Abstract
Three types of cytolethal distending toxin (CDT), namely, CDT-I, CDT-II, and CDT-III, have been described in Escherichia coli. Using primers designed for the detection of sequences common to the cdtB genes, we analyzed by PCR a set of 21 CDT-producing E. coli strains of intestinal and extraintestinal origins isolated from human and different animal species in several European countries and in the United States. On the basis of the existing differences in the cdtB genes, cdt-I-, cdt-II-, and cdt-III-specific primer pairs were designed and used for cdt typing. These new primers successfully differentiated all of the previously described cdt genes. Six strains proved to be cdt-I; eight strains proved to be cdt-III. However, none of the type I-, II-, and III-specific primers generated amplicons from six CDT+ strains, suggesting the existence of a new cdt variant. Sequence analysis of the amplicons from two untypeable genes confirmed the existence of a new cdt variant that we called cdt-IV. Using the new specific primers, cdt-IV was detected in human, porcine, and poultry strains of intestinal and extraintestinal origins. To validate all sets of cdt specific primers, a group of 353 human E. coli strains isolated in Hungary was then investigated for the presence of cdt genes. This included 190 strains isolated from patients with urinary tract infections (UTI), 51 strains isolated from other (nonurinary) extraintestinal infections, and 112 intestinal strains isolated from healthy individuals. Of 190 UTI strains, 15 (7.9%) had cdt genes. Of 51 non-UTI extraintestinal strains 3 (5.9%) contained the cdt gene, and 1 (0.9%) of 112 healthy intestinal strains was PCR positive. Five strains proved to be cdt-I, and fourteen strains proved to be cdt-IV. The CDT-producing extraintestinal strains belonged to a wide variety of serogroups, including O2, O6, O75, and O170. In conclusion, we have developed a new PCR typing system for CDT able to detect a new CDT variant present in pathogenic E. coli strains obtained from animals and humans.
Cytolethal distending toxins (CDT) represent an emerging and unique toxin family (12, 33). CDT was first described in Escherichia coli by Johnson and Lior (22). The culture filtrates of CDT-producing E. coli strains induced characteristic morphological changes, and the distended cells died in 3 to 4 days (23). CDT production was reported from various serotypes of enteropathogenic E. coli (EPEC) (2, 6, 17) and from several bacterial species of medical importance (10, 26, 32, 37). The CDTs induce the formation of giant mononucleated cells of sensitive eukaryotic cell lines and block the cell cycle in G2 phase (3, 30). The block is due to the maintenance of mitosis promoting factor in an inactive form (9, 30).
Molecular genetic studies of CDT-producing E. coli strains were conducted in three different laboratories. Scott and Kaper (34) used an EPEC strain of O86:H34 serotype (15). Pickett et al. (31) used an EPEC O128:H− serotype strain (23), and Pérès et al. (30) used a cytotoxic necrotizing factor type 2 (CNF2)-producing E. coli strain of serogroup O78 isolated from a septicemic calf (28). In all of these cases, it was established that CDT was encoded by three adjacent or slightly overlapping genes, cdtA, cdtB, and cdtC, all of which were required for toxin production (30, 31, 34). The cdtABC genes in EPEC strains were located on the chromosome (31, 34), and in the O78 calf strain the cdtABC genes were located on a transferable large virulence plasmid called pVir that also coded for CNF2 toxin and F17 fimbrial adhesin (30). The function of the proteins encoded by the cdt genes is not fully elucidated, but recent studies have shown that CdtB protein contains an enzymatic motif with homology to several mammalian and bacterial phosphodiesterares, such as the human DNase I and the sphingomyelinase from Bacillus cereus (12, 15, 24). The cdt operon of the O86:H34 EPEC strain E6468/62 was referred to as cdt type I. The operon of the EPEC O128:H− strains 91422-88 and the operon of the O78 calf strain 1404 were referred to as cdt types II and III, respectively (33).
In molecular epidemiological studies, different DNA probes were used for detecting cdt genes in E. coli. By colony hybridization, cdt genes were detected in 3.1% of E. coli strains isolated from Bangladeshi children with diarrhea, while 0.9% of the control strains had cdt genes (2). By using cdtI- and cdtII-specific DNA probes, cdt genes were detected in 1.6% of diarrheagenic E. coli strains isolated in Nigeria (25). It can be assumed that more cdt-bearing strains could have been detected if a PCR system detecting all of the known types of CDT had been available. However, at the beginning of the present study we did not have such a system.
In the present study we designed PCR primers suitable for detecting any type of sequenced cdt genes in E. coli. An additional goal was to use type-specific primers suitable for the detection and identification of known cdt types I, II, and III. In addition, the present study leads to the identification of a novel type (IV) of cdt gene.
MATERIALS AND METHODS
Bacterial strains.
The origins of CDT-producing E. coli strains used in the present study are given in Table 1. The CDT-I-producing prototype strain E6468/62 (O86:H34) was kindly provided by J. B. Kaper, and strain HB101(p2123) containing the cloned cdt-II operon was kindly provided by C. Pickett. A further 353 E. coli strains were isolated and identified by standard bacteriological procedures in three different Hungarian hospitals and in two public health laboratories between 1998 and 2001. A total of 190 E. coli strains were isolated from urine specimens of patients with urinary tract infections (UTI), 51 strains were isolated from patients with different extraintestinal infections, and 112 E. coli strains were isolated from stool samples of healthy individuals.
