Abstract
This work reports results of a systematic molecular analysis involving 113 Burkholderia cepacia complex isolates obtained from 23 cystic fibrosis (CF) patients under surveillance over a 7-year period at the major Portuguese CF center, the Santa Maria Hospital in Lisbon. The majority of the isolates were serial isolates from persistently infected patients (more than one-half of the population examined). In agreement with previous studies, B. cenocepacia (formerly genomovar III) was the most prevalent species; it was isolated from 52% of the patients infected with B. cepacia complex isolates. Contrasting with previous studies, a very significant percentage of the Portuguese CF subpopulation examined was infected with B. cepacia genomovar I (36%) and B. stabilis (18%). B. multivorans was recovered from two of the infected patients. All four of the species or genomovars were associated with poor clinical outcome, including the cepacia syndrome, and gave rise to chronic and transient infections, with the clinical condition depending on the patient and other still-unidentified factors. The B. cepacia epidemic strain marker region was found exclusively in genomovar III strains, while cblA was detected in genomovars I and III, only. There was no clear relation between the presence of these markers and transmissibility. Altogether, our results indicate that the use of these markers or the genomovar status in identifying patients at higher risk for infection is uncertain.
Bacteria belonging to the so called Burkholderia cepacia complex have become important opportunistic pathogens in patients with cystic fibrosis (CF) (12, 13, 14, 21). These bacteria are intrinsically resistant to multiple antibiotics and pose the risk of spread among patients with CF by social contact (11), with hospitalization being a risk factor for acquisition (14, 26, 36). The environment is also a potential source of strains of the B. cepacia complex (13). The rapid decline of lung function observed in CF subpopulations infected with B. cepacia complex, leading to a fatal necrotizing pneumonia accompanied with septicemia (the “cepacia syndrome”), is strongly feared (12, 15). The B. cepacia complex comprises at least nine closely related species or genomovars: B. cepacia genomovar I, B. multivorans (formerly genomovar II), genomovar III (recently denominated B. cenocepacia), B. stabilis (formerly genomovar IV), B. vietnamiensis (formerly genomovar V), B. cepacia genomovar VI, B. ambifaria (genomovar VII), B. anthina (genomovar VIII), and B. pyrrocinia (genomovar IX) (7, 8, 9, 30, 31, 32, 33). Initially, two distinct recA-based phylogenetic subgroups were found within genomovar III (III-A and III-B subgroups) (21). Further recA lineages within genomovar III have also been recently described (10, 33). Although strains from all the species and genomovars are capable of causing human infection (21), B. cepacia genomovar III is the predominant genomovar in the CF populations examined so far in the United States (17), Canada (27), and Italy (1), causing more than 50% of respiratory infections, followed by B. multivorans. Infection with genomovar III bacteria represents a significant clinical risk to patients with CF (20), but B. multivorans, B. stabilis, and the other less-represented genomovars may also be associated with serious clinical outcome (21). Two genetic markers have been associated with transmissibility and virulence of B. cepacia complex strains among CF patients: (i) the cblA gene, coding for the giant cable pili, which is believed to mediate the adherence of the bacterium to the respiratory epithelium (24, 28) and (ii) the B. cepacia epidemic strain marker (BCESM), a conserved 1.4-kb open reading frame (18). However, they cannot be considered absolute markers of virulence and transmissibility since they may be absent in certain strains or genomovars and other factors appear to be important (4, 6, 17, 20).
The first epidemiological survey of B. cepacia complex bacteria involved in pulmonary infections among a Portuguese CF subpopulation under surveillance, from 1995 to the end of 1997, in the major Portuguese CF treatment center at the Santa Maria Hospital in Lisbon was carried out by Richau et al. (22). In this preliminary study, the genomic relatedness of the small number of isolates then examined was characterized by ribotyping, and their ability to produce an extracellular polysaccharide (EPS) was assessed. Results indicated that EPS biosynthesis by B. cepacia complex is not as rare as thought before, suggesting that it may play a role in the colonization and persistence of B. cepacia complex isolates in the lung. The chemical structure of the EPS and its biosynthetic pathway were later characterized (5, 23). With the present work, we extend this preliminary study to isolates obtained until June of 2002 and further the molecular characterization of the B. cepacia complex isolates involved in CF infection in this Portuguese CF treatment center. The reported 7-year molecular analysis involves 113 B. cepacia complex isolates obtained from 23 CF patients, most of them serial isolates from persistently infected patients. The genomovar status of each isolate was determined, the genomic relatedness of the isolates was examined by ribotyping, and the presence of cblA and BCESM genes was investigated. This systematic analysis, focused on a specific and clinically characterized CF population, receiving care for an extended period of time, is a contribution to the understanding of how different strains of distinct species and genomovars of the B. cepacia complex differ in their persistence, epidemiology, and pathogenic potential in CF.
MATERIALS AND METHODS
Bacterial strains and culture.
The 122 putative B. cepacia complex bacteria examined in this study were isolated from the respiratory secretions of 23 patients with CF who attended the Santa Maria Hospital CF Center in Lisbon from January 1995 to June o2002. This CF center is attended by 85% of the CF population living in the Lisbon area, in the south of Portugal, and in the Madeira and Azores islands. Both adult and pediatric patients receive care at this CF treatment center (Tables 1 and 2). Sputum samples are obtained whenever patients are periodically examined to monitor their clinical status. The systematic study was only launched in 1997, but the isolates obtained before and kept by the hospital were also included. The 1996 isolates could not be examined because they were lost due to a prolonged storage period. All the serial isolates obtained from a same patient, during the surveillance period, went through the same molecular analysis. The identification of isolates as members of the B. cepacia complex was carried out by a triphasic analysis: (i) growth on the selective B. cepacia solid medium (Selectatab; Mast Diagnostics, Merseyside, United Kingdom); (ii) positive identification using the commercial system API 20NE (Biomerieux, Marcy L'Etoile, France); (iii) molecular identification by PCR assays based on the rRNA operon (3). Among the 122 isolates tested, only the 113 isolates tested positive for B. cepacia complex are listed in Table 1. Strain J2315 (representative of the highly epidemic genomovar III-A ET12 lineage) (13), kindly provided by J. R. W. Govan, was used as a reference strain.
TABLE 1.
