Abstract
Six independent isolates of an unusual black-pigmented Corynebacterium species (strains CN-1, CN-2, CN-3415, W70124, 91-0032, and 92-0360) were recovered from the human female urogenital tract. Four of the six source patients had complications of pregnancy, including spontaneous abortion, preterm labor, and low amniotic fluid volume at the time of the pathogen isolation. One isolate was recovered from a vaginal ulcer. All six strains yielded black-pigmented colonies on sheep blood agar, chocolate agar, and colistin-nalidixic acid agar after 24 to 48 h of incubation at 35°C. The dry, adherent colonies pitted the agar surface. The cells were coccobacillary to rod-shaped, catalase positive, nonmotile, and nonlipophilic. Only five of six isolates were available for characterization. Biochemical and chemotaxonomic studies revealed that the strains belong to the genus Corynebacterium but differ from known corynebacterial species. Comparative 16S rRNA gene sequence analysis showed that the strains are closely related and form a new subline within the genus Corynebacterium. We propose the name Corynebacterium nigricans sp. nov. for this group of coryneforms. The type strain of Corynebacterium nigricans is CN-1. It is deposited in the American Type Culture Collection (assigned strain number ATCC 700975) and in the Institute Pasteur collection (assigned strain number CIP 107346).
The female urogenital tract is a natural habitat for a multitude of commensal microbes, including diphtheroids. In many instances, these microbes and their metabolites serve as barriers against colonization by harmful pathogens that may cause different diseases. The reported commensal organisms of the female urogenital tract are species of Lactobacillus, Staphylococcus, Streptococcus, Neisseria, Mycoplasma, Ureaplasma, enteric organisms, Acinetobacter, Capnocytophaga, Enterococcus, Bacteroides, Porphyomonas, Prevotella, Fusobacterium, Mobiluncus, Peptostreptococcus, Propionibacterium, Actinomyces, Bifidobacterium, Candida, and coryneform bacteria (8). Like other resident microbes of the human body, some of the vaginal microbiota also are opportunistic pathogens, which become pathogenic when the host immune status is compromised, suppressed, or under immunologic stress. A number of bacterial species are associated with urogenital-related diseases, and among the Corynebacterium species, C. lipophiloflavum, C. amycolatum, C. glucuronolyticum, C. riegelii, and C. urealyticum have been implicated (4, 6-8). The CDC coryneforms group 4 and C. lipophiloflavum are reported to cause urinary tract infections and bacterial vaginosis, respectively (5, 9).
Through the use of nucleic acid-based detection and characterization methods, such as 16S rRNA gene sequence analysis, many new pathogenic organisms are continually being identified, characterized, and linked to diseases of known and unknown etiology. This approach has been successfully applied in our laboratory to identify and characterize a number of fastidious microorganisms (12, 15-17). An unusual black-pigmented species of Corynebacterium that was isolated from a vaginal swab from a 26-year-old woman who experienced a spontaneous abortion was previously reported (18). After detailed biochemical and genetic characterization, it was found that the black-pigmented bacterial isolate was a novel pathogen. Since then, we have identified five additional isolates from the female urogenital tract that have the same phenotypic and genotypic characteristics of the Corynebacterium sp. type strain CN-1. All these strains were also clinically related. Four of the six isolates were recovered following vaginal swab for culture of patients after a report of complications during pregnancy. Based on the phenotypic and genotypic analyses of all six isolates, we propose the name Corynebacterium nigricans sp. nov. for this coryneform, which represents a novel species of the urogenital pathogen.
MATERIALS AND METHODS
Strains used and culture conditions.
Six black-pigmented strains had been isolated, and five of them were characterized in detail. The body sites from which these strains were isolated are as follows: CN-1, CN 3415, W70124, 91-0032 are from vaginal swabs; CN-2 is from a cervical swab; and strain 92-0360 is from a vaginal ulcer (2, 17). These strains have been independently isolated in the following medical centers: CN-1 and CN-2, Marshfield Clinic, Marshfield, Wis.; CN-3415 and W70124, St. John's Hospital, Springfield, Ill.; and 91-0032 and 92-0360, National Microbiology Laboratory (NML) in Canada (2). The strains CDC B8037 and IMMIB D-1488 (C. aurimucosum type strain) were not available for comparison. C. nigricans strains were compared with two strains of black-pigmented, Rothia-like (CDC group 4) bacteria. These isolates (NML identifiers 77-0298 and 83-0250, recovered from sputa) were observed to have characteristics consistent with the genus Rothia, based on phenotypic reactions, cellular fatty acid (CFA) composition analysis, and 16S rRNA gene sequence analysis (K. Bernard, unpublished data).
