Abstract
Repetitive DNA elements have been a part of the genomic fauna of eukaryotes perhaps since their very beginnings. Millions of years of coevolution have given repeats central roles in chromosome maintenance and genetic modulation. Here we review the genomes of parasitic protozoa in the context of the current understanding of repetitive elements. Particular reference is made to repeats in five medically important species with ongoing or completed genome sequencing projects: Plasmodium falciparum, Leishmania major, Trypanosoma brucei, Trypanosoma cruzi, and Giardia lamblia. These organisms are used to illustrate five thematic classes of repeats with different structures and genomic locations. We discuss how these repeat classes may interact with parasitic life-style and also how they can be used as experimental tools. The story which emerges is one of opportunism and upheaval which have been employed to add genetic diversity and genomic flexibility.
INTRODUCTION
The dawn of the genomic age has understandably been dominated by the push for gene discovery. However, to concentrate on the genes at the expense of all else is to miss a wealth of biological information hidden in the repetitive portion of the genome. “Living” (i.e., actively proliferating) repeats are dynamic elements which reshape their host genomes by generating rearrangements, creating and destroying genes, shuffling existing genes, and modulating patterns of expression. Repeats may also accrue functions that are important for the day-to-day maintenance of chromosomes and so become a force for genomic stability as well as instability. “Dead” repeats (i.e., those which are no longer able to proliferate) constitute a palaeontological record, which can be mined for clues about evolutionary events and impetus. The dynamic nature of repeats leads to a rapid evolutionary divergence that can be used in species identification and phylogenetic inference. Repeats can also provide passive markers for studying processes of mutation and selection.
In this review, we focus on repetitive elements in the genomes of five pathogenic protozoa with genome sequencing projects that are either complete or nearing completion. The five parasites are the apicomplexan Plasmodium falciparum (for which the genome sequence is essentially complete [79]); three trypanosomatid Euglenozoa, Leishmania major, Trypanosoma brucei, and Trypanosoma cruzi; and the metamonad (diplomonad) Giardia lamblia. These organisms are of considerable medical interest as pathogens and span three protozoan phyla, each of which seems to have had an intriguing evolutionary history (12, 64, 77, 88, 125, 174).
The genomes of these protozoan parasites, like all eukaryotic genomes, have been colonized by diverse repetitive elements. As the coding and noncoding parts of the genomes have coevolved, some repeats have become bound into nuclear processes. These processes include those that modulate virulence, such as the contingency gene systems of antigenic variation. Others have found genomic niches away from conserved coding regions that could be easily disrupted by their activity. The repeats of a parasite's genome, therefore—their presence and absence, their type, activity, and location—can be a window on the genomic organization that enables parasitism.
CLASSIFICATION OF REPEATS
Repetitive sequences can be artificially divided into two groups: interspersed repeats and tandemly repeated DNA. Interspersed repeats mainly represent inactive copies of presently or historically active transposable elements, which are of three major types (41, 105): elements that transpose through a DNA-based pathway (DNA transposons) and two distinct classes of elements requiring reverse transcription from an RNA intermediate (retroelements). Despite the common link of transposition via RNA, the two classes of retroelements transpose by fundamentally different mechanisms. The long terminal repeat (LTR) retroelements, which include retroviruses and Ty1/Ty3-like retrotransposons, are reverse transcribed from RNA intermediates, duplicated, and then transposed as double-stranded DNA. In contrast, non-LTR retroelements—consisting of short or long interspersed nuclear elements (SINEs or LINEs, respectively [169, 200])—are transposed by reverse transcription of mRNA directly into the site of integration. The critical distinction between the transposition mechanisms of LTR and non-LTR retroelement is often usefully encapsulated by referring to LTR retroelements as retrotransposons and non-LTR retroelements as retroposons (95, 165).
Tandemly repeated DNA satellites are usually confined to specific chromosome locations propagating by replicational slippage and gene conversion (110, 186). The term “satellite DNA”, originally defined by the behavior of DNA in density gradients, has drifted somewhat to include other tandemly repeated DNA elements such as microsatellites and minisatellites. Microsatellites are small (usually <200-bp) clusters of repeats with unit length generally of <5 bp. Minisatellites form larger clusters (several kilobases) from larger repeat units (5 to 25 bp). Macrosatellites are large regions (up to hundreds of kilobases) of repeats of >25 bp; this definition includes the original DNA satellites, although a macrosatellite need not have an abnormal buoyant density.
It is worth noting that the classification of elements as interspersed or tandem is taxonomically not very rigorous. For example, the “interspersed” element Alu, the most abundant SINE in the human genome, forms dense clusters containing many direct (i.e., tandem) repeats (100). It should also be noted that distinction between what is genic and what is repetitive is also somewhat fuzzy. Autonomous transposable elements, at least those that are still active, are obviously coding DNAs and in that sense are genic. Moreover, nontransposable genes can be reiterated by the same mechanisms—replicational slippage and gene conversion—as tandemly repeated noncoding DNAs. Such tandem duplication of protein-coding genes is a particular feature of the Trypanosoma genomes, in which genes expressed at high levels are often multicopy (e.g., tubulin genes and spliced-leader RNA genes). Parasites also frequently contain divergent gene families associated with virulence (e.g., the var, rif, and stevor families of P. falciparum). Multicopy genes (other than transposable elements) are not addressed at length in this review, but it is hoped that some of the parallels between such genes and repeats of other kinds will be clear.
REPETITIVE DNA CONTENT OF GENOMES
The repeat content of the genomes of the protozoan parasites is closely correlated with their haploid genome size (for convenience, all genomic information in this work is given in the context of “haploid” genomes regardless of the typical ploidy, or sexual status, of the various organisms). The small genome of the apicomplexan Theileria parva (10 Mb per haploid), for example, is gene dense and contains virtually no repetitive DNA apart from telomeric repeats and (pseudo)genes of the polymorphic protein family, Tpr (136). Plasmodium berghei, with its intermediate-size genome of ∼25 Mb, contains around 5% repeat sequence (153), whereas in T. cruzi (genome size, ∼40 Mb), DNA reassociation kinetics indicate that 9 to 14% of total cellular DNA is highly repetitive and a further ∼30% is at least moderately reiterated (43). Extrapolating from these data, one might expect the genome of Toxoplasma gondii (a hefty 80 Mb per haploid) to be extremely rich in repetitive DNA. It is interesting then, that at present few repetitive elements have been identified in Toxoplasma (61, 91, 126, 145). This is due at least in part to the initial phase of the T. gondii genome sequencing project being expressed sequence tag-based (7, 182) but could reflect a reiteration of genes rather than noncoding DNA in these organisms.
The existence of genomes containing few or no repetitive elements demonstrates that repeats are not necessary components in basic cellular processes (with the notable exception of chromosome end protection). This can encourage the view of repetitive sequences as purely parasitic or junk elements—the implication being that such sequences are either detrimental or, at best, inconsequential to the fitness of the host organism (96). However, genomes devoid of repeats may equally be invoked to show that such elements are not inevitable in eukaryotic genomes. We should not be too ready to assign organismic fitness costs to genomic elements when we do not know their full consequences. For example, when cyclically transmitted through mosquitoes, the repetitive portion of the genome of P. berghei is ∼5%, but this percentage is drastically reduced during prolonged mechanical propagation in mice (153). Loss of repetitive DNA correlates with decreasing viability of gametocytes (73), suggesting that these repeats are either required for or dependent on some essential process in the parasite life cycle. Our current understanding of the genome organization in T. brucei also indicates that removal of repeats would seriously curtail parasitaemia (see below). Thus, rather than being the freeloaders we might assume, some repeats may be maintained by a positive selection.
DIVERSITY OF REPETITIVE DNA
Table 1 summarizes the major repetitive elements identified in the protozoan parasites P. falciparum, L. major, T. brucei, T. cruzi, and G. lamblia. The most immediate point that can be made from such a list is one of diversity. The genomes differ greatly not only in the amount of repetitive sequence they possess but also in the distribution, type, and unit size of the repeats. No element is common to any two organisms considered. Indeed, with the exception of some of the retroelements of the trypanosomes (T. brucei and T. cruzi), on the basis of sequence alone, no common origin to repeats in any of the organisms listed in Table 1 can be found. For example, the relatives T. brucei and T. cruzi both possess a high-copy-number satellite DNA element with similar repeat sizes: 177 and 195 bp, respectively (170). However, these two trypanosomal repeats have no significant identity and differ greatly in genomic location. Diversity is also the predominant feature when more closely related organisms are considered. Many repeats appear to be entirely species specific, e.g., the TARE-2 and TARE-3 elements of P. falciparum (68). Others are restricted to closely related species, e.g. LST-RB1, specific to the L. major (cutaneous) complex (179), or the Lmet2 repeat, specific to the L. donovani (visceral) complex (93).
TABLE 1.
Major repetitive elements of the parasitic protozoa P. falciparum, L. major, T. brucei, T. cruzi, and G. lamblia
| Repeata | Alternative name(s) | Typeb | STc | Unit size (bp) | Copy no.d | Chromosomal distribution | Comments | Reference(s) |
|---|---|---|---|---|---|---|---|---|
| P. falciparum | ||||||||
| 14-bp repeat | TARE-1, SB-1 | T | • | 14 (28,42) | 2,000e | Most | Directly abuts telomeric repeats; forms suprarepeats | 68, 79, 152, 194 |
| TARE-2f | T | • | 135 | 340e | Most | P. falciparum specific | 68 | |
| TARE-3f | 692-bp repeat, 0.5-kb repeat | T | • | 692 | 60 | Almost all | P. falciparum specific | 56, 68, 198 |
| TARE-4f | T/I | • | ND | Most | Complex repeat structure | 68 | ||
| TARE-5f | 12-bp repeat | T | • | 12 | 4,000e | Most | 68, 146 | |
| 17-bp repeat | T | • | 17 | ND | ND | 146 | ||
| 23/28-bp repeat | T | • | 23/28 | ND | ND | 146 | ||
| Rep11 | T | • | 11 | ND | ND | 33, 151 | ||
| Rep20 | Rep2, 21-bp repeat, TARE-6, SB-3 | T | • | 21 | 12,000 | All | P. falciparum specific; most telomere-distal repeat | 16, 17, 33, 52, 68, 79, 80, 143, 146, 147, 151 |
| L. major | ||||||||
| LCTAS | (TAS-A + TAS-B) | T | • | ∼100 | ND | Many | Directly abuts telomeric repeats | 74, 179 |
| LST-RA | T | • | 58 | ND | Many | 179 | ||
| LST-RB1 | I | • | 23 | ND | Chr 1 and 22 | Specific to L. major (cutaneous) complex | 179 | |
| LST-RB2 | I | • | 25 | ND | Chr 1 and 22 | 179 | ||
| LST-RB3 | T | • | 19 | ND | ND | 179 | ||
| LST-RC | T | • | 55 | ND | Chr 1 and 22 | 179 | ||
| STIR1 | LiSTIR1-like element, LST-RE | T | • | 81 | ND | Chr 1, 5, and 22 | 133, 157, 179 | |
| LST-RI | T | • | 14 | ND | Chr 1 and 22 | 179 | ||
| 272-bp repeat | 274-bp repeat, LST-RJ | T | • | 272 | ND (60 on Chr 1) | Chr 1, 5, and 22 | Most telomere-distal repeat | 74, 133, 179 |
| T. brucei | ||||||||
| 29-bp repeat | T | • | 29 | ND | All | Directly abuts telomeric repeats | 199 | |
| 70-bp repeat | 76-bp repeat | T | 66-81 | ND | All | Upstream of VSG genes | 119, 127 | |
| 50-bp repeat | T | • | 50 | 5,000 | ND | Upstream of VSG-ES | 204 | |
| 177-bp repeat | Alu repeat | T | 177 | 15,000 | MCs and ICs only | Forms large repetitive palindrome on MCs | 170, 171, 199; Wickstead et al., submitted | |
| NR element | T | 36-41 | 0-50,000g | Episomal | Copy number is highly dependent on straing | 10 | ||
| ingi | TRS | I, LINE | 5,200 | 200-500 | Most | Contains a RIME element | 25, 34, 107, 132 | |
| RIME | I, SINE | 494 | 200-600 (incl. ingi) | Most | Related to the LINE ingi | 25, 34, 90 | ||
| SLACS | I, LINE | 6,678 | 9 | ND | Site-specific retroposon of SL-RNA gene | 8, 9, 25 | ||
| T. cruzi | ||||||||
| 189-bp repeat | I | • | 189 | 40 | All | Directly abuts telomeric repeats | 44/PICK> | |
| 195-bp repeat | T | 195 | 20,000 | Several (not all) | Diagnostic tool | 39, 84, 160, 170 | ||
| L1Tc | I, LINE | ∼5,000 | 300h-500 | Most | 25, 35, 124, 141, 142, 160 | |||
| NARTc | I, SINE | 260 | 150h | ND | Related to the LINE L1Tc | 35 | ||
| VIPER | I, LTRi | 2,326 | 70 | ND | Unusual “LTR” elementi; associated with SIRE; dead retrotransposon | 11, 25, 191 | ||
| SIRE | I, SINE | 428 | 700 | All | Associated with VIPER | 25, 39, 160, 191-193 | ||
| CZAR | I, LINE | 7,219 | 6 | One chromosome only | Site-specific retroposon of SL-RNA gene | 25, 195 | ||
| 172-bp repeat | I | 172 | 200-800 | Several (not all) | Associated with rDNA spacer | 54, 156 | ||
| E12 | I | 1,123 | 900 | All | 160-162 | |||
| E13 | RS13Tc | I | 1,025 | 3,000 | All | Diagnostic tool; RS13Tc is a 5′-truncated form of E13 | 142, 159, 160 | |
| E22 | I | ∼1,000 | 1,400 | All | 39, 160, 162 | |||
| C6 | I | 1,433 | 250 | All | 13 | |||
| RS1Tc | I | 1,439 | 700 | Most | 142 | |||
| RLE | I | 317 | ND | ND | 106 | |||
| SRE1-3 | I | 43-145 | 80-200 | ND | Associated with rDNA spacer | 138, 160 | ||
| TcIRE | I | 430-500 | 2,000h | Several (not all) | Two forms: TcIRE(I) and TcIRE(II) | 6 | ||
| G. lamblia | ||||||||
| GiIM | (Genie1 + 771-bp repeat) | T, LINE | • | 5,470 | ∼10 | Most (not all) | Directly abuts telomeric repeats | 15, 25, 37 |
| GiIT | Genie1A | T, LINE | • | 6,000 | ND | ND | Directly abuts telomeric repeats | 15, 25, 37 |
| GiID | Genie2 | I, LINE | 3,019 | ∼30 | ND | Dead retroposon | 15, 25, 37 |
P. falciparum: TARE, telomere-associated repetitive element. L. major: LCTAS, Leishmania conserved telomere-associated sequence; LST-R, Leishmania subtelomeric repeat; STIR, subtelomeric interspersed repeat; TAS, telomere-associated sequence. T. brucei: IC, intermediate-sized chromosome; MC, minichromosome; RIME, ribosomal inserted mobile element; SL, spliced leader; SLACS, spliced leader-associated conserved sequence; TRS, trypanosome repeat sequence; T. cruzi: CZAR, cruzi-associated retrotransposon; NARTc, nonautonomous retroelement in T. cruzi; RLE, retroposon-like element; SL, spliced leader; SIRE, short interspersed repetitive element; SRE, spacer repetitive element; TcIRE, T. cruzi interspersed repeated element; VIPER, vestigial interposed retroelement. G. lamblia: Genie, Giardia early non-LTR insertion element.
