Abstract
We recently identified the genes responsible for the serotype c-specific glucose side chain formation of rhamnose-glucose polysaccharide (RGP) in Streptococcus mutans. These genes were located downstream from the rgpA through rgpF locus that is involved in the synthesis of RGP. In the present study, the corresponding chromosomal regions were isolated from serotype e and f strains and characterized. The rgpA through rgpF homologs were well conserved among the three serotypes. By contrast, the regions downstream from the rgpF homolog differed considerably among the three serotypes. Replacement of these regions in the different serotype strains converted their serotypic phenotypes, suggesting that these regions participated in serotype-specific glucose side chain formation in each serotype strain. Based on the differences among the DNA sequences of these regions, a PCR method was developed to determine serotypes. S. mutans was isolated from 198 of 432 preschool children (3 to 4 years old). The serotypes of all but one S. mutans isolate were identified by serotyping PCR. Serotype c predominated (84.8%), serotype e was the next most common (13.3%), and serotype f occured rarely (1.9%) in Japanese preschool children. Caries experience in the group with a mixed infection by multiple serotypes of S. mutans was significantly higher than that in the group with a monoinfection by a single serotype.
Streptococcus mutans strains are classified into three serotypes (c, e, and f), and the serologic specificity is defined by rhamnose-glucose polysaccharide (RGP) on the cell wall (6). We have characterized the genes involved in RGP synthesis in S. mutans Xc (serotype c) in the course of our previous studies. Four rml genes (rmlA through rmlD) are directly related to the synthesis of dTDP-l-rhamnose (12, 13), and the gluA gene encodes the enzyme producing UDP-d-glucose (18). The rgpG gene is implicated in the initiation of RGP synthesis by transfer of N-acetylglucosamine-1-phosphate to a lipid carrier (16). Furthermore, six other genes (rgpA through rgpF) required for RGP synthesis were identified in the region downstream from rmlD, and these genes are likely to be involved in the transport and assembly of RGP (11, 19).
The RGPs are composed of α1,2- and α1,3-linked rhamnan backbones with glucose side chains linked to alternate rhamnoses. Each serotype-specific polysaccharide has unique linkages of its glucose side chains (serotype c, α1,2-linkage; serotype e, β1,2-linkage; and serotype f, α1,3-linkage) (5, 10). Recently, we identified and characterized the genes required for glucose side chain formation of the serotype c-specific RGP (9). However, the loci responsible for the determination of the other serotypes have not yet been elucidated.
In this study, we identified the loci involved in the glucose side chain formation of RGP in serotypes e and f of S. mutans and confirmed that these regions determine serotype specificities. Furthermore, we designed three pairs of primers from specific DNA sequences within each serotype determinant locus and succeeded in developing a multiplex PCR assay to easily identify serotypes of S. mutans strains. In addition, we evaluated the clinical usefulness of the PCR assay by using epidemiological samples from preschool children.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this study are listed in Table 1. The sources of the bacterial strains and culturte conditions were described previously (7, 13). Antibiotics were used at the following concentrations: 200 μg of erythromycin per ml, 50 μg of ampicillin per ml, 25 μg of kanamycin per ml, or 50 μg spectinomycin per ml for Escherichia coli; 10 μg of erythromycin per ml, 100 μg of kanamycin per ml, or 300 μg spectinomycin per ml for S. mutans.
TABLE 1.
