Abstract
DNA microarrays are an excellent potential tool for clinical microbiology, since this technology allows relatively rapid identification and characterization of microbial and viral pathogens. In the present study, an oligonucleotide microarray was developed and used for the analysis of thermophilic Campylobacter spp., the primary food-borne pathogen in the United States. We analyzed four Campylobacter species: Campylobacter jejuni, C. coli, C. lari, and C. upsaliensis. Our assay relies on the PCR amplification of specific regions in five target genes (fur, glyA, cdtABC, ceuB-C, and fliY) as a first step, followed by microarray-based analysis of amplified DNAs. Alleles of two genes, fur and glyA, which are found in all tested thermophilic Campylobacter spp., were used for identification and discrimination among four bacterial species, the ceuB-C gene was used for discrimination between C. jejuni and C. coli, and the fliY and cdt genes were used as additional genetic markers specific either for C. upsaliensis and C. lari or for C. jejuni. The array was developed and validated by using 51 previously characterized Campylobacter isolates. All isolates were unambiguously identified on the basis of hybridization patterns with 72 individual species-specific oligoprobes. Microarray identification of C. jejuni and C. coli was confirmed by PCR amplification of other genes used for identification (hipO and ask). Our results demonstrate that oligonucleotide microarrays are suitable for rapid and accurate simultaneous differentiation among C. jejuni, C. coli, C. lari, and C. upsaliensis.
Campylobacter is one of the leading causes of bacterial food-borne diarrheal disease throughout the world (2). Campylobacteriosis is estimated to affect over 2.4 million persons every year in the United States. Although Campylobacter does not commonly cause death, the available data suggest that ca. 100 persons with Campylobacter infections die each year (19). The genus Campylobacter comprises 16 closely related species and 6 subspecies of gram-negative bacteria that primarily colonize the gastrointestinal tracts of a wide variety of host species. Epidemiological data show that the most significant food-borne Campylobacter pathogen species is Campylobacter jejuni (19).
Conventional methods for detecting and discriminating between Campylobacter species are tedious and time-consuming procedures. In addition, some of these assays may yield inconsistent results associated with the genetic divergence among the strains of one species and the presence of closely related genes in other Campylobacter species (29, 33). In recent years, numerous molecular diagnostic approaches for detecting and analyzing Campylobacter spp. have been developed, including various PCR-based assays (3, 7, 8, 10-12, 14-17, 20-23, 25-27, 29-32, 34, 35, 37-39). These PCR methods have several advantages. In general, they are faster and have higher sensitivity and specificity. However, as with biochemical tests, genetic variability among the isolates of Campylobacter species, which has been demonstrated previously (9, 18, 28), can reduce the confidence of bacterial identification by using PCR (24, 29, 33).
In previous studies, we demonstrated that oligonucleotide arrays can be used to characterize Shigella spp. and Escherichia coli (4) virulence genes involved in bacterial pathogenesis and to identify Listeria species (36) and clinically relevant rotavirus G genotypes (5). In the present study, an array containing species-specific oligonucleotide probes for four clinically relevant Campylobacter species (C. jejuni, C. coli, C. lari, and C. upsaliensis) was developed by using specific regions of five genes (fur, glyA, cdt, ceuB-C, and fliY). The array readily distinguishes among all four species.
MATERIALS AND METHODS
Bacterial strains.
C. upsalienesis strains were the generous gift of B. Swaminathan and P. Fields of the National Salmonella and Campylobacter Reference Laboratories, Centers for Disease Control and Prevention, Atlanta, Ga. Other strains were obtained from R. Thunberg and T. Tran of the Center for Food Safety and Applied Nutrition, U.S. Food and Drug Administration (FDA), College Park, Md. Bacterial cultures were grown on brain heart infusion plates (Difco, Detroit, Mich.) under microaerophilic conditions. The bacterial strains used in the present study were as follows.
C. coli.
The C. coli strains tested were ATCC 33559 (from porcine feces), ATCC 43473 (from human feces), ATCC 43474 (from human feces), ATCC 43475 (from porcine feces), ATCC 43476 (from sheep feces), ATCC 43481 (from turkey feces), ATCC 49941, ATCC 43480 (from porcine feces), ATCC 43478 (from marmoset feces), ATCC 43485 (from human feces), ATCC 43486, and the clinical isolates 3116, 3117, 5100, 6925, 92B4QA, HB37, 7569, 1420, and USDA11.
C. jejuni.
The C. jejuni strains tested were ATCC 33291 (from human feces), ATCC 35919 (from human feces), ATCC 29428 (from human feces), ATCC 35921 (from human feces), ATCC 35922 (from human feces), ATCC 33560 (from bovine feces), ATCC 43435 (from human feces), ATCC 35918 (from aborted ovine fetus), ATCC 33252 (from human blood), and the clinical isolates DENVER-1, CDC1420, GH18401, GH7493, DENVER-2, and OYSTER-BAY.
C. lari.
The C. lari strains tested were ATCC 35222 (from dog feces), ATCC 35223 (from child with mild diarrhea), ATCC 35221 (from Herring gull cloacal swab), ATCC 43675 (from human feces), and the clinical isolates 3125, 4899, 4902, 4903, 4906, 4907, and BT9.
C. upsaliensis.
The C. upsaliensis strains tested were clinical isolates D1673, D2237, 5613, 5512, and 5502.
Arcobacter butzleri.
The A. butzleri strains tested were ATCC 49616 (from human feces) and clinical isolate 5530.
Non-Campylobacter species.
Listeria monocytogenes, L. innocua, Bacillus subtilis, B. cereus, E. coli, Shigella flexneri, Shigella sonnei, Staphylococcus aureus, Streptococcus pyogenes, and Yersinia enterocolitica were used as negative controls in the present study. These bacteria were grown overnight on brain heart infusion plates (Difco, Detroit, Mass.) at 37°C.
Genomic DNA preparation.
Freshly grown bacteria were boiled in water (ca. 108 cells/ml) for 10 min, followed by centrifugation at 14,000 × g for 10 min to remove denatured proteins and bacterial membranes. The presence of genomic DNA in all prepared samples was confirmed by 1% agarose gel electrophoresis, followed by visualization with ethidium bromide.
PCR amplification.
