determine the linearity and detection limit of the assay, solutions of lysed S. mutans and S. sobrinus were amplified in successive 10-fold dilutions in a series of real-time PCRs (Fig. 1). Based on this approach, correlations were observed between the Ct and the CFU (Fig. 2). Detection and quantification were linear over a range from 1.7 × 101 to 1.7 × 107 CFU per reaction mixture for S. mutans and 1.1 × 101 to 1.1 × 106 CFU per reaction mixture for S. sobrinus. The presence of PCR inhibitors in dental plaque was assessed by the fluorescence levels for serial dilutions of lysed S. mutans and S. sobrinus. Lysates spiked with approximately 10 μg (wet weight) of S. mutans- and S. sobrinus-negative dental plaque in each mixture showed no inhibition (data not shown).
FIG. 1.
Amplification plots of chromosomal DNA from lysed cells. Serial dilutions of chromosomal DNA were from S. mutans (A) or S. sobrinus (B). The log-transformed relative fluorescence [ΔRn(log)] was monitored as the increase in reporter dye intensity relative to the passive internal reference dye. The threshold fluorescence, or level at which the threshold cycle was determined, is shown. The standard curves were generated from the amplification plots in the small panels (correlation coefficient = 0.996 for S. mutans and 1.00 for S. sobrinus). Ct is the cycle number at which the threshold fluorescence was reached.
FIG. 2.
Standard curves generated by known numbers of S. mutans (A) and S. sobrinus (B); linearity is shown for these organisms. The correlation coefficients are 0.982 for S. mutans and 0.958 for S. sobrinus. Ct is the cycle number at which the threshold fluorescence was reached.
Using this assay, we determined the numbers of S. mutans and S. sobrinus bacteria in saliva and dental plaque from 10 individuals (Table 3). The numbers of these organisms in each saliva sample varied by several orders of magnitude, and our results for saliva samples are consistent with a previous report (20). In addition, the relative amounts of these organisms in oral specimens were calculated by the comparative Ct (ΔΔCt) method (21), with a simplification. Briefly, the results, expressed as the fold difference (N) between the number of target gene copies and the number of 16S rRNA gene copies, were determined as follows: N = 2ΔCt = 2(Ct target − Ct 16S rRNA). From a clinical perspective, it is often necessary to analyze the percentage of specific bacteria in a region. Lyons et al. (13) pointed out the importance of the relative quantification of bacteria in clinical specimens. The percentages of these organisms in saliva ranged from 0.0016 to 5.57% for S. mutans and from 0 to 0.19% for S. sobrinus. Note that the percentages of S. mutans and S. sobrinus normalized to the copy number of the 16S rRNA gene may be underestimated, because the copy number of the 16S rRNA gene is higher than that of gtfB or gtfT. Furthermore, we examined the absolute and relative numbers of these organisms in plaque samples on tooth surfaces (Table 3). This is the first report to quantify cariogenic bacteria from dental plaque samples by using a real-time PCR assay. The cell numbers ranged from 0 to 4.82 × 106 per mg (wet weight) of plaque for S. mutans and from 0 to 4.76 × 106 per mg (wet weight) for S. sobrinus. The percentages of these organisms in the plaque ranged from 0 to 7.16% (S. mutans) and from 0 to 6.95% (S. sobrinus). The range of cell numbers and percentages of these bacteria in dental plaque are similar to previous results with cell culture methods (1).
TABLE 3.