Bacterial strains used in this study
| Mutans streptococci (serotype) | Other gram-positive bacteria |
|---|---|
| S. cricetus E49 (a) | Bacillus cereus IFO3131 |
| S. cricetus HS1 (a) | Bacillus megaterium IAH1166 |
| S. ratti BHT (b) | Clostridium bifermentans KZ1012 |
| S. ratti FA1 (b) | Enterococcus faecalis SS499 |
| S. mutans MT8148 (c) | Eubacterium limosum GAI5456 |
| S. mutans Xc (c) | Lactobacillus casei ATCC 393 |
| S. sobrinus MT8145 (d) | Listeria monocytogenes VIU206 |
| S. sobrinus OMZ176 (d) | Micrococcus lylae GIFU9132 |
| S. mutans LM7 (e) | Micrococcus luteus GIFU8717 |
| S. mutans MT703R (e) | Micrococcus varians GIFU9844 |
| S. mutans MT6219 (f) | Mycobacterium smegmatis RIMD 1332001 |
| S. mutans OMZ175 (f) | Staphylococcus aureus IFO 12732 |
| S. sobrinus 6715 (g) | Staphylococcus capitis GIFU9121 |
| S. sobrinus OU8 (g) | Staphylococcus intermedius GIFU3171 |
| S. downei Mfe28 (h) | Staphylococcus simulans GIFU9127 |
| S. downei S28 (h) | Streptococcus gordonii ATCC 10558 |
| Streptococcus gordonii ATCC 12396 | |
| Streptococcus milleri NCTC 10703 | |
| Streptococcus oralis ATCC 10557 | |
| Streptococcus salivarius HT9R | |
| Streptococcus sanguis ATCC 10556 |
DNA manipulation.
Standard DNA recombinant procedures such as DNA isolation, endonuclease restriction, ligation, and agarose gel electrophoresis were carried out as described previously (2). Purification of chromosomal DNA from bacteria was carried out as described previously (13). The nucleotide sequences were determined with a 373 STRETCH automated sequencer (PE Applied Biosystems) as described previously (13). The DNASIS software (Hitachi Software Engineering Co., Yokohama, Japan) was used for sequence analysis. S. mutans and E. coli were transformed as described previously (17).
Confirmation of serotype specificity of RGP.
Serotype specificity of RGP produced in S. mutans transformants was analyzed by immunodiffusion analysis (8). Autoclaved extracts prepared from whole cells of S. mutans strains were examined with serotype c-, e-, and f-specific rabbit antisera and with rhamnan-specific rabbit antiserum in 1% Noble Agar in saline. The serotype c-, e-, and f-specific rabbit antisera were raised by three subcutaneous injections of whole-cell suspensions of MT8148 (serotype c [Table 1]), MT703R (serotype e [Table 1]), or OMZ175 (serotype f [Table 1]) in incomplete Freund's adjuvant at 2-week intervals. These antisera were adsorbed with whole S. mutans cells of another serotype. The rhamnan-specific rabbit antiserum was prepared as described previously (19).
PCR experiments.
PCR experiments designed to discriminate between S. mutans and Streptococcus sobrinus targeted the gene encoding the water-insoluble glucan-synthesizing enzyme (GTF-I) and were done with two sets of primers (GTFB-F plus GTFB-R and GTFI-F plus GTFI-R), as shown in Table 2. Three sets of primers (SC-F plus SC-R, SE-F plus SE-R, and SF-F plus SF-R) were used in the PCR assay to identify S. mutans serotypes (Table 2). Rapidly isolated chromosomal DNA from colonies on strips of Dentocult SM (Orion Diagnostica, Espoo, Finland) was used as a template.
TABLE 2.
