Abstract
We recruited 200 children shortly after birth and collected stool specimens weekly, irrespective of whether the children had diarrhea, until up to 2 years of age. All children were recruited during the first year of the study and were monitored for a median of 18.4 months. To measure pathogenicity, the odds ratio for diarrhea, adjusted for age, sex, and coinfections with other enteropathogens, was determined by logistic regression. Standard estimation of the population attributable risk indicated that rotavirus, enterotoxigenic Escherichia coli that produced only the heat-stable toxin ST, Isospora spp., Cryptosporidium parvum, Shiga toxin (Stx)-producing E. coli (STEC), and Shigella spp. or enteroinvasive E. coli were the most important contributors to diarrhea in this population. Stx2- but not Stx1-producing STEC strains were pathogenic. Enteroaggregative E. coli, diffusely adherent E. coli, and attaching-and-effacing E. coli strains, which were the most commonly isolated microorganisms, were not associated with diarrhea. For most of the microorganisms, primary infections did not confer protection against reinfection with the same organism, but some conferred protection against diarrhea from reinfection.
Diarrheal diseases remain a major cause of childhood mortality and morbidity in developing countries. Although the mortality from diarrheal diseases is declining, diarrheal morbidity is not (27, 48). In order to target preventive measures, it is essential to describe the natural history and the relative importance of the various diarrheagenic agents. Moreover, to guide vaccine development, it is important to estimate the protection against reinfection and disease that natural infections with the various enteropathogens may confer.
Prospective cohort studies that investigate infections with diarrheal pathogens have been carried out for rotavirus (54), Campylobacter spp. (12)., Shigella spp. (20), Giardia lamblia (37), enterotoxigenic Escherichia coli (ETEC) (2), and other diarrheagenic E. coli strains (7, 28, 44). Longitudinal studies that describe a wide range of potential enteropathogens are few and were undertaken before the recognition of more recently described diarrheagenic agents, such as enteroaggregative E. coli (EAggEC) and diffusely adherent E. coli (DAEC) (14, 32).
We used DNA-DNA colony hybridization to identify diarrheagenic E. coli, Salmonella, and Shigella spp. in a cohort of newborn children who were monitored until up to 2 years of age with weekly stool specimen collection. Furthermore, examinations for rotavirus, Yersinia spp., Campylobacter spp., and enteric parasites, including Cryptosporidium parvum, were undertaken. Our objectives were to identify from a wide range of potential enteropathogens those that were associated with diarrhea, to determine the age of primary infection, to quantify any protection that was induced by natural infection, and to estimate their relative contribution to the incidence of diarrhea in children of a developing country.
MATERIALS AND METHODS
Study design and assembly of the cohort.
A total of 603 houses were selected at random in the periurban districts Bandim II and Belem of Bissau, the capital of Guinea-Bissau. A cohort of 200 children born in these houses between 15 January 1996 and 14 January 1997 were recruited within 3 weeks of birth (median, 6 days; interquartile range [IQR], 4 to 9 days) for a community-based cohort study of diarrheal disease, which included weekly morbidity recall visits with stool specimen collection irrespective of whether the children had diarrhea or not. The children were monitored until 2 years of age or until the end of the study on 28 April 1998. The follow-up of the cohort has been described in detail previously (51). The study was approved by the Ministry of Public Health in Guinea-Bissau and the Danish Central Scientific Ethical Committee.
Procedures.
The children were categorized as having diarrhea or no diarrhea according to the information provided by each child's caretaker (who was the mother in most cases) on the day the stool specimen was collected (35, 48). A child was considered to be infected with a given microorganism on the day that the positive specimen was collected. An infection was symptomatic if the child had diarrhea on the day of the infection. In addition, the field-worker asked whether the child had diarrhea or not for each of the 7 days prior to the visit. Information on whether the child was breast-fed was obtained monthly. Stool specimens were collected in plastic containers, and no preservatives were used. When a specimen was unavailable, we used a rectal swab, which afterwards was stored in Cary-Blair transport medium. Within 4 h of collection, the specimens were stored at 4°C until processing—usually within 15 to 18 h.
Microbiological analyses.
Microbiological examination of the stool specimens was undertaken by methods described previously (35), except that a semisolid, blood-free selective motility medium was used to identify Campylobacter spp. (19) because of difficulties in obtaining and storing sheep's blood in Guinea-Bissau. The stool specimens were inoculated and incubated on SSI solid enteric medium (11). Up to 12 (mean, 11.4) individual colonies from each dish were inoculated into meat-extract agar; the selection was made so that morphologically distinct colonies on the dish were represented.
To identify diarrheagenic E. coli, we performed DNA-DNA colony hybridization with nine different DNA probes, including probes specific for EAggEC (6) and DAEC (9). The ipaH probe (55) targets the large virulence plasmids of Shigella spp. and enteroinvasive E. coli (EIEC). The VT1 and VT2 probes (57) target the structural gene of Shiga toxin 1 (Stx1) and Stx2, respectively, of Shiga-like toxin-producing E. coli (STEC). The eae probe (25) targets the intimin structural gene that is common to attaching and effacing E. coli (A/EEC) and most STEC. The STp, STh, and LT probes (49) target the structural gene of the porcine and human heat-stable toxins (STp and STh) and the heat-labile toxin (LT), respectively, of ETEC. We screened the bacterial isolates in hybridization assays with three pools of each three probes: The first pool contained the STp, STh, and the LT probes (50), the second contained the eae, EAggEC, and the DAEC probe, while the third contained the VT1, VT2, and the ipaH probes. When an isolate was positive in a pooled probe assay, it was hybridized with each of the pool's individual probes. Isolates with the characteristics of Salmonella enterica, both isolated from the SSI plate and from desoxycholate citrate agar following selenite enrichment, were examined with the F1217 probe (1). The F1217 probe targets all subspecies of Salmonella enterica. We identified enteric parasites by direct microscopy of a wet mount. For the demonstration of helminth eggs, feces were examined microscopically after sodium chloride flotation. About 1 g of feces was concentrated by the Formol-ether technique (4) and examined for protozoan cysts by microscopy of the iodide-stained sediment. For the demonstration of Cryptosporidium oocysts, a smear was made from the sediment and stained by the modified Ziehl-Neelsen technique (23). We did not examine for enteric parasites in specimens that had been collected with rectal swabs. Stool specimens were tested for rotavirus with the IDEA enzyme-linked immunosorbent assay kit (DAKO, Copenhagen, Denmark) as described by the manufacturer.
Definitions.