Molecular analysis of the 113 B. cepacia complex isolates examined in this worka
| Patient | Gender | Age | Isolate | Isolation date (day/mo/yr) | Ribotype | Genomovar or species | recA | cblA | BCESM | Clinical outcome |
|---|---|---|---|---|---|---|---|---|---|---|
| A | F | 4 mo | IST401 | 28/8/95 | 2 | Genomovar I | K | − | − | |
| 5 mo | IST404 | 5/9/95 | 3 | Genomovar III-B | H | − | − | Era | ||
| B | M | 19 yr | IST402 | 13/4/95 | 1 | B. stabilis | J | − | − | |
| IST409 | 24/4/95 | 1 | B. stabilis | J | − | − | ||||
| IST421 | 3/4/98 | 1 | B. stabilis | J | − | − | ||||
| IST428 | 28/9/98 | 1 | B. stabilis | J | − | − | ||||
| IST437 | 25/1/99 | 1 | B. stabilis | J | − | − | Det | |||
| C | F | 12 yr | IST403 | 14/2/95 | 7 | Genomovar III-A | G | − | + | Unk |
| D | F | 4 mo | IST407 | 6/12/95 | 3 | Genomovar III-B | H | − | + | Era |
| E | M | 3 mo | IST406 | 15/12/95 | 2 | Genomovar I | K | − | − | D/unr |
| F | F | 2 yr | IST408 | 31/1/95 | 2 | Genomovar I | K | − | − | D/unr |
| G | F | 10 yr | IST405 | 7/4/95 | 4 | Genomovar III-B | I | − | + | |
| IST410 | 1/6/95 | 5 | Genomovar I | K | − | − | ||||
| IST411 | 9/12/95 | 6 | Genomovar III-B | I | − | + | D/cs | |||
| H | F | 19 yr | IST412 | 21/1/97 | 1 | B. stabilis | J | − | − | |
| IST413 | 18/3/97 | 1 | B. stabilis | J | − | − | ||||
| IST414 | 13/5/97 | 1 | B. stabilis | J | − | − | ||||
| IST415 | 8/7/97 | 1 | B. stabilis | J | − | − | ||||
| IST420 | 11/3/98 | 1 | B. stabilis | J | − | − | ||||
| IST423 | 6/5/98 | 1 | B. stabilis | J | − | − | ||||
| IST425 | 7/7/98 | 1 | B. stabilis | J | − | − | ||||
| IST427 | 13/8/98 | 1 | B. stabilis | J | − | − | ||||
| IST446 | 13/3/99 | 1 | B. stabilis | J | − | − | ||||
| IST448 | 4/6/99 | 1 | B. stabilis | J | − | − | ||||
| IST451 | 7/1/99 | 1 | B. stabilis | J | − | − | D/cs | |||
| I | F | 4 yr | IST416 | 30/9/97 | 8 | Genomovar III-A | G | − | + | Era |
| J | F | 17 yr | IST418 | 30/1/98 | 1 | B. stabilis | J | − | − | |
| IST419 | 1/2/98 | 9 | B. multivorans | F | − | − | ||||
| IST424 | 4/6/98 | 9 | B. multivorans | F | − | − | ||||
| IST439 | 30/1/99 | 11 | Genomovar III-A | G | − | + | ||||
| IST453 | 19/7/99 | 9 | B. multivorans | F | − | − | ||||
| IST455 | 7/2/00 | 9 | B. multivorans | F | − | − | ||||
| IST461 | 4/4/00 | 9 | B. multivorans | F | − | − | ||||
| IST483 | 29/3/01 | 11 | Genomovar III-A | G | − | + | ||||
| IST495 | 29/5/01 | 9 | B. multivorans | F | − | − | ||||
| IST4103 | 28/7/01 | 11 | Genomovar III-A | G | − | + | ||||
| IST4110 | 1/10/01 | 11 | Genomovar III-A | G | − | + | ||||
| IST4112 | 11/10/01 | 11 | Genomovar III-A | G | − | + | ||||
| IST4113 | 6/11/01 | 11 | Genomovar III-A | G | − | + | ||||
| IST4119 | 22/1/02 | 9 | B. multivorans | F | − | − | ||||
| IST4116 | 11/2/02 | 11 | Genomovar III-A | G | − | − | ||||
| IST4129 | 26/3/02 | 11 | Genomovar III-A | G | − | + | ||||
| IST4130 | 14/5/02 | 11 | Genomovar III-A | G | − | + | ||||
| IST4131 | 26/2/02 | 11 | Genomovar III-A | G | − | + | D/cs | |||
| L | F | 2 mo | IST426 | 22/7/98 | 10 | NT | NT | + | − | Era |
| N | F | 12 yr | IST431 | 30/8/98 | 12 | Genomovar I | AG | − | − | |
| IST443 | 3/3/99 | 12 | Genomovar I | AG | + | − | ||||
| IST444 | 3/3/99 | 12 | Genomovar I | AG | + | − | ||||
| IST449 | 9/6/99 | 12 | Genomovar I | AG | + | − | ||||
| IST457 | 15/3/00 | 12 | Genomovar I | AG | + | − | ||||
| IST463 | 23/5/00 | 12 | Genomovar I | AG | + | − | ||||
| IST472 | 18/10/00 | 12 | Genomovar I | AG | + | − | ||||
| IST485 | 1/4/01 | 12 | Genomovar I | AG | + | − | ||||
| IST491 | 26/4/01 | 12 | Genomovar I | AG | + | − | ||||
| IST4104 | 16/8/01 | 12 | Genomovar I | AG | + | − | ||||
| IST4105 | 31/8/01 | 12 | Genomovar I | AG | + | − | ||||
| IST4106 | 24/9/01 | 12 | Genomovar I | AG | + | − | ||||
| IST4115 | 8/11/01 | 12 | Genomovar I | AG | + | − | ||||
| IST4117 | 3/1/02 | 12 | Genomovar I | AG | + | − | D/r | |||
| O | F | 9 yr | IST430 | 30/9/98 | 11 | Genomovar III-A | G | − | + | |
| IST435 | 1/11/98 | 14 | Genomovar III-B | H | − | − | ||||
| IST440 | 8/2/99 | 14 | Genomovar III-B | H | − | − | ||||
| IST450 | 9/6/99 | 14 | Genomovar III-B | H | − | − | ||||
| IST458 | 17/3/00 | 14 | Genomovar III-B | H | − | − | ||||
| IST462 | 23/5/00 | 11 | Genomovar III-A | G | − | − | ||||
| IST467 | 19/7/00 | 11 | Genomovar III-A | G | − | + | ||||
| IST470 | 18/10/00 | 11 | Genomovar III-A | G | − | + | ||||