Phenotypic characterization.
Biochemical tests, including metabolisms of carbohydrates by these strains, were determined by either the API Coryne kit (bioMerieux, Marcy l'Etolie, France) or standard biochemical tests. The two NML strains were also tested using the API Coryne kit in parallel to conventional methods that included casein, tyrosine, and starch hydrolysis (2). Products of fermentation for 91-0031 and 92-0360 were analyzed as described previously (3).
CFA composition analysis and ancillary tests.
CFAs were determined with the Microbial Identification System (MIDI, Newark, Del.) after 48 h of incubation at 35°C on 5% blood agar plates using version 4.0 of the MIDI operating software. MIDI library generation system software was used to create a library entry for the Canadian strains of the new species. CFA composition data were compared with entries found in the commercial library CLIN (MIDI) version 4.0 (1, 18). The CAMP test was performed as described previously (4). A lipophilic test was performed as described previously (5).
Antimicrobial susceptibility testing.
Antimicrobial susceptibility was determined by E-test on Mueller-Hinton agar plates supplemented with 5% sheep blood. Since the National Committee for Clinical Laboratory Standards has not explicitly set the breakpoints for susceptibility and resistance for Corynebacterium, no interpretation regarding resistance and susceptibility has been made (11). Sensitivity to the vibriostatic disk was assessed (2).
16S rRNA gene sequence analysis.
The 16S rRNA genes of C. nigricans strains (CN-2 and CN-3415) were PCR amplified with the broad-range bacterial primers 27F (5′AGAGTTTGATCCTGGCTCAG3′), 806R(5′GGACTACCAGGGTATCTAAT 3′), 13B (5′AGGCCCGGGAACGTATTCAC 3′), and 1527R(5′AAGGAGGTGATCCAGCC3′) (13, 22). The sequences for the other strains (91-0032, 92-0360, and CN-1) were obtained from GenBank. PCR was performed in a PE7000 thermocycler using the GeneAmp PCR kit and the AmpliTaq DNA polymerase (Perkin Elmer, Branchburg, N.J.). A 100-μl PCR consisted of 10 μl of 10× PCR buffer; 1.4 mM MgCl2; 200 μM dATP, dCTP, dGTP, and dTTP; 2.5 U of Taq polymerase; 20 pmol each of forward and reverse primers; and 5 μl of the template DNA. The PCR products were purified and then sequenced by the cycle sequencing method with a number of nested primers (18). The sequencing reactions were resolved either in an ALF Express DNA sequencer (Amersham Pharmacia Biotech, Piscataway, N.J.) or in an ABI 3100 gene analyzer (Applied Biosystems, Foster City, Calif.). The DNA sequences were aligned by using the DNAStar program (DNAStar Inc., Madison, Wis.) and edited manually.
Phylogenetic analysis.
The rDNA sequences of the C. nigricans strains were aligned with a database of archaeal, bacterial, and eucaryotic small-subunit rRNA sequences (ca. 10,000 sequences in total) by using the ARB software package (19). Both BlastN analysis (http://www.ncbi.nlm.nih.gov) and the parsimony insertion tool of ARB (19) were used to initially determine the phylogenetic group with which these sequences are associated. A subset of the ARB small-subunit rRNA alignment, including the five C. nigricans sequences, other representative corynebacterial sequences, and an M. tuberculosis sequence used as an outgroup, was selected for more detailed phylogenetic analysis. Both full-length datasets and sequence alignments minimized by the use of the Lane mask (10) were analyzed. Multiple sequence datasets, including a variety of taxa and outgroup species, were analyzed by maximum likelihood, maximum parsimony, and evolutionary distance analyses. The presented phylogenetic dendrogram is a consensus tree of 100 maximum likelihood bootstrap replicates (20). The robustness of this tree was assessed by bootstrap resampling (1,000 replicates each) of parsimony and evolutionary distance (weighted least-squares mean with the general time reversal or Kimura two-parameter nucleotide substitution rate corrections) analyses (20). Reported pairwise percent sequence identities are uncorrected.
Nucleotide sequence accession numbers.
GenBank accession numbers for the strains identified in this study are as follows: CN-1, AF220220; CN-2, AY271720; CN-3415, AY227208; 91-0032, AY321294; and 92-0360, AF537608.