Strains that were positive with the eae gene probe and negative for the Stx1 and the Stx2 gene probes were designated A/EEC. Because we did not serotype E. coli strains or examine them for the EAF plasmid or the bundle-forming pilus gene, bfpA, we did not distinguish between typical enteropathogenic E. coli (EPEC) and other A/EEC. ETEC strains that were positive for the STh or the STp gene and negative for the LT gene were designated ST-only ETEC. ETEC strains that were positive for the LT gene and negative for the ST genes were designated LT-only ETEC. ETEC strains that were positive for any of the ST genes and the LT gene were designated STLT ETEC. E. coli strains that were positive for the Stx1 gene and negative for the Stx2 gene were designated Stx1-producing STEC. E. coli strains that were positive for the Stx2 gene and negative for the Stx1 gene were designated Stx2-producing STEC. Strains that were positive for the EAggEC probe or for both the DAEC and the EAggEC probes were designated EAggEC. We considered potential enteropathogens to be the bacterial agents mentioned above plus rotavirus and enteric parasites, including Cryptosporidium parvum, Isospora spp., Blastocystis hominis, Chilomastix mesnilli, Endolimax nana, Entamoeba coli, Entamoeba histolytica/dispar, and Strongyloides stercoralis. The pathogenicity estimates for enteric parasites were calculated both overall (i.e., regardless of whether the parasite was observed in the trophozoitic/larva or the cystic/egg stage) and for the trophozoitic/larva stage alone.
A continuous infection was considered to occur when two or more sequentially collected specimens from the same child were positive for the same species, given that the specimens were collected with less than a 14-day interval. For such continuous infections, we considered the first detection of the enteropathogen to represent the infection and included only this in the analyses.
Data analyses.
The incidence rate of infection with a given potential enteropathogen was calculated as the number of episodes per child-year at risk, in which each stool specimen represented up to 7 days at risk centered on the day of its collection. To estimate the median age of experiencing a primary infection, we used a modification of the Kaplan-Meier estimator that takes into account any gaps in the observation periods (5). We set the median age of a primary infection to be when the estimated cumulative incidence of primary infections reached 50%. Similarly, the method of Kaplan-Meier was used to estimate the median age for ending breast-feeding. The PHREG procedure of the SAS system, version 8.2 (SAS Institute, Cary, N.C.) was used to calculate the Kaplan-Meier estimates.
Pathogenicity (10) was expressed as an odds ratio (OR), i.e., the odds of a pathogen-positive specimen being collected from a child with diarrhea divided by the odds of a pathogen-negative specimen being collected from a child with diarrhea. Because the specimens were collected independently of whether the child had diarrhea, this OR is a direct estimate rather than a biased approximation of the incidence rate ratio for the given enteropathogen (47). A P value of <0.05 was considered to represent statistical significance. A P value between 0.05 and 0.10 indicated borderline significance. We considered an exposure to be a confounder for a pathogenicity estimate if the OR changed by more than 10% when a variable that indicated whether or not the child was exposed was added to the model.
For estimating pathogenicity, we employed the GENMOD procedure of the SAS system to fit multivariable logistic regression models, where the outcome and exposure were diarrhea and infection, respectively. The models were fitted with a generalized estimating equation with a compound symmetry matrix to adjust for both between-child differences in diarrheal burden and differences between the caretakers in the reporting of diarrhea. Other enteropathogens that were isolated significantly more often from the same specimens as the organism of primary interest than from other specimens were considered to be possible confounders for the pathogenicity estimates. We adjusted for the presence of multiple enteropathogens in stool specimens by including variables indicating infections with these enteropathogens in the models. Furthermore, the estimates were adjusted by including variables indicating age in 6-month categories and sex in the models. Interaction with age in 6-month categories, sex, and breast-feeding was explored for each of the pathogens by including the relevant interaction terms in the models. If there was an interaction, defined as a Wald P value of <0.10 for the interaction term, separate estimates for pathogenicity were calculated. For estimating the pathogenicity of primary infections, only specimens collected up to and including the time of the given primary infection were included in the model.
The protection conferred by a primary infection against subsequent infections with the same type of microorganism was expressed as an OR (i.e., as the odds that a child who had already experienced a primary infection would be reinfected divided by the odds that a child who had not previously been infected would be infected). Infection was the outcome, and a time-dependent variable indicating whether the child had experienced a primary infection with the same type of microorganism indicated the exposure. To estimate the protection induced by a given primary infection against diarrhea when the children were reinfected, we calculated an OR (i.e., the ratio between the odds that a reinfection with the same microorganism was symptomatic and the odds that a primary infection would be symptomatic). In this model, only specimens representing infections with the relevant microorganism were included. The outcome was diarrhea, and a time-dependent variable indicating whether the infection was a primary infection or not indicated the exposure. The protection models were adjusted for sex, age, and repeated observations, as described for the models estimating pathogenicity. Percent protection was calculated as (1 − OR) × 100%. For some enteropathogens with which we did not observe any symptomatic reinfections, we calculated protection with the LogXact program, version 4 (Cytel Software Corp., Cambridge, Mass.).
The percent population attributable risk (PAR%) of an enteropathogen, defined as the relative reduction in diarrhea incidence that would be achieved if the population had been entirely unexposed to the enteropathogen compared with its current exposure pattern, was calculated as suggested by Levin (52):
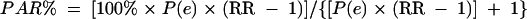 |
where we exchanged RR (the risk ratio) with OR, P(e) = N1/(N1 + N0) is the proportion of exposed children in the entire cohort, N1 is the number of exposed children, and N0 is the number of unexposed children.
RESULTS
Cohort characteristics and monitoring.
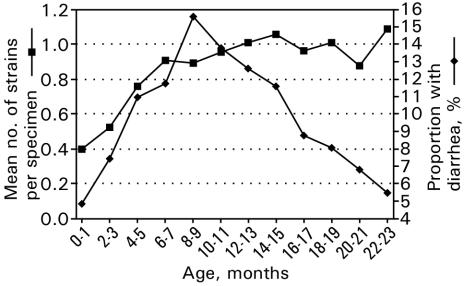
Of the 200 children, who were recruited at an even rate during 1 year, 104 were boys. The observed median age for terminating breast-feeding was 21.5 months (IQR, 18.5 to ≥24.0 months). During the observation period, 18 children died; 10 died within their first year of life. Due to an armed conflict in the spring of 1998, the study was stopped before all children had reached 2 years of age. The children were monitored for a median of 18.4 months (IQR, 13.1 to 22.1 months). In total, we collected 88% (IQR, 73 to 95%) of the scheduled specimens. In our cohort, 10.2% of the stool specimens were from children with diarrhea on the day of the specimen collection. The proportion of children with diarrhea increased steadily from birth to a peak of 16% at 9 to 10 months of age and declined gradually thereafter (Fig. 1).
FIG. 1.
Mean number of enteropathogens per stool specimen by age and proportion of specimens collected from children with diarrhea by age. The results represent a cohort study of 200 children monitored from birth until up to 2 years of age in Guinea-Bissau from 1996 to 1998.
Microbiological analysis.