PCR primers used in this study
| Primer | Sequence (5′ to 3′) | Product size (bp) | Reference |
|---|---|---|---|
| GTFB-F | ACTACACTTTCGGGTGGCTTGG | 517 | 7 |
| GTFB-R | CAGTATAAGCGCCAGTTTCATC | 7 | |
| GTFI-F | GATAACTACCTGACAGCTGACT | 712 | 7 |
| GTFI-R | AAGCTGCCTTAAGGTAATCACT | 7 | |
| SC-F | CGGAGTGCTTTTTACAAGTGCTGG | 727 | This study |
| SC-R | AACCACGGCCAGCAAACCCTTTAT | This study | |
| SE-F | CCTGCTTTTCAAGTACCTTTCGCC | 517 | This study |
| SE-R | CTGCTTGCCAAGCCCTACTAGAAA | This study | |
| SF-F | CCCACAATTGGCTTCAAGAGGAGA | 316 | This study |
| SF-R | TGCGAAACCATAAGCATAGCGAGG | This study | |
| Primer A | ATAGGAAGTCGCGGCTTA | This study | |
| Primer B | AGAGAACGTTTAAGGATA | This study | |
| Primer C | TACAGGCTGAATGGCCTT | This study | |
| Primer D | CTAACAGCCAGCATTCCT | This study | |
| Primer E | TCATCCCCATCAGCCAAT | This study | |
| Primer F | GACCATTCCTACAAAAAT | This study |
The PCR mixture (10 μl) consisted of 0.2 mM each deoxyribonucleoside triphosphate, 10 mM Tris-HCl buffer (pH 8.3), 50 mM KCl, 2 mM MgCl2, 1 U of Ex Taq DNA polymerase (Takara Bio Inc., Tokyo, Japan), a 0.5 μM concentration of each primer, and 1 μl of template DNA. After denaturation at 96°C for 2 min, a total of 25 PCR cycles were performed; each cycle consisted of 15 s of denaturation at 96°C, 30 s of annealing at 61°C, and 1 min of extension at 72°C. To confirm the specificity of PCR for the identification of serotypes of S. mutans, the 16 strains of mutans streptococci and 21 strains of other gram-positive bacteria (Table 1) were examined by multiplex PCR using purified chromosomal DNA samples as templates and the above three sets of primers.
For cloning purposes, PCR was performed with 0.05 U of LA Taq DNA polymerase (Takara Bio Inc.) per μl in 10 mM Tris-HCl buffer (pH 8.3) containing pairs of primers (0.5 μM each primer) drawn from primers A, B, C, D, E, and F, 0.4 mM each deoxyribonucleoside triphosphate, 50 mM KCl, and 2.5 mM MgCl2. LA Taq DNA polymerase was used to enhance the amplification of the large PCR fragment.
Epidemiological procedure.
Caries experience in 3- to 4-year-old children (n = 432, consisting of 237 boys and 195 girls) from five nursery schools in Tokyo, Japan, which is in a nonfluoridated area, was examined by two experienced and calibrated examiners (Cohen's kappa = 0.88) in November 2001. The parents of the children in the study granted permission for dental examination and microbiological sampling. Caries per tooth surface was diagnosed on the basis of visual classification as described by the World Health Organization (14). Oral mutans streptococci were recovered from plaque and saliva by using Dentocult SM Strip Mutans or Site Strip (Orion Diagnostica), as specified by the supplier. Colonies on strips were used for PCR detection.
Statistical analysis.
The differences in the number of tooth surfaces with caries experience (the number of decayed and filled surfaces [dfs] on deciduous teeth per person) between groups were examined using the Kruskal-Wallis test followed by Steel's multiple-comparison test, using the statistical program Excel Toukei version 5.0 (Esumi Co., Ltd., Tokyo, Japan).
Nucleotide sequence accession numbers.
The 3,784-bp (S. mutans Xc), 14,730-bp (S. mutans MT6219), and 16,442-bp (S. mutans LM7) nucleotide sequences determined in this paper have been deposited to the DDBJ data bank (URL, http://www.ddbj.nig.ac.jp) under accession numbers AB108684, AB108685, and AB108686, respectively.
RESULTS
Cloning of regions involved in glucose side chain synthesis of RGP in S. mutans Xc, LM7, and MT6219.
We previously identified new rgp genes (rgpH and rgpI) responsible for the glucose side chain formation in serotype c S. mutans Xc. These genes were located in the region downstream from the rgp locus (rgpA through rgpF) which was involved in rhamnan backbone synthesis (9). Therefore, we designed two primers; forward primer A and reverse primer B, which corresponded to the regions upstream from rgpA and within rgpF, respectively (Fig. 1). A 7.2-kb fragment was amplified by PCR with the two primers, using either LM7 or MT6219 chromosomal DNA as a template. Nucleotide sequence analyses of the PCR fragments from the two templates revealed that the regions from rgpA through rgpF were almost the same in all three serotypes (Fig. 1).