TABLE 1.
E. coli strains used in this study
| Strain | Serogroup and/or serotype | CDT producera | Origin | Country | Source or reference |
|---|---|---|---|---|---|
| H173 | O2 | + | Human, UTI | Hungary | This study |
| H52 | O2 | + | Human, UTI | Hungary | This study |
| 34 | O2 | + | Human, septicemia | Northern Ireland | This study |
| 66LS | O2 | + | Human, septicemia | Northern Ireland | This study |
| 33KH89 | O2 | + | Bovine, septicemia | Belgium | This study |
| AII-40 | O6 | + | Porcine, diarrhea | Austria | 38 |
| S5 | O15:K?:H21 | + | Lamb septicemia | England | 35 |
| B20a | O15 | + | Bovine, diarrhea | Spain | This study |
| 67LS | O18 | + | Human, septicemia | Northern Ireland | This study |
| KS-159 | O23 | + | Food, poultry origin | Hungary | This study |
| H78 | O75 | + | Human, UTI | Hungary | This study |
| 1404 | O78 | + | Bovine, septicemia | France | 28 |
| B26a | O78:K80 | + | Bovine, diarrhea | Spain | This study |
| E6468/62 | O86:H34 | + | Human, EPEC | United States | 34 |
| OS (Outbreak Strain) | O86:K61 | + | Finch, EPEC | Scotland | 16 |
| BM2-10 | O88 | + | Bovine, diarrhea | France | This study |
| E253 | O115 | + | Poultry, septicemia | Hungary | This study |
| 89-201-2/3 | O123 | + | Bovine, diarrhea | France | This study |
| 8 | O123 | + | Human, diarrhea | Northern Ireland | This study |
| B28b | O123:K− | + | Bovine, diarrhea | Spain | This study |
| 28C | O75:K95 | + | Porcine septicemia | Spain | 13 |
| E2348/69 | O127 | − | Human, EPEC | England | 14 |
| HB101(p2123) | K-12 | + | Cloned cdt-II operon | United States | 31 |
| J96 | O4:K6 | − | Human, UTI | Germany | 5 |
| DH5α | K-12 | − | Laboratory strain | 36 |
+, producer; −, nonproducer.
Phenotypic tests.
O-typing was carried out as described by Orskov and Orskov (27). Hemolysin production was examined on Luria-Bertani agar containing 5% sheep red blood cells.
Sequence analysis.
DNA sequences cdt operons sequenced thus far were retrieved from GenBank and included human EPEC strains E6468/62 and 9142-88 and the bovine septicemic strain 1404. Comparison of DNA sequences was made by using the database at the National Center for Biotechnology Information (National Institutes of Health, Bethesda, Md.) with the basic local alignment search tool (BLAST) search algorithm and GCG alignment software and with CLUSTAL W multiple sequence alignment software.
Primers.
On the basis of previously published sequences, two pairs of general PCR primers were designed for the consensus region of all cdtB genes. Furthermore, cdt type-specific primers were designed on the basis of unique cdtB sequences of the different types of cdt genes. All primers used are given in Table 2.
TABLE 2.
Primers used in this study
| PCR | Primer | Sequence | Orientationa | GenBank accession no. | Position | PCR product size (bp) |
|---|---|---|---|---|---|---|
| Multiplex cdt | CDT-s1 | 5′-GAAAGTAAATGGAATATAAATGTCCG | F | U04208 | 1292-1317 | 466 |
| U89305 | 1966-1991 | |||||
| CDT-s2 | 5′-GAAAATAAATGGAACACACATGTCCG | F | U03293 | 1019-1044 | ||
| CDT-as1 | 5′-AAATCACCAAGAATCATCCAGTTA | R | U04208 | 1735-1758 | ||
| U89305 | 2409-2432 | |||||
| CDT-as2 | 5′-AAATCTCCTGCAATCATCCAGTTA | R | U03293 | 1462-1485 | ||
| Multiplex cnf | CNF-s | 5′-TTATATAGTCGTCAAGATGGA | F | U42629 | 1638-1658 | 633 |
| X70670 | 1636-1656 | |||||
| U01097 | 892-912 | |||||
| CNF-as | 5′-CACTAAGCTTTACAATATTGA | R | U42629 | 2251-2271 | ||
| X70670 | 2249-2269 | |||||
| U01097 | 1505-1525 | |||||
| cdt-I | CDT-Is | 5′-CAATAGTCGCCCACAGGA | F | U03293 | 1186-1203 | 411 |
| CDT-IIas | 5′-ATAATCAAGAACACCACCAC | R | U03293 | 1587-1597 | ||
| cdt-II | CDT-IIs | 5′-GAAAGTAAATGGAATATAAATGTCCG | F | U04208 | 1291-1317 | 556 |
| CDT-IIas | 5′-TTTGTGTTGCCGCCGCTGGTGAAA | R | U04208 | 1824-1847 | ||
| cdt-III | CDT-IIIs | 5′-GAAAGTAAATGGAATATAAATGTCCG | F | U89305 | 1966-1991 | 555 |
| CDT-IIIas | 5′-TTTGTGTCGGTGCAGCAGGGAAAA | R | U89305 | 2498-2521 | ||
| cdt-IV | CDT-IVs | 5′-CCTGATGGTTCAGGAGGCTGGTTC | F | U03293 | 1078-1095 | 350 |
| CDT-IVas | 5′-TTGCTCCAGAATCTATACCT | R | AY162217 | 340-360 | ||
| cnf-1 | CNF-1s | 5′-GGGGGAAGTACAGAAGAATTA | F | U42629 | 2700-2720 | 1111 |
| X70670 | 2698-2719 | |||||
| CNF-1as | 5′-TTGCCGTCCACTCTCACCAGT | R | U42629 | 3791-3811 | ||
| X70670 | 3789-3809 | |||||
| cnf-2 | CNF-2s | 5′-TATCATACGGCAGGAGGAAGCACC | F | U01097 | 1942-1965 | 1240 |
| CNF-2as | 5′-GTCACAATAGACAATAATTTTCCG | R | U01097 | 3159-3182 |
F, forward; R, reverse.
