Abstract
Serotype-specific DNA regions involved in the biosynthesis of capsular polysaccharides (cps region) were used to develop a multiplex PCR test for the simultaneous species identification and serotyping of Actinobacillus pleuropneumoniae serotypes 2, 5, and 6. Primers specific for serotypes 2, 5, and 6 were combined with the already existing species-specific primers used in a PCR test based on the omlA gene. The PCR test was evaluated with serotype reference strains of A. pleuropneumoniae as well as 182 Danish field isolates previously serotyped by latex agglutination or immunodiffusion. For all serologically typeable strains, a complete correspondence was found between the results obtained by the multiplex PCR test and the results obtained by the traditional serotyping methods. Six of eight serologically nontypeable strains could be allocated to a serotype on the basis of the multiplex PCR results. The species specificity of the assay was evaluated with a collection of 93 strains representing 29 different species within the family Pasteurellaceae, as well as species normally found in the respiratory tracts of swine. All of these strains were negative by the multiplex PCR test, including 50 field isolates of the phylogenetically closely related species Actinobacillus lignieresii. When the multiplex PCR test was used to test Danish field strains, it was able to identify the serotypes of approximately 94% of all strains isolated from swine with clinical disease. More than 90% of the isolates that cross-reacted by the latex agglutination test were of serotype 2, 5, or 6. Determination of the serotype by PCR represents a convenient and specific method for the serotyping of A. pleuropneumoniae in diagnostic laboratories.
Actinobacillus pleuropneumoniae is an encapsulated respiratory pathogen of swine and the causative agent of porcine pleuropneumonia (28). The disease occurs worldwide and has resulted in large economic losses to the swine industry. At present, 15 different serotypes and 2 biotypes have been described (1, 6, 20, 23). The serotype specificity is predominately due to structural differences in the capsular polysaccharides (22). The presence and prevalence of serotypes vary among countries. In North America, serotypes 1 and 5 are the most commonly isolated, whereas serotypes 2 and 9 are isolated in many European countries (14, 27). In Denmark, serotypes 2 (63%), 5 (5%), and 6 (26%) account for approximately 94% of the strains isolated from swine with clinical disease. Virulence studies indicate considerable differences in virulence between serotypes (5, 11, 25).
A number of serological assays have been developed for the serotyping of A. pleuropneumoniae, but cross-reactions between serotypes are often seen by rapid serological assays such as slide agglutination tests. Definitive typing of such isolates has been achieved by using more time-consuming procedures, for example, immunodiffusion and indirect hemagglutination. Serological cross-reactivity between serotypes 1 and 9, serotypes 3, 6, and 8, and serotypes 4 and 7 has been reported (16, 17, 18). These cross-reactions are most likely due to the presence of similar antigens in the lipopolysaccharides (22). One way to avoid this serological cross-reactivity is to use DNA-based assays for the identification of A. pleuropneumoniae. During the last decade PCR has become a powerful DNA-based tool for the identification of microbes (24). A number of species-specific PCR tests have been developed for identification of A. pleuropneumoniae (8, 19, 26, 29), and a serotype-specific multiplex PCR assay has been designed for the simultaneous identification and serotyping of A. pleuropneumoniae serotype 5 (13).
The aim of the present investigation was to develop a PCR assay for the simultaneous species identification and serotyping of A. pleuropneumoniae serotypes 2, 5, and 6, which will be of great practical importance in diagnostic laboratories where these are the most prevalent serotypes. The PCR test is based on amplification of serotype-specific DNA regions involved in the biosynthesis of the capsular polysaccharides (cps genes). Oligonucleotide primers specific for the cps regions of serotypes 2, 5, and 6 were combined with primers previously used for the species-specific PCR amplification of the omlA gene (8). The specificity and sensitivity of the multiplex PCR test were examined, and the results of the multiplex PCR were compared with those obtained by traditional serological typing methods.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains used for this study are listed in Tables 1 and 2. In addition to reference strains representing serotypes 1 to 15, a total of 182 Danish field isolates of A. pleuropneumoniae from pig lungs representing isolates of serotypes 1 (n = 5), 2 (n = 46), 5 (n = 51), 6 (n = 56), 7 (n = 5), 8 (n = 5), and 12 (n = 6) and nontypeable isolates (n = 8) were included in the study. In addition, 93 strains representing 29 genetically related species, as well as other species normally found in the respiratory tracts of pigs, were used to evaluate the species specificity of the multiplex PCR test. This included 50 field isolates of the phylogenetically closely related species Actinobacillus lignieresii. All V-factor-dependent strains were grown on PPLO agar (Difco, Detroit Mich.), whereas all other strains tested were grown on Columbia agar supplemented with 5% bovine blood. All strains were incubated at 37°C overnight in atmospheric air.