Table 1 lists the primers used to amplify the various Campylobacter genes in the present study. Reverse PCR primers of each pair contained the T7 RNA polymerase promoter sequence (TAATACGACTCACTATAGGG) at the 5′ ends. The standard PCR mixture (30 μl) contained 1.5 U of HotStar Taq DNA polymerase in the recommended buffer supplemented with 2.5 mM MgCl2 (Qiagen, Chatsworth, Calif.), 600 nM concentrations of each forward and reverse primer, 200 μM concentrations of each deoxynucleoside triphosphate (dATP, dGTP, dCTP, and dTTP), and 1 to 2 μl of DNA template (ca. 0.2 μg of genomic bacterial DNA). The PCR was performed by using a GeneAmp PCR system 9600 thermocycler (PE Applied Biosystems, Foster City, Calif.) with the following cycle conditions: initial activation at 95°C for 15 min; 40 cycles of 94°C for 40 s, 50°C for 1 min, and 72°C extension for 1 min for primers S1-S2 and CmpfurF-CmpfurR or for 3 min for primers ceuBCF-ceuBCR and CmpToxF-CmpToxR; and a final extension at 72°C for 10 min. The presence of amplified PCR products was detected by using a 1% agarose gel, followed by UV visualization after ethidium bromide staining.
TABLE 1.
Primers used for amplification of various Campylobacter genes
| Target gene | Primer | Nucleotide sequence (5′-3′) | GenBank accession no. | PCR product size(s) (bp) | Tm (°C) | Source or reference |
|---|---|---|---|---|---|---|
| glyA | S1 | AAYAAATAMGCWGAAGGWTAT | X53816, AF136501, AF136497, AF136496, AF136495, AF136494, AF136493 | 640 | 43-47 | 1 |
| S2 | TAATACGACTCACTATAGGGATGCATYAAWGGWCCWCCTGG | 52-54 | ||||
| ceuB-ceuC | ceuBCF | ATGCTTTAAATTATATACAAAATATCC | X88849, AL139078, NC_002163 | 1,229 | 48 | This study |
| ceuBCR | TAATACGACTCACTATAGGGTAAGTGATATTTACTACTAAAAGTCC | 50 | ||||
| fur | CmpfurF | AGAAAGCTTATATATGGAAATCAAACAAG | NC_002163, L77075 | 362, 366, 367, and 370a | 53 | This study |
| CmpfurR | TAATACGACTCACTATAGGGAATTYTTTGYTGCTCTAAAATATTATCAAAC | 54 | ||||
| cdt cluster | CmpToxF | CAAATATTTGAAAAAGGGTCATAAATTTGTTGC | AL139074 | 2,870 | 56 | This study |
| CmpToxR | TAATACGACTCACTATAGGGTTTCCATGATAGCGATCATAAAAGACAAGC | 58 | ||||
| hipO | VS-15 | GAATGAAATTTTAGAATGGGG | AL139078, X71603 | 358 | 47 | 7 |
| VS-16 | GATATCTATGATTTTATCCTGC | 47 | ||||
| hipO | HIP-F | GAAGAGGGTTTGGGTGGTG | AL139076, Z36940 | 735 | 53 | 7 |
| HIP-R | AGCTAGCTTCGCATAATAACTTG | 52 | ||||
| ask | CC18-F | GGTATGATTTCTACAAAGCGAG | AF017758 | 500 | 51 | 7 |
| CC519-R | ATAAAAGACTATCGTCGCGTG | 50 |
The sizes of the PCR amplicons for C. lari, C. coli, C. upsaliensis, and C. jejuni, respectively.
In vitro transcription and fluorescent chemical labeling of RNA.
Single-stranded RNA (ssRNA) samples for microarray analysis were synthesized by in vitro transcription from the promoter-tagged PCR amplicons by using the MEGAscript T7 high-yield transcription kit (Ambion, Austin, Tex.). The RNA transcription was performed in a 30-μl reaction mixture containing 2 μl of MEGAscript T7 enzyme mix (Ambion); 1× reaction buffer; 5 mM concentrations of ATP, UTP, CTP and GTP; and ca. 0.1 to 0.5 μg of DNA template from the PCR. The reactions were allowed to proceed at 37°C for 1 to 2 h, and then the unincorporated nucleoside triphosphates were removed by purification by using the Centrisep-Spin columns (Princeton Separations, Adelphia, N.J.) according to the manufacturer's protocol.
The Micromax ASAP RNA labeling kit (Perkin-Elmer, Boston, Mass.) was used for Cy5 labeling of the RNA samples for microarray analysis according to the manufacturer's protocol. Fluorescence-labeled ssRNA samples were purified from unincorporated dye by using the Centrisep-Spin columns, dried under vacuum, and solubilized in the Micromax hybridization buffer III at final concentration of 0.3 to 0.5 μM.
Design of oligonucleotide microarray probes.
Basic local alignment search tool (BLAST) searching was used to find and retrieve the sequences of homologous target regions of each of the five genes analyzed (Table 2). The retrieved sequences were aligned by using CLUSTALX software (13). The gene-specific oligonucleotide probes were designed to include species-specific variable regions. The selected oligonucleotides are summarized in Table 2. The 5′ end of each oligonucleotide was modified during the synthesis by using the TFA Aminolink CE reagent (PE Applied Biosystems) for immobilization of the oligonucleotides to silylated slides (CEL Associates, Inc., Houston, Tex.).
TABLE 2.
Oligonucleotide probes for detection and discrimination among Campylobacter spp.