PCR analysis.
All PCRs contained a 0.2 mM mix of each deoxynucleoside triphosphate, 1× Taq DNA polymerase buffer, 0.25 μM concentrations of each primer, and 2.5 U of Taq polymerase. The amplification protocol was as follows: denaturation at 94°C for 5 min, followed by 30 cycles of denaturation (94°C, 1 min), annealing (55°C, 1 min), and extension (72°C, 1 min). A final extension was done at 72°C for 10 min. When the amplicons were produced for sequencing, Pfu DNA polymerase (Promega) was used, and sequence analysis was done by Genome Express, Grenoble, France. The presence of the eae genes was also determined by PCR as described previously (7, 29).
Detection of cytotoxic activity of bacterial lysates and supernatants.
Experiments and the preparation of bacterial lysates were conducted as described previously (11, 30). Briefly, E. coli strains were grown at 37°C in Tryptic soy broth medium with vigorous (200 rpm) shaking for 2 days. Supernatants of bacterial cultures were saved, and bacterial cells were sonicated. Supernatants and sonic lysates were sterile filtered separately by using 0.22-μm-pore-size filters. The amounts of proteins in each extract and supernatant was determined by using a protein assay kit (Bio-Rad). Nonconfluent HeLa cell monolayers were infected in 96-well plates with the culture supernatants and sonic lysates used in twofold dilution, diluted in tissue culture medium. Plates were incubated at 37°C in 5% CO2 atmosphere for 4 days. After 4 days of interaction, the infecting materials were removed by several washings of the HeLa cell monolayers, morphological changes characteristic to CDT were determined, and the cytotoxic activity (expressed as the highest twofold dilution yielding 50% transformed cells after 96 h of incubation [CD50]) values were calculated as the means of the highest twofold dilution of toxic material yielding 50% transformed HeLa cells after 96 h of coincubation. As a negative control, DH5α (36) and the cnf cdt mutant enteropathogenic prototype strain E2348/69 (19) was used. In addition, a CNF1-producing uropathogenic strain (J96 of serotype O4:K6) was used (5). All experiments were conducted in triplicate.
Nucleotide sequence accesion number.
The newly determined cdt-IV sequence reported in the present study has been deposited in the GenBank database under accesion number AY162217.
RESULTS
Detection of cdt genes by PCR with cdt-specific universal primers.
Two pairs of cdtB-specific primers were designed on the basis of available sequences in GenBank (Table 2). A cdt multiplex PCR system containing these primers was used to detect E. coli strains producing CDT in a set of 25 E. coli strains isolated from humans and different animals. Among these strains, 21 have been tested as CDT producer-positive by using the conventional cytotoxic assay (Table 1). The two pairs of cdtB-specific primers generated a product with the expected size (466 bp) of DNA from all of the wild-type strains producing CDT and from the recombinant strains containing the cloned cdt-II operon in a multicopy vector plasmid. On the other hand, no product was detected with templates obtained from strains that did not produce CDT: the uropathogenic E. coli (UPEC) strain J96, the EPEC strain E2348/69, and the laboratory strain DH5α. Further, another set of 50 wild-type E. coli strains that tested as CDT negative by the conventional cytotoxic assay turned out to also be PCR negative (data not shown).
Typing and sequence analysis of the cdt genes by PCR with specific primers.
On the basis of existing sequence differences in the cdtB genes of CDT-I, CDT-II, and CDT-III, type-specific primers were designed and used for typing the cdt genes (Table 2). Seven strains proved to be cdt type I, and eight strains proved to be cdt type III. Our collection did not contain any CDT-II-producing wild-type strains, but the cdt-II operon containing the HB101(p2123) recombinant strain reacted specifically with the type II-specific primers. None of these primers generated amplicons from six CDT+ strains, suggesting the existence of a new cdt variant. To confirm this hypothesis, all of the amplicons were sequenced. The seven cdt-I-specific amplicons had identical sequences. The eight type III-specific sequences were also identical to each other. The sequences of the six untypeable amplicons were identical to each other but were different from the sequences of types I, II, and III, indicating the existence of a new variant of cdt, referred to as type IV, for which E. coli strain 28C was selected as the prototype strain. The alignment of the internal sequences of cdt types I, II, III, and IV is given in Fig. 1. Sequence comparisons revealed that cdt-IV is closely related to cdt-I, with a homology of 84%. The cdt-IV sequence is less related to cdt-II and cdt-III (Table 3). This result suggests the presence of two CDT families represented by types II and III and types I and IV, respectively.