TABLE 1.
A. pleuropneumoniae reference strains and field isolates used for evaluation of the PCR test
| A. pleuropneumoniae species | Strain designation | No. of strains | No. of strains positive by PCR with:
|
|
|---|---|---|---|---|
| A. pleuropneumoniae- specific amplicon (approx 950 bp) | Serotype-specific amplicon | |||
| Reference isolates | ||||
| Serotype 1 | Shope 4074T | 1 | 1 | 0 |
| Serotype 2 | S 1536 | 1 | 1 | 1 (500)a |
| Serotype 3 | S 1421 | 1 | 1 | 0 |
| Serotype 4 | M62 | 1 | 1 | 0 |
| Serotype 5A | K17 | 1 | 1 | 1 (1,100) |
| Serotype 5B | L20 | 1 | 1 | 1 (1,100) |
| Serotype 6 | Femφ | 1 | 1 | 1 (720) |
| Serotype 7 | WF83 | 1 | 1 | 0 |
| Serotype 8 | 405 | 1 | 1 | 0 |
| Serotype 9 | CVJ 13261 | 1 | 1 | 0 |
| Serotype 10 | D 13039 | 1 | 1 | 0 |
| Serotype 11 | 56153 | 1 | 1 | 0 |
| Serotype 12 | 8329 | 1 | 1 | 0 |
| Serotype 13 (biovar 2) | N-273 | 1 | 1 | 0 |
| Serotype 14 (biovar 2) | 3906 | 1 | 1 | 0 |
| Serotype 15 | HS143 | 1 | 1 | 0 |
| Danish field isolates | ||||
| Serotype 1 | 5(5)b | 5 | 0 | |
| Serotype 2 | 46(23) | 46 | 46 (500) | |
| Serotype 5 | 51(36) | 51 | 51 (1,100) | |
| Serotype 6 | 56(46) | 56 | 56 (720) | |
| Serotype 7 | 5(4) | 5 | 0 | |
| Serotype 8 | 5(2) | 5 | 0 | |
| Serotype 12 | 6(2) | 6 | 0 | |
| Nontypeable strains | 8 | 8 | 5 (500) | |
| 1 (720) | ||||
| (−) | ||||
The values in parentheses indicate the approximate amplicon size (in base pairs).
The values in parentheses indicate the number of strains cross-reacting by latex agglutination.
TABLE 2.
