Abstract
The Acinetobacter baumannii 19606 prototype strain produces a 78-kDa iron-regulated outer membrane protein immunologically related to FatA, which is required for iron acquisition by the fish pathogen Vibrio anguillarum via the anguibactin-mediated system. This A. baumannii strain also secretes histamine, a biosynthetic precursor of the siderophore anguibactin. In contrast, the A. baumannii 8399 clinical strain isolated in Oregon produces a siderophore and a putative 73-kDa iron-regulated outer membrane (OM73) receptor that are different from those produced by V. anguillarum and A. baumannii 19606. These observations suggest that different A. baumannii clinical isolates express unrelated iron uptake systems. This hypothesis is supported by differences in outer membrane protein profiles among A. baumannii isolates obtained from Oregon and Europe. The 19606 isolate and some European isolates expressed a FatA-like protein, while neither 19606 nor any of the European isolates expressed proteins related to OM73. Some European isolates failed to express FatA- and OM73-like proteins. All but one of the Oregon isolates expressed OM73-like proteins, while none of them contained a FatA-like protein. The presence of these proteins always correlated with the presence of the om73- and fatA-like genes in the cognate strains. While 19606 and a few European isolates produced histamine, none of the Oregon isolates had this capability. Interestingly, one strain each from the Oregon and European isolates did not express any of these products involved in iron acquisition, indicating that they could acquire iron through siderophore-mediated transport systems different from those expressed by the 19606 and 8399 clinical isolates.
Acinetobacter baumannii is increasingly recognized as an important human pathogen that causes severe infections in hospitalized patients (8) as well as deadly cases of community-acquired pneumonia (5). This bacterium is capable of causing septicemia, endocarditis, meningitis, and skin wound, respiratory tract, and urinary tract infections. Much of the work done with A. baumannii has focused on antibiotic resistance and typing of isolates obtained from different outbreaks, while little is known about the physiology and virulence traits expressed by this opportunistic pathogen. Being a human pathogen, A. baumannii must be able to utilize host resources in order to survive. Iron is an important resource that is not readily available in the human host; rather, it is found complexed with iron-binding molecules such as heme, lactoferrin, and transferrin (18, 23). Bacteria survive and multiply under iron-limiting conditions, such as those found in natural and host environments, by expressing active systems that gather this essential micronutrient. Some systems involve the secretion of low-molecular-mass ferric binding compounds, called siderophores, which can be classified into different categories based on their chemical structures (23, 24). One of these categories of siderophores is the catechols, which use a phenolate group as part of the iron-binding site. Anguibactin is a well-characterized catechol siderophore secreted by the fish pathogen Vibrio anguillarum (1, 20) which has a dihydroxybenzoic acid (DHBA) moiety linked to hydroxyhistamine through one molecule of l-cysteine.
A report from Yamamoto et al. (31) described the structure of acinetobactin, the siderophore secreted by A. baumannii 19606, the prototype strain of this opportunistic human pathogen. The acinetobactin siderophore structure is virtually identical to that of the anguibactin siderophore from V. anguillarum (20). These two siderophores vary in their amino acids linking DHBA to hydroxyhistamine, with acinetobactin using l-threonine instead of l-cysteine. In accordance with the structure of acinetobactin, A. baumannii 19606 is capable of producing histamine by decarboxylation of histidine (3), which is then most likely used for the biosynthesis of this siderophore in a pathway that may be similar to that described for anguibactin in V. anguillarum (11). The almost identical structures of acinetobactin and anguibactin predict that A. baumannii 19606 expresses an outer membrane protein related to FatA, which is the anguibactin receptor produced by V. anguillarum. This prediction was confirmed by immunoblot analysis of A. baumannii 19606 outer membrane proteins with anti-FatA serum (12).
A previous report characterized a high-affinity iron uptake system expressed by A. baumannii 8399, which was isolated during a nosocomial outbreak of respiratory infections at the Oregon Health Sciences University Hospital, Portland (14). This iron uptake system includes a catechol siderophore capable of scavenging iron from high-affinity iron-binding proteins present in the human host. Chemical analyses of a secreted catechol siderophore (14) and the absence of histamine in culture supernatants (3) indicate that the siderophore produced by this isolate is different from acinetobactin. This hypothesis is supported by the recent observation that a chromosomal region of this strain contains a gene cluster that includes a gene encoding a putative outer membrane siderophore receptor protein unrelated to the V. anguillarum and A. baumannii 19606 FatA and FatA-like proteins, respectively (13).