We collected 11,987 specimens, 2,350 (20%) of which were rectal swabs. We found one potential enteropathogen in 37.1% of the specimens, two in 15.6% of the specimens, and three or more in 4.8% of the specimens. The overall probability of detecting any potential enteropathogen was similar whether the child had diarrhea or not (56 versus 58%). Furthermore, if at least one enteropathogen was detected, the numbers of different enteropathogens did not differ between the children who had diarrhea and those who did not (mean, 1.43 versus 1.46 enteropathogens per specimen). The mean number of different enteropathogens per stool specimen increased steadily from birth until reaching a plateau at 12 to 14 months of age (Fig. 1).
Incidence of infection.
EAggEC was the most frequently detected potential enteropathogen, with an incidence rate of almost 10 infections per child-year at risk, followed by DAEC, A/EEC, G. lamblia cysts or trophozoites, LT-only ETEC, G. lamblia in the trophozoitic stage, ST-only ETEC, STLT ETEC, rotavirus, and Salmonella (Table 1).
TABLE 1.
Incidence and OR for diarrhea pathogenicity for potential enteropathogens detected in a cohort study of children monitored from birth until up to 2 years of age in Guinea-Bissau from 1996 to 1998a
| Organism | Incidence (no. of infections/ child-yr at risk)b | Multivariable OR for diarrhea, adjusted for:
|
||
|---|---|---|---|---|
| Age and sex (95% CI) for all infections | Age, sex, and presence of multiple enteropathogens in same stool specimen (95% CI)
|
|||
| All infections | Primary infections | |||
| Rotavirus | 0.57 (116/204.5) | 6.06 (4.06-9.05) | 5.75 (3.77-8.75) | 7.07 (4.30-11.6) |
| EAggEC | 9.66 (1939/200.7) | 0.69 (0.58-0.81) | 0.66 (0.56-0.79) | 0.59 (0.27-1.26) |
| DAEC | 7.32 (1468/200.7) | 0.77 (0.64-0.94) | 0.85 (0.66-1.07) | 0.82 (0.35-1.88) |
| A/EEC | 6.34 (1272/200.7) | 0.88 (0.73-1.06) | 0.91 (0.73-1.13) | 1.45 (0.81-2.59) |
| ST-only ETEC | 1.06 (212/200.8) | 1.79 (1.28-2.51) | 1.87 (1.32-2.65) | 1.51 (0.90-2.53) |
| LT-only ETEC | 2.45 (493/200.8) | 0.84 (0.62-1.13) | 0.84 (0.61-1.15) | 1.52 (0.94-2.46) |
| STLT ETEC | 0.95 (191/200.8) | 1.19 (0.81-1.75) | 0.97 (0.62-1.52) | 1.03 (0.58-1.82) |
| Stx1-producing STEC | 0.05 (10/201.1) | 1.19 (0.24-5.90) | 1.11 (0.21-5.75) | 1.15 (0.17-7.99) |
| Stx2-producing STEC | 0.08 (16/201.1) | 5.34 (2.10-13.6) | 5.09 (1.53-16.9) | 5.41 (1.64-17.9) |
| Shigella spp. or EIEC | 0.29 (61/210.2) | 1.84 (0.95-3.57) | 1.87 (0.85-4.16) | 2.95 (1.31-6.62) |
| S. enterica | 0.57 (105/185.1) | 0.81 (0.43-1.52) | 0.81 (0.43-1.52) | 0.86 (0.39-1.89) |
| Campylobacter spp. | 0.35 (70/198.5) | 0.27 (0.08-0.89) | 0.19 (0.03-1.03) | 0.24 (0.05-1.21) |
| C. parvum | 0.33 (54/161.3) | 2.12 (1.18-3.79) | 2.12 (1.18-3.79) | 2.09 (1.10-3.97) |
| Isospora spp. | 0.07 (11/168.7) | 3.55 (1.15-11.0) | 6.75 (2.15-21.2) | 9.80 (2.45-39.1) |
| G. lamblia | ||||
| Cyst or trophozoite | 3.57 (602/168.7) | 0.65 (0.48-0.89) | 0.64 (0.46-0.89) | 0.83 (0.46-1.50) |
| Trophozoite | 1.85 (312/168.7) | 0.98 (0.69-1.39) | 0.97 (0.67-1.41) | 0.66 (0.34-1.27) |
| Entamoeba coli | ||||
| Cyst or trophozoite | 0.47 (79/168.7) | 0.58 (0.23-1.45) | 0.33 (0.12-0.90) | 0.26 (0.056-1.19) |
| Trophozoite | 0.11 (19/168.7) | 1.01 (0.32-3.20) | 1.10 (0.34-3.58) | 1.39 (0.36-5.38) |
| Entamoeba histolytica or E. dispar | ||||
| Cyst or trophozoitec | 0.08 (13/168.7) | 0.96 (0.12-7.56) | 1.29 (0.11-15.1) | 1.29 (0.06-27.3) |
| E. nana | ||||
| Cyst or trophozoited | 0.13 (22/168.7) | 0.75 (0.19-2.98) | 1.55 (0.35-6.83) | 2.82 (0.62-12.8) |
| C. mesnilli | ||||
| Cyst or trophozoite | 0.20 (33/168.7) | 1.54 (0.61-3.90) | 1.68 (0.64-4.41) | 0.86 (0.21-3.50) |
| Trophozoite | 0.09 (15/168.7) | 0.90 (0.19-4.33) | 0.73 (0.22-2.39) | Not computable |
| B. hominis | ||||
| Cyst | 0.14 (23/168.7) | 1.24 (0.42-3.63) | 2.70 (0.87-8.40) | 3.16 (0.88-11.4) |
| S. stercoralis larvae | 0.21 (35/168.7) | 0.26 (0.04-1.67) | 0.29 (0.07-1.32) | Not computable |
Note that enteropathogens that represented <10 infections were not included in the table.
The number of child-years at risk may vary between potential enteropathogens, mainly because of differences in number of stool specimens analyzed. Approximately 20% of the stool specimens were collected as rectal swabs, which is why detection of parasites was not possible. In contrast, all specimens were examined for rotavirus. Slightly fewer specimens were examined for bacteria due to intermittent and short-lived problems with medium production or accidental drying up of stab cultures.
Only one trophozoite.
Only two trophozoites.
Median age of primary infection.
