Abstract
A multiplex PCR and an improved lead acetate test were developed to discriminate d-tartrate-fermenting and -nonfermenting Salmonella enterica subsp. enterica strains. Both methods showed an accuracy of 100% when 125 Salmonella strains belonging to 15 serovars were tested. Special emphasis was given to S. enterica subsp. enterica serovar Paratyphi B isolates because of the clinical importance of its d-tartrate-nonfermenting variant and the recently increasing numbers of cases of human outbreaks caused by its fermenting variant (formerly Salmonella serovar Java). The lead acetate test described previously (G. A. Alfredsson, R. M. Barker, D. C. Old, and J. P. Duguid, J. Hyg. 70:651-666, 1972) was modified in the inoculation and incubation procedure. The PCR assay was based on the genotypic difference of the presence (d-tartrate-fermenting strains) or absence (d-tartrate-nonfermenting strains) of the ATG start codon for the gene STM 3356, which encodes a putative cation transporter. Sequence data revealed a nucleotide exchange from G to A within the ATG start codon of gene STM 3356 in the d-tartrate-nonfermenting strains. In order to increase the reliability of the PCR assay, a positive control based on a Salmonella genus-specific primer set for the detection of Salmonella DNA was included. The PCR-based discrimination needs only several hours compared to 6 days needed by the improved lead acetate test to obtain reliable results. Consequently, the PCR d-tartrate assay should be the method of choice for the discrimination of d-tartrate-fermenting and -nonfermenting Salmonella strains in the future.
Salmonella enterica subspecies enterica serovar Paratyphi B can be differentiated by the fermentation of dextrorotatory [l(+)]-tartrate (d-tartrate). Kristensen and Kauffmann (7) noted a difference in clinical manifestation of both variants. Whereas d-tartrate-nonfermenting (dT−) strains exhibit an enhanced human pathogenicity causing typhoid-like disease, d-tartrate-fermenting strains (dT+) provoke the less severe gastroenteric disease. Formerly, dT+ strains of this serovar were designated S. enterica subsp. enterica serovar Java by Kauffmann (6). Recently, the Salmonella serovar Paratyphi B dT+ variant has become increasingly important. In The Netherlands, it is the predominating serovar in poultry (14) and has led to human outbreaks in Denmark, Scotland (4), and Canada (13). Therefore, the rapid identification of dT+ and dT− serovar Paratyphi B strains isolated from animals and humans is important for Salmonella risk assessment and adequate surveillance interventions. The discrimination of dT+ and dT− isolates has also been used for the biotyping of other Salmonella serovars (2, 10).
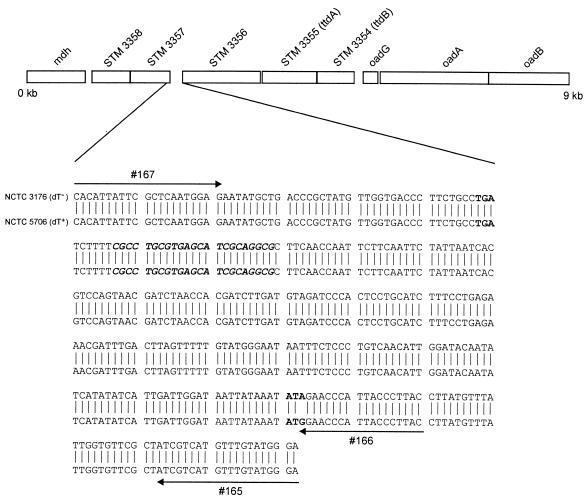
Anaerobic fermentation of tartrate usually proceeds via stereospecific dehydratases to oxaloacetate, which is further converted via pyruvate to acetate, formate, and CO2, thus allowing the synthesis of ATP in the acetate kinase reaction (12). Previously it was shown that anaerobic growth of S. enterica subsp. enterica serovar Typhimurium in the presence of d-tartrate involves an oxaloacetate decarboxylase Na+ pump encoded by the oadGAB genes (15). Upstream of these genes, two open reading frames have been identified; these open reading frames show homology with the ttdB (67%) and ttdA (58%) genes of Escherichia coli (GenBank accession no. L14781), which encode the subunits of the oxygen-labile stereospecific l-tartrate dehydratase (11). These two genes were designated STM 3354 and STM 3355 in the complete genome sequence of Salmonella serovar Typhimurium strain LT2 (GenBank accession no. AE008854). Upstream of the ttdA and ttdB genes, an open reading frame encoding a putative cation transporter (STM 3356), two open reading frames encoding gntR family regulatory proteins (STM 3357 and STM 3358), and the mdh gene encoding the malate dehydrogenase were noted (Fig. 1).