Knowing that there is a difference in siderophore-mediated iron uptake systems between the A. baumannii 19606 and 8399 strains, we decided to compare some components potentially involved in iron acquisition expressed by different clinical A. baumannii isolates obtained from an outbreak at the Oregon Health Sciences University and different European geographical regions. Electrophoretic and immunoblot analyses and thin-layer chromatography (TLC) showed that members of this bacterial species express different iron acquisition systems that allow them to prosper under iron-limiting conditions.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and plasmids.
The A. baumannii 19606 prototype strain was purchased from the American Type Culture Collection. The clinical isolates 9235, 8971, 8143, 8114, 8637, 7133, 7138, 8399, 9606, 7931, 9124, and 9397, which represent the plasmid profiles A to K and W, respectively, were isolated during an outbreak of respiratory tract infections in the Oregon Health Sciences University Hospital (4, 19). The A. baumannii strains BM4420, BM4421, BM4422, BM4424, BM4427, BM4430, BM4432, BM4436, and BM4439, which represent the random amplified polymorphic DNA types A to F, H, I, and K, respectively, were previously described during the characterization of integrons in European isolates of this human pathogen (26). Bacterial strains were maintained on Luria-Bertani agar or broth (27) and grown at 37°C. The plasmid pMU77 is a pUC18 clone that contains a 400-bp HindIII fragment that encompasses an internal region of the A. baumannii 8399 om73 gene (13).
Detection of siderophore activity and catechol compounds produced under iron-rich and iron-limiting conditions.
Bacterial cells were cultured in M9 minimal medium (22) supplemented with either 100 μM FeCl3 (iron rich) or 100 μM ethylenediamine-di-(o-hydroxyphenyl) acetic acid (EDDHA; iron limiting). Production of extracellular compounds with siderophore activity was tested with the chrome azurol S reagent (28). The presence of phenolic (catechol) compounds was detected with the Arnow test (6). Purified anguibactin, acinetobactin, and DHBA obtained from a commercial source (Sigma) were used as standards in chemical and biological assays.
General DNA techniques.
Total DNA was isolated either by using ultracentrifugation in CsCl density gradients (21, 27) or by using the DNeasy tissue kit from Qiagen. Plasmid DNA was isolated according to the method of Birnboim and Doly (9) and further purified by using ultracentrifugation in CsCl-ethidium bromide density gradients (27) or by using the Qiagen mini plasmid spin columns. DNA was digested with restriction enzymes as indicated by the manufacturer (New England Biolabs) and size fractionated by agarose gel electrophoresis (27). Specific restriction fragments were detected by Southern blot analyses which were conducted by using standard protocols (27) under high-stringency conditions (17). The probes to detect the om73 and fatA-like genes were generated by labeling pMU77 and a 750-bp internal fragment of the fatA-like gene (12) that was generated by PCR, respectively. The 32P-labeled probes were prepared by using the oligolabeling method (16), and the radioactive bands were detected with a Storm 860 scanner (Molecular Dynamics).
Isolation and electrophoretic analysis of total and outer membrane proteins.
Bacterial cells used to prepare total and outer membrane fractions were collected, after overnight culture in M9 minimal medium under iron-limiting and iron-rich conditions, by centrifugation and washed once with Tris-buffered saline solution (10 mM Tris-HCl, 0.15 M NaCl [pH 7.5]). Outer membranes were isolated by selective solubilization of total membranes with 1.5% N-lauroylsarcosine (Sarkosyl) and high-speed centrifugation as previously described (2). Outer membrane proteins of all strains were size fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) by using large (14.5-by-15-cm) 12.5% polyacrylamide gels (2). Proteins were visualized by staining with Coomassie brilliant blue. Protein concentration was determined with the Bradford method (10), and 25 μg of proteins was loaded in each lane of polyacrylamide gels.
Detection of the OM73 and FatA-like proteins by immunoblot analyses.
