Abstract
We have evaluated over a period of 18 months the use of 16S ribosomal DNA (rDNA) sequence analysis as a means of identifying aerobic gram-positive rods in the clinical laboratory. Two collections of strains were studied: (i) 37 clinical strains of gram-positive rods well identified by phenotypic tests, and (ii) 136 clinical isolates difficult to identify by standard microbiological investigations, i.e., identification at the species level was impossible. Results of molecular analyses were compared with those of conventional phenotypic identification procedures. Good overall agreement between phenotypic and molecular identification procedures was found for the collection of 37 clinical strains well identified by conventional means. For the 136 clinical strains which were difficult to identify by standard microbiological investigations, phenotypic characterization identified 71 of 136 (52.2%) isolates at the genus level; 65 of 136 (47.8%) isolates could not be discriminated at any taxonomic level. In comparison, 16S rDNA sequencing identified 89 of 136 (65.4%) isolates at the species level, 43 of 136 (31.6%) isolates at the genus level, and 4 of 136 (2.9%) isolates at the family level. We conclude that (i) rDNA sequencing is an effective means for the identification of aerobic gram-positive rods which are difficult to identify by conventional techniques, and (ii) molecular identification procedures are not required for isolates well identified by phenotypic investigations.
Accurate and rapid identification of isolated microorganisms is a key issue in the clinical microbiology laboratory. Identification is traditionally based on phenotypic assessments. However, phenotypic characteristics such as growth factor requirements, fermentation and assimilation of carbohydrates, morphology, and staining behavior are subject to variation and dependent on individual interpretation and expertise.
Over the past few years, genotypic identification procedures have increasingly received attention as an alternative or complement to conventional phenotypic methods (2). Genotypic techniques involve the amplification of a phylogenetically informative target, such as the small-subunit (16S) rRNA gene (28). Broad-range primers that recognize 16S ribosomal DNA (rDNA) sequences conserved among a wide variety of bacteria are used to amplify species-specific variable regions of interest (1, 9). Sequence determination and comparative database searches allow the unknown isolate to be assigned to a group of bacteria (15). 16S rDNA sequencing has been used extensively for bacterial phylogeny (17, 29), for the identification of uncultivated bacterial pathogens (20, 27), and for the identification of cultural isolates (18). However, few studies so far have reported on the use of rDNA sequencing for the identification of bacterial isolates in a more systematic fashion (6, 8, 21, 23, 24).
In this study, we have evaluated the suitability of 16S rDNA sequencing for the identification of aerobic gram-positive rods under routine conditions in a clinical microbiology laboratory. One hundred thirty-six clinical isolates for which conventional phenotypic identification did not result in species identification were investigated by sequence analysis. In addition, 37 randomly selected clinical isolates that had been well-identified by standard microbiological procedures were used to compare the accuracy of molecular identification procedures with those of conventional identification methods.
MATERIALS AND METHODS
Clinical isolates.
From October 2000 to April 2002, we prospectively analyzed all aerobic gram-positive rods (n = 136) that posed problems in identification, i.e., those for which no species identification was achieved by standard microbiological investigations according to the procedure proposed by Von Graevenitz and Funke (25). Thirty-seven strains that were well-identified by standard microbiological investigations, i.e., those identified at the species level, were used to study the concordance of conventional and molecular identification procedures. All strains included in this study were isolated from clinical specimens.
Conventional methods.
Aerobic gram-positive rods were identified according to the method of Von Graevenitz and Funke (25) by means of reactions including the following: catalase; acid production from glucose, maltose, sucrose, mannitol, and xylose in semisolid cystine-Trypticase agar medium; motility; nitrate reduction; hydrolysis of urea; hydrolysis of esculin; CAMP reaction; and a test for lipophilia for catalase-positive isolates.
Sequencing of 16S rDNA.
DNA was extracted using enzymatic lysis and alkaline hydrolysis. A loopful of bacterial cells was lysed in 200 μl of lysis buffer (0.05 M Tris-HCl, 1 mM EDTA, pH 7.5) containing 0.5 mg of lysozyme (Sigma-Aldrich Chemie GmbH, Schnelldorf, Germany) by incubation for 1 h at 37°C. After addition of 10 μl of each of 1 M NaOH and 10% sodium dodecyl sulfate, the mixture was incubated at 95°C for 10 min and neutralized with 10 μl of 1 M HCl. Nucleic acids were then purified using the QIAamp DNA blood mini kit (Qiagen, Basel, Switzerland), resulting in a sample volume of 100 μl.
An 800-bp 16S rDNA fragment, corresponding to Escherichia coli positions 10 to 806 (4), was amplified using primers BAK11w (5′-AGTTTGATC[A/C]TGGCTCAG) and BAK2 (5′-GGACTAC[C/T/A]AGGGTATCTAAT) (13). Cycling parameters included an initial denaturation for 5 min at 95°C, 40 cycles of 1 min at 94°C, 1 min at 48°C, and 1 min at 72°C, and a final extension for 10 min at 72°C. Five microliters of the DNA extract was used for amplification in a total volume of 50 μl containing 1.25 U of AmpliTaq DNA polymerase LD (Applied Biosystems, Rotkreuz, Switzerland), and the appropriate buffer. Amplicons were purified with the QIAquick PCR purification kit (Qiagen AG, Basel, Switzerland) and sequenced with forward primer BAK11w 0010-S-18 using the BigDye kit and an automatic DNA sequencer (ABI Prism 310 Genetic Analyzer; Applied Biosystems).