FIG. 1.
Comparison of the genetic organization of the rgp loci from S. mutans Xc, LM7, and MT6219. The lower part of the diagram indicates regions responsible for glucose side chain formation during RGP synthesis. The identity shown at the bottom indicates the amino acid identity of the corresponding genes between two serotypes or among three serotypes. The rgpA through rgpF genes and ORF12 were common to the three serotypes and showed greater than 98% identity.
Using the S. mutans genome database (http://www.genome.ou.edu/smutans.html), we identified four open reading frames (ORFs) in the region downstream of rgpI, and designated them ORF10, ORF11, ORF12, and ORF13 (Fig. 1). Using database sequences, we designed three reverse primers, primer C, primer D, and primer E, which corresponded to the regions downstream of ORF10, ORF11, and ORF12, respectively, in addition to a forward primer, primer F, within rgpE, (Fig. 1). PCR fragments were amplified from Xc, LM7, or MT6219 chromosomal DNA template by using primer F with either primer C, D, or E. PCR fragments of different sizes were amplified from each serotype with sets of primers F and D and primers F and E, but not from MT6219 with primers F and C. With primers F and E, PCR fragments of 11.7, 11.2, and 9.1 kb were obtained from Xc, LM7, and MT6219 chromosomal DNA templates, respectively. The sequence analyses of these fragments revealed that rgpA through rgpF and ORF12 are common to the three serotypes, with greater than 98% deduced amino acid sequence identity (Fig. 1). ORF3f and ORF2e showed 99 and 81% identity to rgpI and OFR10, respectively, in their deduced amino acid sequences, and both had a conserved glycosyltransferase domain in the N-terminal portion. However, the regions between rgpF and ORF12 differed among the three serotypes. Considering that the region downstream from rgpF is involved in glucose side chain formation in serotype c strain Xc, the regions between rgpF and ORF12 in LM7 and MT6219 seem to be responsible for the serotype-specific glucose side chain formation of RGPs, as is true in Xc.
Conversion of serotype.
To confirm that the regions identified above are determine the serotype, this region of Xc was replaced with the same region from different serotype strains. First, the region (ORF7 through ORF11) responsible for glucose side chain formation of Xc was replaced with the spectinomycin resistance gene (Spcr); the resultant mutant was designated Xc81 and produced only a rhamnan backbone with no glucose side chains. When Spcr of Xc81 was replaced with ORF1e through ORF4e of LM7 plus the erythromycin resistance gene (Emr) or with ORF1f through ORF3f of MT6219 plus Emr, the resultant transformants produced serotype e- or f-specific RGPs, respectively. Previous studies showed that rgpE, which lies upstream from these regions, was also essential in glucose side chain formation of serotype c RGP (11, 19). Therefore, the role of rgpE in serotype e- and f-specific RGP syntheses was examined. rgpE of Xc81 was replaced with the kanamycin resistance gene (Kmr), and the resultant transformant (Xc82) was generated. Even though ORF1e through ORF4e of LM7 or ORF1f through ORF3f of MT6219 were introduced into Xc82, these transformants produced only polysaccharides reacting with rhamnan-specific antiserum but not any serotype-specific RGP.
Next, the region responsible for glucose side chain formation in LM7 (ORF1e through ORF4e) or in MT6129 (ORF1f through ORF3f) was replaced with Spcr to yield LM7-81 and MT6219-81, respectively; the resultant transformants produced only the rhamnan backbone with no glucose side chains. When the region responsible for glucose side chain synthesis in Xc (rgpH through ORF11) was introduced into LM7-81 and MT6219-81, the resultant mutants produced serotype c-specific RGP. These results indicated that the regions between rgpF and ORF12, except for ORF7, were responsible for serotype-specific RGP synthesis in S. mutans, while rgpE has a common function in glucose side chain formation in all serotypes.