RESULTS AND DISCUSSION
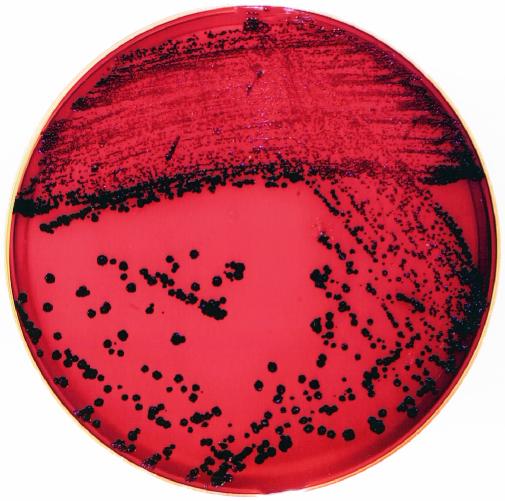
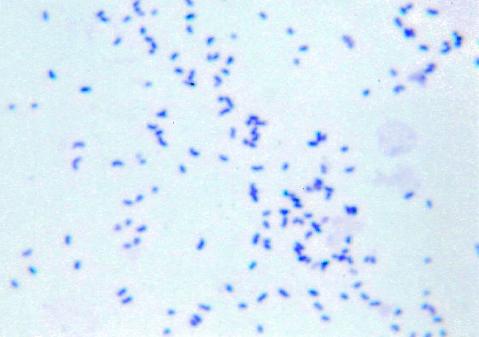
Colonies of C. nigricans are small, round, rough, granular, and 0.5 to 1.5 mm in diameter after 24-h incubation and could grow up to 3.0 mm in diameter upon 48 to 72 h of incubation. The colonies acquire a distinct, dark black color after 24 h of incubation (Fig. 1). The black pigmentation appeared on blood, heme-containing chocolate, and colistin-naladixic acid agar plates incubated at 35°C. The black pigmentation appeared after 24 h on brain heart infusion plates and after 3 to 5 days on trypticase soy agar plates. The pigmentation was not water soluble. The colonies are catalase positive, nonhemolytic, dry, and adherent. Colonies pit the surface of agar and are difficult to remove from the surface. Cells are gram positive, coccoidal or coccobacillary to short rod-shaped, and nonsporulated and are often found in small clusters (Fig. 2) approximately 1.25 to 2.5 microns in size. The common biochemical properties of five C. nigricans strains and a comparison with other urogenital Corynebacterium species, as determined by using the API Coryne strip, are described in Table 1. The C. nigricans strains yielded negative reactions for nitrate reduction, urea, casein, starch, DNA, and esculin hydrolysis. The strains fermented glucose, maltose, sucrose, and fructose. Xylose, lactose, and indole tests were negative for two isolates. Most strains were negative for pyrazinamidase and alkaline phosphatase. CAMP reactions were negative. Two strains, 91-0032 and 92-0360, were characterized by both conventional methods and the API Coryne strip, with equivalent results being obtained, as shown in Table 1. In addition, with conventional means, both strains hydrolyzed tyrosine and fermented fructose and mannose. These were also tested for a range of growth temperatures, and both could grow well (++++) in air and 5% CO2 but poorly under anaerobic conditions (+). Black-pigmented C. nigricans strains can be readily distinguished from similarly pigmented, Rothia-like bacteria based on a number of reactions, including nitrate reductions, esculin hydrolysis, and gelatinase production, and can be distinguished qualitatively by CFA analysis, as summarized in Table 2.
FIG. 1.
Black-pigmented colonies of C. nigricans CN-1 strain on a blood agar plate after 24 h. Reprinted with permission from reference 18.
FIG. 2.
Gram stain of C. nigricans strain CN-3415.
TABLE 1.