Repeats are annotated as to type: T, tandem; I, interspersed. Those identified as LINEs, SINEs, or LTR retrotransposons are indicated.
Elements located exclusively in subtelomeric regions. Repeats of the subtelomere are arranged roughly in order of proximity to the telomeric repeats.
Copy numbers are per haploid nuclear genome (estimated size: 25 Mb, 34 Mb, 25 + 10 Mb, 40 Mb, and 12 Mb for P. falciparum, L. major, T. brucei, T. cruzi, and G. lamblia, respectively). Note that current estimates of T. cruzi genome size are much smaller than earlier estimates (43, 112); hence, copy numbers presented here for repeats of this species are frequently lower than those previously published.
Extrapolated from eight sequenced chromosome ends (68).
TARE-2 to TARE-5 are part of the larger repeat region SB-2 defined in (79).
This element is not detectable in strain TREU927/4, used for the genome-sequencing project (10).
Unusual LTR retrotransposon-like element: reverse transcriptase domain is homologous to LTR retrotransposons from other species, but structure is atypical and terminal repeats have not been identified (191).
As well as diversity of repeats between species, there is diversity within a species: each has been colonized by a variety of repetitive DNAs, most of which show no significant homology to other repeats of the same organism. All of this is indicative of the transitory nature (on an evolutionary timescale) of the repetitive portion of the genome.
There is one obvious exception to this rule of repeat diversity: the telomeric repeats. Constrained by an essential role in chromosome maintenance and interactions with various proteins, the sequence of the telomeric repeats is highly conserved (Table 2). In this way, telomeric repeats are different from the other repetitive DNAs mentioned in this review. There is an extensive literature on the properties of telomeres (47, 108, 155, 167) and they are not discussed at length here. However, it is worth noting that telomeres—with their central role in eukaryotic genome construction—are also promiscuous tandem repeats that most probably had a “selfish” origin. The reverse transcriptase (RT) component of the enzyme telomerase has structural similarities to the RT of non-LTR retroposons, leading to speculations of a common ancestor (96). It should also be remembered that the highly successful short terminal repeat telomeres (such as those in Table 2) are not the only repeats to have filled the niche of chromosome end protection (see below).
TABLE 2.
Telomeric repeats of the parasitic protozoa P. falciparum, L. major, T. brucei, T. cruzi, and G. lamblia
In spite of the lack of sequence conservation, the tangle of repeats identified in the five parasitic species listed in Table 1 can be teased out into five broad thematic classes on the basis of type and location of repeat: (i) subtelomeric satellites consisting of tandemly repeated elements of relatively small unit size (mostly 10 to 100 bp) clustered near telomeres (Plasmodium and Leishmania species contain large numbers of these elements); (ii) subtelomeric retroelements (although tandemly repeated, the subtelomeric repeats of Giardia are different from those of Plasmodium and Leishmania since they are active LINEs); (iii) interspersed retroelements (the two species of Trypanosoma carry these elements in abundance; a number have been identified as LINEs, and it is likely that the others are dependent SINEs); (iv) chromosome internal satellites (such as the macrosatellites of the T. brucei 177-bp repeat and the T. cruzi 195-bp repeat); and (v) microsatellites.
In the menagerie of repetitive elements in the genomes of protozoan parasites, there are some noticeable absences— namely, elements identified as either DNA transposons or retroviruses. In eukaryotes, DNA transposons tend to be less common and have shorter life spans within a species than do retroelements. This can be explained by the inability of the encoded transposase to distinguish between active and inactive elements. As inactive copies accumulate in the genome, transposition activity becomes attenuated, and in due course the transposon will die. LINEs do not experience such severe attenuation since LINE proteins associate predominantly with the RNA from which they were transcribed (but see the discussion of SINEs below), resulting in a selective transposition of functional retroposons. DNA transposons apparently survive extinction by horizontal transfer to virgin genomes (89, 109, 163). Retroviruses, too, propagate by moving between genomes. It is interesting that these parasitic species which now live in such intimate contact with metazoans, and in which metazoan transposons will proliferate if artificially introduced (85), should keep genomes free of DNA transposons and retroviruses. Perhaps this is a result of the tight control these organisms must maintain on traffic at the cell membrane.
LIFE ON THE EDGE: SUBTELOMERIC REPEATS
The subtelomeres of chromosomes are especially turbulent regions prone to nucleotide loss and recombination. Generally, the central core of protozoan chromosomes remains stable while the subtelomeric regions vary. Variability in these regions is responsible for a major part of the large polymorphisms observed between chromosome homologues in parasitic protozoa (72, 73, 113, 129, 179). Because of this variability, DNA elements positioned at chromosomal margins are apt to live short lives unless they can expand rapidly enough to offset the high rate of casualties. The frequent association of tandem repeats with subtelomeric regions in various organisms demonstrates that these elements make particularly good frontiersmen.
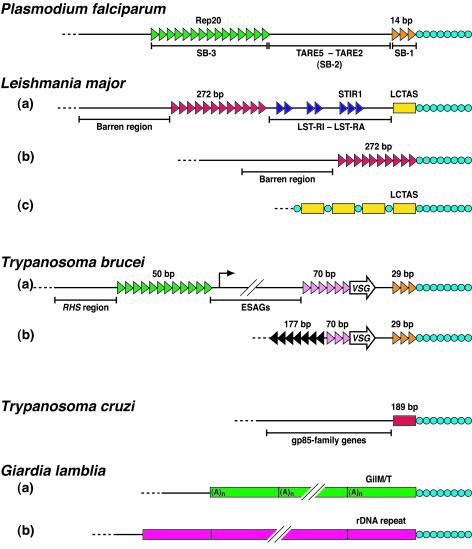
Figure 1 shows schematically the organization of subtelomeres in the parasitic protozoa P. falciparum, L. major, T. brucei, T. cruzi, and G. lamblia. It can be seen that repeats proliferate in the subtelomeres of four of the five organisms. Three of the organisms—P. falciparum, L. major, and T. brucei —contain subtelomeric satellites, while Giardia carries retroelements or tandemly repeated genes at subtelomeres. The clustering of repetitive elements at subtelomeres could be explained by selection against disruption of the central coding regions by repeat insertion. The genomic organization of Leishmania and Plasmodium would certainly fit this paradigm: the major repeat elements of these organisms are found exclusively in subtelomeric regions. Conversely, repetitive subtelomeres could be positively selected—it has already been mentioned that the subtelomeric repeats of P. berghei may be subject to a positive selection during cyclical transmission through a fly vector (153). Alternatively, repeats may flourish at subtelomeres simply because higher rates of recombination allow their proliferation. Most probably, a combination of all these factors is at work.
FIG. 1.
Organization of the subtelomeric regions of the parasitic protozoa P. falciparum, L. major, T. brucei, T. cruzi, and G. lamblia. Telomeric repeats (blue circles) are on the right. For L. major: (a) chr1b, (b) chr1a, and (c) unspecified chromosome. For T. brucei: (a) subtelomere containing a bloodstream form VSG expression site and (b) minichromosomal subtelomere. For G. lamblia: (a) most common subtelomeric organization and (b) rDNA repeat subtelomere. The RHS region contains retrotransposon hot spot (pseudo)genes (34). ESAGs are expression site-associated genes. See Table 1 for other abbreviations and more information on repeats. Not to scale. Adapted from references 15, 37, 40, 68, 74, 79, 80, 114, 133, 146, and 179.
A closer examination of the subtelomeric satellite repeats reveals that they do not colonize the subtelomere randomly. A positional hierarchy exists in which some repeats directly abut the telomeric repeats while others mark the centromere-proximal border of the region. In the first category are the 14-bp repeat of P. falciparum (152, 194), LCTAS of L. major (74, 179), and the 29-bp repeat of T. brucei (199)—their telomere-distal partners being Rep20 (16, 17, 143), the 272-bp repeat (74, 133), and the 50-bp repeat (204), respectively. There are also differences in the chromosomal distribution of the various subtelomeric satellites. For example, the LCTAS element, common to all Leishmania species, is present at almost all chromosome ends whereas elements such as LST-RB1 are much more restricted in terms of chromosomal (and species) distribution (74, 179).
The dynamic relationships between subtelomeric regions means that chromosome end-proximal sequences should be regarded as only a snapshot of subtelomeric organization. A parallel can be drawn with the Y′ elements of Saccharomyces cerevisiae, present in one to four copies at most subtelomeres (196). At any one time, an individual subtelomere may lack Y′ elements, but their mobility is such that virtually any chromosomal extremity is susceptible to Y′ element acquisition (60). A similar mobility has been observed for a 2.3-kb repeat of P. berghei subtelomeres (153). It is likely that many of the subtelomeric repeat satellite elements described are in a similar state of flux. Chromosomal polymorphisms thus reflect not only expansion and contraction of chromosome-specific repeats but also dynamic gain and loss of elements from other subtelomeres.
As mentioned above, the subtelomeric repeats of Giardia are different from the satellite-type repeats of the other species described. A subset of subtelomeres of G. lamblia contain tandem repeats of rRNA genes which show extensive polymorphisms (1, 92, 115) and also apparently jump from telomere to telomere (113, 187), providing an interesting genic parallel to the mobility of the subtelomeric satellites of other species. However, most G. lamblia subtelomeres consist of tandem copies of active LINE retroposons (either GilM or GilT elements), which directly abut the telomeric repeats and are oriented such that reverse transcription would have run toward the chromosome end (15, 37). The organization is suggestive of a possible redundancy between the retroposon and telomerase activities. Such a redundancy was the likely ancestor of the situation now seen in Drosophila, where the role of chromosome end protection has been entirely usurped by the retroposon TART and its dependant, HeT-A (26, 27, 116). This is a prime example of a “parasitic” repetitive element assuming a functional role within a genome.
Unlike the other four organisms, the subtelomeres of T. cruzi do not appear to possess large tracts of subtelomeric repeats. Each chromosome end sequenced to date is capped by a single copy of a 189-bp repeat followed by telomeric repeats (44). Genes can be found immediately centromere-proximal to the 189-bp repeat; no barren (i.e., geneless) region is observed as seen adjacent to Leishmania subtelomeric repeats.
SUBTELOMERIC SATELLITES AND GENE EXPRESSION
The possible function of subtelomeric repetitive regions remains unresolved. Most accounts invoke an idea of a spacer or buffer zone. Subtelomeric tracts may serve to distance coding genes from aberrant expression experienced near the telomeres, whether this be telomeric silencing, as well-documented in yeast (94), or activation, as associated with specialized subtelomeric transcription of contingency genes in T. brucei or Borrelia (19, 83). Alternatively, repeats might insulate conserved chromosome internal regions from the natural volatility of the subtelomere. The gene organization of L. major provides support for such ideas of a buffer zone; the telomeres are separated from coding regions by subtelomeric satellites followed by a further nonrepetitive barren region (133). Moreover, in P. falciparum, deletions of subtelomeric repeats are associated with proximal-gene inactivation (158).