PCR evaluation.
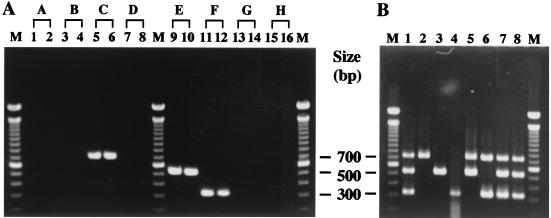
A comparison of the deduced amino acid sequences of the genes newly identified in this study revealed no homology among rgpH, ORF3e, and ORF2f, whereas the other genes showed homology to one another (Fig. 1). Based on this result, serotype-specific primers (SC-F plus SC-R, SE-F plus SE-R, and SF-F plus SF-R) were designed from the DNA sequences of rgpH (serotype c), ORF3e (serotype e), and ORF2f (serotype f), respectively, as listed in Table 2. Each amplified product had a unique size between 316 and 727 bp (Table 2). Of the 16 strains of mutans streptococci, S. mutans strains produced single bands of 727 bp (serotype c), 517 bp (serotype e), or 316 bp (serotype f) (Fig. 2A, lanes 5, 6, and 9 to 12). Other mutans streptococci did not produce an amplified product (lanes 1 to 4, 7, 8, and 13 to 16). Of the 21 other gram-positive strains, no amplified product was detected (data not shown). These results demonstrate that each set of primers can discriminate between specific serotypes of S. mutans strains.
FIG. 2.
Agarose gel electrophoresis of PCR products amplified with multiplex primers. (A) The purified chromosomal DNA samples from strains of mutans streptococci (A through H) were used as the templates. Lanes: 1, S. cricetus E49; 2, S. cricetus HS1; 3, S. ratti BHT; 4, S. ratti FA1; 5, S. mutans MT8148; 6, S. mutans Xc; 7, S. sobrinus MT8145; 8, S. sobrinus OMZ176; 9, S. mutans LM7; 10, S. mutans MT703R; 11, S. mutans MT6219; 12, S. mutans OMZ175; 13, S. sobrinus 6715; 14, S. sobrinus OU8; 15, S. downei Mfe28; 16, S. downei S28; M, molecular size markers. The capital letters above the lanes indicate serotypes, which correspond to those referred to in the text with lowercase letters. (B) The chromosomal DNA extracted from colonies on strips of Dentocult SM was used as the template. Lanes: 1 and 8, a reference marker of mixed PCR products from all three serotypes; 2, subject 1; 3, subject 2; 4, subject 3; 5, subject 4; 6, subject 5; 7, subject 6; M, molecular size markers.
Discrimination of the serotype of S. mutans in clinical samples.
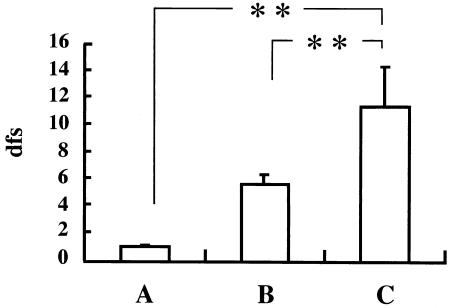
Mutans streptococci were identified from 214 of 432 children by using a Dentocult SM kit, and chromosomal DNA was extracted from the bacteria on the 214 strips. PCR with the extracted DNA and primers that targeted the genes coding for GTF-I amplified a fragment specific for only S. mutans in samples from 170 children, a fragment specific for only S. sobrinus in samples from 9 children, and fragments from both S. mutans and S. sobrinus in samples from 28 children; no fragment bands were produced in samples from 7 children. Serotypes of S. mutans from 197 children who were positive for S. mutans were determined by the serotyping multiplex PCR method developed in this study. Serotypes c, e, and f were identified in 178, 28, and 4 children, respectively. The serotype was not identified in the sample from only one child because no band was amplified. To eliminate any effect of S. sobrinus infection on the analysis of cariogenicity, data from subjects infected with only S. mutans were used to analyze the correlation between caries experience (dfs) and infection with S. mutans serotypes (Fig. 3). The number of children scoring 0 with the Dentocult SM kit was 218, and the dfs for this group was 0.88 ± 0.16 (mean ± standard error). There were 158 children with a monoinfection by a single serotype of S. mutans, with a dfs of 5.58 ± 0.61. Eleven children were infected with more than one serotype, with a dfs of 11.27 ± 2.97. The difference in dfs among these groups was significant using the Kruskal-Wallis test (P < 0.001). In addition, dfs in the group infected with multiple serotypes was significantly higher than dfs in both the group that scored 0 with the Dentocult SM kit (P < 0.01) and the group with a monoinfection (P < 0.01), as shown by an analysis using the Steel's multiple comparison test.