Comparison of C. nigricans with other medically relevant Corynebacterium species which are encountered in the urogenital tract or are phylogenetically closely related taxad
| Speciesa | Pigment and texture | Result of test for:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NO3 | Urea | Esc | PYRa/PYZ | PAL | Gluc | Malt | Suc | Fruc/mannose | Glyc/rib | Other (indicated) | ||
| C. nigricans (n = 5)b | Black, dry, pitting | − | − | − | −/v | v | + | + | (+) | +/+ | v/v | Tyrosine +; O129 sensc, hipp +; prop − |
| C. aurimucosum | Yellowish, sticky or mucoid | − | − | − | −/+ | + | + | + | + | +/− | −/− | Tyrosine and O129, ND; hipp + |
| C. minutissimum | White, creamy, moist | − | − | − | v/+ | + | + | + | v | +/v | −/v | Tyrosine +; O129 sens; hipp − |
| C. amycolatum | White or buff, dry, adherent | v | v | − | −/+ | + | + | v | v | +/+ | −/+ | Mycolates −; prop +; O129, resistant |
| C. glucuronolyticum | White or yellowish | v | v | v | −/+ | + | + | v | + | +/+ | −/+ | β gur +; O129, can be resistant; prop + |
| C. riegelii | White, smooth | − | + | − | −/v | v | − | (+) | − | −/− | −/+ | Glucose − |
| C. urealyticum | Gray-white, fastidious | − | + | − | −/+ | v | − | − | − | −/− | −/− | Lipophilic |
Data according to Bernard (2), Funke and Bernard (4), Shukla et al. (14, 18), Yassin et al. (23), and this study.
CAMP inhibition reaction was not observed for any species. CAMP reaction was always observed for C. glucuronolyticum and occasionally for C. minutissimum.
Based on strain 91-0032 and 92-0360-1.
Abbreviations and symbols: +, positive; −, negative; v, variable; parentheses, delayed or weak reaction; ND, not determined; NO3, nitrate reduction; Esc, esculin hydrolysis; PYRa, pyrrolidonyl arylamidase; PYZ, pyrazinamidase; PAL, alkaline phosphatase; Gluc, glucose; Malt, maltose; Suc, sucrose; Glyc, glycogen; Rib, ribose; sens, sensitive; hipp, hippurate hydrolysis; prop, propionic acid detected as product (otherwise species shown does not produce propionic acid); β gur, beta glucuronidase (using API Coryne or API ZYM [bioMérieux]) or equivalent.
TABLE 2.
Phenotypic and biochemical characteristics comparison between Rothia-like and C. nigricans
| Criterion | Result for organisma
|
|
|---|---|---|
| Rothia-like | C. nigricans | |
| Production of: | ||
| Catalase | Positive | Positive |
| Oxidase | Negative | Negative |
| Citrate | Negative | Negative |
| Urea | Negative | Negative |
| Indol | Negative | Negative |
| Gelatinase | Positive | Negative |
| Fermentation of: | ||
| Glucose | Positive | Positive |
| Sucrose | Positive | Positive |
| Maltose | Positive | Positive |
| Xylose | Negative | Negative |
| Lactose | Negative | Negative |
| Motility | Negative | Negative |
| Nitrate reduction | Positive | Negative |
| Esculin hydrolysis | Positive | Negative |
| CFA | Branched-chain type; similarity index values 0.3 to 0.4 to Rothia dentocariosa | Straight-chained, saturated, monounsaturated; similarity index values 0.4 to 0.6 to Corynebacterium spp. |
Rothia-like organisms are from sputa or respiratory sources; C. nigricans is from vaginal or vulval ulcers.
All five strains were tested against a variety of antimicrobial agents commonly used against gram-positive organisms. The MICs based on the Etest are listed in Table 3. The MICs for the same antimicrobial agent against different strains were very comparable for 8 out of 10 agents. However, the MICs for tetracycline and clindamycin had wide ranges. The tetracycline MIC ranged from 0.5 (91-0032) to 48 μg/ml for CN-3415. The range was much wider in case of clindamycin. The MIC for strain 91-0032 was 0.38 μg/ml, whereas it was >256 μg/ml for strain CN-2. All five C. nigricans strains were negative for beta-lactamase production.
TABLE 3.
