Abstract
A real-time PCR method was developed and used to detect Aspergillus fumigatus mitochondrial DNA (mtDNA) in bronchoalveolar lavage (BAL) fluids and tissue biopsy specimens. The analytical sensitivity of the assay was one A. fumigatus conidium per reaction, and the assay was linear at least over 4 orders of magnitude above the detection limit. BAL fluids from 66 immunocompromised patients at risk of invasive pulmonary aspergillosis (IPA) and 33 immunocompetent controls and tissue biopsy specimens from 10 immunocompromised patients were analyzed. The results were related to the clinical diagnosis established according to recently published consensus criteria. A. fumigatus mtDNA positivity was encountered in 16 of 81 (20%) BAL fluid specimens from patients at risk and 1 of 33 (3%) specimens from immunocompetent controls. PCRs were positive in six of seven, two of four, and four of five of the patients with proven, probable, and possible IPA, respectively, as well as in four patients at risk but without any other evidence of IPA. With qualitative detection, the diagnostic sensitivity of PCR was 73%, specificity was 93%, and predictive values of positive (PPV) and negative (NPV) results were 73 and 95%, respectively. Using a threshold cycle of <35 as a limit for positive PCR, the specificity and PPV of PCR in the diagnosis of invasive aspergillosis were 100%, but its sensitivity was only 45% and NPV was 92%. PCR was positive in tissue biopsy specimens from all patients with invasive aspergillosis caused by A. fumigatus. Semiquantitative detection of A. fumigatus mtDNA in BAL fluid may be helpful in the diagnosis of IPA. PCR is well suited for the verification of the presence of A. fumigatus in tissue biopsy specimens.
Invasive aspergillosis, usually caused by Aspergillus fumigatus, is an important cause of death of patients with hematological malignancy. It may also affect other patients with prolonged neutropenia or impaired function of neutrophils. For a better outcome and optimal use of antifungal treatment, patients developing invasive aspergillosis should be identified at an early stage of the infection. High-resolution computer tomography (HRCT) scanning may reveal typical pulmonary infiltrations, such as the halo sign or air-crescent sign, and contribute to early diagnosis (34). Microbiological diagnosis is difficult to establish, as cultivation of Aspergillus has poor sensitivity and deep tissue biopsy specimens are difficult to obtain. Furthermore, fungal culture is often confounded by antifungal treatment, since initiation of empirical treatment is a common practice in patients with hematological malignancy and fever unresponsive to antibacterial agents. On the other hand, isolation of the fungus from respiratory specimens may also reflect colonization of the airway instead of invasive infection, or even environmental contamination of the culture (13).
Microscopic detection of branching septate hyphae in normally sterile tissue is the most convincing finding for diagnosis of disseminated fungal infection. However, morphology alone is often insufficient for the identification of an invading fungus. Aspergillus is very seldom isolated from the bloodstream, even in fulminant invasive disease (9). However, fungal antigens may be detectable in serum. A commercial sandwich enzyme-linked immunosorbent assay (ELISA) for the detection of galactomannan, a major constituent of the Aspergillus cell wall, has shown high sensitivity (89.7%) and specificity (98.1%) in serial screening of high-risk patients for the development of invasive aspergillosis (24). However, in a recent prospective study of 3,327 sera from 807 patients, the sensitivity of a galactomannan antigen test was only 50% (26). In addition to serum, galactomannan can be detected in bronchoalveolar lavage (BAL) specimens (27, 28, 33) and urine (27, 32), but sensitivity is usually lower than in serum testing.
Many authors have reported detection of Aspergillus nucleic acids by PCR for improved diagnosis of invasive aspergillosis. Target sequences, either panfungal ribosomal DNA (rDNA) (8, 11, 12, 17, 19, 22) or Aspergillus-specific mitochondrial DNA (mtDNA) (2, 3, 6, 15, 33) or rDNA sequences (4, 5, 16, 25, 29) have been amplified from BAL fluids (3-5, 25, 29, 33), serum (2, 6), or whole blood (5, 8, 11, 16, 17, 19, 22, 29). Detection of circulating Aspergillus DNA has shown high sensitivity but poor specificity in the screening of high-risk patients for the development of invasive disease (11). When applied to BAL specimens, the sensitivity of PCR in identifying patients with invasive pulmonary aspergillosis (IPA) (according to the criteria applied in each study) has been almost 100%. False-negative results have been rarely reported, but false-positive results have been obtained in samples from both immunocompromised and immunocompetent patients.
We report the development of a rapid PCR assay for the detection of A. fumigatus DNA in BAL fluids and tissue biopsy specimens. Amplification of A. fumigatus mtDNA is detected by a pair of fluorescing probes in the LightCycler instrument. Since the real-time format of the assay allows quantification of the target DNA in the sample, we also attempted to determine whether the fungal burden in the BAL fluid is higher in IPA than in colonization and whether quantitative or semiquantitative PCR is helpful in differentiating between these conditions.
MATERIALS AND METHODS
Fungi.
The fungal strains used in this study included 20 A. fumigatus strains, 4 related meiosporic Neosartorya strains, 48 other Aspergillus strains representing nine species, as well as 40 strains of other fungal genera representing 32 species and also 20 bacterial strains from 20 species (Table 1). Unless stated otherwise, the experiments used the A. fumigatus type strain CBS 133.61 (Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands). In the experiments using conidia as starting material, a fresh local isolate (TUCH-01-ss-178) was used, since the type strain CBS 133.61 produced little conidia on culture. The fungi were grown on Sabouraud agar at 30°C for 4 to 14 days. Local isolates were identified in a specialized clinical mycology laboratory, on the basis of standard morphological and growth characteristics (7).
TABLE 1.