Total and outer membrane proteins were size fractionated by SDS-PAGE on 12.5% polyacrylamide gels and blotted onto nitrocellulose (2). The presence of OM73 was tested with a rabbit polyclonal antiserum raised as previously described (13). The anti-FatA-like serum was prepared by using the protein isolated from A. baumannii 19606 by SDS-PAGE and electroelution as previously described (15). Specific antibodies were immunopurified with nitrocellulose strips containing the cognate antigen band as previously described (25). The presence of immunocomplexes was detected with horseradish peroxidase-labeled protein A and the SuperSignal CL-HRP substrate (Pierce Chemical).
Detection of histamine by TLC.
The presence of histamine in culture supernatants was analyzed by TLC as previously described (7). Briefly, A. baumannii strains were grown overnight in M9 minimal medium supplemented with 1% histidine. Cells were removed by centrifugation, and the culture supernatants were lyophilized and methanol extracted. Samples were spotted onto silica gel TLC plates and developed with a mobile phase of methanol-ammonium hydroxide (20:1). Ninhydrin reagent was used to visualize histidine and/or histamine.
RESULTS AND DISCUSSION
Iron uptake proficiency and production of catechol compounds.
All A. baumannii strains tested in this study grew well in M9 minimal medium containing either 100 μM EDDHA or 100 μM FeCl3. After overnight incubations with shaking, chrome azurol S assays showed that all the A. baumannii iron-deficient culture supernatants contained iron-chelating activity. In addition, Arnow assays demonstrated that the addition of EDDHA induced the production of catechol compounds that were secreted into the culture supernatant but that the addition of FeCl3 reduced catechol levels to amounts close to or below the detection limit of the test. These results demonstrate that all tested A. baumannii clinical isolates express iron-chelating activity and that all strains tested secrete iron-regulated catechol compounds into the extracellular milieu. These findings are indicative of the type of siderophore that these A. baumannii isolates secrete to scavenge iron from the extracellular environment.
Comparative analysis of outer membrane proteins. (i) Oregon isolates.
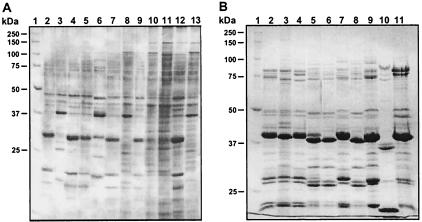
Electrophoretic analysis of outer membrane proteins obtained from the strains isolated during the Oregon Health Sciences University outbreak and cultured under iron limitation showed that these strains can be classified into four groups based on their protein profiles. Group 1 includes the isolates 9235, 8143, 8114, 7133, 8399, and 9124 (Fig. 1A, lanes 2, 4, 5, 7, 9, and 12, respectively), with similar patterns that differ mainly in the presence or absence of relatively large-molecular-size proteins. Group 2 consists of strains 8971, 7138, and 9397 (Fig. 1A, lanes 3, 8, and 13, respectively). The most apparent difference between these two groups is the distinct presence of ca. 30- and 37-kDa proteins in the group 1 and group 2 strains, respectively. The strains 9606 and 7931 (Fig. 1A, lanes 10 and 11, respectively), which are the members of group 3, display protein profiles that are remarkably different from those of group 1 and 2. The strain 8637 (Fig. 1A, lane 6), which is considered the only representative of group 4, displays a pattern that is closer to a combination of the patterns of group 1 and 2 based on the presence of proteins with mobilities corresponding to 30 and 37 kDa. However, this strain displays differences that are apparent when its protein pattern is compared with those shown by members of the first two groups.
FIG. 1.
SDS-PAGE analysis of outer membrane proteins produced by A. baumannii clinical isolates. (A) Oregon strains. Lanes: 2, 9235; 3, 8971; 4, 8143; 5, 8114; 6, 8637; 7, 7133; 8, 7138; 9, 8399; 10, 9606; 11, 7931; 12, 9124; and 13, 9397. (B) European strains. Lanes: 2, BM4420; 3, BM4421; 4, BM4422; 5, BM4424; 6, BM4427; 7, BM4430; 8, BM4432; 9, BM4436; 10, BM4439; and 11, 19606. Lanes 1 in both panels, molecular weight markers. Membrane fractions used for this analysis were isolated from cells cultured under iron-deficient conditions.
