Abstract
This work provides a novel, highly sensitive, hot start PCR method for rapid and specific detection of African swine fever virus (ASFV) that can be used as a routine diagnostic test for ASFV in surveillance, control, and eradication programs. A confirmatory test of the specificity of this method based on restriction endonuclease analysis was also developed.
African swine fever (ASF) is a highly contagious disease of swine caused by a complex DNA virus that has been classified as an Asfivirus (10). It produces great economic losses in the affected countries, due to the high mortality rates of the acute and peracute forms and its potential for extensive and rapid geographical spread. Domestic pigs and European wild boars are very susceptible, showing a wide range of clinical forms (1, 4, 15). African wild boars, bushpigs, and warthogs present unapparent clinical infections, acting as reservoir hosts in Africa (1, 7). Soft ticks, especially Ornithodoros erraticus and Ornithodoros moubata, are reservoir and transmission vectors of ASF virus (ASFV) (2, 13, 14).
At this time, ASF is present in sub-Saharan countries of Africa, where the disease has acquired great importance since 1997, because of the increasing number of outbreaks that have affected many countries (3). Outside Africa, ASF is currently endemic in Sardinia (Italy).
Epidemiological studies have demonstrated that the entrance of ASFV in ASFV-free areas is primarily related to feeding pigs with contaminated garbage from international airports and seaports (15). This fact, together with the extensive commercial trade, keeps ASFV-free countries at constant risk of having the disease introduced in their territory. Since there is no vaccine available at present, control and eradication strategies are mainly based on rapid and accurate laboratory diagnosis of ASFV-positive and carrier animals and on the enforcement of strict sanitary measures. Furthermore, the great similarity of ASF clinical symptoms and lesions with those of other hemorrhagic pig diseases, particularly classical swine fever, makes differential laboratory diagnosis compulsory in order to distinguish ASF from other pig diseases with compatible clinical presentations (15). Therefore, diagnostic laboratories must have rapid and accurate procedures for specific ASFV detection, such as PCR, available.
Only a few PCR-based ASF diagnostic assays have been previously described for detection and identification of ASFV (8, 16, 17). In this paper, a novel, fast, highly sensitive, and specific gel-based hot start PCR test has been specially designed for routine laboratory diagnosis of ASFV.
Different specific primer pairs for ASFV (in the VP73 coding region of the genome) were selected with the aid of Primer Express (Applied Biosystems) computer program. The nucleotide sequences of the VP73 genes of seven different ASFV strains, available in GenBank (5, 9, 11, 12, 18, 19) were aligned using Clustal W software, to assess conservation of the primer sequences in all strains. The primer set PPA1-PPA2 (PPA-1/2) (PPA1, 5′-AGTTATGGGAAACCCGACCC-3′; PPA2, 5′-CCCTGAATCGGAGCATCCT-3′), which delimits an amplicon of 257 bp, was finally selected as providing the best results. The primer set recommended by the Office International des Epizooties (OIE) (Paris, France), primer 1 (5′-ATGGATACCGAGGGAATAGC-3′) and primer 2 (5′-CTTACCGATGAAAATGATAC-3′) (17), were used for comparison.
Different DNA extraction systems were evaluated, and the method that was the most efficient was selected. Briefly, total DNA was purified from 200 μl of a sample (cell culture samples, serum samples, blood samples treated with EDTA, or tissue homogenate samples in 10% phosphate-buffered saline) by using the High Pure PCR template preparation kit (Roche Molecular Biochemicals), following the manufacturer's instructions. The final elution volume was 50 μl.
Total RNA was extracted from 100 μl of a sample with a commercial reagent (Tripure isolation reagent; Roche Molecular Biochemicals) under the conditions recommended by the manufacturer. RNA was resuspended in 10 μl of MilliQ water.
Optimal conditions for the PCR assay were established as follows: 2 μl of sample DNA, 1× PCR buffer II (50 mM KCl, 10 mM Tris-HCl), 2 mM MgCl2, 0.2 mM concentrations of the four deoxynucleoside triphosphates (Roche Molecular Biochemicals), 0.2 μΜ concentrations of both primers (PPA-1/2 or the OIE Manual of Standards for Diagnostic Test and Vaccine primer pair [primers 1 and 2] described previously [17]) and 0.625 U of Taq Gold polymerase (Applied Biosystems), in a total volume of 25 μl. When the PPA-1/2 primer set was used, the reaction mixture was treated as follows: (i) incubated for 10 min at 95°C; (ii) subjected to 40 cycles of PCR, with 1 cycle consisting of 15 s at 95°C, 30 s at 62°C, and 30 s at 72°C; and (iii) incubated for 7 min at 72°C. When the OIE primer set was used, the reaction mixture was treated as follows: (i) incubated for 10 min at 95°C; (ii) subjected to 35 cycles, with 1 cycle consisting of 30 s at 94°C, 30 s at 53°C, and 30 s at 72°C; and (iii) incubated for 7 min at 72°C. Amplification products were analyzed by electrophoresis on a 2% agarose gel containing 0.5 μg of ethidium bromide per ml.