Taming of the subtelomere, however, is only half the story. Some parasitic protozoa may also harness the turbulence of these regions to modulate the activity of genes involved in virulence. The best-characterized example of this is in the antigenic variation of T. brucei (reviewed in references 30, 32, 48, and 49). In this species, expression sites containing the major surface proteins of the bloodstream form are located at subtelomeres (Fig. 1). The most telomere-proximal of the bloodstream expression site genes encodes an immunodominant variable surface glycoprotein (VSG). Silent copies of VSG genes are also found at the subtelomeres of minichromosomes (Fig. 1), as well as at chromosome-internal loci. High recombination rates between subtelomeres ensures a frequent change in the (single) expressed VSG gene and is combined with in situ switching of the transcribed VSG expression site (VSG-ES) to achieve periodic changes in the antigenic character of the parasite.
P. falciparum also undergoes antigenic variation (reviewed in references 31, 111, and 130). Unlike T. brucei VSG genes, the var genes of P. falciparum—encoding immunogenic transmembrane proteins displayed on the surface of schizont-infected erythrocytes—do not require subtelomeric locations for expression (168). Nonetheless, var genes, along with other divergent gene families, rif and stevor, are predominantly subtelomeric (79). This organization promotes recombination between var genes; it has been estimated that recombination rates for subtelomeric var genes are around eight-fold higher than those for the genome as a whole (71, 184). Subtelomeric satellites appear to be central to this process, since they mediate the promiscuous clustering of telomeres, bringing into close association subtelomeric genes which often have little identity (67, 71, 139). The presence or absence of subtelomeric repeats may also modulate the activity of telomere-proximal genes (158). It has been postulated that the presence of the unique Rep20 satellite may be a key reason why P. falciparum is more virulent than other human malarias (139).
From the above discussion, it should not be assumed that involvement of subtelomeric repeats is ubiquitous to antigenic variation in protozoan parasites (or, indeed, to antigenic variation more generally). Trophozoites of G. lamblia undergo antigenic variation both in vivo and in vitro (4, 134). However, genes encoding the variant-specific surface proteins are dispersed throughout the genome and antigenic variation is not associated with DNA rearrangements (2, 3). Interestingly, in the intracellular parasite T. cruzi, which does not undergo antigenic variation (and also lacks subtelomeric satellites), subtelomeric regions are still associated with genes encoding surface antigens from the divergent gp85-sialidase family (44). Moreover, although subtelomeric satellites are absent in T. cruzi, subtelomeric regions in this species are still associated with retroelements (44), as is found for T. brucei subtelomeres (34).
THE QUICK AND THE DEAD: RETROELEMENTS
Unlike Leishmania and Plasmodium species, the genomes of T. brucei and T. cruzi are riddled with interspersed elements (reviewed in reference 25). In these two species, interspersed repeats may be highly reiterated: the elements ingi of T. brucei and L1Tc of T. cruzi both make up ∼6% of their respective genomes (107, 124, 132) (Table 3). It is not yet clear how some of these identified elements have achieved such success, but for many (if not all), retrotransposition has been central. Four of the interspersed elements of Trypanosoma species have the hallmarks of autonomous non-LTR retroposons (i.e., LINEs): ingi and SLACS of T. brucei and L1Tc and CZAR of T. cruzi (Table 1). The mobility of non-LTR retroelements relies on a target-primed reverse transcription reaction in which the cDNA strand is synthesized from an RNA template directly onto a chromosomal target site (120). Thus, LINE cDNA never exists free from the chromosome, unlike cDNAs of LTR retrotransposons. Moreover, complete integration then requires the participation of the cellular DNA repair-replication machinery. These factors may explain the lack of evidence for horizontal transfer of non-LTR retroposons in the last 600 Myr (123).
TABLE 3.
Top 10 repetitive elements of the parasitic protozoa P. falciparum, L. major, T. brucei, T. cruzi, and G. lambliaa
| Rankb | Repetitive element | Organism | Typec | Copy no. | Total amt of DNA (kb) | Proportion of genome (%) |
|---|---|---|---|---|---|---|
| 1 | 195-bp repeat | T. cruzi | T | 20,000 | 3,500 | 9 |
| 2 | 177-bp repeat | T. brucei | T | 15,000 | 3,000 | 8 |
| 3 | E13 | T. cruzi | I | 3,000 | 2,800 | 7 |
| 4 | L1Tc | T. cruzi | I, LINE | 500 | 2,500 | 6 |
| ingi | T. brucei | I, LINE | 400 | 2,000 | 6 | |
| 6 | E22 | T. cruzi | I | 1,400 | 1,400 | 3.5 |
| 7 | E12 | T. cruzi | I | 900 | 1,000 | 2.5 |
| RS1Tc | T. cruzi | I | 700 | 1,000 | 2.5 | |
| 9 | TcIRE | T. cruzi | I | 2,000 | 900 | 2 |
| 10 | Rep20 | P. falciparum | T | 12,000 | 250 | 1 |
See Table 1 for more information and references.
Elements are ranked according to the proportion of the host nuclear genome they are estimated to occupy. The T. brucei circular extrachromosomal NR-element is not ranked, but may occupy up to 5% of nuclear genome in some strains (10).
Abbreviations: I, interspersed; T, tandem.
According to the phylogeny of Malik et al. (123), the elements ingi of T. brucei (107, 132) and L1Tc of T. cruzi (124) belong to the I clade of LINEs, which includes non-site-specific I-elements of Drosophila (reviewed in reference 38). These LINEs encode domains with putative apurinic-apyrimidinic endonuclease, RT, and RNase H activities and display the 3′-oligo(dA) tail and insertion site duplication indicative of retroposons. Transcripts of both ingi and L1Tc have been detected (124, 132). Transcripts of ingi become much more abundant in bloodstream-form cells. This can be attributed to the clustering of ingi retroposons in the subtelomeric VSG expression sites that are active in bloodstream-form cells. Such clustering of ingi at T. brucei subtelomeres may also promote recombination at these specialized loci.
The elements SLACS of T. brucei (8) and CZAR of T. cruzi (195) are LINEs that encode proteins with putative RT and restriction enzyme-like endonuclease activities. Along with retroposons of other parasitic Trypanosomatidae—the two distinct CRE retroposon families of Crithidia fasiculata (CRE1 and CRE2 [78, 185]) and the LINS1 element of Leptomonas seymouri (21)—these elements are members of an ancient clade of LINEs which are site specific for miniexon (spliced leader) arrays. Despite extensive sequence divergence, all these elements have precisely the same insertion site (between nucleotides 11 and 12 of the 39-bp miniexon sequence) and make an apparently frugal living at only a few copies per genome. No transcripts of SLACS, CZAR, CRE1, or CRE2 have been detected, but a transposition frequency of ∼1% per generation has been estimated for CRE1 (78) and the occurrence of intact conserved enzymatic domains suggests that these retroposons are, or have recently been, transpositionally active.
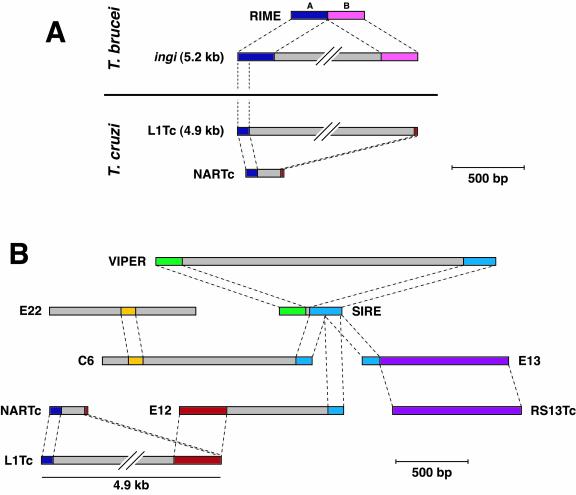
If LINEs are genomic parasites, then SINEs are parasites of parasites (200). Shorter and simpler, SINEs do not possess the necessary genes for autonomous retroactivity but, instead, piggyback on the RT and endonuclease activities encoded by a related LINE. Figure 2A shows the relationship between T. brucei ingi and RIME and T. cruzi L1Tc and NARTc retroposons. Sequence similarity at the 3′-end is typical between a LINE and its dependent SINE since it enables the SINE to recruit the machinery necessary for its retrotransposition. It is easy to see how a SINE can be born from incomplete reverse transcription of its parental LINE. Less explicable from the model of retroposon mobility is the sequence conservation observed at the 5′-extremity of the ingi-RIME and L1Tc-NARTc pairs. However, the canonical LINE-SINE relationship is only one of the many relationships that can be found between the interspersed elements of T. cruzi (Fig. 2B). It is likely that actively proliferating interspersed repeats (whether retroelements or otherwise) may pick up all kinds of passengers. How much of the shared sequence between T. cruzi repeats is due to the likelihood of encountering successful retroposons and how much the modules actually contribute to each individual element's (possible) fertility remains to be seen.
FIG. 2.
Relationships between interspersed repetitive elements in T. brucei and T. cruzi. (A) Homologous regions in T. brucei and T. cruzi LINEs and SINEs. The T. brucei retroposon ingi is bordered by two separate halves of the nonautonomous SINE, RIME (107, 132). The 5′ end of ingi is also conserved in the related T. cruzi LINE, L1Tc (35, 124). The first 77 bp of the T. cruzi SINE, NARTc, are identical to the 5′ extremity of L1Tc, and the two also have a small (13-bp) region of homology preceding the poly(A) tail (35). (B) Identified homologies between some of the interspersed elements of T. cruzi (13, 35, 124, 142, 161, 191, 193). Well-conserved regions (>70% identity at the nucleotide level) are shown in color. Poorly conserved or unrelated DNA is shown in grey.
The composite structure of interspersed elements may help explain the unusual retroelement VIPER of T. cruzi (11, 191). The reconstructed open reading frame of VIPER encodes domains with putative RT and RNase H activities. The encoded RT has significant similarity to LTR retrotransposons and no significant similarity to LINEs. However, its structure is atypical of retrotransposons and it lacks the eponymous LTRs. Instead, the 5′ and 3′ ends of VIPER are composed of parts of SIRE, a T. cruzi SINE element (193). It is unclear who is using whom in this relationship. Has the parental LTR retrotransposon of VIPER incorporated SIRE in place of terminal repeats, or is SIRE the dependant of VIPER?
SINEs exploit LINEs but are dependent on them, just as LINEs exploit the host genome. An infectious agent can (to a certain extent) afford to kill any individual host in the process of infecting others. However, SINEs and LINEs are captives and must live within the means of their host. On the other hand, a silent retroelement cannot remain fertile indefinitely; an element must multiply fast enough to offset the inevitable deaths of elements caused by mutation or recombination. This process leaves the corpses of individuals and whole families of retroelements strewn across the genomes of affected organisms (100). G. lamblia carries a chromosome-internal LINE family, GilD, which has a copy number ca. twofold higher than that of the two active subtelomeric retroposons GilM and GilT combined (15, 37). However, this element has blossomed and died: all copies contain multiple deletions, nucleotide substitutions and frameshifts (15). Similarly, live copies of T. cruzi VIPER have yet to be found (191). Of course, if SIRE is the daughter of another, still-living LINE, then VIPER may prove to be mobile even after death.
RETROELEMENTS AND RNA INTERFERENCE
RNA interference (RNAi) is the homology-dependent ablation of mRNA induced by small interfering RNAs that are produced by the processing of double-stranded RNAs. It has been demonstrated in many organisms, including Drosophila (87), mammals (62), Caenorhabditis (69), and protozoa (20, 137). RNAi is an endogenous mechanism for the posttranscriptional regulation of RNA levels which appears to play a defensive role against viruses (58, 131) and unfettered transposable element activity (103, 104, 180). In C. elegans, genetic mutations which inactivate RNAi are associated with activation of transposon mobility (180). Transposable elements often contain promoters for sense-strand transcription. However, as an element colonizes a genome, it is expected that it will become integrated downstream of external promoters in both sense and antisense orientations. Cotranscription from such loci produces complementary RNA capable of forming double-stranded RNAs and thus initiating an RNAi response. In this way, any reasonably “successful” nucleotide parasite experiences negative feedback that curtails its mobility.
Gene silencing by RNAi has been demonstrated in T. brucei (20, 137) and also T. congolense (99). When small interfering RNAs were recovered from T. brucei, fragments derived from ingi and SLACS were found to be very abundant despite the relatively low abundance of the respective mRNAs (55). This observation fits well with a possible role for RNAi in controlling the mobility of these retroposons. In contrast to the situation in T. brucei, efforts to induce RNAi against target genes in Leishmania have not met with any success, despite many attempts (24, 164). An association of RNAi with transposons and/or retroelements might explain the demonstration of RNAi in trypanosomes but its failure in Leishmania (which has no identified transposable elements). However, the situation may to be more complicated than this simplistic view, given some recent reports of RNAi effects in P. falciparum (122, 128). These experiments are proving hard to reproduce (M. J. Blackman, submitted for publication), but appear to show an active RNAi machinery in an organism with no transposable elements. One could speculate about the relative susceptibilities of sexual versus asexual organisms to colonizing retroelements (14, 23), but in the absence of more experimental data, the situation remains unresolved.
SATELLITES AND SPECIALIZATION
If the success of a repetitive element is measured purely in terms of DNA mass, then the macrosatellite repeats of trypanosomes are the real high flyers (Table 3). The 195-bp repeat of T. cruzi (170) is a chromosome-internal tandem repeat which is estimated to constitute a full 9% of the nuclear genome (84). Large rafts of the repeat occur on several large chromosomes (39, 63). Such repeat regions are presumably propagated by replicational slippage and gene conversion mechanisms, as is the case for other satellites (110, 186); however, unlike for subtelomeric satellites, no function has yet been suggested for these internal repetitive deserts.