FIG. 1.
Diagram of a 9-kb region derived from the Salmonella serovar Typhimurium strain LT2 genome sequence (GenBank accession no. AE008854). A comparison of the DNA sequences of the reference strains NCTC 3176 (dT−) and NCTC 5706 (dT+) between the C terminus of the STM 3357 gene coding region and the N terminus of the STM 3356 gene coding region is shown below the diagram. Relevant primers used for the PCR-based discrimination are marked by arrows. The TGA stop codon of gene STM 3357, a putative palindromic transcriptional terminator, and the ATG start codon of gene STM 3356 are indicated in bold. The start codon of gene STM 3356 in dT− Salmonella strains is absent due to a nucleotide exchange from G to A (marked in bold). The 3′ end of primer 166 fits with the nucleotide G and results in a PCR product of 290 bp, with primer 167 using DNA of strain NCTC 5706 as a template. No PCR signal would be obtained with DNA of strain NCTC 3176 due to the presence of the nucleotide A in the template.
Several biochemical tests, such as the anaerobic plate test, the lead acetate test, and the turbidity test, have been developed to discriminate dT+ and dT− Salmonella strains. A comparison of these methods revealed poor correlation and reproducibility (3). Currently, the WHO Collaborating Centre for Reference and Research on Salmonella located at the Institute Pasteur (Paris, France) uses the lead acetate test as a “gold standard”-like method as described by Alfredsson et al. (2) but with a prolongation of the incubation times to 3 and 6 days. The aim of this study was to develop a rapid PCR-based method for the discrimination of dT+ and dT− S. enterica subsp. enterica isolates. The accuracy of the PCR-based method was compared to that of the lead acetate test as used by the WHO Collaborating Centre. We have found that modifications of the inoculation and incubation conditions of the lead acetate test resulted in 100% accuracy with the PCR-based method.
MATERIALS AND METHODS
Salmonella strains.
Serovars, sources, and countries and years of isolation of the strains used in this study are summarized in Table 1. Most of the strains were isolated from various regions in Germany between the years 1961 and 2002 and collected at the German National Reference Laboratory for Salmonella (NRL-Salm, BfR, Berlin, Germany). The strains originated from animals, foods, and environmental and human sources. The molecular properties of many serovar Paratyphi B dT+ strains used in this study have been described before (9).
TABLE 1.
Results of the discrimination of dT+ and dT− S. enterica subsp. enterica strains by three lead acetate test protocols and a PCR-based method
| Serovar | Total no. of strains tested | Source(s) (no. of strains) | Country of isolation (no. of strains) | Yr(s) of isolation | No. of strains determined to be dT+ bya:
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PCR | Lead acetate testb
|
||||||||||
| Protocol 1
|
Protocol 2
|
Protocol 3
|
|||||||||
| 3 days | 6 days | 3 days | 6 days | 3 days | 6 days | ||||||
| Paratyphi B | 81 | Chicken (37), environment (7), human (7), turkey (3), dog (2), others (25) | Germany (63), England (3), Belgium (5), Australia (3), Austria (2), unknown (2), France (2), The Netherlands (1) | 1961-2002 | 81 | 33c | 71d | 44e | 73 | 68 | 81 |
| Paratyphi B | 21 | Human (10), mussel (2), water (1), sheep (1), sewage (1), others (6) | Germany (14), France (4), The Netherlands (2), unknown (1) | 1964-2002 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Typhimurium | 6 | Minced meat (2), feces (1), sausage (1), ice cream (1), other (1) | Germany (5), unknown (1) | 1996-2000 | 6 | 0 | 6 | 2 | 6 | 2 | 6 |
| Enteritidis | 5 | Minced meat (1), chicken (1), cake (1), cereal (1), egg (1) | Germany (5) | 1999-2000 | 5 | 3e | 4 | 3 | 4 | 3 | 5 |
| Agona | 1 | Pork | Germany | 1999 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| Blockley | 1 | Turkey | Germany | 1998 | 1 | 0 | 1 | 0 | 1 | 0 | 1 |
| Brandenburg | 1 | Sausage | Germany | 2000 | 1 | 0 | 1 | 0 | 1 | 0 | 1 |
| Bredeney | 1 | Chicken | Germany | 2000 | 1 | 0 | 1 | 0 | 1 | 1 | 1 |
| Derby | 1 | Pork | Germany | 1999 | 1 | 0 | 1 | 0 | 1 | 0 | 1 |
| Dublin | 1 | Minced meat | Germany | 1998 | 1 | 0 | 1 | 0 | 1 | 0 | 1 |
| Hadar | 1 | Goose | Germany | 1999 | 1 | 0 | 1 | 0 | 1 | 0 | 1 |
| Heidelberg | 1 | Chicken | Germany | 2000 | 1 | 0 | 1 | 0 | 0 | 1 | 1 |
| Infantis | 1 | Beef | Germany | 1999 | 1 | 0 | 0 | 0 | 1 | 0 | 1 |
| Montevideo | 1 | Feed | Germany | 1998 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Newport | 1 | Duck | Germany | 1999 | 1 | 0 | 1 | 0 | 1 | 0 | 1 |
| Livingstone | 1 | Minced meat | Germany | 2000 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Strains were tested three times each.