Number of S. mutans and S. sobrinus cells detected in oral specimensa
| Specimen type and patient no. |
S. mutans
|
S. sobrinus
|
||
|---|---|---|---|---|
| No. of cellsb | % of bacteriac | No. of cellsb | % of bacteriac | |
| Saliva | ||||
| 1 | 6.33 × 101 ± 0.09 × 101 | 1.33 × 10−1 ± 0.04 × 10−1 | 3.66 ± 0.40d | 2.57 × 10−2 ± 0.38 × 10−2 |
| 2 | 7.41 × 101 ± 0.76 × 101 | 2.50 × 10−2 ± 0.67 × 10−2 | 4.43 × 101 ± 1.84 × 101d | 5.00 × 10−3 ± 1.41 × 10−3 |
| 3 | 1.12 × 102 ± 0.11 × 102 | 6.22 × 10−2 ± 0.57 × 10−2 | NDe | ND |
| 4 | 4.43 × 101 ± 0.24 × 101 | 1.93 × 10−2 ± 0.11 × 10−2 | 5.83 × 101 ± 0.51 × 101d | 1.33 × 10−2 ± 0.10 × 10−2 |
| 5 | 1.26 × 102 ± 0.12 × 102 | 8.62 × 10−2 ± 0.09 × 10−2 | 4.57 × 102 ± 0.13 × 102 | 1.88 × 10−1 ± 0.37 × 10−1 |
| 6 | 1.97 × 103 ± 0.03 × 103 | 1.46 ± 0.02 | ND | ND |
| 7 | 1.34 × 102 ± 0.04 × 102 | 5.57 ± 0.01 | 2.87 ± 0.11d | 3.89 × 10−2 ± 0.14 × 10−2 |
| 8 | 2.15 × 101 ± 0.02 × 101 | 4.60 × 10−3 ± 0.01 × 10−3 | 1.53 × 102 ± 0.16 × 102 | 1.49 × 10−2 ± 0.13 × 10−2 |
| 9 | 5.05 × 102 ± 0.14 × 102 | 1.03 × 10−1 ± 0.04 × 10−1 | 8.76 × 102 ± 0.41 × 102 | 7.40 × 10−2 ± 0.72 × 10−2 |
| 10 | 5.55 ± 0.11d | 1.60 × 10−3 ± 0.00 × 100 | ND | ND |
| Plaque | ||||
| 1 | ND | ND | ND | ND |
| 2 | ND | ND | ND | ND |
| 3 | 3.76 × 101 ± 1.26 × 101 | 2.70 × 10−3 ± 0.70 × 10−3 | ND | ND |
| 4 | 2.44 × 101 ± 0.06 × 101 | 6.37 × 10−3 ± 0.86 × 10−3 | 1.64 × 101 ± 0.30 × 101d | 1.15 × 10−3 ± 0.07 × 10−3 |
| 5 | 3.38 × 102 ± 0.04 × 102 | 2.95 × 10−2 ± 0.04 × 10−2 | 5.17 × 102 ± 0.05 × 102 | 2.21 × 10−2 ± 0.30 × 10−2 |
| 6 | 3.44 × 104 ± 0.06 × 104 | 5.41 ± 0.50 | ND | ND |
| 7 | 3.13 × 101 ± 0.12 × 101 | 1.05 × 10−3 ± 0.07 × 10−3 | 2.55 × 102 ± 0.02 × 102 | 1.03 × 10−2 ± 0.03 × 10−2 |
| 8 | 2.92 × 101 ± 0.12 × 101 | 9.00 × 10−4 ± 2.83 × 10−4 | 2.39 × 102 ± 0.11 × 102 | 5.40 × 10−3 ± 0.14 × 10−3 |
| 9 | 4.82 × 104 ± 0.34 × 104 | 7.16 ± 0.84 | 4.76 × 104 ± 0.19 × 104 | 6.95 ± 0.28 |
| 10 | 4.25 ± 0.19d | 6.00 × 10−4 ± 0.00 × 100 | ND | ND |
Data are means ± standard deviations (n = 3).
CFU per PCR mixture.
Data were calculated by the modified ΔΔCt method.
Theoretical data below the detection limits.
ND, not detected.
This investigation revealed that the TaqMan assay is accurate and useful for the absolute and relative quantification of cariogenic bacteria from oral specimens. Recently, dental plaque has been considered an oral biofilm, and monitoring the absolute or relative amount of cariogenic bacteria in oral biofilm is essential. Moreover, the TaqMan PCR-based quantification system is advantageous, since cell numbers can be monitored in the biofilm as it is. This assay system will be useful for clarifying how these bacteria behave in oral biofilm formation and will contribute to the development of biofilm research.
Acknowledgments
This investigation was supported by research fellowships from the Japan Society for the Promotion of Science for Young Scientists (to N.S.) and by a research grant from the Nakatomi Foundation (to A.Y.).
Footnotes
Respectfully dedicated to the memory of Toshihiko Koga, who passed away 14 October 2001.