| IST471 | 18/10/00 | 14 | Genomovar III-B | H | − | − | ||||
| IST478 | 15/02/01 | 11 | Genomovar III-A | G | − | + | ||||
| IST486 | 3/4/01 | 11 | Genomovar III-A | G | − | + | ||||
| IST490 | 26/4/01 | 11 | Genomovar III-A | G | − | + | ||||
| IST497 | 31/5/01 | 11 | Genomovar III-A | G | − | + | ||||
| IST4107 | 17/9/01 | 11 | Genomovar III-A | G | − | + | ||||
| IST4114 | 8/11/01 | 14 | Genomovar III-B | H | − | − | /PICK> | |||
| IST4118 | 3/1/02 | 14 | Genomovar III-B | H | − | − | ||||
| IST4123 | 28/2/02 | 14 | Genomovar III-B | H | − | − | ||||
| IST4128 | 2/5/02 | 12 | Genomovar I | AG | + | − | Det | |||
| P | F | 6 yr | IST432 | 14/9/98 | 13 | Genomovar III-A | G | − | − | |
| IST468 | 17/10/00 | 13 | Genomovar III-A | G | − | − | ||||
| IST489 | 19/4/01 | 13 | Genomovar III-A | G | − | − | Sta | |||
| Q | F | 11 yr | IST433 | 5/11/98 | 14 | Genomovar III-B | H | − | − | D/unr |
| R | F | 1 yr | IST438 | 25/1/99 | 15 | Genomovar III-B | AN | − | − | |
| IST445 | 8/3/99 | 15 | Genomovar III-B | AN | − | − | ||||
| IST452 | 12/7/99 | 15 | Genomovar III-B | AN | − | − | ||||
| IST454 | 30/12/99 | 15 | Genomovar III-B | AN | − | − | ||||
| IST456 | 15/3/00 | 15 | Genomovar III-B | AN | − | − | ||||
| IST465 | 31/5/00 | 15 | Genomovar III-B | AN | − | − | ||||
| IST474 | 15/1/01 | 15 | Genomovar III-B | AN | − | − | ||||
| IST475 | 20/1/01 | 15 | Genomovar III-B | AN | − | − | ||||
| IST476 | 19/1/01 | 15 | Genomovar III-B | AN | − | − | ||||
| IST484 | 29/3/01 | 15 | Genomovar III-B | AN | − | − | Sta | |||
| T | M | 15 yr | IST466 | 12/6/00 | 16 | Genomovar III-B | H | − | + | |
| IST469 | 18/10/00 | 16 | Genomovar III-B | H | − | + | ||||
| IST473 | 27/12/00 | 16 | Genomovar III-B | H | − | + | ||||
| IST479 | 19/2/01 | 16 | Genomovar III-B | H | − | + | ||||
| IST4102 | 12/7/01 | 16 | Genomovar III-B | H | − | + | ||||
| IST4109 | 4/10/01 | 16 | Genomovar III-B | H | − | + | D/r | |||
| U | F | 6 yr | IST494 | 10/5/01 | 1 | B. stabilis | J | − | − | |
| IST4124 | 28/2/02 | 1 | B. stabilis | J | − | − | Sta | |||
| W | M | <1 yr | IST480 | 21/2/01 | 2 | Genomovar I | K | − | − | |
| IST487 | 6/4/01 | 2 | Genomovar I | K | − | − | ||||
| IST488 | 21/4/01 | 2 | Genomovar I | K | − | − | ||||
| IST492 | 7/5/01 | 2 | Genomovar I | K | − | − | ||||
| IST493 | 10/5/01 | 2 | Genomovar I | K | − | − | ||||
| IST4101 | 4/7/01 | 2 | Genomovar I | K | − | − | Sta | |||
| V | F | 3 yr | IST481 | 28/2/01 | 3 | B. cepacia complex | H2 | − | − | Sta |
| Y | M | 10 yr | IST4108 | 21/9/01 | 9 | B. multivorans | F | − | − | |
| IST4111 | 12/10/01 | 9 | B. multivorans | F | − | − | ||||
| IST4122 | 31/1/02 | 9 | B. multivorans | F | − | − | ||||
| IST4125 | 11/4/02 | 9 | B. multivorans | F | − | − | Sta | |||
| X | M | 5 yr | IST4120 | 24/1/02 | 2 | Genomovar I | K | − | − | |
| IST4126 | 18/4/02 | 2 | Genomovar I | K | − | − | Sta | |||
| AB | M | 14 yr | IST4121 | 31/1/02 | 7 | Genomovar III-A | G | + | − | Det |
All the serial isolates obtained from the same patient (male [M] or female [F]) during the 7-year surveillance period, the date of isolation, the age of the patient at the time of first isolation, and the clinical outcome (at the time of last isolation), are indicated. Abbreviations: NT, not tested; Era, eradicated B. cepacia infection; Det, deteriorated; Unk, unknown; D/unr, deceased, death unrelated to B. cepacia infection; D/cs, Deceased with the cepacia syndrome; Sta, stable; D/r, deceased, death related to B. cepacia infection.
TABLE 2.
Incidence and prevalence rates (calculated from January 1998 to June 2002) of B. cepacia complex respiratory infections in the Portuguese CF patients receiving care at the Santa Maria Hospital in Lisbon
| Yr | No. of CF patients |
B. cepaciab
|
|
|---|---|---|---|
| Incidence | Prevalence | ||
| 1998 | 77 | 6 (7.8) | 8 (10.4) |
| 1999 | 84 | 1 (1.2) | 6 (7.1) |
| 2000 | 78 | 1 (1.3) | 6 (7.6) |
| 2001 | 75 | 4 (5.3) | 10 (13.3) |
| 2002a | 97 | 2 (2.1) | 7 (7.2) |
Values were calculated until June, only.
Incidence and prevalence are reported as number of infected patients, with percentages in parentheses.
Bacterial cultures were maintained on Pseudomonas Isolation Agar plates (Difco, Detroit, Mich.), and liquid cultures were grown overnight in Luria-Bertani broth (Sigma), with orbital agitation, at 30°C.
Ribotyping of B. cepacia isolates.