The lead acetate tests were performed after 3 and 6 days of incubation. The accuracies of the lead acetate tests relative to that of the PCR were as follows (after 3 and 6 days of incubation, respectively): for protocol 1, 47.2 and 89.6%; for protocol 2, 57.6 and 91.2%; for protocol 3,78.4 and 100.0%.
Ten strains showed only two dT+ reactions out of triplicate reactions tested.
Four strains showed only two dT+ reactions out of triplicate reactions tested.
Two strains showed only two dT+ reactions out of triplicate reactions tested.
Molecular methods.
For sequencing of the intergenic region of the STM 3357 and STM 3356 genes, two primers were designed based on the sequence of the serovar Typhimurium LT2 strain (GenBank accession no. AE008854), which amplified a 332-bp DNA fragment. Primer 165 (5′-TCC CAT ACA AAC ATG ACG AT-3′) is located at the N-terminal coding region of the STM 3356 gene, and primer 167 (5′-CAC ATT ATT CGC TCA ATG GAG-3′) is located at the C-terminal coding region of the STM 3357 gene (Fig. 1). All PCRs were carried out in a GeneAmp PCR system 9700 thermocycler (Applied Biosystems, Weiterstadt, Germany). A 50-μl PCR mixture contained 0.4 μM concentrations of primers 167 and 165, a 200 μM concentration of each deoxynucleoside triphosphate (Roche Applied Science, Mannheim, Germany), 1× PCR buffer (20 mM Tris-HCl [pH 8.4], 50 mM KCl), 1.5 mM MgCl2, 1 U of Platinum Taq polymerase (Invitrogen, Karlsruhe, Germany), and a 5-μl aliquot of the sample DNA (approximately 106 CFU). The incubation conditions were 95°C for 1 min, followed by 30 cycles of 95°C for 30 s, 53°C for 30 s, and 72°C for 30 s. A final extension of 72°C for 4 min was employed. The PCR products were sequenced by automated cycle sequencing with an ABI Prism 310 genetic analyzer (Applied Biosystems) according to the manufacturer's instructions. PCR for sequencing the ttdA and ttdB genes of serovar Paratyphi B strains NCTC 3176 and NCTC 5706 was carried out with primers 141 (5′-GGT TAT TAT TAT CGT CAT AAC GTA-3′) and 142 (5′-TCA GCT TGA TCC TGC TGC CA-3′). The PCR conditions used were those described above. The 1,652-bp PCR product was sequenced with primers derived from the published serovar Typhimurium LT2 ttdA (STM 3355) and ttdB (STM 3354) sequences (GenBank accession no. AE008854).