FIG. 3.
Effect of S. mutans infection on caries experience in preschool children with regard to serotype. A, subjects scoring 0 with Dentocult SM kit (n = 218); B, subjects with a monoinfection by a single serotype (n = 158); C, subjects with a mixed infection by multiple serotypes (n = 11). Vertical bars represent standard error. Differences in caries experience among the groups were analyzed by the Kruskal-Wallis test (P < 0.001). **, significant difference according to the Steel multiple-comparison test (P < 0.01).
DISCUSSION
It is known that serotype c S. mutans strains are predominant in the human oral cavity among the serotype c, e, and f strains (4). Bacterial cell wall polysaccharides decorate the cell surface and often play an important role in colonization of their ecological niche. Differences in the binding affinities of the polysaccharide antigens to human oral tissues might have led to this biased distribution. On the other hand, the progenitor of S. mutans is thought to have been serotype c, and serotype f and e strains might have originated through the introduction of a point mutation in the serotype c determinant locus or through deletion of a portion, as is thought to have occurred in the human histo-blood group ABO system (15). Although the sequencing of the entire genome of serotype c S. mutans strain UA159 was completed last year, the data were not sufficient to clarify the genetic factors involved in the generation of serotype specificity.
We previously demonstrated that rgpE, which is located in the middle of the rgp locus responsible for rhamnan backbone synthesis, is involved in glucose side chain formation of serotype c RGP (11, 19). Recently, we identified serotype c-specific genes responsible for α1,2-linked glucose side-chain formation downstream from the rgp locus in S. mutans Xc (9). In this study, it was confirmed that the rgpE homologs participated in a glucose transfer during RGP synthesis in serotypes e and f strains as well. However, the deduced amino acid sequence of the rgpE homologs revealed greater than 99.5% identity among the three serotypes, and a common function was confirmed in all serotypes by the conversion experiment. On the other hand, we recognized considerable discrepancies in the sequences of the region downstream from rgpF among the three serotype strains, although the rgpA through rgpF locus was well conserved among them. These results strongly indicated that the loci responsible for the side chain formation of serotype e and f RGPs were located downstream from rgpF. The conversion analysis confirmed that the regions between rgpF and ORF12 determined the serotype of S. mutans.
On the basis of these results, each serotype strain of S. mutans seems to have acquired its own specific genes for the synthesis of its serotype-specific antigen, and none of the three serotypes could be defined as an ancestral strain. Based on its predominance, the serotype c RGP structure may have advantages for S. mutans colonization of the oral cavity. It is interesting that no S. mutans strain with defective RGP glucose side chains has been isolated from the oral cavity. These findings suggest that the glucose side chains on S. mutans RGP might be important for its colonization of the oral cavity. We need to investigate further the function of RGP in S. mutans colonization.