Fungal and bacterial strains tested by the A. fumigatus PCR
| Species | Type strain(s) or QA strain(s)a | No. of local clinical isolates | PCR result |
|---|---|---|---|
| Fungi | |||
| Aspergillus fumigatus and related meiosporic fungi | |||
| Aspergillus fumigatus | CBS 133.61, NCPF 2078 | 18b | + |
| Neosartorya fennelliae | CBS 598.74 | + | |
| Neosartorya fischeri var. fischeri | CBS 544.65 | + | |
| Neosartorya fischeri var. spinosa | CBS 483.66 | + | |
| Neosartorya pseudofischeri | CBS 208.92 | + | |
| Other Aspergillus spp. | |||
| Aspergillus candidus | NEQAS 4180 | 9 | − |
| Aspergillus glaucus | 1 | − | |
| Aspergillus sydowii | 1 | − | |
| Aspergillus chevalieri | 2 | − | |
| Aspergillus flavus | NEQAS5366, NCPF2617 | 8 | − |
| Aspergillus niger | NCPF 2022 | 15 | − |
| Aspergillus terreus | NCPF 2026 | 1 | − |
| Aspergillus unguis | 1 | − | |
| Aspergillus versicolor | NCPF 2368 | 1 | − |
| Aspergillus sp. | 3 | − | |
| Other fungi | |||
| Acremonium strictum | 1 | − | |
| Alternaria alternata | NEQAS 4522 | − | |
| Alternaria sp. | 1 | − | |
| Aureobasidium pullulans | 1 | − | |
| Candida albicans | ATCC 28366 | − | |
| Candida dubliniensis | NCPF 3949 | 1 | − |
| Candida glabrata | 2 | − | |
| Candida krusei | ATCC 6258 | − | |
| Candida lusitaniae | NCPF 2996 | − | |
| Candida parapsilosis | ATCC 22019 | − | |
| Candida tropicalis | NCPF 3870 | − | |
| Cladosporium sp. | 1 | − | |
| Epidermophyton floccosum | 1 | − | |
| Exophiala dermatitidis | 1 | − | |
| Exophiala sp. | 1 | − | |
| Fusarium solani | 1 | − | |
| Fusarium sp. | 1 | − | |
| Geotrichum penicillatum | NCPF 3520 | − | |
| Microsporum canis | 1 | − | |
| Paecilomyces variotii | 1 | − | |
| Paecilomyces sp. | 1 | − | |
| Penicillium camembertii | 1 | − | |
| Penicillium sp. | 2 | − | |
| Pseudallescheria boydii | 2 | − | |
| Rhizomucor pusillus | NEQAS 4099 | − | |
| Rhizopus oryzae | NCPF 7302 | − | |
| Rhodotorula mucilaginosa | ATCC 9449 | − | |
| Saccharomyces cerevisiae | 2 | − | |
| Scopulariopsis brevicaulis | NEQAS 5649 | − | |
| Syncephalastrum racemosum | 1 | − | |
| Trichophyton meentagrophytes | 1 | − | |
| Trichophyton rubrum | 3 | − | |
| Trichophyton tonsurans | 1 | − | |
| Bacteria | |||
| Acinetobacter calcoaceticus | ATCC 23055 | − | |
| Aeromonas hydrophila | 1 | − | |
| Eikenella corrodens | 1 | − | |
| Escherichia coli | ATCC 25922 | − | |
| Haemophilus influenzae | ATCC 49766 | − | |
| Klebsiella pneumoniae | ATCC 27786 | − | |
| Moraxella catarrhalis | ATCC 25238 | − | |
| Neisseria meningitidis | NCTC 10025 | − | |
| Proteus vulgaris | 1 | − | |
| Pseudomonas aeruginosa | ATCC 27853 | − | |
| Pseudomonas putida | CCUG 264 | − | |
| Pseudomonas stutzerii | CCUG 16590 | − | |
| Salmonella enteritidis | 1 | − | |
| Staphylococcus aureus | ATCC 25923 | − | |
| Stenotrophomonas maltophilia | 1 | − | |
| Streptococcus agalactiae | 1 | − | |
| Streptococcus milleri | 1 | − | |
| Streptococcus mutans | ATCC 25175 | − | |
| Streptococcus oralis | ATCC 35037 | − | |
| Streptococcus pneumoniae | ATCC 49619 | − |
QA, quality assurance strains from UKNEQAS (London, United Kingdom). These strains are referred to either by their UKNEQAS number or by their NCPF (National Collection of Pathogenic Fungi, Bristol, United Kingdom) number, when available; CBS, Centraalbureau voor Schimmelcultures; ATCC, American Type Culture Collection, Manassas, Va.; CCUG, Culture Collection of University of Gothenburg, Gothenburg, Sweden; NCTC, National Collection of Type Cultures, London, United Kingdom.
Two of these were environmental isolates.
DNA purification from fungi.
Fungal hyphae and conidia were harvested by rinsing the cultivation plates with 8 ml of sterile saline, scraping the hyphae, and then collecting them by centrifugation (3,000 × g; 5 min). The supernatant was discarded, and about 400 μl was transferred to sterile Eppendorf tubes, heated (90°C for 10 min), cooled to room temperature, and subjected to lyticase (Sigma) treatment (7 U, +30°C for 30 min for yeasts and 1 h for molds). After proteinase K treatment (0.1 mg/ml; 56°C for 2 to 17 h), DNA was isolated by classic phenol-ether extraction as described previously (14). DNA concentration was determined by measuring absorption at 260 nm and adjusted to 100 μg/ml. If the DNA yield remained low after standard purification, a second isolation was carried out in which the fungal suspension was homogenized for 1 min with 0.3 g of zirconia-silica beads (diameter, 0.1 mm) and a minibeadbeater (Biospec Products, Bartlesville, Okla.) after the lyticase treatment.
PCR.
PCR primers P1 (5′-GAA AGG TCA GGT GTT CGA GTC A-3′) and P2 (5′-CTT GGT TGC GGG TTT AGG GAT T), described by Bretagne et al. (3) and modified by Jones et al. (15), amplify a 134-bp fragment from the A. fumigatus mtDNA sequence encoding mitochondrial tRNA (GenBank accession number L37095). For the LightCycler application, Olfert Landt at Tibmolbiol (Berlin, Germany) designed a pair of probes for online detection of PCR products by fluorescence resonance energy transfer (FRET). The “upper” probe, tRNA FL (5′-TTC TTA TTT ATA TGC GGG TTG ATG TAA TAG TAA CA-3′), contains a fluorescein label in the 3′ end, and the “lower” probe, tRNA LC (5′-AGA TGG CTC ATG ACC ATA ATA TTT AGG TGC p), contains an LC Red 640 label in the 5′ end. When the two probes are hybridized to their targets in close proximity, fluorescein excites LC Red, and its fluorescence was detected by the LightCycler.