REFERENCES
- 1.Bowden, G. H., J. M. Hardie, and G. L. Slack. 1975. Microbial variations in approximal dental plaque. Caries Res. 9:253-277. [DOI] [PubMed] [Google Scholar]
- 2.Cangelosi, G. A., J. M. Iversen, Y. Zuo, T. K. Oswald, and R. J. Lamont. 1994. Oligonucleotide probes for mutans streptococci. Mol. Cell. Probes 8:73-80. [DOI] [PubMed] [Google Scholar]
- 3.de Soet, J. J., P. J. van Dalen, M. J. Pavicic, and J. de Graaff. 1990. Enumeration of mutans streptococci in clinical samples by using monoclonal antibodies. J. Clin. Microbiol. 28:2467-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greisen, K., M. Loeffelholz, A. Purohit, and D. Leong. 1994. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J. Clin. Microbiol. 32:335-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanada, N., Y. Yamashita, Y. Shibata, S. Sato, T. Katayama, T. Takehara, and M. Inoue. 1991. Cloning of a Streptococcus sobrinus gtf gene that encodes a glucosyltransferase which produces a high-molecular-weight water-soluble glucan. Infect. Immun. 59:3434-3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 7.Herzberg, M. C., M. W. Meyer, A. Kilic, and L. Tao. 1997. Host-pathogen interactions in bacterial endocarditis: streptococcal virulence in the host. Adv. Dent. Res. 11:69-74. [DOI] [PubMed] [Google Scholar]
- 8.Holland, P. M., R. D. Abramson, R. Watson, and D. H. Gelfand. 1991. Detection of specific polymerase chain reaction product by utilizing the 5′-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 88:7276-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jensen, B., and D. Bratthall. 1989. A new method for the estimation of mutans streptococci in human saliva. J. Dent. Res. 68:468-471. [DOI] [PubMed] [Google Scholar]
- 10.Koga, T., T. Oho, Y. Shimazaki, and Y. Nakano. 2002. Immunization against dental caries. Vaccine 20:2027-2044. [DOI] [PubMed] [Google Scholar]
- 11.Kuramitsu, H. K. 2001. Virulence properties of oral bacteria: impact of molecular biology. Curr. Issues Mol. Biol. 3:35-36. [PubMed] [Google Scholar]
- 12.Loesche, W. J. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50:353-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyons, S. R., A. L. Griffen, and E. J. Leys. 2000. Quantitative real-time PCR for Porphyromonas gingivalis and total bacteria. J. Clin. Microbiol. 38:2362-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munro, C. L., and F. L. Macrina. 1993. Sucrose-derived exopolysaccharides of Streptococcus mutans V403 contribute to infectivity in endocarditis. Mol. Microbiol. 8:133-142. [DOI] [PubMed] [Google Scholar]
- 15.Oho, T., Y. Yamashita, Y. Shimazaki, M. Kushiyama, and T. Koga. 2000. Simple and rapid detection of Streptococcus mutans and Streptococcus sobrinus in human saliva by polymerase chain reaction. Oral Microbiol. Immunol. 15:258-262. [DOI] [PubMed] [Google Scholar]
- 16.Okada, M., Y. Soda, F. Hayashi, T. Doi, J. Suzuki, K. Miura, and K. Kozai. 2002. PCR detection of Streptococcus mutans and S. sobrinus in dental plaque samples from Japanese pre-school children. J. Med. Microbiol. 51:443-447. [DOI] [PubMed] [Google Scholar]
- 17.Shiroza, T., S. Ueda, and H. K. Kuramitsu. 1987. Sequence analysis of the gtfB gene from Streptococcus mutans. J. Bacteriol. 169:4263-4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki, N., Y. Nakano, Y. Yoshida, D. Ikeda, and T. Koga. 2001. Identification of Actinobacillus actinomycetemcomitans serotypes by multiplex PCR. J. Clin. Microbiol. 39:2002-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tao, L., and M. C. Herzberg. 1999. Identifying in vivo expressed streptococcal genes in endocarditis. Methods Enzymol. 310:109-116. [DOI] [PubMed] [Google Scholar]
- 20.Yano, A., N. Kaneko, H. Ida, T. Yamaguchi, and N. Hanada. 2002. Real-time PCR for quantification of Streptococcus mutans. FEMS Microbiol. Lett. 217:23-30. [DOI] [PubMed] [Google Scholar]
- 21.Yoshida, A., N. Suzuki, Y. Nakano, T. Oho, M. Kawada, and T. Koga. 2003. Development of a 5′ fluorogenic nuclease-based real-time PCR assay for quantitative detection of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. J. Clin. Microbiol. 41:863-866. [DOI] [PMC free article] [PubMed] [Google Scholar]