| Type | Name | Sequence | Length (in nucleotides) | G + C (%) | Tma (°C) |
|---|---|---|---|---|---|
| fur specific | FurCJ1 | GTAACTTCCATTTCTTTTGG | 20 | 35 | 46 |
| FurCJ2 | TGGTTCAGCAGGTAAAAAA | 19 | 37 | 45 | |
| FurCJ3 | GCAAAAGAACATGGTTTTAAA | 21 | 29 | 45 | |
| FurCJ4 | GAACCTGATTTAAATGTAGGA | 21 | 33 | 47 | |
| FurCJ5 | GTGTTTGTGGTGATTGTAATAA | 22 | 32 | 47 | |
| FurCJ6 | CAGGGCATTTGATGCAGC | 18 | 56 | 50 | |
| FurCC1 | GTGTGTGTAATAATTGTAATCA | 22 | 27 | 46 | |
| FurCC2 | TTGCTAAAGAGCATGGATTTA | 21 | 33 | 47 | |
| FurCC3 | ATTGCTACGGTTTATAG | 17 | 35 | 40 | |
| FurCC4 | AAAGAGCATGGATTTAA | 17 | 29 | 37 | |
| FurCC5 | TGTGTGTAATAATTGTAA | 18 | 22 | 37 | |
| FurCC6 | TTCGGCTGGAAAAAAAT | 17 | 35 | 40 | |
| FurCL1 | AAAGTTGCGGCGATATTGTA | 20 | 40 | 48 | |
| FurCL2 | TGCTTCAGGGAAGAAATTTG | 20 | 40 | 48 | |
| FurCL3 | TTATCGAACAACAACAAATGTTA | 23 | 26 | 46 | |
| FurCL4 | GCACAATGCAGTAAAAAATAAGG | 23 | 35 | 50 | |
| FurCL5 | AACAAATGTTAATCGCAAAAGAATATAA | 28 | 21 | 50 | |
| FurCL6 | ATGCAGTAAAAAATAAGGTATGTTTA | 26 | 23 | 49 | |
| FurCU1 | GATTTGCAAAGTCTGTGGAAAA | 22 | 36 | 49 | |
| FurCU2 | AATGAGCATCATTTTAAACTCAC | 23 | 30 | 48 | |
| FurCU3 | TATGGAATTTGTAGCGATTGCAA | 23 | 35 | 50 | |
| FurCU4 | AGCGATTGCAATCATAAAACAAAG | 24 | 33 | 51 | |
| FurCU5 | ACGAACTTTCAAACAAGCCTCA | 22 | 41 | 51 | |
| FurCU6 | TCAAACAAGCCTCACCACGACC | 22 | 55 | 57 | |
| glyA specific | GlyCJ1 | AGATTGAAACTCTAGCTATTGAA | 23 | 30 | 48 |
| GlyCJ2 | TTATGCGGCTTTGATTAATCCAGGT | 25 | 40 | 54 | |
| GlyCJ3 | TGTACGAAAGTTGTTTTTACGGCGTAG | 27 | 41 | 57 | |
| GlyCJ4 | AAATTGCTAAAAAAGAAAAACCAAAACTT | 29 | 21 | 50 | |
| GlyCJ5 | GAAATTGCTAATGAAATAGGTGCCTAT | 27 | 33 | 54 | |
| GlyCJ6 | TTTACGGCGTAGAACTTG | 18 | 44 | 46 | |
| GlyCC1 | AGATGAAATCGAAAATTTAGCTATAG | 26 | 27 | 50 | |
| GlyCC2 | GTTTATGCTGCACTTTTAAATCCAG | 25 | 36 | 53 | |
| GlyCC3 | AGCTCTACAACCCACAAAACC | 21 | 48 | 52 | |
| GlyCC4 | GCGGTATCATCATGACTAATGA | 22 | 41 | 51 | |
| GlyCC5 | CGGATGAAGTTGGAGCTTAT | 20 | 45 | 50 | |
| GlyCC6 | GATGGAAGGATAAACTATGAA | 21 | 33 | 47 | |
| GlyCL1 | AAGGTGTGTATATGGCATTGTTAAAT | 36 | 31 | 52 | |
| GlyCL2 | TGAAACGATTGCTATAGAAAGA | 22 | 32 | 47 | |
| GlyCL3 | ACACTTGACTCATGGTTCTAAA | 22 | 36 | 49 | |
| GlyCL4 | GATAGCAAAAGAGATTAAACCAAAA | 25 | 28 | 49 | |
| GlyCL5 | ACTTATTGTTTGTGGTGCTAG | 21 | 38 | 49 | |
| GlyCL6 | GAAATAGCAGATGAGGTTGGT | 21 | 43 | 50 | |
| GlyCU1 | CTAAGGTTAGTAGCTCGGGTAA | 22 | 45 | 53 | |
| GlyCU2 | AACTCATTGTATGCGGGGCAA | 21 | 48 | 52 | |
| GlyCU3 | TATGCTAGGATTATTGATTTTGC | 23 | 30 | 48 | |
| GlyCU4 | AGATAGCCGATGAAGTGGGG | 20 | 55 | 54 | |
| GlyCU5 | GTGGCTGTGAGATTGTTGAT | 20 | 45 | 50 | |
| GlyCU6 | TTTCCCTCACGCACACATCG | 20 | 55 | 54 | |
| ceuB-C specific | CeuCJ1 | CTTAATGATTGTAAGCATTATCACTAG | 27 | 30 | 52 |
| CeuCJ2 | GCTAATTATCCCAAATTTAGTAGCTCTTT | 29 | 31 | 54 | |
| CeuCJ3 | AGCTCTTTATCTAGGTGATAATCTTAGAAA | 30 | 30 | 55 | |
| CeuCJ4 | TTGGTGCGGCTAATTTAAGTGTTTATAAAAAC | 32 | 31 | 57 | |
| CeuCJ5 | ATGCTAATTTTAAGCTTTTTAACACTTAACA | 31 | 23 | 52 | |
| CeuCJ6 | ATCCTTTAAATTTAGGCAAAGATTTAGCGA | 30 | 30 | 55 | |
| CeuCC1 | CTTAATAATAGTTTCTATTATCACTAG | 27 | 22 | 49 | |
| CeuCC2 | TTATCATCCCTAATCTTGTTGCCATTT | 22 | 33 | 54 | |
| CeuCC3 | TGCCATTTATCGCGGGGATAATCTTAAGA | 29 | 41 | 55 | |
| CeuCC4 | TAGGTTCTGCAAATTTGAGTGTATATAGAAAT | 32 | 28 | 55 | |
| CeuCC5 | TTATATCGTAACTTTGCTTAGTTTTAT | 27 | 22 | 49 | |
| CeuCC6 | ACCCTTATAATGGCACTTGTTTTTGTATT | 29 | 31 | 54 | |
| fliY specific | FliY1 | TTGATCAATTAGCAAATGATCCT | 23 | 30 | 48 |
| FliY2 | AAAGGCATAACTTTTTTTGGCTCTGC | 26 | 38 | 55 | |
| FliY3 | TGATCCTTTAGAAATTTTAATAGGTG | 26 | 27 | 50 | |
| FliY4 | TAGCTGATTTAGAAGAAAAAATTTC | 25 | 24 | 48 | |
| Type | Name | Sequence | Length (in nucleotides) | G + C (%) | Tma (°C) |
| FliY5 | TAAGATTTTAACCAAAACTGACT | 23 | 26 | 46 | |
| FliY6 | TAAGAGTGCGTATAGGTAGTAAA | 23 | 35 | 50 | |
| cdtABC specific | CdtA1 | CCCCAAATCCAATTTCCTTGTGCTAAAGCCCAAAC | 35 | 46 | 64 |
| CdtA2 | CGATTAAAGTATAGCCCCAAATCCAATTTCCTTGT | 35 | 37 | 61 | |
| CdtB1 | GCATATTTGCAAAATGTGCATCTACAGCTGTGATA | 35 | 37 | 61 | |
| CdtB2 | CCGCTTGCTTGAGTTGCGCTAGTTGGAAAAACCAC | 35 | 51 | 67 | |
| CdtC1 | GGATTTGTAAATTGCACATAACCAAAAGGAAGTTC | 35 | 34 | 60 | |
| CdtC2 | CATTCATCAGATTCCAAAACTAAAGAACGAATTTG | 35 | 31 | 59 | |
| QC | QCprb | TGGCAGAAGCTATGAAACGATATGGG | 27 | 44 | 58 |
| Cy3-QC | CCCATATCGTTTCATAGCTTCTGCCA | 26 | 46 | 58 |
The basic melting temperature (Tm) was calculated with the oligonucleotide properties calculator (http://www.basic.nwu.edu/biotools/oligocalc.html).