The PCRs were optimized for MgCl, primer concentrations, and annealing temperature. For this purpose, DNA from A. fumigatus was amplified at three concentrations (20, 2, and 0.2 ng per reaction mixture) with that from Penicillium sp. as a specificity control (20 ng per reaction), and threshold cycle (CT) values determined by the LightCycler instrument under various conditions were compared. The optimized PCR mixture (20 μl) contained each primer at 0.5 μM, each probe at 0.15 μM, MgCl at 3 mM, 1 U of uracil DNA glycosylase (Roche Diagnostics, Mannheim, Germany), and 2 μl of FastStart DNA master hybridization probes (Roche Diagnostics). The FastStart enzyme was activated at 95°C for 10 min prior to the start of cycling. The 45 amplification cycles included denaturation at 95°C for 10 s, annealing of the primers and probes at 60°C for 10 s, and extension at 72°C for 8 s. Fluorescence was measured at channel F2 of the LightCycler at the end of the annealing phase of each cycle. All runs contained distilled water as negative isolation and reaction controls and A. fumigatus DNA in at least three concentrations (20, 2, and 0.2 ng per reaction mixture) as positive amplification controls. Target DNA in the reactions was measured using the instrument's software with the second derivative maximum method. In this method, the CT value, i.e., the cycle in which the beginning of log-linear amplification can be detected, is determined as the first turning point of the fluorescence curve of each sample. Standard CT-versus-concentration plots were constructed using four dilutions of A. fumigatus DNA (20 ng, 2 ng, 200 pg, and 20 pg per reaction mixture). These were amplified in each run from which measurements were required.
Specificity tests.
The specificity of amplification and detection was assessed by amplifying 20 ng of DNA isolated from the fungal strains listed in Table 1. In addition, specificity was tested against DNA from an array of bacteria possibly present in respiratory specimens from immunosuppressed patients (Table 1).
Comparison of analytical sensitivity of different DNA isolation methods.
The efficacy of various DNA isolation systems in releasing aspergillus DNA from the fungal cells for PCR was assessed using suspensions of Aspergillus conidia in saline. After an 11-day culture, conidia from A. fumigatus strain TUCH-01-ss-178 were collected from the young but actively sporulating zone that was 0.5 to 1.5 cm inwards of the edge of the fungal colony and suspended in 1.5% sodium dodecyl sulfate-saline. The number of conidia per milliliter of suspension was determined in a Buerger's calculation chamber under a light microscope and adjusted to 106 conidia per milliliter, and then a series of 10-fold dilutions to a concentration of 1 conidium per ml was prepared. DNA was isolated by using the phenol-ether extraction method described above, with or without bead beating, or by using one of the commercial kits. The kits tested were the DNA-Pure yeast genomic kit (CPG Inc., Lincoln Park, N.J.), the QIAmp DNA mini kit (Qiagen GmbH, Hilden, Germany), the Masterpure DNA purification kit (Epicentre Technologies, Madison, Wis.), and the High Pure PCR template preparation kit (Roche Diagnostics). All samples were treated with lyticase as described above, and then the kits were used according to the manufacturers' instructions. At least two separate runs with two to three replicates of each sample in each run were analyzed. The detection limit was determined as the most dilute sample in which amplification of mtDNA could be constantly detected.
Determination of the precision and linearity of the assay.
The total overall precision of the process was evaluated by analyzing suspensions of A. fumigatus conidia in three concentrations (102, 103, and 104 conidia per ml). The suspensions were concentrated to 200 μl by centrifugation, and DNA was isolated with the lyticase-minibeadbeating-phenol extraction protocol described above. Eight replicates of each template preparation, from two separate DNA extractions, were amplified on three runs performed over 2 days. Average CTs and A. fumigatus DNA concentrations calculated by the software, as well as standard deviations (SD), coefficients of variation, and ±2 SD fidelity regions were determined for each level of the suspensions. Linearity of the amplification was assessed in an experiment in which six replicates of each of the four concentrations (ranging from 102 to 105 conidia per ml of saline) were analyzed.
Efficiency of amplification.
The efficiency of amplification in various sample types was evaluated by analyzing serial dilutions of A. fumigatus conidia in pooled BAL fluids or tissue digests and determining the slope of the target concentration-versus-CT plot, according to the instrument manufacturer's instructions (Technical note no. LC12/2000; Roche Molecular Biochemicals).
Clinical specimens.
Eighty-one BAL fluid specimens were collected from 66 patients at risk of invasive aspergillosis because of various immunosuppressive diseases or treatments and from 33 specimens from 33 immunocompetent patients, 2 of whom were diagnosed with allergic alveolitis. A 100-ml volume of sterile saline was used for bronchoalveolar lavations. A 60- to 70-ml volume was reaspirated, of which 5 to 10 ml was available for PCR. Of the BAL fluid volume, 1.5 ml was concentrated by centrifugation, and DNA was isolated according to the lyticase-beadbeating-phenol extraction protocol described above.
Sixteen tissue biopsy specimens were collected from 10 immunocompromised patients. The biopsy specimens were treated with proteinase K and lyticase and beaten with glass beads. Then, DNA was extracted according to the phenol-ether protocol described above. In each case, 2 μl of template DNA was amplified according to the PCR protocol described above, and the CTs were recorded.
The results of fungal cultures, serum galactomannan antigen tests, and radiological findings from patients at risk of IPA were collected. The patients were designated as having proven, probable, or possible IPA according to the recently published consensus criteria (1). The local ethical committee approved the study, and informed consent was obtained from the patients.
RESULTS
Specificity of the assay.
All 20 A. fumigatus strains tested were amplified in the PCR assay. The four Neosartorya strains known to be sexual (meiosporic) stages of A. fumigatus-related conidiating (mitosporic) fungi also yielded a positive result (7). All other fungal strains, including the pathogenic Aspergillus species A. flavus and A. niger, remained negative, as did all bacteria tested (Table 1).
Analytical sensitivity of the assay.