Collection of strains not giving rise to any amplicons in the multiplex PCR test
| Species | Strain(s) designationa | No. of strains |
|---|---|---|
| Actinobacillus actinomycetemcomitans | CCUG 13227T | 1 |
| Actinobacillus capsulatus | NCTC 11408T, P1364 | 2 |
| Actinobacillus equuli | NCTC 8529T, P1284 | 2 |
| Actinobacillus hominis | NCTC 11529T, P1336 | 2 |
| Actinobacillus indolicus | 46KC2T, 39Cl | 2 |
| Actinobacillus lignieresii | ATCC 49236T, 50 field isolates | 51 |
| Actinobacillus minor | NM 305T | 1 |
| Actinobacillus porcinus | NM 319T, 18765-5(T1), 18870-2(T5), 18870-2(T7), 595 | 5 |
| Actinobacillus rossii | ATCC 27072T | 1 |
| Actinobacillus suis | CAPM 5586T, P1143 | 2 |
| Actinobacillus urea | NCTC 10219T, P1144 | 2 |
| Haemophilus parainfluenzae | CCUG 12836T | 1 |
| Haemophilus paraphrophilus | 3718 | 1 |
| Haemophilus parasuis | NCTC 4557T | 1 |
| Haemophilus segnis | CCUG 10787T | 1 |
| Mannheimia haemolytica | NCTC 9380T | 1 |
| Mannheimia varigena | CCUG 16500, CAPM 5786, 3942/87 | 3 |
| Pasteurella aerogenes | ATCC 27883T | 1 |
| Pasteurella avium biovar 2 | Strain 5 | 1 |
| Pasteurella canis | CCUG 12400T | 1 |
| Pasteurella canis biovar 2 | Strain 25 | 1 |
| Pasteurella langaa | CCUG 15566T | 1 |
| Pasteurella mairii | CCUG 27189T | 1 |
| Pasteurella multocida | NCTC 10320 | 1 |
| Pasteurella multocida subsp. septica | HIM 746-6 | 1 |
| Pasteurella stomatis | CCUG 17979T | 1 |
| Pasteurella sp. strain B | CCUG 19794 | 1 |
| Bordetella bronchiseptica | 1317 | 1 |
| Streptococcus suis | 735, 8074, 24636/74 | 3 |
ATCC, American Type Culture Collection, Manassas, Va.; CAPM, Collection of Animal Pathogenic Microorganisms, Brno, Czech Republic; CCUG, Culture Collection University of Gothenburg, Gothenburg, Sweden; NCTC, National Collection of Type Cultures, London, United Kingdom.
Serotyping of A. pleuropneumoniae.
All the Danish field isolates of A. pleuropneumoniae were serotyped by a latex agglutination test (7). Each isolate was tested with latex particles coated with polyclonal antibodies produced against whole cells of reference strains of A. pleuropneumoniae representing serotypes 1 through 15 (Table 1). If the latex agglutination test showed a cross-reaction between different serotypes, the isolates were also tested by immunodiffusion or indirect hemagglutination to determine the serotype (21).
Oligonucleotide primers for PCR.
The sequences of the oligonucleotide primers used in this study are listed in Table 3. Four different pairs of primers were used in the multiplex PCR test. Serotype-specific primers were designed from the cps genes of serotypes 2, 5, and 6. The primers specific for serotype 2 and serotype 6 were designed in this study, while the primers specific for serotype 5 were designed in an earlier study (13). These three serotype-specific primers were combined with primers used in a previously published species-specific PCR test based on amplification of the omlA gene (8). The DNA primers designed in this study were selected by using the DNASIS sequence analysis program (Hitachi Software Engineering Co., Ltd.).
TABLE 3.
Primers used for PCR amplification
| Name | Function | Sequence (5′ → 3′) | Approx size of amplicon (bp) | DNA amplified |
|---|---|---|---|---|
| HPFa | Forward primer for omlA gene | AAG GTT GAT ATG TCC GCA CC | 950 | A. pleuropneumoniae serotypes 1 to 15 |
| HPRa | Reverse primer for omlA gene | CAC CGA TTA CGC CTT GCC A | ||
| Ap2F | Forward primer for cps region of serotype 2 | ACT ATG GCA ATC AGT CGA TTC AT | 500 | A. pleuropneumoniae serotype 2 |
| Ap2R | Reverse primer for cps region of serotype 2 | CCT AAT CGG AAA CGC CAT TCT G | ||
| Ap5Ab | Forward primer for cps region of serotype 5 | TTT ATC ACT ATC ACC GTC CAC ACC T | 1,100 | A. pleuropneumoniae serotype 5 |
| Ap5Bb | Reverse primer for cps region of serotype 5 | CAT TCG GGT CTT GTG GCT ACT AA | ||
| SGJ14 | Forward primer for cps region of serotype 6 | AAC CAC TCA CTT TCC ACA TTA G | 720 | A. pleuropneumoniae serotype 6 |
| SGJ5 | Reverse primer for cps region of serotype 6 | AAT CGG AAG GTT TTG GTC TCG TG |
Preparation of samples for PCR.