Isolation of total DNA from B. cepacia isolates was carried out using the miniprep procedure described by Ausubel et al. (2). DNA restriction with EcoRI (Gibco BRL), separation of DNA fragments using agarose gel electrophoresis, DNA blotting, and hybridization with [γ-32P]ATP-labeled 16S and 23S rRNA from Escherichia coli were carried out using standard protocols (25). Radioactive labeling of 16S and 23S rRNA was carried out using the 5′-DNA Terminus Labeling system from Amersham following the manufacturer's instructions. Lambda phage DNA restricted with HindIII was used as the molecular mass standard. The sizes of the hybridization restriction fragments were estimated with DNA Simdex software from GenetX. Isolates producing ribopatterns that did not match others within the strain collection were considered to be unique.
Genomovar status identification.
Preliminary identification of genomovar status was performed by a PCR-based procedure for the identification of sequence motifs within the 16S and 23S ribosomal DNA, which uses three sets of primer pairs with different specificities, excluding single species in a stepwise manner (16, 35).
Genomovar status was determined by amplification of the entire recA gene using primers specific for bacteria belonging to the B. cepacia complex (19) and by performing analysis of the recA gene restriction fragments length polymorphisms (RFLPs) with the endonuclease HaeIII (19). Genomovar assignment was confirmed by PCR of the recA gene with genomovar-specific primers (19). Subdivision of genomovar III isolates into distinct recA-based phylogenetic subgroups was performed as described previously (19). For isolates whose species identification remained equivocal, complete recA gene sequences were obtained and analyzed as described elsewhere (19); sequences were aligned and compared with recA sequences from B. cepacia complex strains analyzed in previous studies, using CLUSTAL W (29) and the data analysis software in the Molecular Biology package (DAMBE; http://web.hku.hk/∼xxia/software/software.htm).
Detection of transmissibility markers.
The detection of the two putative transmissibility markers cblA and BCESM on the genome of the B. cepacia isolates was carried out by PCR, according to standard techniques. The 1.4-kb BCESM fragment was amplified with primers BCESM1 (5′-CCACGGACGTGACTAACA-3′) and BCESM2 (5′-CGTCCATCCGAACACGAT-3′) (18); the 664-bp cblA gene was amplified using primers PILI1 (5′-CCAAAGGACTAACCCA-3′) and PILI2 (5′-ACGCGATGTCCATCACA-3′) (24). Amplification reactions were carried out by an initial step of denaturation, for 5 min at 95°C, followed by 30 cycles of 1 min at 95°C for denaturation, 1 min at primer annealing temperature, and 2 min at 72°C for polymerization. Primer annealing was carried out at 63°C for detection of BCESM or at 55°C, for detection of cblA. Isolate J2315 (13) was used as the positive control in both amplification experiments. The amplification mixtures contained, in a total volume of 50 μl, 200 ng of DNA template, a 0.4 mM concentration of each deoxynucleoside triphosphate, 2.5 mM MgCl2, a 4 μM concentration of each primer, 2.5 U of Taq polymerase (Biotaq Taq Polymerase; Bioline), and 5 μl of reaction buffer (10×; supplied by the polymerase manufacturer).
Nucleotide sequence accession data.
The recA gene sequences for isolate IST431 (RFLP type AG) and isolates IST438 and IST445 (both RFLP type AN) have been submitted to GenBank under the following accession numbers, respectively: AF456062, AF456063, and AF456123.
RESULTS
Incidence and prevalence rates of infection with B. cepacia complex bacteria.
From January 1995 to June 2002, 122 putative B. cepacia complex isolates were obtained from the respiratory secretions of 23 CF patients receiving care at the Santa Maria Hospital in Lisbon. The bacteria were isolated at the hospital, during routine bacteriological analysis, onto B. cepacia selective agar and then were subjected to phenotypic analysis, using a commercial identification system. After genotypic analysis based on the ribosomal DNA and recA genes, performed outside the hospital by the partner laboratories in this study, nine of the isolates examined were found to be from outside the complex.
The molecular analysis reported in this work was focused on the 113 isolates (Table 1) confirmed to belong to the B. cepacia complex. Within the 7-year surveying period, the incidence and prevalence of infection with B. cepacia complex bacteria, ranging from 1.2 to 7.8% and from 7.1 to 13.3%, respectively, could only be accurately determined from January 1998 to June 2002 (Table 2). The age of the infected CF patients at the time of first isolation of B. cepacia complex bacteria varied between 2 months and 19 years, with an estimated mean age of 7.7 years. Interestingly, although the CF population examined includes an identical percentage of males and females, during this 7-year study, the B. cepacia complex-infected patients were predominantly (70%) of the female gender. Patients were considered to be persistently infected if at least three positive cultures for B. cepacia complex bacteria persisted for a 6-month period; according to this criterion, more than one-half of the 23 patients infected with B. cepacia complex were persistently infected during the period of surveillance. Eight of the CF patients under study (34.8%) died in the course of the study; five of these patients were infected with B. cepacia complex bacteria at time of death: three of them developed the cepacia syndrome, while two patients also harbored Pseudomonas aeruginosa strains prior to death.
Ribotyping analysis.
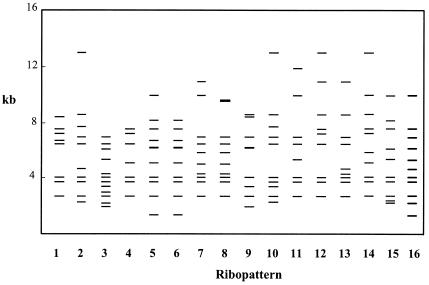
The genomic relatedness of the 113 B. cepacia complex isolates from 23 CF patients was characterized by ribotyping with EcoRI (Fig. 1). A significant percentage of the isolates under study were serial isolates collected from 14 patients, 12 of whom were persistently infected. Sixteen distinct ribopatterns, schematically presented in Fig. 1 and numbered from 1 to 16, were generated, suggesting a wide diversity in the sources of infection. More than one-half of the infected CF patients were found to harbor a unique strain, as would be expected in a CF treatment center with an isolation policy. Strains giving rise to ribopatterns 1 and 2 were the most frequent among the Portuguese CF subpopulation examined, being isolated from four and five patients, respectively. Two small clusters of possible nosocomial infection during 1995 were identified involving the isolates which gave rise to ribopatterns 2 (patients A, E, and F) and 3 (patients A and D), also indistinguishable by RFLP-pulsed-field gel electrophoresis analysis, as reported before (22). Strains exhibiting ribopatterns 2 or 3 were not isolated again until 2001 and 2002, when they were obtained from patients W, V, and X, who were hospitalized but never simultaneously. Strains with ribopatterns 1, 7, 9, 11, or 14 were also isolated from two or more patients, but based on the existing clinical records, the occurrence of any spreading event involving these patients inside the hospital is unlikely. Although social contact outside the hospital is strongly discouraged for these patients, it cannot be excluded.