The lowest amount of purified A. fumigatus DNA constantly detectable by the assay was 20 pg per reaction mixture, which was amplified in 14 consecutive runs. In 10 runs, 2 pg per reaction mixture was detected. The lowest detectable number of conidia in 1 ml of saline varied according to the DNA isolation method. Using phenol extraction, 103 conidia in 1 ml of saline (DNA from 10 conidia per reaction mixture) could be constantly detected. The detection limit was 103 conidia per ml for the High Pure kit, 104 conidia per ml for the Qiagen and Masterpure kits, and 105 conidia per ml for the yeast genomic kit. Beating with glass beads for 1 min before phenol extraction improved sensitivity, so that 102 conidia per ml always yielded detectable amplification, whereas the beating did not affect the detection limit when DNA was purified using the High Pure kit.
Precision and linearity of the assay.
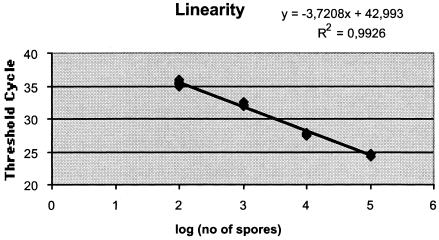
Figure 1 shows the results of the linearity experiment. In this experiment, CTs were plotted against the known concentrations of conidia in the original samples. If the number of conidia in the sample was calculated using three of the six replicates as standards and three as unknowns (modified from the method of Lovatt [23]), all replicates were within 1.5-fold variation at each level (102 to 105 conidia per ml). The overall precision of the PCR assay was assessed by analyzing the CTs determined for the 24 replicates of samples with 104, 103, and 102 conidia. The overall coefficients of variation were 0.59, 1.02, and 2.72%, respectively.
FIG. 1.
Linearity of amplification by the A. fumigatus PCR assay as determined from threshold cycles obtained by amplification of six replicates of each DNA preparation extracted from A. fumigatus conidial suspensions containing 105, 104, 103, or 102 spores per ml.
The ability of the A. fumigatus assay to distinguish between different levels of target cell concentrations was assessed by calculating the ±2 SD (95%) and ±3 SD (99%) fidelity regions for quantification by CT and by the concentration of A. fumigatus DNA in the reaction mixture as calculated using external standards. For samples with 104 conidia per ml, the average CT was 27.91 and the ±2 SD fidelity region was 27.59 to 28.24. The corresponding CTs and fidelity regions for samples with 103 and 102 conidia were 31.40 (±2 SD of 30.76 to 32.05) and 34.31 (±2 SD of 32.45 to 36.18), indicating that the two lowest levels could also be discriminated in >95% of cases. The ±3 SD fidelity regions for CT of the two lowest levels overlapped, as did the ±2 SD fidelity regions for calculated concentrations.
Efficiency of amplification.
Amplification efficiency was equal (1.7) in DNA standards and A. fumigatus-spiked BAL fluids, both of which had been purified according to the phenol protocol. In pooled lung tissue spiked with A. fumigatus conidia, the maximal efficiency of amplification (2.0) was achieved.
Detection of A. fumigatus mtDNA in BAL fluids.
Of the 66 patients at risk, the diagnosis of IPA was proven in 7 (11%), probable in 4 (6%), and possible in 5 (7.5%), according to current consensus criteria (1). Amplification of Aspergillus DNA was detected in 16 of 81 (20%) BAL fluid specimens from patients at risk and in 1 of 33 (3%) samples from immunocompetent controls. PCR was positive in six of seven (86%), two of four (50%), and four of five (80%) patients with proven, probable, and possible IPA, respectively, as well as in 4 of 50 (8%) patients at risk but without any evidence of IPA. All PCR-negative BAL fluid samples from patients with proven, probable, or possible IPA were retested after adding 20 pg of A. fumigatus DNA. None showed inhibition of PCR. Three patients with proven or probable IPA were positive by galactomannan antigen testing but negative by PCR, and two were positive by PCR but negative by antigen testing. Table 2 shows detailed data on patients with positive findings by PCR or antigen testing. At qualitative detection, the diagnostic sensitivity of the PCR per patient was 72%, specificity was 93%, and the predictive values of positive and negative results were 73 and 95%, respectively. Using a CT of <35 as a limit for positive PCR, the specificity and positive predictive value of PCR in the diagnosis of IPA in immunocompromised patients were 100%, but sensitivity was only 45% and negative predictive value was 92%.
TABLE 2.
Patients at risk of aspergillosis with A. fumigatus detected in BAL fluid by PCR or fungal culture or by galactomannan antigen detection in serum or BAL fluid
| No. | Age/sex | Underlying diseasea | Culture | Site of culture |
Aspergillus antigen
|
HRCTb | PCR (CT) | Commentc | Outcome | IPA category | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BAL | Serumd | ||||||||||
| 1 | 27/M | ALL/BMT | A. niger, A. fumigatus | BAL | Pos | Pos (×3) | Pos | Pos (31.61) | Fungal cerebral abscesses | Died | Proven |
| 2 | 47/M | Myeloma/BMT | Aspergillus sp. | Lung | Neg | Neg | Pos | Pos (36.6) | Cured with L-AmB | Survived | Proven |
| 3 | 54/M | Renal transplant | A. fumigatus | Lung and brain | Pos | Pos (×2) | Pos | Pos (25.97) | Pulmonary and cerebral aspergillosis | Died | Proven |
| 4 | 47/M | CML/BMT | Aspergillus sp. | Lung and brain | Not done | Pos (×2) | Pos | Pos (>41) | Pulmonary and cerebral aspergillosis | Died | Proven |
| 5 | 41/F | CLL | A. fumigatus | BAL | Pos | Pos (×3) | Pos | Neg | A. fumigatus pneumonia, cured with L-AmB | Survived | Proven |
| 6 | 66/M | Bronchiectasia | A. fumigatus | Pleural fluid | Neg | Neg | Pos | Pos (25.84) | Empyema, treated with ABLO + Cf + Vc | Survived | Proven |