FIG. 1.
CLUSTAL W-formatted alignments of cdtB type I, II, III, and IV genes. The positions of cdt-IV-specific primers are indicated by arrows starting at positions 34 and 359, respectively.
TABLE 3.
Percentage of nucleotide identities in the cdt internal sequences
| cdt type | % Nucleotide identity with cdt type:
|
|||
|---|---|---|---|---|
| I | II | III | IV | |
| I | 100 | 55 | 54 | 84 |
| II | 100 | 89 | 55 | |
| III | 100 | 54 | ||
| IV | 100 | |||
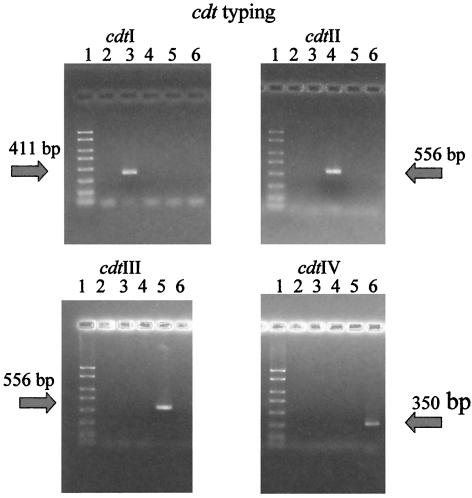
On the basis of the new sequence, type IV-specific primers were designed. By using these primers, a 326-bp product was amplified from all six previously cdt untypeable strains. These six strains were referred to as cdt type IV strains (Table 4). PCR based typing is shown in Fig. 2.
TABLE 4.
Genotypes and phenotypes of E. coli strainsa
| Strains | Sero- group | Genotype
|
Phenotype
|
|||||
|---|---|---|---|---|---|---|---|---|
| cdt | cdt type | cnf | eae | Hly | Lysate | Super- natant | ||
| E6468/62 | O86 | + | I | − | + | − | +++ | +++ |
| 34 | O2 | + | I | 1 | − | − | +++ | +++ |
| 66LS | O2 | + | I | − | − | − | +++ | +++ |
| H52 | O2 | + | I | 1 | − | + | +++ | ++ |
| 67LS | O18 | + | I | − | − | − | +++ | +++ |
| KS159 | O23 | + | I | − | − | − | ++ | ++ |
| OS (outbreak strain) | O86:K61 | + | I | − | + | − | +++ | +++ |
| HB101(p21213) | K-12 | + | II | − | − | − | ND | ND |
| 1404 | O78 | + | III | 2 | − | − | ++ | − |
| 33KH89 | O2 | + | III | 2 | − | + | ++ | ++ |
| B20a | O15 | + | III | 2 | − | − | ++ | − |
| B26a | O78 | + | III | 2 | − | − | ++ | − |
| BM2-10 | O88 | + | III | 2 | − | − | + | − |
| 89-201-2/3 | O123 | + | III | 2 | − | − | + | − |
| B28b | O123 | + | III | 2 | − | − | + | − |
| S5 | O15 | + | III | 2 | − | − | ++ | − |
| 28c | O75 | + | IV | 1 | − | + | ++ | − |
| H173 | O2 | + | IV | − | − | − | + | − |
| AII-40 | O6 | + | IV | 1 | − | + | ++ | − |
| H78 | O75 | + | IV | 1 | − | + | ++ | − |
| E253 | O115 | + | IV | − | − | − | ++ | − |
| 8 | O123 | + | IV | − | − | − | + | − |
| J96 | O4 | − | − | 1 | − | + | * | − |
| DH5α | K-12 | − | − | − | − | − | − | − |
| E2348/49 | O127 | − | − | − | − | − | − | − |
The cytotoxic activity in the sonic lysate and the culture supernatant was titrated as previously described (30) by using as the endpoint the CD50 value. The amounts of proteins in each extract were determined by using a bicinchoninic acid assay (Pierce), and the cytotoxic activity was expressed as the number of CD50 per milligram of protein (+++, ≥1,000; ++, 1,000 to 100; +, <100; −, no specific CDT or CNF effect). If a strain produced CNF and CDT, both cytopathic effects were observed. At the endpoint dilution the transformed HeLa cells were enlarged and polynucleated (CNF effect), and at lower dilutions only the CDT-specific enlargement effect was observed (large mononucleated cells). ND, not done; *, CNF cytopathic effect only.