FIG. 1.
Schematic representation of the ribopatterns of B. cepacia complex clinical isolates examined in this study. Ribopatterns are indicated by numbers across the bottom of the figure, and the molecular sizes of the EcoRI hybridization bands are arranged in a linear scale at left.
Remarkably, two sisters living together (patients N and O), were persistently infected with different strains, as indicated by the different ribopatterns produced by the isolates obtained over 2.5 years. Only in 2002 was the strain with ribotype 12, which persistently infected patient N, isolated from her sister (patient O), strongly suggesting recent transmission. However, since we have only studied one of the colonies grown on B. cepacia selective medium obtained from the sputum of each patient, at a specific date of isolation, the sisters may have been infected with isolates with common genotypes that were not detected. During this 7-year study, persistent colonization with isolates of single ribotypes was the most common situation. Less than 20% of the patients harbored two or more different strains, and among them, only patient A was transiently infected. The five patients whose deaths were linked to B. cepacia infection were infected with single unrelated strains.
Prevalence of B. cepacia complex species and genomovars.
The identification of the isolates recovered from Portuguese CF patients at the species or genomovar level was based on the polymorphisms of the recA gene with HaeIII, using specific primers (19). Nine distinct HaeIII-derived RFLP patterns were obtained (Table 1). The RFLP types F (B. multivorans); G, H, I, and AN (all corresponding to genomovar III); and J (B. stabilis) (Table 1) were previously described (19). RFLP type K was identified by Vermis et al. (34) as genomovar I. However, subsequent results revealed a considerable heterogeneity among type K strains, necessitating further analysis of the homogeneity of this taxon by means of DNA-DNA hybridization experiments (P. Vandamme and E. Mahenthiralingam, unpublished observations). In Table 1, we tentatively classified type K isolates as genomovar I. Two new recA RFLP profiles were obtained and designated AG and H2 (Table 1). Strains corresponding to these new RFLP types failed to react with genomovar-specific primers. Subsequent recA sequence analysis indicated that RFLP type AG corresponds to B. cepacia genomovar I by phylogenetic analysis (data not shown). The taxonomic status of the strain possessing RFLP type H2 was not known, but its reactivity with recA primers BCR1 and BCR2 (19) confirmed that it is a member of the B. cepacia complex.
The 112 B. cepacia complex isolates examined were found to belong to genomovars I and III and to the species B. multivorans and B. stabilis (Table 1). The genomovar status of isolate IST426 could not be determined because this isolate was lost. In agreement with previous reports (1, 17, 27), B. cenocepacia (genomovar III) was the most prevalent species or genomovar in the Portuguese subpopulation examined, being isolated from 52.2% of patients infected with the B. cepacia complex (Table 3). These isolates were separated into subgroups III-A (46.3%) and III-B (53.7%) with no significant differences. B. multivorans, the second most predominant CF pathogen among the three CF populations examined so far (1, 17, 27), was recovered from two patients only (9%). Contrasting with previous studies, a significant percentage of the Portuguese CF subpopulation examined was infected with B. cepacia genomovar I (36%; eight patients) and B. stabilis (18%; four patients) (Table 3).
TABLE 3.
Distribution of species or genomovars and of putative transmissibility factors among the 112 sequential B. cepacia complex isolates recovered from 22 patients with CFe
| Species or genomovar | No. of isolatesa (%) | No. of patientsb (%) |
esmR-positive isolates
|
cblA-positive isolates
|
||
|---|---|---|---|---|---|---|
| No.c (%) | No.d (%) | No.c (%) | No.d (%) | |||
| Genomovar I | 27 (24.1) | 8 (36.4) | 0 | 0 | 14 (12.5) | 14 (51.9) |
| B. multivorans | 11 (9.8) | 2 (9.0) | 0 | 0 | 0 | 0 |
| B. cenocepacia | 54 (48.2) | 12 (52.2) | 28 (25.0) | 28 (51.9) | 1 (0.9) | 1 (1.85) |
| recA subgroup A | 25 (22.3) | 6 (27.3) | 19 (17.0) | 19 (35.2) | 1 (0.9) | 1 (1.85) |
| recA subgroup B | 29 (25.9) | 7 (31.8) | 9 (8.0) | 9 (16.7) | 0 | 0 |
| B. stabilis | 19 (17.0) | 4 (18.2) | 0 | 0 | 0 | 0 |
| B. vietnamiensis | 0 | |||||
| Genomovar VI | 0 | |||||
| B. ambifaria | 0 | |||||
| B. anthina | 0 | |||||
| B. pyrrocinia | 0 | |||||
Number of isolates belonging to a specific genomovar or species (as percentage of the 112 isolates examined).
Number of patients infected with a genomovar/species (as percentage of the 22 patients examined). The sum of the percentage of patients infected with the different genomovars exceeds 100% since three patients harboured strains from more than one genomovar.
Number of isolates belonging to a specific genomovar or species with esmR or cblA genes (as percentage of the 112 isolates examined).
Number of isolates belonging to a specific genomovar or species with esmR or cblA genes (as percentage of number of isolates belonging to this specific genomovar or species).
Calculations were based on results shown in Table 1. (The genomovar status of isolate IST426 from patient L was not determined.)
Among the few patients infected with more than one strain, one patient harbored two different strains, both from B. cenocepacia or genomovar III (recA lineages III-A and III-B); two patients harbored strains of B. cepacia genomovars I and III; and one patient harbored strains belonging to the species B. cenocepacia, B. multivorans, and B. stabilis (Table 1).
Putative transmissibility marker genes.