Microchip design and fabrication.
To increase confidence in the microarray analysis and to overcome potential problems of genetic variability among and within species, each analyzed gene was represented by six individual oligoprobes (Table 2) from different parts of the amplified region. To facilitate interpretation of microarray data, all oligonucleotides specific for each gene were placed on separate rows of the array.
Microchips were printed by using a contact microspotting robotic system PIXSYS 5500 (Cartesian Technologies, Inc., Ann Arbor, Mich.) equipped with a microspotting pin (CMP7; ArrayIt, Sunnyvale, Calif.). The average size of spots was 250 μm. The spotting solution contained a mixture of specific oligonucleotide probe (80 μM) and quality control (QC) oligonucleotide (8 μM) in 50% dimethyl sulfoxide. Printed slides were dried for at least 20 min at 80°C and treated for 15 min with a freshly prepared 0.25% NaBH4 solution in water. Slides were washed once for 5 min with 0.2% sodium dodecyl sulfate in water and five times for 1 min each time with distilled water to remove unbound oligonucleotides. Marker spots for array positioning on the slide were made by using 1× spotting solution (ArrayIt) in 0.25 M acetic acid.
Hybridization conditions.
Hybridization of the fluorescently labeled ssRNA samples to the microarray was performed in the Micromax hybridization buffer III at 45°C for 30 min. Before hybridization, Cy5-labeled ssRNA sample was mixed with a Cy3-QC probe (Table 2) at molar ratio 10 to 1, followed by denaturing at 95°C for 1 min and chilling to 25°C. Each sample was placed on the microchip and covered with a 5- by 5-mm plastic coverslip to prevent evaporation of the probe during incubation. After hybridization, the slides were washed once for 1 min with 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.2% Tween 20, three times for 1 min with 6× SSC buffer, twice with 2× SSC buffer, and once with 1× SSC buffer and then dried in a stream of air.
Microarray scanning.
The fluorescent images of processed microarrays were generated by using ScanArray 5000 (Perkin-Elmer) equipped with two lasers operating at 632 nm (for excitation of Cy5 dye) and 543 nm (for excitation of Cy3 dye). The fluorescent signals from each spot were measured and compared by using QuantArray software (Perkin-Elmer). Fluorescent signals that differed from the average background at a statistically significant level (P < 0.01) were considered positive.
Sequencing.
In some cases, sequences of the genes from some Campylobacter species were determined experimentally. The PCR-amplified DNA fragments were purified by agarose gel electrophoresis, extracted by using the QIAquick gel extraction kit (Qiagen) according to the manufacturer's protocol, and sequenced by using the ABI Prism 310 genetic analyzer system (PE Applied Biosystems).
Nucleotide sequence accession numbers.
The GenBank accession numbers of the deposited sequences are AF545662 (strain ATCC 35221), AF545663 (strain ATCC 35222), AF545664 (strain ATCC 35223), and AF545665 (strain ATCC 43675).
RESULTS
Microarray-based identification of four thermophilic Campylobacter species by using sequence differences in the fur and glyA genes.
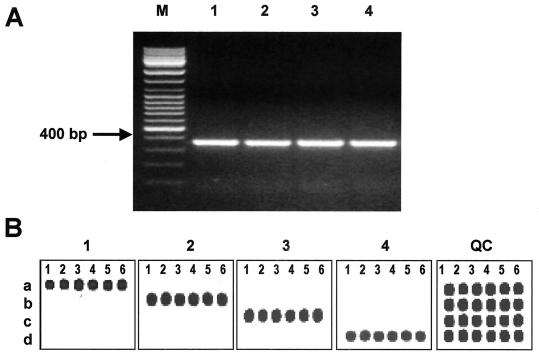
The fur gene sequences from C. jejuni and C. upsaliensis (GenBank AL139075 and L77075) were used to design two primers capable of amplifying any Campylobacter fur allele. Degenerate universal primers, CmpfurF and CmpfurR (Table 1), complementary to the semiconserved regions, were shown to produce a 362- to 370-bp PCR product by using genomic bacterial DNA from isolates of C. jejuni, C. coli, C. lari, and C. upsaliensis as a template (Fig. 1A). However, when DNA from the closely related bacterium A. butzleri, or DNAs from other non-Campylobacter species were used, no PCR products were observed (data not shown). These primers were also tested with all 51 Campylobacter isolates, including C. jejuni (n = 15), C. coli (n = 20), C. lari (n = 11), and C. upsaliensis (n = 5), and a fragment of the expected size was amplified from each.
FIG. 1.
Microarray-based detection of Campylobacter spp. using fur-specific oligoprobes. (A) PCR amplification of fur gene. Genomic DNAs from four reference strains were amplified by using the universal fur gene primers, CmpfurF and CmpfurR (Table 1). The resulting PCR products were separated on a 1.5% agarose gel. Lanes: M, 100-bp DNA Ladder Mix (MBI Fermentas); 1, C. jejuni (ATCC 33560); 2, C. coli (ATCC 43485); 3, C. lari (ATCC 35222); 4, C. upsaliensis (D1673). (B) Microarray-based detection of Campylobacter spp. by using the fur-specific oligoprobes. The fur-derived Cy5-labeled ssRNA transcripts were hybridized to the microchip. Each row of the array contains six individual species-specific probes (Table 2) as follows: a, C. jejuni; B, C. coli; C, C. lari; and D, C. upsaliensis. The image labeled QC is the microarray QC Cy3 image.
Species-specific fur oligoprobes for distinguishing between C. jejuni, C. coli, C. lari, and C. upsaliensis (Table 2) were designed on the basis of comparison of more than 30 different fur gene sequences previously determined in our laboratory. Six individual fur oligoprobes for each Campylobacter species were selected and evaluated in the microarray hybridization with fluorescently labeled ssRNA samples. As shown in Fig. 1B, all species-specific fur gene oligoprobes strongly and specifically hybridized to the sample from their respective Campylobacter species.