Furthermore, we developed a simple, rapid, and reliable PCR method to identify serotypes of S. mutans. Although these serotypes have been hitherto identified by immunological methods, such as immunodiffusion analysis or fluorescent-antibody technique, these techniques can sometimes be ambiguous and time-consuming. There is cross-reactivity among the prepared antigens, and preparation of specific antigens and antibodies for each serotype is not easy. In the present study, we isolated S. mutans from 198 children, and serotypes of all but one isolate could be genetically defined by the multiplex PCR method developed in this study. In fact, of the seven subjects who were not identified by the PCR discriminating between S. mutans and S. sobrinus, serotyping PCR successfully identified two, one with serotype e and the other with serotype f. These findings suggested that the regions used for designing the primers for serotyping PCR were well conserved and that PCR discrimination of the serotype was clinically adequate. Sequence analysis of 16S rRNA revealed that S. sobrinus was present in one of the seven subjects and Streptococcus cricetus was present in two. The remaining two samples could not be identified because heterogeneous PCR products were obtained (data not shown).
Our epidemiological survey revealed that serotype c predominated (84.8%), serotype e was the next most common (13.3%), and serotype f occurred rarely (1.9%) in Japanese preschool children. This result is consistent with the data of Grönroos et al. (4). Further analysis of the worldwide serotype distribution of S. mutans using serotyping PCR will be helpful to understanding the phylogeny of S. mutans. In addition, it is interesting that children with a mixed infection by multiple serotypes seem to have a greater experience of caries than those with a monoinfection by a single serotype. It was reported that children who drank from a nursing bottle were often colonized with more than one clonal type (1). Although the PCR method used in this study could not distinguish ribotypes within S. mutans, a mixed infection by multiple serotypes might be equivalent to that by multiple ribotypes.
Fujiwara et al. (3) recently reported that they had isolated four S. mutans strains from the peripheral blood of patients with bacteremia; two strains were determined to be serotypes e and f by immunodiffusion, and the other two isolates were untypeable isolates with RGP with very low glucose contents. Since serotype c S. mutans strains are most frequently found in oral cavities of Japanese children, we wondered why serotype c S. mutans is not isolated from the blood of patients with bacteremia. In addition, a high incidence of untypeable S. mutans is uncommon. To define the contribution of S. mutans to the pathogenesis of infectious endocarditis, it is necessary to identify the serotypes of many S. mutans strains isolated from patients with infectious endocarditis. The PCR method developed in this study will be a powerful technique for clarifying the clinical importance of serotyping S. mutans.
Acknowledgments
This work was supported in part by the Dental Research Center, Nihon University School of Dentistry, and a Grant To Promote Multi-Disciplinary Research Projects and Grants-in-Aid for Developmental Scientific Research (12557186 and 14571954) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
REFERENCES
- 1.Alaluusua, S., J. Mättö, L. Grönroos, S. Innilä, H. Torkko, S. Asikainen, H. Jousimies-Somer, and M. Saarela. 1996. Oral colonization by more than one clonal type of mutans streptococcus in children with nursing-bottle dental caries. Arch. Oral Biol. 41:167-173. [DOI] [PubMed] [Google Scholar]
- 2.Aoki, H., T. Shiroza, M. Hayakawa, S. Sato, and H. K. Kuramitsu. 1986. Cloning of a Streptococcus mutans glucosyltransferase gene coding for insoluble glucan synthesis. Infect. Immun. 53:587-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujiwara, T., K. Nakano, M. Kawaguchi, T. Ooshima, S. Sobue, S. Kawabata, I. Nakagawa, and S. Hamada. 2001. Biochemical and genetic characterization of serologically untypable Streptococcus mutans strains isolated from patients with bacteremia. Eur. J. Oral Sci. 109:330-334. [DOI] [PubMed] [Google Scholar]
- 4.Grönroos, L., J. Mättö, M. Saarela, A. R. Luoma, H. Luoma, H. Jousimies-Somer, L. Pyhälä, S. Asikainen, and S. Alaluusua. 1995. Chlorhexidine susceptibilities of mutans streptococcal serotypes and ribotypes. Antimicrob. Agents Chemother. 39:894-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Linzer, R., M. S. Reddy, and M. J. Levine. 1986. Immunochemical aspects of serotype carbohydrate antigens of Streptococcus mutans, p. 29-38. In S. Hamada, S. M. Michalek, H. Kiyono, L. Menaker, and J. R. McGhee (ed.), Molecular microbiology and immunology of Streptococcus mutans. Elsevier Science Publishers, Amsterdam, The Netherlands.