Some of the best examples of tandem repeats with putative functions are found in T. brucei. The 177-bp repeat of T. brucei (170), like the 195-bp repeat of T. cruzi, is another extremely populous chromosome-internal satellite repeat (Table 3). However, it has no significant identity to the 195-bp repeat and has a totally different genomic location. The 177-bp repeat is confined to the minichromosomes and intermediate-sized chromosomes of the trypanosome (171). These chromosomal classes are devoid of housekeeping genes, instead carrying a library of subtelomeric VSG genes and VSG-ESs for use during antigenic variation (65) (Fig. 1). The 177-bp repeat forms a central core to minichromosomes that takes up ∼60% of their length (B. Wickstead, K. Ersfeld, and K. Gull, submitted for publication). The minichromosomal core region has an unusual palindromic structure in which direct 177-bp repeats run in from both subtelomeres to an inversion point near the center of the chromosome (Wickstead et al., submitted). The ubiquitous nature of the 177-bp repeat in T. brucei minichromosomes and the association of replication bubbles with the minichromosomal core region (199) suggest a function for this repeat in the maintenance of minichromosomes and intermediate-size chromosomes.
Other tandem repeats associated with particular genomic locations are the 50- and 70-bp repeats of T. brucei. The 50-bp repeat might be described as subtelomeric, although it may be tens of kilobase pairs distal from the telomeric repeats (22). The actual association of the 50-bp repeats is with bloodstream VSG-ESs, and large tracts of the repeat have been found upstream of the promoter in all VSG-ESs investigated to date (22, 118, 203). It could be that the repeats merely insulate the (RNA polymerase I-transcribed) expression site from promiscuous readthrough of RNA polymerase II, in which case they do so at a considerable distance from the immunodominant VSG gene. However, this association is suggestive of a more direct role for 50-bp repeats in the tight transcriptional control exerted on VSG-ESs in bloodstream form cells. It will be interesting to see how 50-bp repeats interact with the VSG expression body, the extranucleolar transcription factory associated with the active expression site (135).
The 70-bp repeat of T. brucei is found immediately upstream of VSG genes. Subtelomeric VSG gene copies have arrays of direct 70-bp repeats of several kilobase pairs, while most copies of the more populous chromosome-internal VSG genes possess a few repeat copies. This conspicuous organization makes 70-bp repeats an obvious site for homologous recombination to instigate antigenic variation, and, indeed, recombination events in these repeats are associated with VSG switching events (53, 119). It was perhaps surprising, then, when McCulloch et al. (127) demonstrated that VSG-ES 70-bp repeats are not essential for switching of the expressed VSG. However, removal or reorientation of the 70-bp repeats in the active VSG-ES alters the proportion of switches occurring via gene conversion events, and the 70-bp repeats supply the homology necessary to access the vast repertoire of VSG genes at minichromosomal subtelomeres or chromosome internal locations (127). Another interesting feature of the 70-bp repeats is that the repeats are rather heterogeneous: very few long stretches of perfect homology exist between arrays (32). Divergence of ES sequences may provide an explanation for the variant-specific switching rates observed in T. brucei, which result in the appearance of VSGs in a statistically preferred order.
CENTROMERIC SATELLITES
No discussion of satellite DNA would be complete without the mention of centromeric DNAs. The centromeres of human, Drosophila, Arabidopsis thaliana, and Schizosaccharomyces pombe are characterized by large tracts of tandem repeats embedded in heterochomatic regions (reviewed in references 45, 177, and 178). In human and S. pombe cells, some repeats are ubiquitous to all centromeres (although not restricted to them), and the same may be true for Drosophila and A. thaliana. What is intriguing about the satellites—both subtelomeric and chromosome internal—of the parasitic protozoa discussed here is that none of them are strong candidates for putative centromeric function: the subtelomeric repeats of plasmodia appear to be dispensable for mitotic function; the 50-bp repeats of T. brucei do not appear to be common to all chromosomes and are adjacent to strong transcriptional units; the 195-bp repeats of T. cruzi are specific to only a subset of chromosomes; and the relatively large chromosomes of G. lamblia (1 to 4 Mb) lack satellite DNA altogether. One possible exception is the 177-bp repeat palindrome of T. brucei, which may have a specialized function in the segregation of the numerous small chromosomes (Wickstead et al., submitted). It has been suggested that telomeres might function as centromeres in T. brucei (148), but in situ hybridization analysis shows some telomeric signal trails behind the majority of DNA during segregation (140). Similarly, in Leishmania, the 272-bp repeat has been implicated in the mitotic stability of artifical chromosomes (59). However, the presence of subtelomeric repeats does not seem to be sufficient for chromosomal stability in this species (181).
Of course, a centromere does not have to be a large repetitive region; the point centromeres of S. cerevisiae are the extensively-studied example (177, 178). However, the centromeres of S. cerevisiae do not assemble kinetochores that are visible by electron microscopy (97), while the mitotic nuclei of P. falciparum, Leishmania, T. brucei, and T. cruzi possess ∼100-nm-long electron-dense laminar structures that are most probably kinetochores (140, 154, 172, 173, 188). The sequences around which these plaques are constructed is unknown, but it is noteworthy that their numbers are significantly smaller than the number of chromosomes in these species. This indicates the presence of diverse mechanisms of chromosome segregation within one nucleus (86).
Alternatively, the centromeres of these species may not be constructed around common high-copy-number sequence elements. The generation of completed sequence for P. falciparum chromosomes 2 and 3 (33, 80) led to the identification of putative centromeres (33). Both putative centromeres were extremely AT-rich regions of DNA (97% AT, compared to ∼82% genomewide), 2 to 3 kb in size, and composed of families of low-copy-number divergent tandem repeats which were not conserved between chromosomes. The recent completion of the P. falciparum genome has led to similar regions being identified on 11 of 14 chromosomes (79); the remaining 3 possess sequencing gaps. It is possible that in these organisms, a variety of noncoding DNA is able to provide necessary centromeric activity if the required epigenetic factors are in place.
USING REPEATS FOR PARASITE IDENTIFICATION, TRAIT MAPPING, AND PHYLOGENETICS
The diversity and dynamism of repetitive DNA can be a valuable asset to the experimentalist. The hyperevolution experienced by repeats means that many are specific to an individual species or a clade of related species. Moreover, some such repeats exist at copy numbers several thousand times that of individual gene markers. These factors have inspired the development of many diagnostic probes based on minisatellite DNA (76, 84, 93, 121, 149). Restriction fragment length polymorphism (RFLP) analysis, combining repeat hybridization with analysis of the locus length, allows an even closer inspection of a parasite's origins, distinguishing between strains and aiding the analysis of genetic crosses (18, 42, 56, 61, 157). The final level of information is accessed by sequencing repetitive elements. Piarroux et al. (150) have used a repeat sequence to investigate the phylogenetic relationships between Old World Leishmania species. The number of informative sites in an alignment of rapidly diverging sequences is much greater for closely related species than for slow-moving phylogenetic standards such as 18S rDNA (190), and even intraspecies relationships can be investigated. Analysis of the divergence of the T. cruzi 195-bp repeat provides strong evidence supporting a hybrid origin for the T. cruzi CL Brener strain (S. Schenkman, personal communication), data backed up by the recent demonstration of genetic exchange in T. cruzi (81). Such work elegantly demonstrates the power of repetitive sequences in close-range phylogenetic inference.
RFLP analysis superseded more time-consuming techniques, such as isoenzyme analysis, as the method of choice for parasite strain identification and genetic analysis (175). In its turn, RFLP is being superseded by PCR-based simple sequence length polymorphism (SSLP) analysis, as has already occurred in the analysis of mammalian systems (66, 102). Simple sequence repeats or microsatellites are a ubiquitous feature of eukaryotic genomes (183), although their density varies between species. Genomically speaking, microsatellites are a local issue and do not exert the long-range influence that can be associated with large satellites. They arise spontaneously by slippage during DNA replication (110, 186). Hence, the identity of a microsatellite depends largely on the base pair bias of a genome and the probability of a seed repeat occurring (which is less likely for longer repeat units). In P. falciparum (82% A+T), the microsatellites (T)n, (TA)n, and (TAA)n predominate (66, 176), while in L. major (37% A+T), (CA)n, (AG)n, (TA)n, (AGG)n, (CAG)n, and (TGG)n have been described at multiple loci (101, 166). SSLPs arise as the microsatellites expand and contract over many generations. Unlike the minisatellites, which are often associated with specific polymorphic loci (e.g., the subtelomeres and the VSG-ES), microsatellite “blooms” are dispersed across the genome. As a result, polymorphic loci in one species are frequently monomorphic in related organisms (101).
One of the most exciting features of SSLP analysis is that it can feed off information from the genome-sequencing projects and expand to be a whole-genome approach. Microsatellite markers can then be used to genetically map DNA sequences that contribute to heritable phenotypes in any organism that undergoes sexual recombination. Although trait mapping is not possible in asexual (or rarely sexual) organisms such as Leishmania and Giardia, a high-resolution linkage map consisting of hundreds of microsatellite markers has been developed for P. falciparum (66, 176).
THE PURPOSE OF REPEATS
The repetitive parts of eukaryotic genomes (in particular the transposable elements) are often referred to as selfish DNA (57, 144). This terminology is rather tautological, since any DNA element, whether genic or otherwise, can be viewed as entirely selfish in an evolutionary sense (50). It is in the nature of evolution that units of replication that best exploit their environmental conditions are most successful. The “purpose” of any unit of replication is to become replicated into more copies than are competing units. Whether this is achieved through phenotypic selection of organisms or reiteration within a genome is largely irrelevant (from the point of view of the unit of replication). Of course, the selfish-DNA theory is more than just a statement of competition between DNAs. Its power lies in the idea that genomic elements may be parasites within their host genomes at the expense of overall genomic fitness (however that may be defined).
Repeats can be the beneficiaries of three types of reproduction: (i) organismic reproduction; (ii) intragenomic replication; and (iii) transmission to virgin genomes. Strict allelic elements are confined to the first of these types of reproduction. Since repeats have additional options for replication, it is possible that the investment of a repeat in organismic survival and reproduction (and, interestingly, sexual status [14, 23]) will differ from that of other parts of the genome (although neither is “altruistic” toward the organism or the rest of the genome). This would make such elements true genomic “outlaws” (51), and modifications may well arise elsewhere in the genome that work to prevent repetitive elements having things entirely their own way, as is suggested for RNAi (103, 104, 180).
Elements such as retroviruses that may kill their host organism and still proliferate are clearly in conflict with the aims of their host genomes and truly deserve the title of genomic parasites. However, the repeats in the genomes of the parasitic protozoa detailed here do not appear capable of transmission between genomes (except via sexual conjugation). In such a closed system, if intragenomic replication is relatively slow or strongly selected against at the organismic level, the interests of “selfish” elements should come to closely resemble those of the host genes. Under these conditions, it is difficult to see how an entirely detrimental repeat could be maintained. There is now a growing acceptance that the status of retroposons with respect to their host genomes may more closely resemble symbiosis than parasitism (36, 46, 82, 105, 117)—“useful parasites” as described by Weiner (200).
Interestingly, nontransposable repeats have often escaped the stigma of selfishness. In contrast to the situation for transposable elements, there can be a tendency to look for a “function” for any satellite DNA. For obvious reasons, few would talk of selfish telomeres or selfish centromeres. Despite this, both may have had a “selfish” origin. Moreover, centromeric elements, such as the α-satellite repeat of humans, often extend farther than necessary for kinetochore assembly (197, 202) and also spill out into noncentromeric regions where their benefit cannot be so easily ascribed (98, 100). Although they do not encode proteins that actively participate in transposition, tandem repeats can still proliferate by encouraging replicational slippage or gene conversion (for instance, through chromosomal clustering, as has been suggested for Rep20 [139]). Hence, it is not necessary to invoke positive selection to explain the appearance of satellite DNAs, even though selective pressure may later be brought to bear.
Repeats play integral parts in ongoing genomic evolution and can play diverse roles at different times (36, 105). Elements which may have proliferated through some “parasitic” path may later prove to be useful and become subject to selective pressure (46, 105, 117). What is clear is that repeats tend to impart a greater changeability to genomes. When an organism faces a changeable environment, the advantages of genomic flexibility (particularly if it can be contained at specific loci) may outweigh the extra cost of replication and any mutagenising tendency exerted on other genomic regions. This may explain the relatively high content of repeats and mobile elements in the genomes of free-living bacteria compared to those of obligate intracellular bacteria whose environments are much more stable (70). Perhaps it is not sufficient to think of repeats in terms of either selfish or altruistic intentions. Repetitive elements may not be essential and may not always have an organism's best interests at heart, but they can be co-opted into specific cellular functions and may add an important genetic flexibility to the genomes they inhabit.
Acknowledgments
We thank Chris Newbold (Institute of Molecular Medicine, John Radcliffe Hospital, Oxford, United Kingdom), John Kelly (Department of Infectious and Tropical Diseases, London School of Hygiene and Tropical Medicine, London, United Kingdom), Frédéric Bringaud (Laboratoire de Parasitologie Moléculaire, Université Victor Segalen Bordeaux II, Bordeaux, France), and Sergio Schenkmann (Disciplina de Biologia Celular, UNIFESP, São Paulo, Brazil) for their helpful comments on early drafts of this review.