FIG. 2.
cdt gene typing. In each panel the cdt type specific amplicons were loaded in the same order. Lanes: 1, PCR marker (Sigma) containing eight defined double-stranded DNA markers from 50 to 2,000 bp; 2, E. coli DH5α; 3, KS159 (cdt-I); 4, HB101(p2123) (cdt-II); 5, 1404 (cdt-III); 6, AII-40 (cdt-IV).
CDT effect of E. coli strains.
Nonconfluent HeLa monolayers were incubated with the supernatants and the sonic lysates of the study strains, and the CD50 values were determined as described in Materials and Methods. All of the 21 CDT-producing strains had cell-associated toxicity, and 8 of them secreted the CDT to a high titer. All but one of them was of the CDT-I type (Table 4). Sonicates and culture supernatants of CDT-I strains showed equally strong toxicity. The sonicates of CDT-III- and CDT-IV-producing strains were less toxic than those of CDT-I-producing strains, and the supernatants of CDT-III and CDT-IV strains were negative, except for one CDT-III strain (Table 4). Eight CDT-producing strains also had the cnf-2 gene, and five had cnf-1. Four strains were hemolytic, and two had the eae gene. Among the CDT- and CNF-producing strains a common tendency was observed. At lower dilutions the morphological transformation typical for CDT could be observed: almost all of the HeLa cells became enlarged and mononucleated. However, at higher dilutions, the CNF-specific polynucleation appeared as well and almost all of the transformed HeLa cells appeared as large polynucleated cells. The cytotoxic activity ranged between 100 and 12,500 CD50/mg of protein. The results of these experiments are shown in Table 4.
Validation of all of the sets of PCR primers.
To validate all sets of the PCR primers, a collection of human wild-type pathogenic extraintestinal and nonpathogenic intestinal strains was tested. Altogether, 353 E. coli strains isolated from humans were examined: 190 strains were isolated from individuals with UTI, 51 strains originated from different extraintestinal infections, and 112 strains were isolated from feces of healthy individuals. All of these strains were tested for both CDT activity on HeLa cells by using the conventional cytotoxic assay and for the presence of the cdtB gene by PCR. The testing of 353 E. coli strains by our PCR protocol (19 CDT-positive strains and 334 CDT-negative strains in HeLa cell culture assay) yielded no false-negative and no false-positive results. The presence of cdt genes was further verified by colony dot blot hybridization (data not shown) (4, 36). As high as 7.9% (15 of 190) of the human UPEC strains and 5.9% (3 of 51) of the other extraintestinal strains possessed cdt genes, and only one (0.9%) strain from a healthy control had a cdt gene. Twelve UPEC strains and two other extraintestinal strains had the cdt-IV gene alone, and three UPEC strains and one extraintestinal strain had the cdt-I gene alone. The cdt-bearing strain isolated from the healthy individual carried the cdt-I gene. No strain had combinations of different types of cdt genes. The CDT-producing strains belonged to a wide variety of serogroups, including O2, O6, O75, O170, and O rough, and most of them also had the cnf-1 gene and were hemolytic (Table 5).
TABLE 5.
Characteristics of cdt-bearing E. coli human strains isolated in Hungarya
| Strain | Source | City | O group | Genotype
|
||
|---|---|---|---|---|---|---|
| cdt | cnf | Hly | ||||
| H52 | Urine | Debrecen | O2 | I | 1 | + |
| H149 | Urine | Budapest | O rough | I | 1 | − |
| H154 | Urine | Budapest | O rough | I | 1 | − |
| H260 | Bile | Budapest | ND | I | − | − |
| H173 | Urine | Budapest | O2 | IV | − | − |
| H4 | Urine | Debrecen | O6 | IV | 1 | + |
| H29 | Urine | Debrecen | O6 | IV | 1 | + |
| H161 | Urine | Budapest | O6 | IV | 1 | + |
| H58 | Urine | Debrecen | O75 | IV | 1 | + |
| H61 | Urine | Debrecen | O75 | IV | 1 | + |
| H62 | Urine | Debrecen | O75 | IV | 1 | + |
| H78 | Urine | Debrecen | O75 | IV | 1 | + |
| H83 | Urine | Debrecen | O75 | IV | 1 | + |
| H104 | Urine | Debrecen | O rough | IV | 1 | + |
| H155 | Urine | Budapest | O rough | IV | − | − |
| H193 | Urine | Pécs | O170 | IV | − | + |
| H327 | Wound | Budapest | ND | IV | − | − |
| H329 | Wound | Budapest | ND | IV | − | − |
| H462* | Feces | Budapest | ND | I | − | − |
ND, not determined; *, only cdt-bearing strain isolated from a healthy person.
DISCUSSION
Since CDT was first identified by Johnson and Lior from E. coli in 1987 (22), several studies reported that CDT can be produced by strains belonging to other intestinal (26) and extraintestinal (10, 37) pathogenic bacteria. Molecular genetic studies have revealed that the E. coli cdt operons sequenced thus far are different (30, 31, 34). Interestingly, the sequenced cdt operons of Campylobacter coli and C. jejuni are much more homologous than the E. coli cdt genes (32, 33). In the present study, we developed PCR-based techniques and successfully applied them for a general and specific identification of the known cdt types in E. coli. Since the cdtB gene is the most conserved gene, we have designed primers for the consensus region of the cdtB genes and developed a general multiplex PCR system that can identify all of the known types of E. coli cdt. Further, we have designed primers specific for cdt types I, II, and III, and we have demonstrated that these type-specific primers can be used for typing cdt genes. The use of these primers suggested the existence of a novel cdt variant. Sequencing results validated all of the PCR typing results, including the existence of a new cdt gene, termed cdt-IV. The cdt-IV gene has 84% homology to cdt-I and less homology to cdt-II and cdt-III.