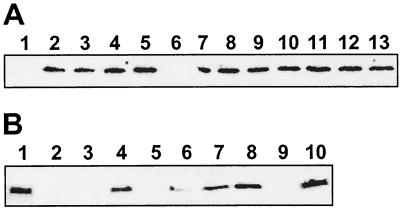
Immunoblot analysis of the same samples with anti-OM73 serum showed that with the exception of strain 8637 (Fig. 2A, lane 6) all the Oregon isolates express the 73-kDa outer membrane protein initially identified in the strain 8399 (Fig. 2A, lanes 2 to 5 and 7 to 13). The fact that the strain 8637 does not produce this protein is consistent with its distinct protein profile (Fig. 1A, lane 6), and when combined, the protein profile and immunoblot data provide a strong basis for the hypothesis that this strain is indeed different from all the other Oregon isolates. None of these strains tested positive when their outer membrane proteins were probed with the anti-FatA-like serum (data not shown).
FIG. 2.
Detection of protein expression by Western blot analysis. (A) Detection of OM73 in the Oregon strains. Lanes: 1, 19606; 2, 9235; 3, 8971; 4, 8143; 5, 8114; 6; 8637; 7, 7133; 8, 7138; 9, 9606; 10, 7931; 11, 9124; 12, 9397; and 13, 8399. (B) Detection of FatA-like protein in the European isolates. Lanes: 1, 19606; 2, BM4439; 3, BM4436; 4, BM4432; 5, BM4430; 6, BM4427; 7, BM4424; 8, BM4422; 9, BM4421; and 10, BM4420.
(ii) European isolates.
A similar analysis of the European strains showed that strains BM4420, BM4421, BM4422, BM4430, BM4436, and the A. baumannii prototype strain 19606 (Fig. 1B, lanes 2, 3, 4, 7, 9, and 11, respectively) can be grouped together based on their similar protein profiles. However, the strain 19606 (Fig. 1B, lane 11) shows a clear difference in the region that includes proteins with apparent molecular sizes ranging from 75 to 100 kDa, a region which also shows some differences in the patterns of the other strains (Fig. 1B, compare lanes 2, 3, 4, 7, and 9). The strains BM4424, BM4427, and BM4432 (Fig. 1B, lanes 5, 6, and 8) constitute the second group, which show differences in the mobilities of apparent major outer membrane proteins, with an apparent molecular size of 37 kDa, and proteins with molecular sizes that range from 25 to 35 kDa. The strain BM4439 is the only member of the third group that displays a protein profile that is clearly different from those shown by the two previous groups of this set of strains (Fig. 1B, lane 10). While OM73 is absent in the outer membranes of these strains, the presence of the FatA-like protein is somewhat variable. Figure 2B shows that the strains BM4432, BM4427, BM4424, BM4422, and BM4420 (lanes 4, 6 to 8, and 10, respectively), in addition to strain 19606 (lane 1), produce the FatA-like protein detected initially in the latter strain with an antiserum raised against the V. anguillarum FatA outer membrane protein. In contrast, the strains BM4439, BM4436, BM4430, and BM4421 (Fig. 2B, lanes 2, 3, 5, and 9, respectively) do not display this protein in their outer membrane fractions. Altogether, the data show that there is not a good correlation between the general outer membrane protein profiles and the production of the FatA-like protein among the European isolates.
Taken together, the results described in this section show that the Oregon isolates tend to display protein profiles that are more related to one another than to those shown by the European strains isolated from different geographical regions. A similar conclusion is obtained when the European isolates are compared among themselves and with those obtained from the Oregon outbreak strains.
Presence of om73 and fatA-like genes.
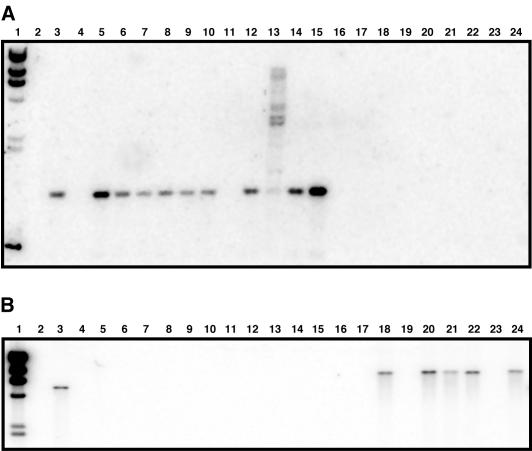
Southern blot analysis of total DNA isolated from each strain was done to test whether the lack of production of either OM73 or the FatA-like protein is due to the absence of the cognate genes, potential mutations, or negative regulatory mechanisms that may impair their expression. Figure 3A shows that with the exception of strain 8637 (lane 11), all the Oregon isolates (lanes 3, 5 to 10, and 12 to 15) contain a 1.2-kb HindIII fragment that hybridizes with the om73 radiolabeled probe. The multiple bands shown in lane 13 are most likely the result of partial digestion of the total DNA isolated from strain 8143. In contrast, 19606 and none of the European isolates produced a detectable signal with this probe (Fig. 3A, lanes 4 and 16 to 24). These results prove that there is a perfect correlation between the production of OM73 and the presence of the gene encoding this protein, which has all the attributes assigned to well-characterized bacterial siderophore receptors (12).
