Abstract
Oral Peptostreptococcus isolates tentatively identified by conventional microbiological culture methods were identified to the species level by a combination of PCR amplification of 16S rRNA genes and restriction enzyme analysis of the amplified products. This method is a reliable and rapid alternative to conventional methods for identification of these bacterial species.
Gram-positive anaerobic cocci (GPAC), which have been isolated from a wide range of body sites, are more commonly known as peptococci and peptostreptococci. Most GPAC clinical isolates have been identified as members of the genus Peptostreptococcus. GPAC are regarded as being part of the normal human flora of the oral cavity, skin, upper respiratory and gastrointestinal tracts, and the female genitourinary system (7). However, they can act as opportunistic pathogens and cause severe infections at distinct body sites. These include infections of the oral cavity, respiratory tract, genitourinary tract, central nervous system, superficial and soft tissues, intra-abdominal sites, and cardiovascular system and septicemia (7). GPAC account for 25 to 30% of all anaerobic isolates and are usually involved in polymicrobial infections, particularly abscesses (7). GPAC species have also been isolated in pure culture from infected sites (7).
Peptostreptococcus species have been identified in a number of oral infections. P. micros and P. anaerobius have been isolated from endodontic abscesses (19). P. micros, P. anaerobius, P. magnus, and P. prevotii are associated with dental root canal infections (3, 16). Peptostreptococcus species have also been implicated in human gingivitis and periodontitis. For example, P. micros is associated with periodontal destruction, especially in sites from disease-active patients (1, 6, 13), while P. anaerobius has been associated with gingivitis and periodontitis (5, 18).
Several methods have been used for the identification of clinical isolates of Peptostreptococcus species. The principal approach has been to use microbiological culture in conjunction with biochemical tests. A selective and differential medium that facilitates the isolation of P. micros, and which is based upon a Columbia agar supplemented with glutathione and lead acetate, has been described (17). The biochemical tests carried out are based upon analysis of bacterial enzymatic activities (Rapid ID 32A) and the capacity to hydrolyze amino acid and phosphate substrates (Rapid ANA II) (11, 13). This approach has been used with limited success in differentiating between Peptostreptococcus species (4, 8). Volatile fatty acid (VFA) production has proved to be a useful identification method, particularly for P. anaerobius, since this is the only gram-positive anaerobic coccus to produce isocaproic acid as the major by-product of metabolism (7, 20).
More recently, molecular-based techniques have been used to identify Peptostreptococcus species in clinical samples. We have developed PCR assays for the direct detection of P. micros (15) and P. anaerobius (14) in clinical samples. In this study, we report the development of a rapid, novel molecular-based method for accurately identifying clinical isolates of oral Peptostreptococcus species. This method is based upon restriction enzyme analysis of PCR-amplified bacterial 16S rRNA genes (PCR-restriction fragment length polymorphism [RFLP]). The method was used in the identification of stored clinical isolates that had been isolated from the oral cavity and assigned to the genus Peptostreptococcus on the basis of microbiological culture alone. A total of 22 isolates were examined by the PCR-RFLP method. Isolates had been originally obtained from the culture of pus aspirates from patients with acute dentoalveolar abscesses or from the subgingival plaque of patients with adult periodontitis. Due to the large number of putative Peptostreptococcus isolates routinely recovered and the prohibitively high cost of carrying out detailed biochemical tests for identification to the species level, identification of clinical isolates had only been carried out to the anaerobic streptococcus level. This was achieved on the basis of colony morphology, Gram staining characteristics, and atmospheric requirements. Samples were cultured by inoculation onto Fastidious Anaerobe agar plates (Life Technologies, Paisley, Scotland) supplemented with 7.5% (vol/vol) sterile defibrinated horse blood and incubated at 37°C for 7 days in an anaerobic chamber (Don Whitley Scientific, Shipley, England). Anaerobic streptococci were identified on the basis of Gram staining and atmospheric requirements: i.e., gram-positive cocci that would not grow at 37°C in an atmosphere of 5% CO2-95% air.
Crude DNA extracts were prepared from each bacterial isolate by inoculation of two loopfuls of bacterial cells into 100 μl of sterile molecular biology-grade water, boiling for 10 min, and removal of cell debris by centrifugation. The supernatant was retained for subsequent PCR analysis.