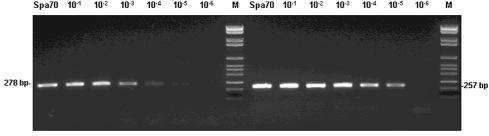
In order to determine the detection limits of the test, PCR assays were performed on serial 10-fold dilutions of a viral suspension of ASFV Spain 70 with a titer of 1.6 × 106 50% hemadsorption units (HADU50)/ml. The sensitivity was consistently observed to be 0.12 HADU50 per PCR mixture (Fig. 1). The amplification efficiency and detection limit of the assay were further examined in a comparative PCR study using PPA-1/2 and the recommended OIE Manual of Standards for Diagnostic Test and Vaccine primer set (17) (Fig. 1). The sensitivity of the assay was increased 10-fold when the PPA-1/2 primer set was used.
FIG. 1.
Sensitivity of the ASFV PCR assay using the OIE primer pair or novel PPA-1/2 primer set. DNAs extracted from serial dilutions, in serum, of a suspension of ASFV strain Spain 70 (Spa70) with a titer of 1.6 × 106 UHAD50/ml were employed as templates in the PCR assays under reaction conditions described in the text using OIE primers (left) or novel PPA-1/2 primers (right). M, molecular weight marker VI (Roche Molecular Biochemicals).
The specificity of the PCR was tested using DNA from 22 different ASFV isolates from different geographical origins and isolation years and DNA or RNA from related viruses (Table 1). In addition, DNA from tissue homogenates (spleen, kidney, and tonsils), blood samples treated with EDTA, and serum samples from healthy pigs, noninfected tissue homogenates from O. erraticus, and several noninfected cell lines (PK15, MDBK, and BHK-21) was employed in the PCR specificity test. PCR gave positive results for all 22 ASFV isolates, but PCR did not give any positive results when other related viruses, cell lines, or noninfected tissue samples were assayed (data not shown).
TABLE 1.
Viruses used to test the specificity of the ASFV PCR assay
| Virus | Virus | |
|---|---|---|
| ASFV | ||
| West Indies and South American strainsa,b | ||
| Brazil 78 | ||
| Haiti 78 | ||
| Dominican Republic 79 | ||
| European strainsa,b | ||
| Spain 70 | ||
| Spain 75 | ||
| Spain 94 | ||
| Italy Ca78/1 | ||
| Italy Nu 84WB | ||
| Italy Nu 90/1 | ||
| Italy Nu 95/2 | ||
| Italy Nu 98 | ||
| Lisbon 57 | ||
| Lisbon 60 | ||
| Malta 78 | ||
| African strainsa | ||
| Uganda 64 | ||
| Mozambique 64 | ||
| Katanga 67 | ||
| Angola 72 | ||
| Cape Verde Islands 97 | ||
| Cape Verde Islands 98 | ||
| Ivory Coast 99 | ||
| Nigeria 2001 | ||
| Classical swine fever virus strainsa,c | ||
| Subgroup 1.1 | ||
| Alfort 187 | ||
| Eystrup | ||
| Subgroup 1.2 | ||
| Brescia | ||
| Subgroup 2.1 | ||
| Spain 97 | ||
| V1240/97 Germany | ||
| Subgroup 2.2 | ||
| Parma 98 | ||
| Subgroup 2.3 | ||
| 2699/Osterode 82 | ||
| Switzerland IV/93 | ||
| 1189/1 Belgium | ||
| VI3837/38 Germany 99 | ||
| Spain 2001 | ||
| Barcelona 2001 | ||
| Visbek/Han 95 | ||
| Disparate strains | ||
| Kanagawa 74 | ||
| Bovine viral diarrhea virus strainsa,c,d | ||
| Type 1 | ||
| Sanders | ||
| NADL | ||
| Oregon | ||
| Type II | ||
| 61/138, Low Saxony, Germany | ||
| 61/120, Low Saxony, Germany | ||
| Border disease virus strainsc,e,f | ||
| X818 | ||
| Moredun | ||
| Fritjers | ||
| 137/4 | ||
| Foot-and-mouth disease virus strainsa | ||
| Serotype A | ||
| A22 Iraq (IRQ24/64) | ||
| Serotype O | ||
| O1-Manisa/Turkey 69 | ||
| Serotype C | ||
| C1-Noville Switzerland/65 | ||
| Reference strains of the serotypes: | ||
| Sat 1 | ||
| Sat 2 | ||
| Sat 3 | ||
| Asia 1 | ||
| Vesicular stomatitis virus serotypesa | ||
| Indiana | ||
| New Jersey | ||
| Swine vesicular disease virusa | ||
| Reference strain UK-72 | ||
| Porcine respiratory and reproductive syndrome strainsa | ||
| European | ||
| 5710 | ||
| American | ||
| VR2332 | ||
| Aujeszky disease virusa | ||
| Phylaxia reference strain |
ASF international reference laboratory (OIE, European Union), Department of Exotic Diseases, Centro de Investigación en Sanidad Animal, Valdeolmos, Madrid, Spain.