FIG. 3.
Southern blot analysis of total DNA obtained from the A. baumannii clinical strains. (A) Detection of om73 in HindIII-digested DNA isolated from the Oregon (lanes 3 and 5 to 15) and European (lanes 16 to 24) isolates. Lanes: 3, 8399; 4, 19606; 5, 9397; 6, 9124; 7, 7931; 8, 9606; 9, 7138; 10, 7133; 11, 8637; 12, 8114; 13, 8143; 14, 8971; 15, 9235; 16, BM4439; 17, BM4436; 18, BM4432; 19, BM4430; 20, BM4427; 21, BM4424; 22, BM4422; 23, BM4421; and 24, BM4420. (B) Detection of fatA-like gene in EcoRV-digested DNA isolated from the Oregon (lanes 4 to 15) and European (lanes 16 to 24) isolates. Lanes: 3, 19606; 4, 9397; 5, 9124; 6, 7931; 7, 9606; 8, 8399; 9, 7138; 10, 7133; 11, 8637; 12, 8114; 13, 8143; 14, 8971; 15, 9235; 16, BM4439; 17, BM4436; 18, BM4432; 19, BM4430; 20, BM4427; 21, BM4424; 22, BM4422; 23, BM4421; and 24, BM4420. Lanes 1, HindIII-digested λ DNA; lanes 2, empty.
Southern analysis of the same isolates showed that European isolates BM4432, BM4427, BM4424, BM4422, and BM4420 (Fig. 3B, lanes 18, 20, 21, 22, and 24, respectively) possess a 6.6-kb EcoRV DNA fragment that hybridizes with the fatA-like probe while the isolates BM4439, BM4436, BM4430, and BM4421 (Fig. 3B, lanes 16, 17, 19, and 23, respectively) all lack any detectable genetic element related to the fatA-like gene. In addition, these results show that all 12 Oregon isolates do not possess a fatA-like homolog (Fig. 3B, lanes 4 to 15). All these findings are in direct correlation with the results of the Western blot analysis shown in Fig. 2B, demonstrating that none of the Oregon isolates express a FatA-like protein because they do not harbor this gene. It is interesting that the fatA-like gene present in the European isolates is located in a 9.6-kb EcoRV fragment while this gene was mapped to a 5.0-kb EcoRV fragment in the 19606 strain (Fig. 3B, compare lanes 3, 18, 20 to 22, and 24). This observation indicates that this gene may be located in different genomic regions in different A. baumannii clinical isolates that harbor it.
Detection of histamine production.
The presence of hydroxyhistamine in the acinetobactin molecule (31) suggests that A. baumannii 19606 expresses histidine decarboxylase activity as described for V. anguillarum (7, 29). This enzymatic activity is encoded in V. anguillarum by angH, and histamine proved to be an essential precursor in the biosynthesis of anguibactin (29). Furthermore, this amine was shown to be secreted into the milieu together with anguibactin (7). Previous work in our lab has shown that A. baumannii 19606 is capable of converting histidine into histamine but that A. baumannii 8399 does not synthesize histamine (3). Based on this information, we tested all European and Oregon isolates for their ability to produce histamine. TLC analysis of methanol extracts of culture supernatants of cells incubated in the presence of 1% histidine showed that none of the Oregon strains secreted detectable amounts of histamine. This observation is in perfect correlation with the results obtained by the SDS-PAGE and immunoblot analyses described above (Table 1). Furthermore, it provides additional support to the hypothesis that these strains acquire iron via a catechol siderophore system that is different from that expressed by V. anguillarum and A. baumannii 19606.
TABLE 1.