Recently, Clark et al. (8) successfully used PCR to detect and identify cdt-I, cdt-II, and cdt-III genes among CDT-producing E. coli strains from humans and animals. Interestingly, that report indicated the existence of a further cdt gene variant, but these authors did not identify a novel cdt.
All of the cdt-bearing strains had only one type of cdt gene; these genes proved to be cdt-I, cdt-III, or cdt-IV types, but our collection did not contain any cdt-II strains. The cdt-I and cdt-IV genes were detected in human, porcine, and poultry strains, and the cnf-1 gene was detected among cdt-I (4 of 11) and cdt-IV (11 of 18) type strains, but none of these strains had cnf-2. In correlation with previous observations, all eight CDT-III-producing strains were isolated from bovine sources, and all of them also had the cnf-2 gene.
We wanted to further characterize the CDT-producing strains and have compared the cytotoxic activity of bacterial lysates and supernatants of the CDT-producing strains from different cdt gene types. Interestingly, the CDT-IV-producing strains seem less toxic than the strains producing CDT-I, whereas the toxic activities of the CDT-IV- and CDT-III-producing strains were almost the same. None of the CDT-IV-producing strains seemed to secrete toxins into the supernatant. However, CDT-I was secreted by CDT-I-positive strains. These in vitro results indicate that CDT-I could be the most potent type of CDT. In our study, all six tested CDT-IV-producing strains had cell-associated CDT activity, and none of these strains' supernatants were toxic in HeLa cell cultures. Clark et al. (8) recently reported that the some CDT-producing strains' supernatants showed a CDT effect in cell culture in the absence of PCR amplification of the cdt-I, cdt-II, or cdt-III genes. Based on these two observations, it is possible that there are other cdt types among pathogenic E. coli, or that in some strains CDT-IV toxin is also secreted, or that there are additional cdt types yet to be described.
As reported earlier, the EPEC strain E6468/62 (34) and the finch outbreak strain OS (16) had the eae gene encoding for intimin, but none of the other study strains had eae. These results clearly show that CDT production is not limited to EPEC strains, as some epidemiological studies have suggested (1, 2, 6, 17).
Since there were several strains of extraintestinal origin among the CDT-producing strains, a collection of extraintestinal E. coli strains was screened for cdt, and the cdt genes were typed. As expected, the cdt gene was found, and the incidence in extraintestinal strains was even higher than that reported earlier for EPEC strains in India (6) and in Brazil (17). Interestingly, most of the of cdt genes proved to be cdt-IV. On the basis of the existence of cdt-IV, we propose that in the former epidemiological reports the incidence of CDT-producing strains was underestimated. In an earlier study (21), cdt genes were detected in urosepsis E. coli isolates when the strains were tested for as many as 29 genes. These genes were virulence factor genes and potential virulence factor genes characteristic for UPEC. The study did not include strains isolated from healthy individuals. Recently, Johnson et al. (20) reported that among E. coli strains isolated from infants with neonatal bacterial meningitis (NBM) the cdtB gene was more prevalent than other well-established NBM-associated virulence genes. We have found that these strains produced CDT-I or CDT-IV.
Our results also highlight that a new CDT type, cdt-IV, could be widely disseminated. We found CDT-IV production among intestinal and extraintestinal strains of human, poultry, and porcine origin. It has been first reported that cdt was encountered among extraintestinal isolates in only two clonal groups: E. coli O2:K5/K7:H1 (21) and E. coli O6:K53:H1 (18, 19). Additional clonal groups (O83:K1 and O18:K1) have been identified among NBM-associated cdt-bearing strains (20). In the present study, the CDT-I-producing strains belonged to the O2, O18, O23, and O28 serogroups; the CDT-III-producing strains to belonged O2, O15, O78, O88, and O123 serogroups; and the CDT-IV-producing strains belonged to several serogroups, including serogroups O2, O6, O75, O115, O123, and O170.
In summary, we describe here a new PCR system that specifically detects all three known types of CDT, leading to the identification of a new member of the CDT family (CDT-IV). Although there are great quantitative differences in toxicity between the CDT-I-, CDT-III-, and CDT-IV-producing strains in vitro, it seems that CDT could be an important virulence factor of intestinal and extraintestinal E. coli in animals and humans.
Acknowledgments
We are grateful to Neil Ledger for editorial assistance. We thank Béla Szabó (Debrecen, Hungary), István Barcs (Budapest, Hungary), Levente Emody (Pécs, Hungary) Gizella Szojka (Budapest, Hungary), and Hywel Ball (Belfast, Ireland) for providing E. coli strains and Michèle Boury (Toulouse, France) for technical assistance. We special thank Béla Nagy for support, helpful discussions, and help with the manuscript.