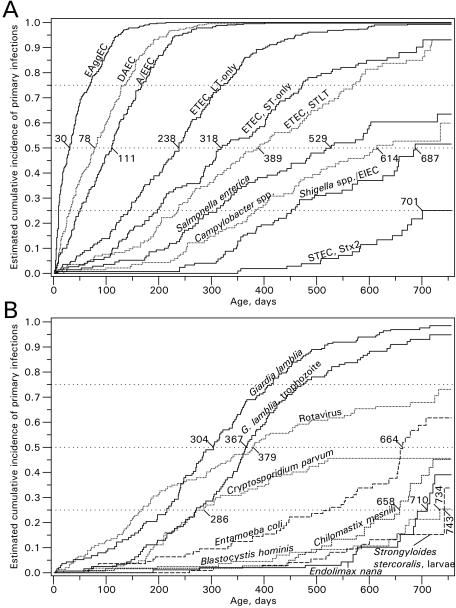
The cumulative incidence of primary infections is shown in Fig. 2A and B. EAggEC was the organism that infected children earliest in life, followed by DAEC, A/EEC, and ETEC. Within 30 days of age, 50% of the children would have experienced an infection with EAggEC.
FIG. 2.
Cumulative incidence of primary enteric infections with bacterial agents (A) and parasites and rotavirus (B) in a cohort study of 200 children monitored from birth until up to 2 years of age in Guinea-Bissau from 1996 to 1998. For the microorganisms that infected <50% of the children, the median age at which 25% of the children are infected is reported. Stx1-producing STEC, Isospora spp., and E. histolytica or E. dispar infected <25% of the children and the results are therefore not shown. The results for G. lamblia, Entamoeba coli, C. mesnilli, and Endolimax nana represent the detection of cysts, trophozoites, or both.
Pathogenicity.
Rotavirus, ST-only ETEC, Stx2-producing STEC, Isospora spp., and C. parvum were significantly associated with diarrhea (Table 1). Shigella spp. or EIEC infections tended to be associated with diarrhea (OR, 1.87; 95% confidence interval [CI], 0.85 to 4.16). The pathogenicity of STEC (OR, 2.33; 95% CI, 0.85 to 6.42) was associated with Stx2 (OR, 5.09; 95% CI, 1.53 to 16.9) rather than with Stx1 (OR, 1.11; 95% CI, 0.21 to 5.75). Eleven of the 16 Stx2 strains were also positive with the STh and the LT probes, a pathogenic subgroup of STLT ETEC (51), and could be classified both as ETEC and STEC. When excluding these from the analysis, the pathogenicity estimate for the remaining five specimens with Stx2 remained high (OR, 4.29; 95% CI, 0.86 to 21.5). When we instead adjusted for SThLT in the model, the pathogenicity estimate for Stx2 was somewhat reduced, but still statistically significant (OR, 3.42; 95% CI, 1.13 to 10.4). No interaction between the pathogenicity for SThLT ETEC and Stx2-producing STEC was observed (P = 0.98). Only one of the Stx2 strains was positive for the eae probe. EAggEC, DAEC, and A/EEC, which were the most commonly isolated organisms, were not associated with diarrhea.
When reclassifying disease status as diarrhea on any day during a 7-day interval centered on the day of the specimen collection, we found a substantially higher pathogenicity of C. parvum (OR, 4.53; 95% CI, 2.75 to 7.46). In contrast, we obtained lower pathogenicity estimates for rotavirus (OR, 5.07; 95% CI, 3.39 to 7.60) and Stx2-producing STEC (OR, 2.67; 95% CI, 0.86 to 8.30).
Effects of age, gender, and breast-feeding on pathogenicity.
For most of the microorganisms, there was no interaction between pathogenicity and age or gender. However, a significant interaction with age was found for DAEC, for which the ORs were 0.62 (95% CI, 0.47 to 0.84) for children below 1 year of age and 1.12 (95% CI, 0.87 to 1.44) for children older than 1 year (P = 0.003). For A/EEC, the pathogenicity estimates were 1.13 (95% CI, 0.90 to 1.42) for children below 1 year of age and 0.76 (95% CI, 0.56 to 1.04) for children older than 1 year (P = 0.04). For C. parvum, the pathogenicity was substantially higher in boys (OR, 5.35; 95% CI, 2.15 to 13.3) than in girls (OR, 1.04; 95% CI, 0.44 to 2.43) (P = 0.01).
Only 6.4% (769 of 11,987) of the stool specimens were from non-breast-fed children. Breast-feeding status did not confound the pathogenicity estimates. For the majority of the organisms, tendencies for an interaction between pathogenicity and breast-feeding were seen, which indicated an increased pathogenicity in non-breast-fed children. Significant interactions were seen for STLT ETEC (P < 0.001) and Salmonella (P = 0.003), which were associated with diarrhea in non-breast-fed children (OR, 4.65; 95% CI, 1.99 to 10.9; and OR, 4.08; 95% CI, 1.33 to 12.6, respectively), but not in breast-fed children (OR, 0.89; 95% CI, 0.57 to 1.41; and OR, 0.53; 95% CI, 0.24 to 1.13, respectively).
Pathogenicity during primary infections.
The pathogenicities of several enteropathogens, including rotavirus, A/EEC, LT-only ETEC, Shigella spp. or EIEC, and Isospora spp., were >20% higher for primary infections than for all infections (Table 1). No marked differences in pathogenicity between primary infection and all infections were seen for, e.g., ST-only ETEC, STLT ETEC, DAEC, and C. parvum.
Protection against reinfection.
We estimated the protection that a primary infection induced against subsequent infections with the same microorganism. For rotavirus, this protection was 52% (95% CI, 16 to 73%), while no substantial protection was apparent for the other enteropathogens. For LT-only ETEC, A/EEC, EAggEC, DAEC, G. lamblia cysts or trophozoites, G. lamblia in the trophozoitic stage, Entamoeba coli cysts or trophozoites, B. hominis, C. mesnilli, and Salmonella, there was, in fact, a statistically significant increase in the rate of subsequent infections after experiencing a primary infection.
Protection against diarrhea from reinfection.
We estimated the degree to which a primary infection protected a subsequent infection from being symptomatic. Such protection was seen for rotavirus (61%; 95% CI, −3 to 86%), LT-only ETEC (55%; 95% CI, 11 to 77%), and A/EEC (49%; 95% CI, 3 to 73%), while no substantial protection was apparent for the other enteropathogens. For Shigella spp. or EIEC, Campylobacter spp., Isospora spp., B. hominis, and the trophozoitic stage of Entamoeba coli, no symptomatic reinfections were observed. Of these, only the protection followed by infection with Shigella spp. or EIEC approached statistical significance (P = 0.11).
PAR%.
Major contributors to diarrheal disease in this population, as identified by PAR%, were, in decreasing order: rotavirus, ST-only ETEC, Isospora spp., C. parvum, Stx2-producing STEC, and Shigella spp. or EIEC (Table 2).
TABLE 2.