PCR was carried out on each crude DNA extract. The PCR primers used targeted conserved regions of the 16S rRNA gene and were designed to amplify DNA from most bacterial species. The primers used were 5′-AGAGTTTGATCMTGGCTCAG-3′ (27f; Escherichia coli positions 8 to 27) and 5′-ACGGGCGGTGTGTRC-3′ (1392r; E. coli positions 1405 to 1391), where M = C + A and R = A + G, and give an expected amplification product of approximately 1,500 bp. PCR amplification was carried out in a total volume of 50 μl, comprising 10 μl of bacterial DNA extract and 40 μl of reaction mixture containing 1× PCR buffer (10 mM Tris-HCl [pH 9.0], 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100), 1.0 U of Taq DNA polymerase (Promega), 0.2 mM each deoxynucleoside triphosphate, and each primer at a concentration of 0.2 μM. For increased sensitivity and specificity, “hot start” PCR was used, whereby the primers are separated from other reaction components by a layer of wax (DyNAwax; Flowgen), therefore preventing the reaction from starting until the wax has melted upon commencement of thermal cycling. PCR amplification was carried out in an OmniGene thermal cycler (Hybaid Ltd., Teddington, England). After an initial denaturation step of 94°C for 5 min, 40 cycles were carried out, comprising denaturation at 94°C for 1 min, annealing at 60°C for 1 min, and extension at 72°C for 1 min, followed by a final extension step at 72°C for 10 min. Ten microliters of each PCR product was electrophoresed on a 2% agarose gel, and amplified DNA was detected by staining with ethidium bromide (0.5 μg/ml) and visualization under UV light.
Ten microliters of each PCR product was separately digested in a total volume of 20 μl with 5 U of each of the restriction enzymes CfoI, HinfI, and RsaI (Promega) and MnlI (Helena Biosciences, Sunderland, England) at 37°C for 3 h. Restriction fragments were visualized by agarose gel electrophoresis as described above.
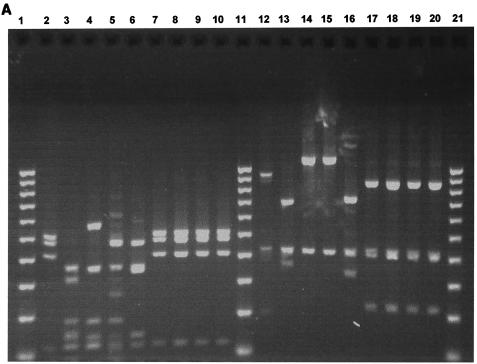
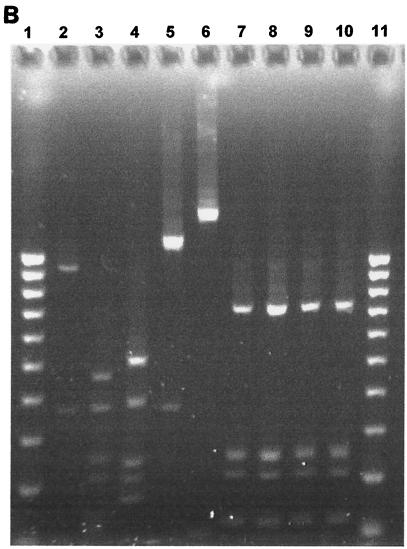
The 16S rRNA gene was successfully amplified from all 22 bacterial isolates and all type strains tested, as demonstrated by the appearance of a PCR product of the expected size. Seven type strains of the Peptostreptococcus species of interest were analyzed by the PCR-RFLP method to generate distinct fingerprints against which those from clinical isolates could be compared for identification purposes. The species selected were P. micros NCTC 11808, P. magnus NCTC 11804, P. asaccharolyticus NCTC 11461, P. productus NCTC 11829, P. indolicus NCTC 11829, P. prevotii NCTC 11806, and P. anaerobius NCTC 11460. Of these species, P. micros, P. anaerobius, P. asaccharolyticus, and P. magnus can be regarded as oral species since they have been detected in the oral cavity by microbiological culture or PCR methods (15, 18). The sizes of the DNA fragments obtained by digestion of the bacterial 16S rRNA PCR product with each of the restriction enzymes CfoI, HinfI, and RsaI for each of the seven type strains tested are summarized in Table 1. Restriction patterns generated by each of these three restriction enzymes for Peptostreptococcus type strains are shown in Fig. 1A and B. Distinct restriction profiles were obtained for all species with the exception of the type strains of P. asaccharolyticus and P. indolicus, whose restriction profiles were identical for all three enzymes. However, these two species could be discriminated by use of an additional restriction enzyme (MnlI), as shown in Fig. 1C, and the sizes of the DNA fragments obtained are shown in Table 1. RsaI and HinfI were the most discriminatory enzymes, sorting the seven type strain species into six distinct RFLP groups, whereas five RFLP groups were obtained for CfoI. Somewhat surprisingly, HinfI did not digest the P. anaerobius 16S rRNA PCR product. Typical restriction patterns obtained for the clinical isolates with each of the restriction enzymes CfoI, HinfI, and RsaI are shown in Fig. 2.