Similar results were observed for the glyA gene-based identification. Regions from the glyA genes were amplified by using previously described primers S1 and S2 (1) and a set of our newly designed oligoprobes (Table 2). A 640-bp amplified DNA fragment was detected with all 51 Campylobacter isolates used in the study, and all species-specific glyA gene oligoprobes strongly and specifically hybridized to the glyA-derived RNA transcripts (data not shown).
Discrimination between C. jejuni and C. coli by using regions of the ceuB-C genes and detection of the C. jejuni cdtABC toxins gene cluster.
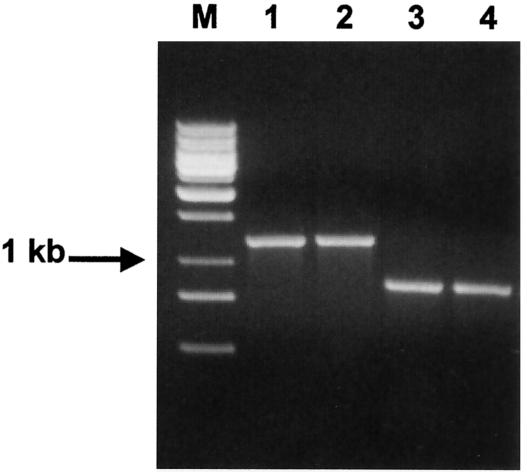
Primers CeuEF and CeuER were designed for PCR amplification of the target region of the ceuB-C genes of C. jejuni and C. coli and were tested with all Campylobacter isolates used. As expected, these primers specifically amplified a 1,229-bp DNA fragment from all C. jejuni and C. coli strains (Fig. 2, lanes 1 and 2). However, an unexpected 866-bp DNA fragment was amplified from C. lari and C. upsaliensis (Fig. 2, lanes 3 and 4). Analysis of the amplicon sequences revealed that although these primers amplified the ceuB-C genes from C. jejuni and C. coli, the DNA amplified from C. lari and C. upsaliensis originated from the putative fliY gene, encoding a protein of the flagellar motor switch complex.
FIG. 2.
PCR amplification of ceuB-C genes. Genomic DNAs from four reference strains were amplified by using the ceuB-C primers (Table 1). The resulting products were separated by using a 1% agarose gel. Lanes: M, 1-kb DNA ladder mix (MBI Fermentas); 1, C. jejuni (ATCC 33560); 2, C. coli (ATCC 43485); 3, C. lari (ATCC 35222); 4, C. upsaliensis (D1673).
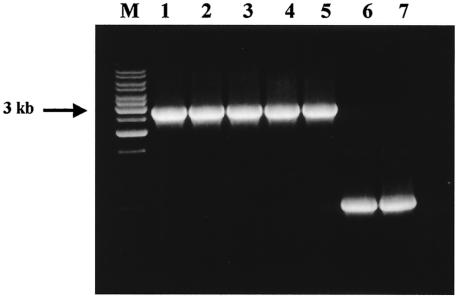
To independently confirm the species identification of C. jejuni, we used the cdtABC gene cluster, since these genes have been found in C. jejuni and C. coli isolates (3, 10). On the basis of the nucleotide sequence of the complete genome of C. jejuni subsp. jejuni NCTC 11168 (GenBank AL139074), we designed the CmpToxF and CmpToxR primers for amplification of the cdtABC gene cluster from C. jejuni (Table 1). The forward and reverse primers include sequences from the flanking lctP and cydA genes, respectively. The oligoprobes for the detection of the C. jejuni cdtABC cluster were designed as described above, and the corresponding sequences are summarized in Table 2. Amplification of a DNA fragment of the predicted size of 2,869 bp (Fig. 3) was observed in all cases when the DNAs from reference ATCC strains and clinical isolates of C. jejuni were used as PCR templates. We also observed amplification of a 700-bp PCR product from all C. coli strains (Fig. 3, lanes 6 and 7). Direct sequencing of this amplicon showed that it resulted from amplification of the lctP-cydA homologous region in the C. coli genome. This amplified C. coli DNA fragment did not hybridize to any of the C. jejuni cdtABC-specific oligoprobes since the lctP-cydA region does not include the cdtABC toxin gene cluster.
FIG. 3.
PCR amplification of the cdtABC gene cluster from the C. jejuni and the lctP-cydA region of C. coli. Genomic DNAs from seven reference strains were amplified by using the cdtABC primers (Table 1). The resulting products were separated by using a 1% agarose gel. Lanes: M, 1-kb DNA ladder mix (MBI Fermentas); 1, C. jejuni (ATCC 33560); 2, C. jejuni (ATCC 35918); 3, C. jejuni (CDC1420); 4, C. jejuni (DENVER-2); 5, C. jejuni (GH18401); 6, C. coli (ATCC 43485); 7, C.coli (ATCC 43473).
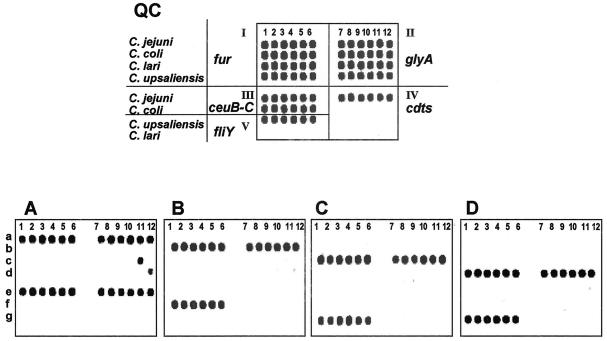
Microarray analysis of four thermophilic Campylobacter spp. by using a composite oligonucleotide microchip.
To create a composite microchip for detection and discrimination of the four thermotolerant Campylobacter spp. by using specific regions of five target genes, we combined all of the oligoprobes described above (Table 2) into a single microarray with five panels (Fig. 4, QC image, panels I to V). All of the Campylobacterisolates listed in Materials and Methods were examined in a four-step procedure: (i) separate amplification of each target gene region, (ii) one-tube synthesis of all ssRNA transcripts from the T7 promoter-tagged PCR amplicons, (iii) fluorescent chemical labeling of ssRNA transcripts, and (iv) hybridization of the RNA probes to the composite microarray.
FIG. 4.
Composite microarray for Campylobacter spp. identification. The QC image shows the layout of the array. The assay was composed of five subarray panels labeled from I to V. Each of four rows (a to d) of the subarray I contains six oligoprobes complementary to species-specific alleles of the fur gene. Subarrays from II to V contain oligoprobes for the glyA, ceuB-C, cdts, and fliY gene alleles, respectively. Microarray hybridization patterns of each of four Campylobacter species—C. jejuni (A), C. coli (B), C. lari (C), and C. upsaliensis (D)—are indicated.