Enteropathogens with adjusted OR larger than unity in a cohort of children monitored from birth and up to 2 years of age in Guinea-Bissau
| Organism | PAR%a |
|---|---|
| Rotavirus | 4.61 |
| ST-only ETEC | 1.65 |
| Isospora spp. | 0.67 |
| C. parvum | 0.66 |
| Stx2-producing STEC | 0.58 |
| Shigella spp. or EIEC | 0.45 |
| B. hominis | 0.41 |
| C. mesnilli | 0.24 |
| Endolimax nana | 0.13 |
| Entamoeba histolytica or E. dispar | 0.04 |
| Entamoeba coli Trophozoite | 0.02 |
| Stx1-producing STEC | 0.01 |
| Total | 9.47 |
Results are listed in descending order of PAR%.
DISCUSSION
Rotavirus was found to be a major diarrheal pathogen. Thus, in agreement with previous studies, it seems that, despite its relatively low incidence and by virtue of its high pathogenicity, rotavirus contributes substantially to the burden of childhood diarrhea; not only in hospital settings (24, 53) but also in the community (45, 54). A primary rotavirus infection conferred protection against infection and tended to protect against diarrhea from reinfection. Results from in-depth analyses of the rotavirus infections and the associated protection are described elsewhere (15).
In line with previous studies (2, 28), ST-only ETEC strains were strongly associated with diarrhea. Whereas primary infections with ST-only ETEC did not confer any protection, primary infections with LT-only ETEC were associated with a substantial protection against diarrhea when the children were reinfected with LT-only ETEC. These observations are consistent with previous findings (13, 36). Overall, infections with LT-only ETEC were not associated with diarrhea in our study. This is in line with most studies (3, 44, 56) and at odds with others (28, 35). Primary LT-only ETEC infection, however, was associated with diarrhea, albeit only with borderline significance, supporting the notion that LT-only ETEC strains are diarrheal agents primarily in immunologically naive individuals. Results from more in-depth analyses of the ETEC infections in our cohort are presented elsewhere (51).
EAggEC strains were not associated with diarrhea in previous investigations (3, 16, 28, 44), albeit such findings are not unanimous (7, 8, 40). In the present investigation, EaggEC infections were not associated with diarrhea and accordingly did not contribute to our PAR% estimate. EAggEC strains show a considerable degree of heterogeneity, and because the pathogenic potential has been confirmed in human volunteer studies and in outbreaks, it seems likely that some but not all strains are pathogenic (41).
Overall, DAEC strains were not associated with diarrhea, and this is in accordance with the findings of other studies (21, 28, 44). Our observation of an age-dependent susceptibility to DAEC-associated diarrhea in children over 12 months of age has been found as well in other community studies (21, 28).
A/EEC strains were not associated with diarrhea overall. Primary A/EEC infections were more strongly associated with diarrhea than were all infections, and a primary infection conferred protection against diarrhea when the child was reinfected with A/EEC. The tendency to cause diarrhea only in children less than 12 months of age has also been observed in other studies (28, 29). These findings suggest that natural infections with A/EEC confer some protection against A/EEC diarrhea. It should be emphasized that, in the present study, we did not subtype A/EEC into the probably more pathogenic typical EPEC (29).
Stx2 is more important than Stx1 for the development of the hemolytic uremic syndrome (38), and Stx2 strains have also been shown to be more diarrheagenic than Stx1 in an animal model (22). The present study is the first to show that the risk of diarrhea in humans was higher when children were infected with Stx2-producing STEC than when they were infected with Stx1-producing STEC. This finding is in accordance with a recent study showing that exposure to Stx2 but not Stx1, even in the absence of STEC bacteria and the Stx receptor, induces structural changes in human gastrointestinal epithelial cells (S. Sculler, G. Frankel, and A. D. Phillips, abstr. from ESPGHAN 36th Annu. Meet., 2003). The combination of enterotoxins and Shiga-like-toxins in the same strain is unusual in humans but has been observed in strains isolated from pigs (42). In the study area, pigs roamed in close surroundings of the houses and often slept in the same room with the humans.
Overall, infections with Shigella spp. or EIEC tended to be associated with diarrhea. Primary infections with Shigella spp. or EIEC, however, were clearly associated with diarrhea and tended to induce protection against diarrhea during reinfection. Infections with Shigella spp. or EIEC were rare during the first 300 days of life and increased steadily thereafter. In a cohort study of infants, Cravioto et al. found no infections with Shigella spp. before 7 months of age (14). Another prospective cohort study in Mexico (20) found, as we did, a very low incidence of Shigella infections during the first 6 months of life. The overall incidence was 0.22 infection/child-year at risk, which is close to the incidence of 0.29 infection/child-year at risk that we estimated in the present study.
Like the coccidian protozoan C. parvum, Isospora spp. are recognized to be pathogenic in immunocomprised individuals (30). Whereas C. parvum is now a recognized diarrheal pathogen also in immunocompetent individuals (34), the pathogenicity of Isospora spp. remains unresolved. A recent study from India detected Isospora spp. in immunocompetent children with diarrhea (33), and Isospora spp. have been described as a cause of traveler's diarrhea (18). We found Isospora spp. to be associated with diarrhea, which is in line with previous findings in our study area (35). Isospora spp. should therefore be recognized as an enteropathogen for healthy children as well. The strong pathogenicity of C. parvum was confirmed in the present study. Our finding that the pathogenicity of C. parvum was higher in boys than in girls has not been described earlier and should be explored further. Because C. parvum infections are seasonal in our study area, with peak prevalences found consistently in the early rainy season of May to July (43), we might have underestimated its incidence because the study was terminated in April 1998.
Different time periods for the definition of diarrhea have been used across different cohort studies, from 1 day (2, 28), to 10 days (54), and up to 14 days (20). We investigated whether a 1-day or 7-day window for defining diarrhea led to different pathogenicity estimates. For C. parvum, which is strongly associated with an increased diarrheal burden (39) and with persistent diarrhea (35, 39), we found substantially higher pathogenicity when using a 7-day (OR, 4.53) rather than a 1-day (OR, 2.12) window. For rotavirus infections, which are associated with diarrheal episodes of relatively short duration (26), the pathogenicity estimate was slightly lower with a 7-day window (OR, 5.07) than with a 1-day (OR, 5.75) window. It therefore seems that, for some enteropathogens, the length of the time periods for defining diarrhea had an effect on the estimated pathogenicity.
Rotavirus was the only pathogen for which primary infection conferred a statistically significant protection against subsequent infections. A possible explanation for this finding is that rotavirus relies on intracellular reproduction to multiply and to be pathogenic (31), in contrast to other microbes, such as LT-only ETEC strains, which colonize the intestine without cellular invasion. From 1 year of age, the mean number of different enteropathogens per specimen remained constant, while the proportion of children with diarrhea declined. This suggests that, with increasing age, the children develop protection against diarrhea but not against infection. This is in agreement with the protection estimates determined in the present study, which showed that most of the enteropathogens did not confer protection against reinfection with the same microorganism, but some, e.g., LT-only ETEC, induced protection against diarrhea. Similar findings were reported from a cohort study of infants in Mexico (14). It is possible that some continuous infections may have been perceived as two separate infections with the same type of microorganism. Such misclassifications would contribute to reducing the estimated protection against infection and possibly also against diarrhea from infection.