Histamine secretion and production of the FatA-like and OM73 proteins in A. baumannii clinical isolates
| Strain | Histaminea | Outer membrane proteinb | Correlationc |
|---|---|---|---|
| 9235 | ND | OM73 | + |
| 8971 | ND | OM73 | + |
| 8143 | ND | OM73 | + |
| 8114 | ND | OM73 | + |
| 8637 | ND | ND | * |
| 7133 | ND | OM73 | + |
| 7138 | ND | OM73 | + |
| 9606 | ND | OM73 | + |
| 7931 | ND | OM73 | + |
| 9124 | ND | OM73 | + |
| 9397 | ND | OM73 | + |
| BM4420 | + | FatA-like protein | + |
| BM4421 | + | ND | NC |
| BM4422 | ND | FatA-like protein | NC |
| BM4424 | + | FatA-like protein | + |
| BM4427 | ND | FatA-like protein | NC |
| BM4430 | + | ND | NC |
| BM4432 | ND | FatA-like protein | NC |
| BM4436 | + | ND | NC |
| BM4439 | ND | ND | * |
| 8399 | ND | OM73 | + |
| 19606 | + | FatA-like protein | + |
The presence (+) or absence (ND) of histamine in culture supernatants was tested by TLC analysis of methanol extracts of culture supernatants and staining with ninhydrin.
The presence of OM73 and the FatA-like protein in outer membrane preparations was tested by immunoblotting. ND, neither OM73 nor FatA-like protein was detected.
+, correlation either between secretion of histamine and production of FatA-like protein or between absence of histamine and presence of OM73. *, strains which did not produce the OM73 and FatA-like proteins and did not secrete histamine; NC, no correlation.
TLC analysis of the culture supernatants of A. baumannii European isolates BM4420, BM4421, BM4424, BM4430, and BM4436 revealed the presence of an amine compound that showed the same Rf value displayed by the histamine standard. In contrast, a similar analysis of the culture supernatants of strains BM4422, BM4427, BM4432, and BM4439 failed to detect the presence of histamine. When these results obtained by TLC analysis are compared with those obtained by protein analyses (Table 1), it is clear that there is straight correlation between the expression of FatA-like protein and the production of histamine in the isolates BM4420 and BM4424. These data indicate that these two A. baumannii strains may acquire iron through the secretion of acinetobactin and the transport of ferric acinetobactin complexes across the bacterial membranes, potentially via the FatA-like protein. In contrast, the strain BM4439 must use a catechol siderophore-mediated iron acquisition system that is unrelated to that based on acinetobactin since it produces neither the FatA-like protein nor histamine. The strains BM4422, BM4427, and BM4432, which produce the FatA-like protein (Fig. 2B), may be able to use acinetobactin for iron acquisition when it is provided from an external source, since the lack of histamine production most likely precludes its biosynthesis in these strains. Finally, the fact that strains BM4421, BM4430, and BM4436 produce histamine but not the FatA-like protein may indicate that these strains produce a catechol siderophore that includes a hydroxyhistamine moiety but that is significantly different from acinetobactin and anguibactin. Another explanation for this phenotype is the potential deletion of the fatA-like gene, a possibility that is supported by the negative results obtained by Southern blotting (Fig. 3B, lanes 17, 19, and 23).
Conclusions.
Our data demonstrate that there are significant variations in the expression of elements involved in iron acquisition among different A. baumannii nosocomial isolates, even within strains obtained during a particular outbreak of nosocomial infections. The variation in the production of a FatA-like protein and the expression of histidine decarboxylase with the concomitant production of histamine are of particular interest. These elements, which are most likely involved in the biosynthesis and utilization of acinetobactin, may play the same function as those described in the biosynthesis of anguibactin and transport of ferric anguibactin in the fish pathogen V. anguillarum. We have recently observed that the disruption of the A. baumannii 19606 fatA-like gene indeed impairs the iron acquisition phenotype of this strain, confirming its role in iron transport (12). The finding of almost identical iron acquisition elements in a human pathogen and a fish pathogen, which are taxonomically and biochemically different from each other, was unexpected and unpredictable. On the other hand, this finding may indicate that the genetic elements encoding the anguibactin- and acinetobactin-mediated systems are mobile and can be transferred among unrelated pathogens. This hypothesis is supported by the fact that the anguibactin-mediated system is mostly encoded by genes located in the plasmid pJM1 present in virulent strains of V. anguillarum (11). Furthermore, the pJM1 genes, particularly those encoding transport functions, are confined to pJM1 regions that contain genes like those encoding transposases and are surrounded by repeated sequences (30). This genetic arrangement, which resembles that of transposable elements, may explain the presence of similar iron uptake systems in unrelated pathogens. The possibility of the acquisition of the acinetobactin transport system via lateral gene transfer is also supported by the observation that the fatA-like gene can be located in different genomic regions in different A. baumannii isolates.