This work was funded by grants from the Wellcome Trust.
REFERENCES
- 1.Adam, R. D. 1992. Chromosome-size variation in Giardia lamblia: the role of rDNA repeats. Nucleic Acids Res. 20:3057-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adam, R. D. 2000. The Giardia lamblia genome. Int. J. Parasitol. 30:475-484. [DOI] [PubMed] [Google Scholar]
- 3.Adam, R. D. 2001. Biology of Giardia lamblia. Clin. Microbiol. Rev. 14:447-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adam, R. D., A. Aggarwal, A. A. Lal, V. F. de la Cruz, T. F. McCutchan, and T. E. Nash. 1988. Antigenic variation of a cysteine-rich protein in Giardia lamblia. J. Exp. Med. 167:109-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adam, R. D., T. E. Nash, and T. E. Wellems. 1991. Telomeric location of Giardia rDNA genes. Mol. Cell. Biol. 11:3326-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agüero, F., R. E. Verdún, A. C. C. Frasch, and D. O. Sánchez. 2000. A random sequencing approach for the analysis of the Trypanosoma cruzi genome: general structure, large gene and repetitive DNA families, and gene discovery. Genome Res. 10:1996-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ajioka, J. W., J. C. Boothroyd, B. P. Brunk, A. Hehl, L. Hillier, I. D. Manger, M. Marra, G. C. Overton, D. S. Roos, K.-L. Wan, R. Waterston, and L. D. Sibley. 1998. Gene discovery by EST sequencing in Toxoplasma gondii reveals sequences restricted to the Apicomplexa. Genome Res. 8:18-28. [DOI] [PubMed] [Google Scholar]
- 8.Aksoy, S., T. M. Lalor, J. Martin, L. H. T. van der Ploeg, and F. F. Richards. 1987. Multiple copies of a retroposon interrupt spliced leader genes in the African trypanosome Trypanosoma brucei gambiense. EMBO J. 6:3819-3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aksoy, S., S. P. Williams, S. Chang, and F. F. Richards. 1990. SLACS retrotransposons form Trypanosoma brucei gambiense is similar to mammalian LINEs. Nucleic Acids Res. 18:785-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alsford, N. S., M. Navarro, H. R. Jamnadass, H. Dunbar, M. Ackroyd, M. Murphy, K. Gull, and K. Ersfeld. 2003. The identification of circular extrachromosomal DNA in the nuclear genome of Trypanosoma brucei. Mol. Microbiol. 47:277-289. [DOI] [PubMed] [Google Scholar]
- 11.Andersson, B., L. Åslund, M. Tammi, A. N. Tran, J. D. Hoheisel, and U. Pettersson. 1998. Complete sequence of a 93.4-kb contig from chromosome 3 of Trypanosoma cruzi containing a strand-switch region. Genome Res. 8:809-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderssson, J. O., A. M. Sjögren, L. A. M. Davis, T. M. Embley, and A. J. Roger. 2003. Phylogenetic analysis of diplomonad genes reveal frequent lateral gene transfers affecting eukaryotes. Curr. Biol. 13:94-104. [DOI] [PubMed] [Google Scholar]
- 13.Araya, J., M. I. Cano, H. B. M. Gomes, E. M. Novak, J. M. Requena, C. Alonso, M. J. Levin, P. Guevara, J. L. Ramirez, and J. F. da Silveira. 1997. Characterization of an interspersed repetitive DNA element in the genome of Trypanosoma cruzi. Parasitology 115:563-570. [DOI] [PubMed] [Google Scholar]
- 14.Arkhipova, I., and M. Meselson. 2000. Transposable elements in sexual and ancient asexual taxa. Proc. Natl. Acad. Sci. USA 97:14473-14477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arkhipova, I. R., and H. G. Morrison. 2001. Three retrotransposon families in the genome of Giardia lamblia: Two telomeric, one dead. Proc. Natl. Acad. Sci. USA 98:14497-14502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Åslund, L., L. Franzén, G. Westin, T. Persson, H. Wigzell, and U. Pettersson. 1985. Highly reiterated non-coding sequence in the genome of Plasmodium falciparum is composed of 21 base-pair tandem repeats. J. Mol. Biol. 185:509-516. [DOI] [PubMed] [Google Scholar]
- 17.Barker, R. H., L. Suebsaeng, W. Rooney, G. C. Alecrim, H. V. Dourado, and D. F. Wirth. 1986. Specific DNA probe for the diagnosis of Plasmodium falciparum malaria. Science 231:1434-1436. [DOI] [PubMed] [Google Scholar]
- 18.Barrett, M. P., A. MacLeod, J. Tovar, J. P. Sweetman, A. Tait, R. W. F. LePage, and S. E. Melville. 1997. A single locus minisatellite sequence which distinguishes between Trypanosoma brucei isolates. Mol. Biochem. Parasitol. 86:95-99. [DOI] [PubMed] [Google Scholar]
- 19.Barry, J. D., M. L. Ginger, and R. McCulloch. 2003. Why are parasite contingency genes often associated with telomeres? Int. J. Parasitol. 33:29-45. [DOI] [PubMed] [Google Scholar]
- 20.Bastin, P., T. Sherwin, and K. Gull. 1998. Paraflagellar rod is vital for trypanosome motility. Nature 391:548-548. [DOI] [PubMed] [Google Scholar]
- 21.Bellofatto, V., R. Cooper, and G. A. M. Cross. 1988. Discontinuous transcription in Leptomonas seymouri: presence of intact and interrupted mini-exon gene families. Nucleic Acids Res. 16:7437-7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berriman, M., N. Hall, K. Sheader, F. Bringaud, B. Tiwari, T. Isobe, S. Bowman, C. Corton, L. Clark, G. A. M. Cross, M. Hoek, T. Zanders, M. Berberof, P. Borst, and G. Rudenko. 2002. The architecture of variant surface glycoprotein gene expression sites in Trypanosoma brucei. Mol. Biochem. Parasitol. 122:131-140. [DOI] [PubMed] [Google Scholar]
- 23.Bestor, T. H. 2000. Sex brings transposons and genomes into conflict. Genetica 107:289-295. [PubMed] [Google Scholar]
- 24.Beverley, S. M. 2003. Protozomics: trypanosomatid parasite genetics comes of age. Nat. Rev. Genet. 4:11-19. [DOI] [PubMed] [Google Scholar]
- 25.Bhattacharya, S., A. Bakre, and A. Bhattacharya. 2002. Mobile genetic elements in protozoan parasites. J. Genet. 81:73-86. [DOI] [PubMed] [Google Scholar]
- 26.Biessmann, H., L. E. Champion, M. O'Hair, K. Ikenaga, B. Kasravi, and J. M. Mason. 1992. Frequent transpositions of Drosophila melanogaster HeT-A transposable elements to receding chromosome ends. EMBO J. 11:4459-4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biessmann, H., K. Valgeirsdottir, A. Lofsky, C. Chin, B. Ginther, R. W. Levis, and M. L. Pardue. 1992. HeT-A, a transposable element specifically involved in ‘healing' broken chromosome ends in Drosophila melanogaster. Mol. Cell. Biol. 12:3910-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blackburn, E. H., and P. B. Challoner. 1984. Identification of a telomeric DNA sequence in Trypanosoma brucei. Cell 36:447-457. [DOI] [PubMed] [Google Scholar]
- 29.Reference deleted.
- 30.Borst, P., W. Bitter, P. A. Blundell, I. Chaves, M. Cross, H. Gerrits, F. van Leeuwen, R. McCulloch, M. Taylor, and G. Rudenko. 1998. Control of VSG gene expression in Trypanosoma brucei. Mol. Biochem. Parasitol. 91:67-76. [DOI] [PubMed] [Google Scholar]
- 31.Borst, P., W. Bitter, R. McCulloch, F. van Leeuwen, and G. Rudenko. 1995. Antigenic variation in malaria. Cell 82:1-4. [DOI] [PubMed] [Google Scholar]
- 32.Borst, P., G. Rudenko, M. C. Taylor, P. A. Blundell, F. van Leeuwen, W. Bitter, M. Cross, and R. McCulloch. 1996. Antigenic variation in trypanosomes. Arch. Med. Res. 27:379-388. [PubMed] [Google Scholar]
- 33.Bowman, S., D. Lawson, D. Basham, D. Brown, T. Chillingworth, C. M. Churcher, A. Craig, R. M. Davies, K. Devlin, T. Feltwell, S. Gentles, R. Gwilliam, N. Hamlin, D. Harris, S. Holroyd, T. Hornsby, P. Horrocks, K. Jagels, B. Jassal, S. Kyes, J. McLean, S. Moule, K. Mungall, L. Murphy, K. Oliver, M. A. Quail, M.-A. Rajandream, S. Rutter, J. Skelton, R. Squares, S. Squares, J. E. Sulston, S. Whitehead, J. R. Woodward, C. Newbold, and B. G. Barrell. 1999. The complete nucleotide sequence of chromosome 3 of Plasmodium falciparum. Nature 400:532-538. [DOI] [PubMed] [Google Scholar]
- 34.Bringaud, F., N. Biteau, S. E. Melville, S. Hez, N. M. El-Sayed, V. Leech, M. Berriman, N. Hall, J. E. Donelson, and T. Baltz. 2002. A new, expressed multigene family containing a hot spot for insertion of retroelements is associated with polymorphic subtelomeric regions of Trypanosoma brucei. Eukaryot. Cell 1:137-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bringaud, F., J. L. García-Pérez, S. R. Heras, E. Ghedlin, N. M. El-Sayed, B. Andersson, T. Baltz, and M. C. López. 2002. Identification of non-autonomous non- LTR retrotransposons in the genome of Trypanosoma cruzi. Mol. Biochem. Parasitol. 124:73-78. [DOI] [PubMed] [Google Scholar]
- 36.Brosius, J. 1999. Genomes were forged by massive bombardments with retroelements and retrosequences. Genetica 107:209-238. [PubMed] [Google Scholar]
- 37.Burke, W. D., H. S. Malik, S. M. Rich, and T. H. Eickbush. 2002. Ancient lineages of non-LTR retrotransposons in the primitive eukaryote, Giardia lamblia. Mol. Biol. Evol. 19:619-630. [DOI] [PubMed] [Google Scholar]
- 38.Busseau, I., M.-C. Chaboissier, A. Pélisson, and A. Bucheton. 1994. I factors in Drosophila melanogaster: transposition under control. Genetica 93:101-116. [DOI] [PubMed] [Google Scholar]
- 39.Cano, M. I., A. Gruber, M. Vazquez, A. Cortés, M. J. Levin, A. González, W. Degrave, E. Rondinelli, B. Zingales, J. L. Ramirez, C. Alonso, J. M. Requena, and J. F. da Silveira. 1995. Molecular karyotype of clone CL Brener chosen for the Trypanosoma cruzi genome project. Mol. Biochem. Parasitol. 71:273-278. [DOI] [PubMed] [Google Scholar]
- 40.Cano, M. I. N. 2001. Telomere biology of trypanosomatids: more questions than answers. Trends Parasitol. 17:425-429. [DOI] [PubMed] [Google Scholar]
- 41.Capy, P. 1998. Classification of transposable elements, p. 37-52. In P. Capy, C. Bazin, D. Hiquet, and T. Langin (ed.), Molecular biology intelligence unit: dynamics and evolution of transposable elements. Landes Bioscience, Georgetown, Tex.
- 42.Carnaby, S., P. D. Butcher, C. D. Summerbell, A. Naeem, and M. J. G. Farthing. 1995. Minisatellites corresponding to the human polycore probe 33.6 and 33.15 in the genome of the most ‘primitive’ known eukaryote Giardia lamblia. Gene 166:167-172. [DOI] [PubMed] [Google Scholar]
- 43.Castro, C., S. P. Craig, and M. Castañeda. 1981. Genome organisation and ploidy number in Trypanosoma cruzi. Mol. Biochem. Parasitol. 4:273-282. [DOI] [PubMed] [Google Scholar]
- 44.Chiurillo, M. A., I. Cano, J. F. da Silveira, and J. L. Ramirez. 1999. Organization of telomeric and sub-telomeric regions of chromosomes from the protozoan parasite Trypanosoma cruzi. Mol. Biochem. Parasitol. 100:173-183. [DOI] [PubMed] [Google Scholar]
- 45.Choo, K. H. A. 1997. Centromere DNA dynamics: latent centromeres and neocentromere formation. Am. J. Hum. Genet. 61:1225-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chu, W. M., R. Ballard, B. W. Carpick, B. R. Williams, and C. W. Schmid. 1998. Potential Alu function: regulation of the activity of double-stranded RNA-activated kinase PKR. Mol. Cell. Biol. 18:58-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Collins, K. 2000. Mammalian telomeres and telomerase. Curr. Opin. Cell Biol. 12:378-383. [DOI] [PubMed] [Google Scholar]
- 48.Cross, G. A. M. 1996. Antigenic variation in trypanosomes: secrets surface slowly. Bioessays 18:283-291. [DOI] [PubMed] [Google Scholar]
- 49.Cross, G. A. M., L. E. Wirtz, and M. Navarro. 1998. Regulation of vsg expression site transcription and switching in Trypanosoma brucei. Mol. Biochem. Parasitol. 91:77-91. [DOI] [PubMed] [Google Scholar]
- 50.Dawkins, R. 1976. The selfish gene. Oxford University Press, Oxford, United Kingdom.