Lysates of pure cultures were prepared for all strains used in this study. A loopful of an overnight culture was taken from the surface of an agar plate and suspended in 200 μl of sterile water. The suspension was boiled for 10 min, and the cells were spun down at 13,000 × g for 2 min. A total of 180 μl of the supernatant was kept for analysis. The lysates were used undiluted as the DNA template in the PCR. The lysates were stored at −20°C until use.
PCR amplification.
PCR was performed in a total volume of 50 μl containing 10 mM Tris-HCl (pH 8.3); 1.5 mM MgCl2; 50 mM KCl; 0.005% Tween 20; 0.005% Nonidet P-40 detergent; 200 μM (each) dATP, dCTP, dGTP, and dTTP; and 1 U of Taq polymerase (Perkin-Elmer). The primers (Table 3) were added at the following final concentrations: 0.4 μM for HPF, HPR, SGJ5, and SGJ14; 2.0 μM for Ap5A and Ap5B; and 0.1 μM for Ap2F and Ap2R. One microliter of undiluted template DNA was added to each reaction mixture. Finally, mineral oil was added to prevent evaporation. The PCRs were performed in a Biometra Trio thermocycler by using 0.5-ml tubes. DNA was amplified for 33 cycles by using the following settings: denaturation at 94°C for 1 min, annealing at 63°C for 1 min, and extension at 72°C for 1 min 20 s. Twelve microliters of each reaction mixture was analyzed by electrophoresis in a 2% agarose gel. The PCR products were stained with ethidium bromide (10 μg/ml) and visualized under UV light.
RESULTS
Latex agglutination of A. pleuropneumoniae strains.
Of the 182 Danish field isolates of A. pleuropneumoniae, only 56 strains could be serotyped by latex agglutination test with polyclonal antibodies to whole cells. The remaining 126 isolates showed cross-reactions between two or more serotypes by the latex agglutination test. The serotypes of these 126 isolates were identified by immunodiffusion or indirect hemagglutination. Eight A. pleuropneumoniae strains could not be allocated to a serotype by any of these methods (Table 1).
Optimization of multiplex PCR test.
Two pairs of oligonucleotide primers were designed to amplify a part of the serotype-specific cps region from serotype 2 and 6 isolates, respectively. Primers Ap2F and Ap2R were designed to produce an approximately 500-bp PCR fragment from serotype 2 isolates, whereas primers SGJ14 and SGJ5 were designed to produce an approximately 720-bp PCR fragment from serotype 6 isolates. The Ap5A-Ap5B primer pair amplifies an approximately 1,100-bp portion of the serotype-specific cps region from serotype 5 isolates (13). These three primer pairs were combined with the primers used in an existing species-specific A. pleuropneumoniae PCR test that produce an approximately 950-bp amplicon from isolates of all A. pleuropneumoniae serotypes (8).
For optimization of the multiplex PCR test, serotype reference strains of A. pleuropneumoniae serotypes 1 through 15 were used. The annealing temperature and the reaction buffer were the same as those used for the species-specific test based on amplification of omlA (8). The magnesium concentration was varied from 1 to 2.5 mM. With low magnesium concentrations some of the expected PCR fragments were faint or nondetectable, while with high magnesium concentrations nonspecific PCR products of various sizes were amplified. From these results the optimum magnesium concentration for the assay was determined to be 1.5 mM. The concentrations of all primers were set from the beginning to be 0.4 μM, which resulted in differences in the intensities of the amplified DNA fragments. Initially, the intensity of the serotype 2-specific amplicon was much more intense than those of the other PCR products produced. The intensities of the amplicons became almost identical when the concentration of the serotype 2 primers was lowered to 0.1 μM and the concentration of the serotype 5 primers was increased to 2.0 μM.
Serotype specificity of multiplex PCR test.