| 7 | 61/F | Wegener's disease | A. fumigatus, A. niger | BAL | Pos | Neg | Not done | Pos (32.09) | Post mortem: necrotizing foci of aspergillosis | Died | Proven |
| 8 | 30/M | AML/BMT | A. fumigatus | BAL | Neg | Neg | Pos | Pos (31.68) | Cured with L-AmB | Survived | Probable |
| 9 | 43/M | NHL/APBSCT | No fungal growth | BAL liver | Neg | Pos (×5) | Pos | Neg | Pulmonary and hepatic aspergillosis | Survived | Probable |
| 7 | 55/F | NHL/APBSCT | No fungal growth | BAL | Pos | Pos (×2) | Pos | Pos (37.73) | Cured with L-AmB | Survived | Probable |
| 11 | 72/F | CLL | Paecilomyces sp. | BAL | Pos | Pos (×2) | Pos | Neg | Cured with L-AmB | Survived | Probable |
| 12 | 68/M | Myeloma | No fungal growth | BAL | Neg | Pos | Not done | Pos (>41) | HSV + BAL | Survived | Possible |
| 13 | 52/M | NHL/BMT | S. cerevisiae | BAL | Neg | Neg | Pos | Pos (36.83) | The etiology of pulmonary infiltration? | Died | Possible |
| 14 | 63/M | Renal transplant | A. fumigatus, A. flavus | BAL | Not done | Neg | Not done | Pos (38.37) | Survived | Possible | |
| 15 | 55/M | AML/BMT | S. cerevisiae | BAL | Pos | Pos | Pos | Neg | Post mortem: no findings related to IPA | Died | Possible |
| 16 | 44/M | Hodgkin's lymphoma | No fungal growth | BAL | Neg | Neg | Pos | Pos (35.05) | Survived | Possible | |
| 17 | 17/M | ALL/BMT | No fungal growth | BAL | Not done | Neg | Neg | Pos (36.71) | Post mortem: no findings related to IPA | Died | Not probable |
| 18 | 52/M | CML | No fungal growth | BAL | Neg | Neg | Neg | Pos (36.84) | Survived | Not probable | |
| 19 | 75/F | CLL | No fungal growth | BAL | Neg | Not done | Not done | Pos (40.72) | Died | Not probable | |
| 20 | 78/M | MDS | No fungal growth | BAL | Neg | Neg | Not done | Pos (>41) | Survived | Not probable | |
ALL, acute lymphoblastic leukemia; BMT, bone marrow transplantation; CML, chronic myelogenous leukemia; CLL, chronic lymphocytic leukemia; NHL, non-Hodgkin's lymphoma; APBSCT, autologous peripheral blood stem cell transplantation; MDS, myelodysplastic syndrome.
A positive HRCT indicates findings consistent with invasive pulmonary fungal infection.
L-AmB, liposomal amphotericin B; ABLC, amphotericin B lipid complex; Cf, caspofungin; Vc, voriconazole. HSV, herpes simplex virus.
Values in parentheses are numbers of separate serum samples in which galactomannan antigen was detected.
Detection of A. fumigatus DNA in tissue biopsy specimens.
Table 3 presents the results of PCR and fungal cultures from 16 tissue biopsy specimens of immunocompromised patients as well as the results of galactomannan antigen testing and clinical data on the patients. Six of the patients had invasive aspergillosis, one of which was caused by A. flavus instead of A. fumigatus. PCR was positive in all patients with culture-confirmed IPA caused by A. fumigatus. Both PCR and culture on lung biopsy specimens remained negative in a patient with cerebral aspergillosis diagnosed post mortem. The mean CT value of the PCR-positive biopsy specimens was 28.39.
TABLE 3.
Tissue biopsy specimens from patients at risk of aspergillosis
| No. | Age/sex | Underlying diseasea | Tissue | Culture | PCR | S-agc | Diagnosis |
|---|---|---|---|---|---|---|---|
| 1 | 25/M | ALL | Lung | A. flavus | Negative | Positive (×3) | IPA (proven) |
| 2 | 27/M | ALL | Brain | Negative (hyphae seen) | Positive | Positive (×3) | IA (proven) |
| Kidney | A. fumigatus | Positive | |||||
| Brain | Not done | Negative | |||||
| Liver biopsy | Not done | Positive | |||||
| Spleen | No fungal growth | Positive | |||||
| 3 | 53/M | Collagenosis, DM | Brain | A. fumigatus | Positive | Positive | Rhinocerebral aspergillosis |
| 4 | 31/M | MDS, MUD transplantation | Lung | No fungal growth | Negative | Positive (X2) | IA (CNS, post mortem) |
| 5 | 66/M | Aplastic anemia/BMT | Liver | No fungal growth | Negative | Positive | IA (probable)b |
| 6 | 57/M | CLL | Lung | A. fumigatus | Positive | Positive (×4) | IPA (proven) |
| Liver | Not done | Negative | |||||
| 7 | 54/M | AML | Lung | C. tropicalis | Negative | Negative | Sepsis (C. tropicalis) |
| Liver | C. albidus, Acremonium sp. | Negative | |||||
| 8 | 29/F | ALL | Lung | No fungal growth | Negative | Negative | Mucormycosis (histology) |
| 9 | 49/M | NHL | Lung | C. albicans | Negative | Negative | Pneumonia |
| 10 | 70/F | MDS-AML | Lung | C. albicans | Negative | Negative | Pneumonia |
ALL, acute lymphoblastic leukemia; DM, diabetes mellitus; MDS, myelodysplastic syndrome; MUD, matched unrelated donor; BMT, bone marrow transplantation; CLL, chronic lymphocytic leukemia; AML, acute myelogenous leukemia; NHL, non-Hodgkin's lymphoma.
RCT of liver was consistent with invasive fungal infection.
S-ag, galactomannan antigen in serum. Values in parentheses are the numbers of separate serum samples in which galactomannan antigen was detected.
Patient 3 (Table 3) with rhinocerebral aspergillosis exemplifies the value of PCR in the diagnosis of fungal infections. The patient had diabetes mellitus and was receiving high-dose corticosteroids and intravenous cyclophosphamide for collagenosis. He experienced visual loss in one eye and was operated on because a tumor compressing the optical nerve was suspected. As the patient had no symptoms suggestive of infection, microbiological samples were not obtained. Surprisingly, invasive fungal hyphae were detected in biopsy specimens taken for histopathological diagnosis. PCR on sections of paraffinized tissue showed the presence of A. fumigatus DNA. Later on, the mold could also be cultured from biopsy specimens obtained at a reoperation.