- 51.Dawkins, R. 1982. The extended phenotype, p.133-155. Oxford University Press, Oxford, United Kingdom.
- 52.de Bruin, D., M. Lanzer, and J. V. Ravetch. 1994. The polymorphic subtelomeric regions of Plasmodium falciparum chromosomes contain arrays of repetitive sequence elements. Proc. Natl. Acad. Sci. USA 91:619-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Lange, T., J. M. Kooter, J. Luirink, and P. Borst. 1985. Transcription of a transposed trypanosome surface antigen gene starts upstream of the transposed segment. EMBO J. 4:3299-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dietrich, P., M. B. Soares, M. H. T. Affonso, and L. M. Floeter-Winter. 1993. The Trypanosoma cruzi ribosomal RNA-encoding gene: analysis of promoter and upstream intergenic spacer sequences. Gene 125:103-107. [DOI] [PubMed] [Google Scholar]
- 55.Djikeng, A., H. Shi, C. Tschudi, and E. Ullu. 2001. RNA interference in Trypansoma brucei: cloning of small interfering RNAs provides evidence for retroposon-derived 24- 26 nucleotide RNAs. RNA 7:1522-1530. [PMC free article] [PubMed] [Google Scholar]
- 56.Dolan, S. A., J. A. Herrfeldt, and T. E. Wellems. 1993. Restriction polymorphisms and fingerprint patterns from an interspersed repetitive element of Plasmodium falciparum DNA. Mol. Biochem. Parasitol. 61:137-142. [DOI] [PubMed] [Google Scholar]
- 57.Doolittle, W. F., and C. Sapienza. 1980. Selfish genes, the phenotype paradigm and genome evolution. Nature 284:601-603. [DOI] [PubMed] [Google Scholar]
- 58.Dougherty, W. G., J. A. Lindbo, H. A. Smith, T. D. Parks, S. Swaney, and W. M. Proebsting. 1994. RNA-mediated virus resistance in transgenic plants: exploitation of a cellular pathway possibly involved in RNA degradation. Mol. Plant-Microbe Interact. 7:544-552. [PubMed] [Google Scholar]
- 59.Dubessay, P., C. Ravel, P. Bastien, K. Stuart, J.-P. Dedet, C. Blaineau, and M. Pagès. 2002. Mitotic stability of a coding DNA sequence-free version of Leishmania major chromosome 1 generated by targeted chromosome fragmentation. Gene 289:151-159. [DOI] [PubMed] [Google Scholar]
- 60.Dunn, B., P. Szauter, M. L. Pardue, and J. W. Szostak. 1984. Transfer of yeast telomeres to linear plasmid by recombination. Cell 39:191-201. [DOI] [PubMed] [Google Scholar]
- 61.Echeverria, P. C., P. A. Rojas, V. Martin, E. A. Guarnera, V. Pszenny, and S. O. Angel. 2000. Characterisation of a novel interspersed Toxoplasma gondii DNA repeat with potential uses for PCR diagnosis and PCR-RFLP analysis. FEMS Microbiol. Lett. 184:23-27. [DOI] [PubMed] [Google Scholar]
- 62.Elbashir, S. M., W. Lendeckel, and T. Tuschl. 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15:188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Elias, M. C. Q. B., N. S. Vargas, B. Zingales, and S. Schenkman. 2003. Organization of satellite DNA in the genome of Trypanosoma cruzi. Mol. Biochem. Parasitol. 129:1-9. [DOI] [PubMed] [Google Scholar]
- 64.Embley, T. M., and R. P. Hirt. 1998. Early branching eukaryotes? Curr. Opin. Genet. Dev. 8:624-629. [DOI] [PubMed] [Google Scholar]
- 65.Ersfeld, K., S. E. Melville, and K. Gull. 1999. Nuclear and genome organization of Trypanosoma brucei. Parasitol. Today 15:58-63. [DOI] [PubMed] [Google Scholar]
- 66.Ferdig, M. T., and X.-Z. Su. 2000. Microsatellite markers and genetic mapping in Plasmodium falciparum. Parasitol. Today 16:307-312. [DOI] [PubMed] [Google Scholar]
- 67.Figueiredo, L. M., L. H. Freitas-Junior, E. Bottius, J.-C. Olivo-Marin, and A. Scherf. 2002. A central role for Plasmodium falciparum subtelomeric regions in spatial positioning and telomere length regulation. EMBO J. 21:815-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Figueiredo, L. M., L. A. Pirritt, and A. Scherf. 2000. Genomic organisation and chromatin structure of Plasmodium falciparum chromosome ends. Mol. Biochem. Parasitol. 106:169-174. [DOI] [PubMed] [Google Scholar]
- 69.Fire, A., S. Xu, M. K. Mongomery, S. A. Kostas, S. E. Driver, and C. C. Mello. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806-811. [DOI] [PubMed] [Google Scholar]
- 70.Frank, A. C., H. Amiri, and S. G. E. Andersson. 2002. Genome deterioration: loss of repeated sequences and accumulation of junk DNA. Genetica 115:1-12. [DOI] [PubMed] [Google Scholar]
- 71.Freitas-Junior, L. H., E. Bottius, L. A. Pirrit, K. W. Deitsch, C. Scheidig, F. Guinet, U. Nehrbass, T. E. Wellems, and A. Scherf. 2000. Frequent ectopic recombination of virulence factor genes in telomeric clusters of P. falciparum. Nature 407:1018-1022. [DOI] [PubMed] [Google Scholar]
- 72.Freitas-Junior, L. H. G., R. M. Porto, L. A. Pirrit, S. Schenkman, and A. Scherf. 1999. Identification of the telomere in Trypanosoma cruzi reveals highly heterogeneous telomere lengths in different parasite strains. Nucleic Acids Res. 27:2451-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Frontali, C. 1994. Genome plasticity in Plasmodium. Genetica 94:91-100. [DOI] [PubMed] [Google Scholar]
- 74.Fu, G. L., and D. C. Barker. 1998. Characterisation of Leishmania telomeres reveals unusual telomeric repeats and conserved telomere-associated sequence. Nucleic Acids Res. 26:2161-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fu, G. L., and D. C. Barker. 1998. Rapid cloning of telomere-associated sequence using primer-tagged amplification. BioTechniques 24:386-390. [DOI] [PubMed] [Google Scholar]
- 76.Fu, G. L., G. Perona-Wright, and D. C. Barker. 1998. Leishmania braziliensis: Characterisation of a complex specific subtelomeric repeat sequence and its use in the detection of parasites. Exp. Parasitol. 90:236-243. [DOI] [PubMed] [Google Scholar]
- 77.Funes, S., E. Davidson, A. Reyes-Prieto, S. Magallón, P. Herion, M. P. King, and D. González-Halphen. 2002. A green algal apicoplast ancestor. Science 298:2155. [DOI] [PubMed] [Google Scholar]
- 78.Gabriel, A., T. J. Yen, D. C. Schwartz, C. L. Smith, J. D. Boeke, B. Sollner-Webb, and D. W. Cleveland. 1990. A rapidly rearranging retrotransposon within the mini-exon gene locus of Crithidia fasciculata. Mol. Cell. Biol. 10:615-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gardner, M. J., N. Hall, E. Fung, O. White, M. Berriman, R. W. Hyman, J. M. Carlton, A. Pain, K. E. Nelson, S. Bowman, I. T. Paulsen, K. James, J. A. Eisen, K. Rutherford, S. Salzberg, A. Craig, S. Kyes, M.-S. Chan, V. Nene, S. Shallom, B. Suh, J. Peterson, S. Angiuoli, M. Pertea, J. Allen, J. Selengut, D. Haft, M. W. Mather, A. B. Vaidya, D. M. A. Martin, A. H. Fairlamb, M. J. Fraunholz, D. S. Roos, S. A. Ralph, G. I. McFadden, L. M. Cummings, G. M. Subramanian, C. Mungall, J. C. Venter, D. J. Carucci, S. L. Hoffman, C. Newbold, R. W. Davies, C. M. Fraser, and B. Barrell. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419:498-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gardner, M. J., H. Tettelin, D. J. Carucci, L. M. Cummings, L. Aravind, E. V. Koonin, S. Shallom, T. Mason, K. Yu, C. Fujii, J. Pederson, K. Shen, J. P. Jing, C. Aston, Z. W. Lai, D. C. Schwartz, M. Pertea, S. Salzberg, L. X. Zhou, G. G. Sutton, R. Clayton, O. White, H. O. Smith, C. M. Fraser, M. D. Adams, J. C. Venter, and S. L. Hoffman. 1998. Chromosome 2 sequence of the human malaria parasite Plasmodium falciparum. Science 282:1126-1132. [DOI] [PubMed] [Google Scholar]
- 81.Gaunt, M. W., M. Yeo, I. A. Frame, J. R. Stothard, H. J. Carrasco, M. C. Taylor, S. S. Mena, P. Veazey, G. A. J. Miles, N. Acosta, A. R. de Arias, and M. A. Miles. 2003. Mechanisms of genetic exchange in American trypanosomes. Nature 421:936-939. [DOI] [PubMed] [Google Scholar]
- 82.Georgiev, G. P. 1984. Mobile genetic elements in animal cells and their biological significance. Eur. J. Biochem. 145:203-220. [DOI] [PubMed] [Google Scholar]
- 83.Gilson, E., T. Laroche, and S. M. Gasser. 1993. Telomeres and the functional architecture of the nucleus. Trends Cell Biol. 3:128-134. [DOI] [PubMed] [Google Scholar]
- 84.González, A., E. Prediger, M. E. Huecas, N. Nogueira, and P. M. Lizardi. 1984. Minichromosomal repetitive DNA in Trypanosoma cruzi: its use in a high-sensitivity parasite detection assay. Proc. Natl. Acad. Sci. USA 81:3356-3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gueiros-Filho, F. J., and S. M. Beverley. 1997. Trans-kingdom transposition of the Drosophila element mariner within the protozoan Leishmania. Science 276:1716-1719. [DOI] [PubMed] [Google Scholar]
- 86.Gull, K., S. Alsford, and K. Ersfeld. 1998. Segregation of minichromosomes in trypanosomes: implications for mitotic mechanisms. Trends Microbiol. 6:319-323. [DOI] [PubMed] [Google Scholar]
- 87.Hammond, S. M., E. Bernstein, D. Beach, and G. J. Hannon. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404:293-296. [DOI] [PubMed] [Google Scholar]
- 88.Hannaert, V., E. Saavedra, F. Duffieux, J.-P. Szikora, D. J. Rigden, P. A. M. Michels, and F. R. Opperdoes. 2003. Plant-like traits associated with metabolism of Trypanosoma parasites. Proc. Natl. Acad. Sci. USA 100:1067-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Haring, E., S. Hagemann, and W. Pinsker. 2000. Ancient and recent horizontal invasions of Drosophilids by P elements. J. Mol. Evol. 51:577-586. [DOI] [PubMed] [Google Scholar]
- 90.Hasan, G., M. Turner, and J. S. Cordingley. 1984. Complete nucleotide sequence of an unusual mobile element from Trypanosoma brucei. Cell 37:333-341. [DOI] [PubMed] [Google Scholar]
- 91.Homan, W. L., M. Vercammen, J. De Braekeleer, and H. Verschueren. 2000. Identification of a 200-to 300-fold repetitive 529 bp DNA fragment in Toxoplasma gondii, and its use for diagnostic and quantitative PCR. Int. J. Parasitol. 30:69-75. [DOI] [PubMed] [Google Scholar]
- 92.Hou, G., S. M. le Blancq, Y. E, H. Zhu, and M. G.-S. Lee. 1995. Structure of a frequently rearranged rRNA-encoding chromosome in Giardia lamblia. Nucleic Acids Res. 23:3310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Howard, M. K., J. M. Kelly, R. P. Lane, and M. A. Miles. 1991. A sensitive repetitive DNA probe that is specific to the Leishmania donovani complex and its use as an epidemiologic and diagnostic reagent. Mol. Biochem. Parasitol. 44:63-72. [DOI] [PubMed] [Google Scholar]
- 94.Huang, Y. 2002. Transcriptional silencing in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Nucleic Acids Res. 30:1465-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hull, R., and H. Will. 1989. Molecular biology of viral and non-viral retroelements. Trends Genet. 5:357-359. [DOI] [PubMed] [Google Scholar]
- 96.Hurst, G. D. D., and J. H. Werren. 2001. The role of selfish genetic elements in eukaryotic evolution. Nat. Rev. Genet. 2:597-606. [DOI] [PubMed] [Google Scholar]
- 97.Hyman, A. A., and P. K. Sorger. 1995. Structure and function of kinetochores in budding yeast. Annu. Rev. Cell Dev. Biol. 11:471-495. [DOI] [PubMed] [Google Scholar]
- 98.Ikeno, M., H. Masumoto, and T. Okazaki. 1994. Distribution of CENP-B boxes reflected in CREST centromere antigenic sites on long-range α-satellite DNA arrays of human chromosome 21. Hum. Mol. Genet. 3:1245-1257. [DOI] [PubMed] [Google Scholar]
- 99.Inoue, N., K. Otsu, D. M. Ferraro, and J. E. Donelson. 2002. Tetracycline-regulated RNA interference in Trypanosoma congolense. Mol. Biochem. Parasitol. 120:309-313. [DOI] [PubMed] [Google Scholar]
- 100.International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 101.Jamjoom, M. B., R. W. Ashford, P. A. Bates, S. J. Kemp, and H. A. Noyes. 2002. Polymorphic microsatellite repeats are not conserved between Leishmania donovani and Leishmania major. Mol. Ecol. Notes 2:104-106. [Google Scholar]
- 102.Jeffrey, A. J., V. Wilson, and S. L. Thein. 1985. Hypervariable minisatellite regions in human DNA. Nature 314:67-73. [DOI] [PubMed] [Google Scholar]
- 103.Jensen, S., M. P. Gassama, and T. Heidmann. 1999. Cosuppression of I transposon activity in Drosophila by I-containing sense and antisense transgenes. Genetics 153:1767-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jensen, S., M. P. Gassama, and T. Heidmann. 1999. Taming of transposable elements by homology-dependent gene silencing. Nat. Genet. 21:209-212. [DOI] [PubMed] [Google Scholar]
- 105.Jurka, J. 1998. Repeats in genomic DNA: mining and meaning. Curr. Opin. Struct. Biol. 8:333-337. [DOI] [PubMed] [Google Scholar]
- 106.Kendall, G., A. F. Wilderspin, F. Ashall, M. A. Miles, and J. M. Kelly. 1990. Trypanosoma cruzi glycosomal glyceraldehyde-3-phosphate dehydrogenase does not conform to the ‘hotspot' topogenic signal model. EMBO J. 9:2751-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kimmel, B. E., O. K. Olemoiyoi, and J. R. Young. 1987. Ingi, a 5.2-kb dispersed sequence element from Trypanosoma brucei that carries half of a smaller mobile element at either end and has homology with mammalian LINEs. Mol. Cell. Biol. 7:1465-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kipling, D. 1995. The telomere. Oxford University Press, Oxford, United Kingdom.