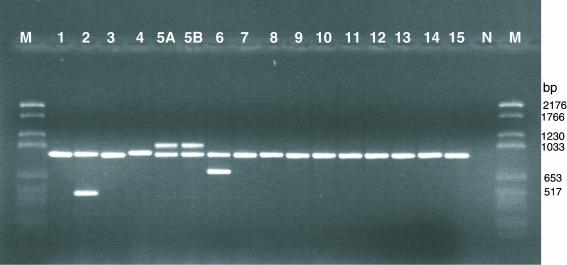
The serotype specificity of the multiplex PCR was determined by applying the test to a collection of 198 A. pleuropneumoniae isolates, including the serotype reference strains (Table 1). The species-specific fragment of approximately 950 bp was amplified from all A. pleuropneumoniae strains tested. An additional serotype-specific fragment was amplified from strains belonging to serotypes 2, 5, and 6 (Fig. 1). All Danish field isolates of A. pleuropneumoniae typed as serotype 2, 5, or 6 by one of the traditional serotyping methods were allocated to the same serotype by the multiplex PCR test. Eight strains of A. pleuropneumoniae which could not be serotyped by latex agglutination, immunodiffusion, or indirect hemagglutination were tested. Five of these strains gave rise to an amplicon of the same size as that specific for serotype 2 (approximately 500 bp), and one of the strains gave rise to an amplicon of the same size as that specific for serotype 6 (approximately 720 bp). Two of the nontypeable strains gave rise only to the A. pleuropneumoniae species-specific fragment of approximately 950 bp, indicating that they did not belong to serotype 2, 5, or 6. Nonspecific PCR products were not observed from any of the strains when the optimized conditions for the multiplex PCR assay were used.
FIG. 1.
Agarose gel electrophoresis of DNA fragments generated by multiplex PCR with the reference A. pleuropneumoniae strains of serotypes 1 to 15 (as indicated above the lanes) (Table 1). All strains gave rise to a species-specific band of approximately 950 bp, in addition to serotype-specific bands: approximately 500 bp (serotype 2), approximately 1,100 bp (serotypes 5A and 5B), and approximately 720 bp (serotype 6). Lane N, nontemplate control; lanes M, DNA molecular weight marker VI (Boehringer Mannheim).
Species specificity of multiplex PCR test.
The species specificity of the PCR assay was evaluated with a collection of 93 strains representing 29 different species within the family Pasteurellaceae as well as other species normally found in the upper respiratory tracts of swine (Table 2). The collection also included 50 field isolates of the phylogenetically closely related species A. lignieresii. The species-specific primers did not amplify the A. pleuropneumoniae species-specific amplicon of approximately 950 bp, and the serotype-specific primers did not amplify any product from any of the 93 heterologous strains tested in this study. Nonspecific PCR products were not observed from any of the related species tested when the optimized conditions for the multiplex PCR assay were used.
DISCUSSION
This is the first report of a PCR assay that simultaneously identifies A. pleuropneumoniae and three different serotypes. Cross-reactions between serotypes often occur when traditional serological assays are used for the serotyping of A. pleuropneumoniae. One way to avoid this serological cross-reactivity is to use DNA-based methods for serotyping, such as PCR, which does not involve antigens or antibodies. The present study shows that the use of multiplex PCR can be of great practical importance in the serotyping of A. pleuropneumoniae. A multiplex PCR test for the simultaneous detection of both the species A. pleuropneumoniae and serotypes 2, 5, and 6 was evaluated. The assay uses the advantage of multiplex PCR, in which more than one set of primers leads to the amplification of DNA from two or more target sequences in the same reaction under a single set of reaction conditions (15).
The design of the multiplex PCR test was determined by the distribution of A. pleuropneumoniae serotypes in Denmark. Almost 94% of the field strains isolated belong to serotype 2, 5, or 6. In Denmark, serotype 6 has been regarded as less virulent than serotype 2 and 5 in specific-pathogen-free animals. A quick and reliable means for the serotype designation of an isolate has therefore had a high priority in diagnostic bacteriology. Approximately 10% of the field isolates received at the Danish Veterinary Institute cross-react in the routinely performed latex agglutination tests, making it necessary to use more time-consuming serological serotyping procedures, such as immunodiffusion or indirect hemagglutination. More than 90% of these cross-reacting isolates represent serotype 2, 5, or 6. After optimization of the multiplex PCR assay, nonspecific amplicons were not observed from any of the isolates tested. All 182 Danish field isolates tested by the assay were identified as A. pleuropneumoniae; and all isolates identified as serotypes 2, 5, and 6 by serological assays were also identified as serotype 2, 5, and 6, respectively, by the multiplex PCR test. Furthermore, the multiplex PCR test was able to allocate six of the eight nonserotypeable isolates of A. pleuropneumoniae to either serotype 2 or serotype 6. These results show that the multiplex PCR test is a highly specific method for the identification and serotyping of A. pleuropneumoniae isolates and will be of great practical importance in the diagnostic laboratory.