Although more than half of the specimens were positive for potential enteropathogens, it is striking that we were able to explain less than 10% of the diarrheal episodes by using Levin's formula for calculating PAR%. This method, which initially was developed for studies of chronic diseases, assumes that all individuals in the study are at risk of developing disease. However, in our study population, many children were likely to be protected from symptomatic infection by passively transferred immunity from maternal antibodies and from acquired immunity, thus underestimating the pathogenicity and, therefore, the PAR%. For LT-only ETEC and other highly incident organisms, such as EAggEC, DAEC, and A/EEC, there may be a certain analogy to the thoughts of Rose, who argues that if exposure to a certain agent is homogenous within a population, even epidemiological studies will fail to detect its association with the disease (46). Hence, disease is related to other factors, such as individual susceptibility, which may depend on immunity, micronutrient status, and several unknown factors. This predicament underscores the limitations of observational studies carried out in settings with hyperendemicity of infectious agents as well as the limitations of currently available epidemiological methodology. It is likely that we underestimate the contribution of the highly incident pathogens for diarrheal burden and that the estimated overall PAR% is highly conservative.
Other factors may in addition contribute to reducing the overall PAR%. For some of the seemingly nonpathogenic organisms, important pathogenic subgroups exist, the pathogenicity of which is masked by other nonpathogenic subgroups. This is the case for LT-only ETEC, where we have recently shown that there are substantial differences in pathogenicity between different subgroups (51). Such pathogenic subgroups, although clearly of importance for diarrheal burden, are not reflected in the estimated PAR%. Although our study included the identification of a large number of potential enteropathogens, we did not include all. Several enteric viruses other than rotavirus, such as adeno, astro-, and caliciviruses, are thought to cause diarrhea in children (17). Furthermore, the weekly specimen collection schedule probably failed to identify some infections of short duration.
Even though detection of the fragile trophozoitic stage of, e.g., G. lamblia was very common, it is possible that we have underestimated its prevalence and the estimated cumulative incidence of primary infections in the present study, because it was not logistically feasible to examine freshly passed stools.
In this cohort study of children monitored from birth and up to 2 years of age, we have estimated the pathogenicity and incidence of a wide range of potential enteropathogens. Major contributors to childhood diarrhea were rotavirus, ST-only ETEC, Isospora spp., C. parvum, STEC, and Shigella spp. or EIEC. The pathogenicity of STEC was limited to Stx2, a finding that should be further explored.
Acknowledgments
We thank Gina Santos for support in the field. We gratefully acknowledge the extraordinary effort of the laboratory personnel at The National Public Health Laboratory; fieldwork supervisor Queba Djana; and the field-workers Francisco Da Silva, Carlos Sá, Adelino Bassam, Leontino Da Silva, and Domingos Pereira at the Bandim Health Project in Guinea-Bissau. We thank Henrik Jensen and Per Kragh Andersen for advice during the statistical analysis.
Financial support was provided by the European Commission-DG Research, FP3-International Cooperation STD programme (TC*-CT94-0311), Danish Council for Development Research (104.Dan.8/717, 9501931, and 91010), DANIDA, The Danish Medical Research Council, The Danish National Research Foundation, L. Meltzers Høyskolefond, and The Research Council of Norway.
REFERENCES
- 1.Aabo, S., A. Thomas, M. L. M. Hall, H. R. Smith, and J. E. Olsen. 1992. Evaluation of a Salmonella-specific DNA probe by colony hybridization using non-isotopic and isotopic labelling. APMIS 100:623-628. [DOI] [PubMed] [Google Scholar]
- 2.Abu-Elyazeed, R., T. F. Wierzba, A. S. Mourad, L. F. Peruski, B. A. Kay, M. Rao, A. M. Churilla, A. L. Bourgeois, A. K. Mortagy, S. M. Kamal, S. J. Savarino, J. R. Campbell, J. R. Murphy, A. Naficy, and J. D. Clemens. 1999. Epidemiology of enterotoxigenic Escherichia coli diarrhea in a pediatric cohort in a periurban area of lower Egypt. J. Infect. Dis. 179:382-389. [DOI] [PubMed] [Google Scholar]
- 3.Albert, M. J., S. M. Faruque, A. S. G. Faruque, P. K. B. Neogi, M. Ansaruzzaman, N. A. Bhuiyan, K. Alam, and M. S. Akbar. 1995. Controlled study of Escherichia coli diarrhoeal infections in Bangladeshi children. J. Clin. Microbiol. 33:973-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen, A. V., and D. S. Ridley. 1970. Further observations on the Formol-ether concentration technique for faecal parasites. J. Clin. Pathol. 23:545-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersen, P. K. 1993. The Kaplan-Meier estimator, p. 255-286. In Ø. Borgan, R. D. Gill, and N. Keiding (ed.), Statistical models based on counting processes. Springer-Verlag, New York, N.Y.