- 109.Koga, A., A. Shimada, A. Shima, M. Sakaizumi, H. Tachida, and H. Hori. 2000. Evidence for recent invasion of the medaka fish genome by the To12 transposable element. Genetics 155:273-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kruglyak, S., R. T. Durrett, M. D. Schug, and C. F. Aquadro. 1998. Equilibrium distribution of microsatellite repeat length resulting from a balance between slippage events and point mutations. Proc. Natl. Acad. Sci. USA 95:10774-10778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kyes, S., P. Horrocks, and C. Newbold. 2001. Antigenic variation at the infected red cell surface in malaria. Annu. Rev. Microbiol. 55:673-707. [DOI] [PubMed] [Google Scholar]
- 112.Lanar, D. E., L. S. Levy, and J. E. Manning. 1981. Complexity and content of the DNA and RNA in Trypanosoma cruzi. Mol. Biochem. Parasitol. 3:327-341. [DOI] [PubMed] [Google Scholar]
- 113.le Blancq, S. M., and R. D. Adam. 1998. Structural basis of karyotype heterogeneity in Giardia lamblia. Mol. Biochem. Parasitol. 97:199-208. [DOI] [PubMed] [Google Scholar]
- 114.le Blancq, S. M., R. S. Kase, and L. H. T. Van der Ploeg. 1991. Analysis of a Giardia lamblia rRNA encoding telomere with (TAGGG)n as the telomere repeat. Nucleic Acids Res. 19:5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.le Blancq, S. M., M. H. Korman, and L. H. T. Van der Ploeg. 1991. Frequent rearrangements of rRNA-encoding chromosomes in Giardia lamblia. Nucleic Acids Res. 19:4405-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Levis, R. W., R. Ganesan, K. Houtchens, L. A. Tolar, and F.-M. Sheen. 1993. Transposons in place of telomeric repeats at the Drosophila telomere. Cell 75:1083-1093. [DOI] [PubMed] [Google Scholar]
- 117.Li, T. H., and C. W. Schmid. 2001. Differential stress induction of individual Alu loci: implications for transcription and retrotransposition. Gene 276:135-141. [DOI] [PubMed] [Google Scholar]
- 118.Lips, S., P. Revelard, and E. Pays. 1993. Identification of a new expression site- associated gene in the complete 30.5 kb sequence from the AnTat 1.3a variant surface protein gene-expression site of Trypanosoma brucei. Mol. Biochem. Parasitol. 62:135-137. [DOI] [PubMed] [Google Scholar]
- 119.Liu, A. Y. C., L. H. T. Van der Ploeg, F. A. M. Rijsewijk, and P. Borst. 1983. The transposition unit of variant surface glycoprotein gene 118 of Trypanosoma brucei. J. Mol. Biol. 167:57-75. [DOI] [PubMed] [Google Scholar]
- 120.Luan, D. D., M. H. Korman, J. L. Jakubczak, and T. H. Eickbush. 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72:595-605. [DOI] [PubMed] [Google Scholar]
- 121.Majiwa, P. A. O., and P. Webster. 1987. A repetitive deoxyribonucleic acid sequence distinguishes Trypanosoma simiae form T. congolense. Parasitology 95:543-598. [DOI] [PubMed] [Google Scholar]
- 122.Malhotra, P., P. V. N. Dasaradhi, A. Kumar, A. Mohammed, N. Agrawal, R. K. Bhatnagar, and V. S. Chauhan. 2002. Double-stranded RNA-mediated gene silencing of cysteine proteases (falcipain-1 and -2) of Plasmodium falciparum. Mol. Microbiol. 45:1245-1254. [DOI] [PubMed] [Google Scholar]
- 123.Malik, H. S., W. D. Burke, and T. H. Eickbush. 1999. The age and evolution of non- LTR retrotransposable elements. Mol. Biol. Evol. 16:793-805. [DOI] [PubMed] [Google Scholar]
- 124.Martín, F., C. Marañón, M. Olivares, C. Alonso, and M. C. López. 1995. Characterization of a non-long terminal repeat retrotransposon cDNA (L1Tc) from Trypanosoma cruzi: homology of the first ORF with the Ape family of DNA repair enzymes. J. Mol. Biol. 247:49-59. [DOI] [PubMed] [Google Scholar]
- 125.Martin, W., and P. Borst. 2003. Secondary loss of chloroplasts in trypanosomes. Proc. Natl. Acad. Sci. USA 100:765-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Matrajt, M., S. O. Angel, V. Pszenny, E. Guarnera, D. S. Roos, and J. C. Garberi. 1999. Arrays of repetitive DNA elements in the largest chromosomes of Toxoplasma gondii. Genome 42:265-269. [DOI] [PubMed] [Google Scholar]
- 127.McCulloch, R., G. Rudenko, and P. Borst. 1997. Gene conversions mediating antigenic variation in Trypanosoma brucei can occur in variant surface glycoprotein expression sites lacking 70-base-pair repeat sequences. Mol. Cell. Biol. 17:833-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.McRobert, L., and G. A. McConkey. 2002. RNA interference (RNAi) inhibits growth of Plasmodium falciparum. Mol. Biochem. Parasitol. 119:273-278. [DOI] [PubMed] [Google Scholar]
- 129.Melville, S. E., C. S. Gerrard, and J. M. Blackwell. 1999. Multiple causes of size variation in the diploid megabase chromosomes of African trypanosomes. Chromosome Res. 7:191-203. [DOI] [PubMed] [Google Scholar]
- 130.Miller, L. H., M. P. Good, and G. Milon. 1994. Malaria pathogenesis. Science 264:1878-1883. [DOI] [PubMed] [Google Scholar]
- 131.Mourrain, P., C. Beclin, T. Elmayan, F. Feuerbach, C. Godon, J. B. Morel, D. Jouette, A. M. Lacombe, S. Nikic, N. Picault, et al. 2000. Arabidopsis SGS2 and SGS3 genes are required for post-transcriptional gene silencing and natural virus resistance. Cell 101:533-542. [DOI] [PubMed] [Google Scholar]
- 132.Murphy, N. B., A. Pays, P. Tebabi, H. Coquelet, M. Guyaux, M. Steinert, and E. Pays. 1987. Trypanosoma brucei repeated element with unusual structural and transcriptional properties. J. Mol. Biol. 195:855-871. [DOI] [PubMed] [Google Scholar]
- 133.Myler, P. J., L. Audleman, T. DeVos, G. Hixson, P. Kiser, C. Lemley, C. Magness, E. Rickel, E. Sisk, S. Sunkin, S. Swartzell, T. Westlake, P. Bastien, G. L. Fu, A. Ivens, and K. Stuart. 1999. Leishmania major Friedlin chromosome 1 has an unusual distribution of protein-coding genes. Proc. Natl. Acad. Sci. USA 96:2902-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nash, T. E., A. Aggarwal, R. D. Adam, J. T. Conrad, and J. W. Merritt. 1988. Antigenic variation in Giardia lamblia. J. Immunol. 141:636-641. [PubMed] [Google Scholar]
- 135.Navarro, M., and K. Gull. 2001. A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature 414:759-763. [DOI] [PubMed] [Google Scholar]
- 136.Nene, V., S. Morzaria, and R. Bishop. 1998. Organisation and informational content of the Theileria parva genome. Mol. Biochem. Parasitol. 95:1-8. [DOI] [PubMed] [Google Scholar]
- 137.Ngo, H., C. Tschudi, K. Gull, and E. Ullu. 1998. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 95:14687-14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Novak, E. M., M. P. de Mello, H. B. Gomes, I. Galindo, P. Guevara, A. H. Ramirez, and J. F. da Silveira. 1993. Repetitive sequences in the ribosomal intergenic spacer of Trypanosoma cruzi. Mol. Biochem. Parasitol. 60:273-280. [DOI] [PubMed] [Google Scholar]
- 139.O'Donnell, R. A., L. H. Freitas-Junior, P. R. Preiser, D. H. Williamson, M. Duraisingh, T. F. McElwain, A. Scherf, A. F. Cowman, and B. S. Crabb. 2002. A genetic screen for improved plasmid segregation reveals a role for Rep20 in the interaction of Plasmodium falciparum chromosomes. EMBO J. 21:1231-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ogbadoyi, E., K. Ersfeld, D. Robinson, T. Sherwin, and K. Gull. 2000. Architecture of the Trypanosoma brucei nucleus during interphase and mitosis. Chromosoma 108:501-513. [DOI] [PubMed] [Google Scholar]
- 141.Olivares, M., J. L. García-Pérez, M. C. Thomas, S. R. Heras, and M. C. López. 2002. The non-LTR (long terminal repeat) retrotransposon L1Tc from Trypanosoma cruzi codes for a protein with RNase H activity. J. Biol. Chem. 277:28025-28030. [DOI] [PubMed] [Google Scholar]
- 142.Olivares, M., M. D. Thomas, A. López-Barajas, J. M. Requena, J. L. García-Pérez, S. Angel, C. Alonso, and M. C. López. 2000. Genomic clustering of the Trypanosoma cruzi nonlong terminal L1Tc retrotransposon with defined interspersed repeated DNA elements. Electrophoresis 21:2973-2982. [DOI] [PubMed] [Google Scholar]
- 143.Oquendo, P., M. Goman, M. Mackay, G. Langsley, D. Walliker, and J. Scaife. 1986. Characterization of a repetitive DNA sequence from the malaria parasite, Plasmodium falciparum. Mol. Biochem. Parasitol. 18:89-101. [DOI] [PubMed] [Google Scholar]
- 144.Orgel, L. E., and F. H. Crick. 1980. Selfish DNA: the ultimate parasite. Nature 284:604-607. [DOI] [PubMed] [Google Scholar]
- 145.Ossorio, P. N., L. D. Sibley, and J. C. Boothroyd. 1991. Mitochondrial-like DNA sequences flanked by direct and inverted repeats in the nuclear genome of Toxoplasma gondii. J. Mol. Biol. 222:525-536. [DOI] [PubMed] [Google Scholar]
- 146.Pace, T., M. Ponzi, R. Scotti, and C. Frontali. 1995. Structure and superstructure of Plasmodium falciparum subtelomeric regions. Mol. Biochem. Parasitol. 69:257-268. [DOI] [PubMed] [Google Scholar]
- 147.Patarapotikul, J., and G. Langsley. 1988. Chromosome size polymorphism in Plasmodium falciparum can involve deletion of the subtelomeric pPfRep20 sequence. Nucleic Acids Res. 16:4331-4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Perez-Morga, D., A. Amiguet-Vercher, D. Vermijlen, and E. Pays. 2001. Organization of telomeres during the cell and life cycles of Trypanosoma brucei. J. Eukaryot. Microbiol. 48:221-226. [DOI] [PubMed] [Google Scholar]
- 149.Piarroux, R., R. Azaiez, A. M. Lossi, P. Reynier, F. Muscatelli, F. Gambarelli, M. Fontes, H. Dumon, and M. Quilici. 1993. Isolation and characterization of a repetitive DNA sequence from Leishmania infantum: development of a visceral leishmaniasis polymerase chain reaction. Am. J. Trop. Med. Hyg. 49:364-369. [DOI] [PubMed] [Google Scholar]
- 150.Piarroux, R., M. Fontes, R. Perasso, F. Gambarelli, C. Joblet, H. Dumon, and M. Quilici. 1995. Phylogenetic-relationships between Old-World Leishmania strains revealed by analysis of a repetitive DNA-sequence. Mol. Biochem. Parasitol. 73:249-252. [DOI] [PubMed] [Google Scholar]
- 151.Pizzi, E., and C. Frontali. 2001. Fine structure of Plasmodium falciparum subtelomeric sequences. Mol. Biochem. Parasitol. 118:253-258. [DOI] [PubMed] [Google Scholar]
- 152.Pizzi, E., S. Liuni, and C. Frontali. 1990. Detection of latent sequence periodicities. Nucleic Acids Res. 18:3745-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ponzi, M., C. J. Janse, E. Dore, R. Scotti, T. Pace, T. J. F. Reterink, F. M. van der Berg, and B. Mons. 1990. Generation of chromosome size polymorphism during in vivo mitotic multiplication of P. berghei involves both loss and addition of subtelomeric repeat sequences. Mol. Biochem. Parasitol. 41:73-82. [DOI] [PubMed] [Google Scholar]
- 154.Prensier, G., and C. Slomianny. 1986. The karyotype of Plasmodium falciparum determined by ultrastructural serial sectioning and 3D reconstruction. J. Parasitol. 72:731-736. [PubMed] [Google Scholar]