Istituto Zooprofilacttico della Sardegna, Sardegna, Italy.
Classical swine fever virus European reference laboratory, Institute of Virology, Hannover, Germany.
Veterinary Laboratories Agency, Weybridge, United Kingdom.
Elizabeth Macarthur Agricultural Institute, Camden, Australia.
Institute for Animal Science and Health-Lelystad, Lelystad, The Netherlands.
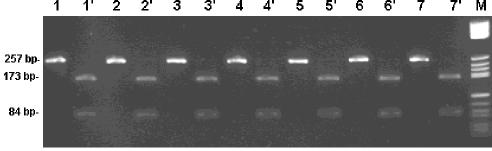
Sequence analysis of the genome region delimited by the PPA-1/2 primer set enabled us to determine the location of a BsmAI restriction endonuclease site, splitting the amplicon into two fragments of 173 and 84 bp in length. The BsmAI site was conserved in the sequences of the seven isolates available in GenBank.
BsmAI digestion of the amplification products were performed in 20-μl reaction mixtures, using 5 μl of the PCR amplification product and 5 U of the enzyme. The reaction mixtures were incubated for at least 2 h at 55°C, and restriction fragments were analyzed by electrophoresis on a 3% agarose gel. BsmAI digestion of the corresponding amplicon from the 22 ASFV isolates resulted in the two expected specific restriction fragments in all cases. The results obtained after analysis of seven representative isolates of the 22 ASFV isolates are shown in Fig. 2.
FIG. 2.
BsmAI restriction endonuclease analysis of amplification products of different ASFV strains. Lanes 1 and 1′, Brazil 78; 2 and 2′, Haiti 78; 3 and 3′, Spain 70; 4 and 4′, Lisbon 60; 5 and 5′, Cape Verde Islands 97; 6 and 6′, Ivory Coast 99; 7 and 7′, Nigeria 2001. Lanes 1, 2, 3, 4, 5, 6, and 7 are amplification products. Lanes 1′, 2′, 3′, 4′, 5′, 6′, and 7′ are amplification products after digestion with BsmA. Reaction conditions are described in the text. M, molecular weight marker V (Roche Molecular Biochemicals).
In order to assess the validity of the method in clinical specimens, samples from pigs experimentally inoculated with two different ASFV isolates were tested. Serum and blood samples obtained at 0, 1, 2, 3, 4, and 7 days postinoculation (dpi) from two pigs (1-year-old) intramuscularly inoculated with 104 HADU50 of the Spain 75 strain, which has low virulence, were analyzed by PCR using the novel PPA-1/2 primer set and the OIE primer set. The results are shown in Fig. 3. The ASFV genome was detected 2 dpi in blood samples treated with EDTA when the PPA-1/2 primers were employed, 2 days before the first clinical signs (fever) were observed, which indicates the power of this procedure for early diagnosis. When the OIE primers were used, positive results were detected by PCR at 3 dpi. Similarly, blood, kidney, liver, lung, lymph node, spleen, and tonsil samples obtained at 5 dpi from a 3-month-old pig experimentally inoculated with 107 UHAD50 of Lisbon 60 strain were analyzed by PCR using the PPA-1/2 primer set (Fig. 4) or the OIE primer set (data not shown). All the samples analyzed gave positive results by both tests.
FIG. 3.
ASFV detection by PCR in blood (B) or serum (S) samples from experimentally infected animals using OIE (a) or PPA-1/2 (b) primer pairs. Samples of pigs were obtained at different dpi, as described in the text. M, molecular weight marker VI (Roche Molecular Biochemicals).
FIG. 4.