TABLE 1.
Sizes of DNA fragments obtained following PCR-RFLP analysis of 16S rRNA genes of Peptostreptococcus type strains with the restriction enzymes CfoI, HinfI, RsaI, and MnlI
| Bacterial species | Size (bp) of fragment obtaineda
|
|||
|---|---|---|---|---|
| CfoI | HinfI | RsaI | MnlI | |
| P. anaerobius | 700, 430, 340 | 1,500 | 460, 360, 350, 150, 120 | ND |
| P. asaccharolyticus | 870, 430, 220 | 750, 260, 220, 120 | 530, 490, 420, 120 | 420, 350, 270, 180, 130 |
| P. indolicus | 870, 430, 220 | 750, 260, 220, 120 | 530, 490, 420, 120 | 350, 270, 190, 170, 130 |
| P. magnus | 1,100, 430 | 1,100, 380 | 460, 360, 180, 150, 120 | ND |
| P. micros | 1,100, 430 | 520, 390, 240, 200, 180 | 560, 360, 180, 150, 120 | ND |
| P. prevotii | 700, 430, 380 | 470, 380, 250, 220, 150, 125 | 370, 330, 180, 150, 120 | ND |
| P. productus | 940, 430, 200 | 950, 380, 100 | 530, 490, 420, 120 | ND |
Only DNA fragments at least 100 bp in size are shown. ND, not determined.
FIG. 1.
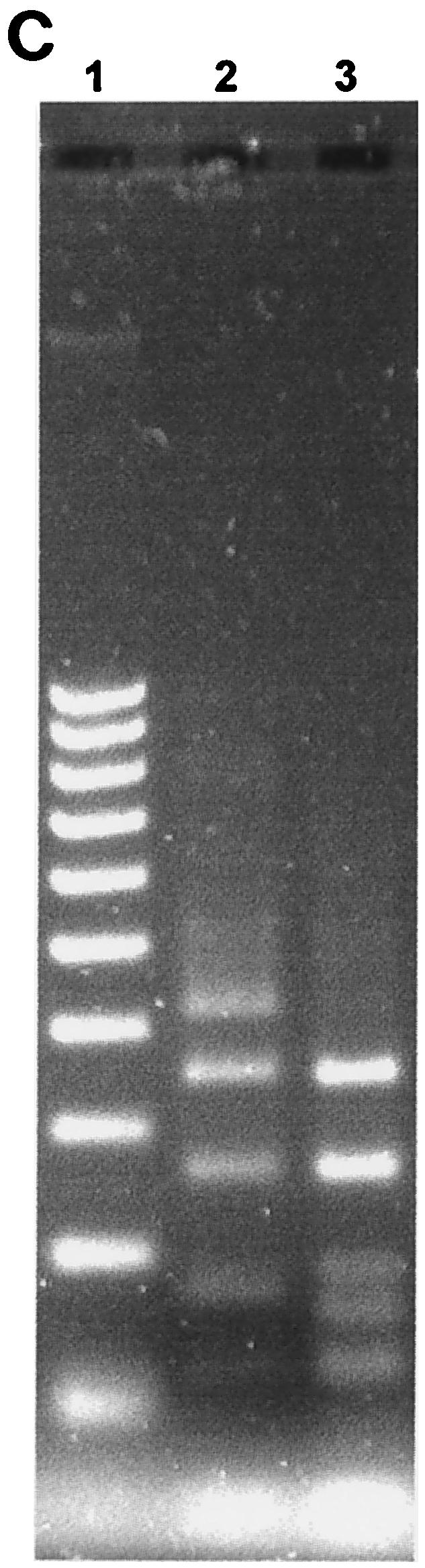
(A) Two percent agarose gel electrophoresis of restriction fragments obtained by digestion of 16S rRNA gene products of Peptostreptococcus type strains with the restriction enzymes RsaI and CfoI. Lanes: 1, 11, and 21, 100-bp DNA ladder; 2, P. productus (RsaI); 3, P. prevotii (RsaI); 4, P. micros (RsaI); 5, P. magnus (RsaI); 6, P. anaerobius (RsaI); 7, P. asaccharolyticus (RsaI); 8, P. asaccharolyticus (RsaI); 9, P. indolicus (RsaI); 10, P. indolicus (RsaI); 12 to 20, same as lanes 2 to 10 but digested with CfoI. (B) Two percent agarose gel electrophoresis of restriction fragments obtained by digestion of 16S rRNA gene products of Peptostreptococcus type strains with the restriction enzyme HinfI. Lanes: 1 and 11, 100-bp DNA ladder; 2 to 10, as in Fig. 1A (lanes 2 to 10) but digested with HinfI. (C) Two percent agarose gel electrophoresis of restriction fragments obtained by digestion of 16S rRNA gene products of P. asaccharolyticus and P. indolicus type strains with the restriction enzyme MnlI. Lanes: 1, 100-bp DNA ladder; 2, P. asaccharolyticus; 3, P. indolicus.