Sequence analysis.
16S rDNA sequences were compared with those available in the GenBank, EMBL, and DDBJ databases using a two-step procedure. A first search was performed with the FASTA algorithm of the Wisconsin GCG program package (7). All positions showing differences to the best scoring reference sequence were visually inspected in the electropherogram, and the sequence was corrected if necessary. Thereafter, a second search was done using BLASTN. Undetermined nucleotides (designated with an N) in either the determined sequence or the reference sequence were counted as matches. The mean length of the sequences (± the standard deviation) after manual editing was 398 ± 89 nucleotides containing 1.6 ± 2.4 undetermined (N) positions.
Criteria for identification.
For identification at the genus or species level, the following criteria were used: (i) when the comparison of the determined sequence with a reference sequence of a classified species yielded a similarity score that was ⩾99%, the unknown isolate was assigned to this species; (ii) when the score was <99% and ⩾95%, the unknown isolate was assigned to the corresponding genus; and (iii) when the score was <95%, the unknown isolate was assigned to a family. If the unknown isolate was assigned to a species (more than 99% sequence similarity to a classified species) and the second classified species in the scoring list showed less than 0.5% additional sequence divergence, this was marked as a “species with low demarcation to next species.”
RESULTS
Comparison of molecular and phenotypic identification procedures: conventionally well-identified isolates.
A collection of 37 strains, well identified at the species level by phenotypic investigations, was used for comparison and evaluation of the methods. A total of 33 of 37 strains were assigned to the same species by phenotypic and molecular identification procedures (Tables 1 and 2); of these, 6 were identified as a species with low demarcation to the next species, i.e., less than 0.5% additional sequence difference to another sequence entry.
TABLE 1.
Molecular versus conventional identification
| Conventional identification | No. (%) identified by molecular methods
|
|||
|---|---|---|---|---|
| Taxonomic level | No. inves- tigated | Iden- tical | Molecular methods more discriminativea | Conventional methods more discriminativea |
| Conventionally well- identified isolates | ||||
| Species | 37 | 33 (89.2) | 2 (5.4) | 2 (5.4)b |
| Prospective study (con- ventionally difficult to identify isolates) | ||||
| Genus | 71 | 21 (29.6) | 48 (67.6) | 2 (2.8)c |
| No identification | 65 | 0 (0.0) | 65 (100.0)d | 0 (0.0) |
Sequence analysis identified one strain as Actinomyces sp. and one strain as Rothia sp.
Sequence analysis did not identify these strains at the genus level.
By 16S rDNA sequencing 43 isolates were assigned to a species, 20 isolates were assigned to a genus, and 2 isolates were assigned to a family.
TABLE 2.
Molecular versus conventional identification for 37 isolates identified at the species level by conventional means
| Conventional identification | No. | Molecular identification
|
||
|---|---|---|---|---|
| Result | Difference (%) from reference sequence | Reference sequence | ||
| Identical species assignment by phenotypic and molecular identification | ||||
| Actinomyces israelii | Actinomyces israelii | 0.5 | A. israelii | |
| Actinomyces meyeri | Actinomyces meyeri | 0.0 | A. meyeri | |
| Actinomyces neuii subsp. neuii | Actinomyces neuii | 0.0 | A. neuii | |
| Actinomyces turicensis | Actinomyces turicensis | 0.8 | A. turicensis | |
| Corynebacterium accolens | Corynebacterium accolens | 0.2 | C. accolens | |
| Corynebacterium afermentans | Corynebacterium afermentans | 0.2 | C. afermentans | |
| Corynebacterium diphtheriae | 8 | Corynebacterium diphtheriae | 0.0-0.3 | C. diphtheriae |
| Corynebacterium group G | Corynebacterium group G-2 | 0.0 | Corynebacterium group G-2 | |
| Corynebacterium jeikeium | Corynebacterium jeikeium | 0.0 | C. jeikeium | |
| C. pseudodiphtheriticum | 2 | C. pseudodiphtheriticum | 0.0 | C. pseudodiphtheriticum |
| C. pseudotuberculosis | C. pseudotuberculosis | 0.0 | C. pseudotuberculosis | |
| Corynebacterium striatum | 2 | Corynebacterium striatum | 0.0 | C. striatum |
| Corynebacterium ulcerans | 2 | Corynebacterium ulcerans | 0.2 | C. ulcerans |
| Dermabacter hominis | Dermabacter hominis | 0.0 | D. hominis | |
| Gardnerella vaginalis | Gardnerella vaginalis | 0.0 | G. vaginalis | |
| Listeria monocytogenes | Listeria monocytogenes | 0.3 | L. monocytogenes | |