A multiplex PCR for the discrimination of dT+ and dT− Salmonella strains contained 0.4 μM concentrations of primers 166 (5′-GTA AGG GTA ATG GGT TCC-3′) and 167, 0.2 μM concentrations of primers ST11 and ST15 (1), a 200 μM concentration of each deoxynucleoside triphosphate (Roche Applied Science), 1× PCR buffer, 2.5 mM MgCl2, 1 U of Platinum Taq polymerase (Invitrogen), and a 5-μl aliquot of the sample DNA (approximately 106 CFU). The total volume was 25 μl. The incubation conditions were 95°C for 1 min, followed by three cycles of 95°C for 30 s, 61°C for 30 s, and 72°C for 30 s and 27 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 30 s. A final extension of 72°C for 4 min was employed. Sample DNA for PCRs was prepared from an aerobically grown Salmonella culture (16 h in Luria-Bertani medium with shaking at 37°C) (approximately 2 × 109 CFU). A 1-ml aliquot of the culture was centrifuged in a tube for 5 min at 10,000 × g at 4°C. The supernatant was carefully discarded, and the cell pellet was suspended in 300 μl of Tris-EDTA buffer (10 mM Tris-HCl, 0.1 mM EDTA [pH 8.0]). The tube was incubated for 10 min at 100°C in a water bath and immediately chilled on ice. After centrifugation for 5 min at 14,000 × g at 4°C, the DNA-containing supernatant was carefully transferred to a new tube. A 5-μl aliquot was used as template DNA for the PCR. A 7-μl aliquot of a PCR product was loaded onto a 1.6% agarose gel, and electrophoresis was performed in 1 × Tris-borate-EDTA buffer at 6 V/cm for 90 min. After electrophoresis, the gel was stained for 10 min in 1 × Tris-borate-EDTA buffer containing 0.5 μg of ethidium bromide/ml. The gel results were documented with a video camera. The PCR was used for all 125 strains and performed in triplicate (see Table 2).
TABLE 2.
Properties of the three lead acetate test protocols used in this study
| Parameter | Protocol 1 (current WHO method)a | Protocol 2 | Protocol 3 |
|---|---|---|---|
| Broth | 8 ml, pH 7.4; 1% Difco Bacto Peptone, 1% d-tartrate, 0.0023% bromothymol blue | 8 ml, pH 7.4; 1% Difco Bacto Peptone, 1% d-tartrate, 0.0023% bromothymol blue | 8 ml, pH 7.4; 1% Difco Bacto Peptone, 1% d-tartrate, 0.0023% bromothymol blue |
| Inoculate | 5 × 107 bacteria in 0.85% NaCl | 5 × 107 bacteria in 0.85% NaCl | Loopful of bacteria from plate |
| Incubation atmosphere | Air, 37°C | 10% CO2, 37°C | 10% CO2, 37°C |
| Incubation time | 3 and 6 days | 3 and 6 days | 3 and 6 days |
Protocol 1 is that described by Alfredsson et al. (2). WHO, World Health Organization.
The detection limit of the multiplex PCR was determined with DNA prepared from strains NCTC 5706 and NCTC 3176 with the Genomic-tip 100 columns (Qiagen, Hilden, Germany) according to the manufacturer's manual. The DNA was serially diluted 10-fold in Tris-EDTA buffer and cycled according to the incubation conditions described above. The experiment was repeated three times.
Lead acetate test protocols.
Three different protocols of the test were used for all 125 strains and performed three times each (Table 2).
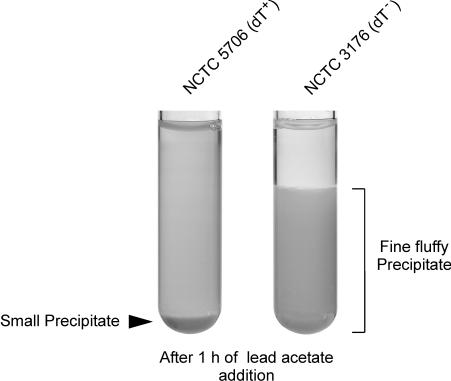
Protocol 1 was performed as described by Alfredsson et al. (2) with some minor modifications and is currently the gold standard-like method used by the WHO Collaborating Centre. Ten grams of Difco Bacto Peptone (Becton Dickinson, Heidelberg, Germany) per liter was autoclaved at 121°C for 15 min. Potassium sodium tartrate tetrahydrate (Merck Eurolab, Darmstadt, Germany; catalog no. 1.08087) was added to a final concentration of 1%. The pH was adjusted with NaOH to pH 7.4. A solution of bromothymol blue sodium salt was added as an indicator to a final concentration of 0.0023%. The broth was dispensed in 8-ml volumes in cotton-wool-stoppered test tubes and sterilized two times for 15 min at 100°C each time. The inoculate was prepared by suspension of several colonies in saline (0.85% NaCl) to a density of about 109 bacteria per ml. A 50-μl aliquot of this suspension was inoculated into the test tubes. The cultures were incubated at 37°C for 3 and 6 days aerobically without shaking. After incubation, the cultures were tested for d-tartrate utilization by the addition of a saturated aqueous lead acetate solution in the proportion of 0.1 ml per 1 ml of culture. The immediately produced precipitate was homogenized by brief mixing. dT+ strains were detected by the formation of a small precipitate after 1 to 2 h of lead acetate addition. The presence of a fluffy fine precipitate after the 1 to 2 h of lead acetate addition indicated a dT− strain (Fig. 2).