ASFV detection by PCR in blood and pig tissue samples. Samples of blood (treated with EDTA), kidney, liver, lung, lymph nodes, spleen, and tonsils were obtained from a pig experimentally inoculated with Lisbon 60 at 5 dpi and analyzed by PCR using PPA-1/2 primers. M, molecular weight marker VI (Roche Molecular Biochemicals).
To determine the sensitivity of the PCR method on badly preserved samples, pieces of kidney tissue obtained from a pig experimentally infected with ASFV strain Lisbon 60 were stored at room temperature for 0, 14, or 28 days, homogenized, and analyzed by PCR using PPA-1/2 and OIE primer sets. The viral genome was detected when both primer sets were used, even after 28 days of tissue sample storage at room temperature when the samples were putrefied (Fig. 5). However, a stronger signal was obtained when the PPA-1/2 primer set was employed.
FIG. 5.
PCR detection of ASFV on badly preserved samples. Pieces of kidney from a pig infected with ASFV isolate Lisbon 60 were homogenized and analyzed by PCR using either the PPA-1/2 or OIE primer set after 0, 14, or 28 days of storage at room temperature. M, molecular weight marker VI (Roche Molecular Biochemicals).
Finally, a retrospective study was also performed by analysis of a collection of 70 frozen samples, including 18 serum samples, 4 blood samples treated with EDTA, and 48 tissue samples recovered from ASF outbreaks that occurred in Spain from 1960 to 1994 and, more recently, in Cape Verde Islands (1997 and 1998), Ivory Coast (1999), and Nigeria (2001). These samples were previously confirmed to be ASFV positive by virus isolation or hemadsorption or by FAT reference tests (and for the samples obtained more recently, by the OIE PCR protocol [17]). All samples gave positive results by PCR when the PPA-1/2 primer pair was used (data not shown).
In summary, the novel ASFV PCR assay possesses a higher sensitivity (0.12 HADU50) than that recommended by the OIE Manual of Standards for Diagnostic Test and Vaccine (17) and allows the detection of ASFV strains worldwide, as it was established after the analysis of 22 ASFV strains from different years and geographical locations. This method is both rapid (the results can be obtained in less than 5 h) and specific (no false-positive reactions were observed when samples from healthy pigs, noninfected ticks, uninfected cell lines, and related porcine viruses were assayed). Other advantages of this method is that it has been standardized so that it can be used on tissue samples, blood samples treated with EDTA, and serum samples, and it can be performed even on poorly preserved or putrefied tissues, while previous PCR methods did not have such wide applications (8, 16, 17). The ability to check the specificity of the amplicons by simple restriction endonuclease digestion, instead of sequencing, broadens the range of laboratories in which this diagnostic technique can be used with confidence.
In addition, the results of this study show that the DNA extraction method used efficiently removes potential inhibitors that are present in the clinical samples, since all the ASFV-positive tissue, serum, and blood samples analyzed were confirmed by PCR. A simpler DNA preparation procedure has been described previously (16, 17) which sometimes gives false-negative results, as recently demonstrated by Gonzague et al. (6), probably due to the presence of inhibitors which are not removed during sample preparation.
Acknowledgments
We thank U. Moennig and I. Greiser-Wilke of the Institute of Virology (Hannover, Germany), D. Paton of the Veterinary Laboratories Agency (Weybridge, United Kingdom), A. D. Shannon of the Elizabeth Macarthur Agricultural Institute (Camden, Australia), and A. J. De Smit of the Institute for Animal Science and Health—Lelystad, Lelystad, The Netherlands for supplying some of the pestivirus isolates. We thank C. Patta of the Istituto Zooprofilacttico della Sardegna (Sardegna, Italy) for supplying the Italian ASFV isolates and M. Moyano and A. Robles for their skillful technical assistance.
This work has been funded in part by governmental grants (agreement INIA and Ministry of Agriculture of Spain) and the EU project QLK2-CT-2000-00486.
REFERENCES
- 1.Arias, M., C. Sánchez, M. A. González, L. Carrasco, and J. M. Sánchez-Vizcaíno. 2002. Peste porcina Africana. In Curso digital de enfermedades infecciosas porcinas. [Online.] http:www.sanidadanimal.info.
- 2.Arias, M., and J. M. Sánchez-Vizcaíno. 2002. African swine fever (ASF), p. 119-124. In A. Morilla, K.-J. Yoon, and J. Zimmerman (ed.), Trends in emerging viral infections of swine. Iowa State University Press, Ames.
- 3.Arias, M., and J. M. Sánchez-Vizcaíno. 2002. African swine fever eradication: the Spanish model, p. 133-139. In A. Morilla, K.-J. Yoon, and J. Zimmerman (ed.), Trends in emerging viral infections of swine. Iowa State University Press, Ames.