| Nocardia asteroides | Nocardia asteroides | 0.0 | N. asteroides | |
| Nocardia asteroides | N. asteroides/cyriacigeorgici | 0.0 | N. asteroides/cyriacigeorgici | |
| Propionibacterium acnes | 3 | Propionibacterium acnes | 0.0 | P. acnes |
| Rothia dentocariosa | 2 | Rothia dentocariosa | 0.0-0.2 | R. dentocariosa |
| Conventional identification more discriminative | ||||
| Actinomyces odontolyticus | Actinomyces sp. | 0.3/1.5 | Actinomyces sp./A. odontolyticus | |
| Rothia dentocariosa | Rothia sp. | 0.0/1.5 | Rothia sp./R. dentocariosa | |
| Assignment to different taxa by phenotypic and molecular identification | ||||
| Corynebacterium macginleyi | Corynebacterium group G-2 | 0.2 | Corynebacterium group G-2 | |
| Nocardia brasiliensis | Streptomyces albidoflavus | 0.0 | S. albidoflavus | |
Discrepant results were found in 2 of 37 strains (Tables 1 and 2). In one case, conventional identification resulted in Corynebacterium macginleyi, whereas sequence comparison with public databases resulted in 99.8% similarity with Corynebacterium sp. CDC group G2 and 96.6% similarity with C. macginleyi. According to Stackebrandt and Goebel (22), 16S rDNA similarities of less than 97% indicate that isolates belong to different species. Although only partial sequences were used here, it was thus assumed that this isolate does not belong to C. macginleyi (sequence similarity below 97%); further investigations would be necessary to analyze this minor discrepancy between phenotypic and molecular identification. In the other case, conventional identification resulted in Nocardia brasiliensis, whereas sequence comparison with public databases resulted in 100% sequence identity with Streptomyces albidoflavus (major discrepancy). Upon reanalysis, it was found that conventional identification was incorrect due to false-positive acid-fast staining.
In 2 of 37 cases, molecular identification was less discriminative than conventional identification (Tables 1 and 2). In one case, conventional identification resulted in Actinomyces odontolyticus; the 16S rDNA sequence determined showed 99.7% similarity with an unclassified Actinomyces sp. and 98.5% with A. odontolyticus. In the other case, the result of conventional identification was Rothia dentocariosa; the 16S rDNA sequence determined showed 100% identity with an unclassified Rothia sp. and 98.5% identity with R. dentocariosa. According to the criteria defined for molecular identification, these two isolates were not assigned to an established species by sequence analysis because the similarity values obtained were below the defined threshold. It was assumed that the phenotypic approach correctly identified the isolates at the species level.
Comparison of molecular and phenotypic identification procedures: a prospective study for isolates difficult to identify by conventional investigations.
By conventional identification methods, 71 of the 136 gram-positive rods investigated were identified at the genus level, and 65 gram-positive rods could not be further identified (Table 1).
In 44 of 71 cases identified at the genus level by phenotypic methods, 16S rDNA sequencing allowed species assignment; in all of these 44 cases the species assignment did not contradict the genus determined conventionally (Tables 1 and 3). As an example, conventional procedures identified an isolate as Actinomyces sp., whereas molecular methods resulted in a sequence that was identical with that of A. naeslundii. In 4 of 71 cases, conventional identification was incorrect. In the first of these four cases, phenotypic identification resulted in Bacillus sp., whereas sequencing revealed Paenibacillus sp.; by conventional methods, Bacillus sp. is not differentiated from Paenibacillus sp. (minor discrepancy). In the second case, phenotypic identification resulted in Actinomyces sp., whereas sequencing revealed Actinobaculum sp.; by conventional methods, Actinomyces sp. is not differentiated from Actinobaculum sp. (minor discrepancy). In the third case, conventional methods misidentified an isolate of Corynebacterium mucefaciens as Microbacterium sp. due to misinterpretation of the carbohydrate metabolism (major discrepancy). In the fourth case, phenotypic identification resulted in Corynebacterium sp., whereas sequencing revealed Propionibacterium acnes, which by the production of propionic acid can be differentiated from Corynebacterium sp. (in this case, the search for propionic acid had been neglected; major discrepancy).
TABLE 3.