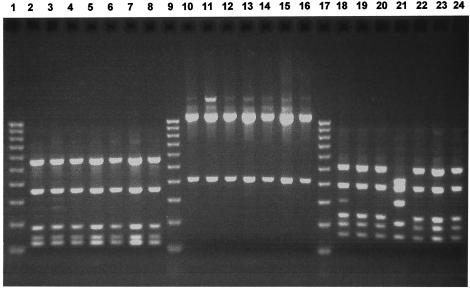
FIG. 2.
Two percent agarose gel electrophoresis of restriction fragments obtained by digestion of 16S rRNA gene products of seven selected Peptostreptococcus clinical isolates with the restriction enzymes RsaI, CfoI, and HinfI. Lanes: 1, 9, and 17, 100-bp DNA ladder; 2 to 8, RsaI digests; 10 to 16, CfoI digests; 18 to 24, HinfI digests. The atypical HinfI restriction profile of the clinical isolate that possesses RsaI and CfoI restriction profiles corresponding to those of P. micros is shown in lane 21.
Of the 22 bacterial isolates examined, 15 had restriction patterns for CfoI, HinfI, and RsaI that perfectly matched that of P. micros NCTC 11808, thus confirming the identity of these clinical isolates as P. micros. Of the remaining seven isolates, four gave restriction patterns for CfoI and RsaI that were identical to that for P. micros, but the restriction profile for HinfI was distinct from that of P. micros and did not match any of the profiles for the other six species examined. However, the HinfI restriction profiles were identical for all four clinical isolates. This suggests that these four isolates may be strain variants of P. micros or perhaps represent another, possibly unidentified, bacterial species that is very closely related to P. micros. Three of the 22 isolates examined did not match any of the restriction profiles for the seven species under investigation and as such probably represent species that have been misidentified by culture methods as belonging to the genus Peptostreptococcus. Since none of the restriction profiles generated with CfoI, HinfI, and RsaI matched those expected for P. asaccharolyticus or P. indolicus, it was unnecessary to digest the PCR products of any bacterial isolates with MnlI in this instance.
Conventional methods for identifying Peptostreptococcus species are reliant upon microbiological culture coupled to biochemical tests for phenotypic identification to the species level. This identification method has been useful, as demonstrated in this study by the observation that 19 of 22 bacterial isolates tentatively identified as members of the genus Peptostreptococcus by microbiological culture alone were confirmed as such by the PCR-RFLP identification method. However, biochemical test kits that detect enzymatic activities, such as the Rapid ID32A system, are expensive and not totally reliable for species identification, particularly when large numbers of clinical isolates are to be analyzed. The inaccuracy of this identification system was highlighted previously by Ng et al. (11). In that study, it was demonstrated that although the system was particularly well suited to the identification of P. anaerobius and P. asaccharolyticus, it was far less reliable for the identification of the three oral species P. magnus, P. micros, and, in particular, P. prevotii. These methods are both time-consuming and labor intensive, and the results obtained are confounded by the existence of phenotypically variable strains that exhibit different enzymatic activities from the species type strains.