FIG. 2.
Lead acetate test. The test tubes inoculated with reference strains NCTC 5706 (dT+) and NCTC 3176 (dT−) are shown after 3 days of incubation under a 10% CO2 atmosphere and after 1 h of the addition of a saturated lead acetate solution. A fluffy precipitate, which is visible for strain NCTC 3176, indicates a dT− Salmonella strain, whereas a small precipitate indicates a dT+ Salmonella strain, as shown for strain NCTC 5706.
Protocol 2 was identical to protocol 1 except that incubation was in the presence of a 10% CO2 atmosphere instead of air. Identical inoculates were used.
Protocol 3 differed from protocol 1 in the incubation conditions (protocol 3 was in the presence of a 10% CO2 atmosphere) and the type of inoculate. Inoculation was performed with a loopful of bacteria grown aerobically overnight on Luria-Bertani plates (Merck Eurolab). Before use, the test tubes were preincubated at 37°C in a 10% CO2 atmosphere for 30 min.
Statistics.
The accuracy of the assay was determined as described by Gardner and Greiner (5). It describes the closeness of agreement between a test result and the accepted reference value.
Nucleotide sequence accession numbers.
The sequences of strains NCTC 5706 (dT+) and NCTC 3176 (dT−) have been deposited in GenBank (accession no. AY211490 and AY211491).
RESULTS AND DISCUSSION
Development of a PCR assay for the discrimination of dT+ and dT− Salmonella strains.
PCR amplification and sequencing showed the presence of the ttdA and ttdB genes in the serovar Paratyphi B dT+ reference strain NCTC 5706 and the dT− reference strain NCTC 3176 (GenBank accession no. AY211492 and AY211493). The translated protein sequences of TtdA and TtdB in both strains were identical to those in the serovar Typhimurium strain LT2. To identify possible sequence differences of both strains in potential regulatory binding sites next to the ttdA and ttdB genes, the intergenic region of the STM 3357 and STM 3356 genes was amplified and sequenced (Fig. 1). The sequences showed that only a single nucleotide located in the ATG start codon of gene STM 3356 was different. The serovar Paratyphi B dT+ strain possessed a regular ATG start codon, whereas the serovar Paratyphi B dT− strain possessed an A nucleotide instead of a G within the start codon. Consequently, a gene transcript cannot be translated into the essential putative cation transporter protein. This affects the anaerobic growth of Salmonella in the presence of d-tartrate. The role of the protein encoded by the STM 3356 gene in the fermentation pathway for d-tartrate remains unknown and has yet to be elucidated.
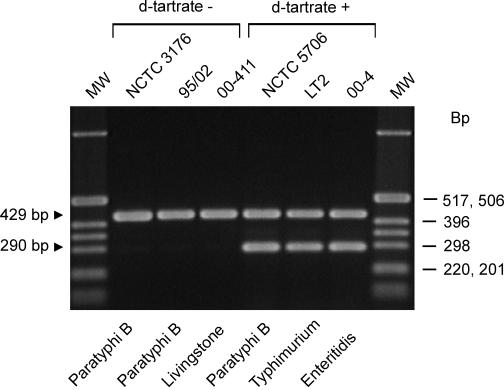
A primer set (primers 167 and 166) was designed to give a positive PCR signal (290-bp PCR product) when the ATG start codon is present in the STM 3356 gene and no PCR amplicon when the start codon is absent (Fig. 3). The forward primer, 167, is located at the C-terminal coding region of the STM 3357 gene and anneals to both variants. The backward primer, 166, is located at the N-terminal coding region of the STM 3356 gene. The 3′ primer end fits exactly with the G located in the ATG codon in dT+ strains (Fig. 1). This primer set is combined with a second primer set (ST11-ST15) generating a 429-bp PCR fragment when Salmonella DNA is present (1). This primer set serves as an internal positive control and detects the presence of amplifiable Salmonella DNA in the PCR. Using purified chromosomal DNA of strains NCTC 5706 (dT+) and NCTC 3176 (dT−) as a template, the detection limit was determined to be 10 to 1 pg of DNA (approximately 104 to 103 Salmonella genome equivalents (8) for both strains.