- 4.Arias, M., J. A. Martínez Escribano, A. Rueda, and J. M. Sánchez-Vizcaíno. 1986. La peste porcina Africana. Med. Vet. 3:333-359. [Google Scholar]
- 5.Cistue, C., and E. Tabarés. 1992. Expression in vivo and in vitro of the major structural protein (VP73) of African swine fever virus. Arch. Virol. 123:111-124. [DOI] [PubMed] [Google Scholar]
- 6.Gonzague, M., C. Plin, L. Bakkali-Kassimi, A. Boutrouille, and C. Cruciere. 2002. Development of an internal control for the detection of the African swine fever virus by PCR. Mol. Cell. Probes 16:237-242. [DOI] [PubMed] [Google Scholar]
- 7.Heuschele, W. P., and L. Coggins. 1965. Isolation of African swine fever virus from a giant forest hog. Bull. Epizoot. Dis. Afric. 13:255-256. [PubMed] [Google Scholar]
- 8.King, D. P., S. M. Reid, G. H. Hutchings, S. Grierson, P. J. Wilkinson, L. K. Dixon, D. S. Bastos, and T. W. Drew. 2003. Development of a TaqMan PCR assay with internal amplification control for the detection of African swine fever virus. J. Virol. Methods 107:53-61. [DOI] [PubMed] [Google Scholar]
- 9.Lopez-Otin, C., J. M. Freije, F. Parra, E. Mendez, and E. Viñuela. 1990. Mapping and sequence of the gene coding for protein p72, the major capsid protein of African swine fever virus. Virology 175:477-484. [DOI] [PubMed] [Google Scholar]
- 10.Murphy, F. A., E. P. J. Gibbs, M. A. Horzinek, and M. J. Studdert. 1999. Asfarviridae and Iridoviridae, p. 293-300. In F. A. Murphy, E. P. J. Gibbs, M. A. Horzinek, and M. J. Studdert (ed.), Veterinary virology, 3rd ed. Academic Press, San Diego, Calif.
- 11.Odemuyiwa, S. O., I. A. Adebayo, W. Ammerlaan, A. T. Ajuwape, O. O. Alaka, O. I. Oyedele, K. O. Soyelu, D. O. Olaleye, E. B. Otesile, and C. P. Muller. 2000. An outbreak of African swine fever in Nigeria: virus isolation and molecular characterization of the VP72 gene of a first isolate from West Africa. Virus Genes 20:139-142. [DOI] [PubMed] [Google Scholar]
- 12.Pitcher, L. J., L. K. Dixon, and P. C. Turner. 1992. A sequence comparison of the VP72 gene of African swine fever virus. Biochem. Soc. Trans. 20:30S. [DOI] [PubMed] [Google Scholar]
- 13.Plowright, W., C. T. Perry, and M. A. Peirce. 1970. Experimental infection of the argasid tick, Ornithodoros moubata porcinus, with African swine fever virus. Arch. Ges. Virusforsch. 31:33-50. [DOI] [PubMed] [Google Scholar]
- 14.Sánchez Botija, C. 1963. Reservorios del virus de la peste porcina Africana. Investigación del virus de la PPA en los artrópodos mediante la prueba de la hemoadsorción. Bull. Office Int. Epizoot. 60:895-899. [Google Scholar]
- 15.Sánchez-Vizcaíno, J. M. 1999. African swine fever, p. 93-102. In A. D. Leman, B. E. Straw, W. L. Mengeling, S. Dallaire, and D. J. Taylor (ed.), Diseases of swine, 8th ed. Iowa State University Press, Ames.
- 16.Steiger, Y., M. Ackermann, C. Mettraux, and U. Kihm. 1992. Rapid and biologically safe diagnosis of African swine fever virus infection by using polymerase chain reaction. J. Clin. Microbiol. 30:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilkinson, P. J. 2000. African swine fever, p. 189-198. In Manual of standards for diagnostic test and vaccines, 4th ed. Office International des Epizooties, Paris, France.
- 18.Yanez, R. J., J. M. Rodriguez, M. L. Nogal, L. Yuste, C. Enriquez, J. F. Rodriguez, and E. Viñuela. 1995. Analysis of the complete nucleotide sequence of African swine fever virus. Virology 208:249-278. [DOI] [PubMed] [Google Scholar]
- 19.Yu, M., C. J. Morrissy, and H. A. Westbury. 1996. Strong sequence conservation of African swine fever virus p72 protein provides the molecular basis for its antigenic stability. Arch. Virol. 141:1795-1802. [DOI] [PubMed] [Google Scholar]