Using the optimized protocol conditions, we observed efficient and specific hybridization of species-specific oligoprobes with the corresponding Campylobacter species (Fig. 4A to D). Some cross-hybridization was observed for the glyA probes of C. jejuni and C. lari (Fig. 4A, spot c-11) and C. upsaliensis (Fig. 4A, spot d-12). However, since the Campylobacter sp. identification relied on the results of hybridization with six independent oligoprobes for each gene, the cross-hybridization of one or two spots did not affect the species identification.
The specificity of the composite microarray assay was evaluated by analyzing the collection of 51 Campylobacter isolates. All of the isolates were unambiguously identified; the results of 16 of these analyses are shown in Fig. 5. The results for C. jejuni and C. coli were confirmed by a PCR-based species detection method based on the hipO and ask genes (6). The results of the PCR assays were concordant with those of the microarray-based identification (data not shown).
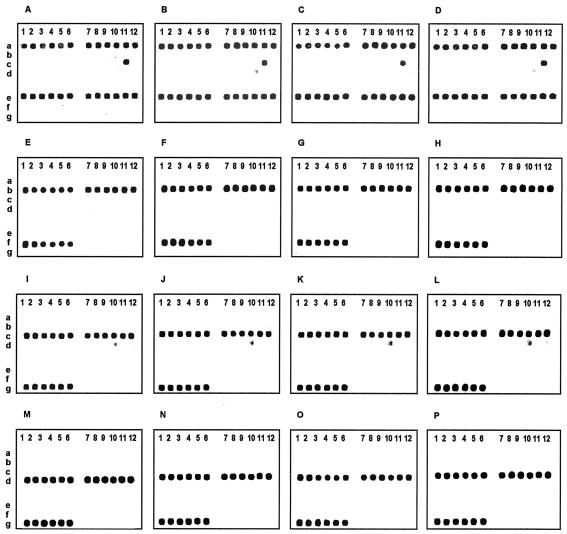
FIG. 5.
Hybridization patterns of sixteen Campylobacter isolates. The composite microarray (Fig. 4) was used for the analysis of 16 Campylobacter isolates: C. jejuni (ATCC 35919, ATCC 29428, ATCC 33560, and DENVER-1) (A to D, respectively); C. coli (ATCC 33559, ATCC 43481, ATCC 43478, and 92B4QA) (E to H, respectively); C. lari (ATCC 35222, ATCC 35221, ATCC 43675, and 3125) (I to L, respectively); and C. upsaliensis (D2237, 5613, 5512, and 5502) (M to P, respectively).
Microarray identification of Campylobacter spp. from mixed samples.
To determine the ability of microarray identification to determine Campylobacter species in mixed bacterial populations, we prepared and analyzed artificial DNA mixtures of different Campylobacter spp. The results showed that the microarray allowed efficient and highly specific detection of each Campylobacter species present in the mixtures (Fig. 6). Moreover, this microarray system allowed unambiguous species identification in the presence of DNA from various non-Campylobacter species (data not shown).
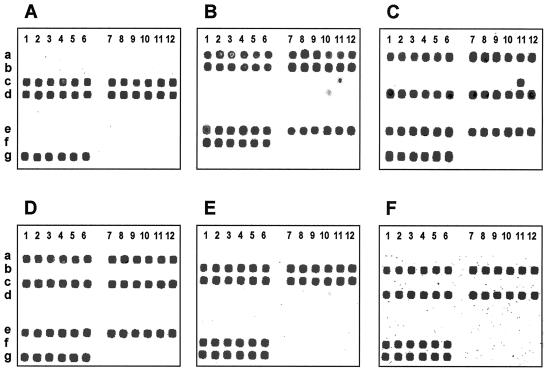
FIG. 6.
Microarray hybridization patterns of bacterial samples containing mixtures of different Campylobacter species are shown. The composite microarray (Fig. 4) was used for six analyses of mixed Campylobacter isolates. Panels A to F show hybridization patterns for mixtures of C. lari and C. upsaliensis (A), C. jejuni and C. coli (B), C. jejuni and C. upsaliensis (C), C. jejuni and C. lari (D), C. coli and C. lari (E), and C. coli and C. upsaliensis (F).
DISCUSSION
We describe here an oligonucleotide microarray assay for rapid detection and identification of four Campylobacter species of clinical relevance (C. jejuni, C. coli, C. lari, and C. upsaliensis). The approach uses the target regions of five genes: fur, glyA, cdtABC cluster, ceuB-C, and fliY. The initial microarray identification of each Campylobacter species is based on analysis of the fur and glyA genes, since these genes are well characterized and are found in all Campylobacter species and some of them have been used in previous PCR-based analysis methods.
Although the fur and glyA genes were used to unambiguously identify the four Campylobacter species, the ceuB-C genes were used to discriminate between C. jejuni and C. coli, the fliY gene to identify both C. lari and C. upsaliensis, and the cdtABC cluster to identify C. jejuni. The use of only one set of primers for simultaneous amplification of alleles of the ceuB-C genes of C. jejuni and C. coli and the fliY gene of C. lari and C. upsaliensis allows us to reduce the number of PCRs required for the analysis. The presence of the cdtABC gene cluster was used to confirm the identification of C. jejuni. Although homologues of these genes are found in some other diarrheagenic bacterial species and some closely related Campylobacter spp. such as C. coli, the oligonucleotide probes on the array were specific to C. jejuni and did not cross-react with other species (Fig. 4).
In our microarray system, we used relatively short oligonucleotides (17 to 35 nucleotides) for two reasons. First, shorter oligoprobe sequences (<25 bp) are often capable of detecting a singe nucleotide mismatch between the template ssRNA and the oligoprobe, thus detecting minor genetic variants in target genes in a bacterial population. Second, the use of multiple oligoprobes allows independent testing of several species-specific regions of each gene. This reduces the probability of misidentification.
We took advantage of the high-density capabilities of the array by analyzing 10 different species on one slide using several sequences per strain, and we performed this analysis simultaneously.
The genetic variability of Campylobacter spp., which has been demonstrated previously (9, 18, 28), may be problematic for PCR methods that rely on species-specific primers to identify the bacterial species. To avoid this problem, we deliberately designed degenerate primers for the PCR amplification and replaced the gel-based characterization of PCR products with a sequence-based hybridization method.
By using six spots representing six different sequences of the same gene, we assured detection despite sequence divergence. In addition we used several genes for analysis. This redundancy of sequences within genes and of genes within species will help to overcome the potential problem of sequence divergence and hybridization specificity. However, the aim of this array was not to distinguish among strains of the same species. Indeed, we deliberately chose conserved sequences found in all strains of a specific species.