FIG. 3.
PCR-based discrimination of dT− and dT+ Salmonella strains. The agarose gel shows the multiplex PCR profiles from dT− and dT+ Salmonella strains. The original strain numbers are indicated above the gel, and the serovars are indicated below the gel. The Salmonella-positive control for each PCR, a 429-bp fragment, is indicated. A 290-bp fragment is present when dT+ Salmonella strain DNA has been detected. The molecular size standard markers (Marker X; Roche Applied Science) are indicated on the right in base pairs.
Comparison of the PCR assay with three lead acetate test protocols.
One hundred two Salmonella serovar Paratyphi B strains isolated between 1961 and 2002 in various countries worldwide, 6 serovar Typhimurium strains, 5 serovar Enteritidis strains, and 12 Salmonella strains belonging to various other serovars were selected to determine the accuracy between the genotypic PCR-based method and three phenotypic lead acetate test protocols (Table 1). Lead acetate test protocol 1 is currently used as the gold standard-like method for the discrimination of dT+ and dT− Salmonella strains at the WHO Collaborating Centre for Reference and Research on Salmonella (M. Popoff, personal communication). The accuracy of this protocol compared to that of the PCR-based method was 47% after 3 days of incubation and 90% after 6 days of incubation. When the incubation was performed under a 10% CO2 atmosphere (protocol 2), the accuracy increased to 58% (3 days of incubation) and 91% (6 days of incubation). A change in inoculation method from use of a sodium chloride suspension to a loopful of bacteria from a plate (protocol 3) led to an accuracy of 78% after 3 days of incubation and 100% after 6 days of incubation.
These data indicate that obviously poorly aerated conditions and the inoculation of approximately 5 × 107 bacterial cells per culture suspended in 0.85% NaCl are not sufficient to activate the oxygen-labile stereospecific l-tartrate dehydratase in all strains. A loopful of bacteria contains approximately 10- to 50-fold more cells.
An improved performance of the lead acetate assay was also shown when a 10% CO2 atmosphere was used during incubation. Fifty percent of the results were false negative when reference strain NCTC 5706 (dT+) was subcultured 20 times under the conditions of protocol 1 (2) for 2 days of incubation. In the presence of a 10% CO2 atmosphere, 17 of 20 (85%) subcultures reacted positively.
It is interesting that various strains showed a delay in fermenting d-tartrate, which can lead to a false-negative result depending on the test conditions. Barker (3) described eight strains which were d-tartrate negative after 24 h of incubation but positive after 48 h. It was speculated that the delay in fermenting d-tartrate was due to a deficient permease system responsible for the passage of d-tartrate into the cell or for the export of d-tartrate dehydrase. However, the reasons for such a delay remain to be elucidated. To our knowledge, nothing is known about the frequency with which dT+ strains become dT− and if the DNA region of the genome is located within a mutational hot spot. Our data indicate a stable phenotype because triplicate determinations of nonfermenting strains showed no phenotypic or genotypic exceptions.
The PCR and lead acetate test results presented here indicate quite clearly that the incubation times and conditions used by Alfredsson et al. (2) and Barker (3) should give a high degree of false-negative results and were insufficient to generate correct and reliable results.
In conclusion, despite the improved lead acetate test (protocol 3), the PCR assay described here should be the method of choice to discriminate between dT+ and dT− Salmonella strains in the future. The assay is rapid and reliable. An interlaboratory validation study is in progress to evaluate the reproducibility of the PCR method.
Acknowledgments
B.M. was supported by the Bundesministerium für Verbraucherschutz, Ernährung und Landwirtschaft (BMVEL).
We thank C. Kornschober (BBSUA, Graz, Austria), H. Imberechts (CODA, Brussels, Belgium), Philippe Bouvet (Institut Pasteur, Paris, France), W. Rabsch (RKI, Wernigerode, Germany), D. Lightfoot (University of Melbourne, Australia), S. Chappell (Veterinary Laboratory Agency, Weybridge, United Kingdom), W. Wannett (RIVM, Bilthoven, The Netherlands) for kindly providing Salmonella serovar Paratyphi B strains.