- 155.Promisel-Cooper, J. 2000. Telomere transitions in yeast: the end of the chromosome as we know it. Curr. Opin. Genet. Dev. 10:169-177. [DOI] [PubMed] [Google Scholar]
- 156.Pulido, M., S. Martínez-Calvillo, and R. Hernández. 1996. Trypanosoma cruzi rRNA genes: a repeated element from the non- transcribed spacer is locus specific. Acta Trop. 62:163-170. [DOI] [PubMed] [Google Scholar]
- 157.Ravel, C., P. Wincker, P. Bastien, C. Blaineau, and M. Pagés. 1995. A polymorphic minisatellite sequence in the subtelomeric regions of chromosomes I and V in Leishmania infantum. Mol. Biochem. Parasitol. 74:31-41. [DOI] [PubMed] [Google Scholar]
- 158.Ravetch, J. V. 1989. Chromosomal polymorphisms and gene expression in P. falciparum. Exp. Parasitol. 68:121-125. [DOI] [PubMed] [Google Scholar]
- 159.Requena, J. M., A. Jimenez-Ruiz, M. Soto, M. C. López, and C. Alonso. 1992. Characterization of a highly repeated interspersed DNA sequence of Trypanosoma cruzi: its potential use in diagnosis and strain classification. Mol. Biochem. Parasitol. 51:271-280. [DOI] [PubMed] [Google Scholar]
- 160.Requena, J. M., M. C. López, and C. Alonso. 1996. Genomic repetitive DNA elements of Trypanosoma cruzi. Parasitol. Today 12:279-283. [DOI] [PubMed] [Google Scholar]
- 161.Requena, J. M., F. Martín, M. Soto, M. C. López, and C. Alonso. 1994. Characterization of a short interspersed reiterated DNA sequence of Trypanosoma cruzi located at the 3′-end of a poly(A)+ transcript. Gene 146:245-250. [DOI] [PubMed] [Google Scholar]
- 162.Requena, J. M., M. Soto, and C. Alonso. 1993. Isolation of Trypansoma cruzi specific nuclear repeated DNA sequences. Biol. Res. 26:11-18. [PubMed] [Google Scholar]
- 163.Robertson, H. M., and D. J. Lampe. 1995. Recent horizontal transfer of a mariner transposable element among and between diptera and neuroptera. Mol. Biol. Evol. 12:850-862. [DOI] [PubMed] [Google Scholar]
- 164.Robinson, K. A., and S. M. Beverley. 2003. Improvements in transfection efficiency and tests of RNA interference (RNAi) approaches in the protozoan parasite Leishmania. Mol. Biochem. Parasitol. 128:217-228. [DOI] [PubMed] [Google Scholar]
- 165.Rogers, J. H. 1985. The origin and evoluation of retroposons. Int. Rev. Cytol. 93:187-279. [DOI] [PubMed] [Google Scholar]
- 166.Rossi, V., P. Wincker, C. Ravel, C. Blaineau, M. Pagés, and P. Bastien. 1994. Structural organisation of microsatellite families in the Leishmania genome and polymorphisms at two (CA)n loci. Mol. Biochem. Parasitol. 65:271-282. [DOI] [PubMed] [Google Scholar]
- 167.Scherf, A., L. M. Figueiredo, and L. H. Freitas-Junior. 2001. Plasmodium telomeres: A pathogen's perspective. Curr. Opin. Microbiol. 4:409-414. [DOI] [PubMed] [Google Scholar]
- 168.Scherf, A., R. Hernandez-Rivas, P. Buffet, E. Bottius, C. Benatar, B. Pouvelle, J. Gysin, and M. Lanzer. 1998. Antigenic variation in malaria: in situ switching, relaxed and mutually exclusive transcription of var genes during intra-erythrocytic development in Plasmodium falciparum. EMBO J. 17:5418-5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Singer, M. F. 1982. SINEs and LINEs: highly repeated short and long interspersed sequences in mammalian genomes. Cell 28:433-434. [DOI] [PubMed] [Google Scholar]
- 170.Sloof, P., J. L. Bos, A. F. J. M. Konings, H. H. Menke, P. Borst, W. E. Gutteridge, and W. Leon. 1983. Characterization of satellite DNA in Trypanosoma brucei and Trypanosoma cruzi. J. Mol. Biol. 167:1-21. [DOI] [PubMed] [Google Scholar]
- 171.Sloof, P., H. H. Menke, M. P. M. Caspers, and P. Borst. 1983. Size fractionation of Trypanosoma brucei DNA: localization of the 177 bp repeat satellite DNA and a variant surface glycoprotein gene in a minichromosomal DNA fraction. Nucleic Acids Res. 11:3889-3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Solari, A. J. 1980. The 3-dimensional fine structure of the mitotic spindle in Trypanosoma cruzi. Chromosoma 78:239-255. [DOI] [PubMed] [Google Scholar]
- 173.Solari, A. J. 1980. Function of the dense plaques during mitosis in Trypanosoma cruzi. Exp. Cell Res. 127:457-460. [DOI] [PubMed] [Google Scholar]
- 174.Stechmann, A., and T. Cavalier-Smith. 2002. Rooting the eukaryotic tree by using a derived gene fusion. Science 297:89-91. [DOI] [PubMed] [Google Scholar]
- 175.Sternberg, J., and A. Tait. 1990. Genetic exchange in African trypanosomes. Trends Genet. 6:317-322. [DOI] [PubMed] [Google Scholar]
- 176.Su, X.-Z., and T. E. Wellems. 1996. Toward a high-resolution Plasmodium falciparum linkage map: polymorphic markers from hundreds of simple sequence repeats. Genomics 33:430-444. [DOI] [PubMed] [Google Scholar]
- 177.Sullivan, B. A., M. D. Blower, and G. H. Karpen. 2001. Determining centromere identity: cyclical stories and forking paths. Nat. Rev. Genet. 2:584-596. [DOI] [PubMed] [Google Scholar]
- 178.Sunkel, C., and P. A. Coelho. 1995. The elusive centromere: sequence divergence and functional conservation. Curr. Opin. Genet. Dev. 5:756-767. [DOI] [PubMed] [Google Scholar]
- 179.Sunkin, S. M., P. Kiser, P. J. Myler, and K. Stuart. 2000. The size difference between Leishmania major Friedlin chromosome one homologues is localized to sub-telomeric repeats at one chromosomal end. Mol. Biochem. Parasitol. 109:1-15. [DOI] [PubMed] [Google Scholar]
- 180.Tabara, H., M. Sarkissian, W. G. Kelly, J. Fleenor, A. Grishok, L. Timmons, A. Fire, and C. C. Mello. 1999. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99:123-132. [DOI] [PubMed] [Google Scholar]
- 181.Tamar, S., and B. Papadopoulou. 2001. A telomere-mediated chromosome fragmentation approach to assess mitotic stability and ploidy alterations of Leishmania chromosomes. J. Biol. Chem. 276:11662-11673. [DOI] [PubMed] [Google Scholar]
- 182.Tarleton, R. L., and J. Kissinger. 2001. Parasite genomics: current status and future prospects. Curr. Opin. Immunol. 13:395-402. [DOI] [PubMed] [Google Scholar]
- 183.Tautz, D., and M. Renz. 1984. Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res. 12:4127-4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Taylor, H. M., S. Kyes, and C. Newbold. 2000. Var gene diversity in Plasmodium falciparum is generated by frequent recombination events. Mol. Biochem. Parasitol. 110:391-397. [DOI] [PubMed] [Google Scholar]
- 185.Teng, S. C., S. X. Wang, and A. Gabriel. 1995. A new non-LTR retrotransposon provides evidence for multiple distinct site-specific elements in Crithidia fasciculata miniexon arrays. Nucleic Acids Res. 23:2929-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Toth, G., Z. Gaspari, and J. Jurka. 2000. Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res. 10:967-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Upcroft, P., and J. A. Upcroft. 1999. Organization and structure of the Giardia genome. Protist 150:17-23. [DOI] [PubMed] [Google Scholar]
- 188.Urena, F. 1986. 3-dimensional reconstructions of the mitotic spindle and dense plaques in 3 species of Leishmania. Parasitol. Res. 72:299-306. [DOI] [PubMed] [Google Scholar]
- 189.Van der Ploeg, L. H. T., A. Y. C. Liu, and P. Borst. 1984. Structure of the growing telomeres of trypanosomes. Cell 36:459-468. [DOI] [PubMed] [Google Scholar]
- 190.van Eys, G. J. J. M., L. F. Schnur, N. C. M. Kroon, and S. B. Ebeling. 1992. Sequence analysis of small subunit ribosomal RNA genes and its use for detection of Leishmania parasites. Mol. Biochem. Parasitol. 51:133-142. [DOI] [PubMed] [Google Scholar]
- 191.Vázquez, M., C. Ben-Dov, H. Lorenzi, T. Moore, A. Schijman, and M. J. Levin. 2000. The short interspersed repetitive element of Trypanosoma cruzi, SIRE, is part of VIPER, an unusual retroelement related to long terminal repeat retrotransposons. Proc. Natl. Acad. Sci. USA 97:2128-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Vázquez, M., H. Lorenzi, A. G. Schijman, C. Ben-Dov, and M. J. Levin. 1999. Analysis of the distribution of SIRE in the nuclear genome of Trypanosoma cruzi. Gene 239:207-216. [DOI] [PubMed] [Google Scholar]
- 193.Vázquez, M. P., A. G. Schijman, and M. J. Levin. 1994. A short interspersed repetitive element provides a new 3′ acceptor site for trans-splicing in certain ribosomal P2β protein genes of Trypanosoma cruzi. Mol. Biochem. Parasitol. 64:327-336. [DOI] [PubMed] [Google Scholar]
- 194.Vernick, K. D., and T. F. McCutchan. 1988. Sequence and structure of a Plasmodium falciparum telomere. Mol. Biochem. Parasitol. 28:85-94. [DOI] [PubMed] [Google Scholar]
- 195.Villanueva, M. S., S. P. Williams, C. B. Beard, F. F. Richards, and S. Aksoy. 1991. A new member of a family of site-specific retrotransposons is present in the spliced leader RNA genes of Trypanosoma cruzi. Mol. Cell. Biol. 11:6139-6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Walmsley, R. M., C. S. M. Chan, B. K. Tye, and T. D. Petes. 1984. Unusual DNA sequences associated with the ends of yeast chromosomes. Nature 310:157-160. [DOI] [PubMed] [Google Scholar]
- 197.Warburton, P. E., C. A. Cooke, S. Bourassa, O. Vafa, B. A. Sullivan, G. Stetten, G. Gimelli, D. Warburton, C. TylerSmith, K. F. Sullivan, G. G. Poirier, and W. C. Earnshaw. 1997. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol. 7:901-904. [DOI] [PubMed] [Google Scholar]
- 198.Wasserman, M., J. Contreras, G. Pinilla, M. O. Rojas, A. Paez, and E. Caminos. 1995. Plasmodium falciparum: characterization of a 0.7 kbp, moderately repetitive sequence. Exp. Parasitol. 81:165-171. [DOI] [PubMed] [Google Scholar]
- 199.Weiden, M., Y. N. Osheim, A. L. Beyer, and L. H. T. van der Ploeg. 1991. Chromosome structure: DNA nucleotide sequence elements of a subset of the minichromosomes of the protozoan Trypanosoma brucei. Mol. Cell. Biol. 11:3823-3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Weiner, A. M. 2002. SINEs and LINEs: the art of biting the hand that feeds you. Curr. Opin. Cell Biol. 14:343-350. [DOI] [PubMed] [Google Scholar]
- 201.Reference deleted.
- 202.Yang, J. W., C. Pendon, J. Yang, N. Haywood, A. Chand, and W. R. A. Brown. 2000. Human mini-chromosomes with minimal centromeres. Hum. Mol. Genet. 9:1891-1902. [DOI] [PubMed] [Google Scholar]
- 203.Zomerdijk, J. C. B. M., R. Kieft, M. Duyndam, P. G. Shiels, and P. Borst. 1991. Antigenic variation in Trypanosoma brucei: a telomeric expression site for variant- specific surface glycoprotein genes with novel features. Nucleic Acids Res. 19:1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zomerdijk, J. C. B. M., M. Ouellette, A. L. M. A. Ten Asbroek, R. Kieft, A. M. M. Bommer, C. E. Clayton, and P. Borst. 1990. The promoter for a variant surface glycoprotein gene expression site in Trypanosoma brucei. EMBO J. 9:2791-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]