MIC values of 12 commonly used antibiotics against gram-positive bacteriaa and test for β-lactamase production
| Strain | MIC (μg/ml) of:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TX | EM | FX | CT | PG | LE | LZ | CI | TC | CM | VA | TS | β-lactamase | |
| CN-1 | 2 | 0.016 | 1 | 1 | 0.25 | 0.125 | 1 | 0.125 | 1.5 | 1.5 | 0.5 | 0.75 | Negative |
| CN-2 | 1.5 | 0.75 | 3 | 1 | 0.25 | 0.19 | 0.75 | 0.25 | 32 | >256 | 0.38 | 0.032 | Negative |
| CN-3415 | 2 | 1.5 | 1.5 | 1 | 0.38 | 0.94 | 0.75 | 0.064 | 48 | 2 | 0.5 | 0.5 | Negative |
| NML 91-0032 | 0.75 | <0.016 | 0.75 | 0.75 | 0.094 | 0.094 | 0.75 | 0.094 | 0.5 | 0.38 | 0.75 | 0.75 | Negative |
| NML 92-0360 | 1.5 | <0.016 | 0.25 | 0.75 | 0.064 | 0.19 | 0.75 | 0.094 | 1 | 2 | 0.75 | 0.75 | Negative |
Abbreviations: TX, ceftriaxone; EM, erythromycin; FX, cefoxitin; CT, cefotaxime; PG, penicillin; LE, levofloxacin; LZ, linezolid; CI, ciprofloxacin; TC, tetracycline; CM, clindamycin; VA, vancomycin; TS, sulfamethoxazole trimethoprim.
The API Coryne kit was employed to identify these coryneforms. The API codes for three out of five strains tested was 0000125. Specifically, the API Coryne codes for strains CN-3415 and CN-2 were 2000125 and 0000125, respectively. The code for strain CN-1 was 2100327, whereas the code for strains 91-0032 and 92-0360 were 0000125, as described previously (2, 18). Codes for this newly named species generated poor confidence levels towards CDC group F-1, C. striatum, C. amycolatum, C. minutissimum, and CDC group G when compared with API Coryne database version 2.
Total CFA composition was determined for strains 91-0030 and 92-0360, and a library entry was generated. CFAs were found to be consistent with those described for the genus Corynebacterium, where the majority of the CFAs are of the straight-chained or monounsaturated types (1). A significant percentage of hexadecanoic acid (C16:0) (35%), cis-9 octadecanoic acid (C18:1w9c), cis-9,12 octadecadienoic acid (C18:2), and octadecanoic acid (C18:0) and tuberculosteric acid (1%) were observed. Peaks thought to represent coelution of corynemycolates at or near equivalent chain lengths 14.958 and 16.764 were present (1). The CFA composition of CN-1 was described previously (18) and was qualitatively and qualitatively consistent with these strains. Prior to creation of a library entry, each strain was found to generate good to excellent similarity index values (0.48 to 0.76) to the entry in CLIN for C. minutissimum, with no other significant matches. Propionic acid was not detected as a product of fermentation (2). Based on the unique colony phenotype, biochemical properties, and chemotaxonomic data presented here, this bacterium represents a new species in the genus Corynebacterium.
Phylogenetic analysis.
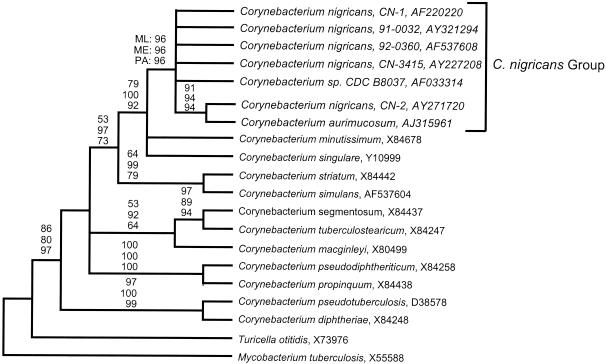
In order to do a comprehensive phylogenetic analysis of C. nigricans, 16S rRNA gene sequences were obtained for four additional C. nigricans strains, namely, CN-2, CN-3415, 91-0032, and 92-0360 (2, 18; this study). A detailed 16S rDNA-based phylogenetic analysis of the first reported black-pigmented Corynebacterium species, strain CN-1, by Shukla et al. (18) revealed that it was a novel species (GenBank accession no. AF220220) related to another corynebacterial isolate, CDC B8037 (GenBank accession no. AF033314). Initial phylogenetic analyses of these sequences with BlastN and ARB software (19) clearly placed these strains within the genus Corynebacterium and suggested a close phylogenetic relationship of these strains with C. nigricans strain CN-1 and CDC B8037. The C. aurimucosum strain that was reported after the initial description of the strain CN-1 had 16S rDNA sequences that were closely related to strain CN-1 by this preliminary analysis (23). Therefore, more precise phylogenetic relationships among the C. nigricans strains and other corynebacterial species were inferred by maximum likelihood, parsimony, and