Molecular identification for 71 isolates identified by conventional procedures at the genus level
| Conventional identification | No. | Molecular identification
|
||
|---|---|---|---|---|
| Result | Difference (%) from reference sequence | Reference sequence | ||
| Molecular identification at the species level | ||||
| Actinomyces sp. | Actinomyces europaeus | 0.4 | A. europaeus | |
| Actinomyces sp. | Actinomyces georgiae | 0.5 | A. georgiae | |
| Actinomyces sp. | 2 | Actinomyces naeslundii | 0.0 | A. naeslundii |
| Actinomyces sp. | 3 | Actinomyces odontolyticus | 0.3-0.9 | A. odontolyticus |
| Actinomyces sp. | 4 | Actinomyces radingae | 0.8-0.9 | A. radingae |
| Actinomyces sp. | Actinomyces turicensis | 0.6 | A. turicensis | |
| Actinomyces sp. | 2 | Actinomyces urogenitalis | 0.2-0.3 | A. urogenitalis |
| Arcanobacterium sp. | Arcanobacterium bernardiae | 0.2 | A. bernardiae | |
| Corynebacterium sp. | C. pseudotuberculosis | 0.0 | C. pseudotuberculosis | |
| Corynebacterium sp. | Corynebacterium amycolatum | 0.5 | C. amycolatum | |
| Corynebacterium sp. | 3 | Corynebacterium coyleiae | 0.0-0.7 | C. coyleiae |
| Corynebacterium sp. | Corynebacterium group G-2 | 0.4 | Corynebacterium group G-2 | |
| Corynebacterium sp. | Corynebacterium mucifaciens | 0.0 | C. mucifaciens | |
| Corynebacterium sp. | 2 | C. pseudogenitalium | 0.0 | C. pseudogenitalium |
| Corynebacterium sp. | Corynebacterium striatum | 0.0 | C. striatum | |
| Corynebacterium sp. | Corynebacterium minutissimum | 0.5 | C. minutissimum | |
| Lactobacillus sp. | Lactobacillus gasseri | 0.0 | L. gasseri | |
| Lactobacillus sp. | 2 | Lactobacillus rhamnosus | 0.0 | L. rhamnosus |
| Listeria sp. | Listeria ivanovii | 0.0 | L. ivanovii | |
| Nocardia asteroides complex | 2 | Nocardia asteroides | 0.0 | N. asteroides |
| Nocardia asteroides complex | Nocardia brasiliensis | 0.6 | N. brasiliensis | |
| Nocardia asteroides complex | Nocardia farcinica | 0.0 | N. farcinica | |
| Nocardia asteroides complex | Nocardia pseudosporangifera | 0.0 | N. pseudosporangifera | |
| Nocardia sp. | N. asteroides/translavensis | 0.2 | N. asteroides/translavensis | |
| Nocardia sp. | Nocardia beijingensis | 0.5 | N. beijingensis | |
| Nocardia sp. | 3 | Nocardia farcinica | 0.0 | N. farcinica |
| Nocardia sp. | Nocardia otitidiscaviarum | 0.5 | N. otitidiscaviarum | |
| Rhodococcus sp. | R. erythropolis/erythrehus | 0.0 | R. erythropolis/erythrehus | |
| Rothia sp. | Rothia dentocariosa | 0.2 | R. dentocariosa | |
| Streptomyces sp. | S. caviscabies/setonii/lavendulae | 0.0 | S. caviscabies/setonii/lavendulae | |
| Molecular identification at the genus level | ||||
| Actinomyces sp. | Actinomyces sp. | 3.5 | A. graevenitzii | |
| Actinomyces sp. | Actinomyces sp. | 1.3 | A. odontolyticus | |
| Actinomyces sp. | 5 | Actinomyces sp. | 1.1-2.9 | A. radingae |
| Actinomyces sp. | Actinomyces sp. | 1.1 | A. viscosus | |
| Actinomyces sp. | 2 | Actinomyces sp. | 0.0/1.9 | Actinomyces sp./A. viscosus |
| Actinomyces sp. | Actinomyces sp. | 0.0/3.7 | Actinomyces sp./A. naeslundii | |
| Corynebacterium sp. | 2 | Corynebacterium sp. | 1.7-2.0 | C. jeikeium |
| Corynebacterium sp. | Corynebacterium sp. | 1.2 | C. kroppenstedtii | |
| Corynebacterium sp. | Corynebacterium sp. | 0.0/6.0 | Corynebacterium sp./C. thomssenii | |
| Corynebacterium sp. | Corynebacterium sp. | 2.8/6.0 | Corynebacterium sp./C. thomssenii | |
| Corynebacterium sp. | Corynebacterium sp. | 1.4/2.6 | Corynebacterium sp./C. variabilis | |
| Nocardia sp. | Nocardia sp. | 1.0 | N. flavorosea | |
| Propionibacterium sp. | Propionibacterium sp. | 1.9 | P. propionicus | |
| Streptomyces sp. | 2 | Streptomyces sp. | 0.0 | S. albidoflavus/griseus/somaliensis/albus |
| Molecular identification at the family level | ||||
| Actinomyces sp. | Actinobacteriaceae | 5.4 | Actinomyces sp. | |
| Actinomyces sp. | Actinobacteriaceae | 5.6 | Actinomyces bovis | |
| Assignment to different taxa by phenotypic and molecular identification | ||||
| Actinomyces sp. | Actinobaculum sp. | 4.6 | A. schalii | |
| Bacillus sp. | Paenibacillus sp. | 0.0/7.3 | Paenibacillus sp./P. lautus | |
| Corynebacterium sp. | Propionibacterium acnes | 0.0 | P. acnes | |
| Microbacterium sp. | Corynebacterium mucifaciens | 0.0 | C. mucifaciens | |
In 2 of 71 cases conventional methods were more discriminative. In these cases the isolates were identified as Actinomyces sp. by phenotypic methods; 16S rDNA sequence determination showed 94.4% similarity to A. bovis in one case and 94.6% similarity to an unclassified Actinomyces sp. in the other case. Based on our defined criteria for molecular identification, these values are below the threshold value for genus assignment; these two isolates were thus reported as belonging to the family Actinobacteriaceae.