- 6.Loesche, W. J. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50:353-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oho, T., Y. Yamashita, Y. Shimazaki, M. Kushiyama, and T. Koga. 2000. Simple and rapid detection of Streptococcus mutans and Streptococcus sobrinus in human saliva by polymerase chain reaction. Oral Microbiol. Immunol. 15:258-262. [DOI] [PubMed] [Google Scholar]
- 8.Ouchterlony, Ö. 1958. Diffusion-in-gel methods for immunological analysis. Prog. Allergy 5:1-78. [PubMed] [Google Scholar]
- 9.Ozaki, K., Y. Shibata, Y. Yamashita, Y. Nakano, H. Tsuda, and T. Koga. 2002. A novel mechanism for glucose side-chain formation in rhamnose-glucose polysaccharide synthesis. FEBS Lett. 532:159-163. [DOI] [PubMed] [Google Scholar]
- 10.Pritchard, D. G., R. L. Gregory, S. M. Michalek, and J. R. McGhee. 1986. Biochemical aspects of serotype carbohydrate antigens of Streptococcus mutans, p. 39-49. In S. Hamada, S. M. Michalek, H. Kiyono, L. Menaker, and J. R. McGhee (ed.), Molecular microbiology and immunology of Streptococcus mutans. Elsevier Science Publishers, Amsterdam, The Netherlands.
- 11.Shibata, Y., Y. Yamashita, K. Ozaki, Y. Nakano, and T. Koga. 2002. Expression and characterization of streptococcal rgp genes required for rhamnan synthesis in Escherichia coli. Infect. Immun. 70:2891-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsukioka, Y., Y. Yamashita, Y. Nakano, T. Oho, and T. Koga. 1997. Identification of a fourth gene involved in dTDP-rhamnose synthesis in Streptococcus mutans. J. Bacteriol. 179:4411-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsukioka, Y., Y. Yamashita, T. Oho, Y. Nakano, and T. Koga. 1997. Biological function of the dTDP-rhamnose synthesis pathway in Streptococcus mutans. J. Bacteriol. 179:1126-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.World Health Organization. 1997. Oral health survey-basic methods, 4th ed. World Health Organization, Geneva, Switzerland.
- 15.Yamamoto, F., H. Clausen, T. White, J. Marken, and S. Hakomori. 1990. Molecular genetic basis of the histo-blood group ABO system. Nature 345:229-233. [DOI] [PubMed] [Google Scholar]
- 16.Yamashita, Y., Y. Shibata, Y. Nakano, H. Tsuda, N. Kido, M. Ohta, and T. Koga. 1999. A novel gene required for rhamnose-glucose polysaccharide synthesis in Streptococcus mutans. J. Bacteriol. 181:6556-6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamashita, Y., T. Takehara, and H. K. Kuramitsu. 1993. Molecular characterization of a Streptococcus mutans mutant altered in environmental stress responses. J. Bacteriol. 175:6220-6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamashita, Y., Y. Tsukioka, Y. Nakano, K. Tomihisa, T. Oho, and T. Koga. 1998. Biological functions of UDP-glucose synthesis in Streptococcus mutans. Microbiology 144:1235-1245. [DOI] [PubMed] [Google Scholar]
- 19.Yamashita, Y., Y. Tsukioka, K. Tomihisa, Y. Nakano, and T. Koga. 1998. Genes involved in cell wall localization and side chain formation of rhamnose-glucose polysaccharide in Streptococcus mutans. J. Bacteriol. 180:5803-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]