The presence of the cblA and BCESM genetic markers in the 112 B. cepacia isolates was investigated by PCR. The cblA gene was detected in 13.4% of the isolates (Table 3), corresponding to 2 of the 16 strains examined (12.5%). The cblA-positive isolates were recovered from three infected patients and belong to genomovars I and III (12.5 and 0.9% of the 112 isolates examined, respectively) (Table 3). The seven strains found positive for the presence of the BCESM marker (25% of the 112 isolates examined) belong exclusively to B. cepacia genomovar III (Table 3). The esmR gene was found in one-half of the genomovar III isolates, being significantly more frequent in isolates from genomovar III subgroup III-A (35.2%) than those from subgroup III-B (16.7%) (Tables 1 and 3). Among the 113 isolates examined, no isolate was found to exhibit both cblA and esmR genes. The study of several isolates, most of them serial isolates of the same strain (with a common ribopattern), indicates that the presence of the BCESM region is not a stable trait (Table 1). The instability of the BCESM region was registered in both genomovar III subgroups (Tables 1 and 3), but no significant differences were detected in the two subgroups, as reported before (21).
Clinical outcome and epidemiology in relation to genomovar and transmissibility markers.
During the 7-year surveillance period, three patients died with symptoms of the cepacia syndrome. From one of these patients (patient G), three different strains were isolated over 8 months: two of them belonging to genomovar III-B, both being BCESM-positive, and the other isolate belonging to genomovar I; close to the time of death, an isolate belonging to genomovar III-B was isolated from this patient. Another patient deceased from the cepacia syndrome (patient J) simultaneously harbored B. multivorans and genomovar III-A strains. Notably, the third patient (patient H) who developed the cepacia syndrome was persistently infected with B. stabilis. Interestingly, this same B. stabilis strain (ribopattern 1) chronically infected another patient (patient B) for 5 years and was apparently eradicated. The same B. stabilis strain was once transiently isolated from patient J, who is now deceased, and more recently was isolated from patient U, who was infected over 7 months with no apparent impact on the clinical status. The high prevalence of B. stabilis among the Portuguese CF subpopulation under study results from the presence of this same strain in 4 of the 23 CF patients under surveillance. Although this may suggest its spread among the patients, there is no evidence of patient contact within the hospital or of any other way of nosocomial infection.
Two other deceases (patients N and T) were registered during the surveillance period, following progressive deterioration during persistent infection with a unique strain of genomovar I (during 5 years) or with a BCESM-positive genomovar III-B strain (for more than 1 year). The ribopatterns exhibited by these two strains were not detected in any other isolate recovered from other patients, with one exception. The isolate IST4128 (genomovar I, cblA-positive), which chronically infected patient N over a 5-year period, has recently been recovered from her sister (patient O). Notably, although these sisters lived together and most of the isolates recovered from patient N harbor the giant cable pilus subunit gene, cross-infection with this strain was never detected previously. During this period, two other strains of genomovar III-A (BCESM positive) and III-B were isolated from patient O, whose clinical situation has currently deteriorated. During the surveying period, other patients (patients E, F, and Q), who had been transiently infected with strains of genomovar I and III-A, died, but these deceases were not related to B. cepacia complex infection.
B. multivorans and genomovar III-A strains were alternately isolated from patient J, throughout a 4-year period, indicating that during such an extended period the replacement of the B. multivorans strain with the genomovar III-A strain did not occur, as suggested by other studies (20).
Patients sharing the same strains (with a common ribopattern), persistently infected with the same strain, or exhibiting lung deterioration were observed for all the four B. cepacia complex genomovars and species represented in the collection examined. There were also cases of transient infection with antibiotic clearance with strains belonging to the four genomovars, including genomovar III (subgroups III-A, III-B, and BCESM-positive strains).
The eight patients infected with strains from genomovar I were children (ages ranging from 3 months to 12 years), while three of the four patients infected with B. stabilis were adults (>16 years old). Due to the very small CF population examined, results do not support or reject a previous observation that patients with genomovar III infection are significantly older than those infected with B. multivorans (20).
DISCUSSION
Results of the 7-year systematic study reported here, focused on a defined Portuguese subpopulation infected with strains of the B. cepacia complex, may contribute to a better definition of the capacity of the genomovars and species infrequently recovered from the sputum of CF patients to spread and to cause persistent infection and severe disease. In fact, isolates from the less represented B. stabilis (ranging from 0.2 to 3.2% in the studied CF populations) (21) and genomovar I species (ranging from 0.2 to 4.8% in the studied CF populations) (21) were obtained from a significant percentage of the Portuguese CF patients under surveillance (18 and 36%, respectively). A strain of the rarely found species B. stabilis was in the origin of the persistent infection of one patient over 3 years, leading to one of the three cases of cepacia syndrome registered, also associated with genomovar III and B. multivorans, indicating that B. stabilis may be associated with serious clinical deterioration. However, this same strain (as assessed by a common ribopattern) was eradicated or had no apparent impact on the clinical state of other patients. The unusual prevalence of B. stabilis among this Portuguese CF subpopulation was related to the fact that a same strain (ribopattern 1) infected four patients, even though the hospital records are not consistent with nosocomial, patient-to-patient spread. This Portuguese CF center follows the recommended control measures; CF patients do not share hospital rooms as inpatients, and contact between outpatients in the clinic is limited. It should be noted that in contrast with other B. cepacia complex species, the B. stabilis genome was reported to be unusually stable and identical or very similar pulsed-field gel electrophoresis and random amplified polymorphic DNA fingerprints have been reported in a collection of 21 B. stabilis isolates obtained from CF patients and environmental sources over a period of nearly 30 years and derived from very diverse geographical origins (31). With respect to genomovar I, eight patients were infected with three different strains. One of the strains (ribopattern 2) was isolated from five of these eight patients. During 1995, this strain was isolated from three patients, probably due to nosocomial transmission (22). During 2001 and 2002 it was again isolated from two other patients, who were hospitalized but not simultaneously. Two other genomovar I unique strains were isolated, suggesting that different sources accounted for infection with genomovar I in the Portuguese CF subpopulation examined. Since genomovar I was isolated exclusively from children (<12 years old) and the Portuguese population under study predominantly comprised children, this characteristic may have contributed to the high prevalence registered for genomovar I. A follow-up study, extended to other Portuguese CF centers, is needed to evaluate if this prevalence is specific to the particular subpopulation and CF center examined or if it reflects a general trait of B. cepacia complex infections in the Portuguese CF population. Distinct differences between the U.S., Canadian. and Italian CF populations concerning the prevalence of B. cepacia complex genomovars and species were also observed (1, 17, 27), the basis for this being unclear.