evolutionary distance methods of phylogenetic reconstruction. Figure 3 shows a representative consensus phylogenetic tree which was calculated by maximum likelihood analysis (20). Parsimony and evolutionary distance analyses with a variety of sequence data sets and outgroup species provided results substantially similar to those depicted in Fig. 3. Multiple bootstrap replicates were analyzed for each phylogenetic method in order to assess the robustness of any particular tree. In the case of the phylogenetic tree shown in Fig. 3, the branch points for which high bootstrap values (i.e., 75%) were determined by at least one method of phylogenetic reconstruction are labeled. Near-full-length 16S rDNA sequences from five C. nigricans strains and sequences obtained from GenBank for strains D-1488 (type strain C. aurimucosum) and CDC B8037 consistently formed a monophyletic relatedness group exclusive of all other known corynebacterial isolates, with high bootstrap values (96, 96, and 96% for maximum likelihood, evolutionary distance, and parsimony analyses, respectively). This clade has been designated “C. nigricans group” in Fig. 3. Pairwise percent sequence identities of the 16S rDNA sequence of the five C. nigricans strains and C. aurimucosum are presented in Table 4. Although no rigid criterion exists for defining species based on 16S rRNA sequence similarities, the high pairwise percent sequence identities evident within this group (99.5% mean identity; range, 99.0 to 100% identity) are in the range observed for interspecific variation in the 16S rRNA sequences of other well-established species (e.g., Escherichia coli). Thus, the molecular-phylogenetic analyses reported here are highly suggestive that the members of the C. nigricans group represent strains of the same species. However, C. aurimucosum, a recently described species, did not include dryness, pitting, and black pigmentation as a colony phenotype. Its ecological niche is yet to be defined. The single reported strain of C. aurimucosum was a bronchial isolate, whereas, remarkably, all C. nigricans strains were of urogenital origin and four of the six source patients had complications of pregnancy. In the case of C. nigricans, there seems to be a correlation between its urogenital ecological niche and clinical syndromes associated with it. However, additional isolates of C. aurimucosum and a critical phylogenetic analysis of them by using genes in addition to the 16S rRNA gene should clarify the true relationship between C. nigricans and C. aurimucosum.
FIG. 3.
Maximum-likelihood cladogram of selected corynebacterial 16S rRNA sequences, including those of newly isolated C. nigricans strains. The 16S rRNA sequence of Mycobacterium tuberculosis (X55588) was chosen as an outgroup for this phylogenetic analysis. Sequences are identified by species name and GenBank accession number, if available. The presented tree is a consensus of 100 bootstrap replicates. Bootstrap values of maximum likelihood (ML) (100 replicates), minimum evolutionary distance (ME) (1,000 replicates), and parsimony analyses (PA) (1,000 replicates) are indicated for strongly supported branch points (i.e., bootstrap value of >75%). Taxa representative of the proposed C. nigricans species are labeled “C. nigricans group.” C. segmentosum and C. tuberculostearicum are not valid species names.
TABLE 4.
Pairwise percent sequence identity of the 16S rDNA sequences of the five C. nigricans strains and C. aurimucosum
| Species or strain (no. of nucleotides) | CN-1 | CN-2 | 91-0032 | 92-0360 | CN-3415 | CDC B8037 |
|---|---|---|---|---|---|---|
| CN-1 (1,437) | ||||||
| CN-2 (1,414) | 99.2 | |||||
| 91-0032 (1,414) | 99.9 | 99.4 | ||||
| 92-0360 (1,443) | 99.9 | 99.4 | 100 | |||
| CN-3415 (1,441) | 99.9 | 99.3 | 100 | 100 | ||
| CDC B8037 (1,428) | 99.2 | 99.0 | 99.3 | 99.3 | 99.3 | |
| C. aurimucosum (1,428) | 99.4 | 99.8 | 99.4 | 99.4 | 99.3 | 99.0 |
Clinical significance.