- 6.Baudry, B., S. J. Savarino, P. Vial, J. B. Kaper, and M. M. Levine. 1990. A sensitive and specific DNA probe to identify enteroaggregative Escherichia coli, a recently discovered diarrhoeal pathogen. J. Infect. Dis. 161:1249-1251. [DOI] [PubMed] [Google Scholar]
- 7.Bhan, M. K., P. Raj, M. M. Levine, J. B. Kaper, N. Bhandari, R. Srivastava, R. Kumar, and S. Sazawal. 1989. Enteroaggregative Escherichia coli associated with persistent diarrhoea in a cohort of rural children in India. J. Infect. Dis. 159:1061-1064. [DOI] [PubMed] [Google Scholar]
- 8.Bhatnagar, S., M. K. Bhan, H. Sommerfelt, S. Sazawal, R. Kumar, and S. Saini. 1993. Enteroaggregative Escherichia coli may be a new pathogen causing acute and persistent diarrhoea. Scand. J. Infect. Dis. 25:579-583. [DOI] [PubMed] [Google Scholar]
- 9.Bilge, S. S., C. R. Clausen, W. Lau, and S. L. Moseley. 1989. Molecular characterization of a fimbrial adhesin, F1845, mediating diffuse adherence of diarrhea-associated Escherichia coli to HEp-2 cells. J. Bacteriol. 171:4281-4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Black, R. E. 1993. Epidemiology of diarrhoeal disease: implications for control by vaccines. Vaccine 11:100-106. [DOI] [PubMed] [Google Scholar]
- 11.Blom, M., A. Meyer, P. Gerner-Smidt, K. Gaarslev, and F. Espersen. 1999. Evaluation of Statens Serum Institut enteric medium for detection of enteric pathogens. J. Clin. Microbiol. 37:2312-2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calva, J. J., G. M. Ruiz-Palacios, A. B. Lopez-Vidal, A. Ramos, and R. Bojalil. 1988. Cohort study of intestinal infection with Campylobacter in Mexican children. Lancet i:503-506. [DOI] [PubMed]
- 13.Clemens, J. D., D. A. Sack, J. R. Harris, J. Chakraborty, P. K. Neogy, B. Stanton, N. Huda, M. U. Khan, B. A. Kay, and M. R. Khan. 1988. Cross-protection by B subunit-whole cell cholera vaccine against diarrhea associated with heat-labile toxin-producing enterotoxigenic Escherichia coli: results of a large-scale field trial. J. Infect. Dis. 158:372-377. [DOI] [PubMed] [Google Scholar]
- 14.Cravioto, A., R. A. Reyes, F. Trujillo, F. Uribe, A. Navarro, J. de la Roca, J. Hernandez, G. Perez, and V. Vazquez. 1990. Risk of diarrhea during the first year of life associated with initial and subsequent colonization by specific enteropathogens. Am. J. Epidemiol. 131:886-904. [DOI] [PubMed] [Google Scholar]
- 15.Fischer, T. K., P. Valentiner-Branth, H. Steinsland, M. Perch, G. Santos, P. Aaby, K. Mølbak, and H. Sommerfelt. 2002. Protective immunity after natural rotavirus infection. A community cohort study of newborn children in Guinea-Bissau, West Africa. J. Infect. Dis. 186:593-597. [DOI] [PubMed] [Google Scholar]
- 16.Gascón, J., M. Vargas, D. Schellenberg, H. Urassa, C. Casals, E. Kahigwa, J. J. Aponte, H. Mshinda, and J. Vila. 2000. Diarrhea in children under 5 years of age from Ifakara, Tanzania: a case-control study. J. Clin. Microbiol. 38:4459-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glass, R. I., J. Bresee, B. Jiang, J. Gentsch, T. Ando, R. Fankhauser, J. Noel, U. Parashar, B. Rosen, and S. S. Monroe. 2001. Gastroenteritis viruses: an overview. Novartis Found. Symp. 238:5-19. [DOI] [PubMed] [Google Scholar]
- 18.Goodgame, R. 2003. Emerging causes of traveler's diarrhea: Cryptosporidium, Cyclospora, Isospora, and microsporidia. Curr. Infect. Dis. Rep. 5:66-73. [DOI] [PubMed] [Google Scholar]
- 19.Goossens, H., L. Vlaes, L. Galand, C. Van den Borre, and J.-P. Butzler. 1989. Semisolid blood-free selective-motility medium for the isolation of campylobacters from stool specimens. J. Clin. Microbiol. 27:1077-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guerrero, L., J. J. Calva, A. L. Morrow, F. R. Velazquez, F. Tuz-Dzib, Y. Lopez-Vidal, H. Ortega, H. Arroyo, T. G. Cleary, and L. K. Pickering. 1994. Asymptomatic Shigella infections in a cohort of Mexican children younger than two years of age. Pediatr. Infect. Dis. J. 13:597-602. [DOI] [PubMed] [Google Scholar]
- 21.Gunzburg, S. T., B. J. Chang, S. J. Elliott, V. Burke, and M. Gracey. 1993. Diffuse and enteroaggregative patterns of adherence of enteric Escherichia coli isolated from aboriginal children from the Kimberley region of Western Australia. J. Infect. Dis. 167:755-758. [DOI] [PubMed] [Google Scholar]
- 22.Hammermueller, J., S. Kruth, J. Prescott, and C. Gyles. 1995. Detection of toxin genes in Escherichia coli isolated from normal dogs and dogs with diarrhea. Can. J. Vet. Res. 59:265-270. [PMC free article] [PubMed] [Google Scholar]
- 23.Henriksen, S. A., and J. F. L. Pohlenz. 1981. Staining of cryptosporidia by a modified Ziehl-Neelsen technique. Acta Vet. Scand. 22:594-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jain, V., U. D. Parashar, R. I. Glass, and M. K. Bhan. 2001. Epidemiology of rotavirus in India. Indian J. Pediatr. 68:855-862. [DOI] [PubMed] [Google Scholar]
- 25.Jerse, A. E., and J. B. Kaper. 1991. The eae gene of enteropathogenic Escherichia coli encodes a 94-kilodalton membrane protein, the expression of which is influenced by the EAF plasmid. Infect. Immun. 59:4302-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kapikian, A. Z., Y. Hoshino, and R. M. Chanock. 2001. Rotaviruses, p. 1788-1834. In Fields virology, 4th ed. Lippincott-Williams & Wilkins, Philadelphia, Pa.
- 27.Kosek, M., C. Bern, and R. L. Guerrant. 2003. The global burden of diarrhoeal disease, as estimated from studies published between 1992 and 2000. Bull. W. H. O. 81:197-204. [PMC free article] [PubMed] [Google Scholar]
- 28.Levine, M., C. Ferrecoccio, V. Prado, M. Cayazzo, P. Abrego, J. Martinez, L. Maggi, M. M. Baldini, W. Martin, D. Maneval, B. Kay, L. Guers, H. Lior, S. Wasserman, and P. Nataro. 1993. Epidemiologic studies of Escherichia coli diarrheal infections in a low socioeconomic level peri-urban community in Santiago, Chile. Am. J. Epidemiol. 138:849-869. [DOI] [PubMed] [Google Scholar]
- 29.Levine, M. M., and R. Edelman. 1984. Enteropathogenic Escherichia coli of classic serotypes associated with infant diarrhea: epidemiology and pathogenesis. Epidemiol. Rev. 6:31-51. [DOI] [PubMed] [Google Scholar]