Despite the high prevalence of B. stabilis and genomovar I among the Portuguese patients monitored over 7 years, the BCESM region was found exclusively in genomovar III strains, being more frequent among subgroup III-A strains, while the cblA gene was only detected in genomovars I and III strains, as reported before (21). The molecular analysis of isolates with a common ribopattern and therefore considered to be the same strain, confirms the instability of the BCESM region on the genome (21). Notably, none of the putative transmissibility markers cblA and BCESM was present in the strain (ribopattern 2) that was found to be capable of nosocomial transmission. In contrast, the cblA-positive genomovar I strain (with ribopattern 12) that persistently infected patient N over a 5-year period, leading to her death, was apparently not transmitted, until very recently, to her sister, also chronically infected with genomovar III-A and III-B strains. Although there are BCESM-positive strains of genomovar III-B related to the death of persistently infected patients, other BCESM-positive strains, from either subgroup III-A or III-B, were cleared from other patients. The lack of correlation between the presence of the putative transmissibility markers in the genome of the isolates and epidemiological evidence for spread suggests that its use in predicting epidemic strains and in identifying patients at higher risk for infection is uncertain.
Chronic or transient infections with strains of the three more-represented species and genomovars were observed. The few cases involving the less represented B. multivorans limit firm conclusions. There are examples that indicate that a same strain (same ribopattern) may persistently infect a patient for several years while it can be cleared by antibiotic therapy from another patient. The reasons why strains of distinct species and genomovars of the B. cepacia complex differ in their persistence, epidemiology, and pathogenic potential in CF and why the same strain may be associated with very different clinical outcomes in different patients are still unclear, and further research is required. Due to the unusual predominance of genomovar I and B. stabilis isolates in the Portuguese CF population receiving care at the Santa Maria Hospital in Lisbon, this study supports the concept that strains of these genomovars and species are both capable of causing devastating infections and can give rise to chronic or transient infections, with the clinical outcome depending on the patient and on other still unidentified factors.
Acknowledgments
M.V.C. acknowledges a Ph.D. scholarship (PRAXIS XXI/BD/19809/99) from Fundação para a Ciência e a Tecnologia. E.M. acknowledges grant funding from the U.K. Cystic Fibrosis Trust (grant PJ472).
E.M. is is grateful to Julie Fadden for technical assistance.
REFERENCES
- 1.Agodi, A., E. Mahenthiralingam, M. Barchitta, V. Giannino, A. Sciacca, and S. Stefani. 2001. Burkholderia cepacia complex infection in Italian patients with cystic fibrosis: prevalence, epidemiology, and genomovar status. J. Clin. Microbiol. 39:2891-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., M. Brent, R. E. Kingston, D. E. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1989. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Bauernfeind, A., I. Schneider, R. Jungwirth, and C. Roller. 1998. Discrimination of Burkholderia gladioli from other Burkholderia species detectable in cystic fibrosis patients by PCR. J. Clin. Microbiol. 36:2748-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bevivino, A., C. Dalmastri, S. Tabacchioni, L. Chiarini, M. L. Belli, S. Piana, A. Materazzo, P. Vandamme, and G. Manno. 2002. Burkholderia cepacia complex bacteria from clinical and environmental sources in Italy: genomovar status and distribution of traits related to virulence and transmissibility. J. Clin. Microbiol. 40:846-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cescutti, P., M. Bosco, F. Picotti, G. Impallomeni, J. H. Leitão, J. A. Richau, and I. Sá-Correia. 2000. Structural study of the exopolysaccharide produced by a clinical isolate of Burkholderia cepacia. Biochem. Biophys. Res. Commun. 273:1088-1094. [DOI] [PubMed] [Google Scholar]
- 6.Clode, F. E., M. E. Kaufmann, H. Malnick, and T. L. Pitt. 2000. Distribution of genes encoding putative transmissibility factors among epidemic and nonepidemic strains of Burkholderia cepacia from cystic fibrosis patients in the United Kingdom. J. Clin. Microbiol. 38:1763-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coenye, T., J. J. LiPuma, D. Henry, B. Hoste, K. Vandemeulebroecke, M. Gillis, D. P. Speert, and P. Vandamme. 2001. Burkholderia cepacia genomovar VI, a new member of the Burkholderia cepacia complex isolated from cystic fibrosis patients. Int. J. Syst. E vol. Microbiol. 51:271-279. [DOI] [PubMed] [Google Scholar]
- 8.Coenye, T., P. Vandamme, J. R. W. Govan, and J. J. LiPuma. 2001. Taxonomy and identification of the Burkholderia cepacia complex. J. Clin. Microbiol. 39:3427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coenye, T., E. Mahenthiralingam, D. Henry, J. J. LiPuma, S. Laevens, M. Gillis, D. P. Speert, and P. Vandamme. 2001. Burkholderia ambifaria sp. nov., a novel member of the Burkholderia cepacia complex including biocontrol and cystic fibrosis-related isolates. Int. J. Syst. E vol. Microbiol. 51:1481-1490. [DOI] [PubMed] [Google Scholar]
- 10.Coenye, T., and J. J. LiPuma. 2002. Multilocus restriction typing: a novel tool for studying global epidemiology of Burkholderia cepacia complex infection in cystic fibrosis. J. Infect. Dis. 185:1454-1462. [DOI] [PubMed] [Google Scholar]
- 11.Govan, J. R. W., P. H. Brown, J. Maddison, C. J. Doherty, J. W. Nelson, M. Dodd, A. P. Greening, and A. K. Webb. 1993. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis. Lancet 342:15-19. [DOI] [PubMed] [Google Scholar]
- 12.Govan, J. R. W., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Govan, J. R. W., J. E. Hughes, and P. Vandamme. 1996. Burkholderia cepacia: medical, taxonomic and ecological issues. J. Med. Microbiol. 45:395-407. [DOI] [PubMed] [Google Scholar]