DISCUSSION
We report the development of a semiquantitative real-time PCR assay for detection of A. fumigatus DNA in BAL fluids and tissue biopsy specimens. The assay was carefully validated and applied to an analysis of 81 BAL fluid specimens from 66 patients and 16 tissue biopsy specimens from 10 patients at risk of invasive aspergillosis owing to their underlying disease and/or immunosuppressive treatment.
The specificity of amplification was tested against a panel of fungal strains. All 20 A. fumigatus strains tested were amplified in the PCR assay with FRET detection. As we expected on the basis of morphological taxonomy, the four Neosartorya strains also yielded a positive result. All known Aspergillus species are asexual (mitosporic) stages of fungi, and their full life cycles include a sexual (meiosporic) stage in the various genera of the Ascomycetes class. The sexual stage of aspergilli morphologically similar to the A. fumigatus group, as far as the full life cycles are known, belongs to the ascomycetous genus Neosartorya. Neosartorya species have been very seldom isolated from clinical specimens (7).
All other fungal strains, including the pathogenic Aspergillus species A. flavus and A. niger, remained negative, as did all bacteria tested. Our assay was designed to be specific for A. fumigatus, which is the most common Aspergillus species causing invasive disease. Indeed, our assay did not detect the one invasive disease caused by A. flavus. According to the original description of the specificity of the primers (15) and our preliminary tests with SYBR Green I detection, the primers also amplified A. flavus mtDNA, but that amplification was not visualized with the FRET probes used. An additional probe pair for detection of A. flavus might be useful.
According to the recently published international consensus on diagnostic criteria (1), a diagnosis of “proven” invasive aspergillosis can be established by culture from a sterile tissue biopsy specimen or by needle aspiration, or by microscopic detection of branching septate hyphae in such samples with histopathological evidence of associated tissue damage. Isolation of Aspergillus from respiratory specimen or sinus aspirate is regarded as evidence for “probable” infection in a high-risk patient with relevant clinical and radiological findings, and so is detection of Aspergillus antigen in BAL or cerebrospinal fluid or its repeated detection in serum. The radiological findings include typical pulmonary HRCT findings, such as the halo sign, air-crescent sign, or a cavity with a consolidated area, but also findings in other tissues suggestive of fungal infection. If both radiological and mycological evidence for invasive aspergillosis is not obtained, the diagnosis can only be classified as “possible.”
In recent studies, the sensitivity of PCR when applied to BAL specimens has been almost 100% in patients having IPA according to the consensus criteria, but false-positive results have occurred with BAL fluids from both immunocompromised and immunocompetent patients at a rate of 3 to 11% (5, 10). Detection of Aspergillus DNA in the circulation as a marker of invasive disease allows repeated noninvasive testing and thus serial screening of patients at risk of invasive disease. Analyzing both blood and BAL samples of 45 patients, 8 of them with invasive aspergillosis, Buchheidt et al. (5) had slightly better results from BAL than from the blood (sensitivity of 100% versus 91.7%, specificity of 92.6% versus 81.3% for BAL and blood, respectively). Recently, they developed a LightCycler assay which successfully detected A. fumigatus DNA in all 12 tested BAL fluids but only in 6 of 14 blood samples from neutropenic patients with proven or probable IPA, which had all been positive by the previously described nested PCR assay (30). Another German group has been concentrating on the detection of Aspergillus DNA in whole blood by 18S rDNA PCR with panfungal primers and species-specific probes (8, 19, 21, 22) or, more recently, on the detection of rRNA by nucleic acid sequence-based amplification (20). Two recent studies evaluated a PCR-ELISA in screening patients at high risk for invasive aspergillosis. Prospective screening of blood samples was reported to have a diagnostic sensitivity of 100% and a specificity of 65% when single positive results were considered (11) and a sensitivity of 75% and specificity of 96% when at least two consecutive positive results were regarded as indicative of invasive disease (17). To date, no prospective comparison of antigen detection by ELISA and DNA detection by PCR in screening patients at risk of IPA has been published. Results from retrospective comparisons suggest that detection of galactomannan by ELISA is more sensitive than amplification of Aspergillus mtDNA from serum (2, 6). According to the results obtained by quantitative PCR, the concentration of Aspergillus DNA in patient sera is less than 30 fg/ml (6) and that in whole blood is 10 to 100 fg/ml (21). The concentration of galactomannan is in nanograms per milliliter of serum. This is not surprising, since galactomannan is a major component of the Aspergillus cell wall and is released to the medium in large quantities in fungal cultures (18), whereas circulating DNA may be derived from a few hyphae that enter the bloodstream from the site of invasive infection.
Exact measurement of fungal DNA in BAL fluids would be difficult. Although the linearity of our assay was good and the reproducibility of CT values was as good as is achievable, according to the manufacturer, using the LightCycler instrument, our results showed that just above the detection limit, samples with 102 or 103 conidia per ml (1 to 10 conidia per reaction mixture) could be discriminated from each other, but more precise quantification would have been impossible. Most A. fumigatus-positive BAL specimens had CT values close to those of these simulated specimens with the lowest concentrations of conidia. According to a Poisson distribution, the sampling variation alone increases rapidly when less than 1,000 target gene copies are present in the reaction (31). Estimation of target gene copy numbers in our samples is difficult, as the number of mitochondria per fungal cell varies. In addition, fungal conidia may be more resistant to the DNA purification procedure than fungal hyphae, which are the cell form present in clinical specimens. Further inaccuracy is brought about by using bronchoalveolar lavation as the sampling method. IPA is a focal disease, and the alveoli that are sampled and the amount of fluid that can be reaspirated affect the concentration of fungal cells in the BAL fluid specimen. Because of these inaccuracies, a semiquantitative approach based on CTs was selected to determine the amount of fungal mtDNA in the specimens. Quantification of A. fumigatus DNA in tissue biopsy specimens would not have been possible, because amplification efficiency in them differed from that in the standards used. Neither would quantification be necessary, because qualitative detection of the fungus in deep tissues is diagnostic (1).