Several methods exist for analysis of Campylobacter including: nucleic acid hybridization, biochemical reactions, enzyme-linked immunosorbent assay, the combination of enzyme-linked immunosorbent assay and immunomagnetic separation, enzyme-linked fluorescent assay, and PCRs. The combined PCR and microarray analysis we present here has important advantages over these methods. First, it takes advantage of the sensitivity and simplicity of PCR amplification for analyzing even low levels of bacterial contamination in many different samples, including food products, while overcoming the problems of nonspecific products that are often produced in highly sensitive PCR assays. Second, the microarray method enables simultaneous analysis of multiple genetic characteristics of target organism in one experiment. Unlike other nucleic acid hybridization methods, the glass microarray chips analyze several genes, and several sequences for each gene, simultaneously. Thus, identification is made on the basis of multiple genetic characteristics, which limits the probability of both false-positive and false-negative results. In the experiments reported here, the species determination was made based on 72 parameters (the number of spots), increasing the reliability of the results. Third, this method can be used to carry out many analyses simultaneously. We demonstrated that as few as 10 different Campylobacter strains could be analyzed on one slide. The PCR-microarray assay can also be scaled up through the use of universal primers for amplification, which reduces the number of primers and the number of reactions needed for analysis of several genes from several species. Finally, microarray analysis can be viewed as a spot pattern recognition assay, which can now be carried out automatically by an increasing number of computerized devices. Thus, the data presented here suggest that microarray analysis is a valuable tool for the identification and characterization of bacterial pathogens and other organisms.
Acknowledgments
We thank P. Fields and B. Swaminathan of the National Salmonella and Campylobacter Reference Laboratories, Centers for Disease Control and Prevention, and R. Thunberg and T. Tran of the FDA Center of Food Safety and Applied Nutrition for providing the strains used in the present study. We thank R. Thunberg for technical assistance.
This work was supported in part by USDA grant 0013000 and funding provided by the FDA Office of Science to A.R. and V.C. and by a grant from the U.S. Defense Advanced Research Project Agency to K.C.
REFERENCES
- 1.Al Rashid, S. T., I. Dakuna, H. Louie, D. Ng, P. Vandamme, W. Johnson, and V. L. Chan. 2000. Identification of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis, Arcobacter butzleri, and A. butzleri-like species based on the glyA gene. J. Clin. Microbiol. 38:1488-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allos, B. M. 2001. Campylobacter jejuni Infections: update on emerging issues and trends. Clin. Infect. Dis. 32:1201-1206. [DOI] [PubMed] [Google Scholar]
- 3.Bang, D. D., F. Scheutz, P. Ahrens, K. Pedersen, J. Blom, and M. Madsen. 2001. Prevalence of cytolethal distending toxin (cdt) genes and CDT production in Campylobacter spp. isolated from Danish broilers. J. Med. Microbiol. 50:1087-1094. [DOI] [PubMed] [Google Scholar]
- 4.Chizhikov, V., A. Rasooly, K. Chumakov, and D. D. Levy. 2001. Microarray analysis of microbial virulence factors. Appl. Environ. Microbiol. 67:3258-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chizhikov, V., M. Wagner, A. Ivshina, Y. Hoshino, A. Z. Kapikian, and K. Chumakov. 2002. Detection and genotyping of human group A rotaviruses by oligonucleotide microarray hybridization. J. Clin. Microbiol. 40:2398-2407. [DOI] [PMC free article] [PubMed]
- 6.Chuma, T., S. Hashimoto, and K. Okamoto. 2000. Detection of thermophilic Campylobacter from sparrows by multiplex PCR: the role of sparrows as a source of contamination of broilers with Campylobacter. J. Vet. Med. Sci. 62:1291-1295. [DOI] [PubMed] [Google Scholar]
- 7.Chuma, T., K. Yano, H. Omori, K. Okamoto, and H. Yugi. 1997. Direct detection of Campylobacter jejuni in chicken cecal contents by PCR. J. Vet. Med. Sci. 59:85-87. [DOI] [PubMed] [Google Scholar]
- 8.Cloak, O. M., and P. M. Fratamico. 2002. A multiplex polymerase chain reaction for the differentiation of Campylobacter jejuni and Campylobacter coli from a swine processing facility and characterization of isolates by pulsed-field gel electrophoresis and antibiotic resistance profiles. J. Food Prot. 65:266-273. [DOI] [PubMed] [Google Scholar]
- 9.Dickins, M. A., S. Franklin, R. Stefanova, G. E. Schutze, K. D. Eisenach, I. Wesley, and M. D. Cave. 2002. Diversity of Campylobacter isolates from retail poultry carcasses and from humans as demonstrated by pulsed-field gel electrophoresis. J. Food Prot. 65:957-962. [DOI] [PubMed] [Google Scholar]
- 10.Eyigor, A., K. A. Dawson, B. E. Langlois, and C. L. Pickett. 1999. Cytolethal distending toxin genes in Campylobacter jejuni and Campylobacter coli isolates: detection and analysis by PCR. J. Clin. Microbiol. 37:1646-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fermer, C., and E. O. Engvall. 1999. Specific PCR identification and differentiation of the thermophilic campylobacters, Campylobacter jejuni, C. coli, C. lari, and C. upsaliensis. J. Clin. Microbiol. 37:3370-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez, I., T. Garcia, A. Antolin, P. E. Hernandez, and R. Martin. 2000. Development of a combined PCR-culture technique for the rapid detection of Arcobacter spp. in chicken meat. Lett. Appl. Microbiol. 30:207-212. [DOI] [PubMed] [Google Scholar]
- 13.Heringa, J. 1999. Two strategies for sequence comparison: profile-preprocessed and secondary structure-induced multiple alignment. Comput. Chem. 23:341-364. [DOI] [PubMed] [Google Scholar]
- 14.Houng, H. S., O. Sethabutr, W. Nirdnoy, D. E. Katz, and L. W. Pang. 2001. Development of a ceuE-based multiplex polymerase chain reaction (PCR) assay for direct detection and differentiation of Campylobacter jejuni and Campylobacter coli in Thailand. Diagn. Microbiol. Infect. Dis. 40:11-19. [DOI] [PubMed] [Google Scholar]