The use of gas-liquid chromatography (GLC) for detecting VFA by-products of bacterial metabolism has been touted as an additional method for identifying Peptostreptococcus species (7, 8, 20). GLC can be used to classify Peptostreptococcus species into three groups on the basis of the major terminal VFA produced: an acetate group that produces acetic acid or no VFAs at all (e.g., P. magnus and P. micros); the butyrate group (which is the largest group), which produces butyric acid as its major VFA (e.g., P. asaccharolyticus and P. prevotii); and a caproate group, which produces longer-chain VFAs (e.g., P. anaerobius). Taken on its own, this method lends itself only to the specific identification of P. anaerobius, since this is the only species in this group to produce isocaproic acid as its major terminal VFA. GLC results have been shown to closely correlate with those produced by conventional biochemical methods (8). However, the method is time-consuming, costly, and not suitable for use in diagnostic laboratories.
The PCR-RFLP method presented in this study overcomes the limitations of biochemical and GLC-based methods. On the basis of the PCR-RFLP scheme used in this study for the identification of seven major Peptostreptococcus species, including four oral species, the greatest discriminatory capacity was conferred by the enzyme RsaI, which could distinguish between all species except P. asaccharolyticus or P. indolicus and P. productus. The additional use of CfoI or HinfI could readily discriminate P. productus from P. asaccharolyticus or P. indolicus. However, an additional restriction enzyme (MnlI) had to be used in order to distinguish between P. asaccharolyticus and P. indolicus, which possessed identical restriction profiles for CfoI, HinfI, and RsaI.
Several recent studies have questioned the accuracy of current classification systems for members of the genus Peptostreptococcus. For example, Rajendram et al. (12) suggested reclassification of P. ascaccharolyticus as Schleiferella asaccharolyticus, while it has been recommended that P. magnus and P. micros be reclassified as Finegoldia magna and Micromonas micros, respectively (9). Three new genera have been proposed for members of the genus Peptostreptococcus, namely Peptoniphilus (which includes P. asaccharolyticus, P. indolicus, and three other species), Anaerococcus (which includes P. prevotii and five other species), and Gallicola (which comprises solely P. barnesae) (2). P. anaerobius has been proposed as the only remaining member of the genus Peptostreptococcus (10).
In conclusion, we have developed a rapid, accurate, and specific method for identifying oral Peptostreptococcus species and which can also be applied to the identification of nonoral species. We propose the use of this PCR-RFLP method as an alternative and more accurate method than phenotypic methods for species-level identification of clinical isolates tentatively identified by culture methods as Peptostreptococcus species in the clinical microbiology laboratory. The method is cheaper, simpler, and more rapid than conventional identification methods and provides definitive results, since it can also correctly identify phenotypically variable strains. The simplicity of this PCR-RFLP identification method makes it suitable for use in both clinical and reference laboratory settings.
REFERENCES
- 1.Dzink, J. L., S. S. Socransky, and A. D. Haffajee. 1988. The predominant cultivable microbiota of active and inactive lesions of destructive periodontal diseases. J. Clin. Periodontol. 15:316-323. [DOI] [PubMed] [Google Scholar]
- 2.Ezaki, T., Y. Kawamura, N. Li, Z. Y. Li, L. C. Zhao, and S. E. Shu. 2001. Proposal of the genera Anaerococcus gen. nov., Peptoniphilus gen. nov. and Gallicola gen. nov. for members of the genus Peptostreptococcus. Int. J. Syst. E vol. Microbiol. 51:1521-1528. [DOI] [PubMed] [Google Scholar]
- 3.Gomes, B. P. F. A., J. D. Lilley, and D. B. Drucker. 1996. Clinical significance of dental root canal microflora. J. Dent. 24:47-55. [DOI] [PubMed] [Google Scholar]