The fact that some of the European isolates produce histamine but not the FatA-like protein, and vice versa, may reflect either some instability of the genetic elements encoding these iron acquisition components or remnants of genetic processes that were not resolved properly. Another possibility that should be considered, particularly in the situation in which histamine is produced in the absence of detectable FatA-like protein, is that some of these elements like histamine may be used for the biosynthesis of siderophores structurally different from acinetobactin and anguibactin. The data also demonstrate that some isolates, independent of their origin, must express iron acquisition systems unrelated to those mediated by acinetobactin and anguibactin, and even different from that expressed by most of the Oregon isolates, which involve a novel iron chelator and may require the expression of the OM73 outer membrane protein.
All the data together demonstrate the complexity and variability of the iron uptake systems expressed by this opportunistic human pathogen. However, all these variations in the production of iron acquisition elements, either within members of a particular set of isolates related by their nosocomial origin or among strains classified by their geographical origins, may be an accurate reflection of the taxonomic and metabolic variability already described for this bacterial group. They may also reflect the ubiquitous nature of Acinetobacter and its ability to survive under significantly different environmental conditions.
Acknowledgments
We thank M.-C. Ploy (Laboratoire de Bactériologie-Virologie-Hygiène, CHU Dupuytren, France) for providing the A. baumannii strains BM4420, BM4421, BM4422, BM4424, BM4427, BM4430, BM4432, BM4436, and BM4439. M. S. Beglin was a recipient of an Undergraduate Summer Scholars scholarship from Miami University.
REFERENCES
- 1.Actis, L. A., W. Fish, J. H. Crosa, K. Kellerman, S. Ellenberger, F. Hauser, and J. Sanders-Loehr. 1986. Characterization of anguibactin, a novel siderophore from Vibrio anguillarum 775(pJM1). J. Bacteriol. 167:57-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Actis, L. A., S. Potter, and J. H. Crosa. 1985. Iron-regulated outer membrane protein OM2 of Vibrio anguillarum is encoded by virulence plasmid pJM1. J. Bacteriol. 161:736-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Actis, L. A., J. C. Smoot, C. E. Barancin, and R. H. Findlay. 1999. Comparison of differential plating media and two chromatography techniques for the detection of histamine production in bacteria. J. Microbiol. Methods 39:79-90. [DOI] [PubMed] [Google Scholar]
- 4.Actis, L. A., M. E. Tolmasky, L. M. Crosa, and J. H. Crosa. 1993. Effect of iron-limiting conditions on growth of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 31:2812-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anstey, N. M., B. J. Currie, and K. M. Withnall. 1991. Community-acquired Acinetobacter pneumonia in the northern territory of Australia. Clin. Infect. Dis. 14:83-91. [DOI] [PubMed] [Google Scholar]
- 6.Arnow, L. 1937. Colorimetric determination of the components of 3,4-dihydroxyphenylalanine-tyrosine mixtures. J. Biol. Chem. 118:531-537. [Google Scholar]
- 7.Barancin, C. E., J. C. Smoot, R. H. Findlay, and L. A. Actis. 1998. Plasmid-mediated histamine biosynthesis in the bacterial fish pathogen Vibrio anguillarum. Plasmid 39:235-244. [DOI] [PubMed] [Google Scholar]
- 8.Bergogne-Berenzin, E., and K. J. Towner. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 9:148-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradford, M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of proteins utilizing the principle of protein-dye binding. Anal. Biochem. 72:249-252. [DOI] [PubMed] [Google Scholar]
- 11.Crosa, J. H., and C. T. Walsh. 2002. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 66:223-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorsey, C. W. 2002. Molecular and genetic analysis of the iron regulon in different clinical isolates of Acinetobacter baumannii. Ph.D. dissertation. Miami University, Oxford, Ohio.