In 21 of 71 strains identified at the genus level by phenotypic methods, 16S rDNA sequencing did not yield more-discriminative results, i.e., the isolate was assigned to the same genus without further species assignment (Tables 1 and 3).
Of the 65 strains which could not be assigned to a genus by conventional identification procedures, molecular methods allowed identification in 63 cases; 43 strains were identified at the species level, and 20 strains were identified at the genus level (Tables 1 and 4). In 2 of 65 cases a genus assignment was not possible by molecular methods. In one of these cases, the sequence determined showed 93.0% sequence similarity to Corynebacterium bovis, and the isolate was thus reported as belonging to the family Corynebacteriaceae. In the other case the sequence determined showed 98.7% sequence similarity to an unclassified Microbacterium sp., 98.2% sequence similarity to Leifsonia poae, and 96.9% sequence similarity to Clavibacter xyli; the isolate was reported as belonging to the family Microbacteriaceae.
TABLE 4.
Molecular identification for 65 isolates identified by conventional procedures as gram-positive rods
| Result | No. | Difference (%) from reference sequence | Reference sequence |
|---|---|---|---|
| Molecular identification at the species level | |||
| Actinomyces biruadii | 0.7 | Actinomyces biruadii | |
| Actinomyces europaeus | 0.0 | Actinomyces europaeus | |
| Actinomyces hordeovulneris | 0.4 | Actinomyces hordeovulneris | |
| Actinomyces odontolyticus | 0.3 | Actinomyces odontolyticus | |
| Actinomyces turicensis | 4 | 0.0-0.6 | Actinomyces turicensis |
| Arcanobacterium bernardiae | 0.3 | Arcanobacterium bernardiae | |
| Atopobium parvulum | 0.0 | Atopobium parvulum | |
| Bifidobacterium breve | 0.2 | Bifidobacterium breve | |
| Bifidobacterium infantis/longus | 0.6 | Bifidobacterium infantis/longus | |
| Corynebacterium afermentans | 0.5 | Corynebacterium afermentans | |
| Corynebacterium amycolatum | 0.2 | Corynebacterium amycolatum | |
| Corynebacterium asperum | 0.0 | Corynebacterium asperum | |
| Corynebacterium coyleiae | 0.0 | Corynebacterium coyleiae | |
| Corynebacterium kroppenstedtii | 0.0 | C. kroppenstedtii | |
| Corynebacterium pseudodiphtheriticum | 0.0 | C. pseudodiphtheriticum | |
| Corynebacterium riegelii | 2 | 0.0-0.6 | Corynebacterium riegelii |
| Dermabacter hominis | 0.0 | Dermabacter hominis | |
| Dietzia maris | 0.2 | Dietzia maris | |
| Eggerthella lenta | 0.0 | Eggerthella lenta | |
| Gardnerella vaginalis | 4 | 0.0-0.9 | Gardnerella vaginalis |
| Gordonia terrae | 0.7 | Gordonia terrae | |
| Gordonia rubripertinctus | 0.2 | Gordonia rubripertinctus | |
| Lactobacillus fermentans | 0.2 | Lactobacillus fermentans | |
| Lactobacillus gasseri | 0.3 | Lactobacillus gasseri | |
| Lactobacillus sake | 0.5 | Lactobacillus sake | |
| Microbacterium lacticum/aurum | 0.8 | Microbacterium lacticum/aurum | |
| Mycobacterium fortuitum group | 0.0 | M. fortuitum/septicum/peregrinum | |
| Nocardia farcinica | 0.0 | Nocardia farcinica | |
| Paenibacillus lautus | 0.2 | Paenibacillus lautus | |
| Propionibacterium acnes | 0.0 | Propionibacterium acnes | |
| Propionibacterium propionicum | 0.0 | Propionibacterium propionicum | |
| Rhodococcus corynebacteroides | 0.2 | Rhodococcus corynebacteroides | |
| Streptomyces griseus | 0.0 | Streptomyces griseus | |
| Streptomyces lincolnensis | 0.0 | Streptomyces lincolnensis | |
| Streptomyces tendae | 1.0 | Streptomyces tendae | |
| Terrabacter tumescens | 0.2 | Terrabacter tumescens | |
| Molecular identification at the genus level | |||
| Actinobaculum sp. | 3 | 1.0-4.7 | Actinobaculum sp. |
| Actinomyces sp. | 4 | 1.0-4.3 | Actinomyces sp. |
| Actinomyces sp. | 0.0/5.0 | Actinomyces sp./A. viscosus | |
| Bifidobacterium sp. | 1.6 | Bifidobacterium sp. | |
| Corynebacterium sp. | 2 | 0.2-0.7/1.4-1.7 | Corynebacterium sp./C. pseudogenitalium |
| Corynebacterium sp. | 2 | 2.3-2.5 | Corynebacterium sp. |
| Dietzia sp. | 0.0/1.1 | Dietzia sp./D. maris | |
| Lactobacillus sp. | 0.2/7.4 | Lactobacillus sp./L. acidophilus | |
| Nocardiopsis sp. | 3.0 | Nocardiopsis sp. | |
| Rhodococcus sp. | 1.6 | Rhodococcus sp. | |
| Rothia sp. | 3 | 0.0/1.6-2.2 | Rothia sp./R. dentocariosa |
| Molecular identification at the family level | |||
| Corynebacteriaceae | 7.1 | Corynebacterium confusum | |
| Microbacteriaceae | 1.3 | Microbacteriaceae |
DISCUSSION
Accurate and rapid identification of microorganisms is a prerequisite for the generation of high-quality data in the clinical laboratory and is necessary for decisions concerning the installment of antibiotic therapy. In this study, we have shown that 16S rDNA sequence analysis improves the identification of aerobic gram-positive rods compared to conventional phenotypic methods.