REFERENCES
- 1.Aabo, S., O. F. Rasmussen, L. Rossen, P. D. Sorensen, and J. E. Olsen. 1993. Salmonella identification by the polymerase chain reaction. Mol. Cell. Probes. 7:171-178. [DOI] [PubMed] [Google Scholar]
- 2.Alfredsson, G. A., R. M. Barker, D. C. Old, and J. P. Duguid. 1972. Use of tartaric acid isomers and citric acid in the biotyping of Salmonella typhimurium. J. Hyg. 70:651-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker, R. M. 1985. Utilization of d-tartaric acid by Salmonella paratyphi B and Salmonella java: comparison of anaerobic plate test, lead acetate test and turbidity test. J. Hyg. 95:107-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, D. J., L. M. Browning, and J. E. Coica. 2003. Investigation of human infections with Salmonella enterica serovar Java in Scotland and possible association with imported poultry. Eurosurveillance 8:35-40. [DOI] [PubMed] [Google Scholar]
- 5.Gardner, I. A., and M. Greiner. 1999. Advanced methods for test validation and interpretation in veterinary medicine. Joint Cooperation between the Freie Universität Berlin and the University of California, Davis. Freie Universität Berlin, Berlin, Germany.
- 6.Kauffmann, F. 1955. Zur Differentialdiagnose und Pathogenität von Salmonella java und Salmonella paratyphi B. B. Z. Hyg. 141:546-550. [PubMed] [Google Scholar]
- 7.Kristensen, M., and F. Kauffmann. 1937. Bakteriologische und klinische Erfahrungen über Infektionen mit d-weinsäurevergärenden Paratyphus B-bacillen. Z. Hyg. Infektionskr. 120:149-154. [Google Scholar]
- 8.Liu, S. L., A. Hessel, and K. E. Sanderson. 1993. The XbaI-BlnI-CeuI genomic cleavage map of Salmonella typhimurium LT2 determined by double digestion, end labelling, and pulsed-field gel electrophoresis. J. Bacteriol. 175:4104-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miko, A., B. Guerra, A. Schroeter, C. Dorn, and R. Helmuth. 2002. Molecular characterization of multiresistant d-tartrate-positive Salmonella enterica serovar Paratyphi B isolates. J. Clin. Microbiol. 40:3184-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Odongo, M. O., I. M. McLaren, J. E. Smith, and C. Wray. 1990. A biotyping scheme for Salmonella livingstone. Br. Vet. J. 146:75-79. [DOI] [PubMed] [Google Scholar]
- 11.Reaney, S. K., C. Begg, S. J. Bungard, and J. R. Guest. 1993. Identification of the L-tartrate dehydratase genes (ttdA and ttdB) of Escherichia coli and evolutionary relationship with the class I fumarase genes. J. Gen. Microbiol. 139:1523-1530. [DOI] [PubMed] [Google Scholar]
- 12.Schink, B. 1984. Fermentation of tartrate enantiomers by anaerobic bacteria, and description of two new species of strict anaerobes, Ruminococcus pasteurii and Ilyobacter tartaricus. Arch. Microbiol. 139:409-414. [Google Scholar]
- 13.Stratton, J., L. Stefaniw, K. Grimsrud, D. H. Werker, A. Ellis, E. Ashton, L. Chui, E. Blewett, R. Ahmed, C. Clark, F. Rodgers, L. Trottier, and B. Jensen. 2001. Outbreak of Salmonella paratyphi B var java due to contaminated alfalfa sprouts in Alberta, British Columbia and Saskatchewan. Can. Commun. Dis. Rep. 27:133-137. [PubMed] [Google Scholar]
- 14.Van Pelt, W., H. van der Zee, W. J. B. Wannet, A. W. van de Giessen, D. J. Mevius, N. M. Bolder, R. E. Komijn, and Y. T. H. P. van Duynhoven. 2003. Explosive increase of Salmonella Java in poultry in The Netherlands: consequences for public health. Eurosurveillance 8:31-35. [DOI] [PubMed] [Google Scholar]
- 15.Woehlke, G., and P. Dimroth. 1994. Anaerobic growth of Salmonella typhimurium on L(+)- and D(−)-tartrate involves an oxaloacetate decarboxylase Na+ pump. Arch. Microbiol. 162:233-237. [DOI] [PubMed] [Google Scholar]