Compared to the results presented by other authors, our assay had modest analytical sensitivity, considering the minimal amount of DNA constantly detected (picograms), and it also showed modest clinical sensitivity in analyzing BAL specimens. Its clinical sensitivity might be somewhat improved by increasing the template volume, since the amount of fungal DNA in the specimens was low, as judged from the late CTs. Implementing an internal control for the detection of any inhibition could also be useful, although inhibition was not detected in the false-negative specimens and the use of an internal control might compromise the detection of small amounts of target DNA. Semiquantitative detection of A. fumigatus DNA could not discriminate between colonization and invasion very well, but reporting of samples with a CT of <35 as positive and those with a CT of >35 as borderline might be useful for clinical purposes. In comparing PCRs to galactomannan antigen tests, three patients with proven or probable IPA were positive in antigen testing but negative in PCR, and two were positive in PCR but negative in antigen testing. Thus, the combination of semiquantitative PCR and antigen detection may help confirm the microbiological diagnosis of IPA. PCR can also complement HRCT findings suggestive of fungal infections, as many molds can produce indistinguishable radiological changes. A further application area is the confirmation of histological findings, as identification of the invading fungus may be possible by molecular methods, when culture has not been done or is inhibited by empirical antifungal treatment.
Acknowledgments
Elizabeth Johnson, Head of the PHLS Mycology Reference Laboratory, Bristol, United Kingdom, is acknowledged for help with identification of UKNEQAS fungal strains.
Tiina Haarala, Kaisa Leppänen, and Tarja Laine, as well as the personnel of the Mycology and Parasitology Laboratory of the Turku University Central Hospital, are thanked for excellent technical assistance, and Simo Merne is thanked for revising the language of the manuscript. This study was supported by an EVO grant of the Turku University Central Hospital.
REFERENCES
- 1.Ascioglu, S., J. H. Rex, B. de Pauw, J. E. Bennett, J. Bille, F. Crokaert, D. W. Denning, J. P. Donnelly, J. E. Edwards, Z. Erjavec, D. Fiere, O. Lortholary, J. Maertens, J. F. Meis, T. F. Patterson, J. Ritter, D. Selleslag, P. M. Shah, D. A. Stevens, and T. J. Walsh. 2002. Defining opportunistic invasive fungal infections in immunocompromised patients with cancer and hematopoietic stem cell transplants: an international consensus. Clin. Infect. Dis. 34:7-14. [DOI] [PubMed] [Google Scholar]
- 2.Bretagne, S., J. M. Costa, E. Bart-Delabesse, N. Dhedin, C. Rieux, and C. Cordonnier. 1998. Comparison of serum galactomannan antigen detection and competitive polymerase chain reaction for diagnosing invasive aspergillosis. Clin. Infect. Dis. 26:1407-1412. [DOI] [PubMed] [Google Scholar]
- 3.Bretagne, S., J. M. Costa, A. Marmorat-Khuong, F. Poron, C. Cordonnier, M. Vidaud, and J. Fleury-Feith. 1995. Detection of Aspergillus species DNA in bronchoalveolar lavage samples by competitive PCR. J. Clin. Microbiol. 33:1164-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchheidt, D., C. Baust, H. Skladny, M. Baldus, S. Brauninger, and R. Hehlmann. 2002. Clinical evaluation of a polymerase chain reaction assay to detect Aspergillus species in bronchoalveolar lavage samples of neutropenic patients. Br. J. Haematol. 116:803-811. [DOI] [PubMed] [Google Scholar]
- 5.Buchheidt, D., C. Baust, H. Skladny, J. Ritter, T. Suedhoff, M. Baldus, W. Seifarth, C. Leib-Moesch, and R. Hehlmann. 2001. Detection of Aspergillus species in blood and bronchoalveolar lavage samples from immunocompromised patients by means of 2-step polymerase chain reaction: clinical results. Clin. Infect. Dis. 33:428-435. [DOI] [PubMed] [Google Scholar]
- 6.Costa, C., J. M. Costa, C. Desterke, F. Botterel, C. Cordonnier, and S. Bretagne. 2002. Real-time PCR coupled with automated DNA extraction and detection of galactomannan antigen in serum by enzyme-linked immunosorbent assay for diagnosis of invasive aspergillosis. J. Clin. Microbiol. 40:2224-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Hoog, G. S., J. Guarro, and M. J. Figueras. 2000. Atlas of clinical fungi, 2nd ed. Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands.
- 8.Einsele, H., H. Hebart, G. Roller, J. Loffler, I. Rothenhofer, C. A. Muller, R. A. Bowden, J. van Burik, D. Engelhard, L. Kanz, and U. Schumacher. 1997. Detection and identification of fungal pathogens in blood by using molecular probes. J. Clin. Microbiol. 35:1353-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Girmenia, C., M. Nucci, and P. Martino. 2001. Clinical significance of Aspergillus fungaemia in patients with haematological malignancies and invasive aspergillosis. Br. J. Haematol. 114:93-98. [DOI] [PubMed] [Google Scholar]
- 10.Hayette, M. P., D. Vaira, F. Susin, P. Boland, G. Christiaens, P. Melin, and P. De Mol. 2001. Detection of Aspergillus species DNA by PCR in bronchoalveolar lavage fluid. J. Clin. Microbiol. 39:2338-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hebart, H., J. Loffler, H. Reitze, A. Engel, U. Schumacher, T. Klingebiel, P. Bader, A. Bohme, H. Martin, D. Bunjes, W. V. Kern, L. Kanz, and H. Einsele. 2000. Prospective screening by a panfungal polymerase chain reaction assay in patients at risk for fungal infections: implications for the management of febrile neutropenia. Br. J. Haematol. 111:635-640. [DOI] [PubMed] [Google Scholar]
- 12.Hendolin, P. H., L. Paulin, P. Koukila-Kahkola, V. J. Anttila, H. Malmberg, M. Richardson, and J. Ylikoski. 2000. Panfungal PCR and multiplex liquid hybridization for detection of fungi in tissue specimens. J. Clin. Microbiol. 38:4186-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horvath, J. A., and S. Dummer. 1996. The use of respiratory-tract cultures in the diagnosis of invasive pulmonary aspergillosis. Am. J. Med. 100:171-178. [DOI] [PubMed] [Google Scholar]