Isolates well identified at the species level by conventional phenotypic methods served as a control for both conventional and molecular identification procedures. For this collection of isolates (n = 37), the two methods showed good overall agreement. In rare instances (n = 4), 16S rDNA sequencing and conventional identification gave different results, three of which were minor, i.e., a higher resolution was obtained with one or the other method. In one case a conflicting result was obtained, i.e., conventional identification resulted in N. brasiliensis, whereas sequence analysis resulted in Streptomyces albidoflavus (conventional identification was incorrect due to false acid-fast staining). In 16.2% of cases, sequencing resulted in recovery of a sequence with low demarcation to the next species. In all of these cases, the results of the conventional identification procedures supported the interpretation of the sequencing data.
Our study demonstrates that 16S rDNA analysis has a great potential for the identification of gram-positive rods that are difficult to identify by conventional means: (i) in 64.8% (46 of 71) of isolates which could only be identified at the genus level by conventional procedures, sequence analysis allowed species identification; (ii) in the case of isolates that could not be discriminated at any taxonomic level conventionally (n = 65), 66.2% and 30.8% of the isolated bacteria could be assigned to a species and genus, respectively.
The two-step procedure for sequence analysis used in this study proved to be very helpful in generating high-quality sequence data. The FASTA search rapidly detected sequencing problems at either end of or within the sequence and allowed for correction if necessary. Ambiguous ends of the sequence were deleted before performing the BLASTN search. The resulting sequences often showed no or only a few mismatches to database entries, allowing the successful use of the defined criteria for species and genus assignment.
The conventional identification system used in our routine laboratory (25) is an elaborate procedure that forms the basis for identification of coryneform gram-positive rods as published in the Manual of Clinical Microbiology (12). Most aerobic gram-positive rods are readily identified by this system. However, frequently strains are isolated which cannot be identified using this approach. Approximately 14% of all aerobic gram-positive rods (not including Gardnerella vaginalis and Lactobacillus sp.) isolated during the course of this study belonged to this category, all of which were included in this analysis. For such isolates, it has been suggested to supplement the system by further phenotypic tests (25), e.g., analysis of carbon source utilization, cellular fatty acid analysis, and cell wall analyses (26). We intentionally abandoned additional phenotypic investigations but used rDNA sequencing as a complement to conventional identification, as molecular identification procedures provide results in 1 to 2 working days.
Very few studies so far have reported on the use of rDNA sequencing for the identification of clinical isolates in a systematic fashion. Most of these studies focused on mycobacteria (6, 10, 19, 21), and a few reports investigated other groups of bacteria (8, 23, 24). Using 16S rDNA sequencing, Drancourt et al. (8) analyzed 177 gram-positive and gram-negative isolates from environmental, veterinary, and clinical sources that could not be identified by conventional means; 78.5% and 89.8% of the isolates could be identified at the species and genus level, respectively. Tang and coworkers (24) used the MicroSeq 500 16S bacterial sequencing kit (Applied Biosystems) for the identification of 52 coryneform bacilli and concluded that the MicroSeq system is superior to conventional identification.
The present study is unique in that (i) we prospectively evaluated the value of 16S rDNA sequencing under routine conditions in a clinical laboratory, and (ii) we investigated various groups of aerobic gram-positive rods. The part of the 16S rRNA gene chosen for analysis covers the most discriminating regions within the 16S rDNA and is therefore suitable for identification purposes (16). The present data suggest that 16S rDNA sequence analysis not only is more accurate and more objective than conventional identification, which relies on individual interpretation and expertise, but also offers the possibility of rapidly recognizing yet undescribed taxa. This is because 16S rDNA similarity reflects phylogenetic relationships (28).