Until now, most of the PCR assays reported for A. pleuropneumoniae have been used either for species identification or for subtyping of an isolate and do not simultaneously identify the species and the serotype. Various PCR tests have been developed for species identification of A. pleuropneumoniae (2, 8, 19, 26, 29). A common feature of these PCR tests is that the primers used for amplification are intended to be specific for A. pleuropneumoniae and not for any of the A. pleuropneumoniae serotypes. These assays are used only for identification of the organism, and therefore, other methods are still needed for serotyping. Furthermore, in these assays primers specific for the target gene(s) often amplify DNA from closely related species, such as Actinobacillus equuli and A. lignieresii (2, 19, 29). In the present multiplex PCR test, the species-specific amplicon was observed only when strains of A. pleuropneumoniae were tested and not when other species were tested (Table 2). Other PCR-based assays have been designed for the subtyping of A. pleuropneumoniae. However, most of the PCR-based typing methods do not use primers for genes directly involved in capsule production, which results in a subtype that at best is associated with the serotype. Hennessy et al. (10) described the use of arbitrarily primed PCR for serotyping. This method is able to differentiate between serotypes 1 through 12 by using a combination of two tests, although some of the serotypes can be distinguished only by minor differences in band migration. Gram et al. (9) developed an A. pleuropneumoniae-specific PCR typing system based on the apx and omlA genes. This PCR typing system could discriminate the majority of A. pleuropneumoniae serotypes of biotype 1 except serotypes 1, 9, and 11 and serotypes 2 and 8. Furthermore, the PCR typing system failed to discriminate between serotype 6 isolates and field isolates of serotype 8.
A more optimal design for DNA-based serotyping would be an assay with genes that are unique to the different serotypes, which can make the test independent of contaminating DNA. Lo et al. (13) were the first to describe the use of a multiplex PCR to amplify conserved and serotype-specific DNA regions involved in encapsulation to simultaneously detect both A. pleuropneumoniae and serotype 5 isolates. The species-specific PCR included in that assay failed to amplify a DNA fragment from A. pleuropneumoniae serotype 4. Furthermore, the species specificity of the PCR test was evaluated with only a few species; the closely related species A. lignieresii was not included in that study.
The present multiplex PCR test did not amplify DNA fragments from any of the 29 different species related to A. pleuropneumoniae or species normally found in the respiratory tracts of swine (Table 2). These results indicate a 100% species specificity of the multiplex PCR assay. Furthermore, DNA from 50 field isolates of the closely related species A. lignieresii did not provide a template for the multiplex PCR. A. lignieresii is the species that is the most phylogenetically related to A. pleuropneumoniae, as determined by comparison of 16S rRNA sequences, and serological cross-reactions between the two species have been observed (3, 4, 12). These serological cross-reactions have been reported for serotypes 3, 4, and 7 and consequently do not involve any of the serotypes for which the present multiplex PCR test is specific. Furthermore, the nature of these serological cross-reactions might rely on lipopolysaccharide or protein antigens. Future sequencing of the cps genes from more A. pleuropneumoniae serotypes might help to elucidate these cross-reactions.
In conclusion, the results obtained in this study indicate that the multiplex PCR test is a sensitive, specific, and highly effective diagnostic tool for the simultaneous identification and serotyping of A. pleuropneumoniae serotypes 2, 5, and 6. Moreover, problems with serological cross-reactions can be avoided in diagnostic laboratories by using multiplex PCR. The results also confirm that the genes involved in the biosynthesis of the capsular polysaccharides contain serotype-specific regions in A. pleuropneumoniae serotypes 2, 5, and 6. A future aim is to develop multiplex PCR tests based on the serotype-specific cps region that can allocate all isolates of A. pleuropneumoniae to a serotype.
Acknowledgments
We thank Jannie Jensen for competent technical assistance.