- 14.Jalava, J., P. Kotilainen, S. Nikkari, M. Skurnik, E. Vanttinen, O. P. Lehtonen, E. Eerola, and P. Toivanen. 1995. Use of the polymerase chain reaction and DNA sequencing for detection of Bartonella quintana in the aortic valve of a patient with culture-negative infective endocarditis. Clin. Infect. Dis. 21:891-896. [DOI] [PubMed] [Google Scholar]
- 15.Jones, M. E., A. J. Fox, A. J. Barnes, B. A. Oppenheim, P. Balagopal, G. R. Morgenstern, and J. H. Scarffe. 1998. PCR-ELISA for the early diagnosis of invasive pulmonary aspergillus infection in neutropenic patients. J. Clin. Pathol. 51:652-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kami, M., T. Fukui, S. Ogawa, Y. Kazuyama, U. Machida, Y. Tanaka, Y. Kanda, T. Kashima, Y. Yamazaki, T. Hamaki, S. Mori, H. Akiyama, Y. Mutou, H. Sakamaki, K. Osumi, S. Kimura, and H. Hirai. 2001. Use of real-time PCR on blood samples for diagnosis of invasive aspergillosis. Clin. Infect. Dis. 33:1504-1512. [DOI] [PubMed] [Google Scholar]
- 17.Lass-Florl, C., J. Aigner, E. Gunsilius, A. Petzer, D. Nachbaur, G. Gastl, H. Einsele, J. Loffler, M. P. Dierich, and R. Wurzner. 2001. Screening for Aspergillus spp. using polymerase chain reaction of whole blood samples from patients with haematological malignancies. Br. J. Haematol. 113:180-184. [DOI] [PubMed] [Google Scholar]
- 18.Latge, J. P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loeffler, J., H. Hebart, U. Brauchle, U. Schumacher, and H. Einsele. 2000. Comparison between plasma and whole blood specimens for detection of Aspergillus DNA by PCR. J. Clin. Microbiol. 38:3830-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loeffler, J., H. Hebart, P. Cox, N. Flues, U. Schumacher, and H. Einsele. 2001. Nucleic acid sequence-based amplification of Aspergillus RNA in blood samples. J. Clin. Microbiol. 39:1626-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loeffler, J., N. Henke, H. Hebart, D. Schmidt, L. Hagmeyer, U. Schumacher, and H. Einsele. 2000. Quantification of fungal DNA by using fluorescence resonance energy transfer and the LightCycler system. J. Clin. Microbiol. 38:586-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loffler, J., H. Hebart, S. Sepe, U. Schumcher, T. Klingebiel, and H. Einsele. 1998. Detection of PCR-amplified fungal DNA by using a PCR-ELISA system. Med. Mycol. 36:275-279. [DOI] [PubMed] [Google Scholar]
- 23.Lovatt, A. 2002. Applications of quantitative PCR in the biosafety and genetic stability assessment of biotechnology products. J. Biotechnol. 82:279-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maertens, J., J. Verhaegen, K. Lagrou, J. Van Eldere, and M. Boogaerts. 2001. Screening for circulating galactomannan as a noninvasive diagnostic tool for invasive aspergillosis in prolonged neutropenic patients and stem cell transplantation recipients: a prospective validation. Blood 97:1604-1610. [DOI] [PubMed] [Google Scholar]
- 25.Melchers, W. J., P. E. Verweij, P. van den Hurk, A. van Belkum, B. E. De Pauw, J. A. Hoogkamp-Korstanje, and J. F. Meis. 1994. General primer-mediated PCR for detection of Aspergillus species. J. Clin. Microbiol. 32:1710-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinel, C., H. Fricker-Hidalgo, B. Lebeau, F. Garban, R. Hamidfar, P. Ambroise-Thomas, and R. Grillot. 2003. Detection of circulating Aspergillus fumigatus galactomannan: value and limits of the Platelia test for diagnosing invasive aspergillosis. J. Clin. Microbiol. 41:2184-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salonen, J., O. P. Lehtonen, M. R. Terasjarvi, and J. Nikoskelainen. 2000. Aspergillus antigen in serum, urine and bronchoalveolar lavage specimens of neutropenic patients in relation to clinical outcome. Scand. J. Infect. Dis. 32:485-490. [DOI] [PubMed] [Google Scholar]
- 28.Seyfarth, H.-J., P. Nenoff, J. Winkler, R. Krahl, U. F. Haustein, and J. Schauer. 2001. Aspergillus detection in bronchoscopically acquired material. Significance and interpretation. Mycoses 44:356-360. [DOI] [PubMed] [Google Scholar]
- 29.Skladny, H., D. Buchheidt, C. Baust, F. Krieg-Schneider, W. Seifarth, C. Leib-Mosch, and R. Hehlmann. 1999. Specific detection of Aspergillus species in blood and bronchoalveolar lavage samples of immunocompromised patients by two-step PCR. J. Clin. Microbiol. 37:3865-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spiess, B., D. Buchheidt, C. Baust, H. Skladny, W. Seifarth, U. Zeilfelder, C. Leib-Mosch, H. Morz, and R. Hehlmann. 2003. Development of a LightCycler PCR assay for detection and quantification of Aspergillus fumigatus DNA in clinical samples from neutropenic patients. J. Clin. Microbiol. 41:1811-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stenman, J., and A. Orpana. 2001. Accuracy in amplification. Nat. Biotechnol. 19:1011-1012. [DOI] [PubMed] [Google Scholar]
- 32.Stynen, D., A. Goris, J. Sarfati, and J. P. Latge. 1995. A new sensitive sandwich enzyme-linked immunosorbent assay to detect galactofuran in patients with invasive aspergillosis. J. Clin. Microbiol. 33:497-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verweij, P. E., J. P. Latge, A. J. Rijs, W. J. Melchers, B. E. De Pauw, J. A. Hoogkamp-Korstanje, and J. F. Meis. 1995. Comparison of antigen detection and PCR assay using bronchoalveolar lavage fluid for diagnosing invasive pulmonary aspergillosis in patients receiving treatment for hematological malignancies. J. Clin. Microbiol. 33:3150-3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.von Eiff, M., N. Roos, R. Schulten, M. Hesse, M. Zuhlsdorf, and J. van de Loo. 1995. Pulmonary aspergillosis: early diagnosis improves survival. Respiration 62:341-347. [DOI] [PubMed] [Google Scholar]