In general, 16S rDNA similarities below 97% are indicative of different species. In the present study, 21 (11.9%) isolates showed 16S rDNA similarities below 97% with sequences of established species. Although only partial sequences were used, it is likely that some of these isolates are indicative of a new species (22). Sequence homology values above 97% similarity, however, do not guarantee species identity; in the extreme, two species may have identical 16S rDNA sequences, as reported for Bacillus psychrophilus and Bacillus globisporus (11). In practice, these rare exceptions of 16S rDNA sequence identity despite different species assignment do not pose problems, as the systematic sequence comparison by searches of public databases will reveal the identity with both species. Identification by sequencing may be complicated by significant sequence variability within and between strains of the same species (3, 5), e.g., Clayton et al. (5) found that 28% of prokaryotic species with two 16S rRNA sequences deposited in public databases had more than 1.0% variable sequence positions. They concluded that some of these variations have to be attributed to intraspecies heterogeneity, whereas others are likely to be the result of sequencing errors. With the above in mind, we decided to use stringent criteria for species identification; i.e., 99% sequence similarity or higher for species assignment and 95% sequence similarity or higher for genus assignment. Other authors have used cutoff values of 99% (8), 99.2% (19), or 98.6% (14) for species identification.
An important issue is the quality of public databases such as GenBank, EMBL, and DDBJ. Sequences can be deposited in these databases largely independent of their quality, e.g., the number of ambiguous nucleotides, the length of the sequence, or the correct assignment of the investigated strain. Among the deposited sequences of aerobic gram-positive rods, we found some which we believe are misidentified; e.g., M29560 “Mycobacterium chitae” shows >98% total 16S rDNA similarity to P. acnes but <86% to other sequences of Mycobacterium chitae. We therefore assume that M29560 is the sequence of a Propionibacterium sp. strain. Similar findings apply to X81906 “Corynebacterium xerosis” (>99% with C. striatum, <95% with other sequences of C. xerosis), X80611 “Nocardia otitidiscaviarum” (>99% with N. farcinica, <96% with other sequences of N. otitidiscaviarum), Y13025 “Actinomyces sp.” (100% with Rothia sp., <94% with other sequences of Actinomyces sp.).
For sequence analysis, we suggest threshold values of sequence similarities of 99% and 95% for species and genus assignment, respectively. If, in the case of a species assignment, the second scoring reference species shows less than 0.5% additional sequence divergence, this should be noted (our category of “species with low demarcation to next species”). Efforts should be undertaken to provide 16S rDNA databases with high-quality sequences. First steps in this direction have already been made (18) with the Ribosomal Database Project (rdp.cme.msu.edu), Ribosomal Differentiation of Medical Microorganisms (www.ridom.de), SmartGene IDNS (www.smartgene.ch), and MicroSeq (Applied Biosystems).
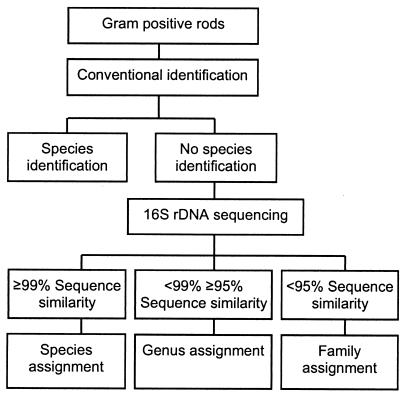
Despite some unresolved issues in molecular identification procedures, e.g., interspecies sequence homology and quality of public databases, our study demonstrates that molecular identification even at this stage is vastly superior to standard identification procedures and is ready to be implemented in the clinical laboratory. For the identification of aerobic gram-positive rods in routine practice, we suggest a two-step procedure that combines phenotypic methods with molecular methods, as shown in the algorithm in Fig. 1. (i) For isolates which give a reliable identification result by phenotypic identification procedures, sequence analyses are not required; and (ii) for isolates which are difficult to identify by conventional techniques, rDNA sequencing is an effective means for identification. As the majority of isolates (approximately 86%) fall in the first category, costs will be kept to a minimum, allowing this technology to be within the reach of many microbiological laboratories.
FIG. 1.
Algorithm for the identification of gram-positive rods.
Acknowledgments
We thank the technicians of the Institute of Medical Microbiology for their excellent technical assistance.
This study was supported by the University of Zurich.