The present work was financed by the Danish Agricultural and Veterinary Research Council (grant 9702797).
REFERENCES
- 1.Blackall, P. J., H. L. Klaasen, H. Van Den Bosch, P. Kuhnert, and J. Frey. 2002. Proposal of a new serovar of Actinobacillus pleuropneumoniae: serovar 15. Vet. Microbiol. 84:47-52. [DOI] [PubMed] [Google Scholar]
- 2.Chiers, K., O. Van, E. Donné, M. Baele, R. Ducatelle, B. De, and F. Haesebrouck. 2001. Detection of Actinobacillus pleuropneumoniae in cultures from nasal and tonsillar swabs of pigs by a PCR assay based on the nucleotide sequence of a dsbE-like gene. Vet. Microbiol. 83:147-159. [DOI] [PubMed] [Google Scholar]
- 3.Christensen, H., M. Bisgaard, Ø. Angen, and J. E. Olsen. 2002. Final classification of Bisgaard taxon 9 as Actinobacillus arthritidis sp. nov. and recognition of a novel genomospecies for equine strains of Actinobacillus lignieresii. Int. J. Syst. E vol. Microbiol. 52:1239-1246. [DOI] [PubMed] [Google Scholar]
- 4.Dewhirst, F. E., B. J. Paster, I. Olsen, and G. J. Fraser. 1992. Phylogeny of 54 representative strains of species in the family Pasteurellaceae as determined by comparison of 16S rRNA sequences. J. Bacteriol. 174:2002-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dom, P., and F. Haesebrouck. 1992. Comparative virulence of NAD-dependent and NAD-independent Actinobacillus pleuropneumoniae strains. Zentbl. Vet. Med. Ser. B 39:303-306. [DOI] [PubMed] [Google Scholar]
- 6.Fodor, L., J. Varga, É. Molnár, and I. Hajtós. 1989. Biochemical and serological properties of Actinobacillus pleuropneumoniae biotype 2 strains isolated from swine. Vet. Microbiol. 20:173-180. [DOI] [PubMed] [Google Scholar]
- 7.Giese, S. B., E. Stenbaek, and R. Nielsen. 1993. Identification of Actinobacillus pleuropneumoniae serotype 2 by monoclonal or polyclonal antibodies in latex agglutination tests. Acta Vet. Scand. 34:223-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gram, T., and P. Ahrens. 1998. Improved diagnostic PCR assay for Actinobacillus pleuropneumoniae based on the nucleotide sequence of an outer membrane lipoprotein. J. Clin. Microbiol. 36:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gram, T., P. Ahrens, M. Andreasen, and J. P. Nielsen. 2000. An Actinobacillus pleuropneumoniae PCR typing system based on the apx and omlA genes—evaluation of isolates from lungs and tonsils of pigs. Vet. Microbiol. 75:43-57. [DOI] [PubMed] [Google Scholar]
- 10.Hennessy, K. J., J. J. Iandolo, and B. W. Fenwick. 1993. Serotype identification of Actinobacillus pleuropneumoniae by arbitrarily primed polymerase chain reaction. J. Clin. Microbiol. 31:1155-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobsen, M. J., J. P. Nielsen, and R. Nielsen. 1996. Comparison of virulence of different Actinobacillus pleuropneumoniae serotypes and biotypes using an aerosol infection model. Vet. Microbiol. 49:159-168. [DOI] [PubMed] [Google Scholar]
- 12.Lebrun, A., S. Lacouture, D. Côté, K. R. Mittal, and M. Gottschalk. 1999. Identification of Actinobacillus pleuropneumoniae strains of serotypes 7 and 4 using monoclonal antibodies: demonstration of common LPS O-chain epitopes with Actinobacillus lignieresii. Vet. Microbiol. 65:271-282. [DOI] [PubMed] [Google Scholar]
- 13.Lo, T. M., C. K. Ward, and T. J. Inzana. 1998. Detection and identification of Actinobacillus pleuropneumoniae serotype 5 by multiplex PCR. J. Clin. Microbiol. 36:1704-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macinnes, J. I., and N. L. Smart. 1993. Actinobacillus and Haemophilus, p. 188-200. In C. L. Gyles and C. O. Thoen (ed.), Pathogenesis of bacterial infections in animals. Iowa State University Press, Ames.