- 14.Holmes, A., R. Nolan, R. Taylor, R. Finley, M. Riley, R.-Z. Jiang, S. Steinbach, and R. Goldstein. 1999. An epidemic of Burkholderia cepacia transmitted between patients with and without cystic fibrosis. J. Infect. Dis. 179:1197-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones, A. M., M. E. Dodd, and A. K. Webb. 2001. Burkholderia cepacia: current clinical issues, environmental controversies and ethical dilemmas. Eur. Respir. J. 17:295-301. [DOI] [PubMed] [Google Scholar]
- 16.LiPuma, J. J., B. J. Dulaney, J. D. McMenamin, P. W. Whitby, T. L. Stull, T. Coenye, and P. Vandamme. 2000. Development of rRNA-based PCR assays for identification of Burkholderia cepacia complex isolates recovered from cystic fibrosis patients. J. Clin. Microbiol. 37:3167-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LiPuma, J. J., T. Spilker, L. H. Gill, P. W. Campbell III, L. Liu, and E. Mahenthiralingam. 2001. Disproportionate distribution of Burkholderia cepacia complex species and transmissibility markers in cystic fibrosis. Am. J. Respir. Crit. Care Med. 164:92-96. [DOI] [PubMed] [Google Scholar]
- 18.Mahenthiralingam, E., D. A. Simpson, and D. P. Speert. 1997. Identification and characterization of a novel DNA marker associated with epidemic Burkholderia cepacia strains recovered from patients with cystic fibrosis. J. Clin. Microbiol. 35:808-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahenthiralingam, E., J. Bischof, S. K. Byrne, C. Radomski, J. E. Davies, Y. Av-Gay, and P. Vandamme. 2000. DNA-based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J. Clin. Microbiol. 38:3165-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahenthiralingam, E., P. Vandamme, M. E. Campbell, D. A. Henry, A. M. Gravelle, L. T. Wong, A. G. Davidson, P. G. Wilcox, B. Nakielna, and D. P. Speert. 2001. Infection with Burkholderia cepacia complex genomovars in patients with cystic fibrosis: virulent transmissible strains of genomovar III can replace Burkholderia multivorans. Clin. Infect. Dis. 33:1469-1475. [DOI] [PubMed] [Google Scholar]
- 21.Mahenthiralingam, E., A. Baldwin, and P. Vandamme. 2002. Burkholderia cepacia complex infection in patients with cystic fibrosis. J. Med. Microbiol. 51:533-538. [DOI] [PubMed] [Google Scholar]
- 22.Richau, J. A., J. H. Leitão, M. Correia, L. Lito, M. J. Salgado, C. Barreto, P. Cescutti, and I. Sá-Correia. 2000. Molecular typing and exopolysaccharide biosynthesis of Burkholderia cepacia isolates from a Portuguese cystic fibrosis center. J. Clin. Microbiol. 38:1651-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richau, J. A., J. H. Leitão, and I. Sá-Correia. 2000. Enzymes leading to the nucleotide sugar precursors for exopolysaccharide synthesis in Burkholderia cepacia. Biochem. Biophys. Res. Commun. 276:71-76. [DOI] [PubMed] [Google Scholar]
- 24.Sajjan, U. S., L. Sun, R. Goldstein, and J. F. Forstner. 1995. Cable (cbl) type II pili of cystic fibrosis-associated Burkholderia (Pseudomonas) cepacia: nucleotide sequence of the cblA major subunit pilin gene and novel morphology of the assembled appendage fibers. J. Bacteriol. 177:1030-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Smith, D. L., L. B. Gumery, E. G. Smith, D. E. Stableforth, M. E. Kaufmann, and T. L. Pitt. 1993. Epidemic of Pseudomonas cepacia in an adult cystic fibrosis unit: evidence of person-to-person transmission. J. Clin. Microbiol. 31:3017-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Speert, D. P., D. Henry, P. Vandamme, M. Corey, and E. Mahenthiralingam. 2002. Epidemiology of Burkholderia cepacia complex in patients with cystic fibrosis in Canada: geographical distribution and clustering of strains. Emerg. Infect. Dis. 8:181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun, L., R.-Z. Jiang, S. Steinbach, A. Holmes, C. Campanelli, J. Forstner, U. Sajjan, Y. Tan, M. Riley, and R. Goldstein. 1995. The emergence of a highly transmissible lineage of cblA+ Pseudomonas (Burkholderia) cepacia causing epidemics in North America and Britain. Nat. Med. 1:661-666. [DOI] [PubMed] [Google Scholar]
- 29.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vandamme, P., B. Holmes, M. Vancanneyt, T. Coenye, B. Hoste, R. Coopman, H. Revets, S. Lauwers, M. Gillis, K. Kersters, and J. R. W. Govan. 1997. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int. J. Syst. Bacteriol. 47:1188-1200. [DOI] [PubMed] [Google Scholar]
- 31.Vandamme, P., E. Mahenthiralingam, B. Holmes, T. Coenye, B. Hoste, P. De Vos, D. Henry, and D. P. Speert. 2000. Identification and population structure of Burkholderia stabilis sp. nov. (formerly Burkholderia cepacia genomovar IV). J. Clin. Microbiol. 38:1042-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vandamme, P., D. Henry, T. Coenye, S. Nzula, M. Vancanneyt, J. J. LiPuma, D. P. Speert, J. R. W. Govan, and E. Mahenthiralingam. 2002. Burkholderia anthina sp. nov. and Burkholderia pyrrocinia, two additional Burkholderia cepacia complex bacteria, may confound results of new molecular diagnostic tools. FEMS Immunol. Med. Microbiol. 33:143-149. [DOI] [PubMed] [Google Scholar]
- 33.Vandamme, P., B. Holmes, T. Coenye, J. Goris, E. Mahenthiralingam, and J. R. W. Govan. 2003. Burkholderia cenocepacia sp. nov., a new twist of an old story. Res. Microbiol. 154:91-96. [DOI] [PubMed] [Google Scholar]
- 34.Vermis, K., T. Coenye, E. Mahenthiralingam, H. J. Nelis, and P. Vandamme. 2002. Evaluation of species-specific recA-based PCR tests for genomovar level identification within the Burkholderia cepacia complex. J. Med. Microbiol. 51:937-940. [DOI] [PubMed] [Google Scholar]
- 35.Whitby, P. W., K. B. Carter, K. L. Hatter, J. J. LiPuma, and T. L. Stull. 2000. Identification of members of the Burkholderia cepacia complex by species-specific PCR. J. Clin. Microbiol. 38:2962-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whiteford, M. L., J. D. Wilkinson, J. H. McColl, F. M. Conlon, J. R. Michie, T. J. Evans, and J. Y. Paton. 1995. Outcome of Burkholderia (Pseudomonas) cepacia colonisation in children with cystic fibrosis following a hospital outbreak. Thorax 50:1194-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]