This work was supported by the Hungarian Research Fund OTKA (T37890) and Hungarian-French bilateral grants.
REFERENCES
- 1.Albert, M. J., S. M. Faruque, A. S. G. Faruque, K. A. Bettelheim, P. K. B. Neogi, N. A. Bhuiyan, and J. B. Kaper. 1996. Controlled study of cytolethal distending toxin-producing Escherichia coli infections in Bangladeshi children. J. Clin. Microbiol. 34:717-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansaruzzaman, M., M. J. Albert, S. Nahar, R. Byun, M. Kataouli, I. Kuhn, and R. Molby. 2000. Clonal groups of enteropathogenic Escherichia coli isolated in case-control studies of diarrhoea in Bangladesh. J. Med. Microbiol. 49:177-185. [DOI] [PubMed] [Google Scholar]
- 3.Aragon, V., K. Chao, and L. A. Dreyfus. 1997. Effect of cytolethal distending toxin on F-actin assembly and cell division in Chinese hamster ovary cells. Infect. Immun. 65:3774-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birnboim, H. C., and J. Dolly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed]
- 5.Blum, G., V. Falbo, A. Caprioli, and J. Hacker. 1995. Gene clusters encoding the cytotoxic necrotizing factor type 1, Prs-fimbriae and -hemolysin from the pathogenicity island II of uropathogenic Escherichia coli strain J96. FEMS Microbiol. Lett. 126:189-196. [DOI] [PubMed] [Google Scholar]
- 6.Bouzari, S., and A. Varghese. 1990. Cytolethal distending toxin (CLDT) production by enteropathogenic Escherichia coli (EPEC). FEMS Microbiol. Lett. 59:193-198. [DOI] [PubMed] [Google Scholar]
- 7.China, B., V. Pirson, and J. Mainil. 1996. Typing of bovine attaching and effacing Escherichia coli by multiplex in vitro amplification of virulence-associated genes. Appl. Environ. Microbiol. 62:3462-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark, C. G., S. T. Johnson, R. H. Easy, J. L. Campbell, and F. G. Rodgers. 2002. PCR for detection of cdt-III and the relative frequencies of cytolethal distending toxin variant-producing Escherichia coli isolates from humans and cattle. J. Clin. Microbiol. 40:2671-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Comayras, C., C. Tasca, S. Y. Peres, B. Ducommun, E. Oswald, and J. De Rycke. 1997. Escherichia coli cytolethal distending toxin blocks the HeLa cell cycle at the G2/M transition by preventing cdc2 protein kinase dephosphorylation and activation. Infect. Immun. 65:5088-5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cope, L., S. Lumbley, L. Latimer, J. Klesney-Tait, M. K. Stevensen, L. S. Johnson, M. Purven, M. Munson, J. R. S. Munson, T. Langerpard, J. D. Radolf, and E. J. Hansen. A diffuse cytotoxin of Haemophilus ducreyi. Proc. Natl. Acad. Sci. USA 94:4056-4406. [DOI] [PMC free article] [PubMed]
- 11.De Rycke, J., P. Mazars, J.-P. Nougayrede, C. Tasca, M. Boury, F. Herault, A. Valette, and E. Oswald. 1996. Miotic block and delayed lethality in HeLa epitheilial cells exposed to Escherichia coli BM2-1 producing cytotoxic necrotizing factor type 1. Infect. Immun. 64:1694-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Rycke, J., and E. Oswald. 2001. Cytolethal distending toxin (CDT): a bacterial weapon to control host cell proliferation? FEMS Microbiol. Lett. 203:141-148. [DOI] [PubMed] [Google Scholar]
- 13.Dozois, C. M., S. Clement, C. Desautels, E. Oswald, and J. M. Fairbrother. 1997. Expression of P, S, and F1C adhesins by cytotoxic necrotizing factor 1-producing Escherichia coli from septicemic and diarrheic pigs. FEMS Microbiol. Lett. 152:307-312. [DOI] [PubMed] [Google Scholar]
- 14.Elliott, S. J., L. A. Wainwright, T. K. McDaniel, K. G. Jarvis, Y. K. Deng, L. C. Lai, B. P. McNamara, M. S. Donnenberg, and J. B. Kaper. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28:1-4. [DOI] [PubMed] [Google Scholar]
- 15.Elwell, C. A., and L. A. Dreyfus. 2000. DNase I homologous residues in CdtB are critical for cytolethal distending toxin-mediated cell cycle arrest. Mol. Microbiol. 37:952-963. [DOI] [PubMed] [Google Scholar]
- 16.Foster, G., H. M. Ross, T. W. Pennycott, G. F. Hopkins, and I. M. McLaren. 1998. Isolation of Escherichia coli O86:K61 producing cyto-lethal distending toxin from wild birds of the finch family. Lett. Appl. Microbiol. 26:395-398. [DOI] [PubMed] [Google Scholar]