- 4.Marler, L. M., J. A. Siders, L. C. Wolters, Y. Pettigrew, B. L. Skitt, and S. D. Allen. 1991. Evaluation of the new RapID-ANA II System for the identification of clinical anaerobic isolates. J. Clin. Microbiol. 29:874-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moore, L. V. H., W. E. C. Moore, E. P. Cato, R. M. Smibert, J. A. Burmeister, A. M. Best, and R. R. Ranney. 1987. Bacteriology of human gingivitis. J. Dent. Res. 66:989-995. [DOI] [PubMed] [Google Scholar]
- 6.Moore, W. E. C., L. H. Moore, R. R. Ranney, R. M. Smibert, J. A. Burmeister, and H. A. Schenkein. 1991. The microflora of periodontal sites showing active destructive progression. J. Clin. Periodontol. 18:729-739. [DOI] [PubMed] [Google Scholar]
- 7.Murdoch, D. A. 1998. Gram-positive anaerobic cocci. Clin. Microbiol. Rev. 11:81-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murdoch, D. A., and I. J. Mitchelmore. 1991. The laboratory identification of gram-positive anaerobic cocci. J. Med. Microbiol. 34:295-308. [DOI] [PubMed] [Google Scholar]
- 9.Murdoch, D. A., and H. N. Shah. 1999. Reclassification of Peptostreptococcus magnus (Prevot 1933) Holdeman and Moore 1972 as Finegoldia magna comb. nov. and Peptostreptococcus micros (Prevot 1933) Smith 1957 as Micromonas micros comb. nov. Anaerobe 5:555-559. [Google Scholar]
- 10.Murdoch, D. A., H. N. Shah, S. E. Gharbia, and D. Rajendram. 2000. Proposal to restrict the genus Peptostreptococcus (Kluyver & van Niel 1936) to Peptostreptococcus anaerobius. Anaerobe 6:257-260. [Google Scholar]
- 11.Ng, J., L.-K. Ng, A. W. Chow, and J.-A. R. Dillon. 1994. Identification of five Peptostreptococcus species isolated predominantly from the female genital tract by using the Rapid ID32A System. J. Clin. Microbiol. 32:1302-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajendram, D., H. N. Shah, S. E. Gharbia, and D. A. Murdoch. 2001. Reclassification of Peptostreptococcus asaccharolyticus (Distaso 1912) Ezaki, Yamamoto, Ninomiya, Suzuki and Yabuuchi 1983 as Schleiferella asaccharolytica comb. nov., Peptostreptococcus indolicus (Christiansen 1934) Ezaki, Yamamoto, Ninomiya, Suzuki and Yabuuchi 1983 as Schleiferella indolica comb. nov., Peptostreptococcus lacrimalis Li, Hashimoto, Adnan, Miura, Yamamoto and Ezaki 1992 as Schleiferella lacrimalis comb. nov. and Peptostreptococcus harei (Murdoch, Collins, Willems, Hardie, Young and Magee 1997) as Schleiferella harei comb. nov. Anaerobe 7:93-101. [Google Scholar]
- 13.Rams, T. E., D. Feik, M. A. Listgarten, and J. Slots. 1992. Peptostreptococcus micros in human periodontitis. Oral Microbiol. Immunol. 7:1-6. [DOI] [PubMed] [Google Scholar]
- 14.Riggio, M. P., and A. Lennon. 2002. Development of a PCR assay specific for Peptostreptococcus anaerobius. J. Med. Microbiol. 51:1097-1101. [DOI] [PubMed] [Google Scholar]
- 15.Riggio, M. P., A. Lennon, and A. Smith. 2001. Detection of Peptostreptococcus micros DNA in clinical samples by PCR. J. Med. Microbiol. 50:249-254. [DOI] [PubMed] [Google Scholar]
- 16.Sundqvist, G. 1992. Associations between microbial species in dental root canal infections. Oral Microbiol. Immunol. 7:257-262. [DOI] [PubMed] [Google Scholar]
- 17.Turng, B.-F., J. M. Guthmiller, G. E. Minah, and W. A. Falkler. 1996. Development and evaluation of a selective and differential medium for the primary isolation of Peptostreptococcus micros. Oral Microbiol. Immunol. 5:356-361. [DOI] [PubMed] [Google Scholar]
- 18.Wade, W. G., J. Moran, J. R. Morgan, R. Newcombe, and M. Addy. 1992. The effects of antimicrobial acrylic strips on the subgingival microflora in chronic periodontitis. J. Clin. Periodontol. 19:127-134. [DOI] [PubMed] [Google Scholar]
- 19.Williams, B. L., G. F. McCann, and F. D. Schoenknecht. 1983. Bacteriology of dental abscesses of endodontic origin. J. Clin. Microbiol. 18:770-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson, M. J., V. Hall, J. Brazier, and M. A. O. Lewis. 2000. Evaluation of a phenotypic scheme for the identification of ‘butyrate-producing’ Peptostreptococcus species. J. Med. Microbiol. 49:747-751. [DOI] [PubMed] [Google Scholar]