- 15.Markoulatos, P., N. Siafakas, and M. Moncany. 2002. Multiplex polymerase chain reaction: a practical approach. J. Clin. Lab. Anal. 16:47-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mittal, K. R. 1990. Cross-reactions between Actinobacillus (Haemophilus) pleuropneumoniae strains of serotypes 1 and 9. J. Clin. Microbiol. 28:535-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mittal, K. R., and S. Bourdon. 1991. Cross-reactivity and antigenic heterogeneity among Actinobacillus pleuropneumoniae strains of serotypes 4 and 7. J. Clin. Microbiol. 29:1344-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mittal, K. R., R. Higgins, and S. Lariviére. 1988. Serologic studies of Actinobacillus (Haemophilus) pleuropneumoniae strains of serotype-3 and their antigenic relationships with other A. pleuropneumoniae serotypes in swine. Am. J. Vet. Res. 49:152-155. [PubMed] [Google Scholar]
- 19.Moral, C. H., A. C. Soriano, M. S. Salazar, J. Y. Marcos, S. S. Ramos, and G. N. Carrasco. 1999. Molecular cloning and sequencing of the aroA gene from Actinobacillus pleuropneumoniae and its use in a PCR assay for rapid identification. J. Clin. Microbiol. 37:1575-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nielsen, R. 1990. New diagnostic techniques: a review of the HAP group of bacteria. Can. J. Vet. Res. 54(Suppl.):S68-S72. [PubMed] [Google Scholar]
- 21.Nielsen, R., and P. J. O'Connor. 1984. Serological characterization of 8 Haemophilus pleuropneumoniae strains and proposal of a new serotype: serotype 8. Acta Vet. Scand. 25:96-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perry, M. B., E. Altman, J.-R. Brisson, L. M. Beynon, and J. C. Richards. 1990. Structural characteristics of the antigenic capsular polysaccharides and lipopolysaccharides involved in the serological classification of Actinobacillus (Haemophilus) pleuropneumoniae. Serodiagn. Immunother. Infect. Dis. 4:299-308. [Google Scholar]
- 23.Pohl, S., H. U. Bertschinger, W. Frederiksen, and W. Mannheim. 1983. Transfer of Haemophilus pleuropneumoniae and the Pasteurella haemolytica-like organism causing porcine necrotic pleuropneumoniae to the genus Actinobacillus (Actinobacillus pleuropneumoniae comb. nov.) on the basis of phenotypic and deoxyribonucleic acid relatedness. Int. J. Syst. Bacteriol. 33:510-514. [Google Scholar]
- 24.Rodu, B. 1990. The polymerase chain reaction: the revolution within. Am. J. Med. Sci. 299:210-216. [DOI] [PubMed] [Google Scholar]
- 25.Rosendal, S., D. A. Boyd, and K. A. Gilbride. 1985. Comparative virulence of porcine Haemophilus bacteria. Can. J. Comp. Med. 49:68-74. [PMC free article] [PubMed] [Google Scholar]
- 26.Schaller, A., S. P. Djordjevic, G. J. Eamens, W. A. Forbes, R. Kuhn, P. Kuhnert, M. Gottschalk, J. Nicolet, and J. Frey. 2001. Identification and detection of Actinobacillus pleuropneumoniae by PCR based on the gene apxIVA. Vet. Microbiol. 79:47-62. [DOI] [PubMed] [Google Scholar]
- 27.Sebunya, T. N., and J. R. Saunders. 1983. Haemophilus pleuropneumoniae infection in swine: a review. J. Am. Vet. Med. Assoc. 182:1331-1337. [PubMed] [Google Scholar]
- 28.Shope, R. E. 1964. Porcine contagious pleuropneumoniae. I. Experimental transmission, etiology, and pathology. J. Exp. Med. 119:357-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sirois, M., E. G. Lemire, and R. C. Levesque. 1991. Construction of a DNA probe and detection of Actinobacillus pleuropneumoniae by using polymerase chain reaction. J. Clin. Microbiol. 29:1183-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]