- 17.Guth, B. E., R. Giraldi, T. A. Gomes, and L. R. Marques. 1994. Survey of cytotoxin production among Escherichia coli strains characterized as enteropathogenic (EPEC) by serotyping and presence of EPEC adherence factor (EAF) sequences. Can. J. Microbiol. 40:341-344. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, J. R., P. Delavari, A. L. Stell, T. S. Whittam, U. Carlino, and T. A. Russo. 2001. Molecular comparison of extraintestinal Escherichia coli isolates from the same electrophoretic lineages from humans and domestic animals. J. Infect. Dis. 183:154-159. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, J. R., T. T. O'Bryan, M. Kuskowski, and J. N. Maslow. 2001. Ongoing horizontal and vertical transmission of virulence genes and papA alleles among Escherichia coli blood isolates from patients with diverse-source bacteremia. Infect. Immun. 69:5363-5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson, J. R., E. Oswald, T. T. O'Bryan, M. A. Kuskowski, and L. Spanjaard. 2002. Phylogenetic distribution of virulence-associated genes among Escherichia coli isolates associated with neonatal bacterial meningitis in The Netherlands. J. Infect. Dis. 185:774-784. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, J. R., and A. L. Stell. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261-272. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, W. M., and H. Lior. 1987. Response of chinese hamster ovary cells to a cytolethal distending toxin (CDT) of Escherichia coli possible misinterpretation as a heat-labile (LT) enterotoxin. FEMS Microbiol. Lett. 43:19-23. [Google Scholar]
- 23.Johnson, W. M., and H. Lior. 1988. A new heat-labile cytolethal distending toxin (CLDT) produced by Escherichia coli isolates from clinical matherial. Microb. Pathog. 4:103-113. [DOI] [PubMed] [Google Scholar]
- 24.Lara-Tejero, M., and J. E. Galan. 2000. A bacterial toxin that controls cell cycle progression as a deoxyribonuclease I-like protein. Science 290:354-357. [DOI] [PubMed] [Google Scholar]
- 25.Okeke, I. N., A. Lamikanra, H. Steinruck, and J. B. Kaper. 2000. Characterization of Escherichia coli strains from cases of childhood diarrhea in provincial southwestern Nigeria. J. Clin. Microbiol. 38:7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okuda, J., H. Kurazono, and Y. Takeda. 1995. Distribution of the cytolethal distending toxin A gene (cdtA) among species of Shigella and Vibrio, and cloning and sequencing of the cdt gene from Shigella dysenteriae. Microb. Pathog. 18:167-172. [DOI] [PubMed] [Google Scholar]
- 27.Orskov, I., and F. Orskov. 1984. Serotyping of Escherichia coli. Methods Microbiol. 14:43-112. [Google Scholar]
- 28.Oswald, E., J. De Rycke, P. Lintermans, K. Van Muylen, J. Mainil, G. Daube, and P. Pohl. 1991. Virulence factors associated with CNF2 in bovine diarrheic and septicemic strains of Escherichia coli. J. Clin. Microbiol. 29:2522-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oswald, E., H. Schmidt, S. Morabito, H. Karch, O. Marches, and A. Caprioli. 2000. Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: characterization of a new intimin variant. Infect. Immun. 68:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pérès, S. Y., O. Marchès, F. Daigle, J.-P. Nougayrede, F. Herault, C. Tasca, J. DeRycke, and E. Oswald. 1997. A new cytolethal distending toxin (CDT) from Escherichia coli producing CNF2 blocks HeLa cell division in G2/M phase. Mol. Microbiol. 24:1095-1107. [DOI] [PubMed] [Google Scholar]
- 31.Pickett, C. L., D. L. Cottle, E. C. Pesci, and G. Bikah. 1994. Cloning, sequencing and expression of the Escherichia coli cytolethal distending toxin production genes. Infect. Immun. 62:1046-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pickett, C. L., E. C. Pesci, L. Cottle, G. Russell, A. N. Erdem, and H. Zeytin. 1996. Prevalence of cytolethal distending toxin production in Campylobacter jejuni and relatedness of Campylobacter sp. cdtB genes. Infect. Immun. 64:2070-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pickett, C. A., and C. A. Whitehouse. 1999. The cytolethal distending toxin family. Trends Microbiol. 7:292-297. [DOI] [PubMed] [Google Scholar]
- 34.Scott, D. A., and J. Kaper. 1994. Cloning and sequencing of the genes encoding Escherichia coli cytolethal distending toxin. Infect. Immun. 62:244-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith, H. W. 1974. Seach for transmissible pathogenic characters in invasive strains of Escherichia coli: the discovery of a plasmid toxin and a plasmid lethal character closely associated, or identical, with colicine V. J. Gen. Microbiol. 83:95-111. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Sugai, M., T. Kawamoto, S. Y. Peres, Y. Ueno, H. Komatsuzawa, T. Fujiwara, H. Kurihara, H. Suginaka, and E. Oswald. 1998. The cell cycle-specific growth-inhibitory factor produced by Actinobaccillus actinomycetemcomitans is a cytolethal distending toxin. Infect. Immun. 66:5008-5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tóth, I., E. Oswald, J. G. Mainil, M. Awad-Masalmeh, and B. Nagy. 2000. Characterization of intestinal cnf1+ Escherichia coli from weaned pigs. Int. J. Med. Microbiol. 290:539-542. [DOI] [PubMed] [Google Scholar]