- 13.Dorsey, C. W., M. E. Tolmasky, J. H. Crosa, and L. A. Actis. 2003. Genetic organization of an Acinetobacter baumannii chromosomal region harbouring genes related to siderophore biosynthesis and transport. Microbiology 149:1227-1238. [DOI] [PubMed] [Google Scholar]
- 14.Echenique, J. R., H. Arienti, M. E. Tolmasky, R. Read, J. Staneloni, J. H. Crosa, and L. A. Actis. 1992. Characterization of a high-affinity iron transport system in Acinetobacter baumannii. J. Bacteriol. 174:7670-7679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Echenique, J. R., C. W. Dorsey, L. C. Patrito, A. Petroni, M. E. Tolmasky, and L. A. Actis. 2001. Acinetobacter baumannii has two genes encoding glutathione-dependent formaldehyde dehydrogenase: evidence for differential regulation in response to iron. Microbiology 147:2805-2815. [DOI] [PubMed] [Google Scholar]
- 16.Feinberg, A. P., and B. Vogelstein. 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132:6-13. [DOI] [PubMed] [Google Scholar]
- 17.Graber, K., L. M. Smoot, and L. A. Actis. 1998. Expression of iron binding proteins and hemin binding activity in the dental pathogen Actinobacillus actinomycetemcomitans. FEMS Microbiol. Lett. 163:135-142. [DOI] [PubMed] [Google Scholar]
- 18.Gray-Owen, S. D., and A. B. Schryvers. 1996. Bacterial transferrin and lactoferrin receptors. Trends Microbiol. 4:185-191. [DOI] [PubMed] [Google Scholar]
- 19.Hartstein, A. I., A. L. Rashad, J. M. Liebler, L. A. Actis, J. Freeman, J. W. Rourke, Jr., T. B. Stibolt, M. E. Tolmasky, G. R. Ellis, and J. H. Crosa. 1988. Multiple intensive care unit outbreak of Acinetobacter calcoaceticus subspecies anitratus respiratory infection and colonization associated with contaminated, reusable ventilators and resuscitation bags. Am. J. Med. 85:624-631. [DOI] [PubMed] [Google Scholar]
- 20.Jalal, M., D. Hossain, J. van der Helm, J. Sanders-Loerh, L. A. Actis, and J. H. Crosa. 1989. Structure of anguibactin, a unique plasmid-related bacterial siderophore from the fish pathogen Vibrio anguillarum. J. Am. Chem. Soc. 111:292-296. [Google Scholar]
- 21.Meade, H. M., S. R. Long, S. E. Ruvkum, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.Neilands, J. 1981. Microbial iron compounds. Annu. Rev. Biochem. 50:715-731. [DOI] [PubMed] [Google Scholar]
- 24.Neilands, J. 1995. Siderophores: structure and function of microbial iron transport compounds. J. Biol. Chem. 270:26723-26726. [DOI] [PubMed] [Google Scholar]
- 25.Olmsted, J. B. 1981. Affinity purification of antibodies from diazotized paper blots of heterogeneous protein samples. J. Biol. Chem. 256:11955-11957. [PubMed] [Google Scholar]
- 26.Ploy, M. C., F. Denis, P. Courvalin, and T. Lambert. 2000. Molecular characterization of integrons in Acinetobacter baumannii: description of a hybrid class 2 integron. Antimicrob. Agents Chemother. 44:2684-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 29.Tolmasky, M. E., L. A. Actis, and J. H. Crosa. 1995. A histidine decarboxylase gene encoded by the Vibrio anguillarum plasmid pJM1 is essential for virulence: histamine is a precursor in the biosynthesis of anguibactin. Mol. Microbiol. 15:87-95. [DOI] [PubMed] [Google Scholar]
- 30.Tolmasky, M. E., and J. H. Crosa. 1995. Iron transport genes of the pJM1-mediated iron uptake system of Vibrio anguillarum are included in a transposonlike structure. Plasmid 33:180-190. [DOI] [PubMed] [Google Scholar]
- 31.Yamamoto, S., N. Okujo, and Y. Sakakibara. 1994. Isolation and structure elucidation of acinetobactin, a novel siderophore from Acinetobacter baumannii. Arch. Microbiol. 162:249-252. [DOI] [PubMed] [Google Scholar]