This pathogen appears to be a clinically significant pathogen even though only a limited number of published reports are available. The first reported isolate came from a 34-year-old woman who presented with sudden onset of premature labor during her sixth month of pregnancy (18). The patient had a spontaneous abortion after hospitalization. The second and third isolates were recovered from vaginal swabs, one of them being from a vaginal ulcer (2). No medical histories were available for these patients. The case histories of the fourth, fifth, and the sixth isolates have been reported recently (14). The fourth isolate, from a vaginal-rectal swab, was recovered from a 25-year-old woman with preterm labor at 23 weeks of gestation. The patient had a history of preterm, as well as caesarian-section, deliveries. The fifth patient was a 21-year-old admitted at 38 weeks for induction of labor. This patient had a history of two stillbirths. The patient was positive for anti-cardiolipin antibodies and a lupus anticoagulant. The sixth isolate was collected from a 24-year-old woman who had a history of four prior spontaneous miscarriages (14). The patient also had unexplained low amniotic fluid. Remarkably, all six isolates were of urogenital origin, and four of them came from women who had complications of pregnancy. Incidentally, no other significant sexually transmitted agents were recovered. These case histories suggested, at least, a case for an opportunistic infection by this pathogen (14). Interestingly, even though no similar isolate has been recovered from the male genital tract, molecular evidence based on the 16S rDNA sequence suggests that a similar pathogen is present in prostatitis tissue (21). This finding could infer sexual activity as a means to transmit this bacterium to women. Additional isolates from human clinical samples will help establish its ecological niche and pathogenic potential.
Description of Corynebacterium nigricans sp. nov.
This description is based on the biochemical and genetic characteristics of five strains characterized so far. The name Corynebacterium nigricans (ni' gri-kanz) is derived from the word “niger” (L. fr. niger, black). The bacterial cells are gram positive, catalase positive, coccobacillary to rod-shaped, nonmotile, nonsporulated, and 1.25 to 2.5 micron in size. Colonies are small, dry, nonhemolytic, and granular. They are pitted, with a strong adhesion to agar. The colonies grow up to 2 to 3 mm in size after 48 h. The colonies produce black pigmentation on heme-containing sheep blood agar, colistin-naladixic acid agar, chocolate agar, and non-heme-containing brain heart infusion agar after 24 to 48 h of incubation. In a laboratory, these could be the criteria to distinguish this coryneform from other coryneforms. The pigment is not water soluble. Colonies also turn black on trypticase soy agar after 4 to 5 days. The organism grows well in ambient air or in air saturated with 5% CO2. The C. nigricans strain fermented glucose, maltose, and sucrose. Ribose and glycogen reactions were variable. The fermentation of glycogen, ribose, and sucrose was weak at 24 h of incubation. The tests for nitrate reduction, urea, casein, starch, xanthine, and esculin hydrolysis are negative. The C. nigricans strains produce variable reactions for pyrazinamidase and alkaline phosphatase as well. For example, CN-2 is negative for pyrazinamidase and weakly positive for alkaline phosphatase. Interestingly, CN 3415 is negative for alkaline phosphatase and positive for pyrazinamidase (Table 1).
Strain CN-1 is CAMP negative. Its major CFAs are hexadecanoic acid and cis-9 octadecanoic acid, and its minor CFAs are tetradecanoic acid, octadecanoic acid, cis-9 hexadecanoic acid, cis-9,12 octadecadienoic acid, and octadecanoic acid. Finally, phylogenetic analysis based on 16S rRNA sequence data indicates that all the proposed C. nigricans strains form a closely related group of organisms that exhibit greater than 99% pairwise sequence identity. The C. nigricans type strain CN-1 has been deposited into the ATCC as accession ATCC 700795.
Acknowledgments
We acknowledge the excellent technical assistance of Deborah Wiebe, Emma Ongsansoy, Cindy Munro, Teresa Aspeslet, Lisa Baeten, Mary Stemper, and Jennifer Conradt. We also thank Alice Stargardt for assistance in preparing the manuscript for submission.
REFERENCES
- 1.Bernard, K. A., M. Bellefeuille, and E. P. Ewan. 1991. Cellular fatty acid composition as an adjunct to the identification of asporogenous, aerobic gram-positive rods. J. Clin. Microbiol. 29:83-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernard, K. A., C. Munro, D. Wiebe, and E. Ongsansoy. 2002. Characteristics of rare or recently described Corynebacterium species recovered from human clinical material in Canada. J. Clin. Microbiol. 40:4375-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernard, K. A., L. Shuttleworth, C. Munro, J. C. Forbes-Faulkner, D. Pitt, J. H. Norton, and A. D. Thomas. 2002. Propionibacterium australiense sp. nov. derived from granulomatous bovine lesions. Anaerobe 8:41-47. [Google Scholar]
- 4.Funke, G., and K. A. Bernard. 2003. Coryneform gram-positive rods, p. 472-501. In P. R. Murray, E. J. Baron, M. J. Pfaller, F. C. Tenover, and R. H. Holden (ed.), Manual of clinical microbiology, 8th ed. ASM Press, Washington, D.C.