REFERENCES
- 1.Boettger, E. C. 1989. Rapid determination of bacterial ribosomal RNA sequences by direct sequencing of enzymatically amplified DNA. FEMS Microbiol. Lett. 65:171-176. [DOI] [PubMed] [Google Scholar]
- 2.Boettger, E. C. 1996. Approaches for identification of microorganisms. ASM News 62:247-250. [Google Scholar]
- 3.Bosshard, P. P., R. Zbinden, and M. Alwegg. 2002. Paenibacillus turicensis sp. nov., a novel bacterium harboring heterogeneities between 16S rRNA genes. Int. J. Syst. E vol. Microbiol. 52:2241-2249. [DOI] [PubMed] [Google Scholar]
- 4.Brosius, J., M. L. Palmer, P. J. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clayton, R. A., G. Sutton, P. S. Hinkle, C. Bult, and C. Fields. 1995. Intraspecific variation in small-subunit rRNA sequences in GenBank: why single sequences may not adequately represent prokaryotic taxa. Int. J. Syst. Bacteriol. 45:595-599. [DOI] [PubMed] [Google Scholar]
- 6.Cloud, J. L., H. Neal, R. Rosenberry, C. Y. Turenne, M. Jama, D. R. Hillyard, and K. C. Carroll. 2002. Identification of Mycobacterium spp. by using a commercial 16S ribosomal DNA sequencing kit and additional sequencing libraries. J. Clin. Microbiol. 40:400-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drancourt, M., C. Bollet, A. Carlioz, R. Martelin, J. P. Gayral, and D. Raoult. 2000. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J. Clin. Microbiol. 38:3623-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwards, U., T. Rogalt, H. Bloecker, M. Emde, and E. C. Boettger. 1989. Isolation and direct complete nucleotide determination of entire genes characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El Amin, N. M., H. S. Hanson, B. Pettersson, B. Petrini, and L. V. Von Stedingk. 2000. Identification of non-tuberculous mycobacteria: 16S rRNA gene sequence analysis vs. conventional methods. Scand. J. Infect. Dis. 32:47-50. [DOI] [PubMed] [Google Scholar]
- 11.Fox, G. E., J. D. Wisotzkey, and P. Jurtshuk. 1992. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol. 42:166-170. [DOI] [PubMed] [Google Scholar]
- 12.Funke, G., and K. A. Bernard. 1999. Coryneform gram-positive rods, p. 319-345. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology. American Society for Microbiology, Washington, D.C.
- 13.Goldenberger, D., A. Kuenzli, P. Vogt, R. Zbinden, and M. Altwegg. 1997. Molecular diagnosis of bacterial endocarditis by broad-range PCR amplification and direct sequencing. J. Clin. Microbiol. 35:2733-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keswani, J., and W. B. Whitman. 2001. Relationship of 16S rRNA sequence similarity to DNA hybridization in prokaryotes. Int. J. Syst. Evol. Microbiol. 51:667-678. [DOI] [PubMed] [Google Scholar]
- 15.Kolbert, C. P., and D. H. Persing. 1999. Ribosomal DNA sequencing as a tool for identification of bacterial pathogens. Curr. Opin. Microbiol. 2:299-305. [DOI] [PubMed] [Google Scholar]
- 16.Ludwig, W., O. Strunk, S. Klugbauer, N. Klugbauer, M. Weizenegger, J. Neumaier, M. Bachleitner, and K. H. Schleifer. 1998. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19:554-568. [DOI] [PubMed] [Google Scholar]
- 17.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734-740. [DOI] [PubMed] [Google Scholar]
- 18.Patel, J. B. 2001. 16S rRNA gene sequencing for bacterial pathogen identification in the clinical laboratory. Mol. Diagn. 6:313-321. [DOI] [PubMed] [Google Scholar]
- 19.Patel, J. B., D. G. Leonard, X. Pan, J. M. Musser, R. E. Berman, and I. Nachamkin. 2000. Sequence-based identification of Mycobacterium species using the MicroSeq 500 16S rDNA bacterial identification system. J. Clin. Microbiol. 38:246-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Relman, D. A., J. S. Loutit, T. M. Schmidt, S. Falkow, and L. S. Tompkins. 1990. The agent of bacillary angiomatosis. An approach to the identification of uncultured pathogens. N. Engl. J. Med. 323:1573-1580. [DOI] [PubMed] [Google Scholar]
- 21.Springer, B., L. Stockman, K. Teschner, G. D. Roberts, and E. C. Boettger. 1996. Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J. Clin. Microbiol. 34:296-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stackebrandt, E., and B. M. Goebel. 1994. A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 23.Tang, Y. W., N. M. Ellis, M. K. Hopkins, D. H. Smith, D. E. Dodge, and D. H. Persing. 1998. Comparison of phenotypic and genotypic techniques for identification of unusual aerobic pathogenic gram-negative bacilli. J. Clin. Microbiol. 36:3674-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang, Y. W., A. Von Graevenitz, M. G. Waddington, M. K. Hopkins, D. H. Smith, H. Li, C. P. Kolbert, S. O. Montgomery, and D. H. Persing. 2000. Identification of coryneform bacterial isolates by ribosomal DNA sequence analysis. J. Clin. Microbiol. 38:1676-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Von Graevenitz, A., and G. Funke. 1996. An identification scheme for rapidly and aerobically growing gram-positive rods. Zentbl. Bakteriol. 284:246-254. [DOI] [PubMed] [Google Scholar]
- 26.Von Graevenitz, A., G. Osterhout, and J. Dick. 1991. Grouping of some clinically relevant gram-positive rods by automated fatty acid analysis. Diagnostic implications. APMIS 99:147-154. [DOI] [PubMed] [Google Scholar]
- 27.Wilson, K. H., R. Blitchington, R. Frothingham, and J. A. Wilson. 1991. Phylogeny of the Whipple's-disease-associated bacterium. Lancet 338:474-475. [DOI] [PubMed] [Google Scholar]
- 28.Woese, C. R., and G. E. Fox. 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. USA 74:5088-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woese, C. R., O. Kandler, and M. L. Wheelis. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA 87:4576-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]