- 30.Lindsay, D. S., J. P. Dubey, and B. L. Blagburn. 1997. Biology of Isospora spp. from humans, nonhuman primates, and domestic animals. Clin. Microbiol. Rev. 10:19-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lundgren, O., and L. Svensson. 2001. Pathogenesis of rotavirus diarrhea. Microbes Infect. 3:1145-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mata, L. J., J. J. Urrutia, and J. E. Gordon. 1967. Diarrhoeal disease in a cohort of Guatemala village children observed from birth to age two years. Trop. Geogr. Med. 19:247-257. [PubMed] [Google Scholar]
- 33.Mirdha, B. R., S. K. Kabra, and J. C. Samantray. 2002. Isosporiasis in children. Indian Pediatr. 39:941-944. [PubMed] [Google Scholar]
- 34.Mølbak, K., I. M. Lisse, N. Højlyng, and P. Aaby. 1994. Severe cryptosporidiosis in children with normal T-cell subsets. Parasite Immunol. 16:275-277. [DOI] [PubMed] [Google Scholar]
- 35.Mølbak, K., N. Wested, N. Højlyng, F. Scheutz, A. Gottschau, P. Aaby, and A. P. J. da Silva. 1994. The etiology of early childhood diarrhea: a community study from Guinea-Bissau. J. Infect. Dis. 169:581-587. [DOI] [PubMed] [Google Scholar]
- 36.Moon, H. W., A. L. Baetz, and R. A. Giannella. 1983. Immunization of swine with heat-stable Escherichia coli enterotoxin coupled to a carrier protein does not protect suckling pigs against an Escherichia coli strain that produces heat-stable enterotoxin. Infect. Immun. 39:990-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morrow, A. L., R. R. Reves, M. S. West, M. L. Guerrero, G. M. Ruiz-Palacios, and L. K. Pickering. 1992. Protection against infection with Giardia lamblia by breast-feeding in a cohort of Mexican infants. J. Pediatr. 121:363-370. [DOI] [PubMed] [Google Scholar]
- 38.Nakao, H., and T. Takeda. 2000. Escherichia coli Shiga toxin. J. Nat. Toxins 9:299-313. [PubMed] [Google Scholar]
- 39.Newman, R. D., C. L. Sears, S. R. Moore, J. P. Nataro, T. Wuhib, D. A. Agnew, R. L. Guerrant, and A. A. Lima. 1999. Longitudinal study of Cryptosporidium infection in children in northeastern Brazil. J. Infect. Dis. 180:167-175. [DOI] [PubMed] [Google Scholar]
- 40.Okeke, I. N., A. Lamikanra, H. Steinrück, and J. B. Kaper. 2000. Characterization of Escherichia coli strains from cases of childhood diarrhea in provincial southwestern Nigeria. J. Clin. Microbiol. 38:7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okeke, I. N., and J. P. Nataro. 2001. Enteroaggregative Escherichia coli. Lancet Infect. Dis. 1:304-312. [DOI] [PubMed] [Google Scholar]
- 42.Parma, A. E., M. E. Sanz, M. R. Vinas, M. E. Cicuta, J. E. Blanco, S. I. Boehringer, M. M. Vena, W. R. Roibon, M. C. Benitez, J. Blanco, and M. Blanco. 2000. Toxigenic Escherichia coli isolated from pigs in Argentina. Vet. Microbiol. 72:269-276. [DOI] [PubMed] [Google Scholar]
- 43.Perch, M., M. Sodemann, M. S. Jakobsen, P. Valentiner-Branth, H. Steinsland, T. K. Fischer, D. D. Lopes, P. Aaby, and K. Mølbak. 2001. Seven years' experience with Cryptosporidium parvum in Guinea-Bissau, West Africa. Ann. Trop. Paediatr. 21:313-318. [DOI] [PubMed] [Google Scholar]
- 44.Porat, N., A. Levy, D. Fraser, R. J. Deckelbaum, and R. Dagan. 1998. Prevalence of intestinal infections caused by diarrheagenic Escherichia coli in Bedouin infants and young children in Southern Israel. Pediatr. Infect. Dis. J. 17:482-488. [DOI] [PubMed] [Google Scholar]
- 45.Raul, V. F., J. J. Calva, G. M. Lourdes, D. Mass, R. I. Glass, L. K. Pickering, and G. M. Ruiz-Palacios. 1993. Cohort study of rotavirus serotype patterns in symptomatic and asymptomatic infections in Mexican children. Pediatr. Infect. Dis. J. 12:54-61. [DOI] [PubMed] [Google Scholar]
- 46.Rose, G. 2001. Sick individuals and sick populations. Int. J. Epidemiol. 30:427-432. [DOI] [PubMed] [Google Scholar]
- 47.Rothman, K., and S. Greenland. 1998. Modern epidemiology, p. 29-47. Lippincott-Raven, Philadelphia, Pa.
- 48.Snyder, J. D., and M. H. Merson. 1982. The magnitude of the global problem of acute diarrhoeal disease: a review of active surveillance data. Bull. W. H. O. 60:605-613. [PMC free article] [PubMed] [Google Scholar]
- 49.Sommerfelt, H., H. M. S. Grewal, and M. K. Bhan. 1990. Simplified and accurate nonradioactive polynucleotide gene probe assay for identification of enterotoxigenic Escherichia coli. J. Clin. Microbiol. 28:49-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steinsland, H., P. Valentiner-Branth, H. M. S. Grewal, W. Gaastra, K. Mølbak, and H. Sommerfelt. 2003. Development and evaluation of genotypic assays for the detection and characterization of enterotoxigenic Escherichia coli. Diagn. Microbiol. Infect. Dis. 45:97-105. [DOI] [PubMed] [Google Scholar]
- 51.Steinsland, H., P. Valentiner-Branth, M. Perch, F. Dias, T. K. Fischer, P. Aaby, K. Mølbak, and H. Sommerfelt. 2002. Enterotoxigenic Escherichia coli infections and diarrhea in a cohort of young children in Guinea-Bissau. J. Infect. Dis. 186:1740-1747. [DOI] [PubMed] [Google Scholar]
- 52.Szklo, M., and F. J. Nieto. 2000. Epidemiology beyond the basics. Aspen Publishers, Inc, Gaithersburg, Md.
- 53.Unicomb, L. E., P. E. Kilgore, S. G. Faruque, J. D. Hamadani, G. J. Fuchs, M. J. Albert, and R. I. Glass. 1997. Anticipating rotavirus vaccines: hospital-based surveillance for rotavirus diarrhea and estimates of disease burden in Bangladesh. Pediatr. Infect. Dis. J. 16:947-951. [DOI] [PubMed] [Google Scholar]
- 54.Velazquez, F. R., D. O. Matson, J. J. Calva, L. Guerrero, A. L. Morrow, S. Carter-Campbell, R. I. Glass, M. K. Estes, L. K. Pickering, and G. M. Ruiz-Palacios. 1996. Rotavirus infections in infants as protection against subsequent infections. N. Engl. J. Med. 335:1022-1028. [DOI] [PubMed] [Google Scholar]
- 55.Venkatesan, M. M., J. M. Buysse, and D. J. Kopecko. 1989. Use of Shigella flexneri ipaC and ipaH gene sequences for the general identification of Shigella spp. and enteroinvasive Escherichia coli. J. Clin. Microbiol. 27:2687-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Viboud, G. I., M. J. Jouve, N. Binzstein, M. Vergara, M. Rivas, M. Quiroga, and A. Svennerholm. 1999. Prospective cohort study of enterotoxigenic Escherichia coli infections in Argentinian children. J. Clin. Microbiol. 37:2829-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Willshaw, G. A., H. R. Smith, S. M. Scotland, A. M. Field, and B. Rowe. 1987. Heterogeneity of Escherichia coli phages encoding Vero cytotoxins: comparison of cloned sequences determining VT1 and VT2 and development of specific gene probes. J. Gen. Microbiol. 133:1309-1317. [DOI] [PubMed] [Google Scholar]