- 5.Funke, G., R. A. Hutson, M. Hilleringmann, W. R Heizmann, and M. D. Collins. 1997. Corynebacterium lipophiloflavum sp. nov. isolated from a patient with bacterial vaginosis. FEMS Microbiol. Lett. 150:219-224. [DOI] [PubMed] [Google Scholar]
- 6.Funke, G., P. A. Lawson, K. A. Bernard, and M. D. Collins. 1996. Most Corynebacterium xerosis strains identified in the routine clinical laboratory correspond to Corynebacterium amycolatum. J. Clin. Microbiol. 34:1124-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Funke, G., P. A. Lawson, and M. D. Collins. 1998. Corynebacterium riegelii sp. nov., an unusual species isolated from female patients with urinary tract infections. J. Clin. Microbiol. 36:624-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janda, W. M. 1998. Corynebacterium species and the coryneform bacteria. Part I: new and emerging species in the genus Corynebacterium. Clin. Microbiol. Newslett. 20:41-52. [Google Scholar]
- 9.Janda, W. M. 1998. Corynebacterium species and the coryneform bacteria. Part II: current status of the CDC coryneform groups. Clin. Microbiol. Newslett. 20:53-66. [Google Scholar]
- 10.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, N.Y.
- 11.National Committee for Clinical Laboratory Standards. 1997. Minimum inhibitory concentration (MIC) interpretive standards (μg/ml) for organisms other than Haemophilus spp., Neisseria gonorrhoeae, and Streptococcus spp. NCCLS document M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 12.Reed, K. D., G. R. Ruth, J. A. Meyer, and S. K. Shukla. 2000. Chlamydia pneumoniae infection in a breeding colony of African clawed frogs (Xenopus tropicalis). Emerg. Infect. Dis. 6:196-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Relman, D. A., T. M. Schmidt, R. P. MacDermott, and S. Falkow. 1992. Identification of the uncultured bacillus of Whipple's disease. N. Engl. J. Med. 327:293-301. [DOI] [PubMed] [Google Scholar]
- 14.Shukla, S. K., M. Harney, M. Jhaveri, A. Andrews, and K. D. Reed. Is a black-pigmented Corynebacterium species an opportunistic pathogen during pregnancy? Literature review and report of three new cases. Clin. Infect. Dis., in press. [DOI] [PubMed]
- 15.Shukla, S. K., P. R. Meier, P. D. Mitchell, D. N. Frank, and K. D. Reed. 2002. Leptotrichia amnionii sp. nov., a novel bacterium isolated from the amniotic fluid of a woman after intrauterine fetal demise. J. Clin. Microbiol. 40:3346-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shukla, S. K., and K. D. Reed. 2000. Desulfovibrio desulfuricans bacteremia in a dog. J. Clin. Microbiol. 6:1701-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shukla, S. K., T. Tak, R. C. Haselby, C. S. McCauley Juniorperiod, and K. D. Reed. 2002. Second case of infective endocarditis caused by Gemella sanguinis. WMJ 101:37-39. [PubMed] [Google Scholar]
- 18.Shukla, S. K., D. N. Vevea, D. N. Frank, N. R. Pace, and K. D. Reed. 2001. Isolation and characterization of a black-pigmented Corynebacterium sp. from a woman with spontaneous abortion. J. Clin. Microbiol. 39:1109-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strunk, O., O. Gross, B. Reichel, M. May, S. Hermann, N. Stuckmann, B. Nonhoff, T. Ginhart, A. Vibig, M. Lenke, T. Ludwig, A. Bode, K.-H. Schleifer, and W. Ludwig. 1996. ARB: a software environment for sequence data. [Online.] http://www.mikro.biologie.tu-muenchen.de/.
- 20.Swofford, D. L. 1999. PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, Mass.
- 21.Tanner, M. A., D. Shoskes, A. Shahed, and N. R. Pace. 1999. Prevalence of corynebacterial 16S rRNA sequences in patients with bacterial and “nonbacterial” prostatitis. J. Clin. Microbiol. 37:1863-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yassin, A. F., U. Steiner, and W. Ludwig. 2002. Corynebacterium aurimucosum sp. nov. and emended description of Corynebacterium minutissimum Collins and Jones (1983). Int. J. Syst. E vol. Microbiol. 52:1001-1005. [DOI] [PubMed] [Google Scholar]