- 15.Kirk, R., and M. T. Rowe. 1994. A PCR assay for the detection of Campylobacter jejuni and Campylobacter coli in water. Lett. Appl. Microbiol. 19:301-303. [DOI] [PubMed] [Google Scholar]
- 16.Lawson, A. J., M. S. Shafi, K. Pathak, and J. Stanley. 1998. Detection of campylobacter in gastroenteritis: comparison of direct PCR assay of faecal samples with selective culture. Epidemiol. Infect. 121:547-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linton, D., R. J. Owen, and J. Stanley. 1996. Rapid identification by PCR of the genus Campylobacter and of five Campylobacter species enteropathogenic for man and animals. Res. Microbiol. 147:707-718. [DOI] [PubMed] [Google Scholar]
- 18.Matsuda, M., M. Tsukada, M. Fukuyama, Y. Kato, Y. Ishida, M. Honda, and C. Kaneuchi. 1995. Detection of genomic variability among isolates of Campylobacter jejuni from chickens by crossed-field gel electrophoresis. Cytobios 82:73-79. [PubMed] [Google Scholar]
- 19.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metherell, L. A., J. M. Logan, and J. Stanley. 1999. PCR-enzyme-linked immunosorbent assay for detection and identification of Campylobacter species: application to isolates and stool samples. J. Clin. Microbiol. 37:433-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moreno, Y., M. Hernandez, M. A. Ferrus, J. L. Alonso, S. Botella, R. Montes, and J. Hernandez. 2001. Direct detection of thermotolerant campylobacters in chicken products by PCR and in situ hybridization. Res. Microbiol. 152:577-582. [DOI] [PubMed] [Google Scholar]
- 22.Nesbit, E. G., P. Gibbs, D. W. Dreesen, and M. D. Lee. 2001. Epidemiologic features of Campylobacter jejuni isolated from poultry broiler houses and surrounding environments as determined by use of molecular strain typing. Am. J. Vet. Res. 62:190-194. [DOI] [PubMed] [Google Scholar]
- 23.Okada, M., F. Hayashi, and N. Nagasaka. 2001. PCR detection of 5 putative periodontal pathogens in dental plaque samples from children 2 to 12 years of age. J. Clin. Periodontol. 28:576-582. [DOI] [PubMed] [Google Scholar]
- 24.Opfer, C., J. Kleer, and G. Hildebrandt. 2001. Comparison of the two different PCR assays for the detection of thermotolerant Campylobacter in poultry. Berl. Munch. Tierarztl. Wochenschr. 114:470-472. (In German.) [PubMed]
- 25.O'Sullivan, N. A., R. Fallon, C. Carroll, T. Smith, and M. Maher. 2000. Detection and differentiation of Campylobacter jejuni and Campylobacter coli in broiler chicken samples using a PCR/DNA probe membrane based colorimetric detection assay. Mol. Cell Probes 14:7-16. [DOI] [PubMed] [Google Scholar]
- 26.Oyofo, B. A., S. M. Abd el Salam, A. M. Churilla, and M. O. Wasfy. 1997. Rapid and sensitive detection of Campylobacter spp. from chicken using the polymerase chain reaction. Zentbl. Bakteriol. 285:480-485. [DOI] [PubMed] [Google Scholar]
- 27.Oyofo, B. A., S. A. Thornton, D. H. Burr, T. J. Trust, O. R. Pavlovskis, and P. Guerry. 1992. Specific detection of Campylobacter jejuni and Campylobacter coli by using polymerase chain reaction. J. Clin. Microbiol. 30:2613-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ragimbeau, C., G. Salvat, P. Colin, and G. Ermel. 1998. Development of a multiplex PCR gene fingerprinting method using gyrA and pflA polymorphisms to identify genotypic relatedness within Campylobacter jejuni species. J. Appl. Microbiol. 85:829-838. [DOI] [PubMed] [Google Scholar]
- 29.Rautelin, H., J. Jusufovic, and M. L. Hanninen. 1999. Identification of hippurate-negative thermophilic campylobacters. Diagn. Microbiol. Infect. Dis. 35:9-12. [DOI] [PubMed] [Google Scholar]
- 30.Sails, A. D., F. J. Bolton, A. J. Fox, D. R. Wareing, and D. L. Greenway. 2002. Detection of Campylobacter jejuni and Campylobacter coli in environmental waters by PCR enzyme-linked immunosorbent assay. Appl. Environ. Microbiol. 68:1319-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sails, A. D., A. J. Fox, F. J. Bolton, D. R. Wareing, D. L. Greenway, and R. Borrow. 2001. Development of a PCR ELISA assay for the identification of Campylobacter jejuni and Campylobacter coli. Mol. Cell Probes 15:291-300. [DOI] [PubMed] [Google Scholar]
- 32.Thunberg, R. L., T. T. Tran, and M. O. Walderhaug. 2000. Detection of thermophilic Campylobacter spp. in blood-free enriched samples of inoculated foods by the polymerase chain reaction. J. Food Prot. 63:299-303. [DOI] [PubMed] [Google Scholar]
- 33.Totten, P. A., C. M. Patton, F. C. Tenover, T. J. Barrett, W. E. Stamm, A. G. Steigerwalt, J. Y. Lin, K. K. Holmes, and D. J. Brenner. 1987. Prevalence and characterization of hippurate-negative Campylobacter jejuni in King County, Washington. J. Clin. Microbiol. 25:1747-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Doorn, L. J., B. A. Giesendorf, R. Bax, B. A. van der Zeijst, P. Vandamme, and W. G. Quint. 1997. Molecular discrimination between Campylobacter jejuni, Campylobacter coli, Campylobacter lari, and Campylobacter upsaliensis by polymerase chain reaction based on a novel putative GTPase gene. Mol. Cell Probes 11:177-185. [DOI] [PubMed] [Google Scholar]
- 35.Vanniasinkam, T., J. A. Lanser, and M. D. Barton. 1999. PCR for the detection of Campylobacter spp. in clinical specimens. Lett. Appl. Microbiol. 28:52-56. [DOI] [PubMed] [Google Scholar]
- 36.Volokhov, D., A. Rasooly, K. Chumakov, and V. Chizhikov. 2002. Identification of listeria species by microarray-based assay. J. Clin. Microbiol. 40:4720-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waage, A. S., T. Vardund, V. Lund, and G. Kapperud. 1999. Detection of small numbers of Campylobacter jejuni and Campylobacter coli cells in environmental water, sewage, and food samples by a seminested PCR assay. Appl. Environ. Microbiol. 65:1636-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waller, D. F., and S. A. Ogata. 2000. Quantitative immunocapture PCR assay for detection of Campylobacter jejuni in foods. Appl. Environ. Microbiol. 66:4115-4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winters, D. K., and M. F. Slavik. 2000. Multiplex PCR detection of Campylobacter jejuni and Arcobacter butzleri in food products. Mol. Cell Probes 14:95-99. [DOI] [PubMed] [Google Scholar]