Abstract
Gram-positive bacteria possess a myriad of acid resistance systems that can help them to overcome the challenge posed by different acidic environments. In this review the most common mechanisms are described: i.e., the use of proton pumps, the protection or repair of macromolecules, cell membrane changes, production of alkali, induction of pathways by transcriptional regulators, alteration of metabolism, and the role of cell density and cell signaling. We also discuss the reponses of Listeria monocytogenes, Rhodococcus, Mycobacterium, Clostridium perfringens, Staphylococcus aureus, Bacillus cereus, oral streptococci, and lactic acid bacteria to acidic environments and outline ways in which this knowledge has been or may be used to either aid or prevent bacterial survival in low-pH environments.
INTRODUCTION
Gram-positive bacteria play vital roles in food and health, although the nature of this role can vary greatly. Gram-positive bacteria include generally-regarded-as-safe organisms used in food fermentations, ubiquitous gastric commensals, and potentially beneficial probiotic bacteria. However, gram-positive pathogens can also cause diseases ranging from dental caries to potentially fatal gastrointestinal infections. The acidic environments encountered in food and in the gastrointestinal tract provide a significant survival challenge for these diverse organisms. For example, the preferred delivery vehicles for probiotic cultures are yogurts and fermented milks, both of which present an acid challenge. Subsequently the bacteria will need to survive the highly acidic gastric juice if they are to reach the small intestine in a viable state (113, 140). Hence, acid tolerance is accepted as one of the desirable properties used to select potentially probiotic strains. In striking contrast, the acid tolerance of a pathogen such as Listeria monocytogenes, again assisting survival in the stomach and also in the macrophage phagosome, could be considered a virulence factor. Thus, it is timely to consider the mechanisms used by gram-positive organisms to protect themselves from the challenge posed by low-pH environments such as food and gastric juice, and comment on the strategies by which they can be aided or impeded.
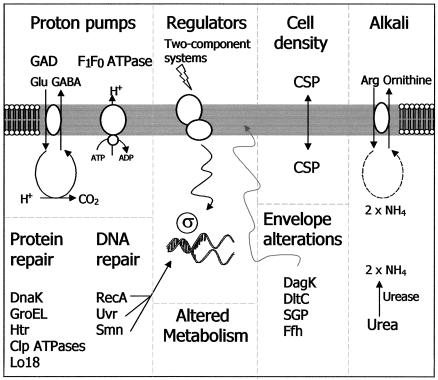
While many excellent reviews have outlined the numerous systems which have been characterized in gram-negative bacteria (7, 196, 215) and, to a lesser extent, in oral streptococci (27, 186), until recently the relative lack of information pertaining to acid resistance systems in gram-positive bacteria has militated against a review of this area. This may reflect the perception that the F1F0-ATPase proton pump was solely responsible for the survival of some of these organisms in acidic environments. It is now apparent that a combination of constitutive and inducible strategies which result in the removal of protons (H+), alkanization of the external environment, changes in the composition of the cell envelope, production of general shock proteins and chaperones, expression of transcriptional regulators, and responses to changes in cell density can all contribute to survival (Fig. 1). These mechanisms counter the negative impact of a reduction in cytoplasmic pH, which can include loss of activity of the relatively acid-sensitive glycolytic enzymes (which severely affects the ability to produce ATP) and structural damage to the cell membrane and macromolecules such as DNA and proteins.
FIG. 1.
Graphical presentation of the mechanisms of resistance available to gram-positive bacteria. These have been divided into eight categories, and a number of examples are demonstrated. (i) Proton pumps such as the F1F0ATPase or that utilized by the GAD system bring about an increase in internal pH. (ii) Proton repair involving chaperones, proteases, and heat shock proteins results in the protection of proteins or their degradation if damaged. (iii) DNA damaged as a consequence of a low internal pH can be repaired through the excision of errors or the restarting of stalled replication forks. (iv) The involvement of regulators such as 2CSs and sigma factors can induce minor or global responses. (v) Cell density affects cell-to-cell communication. (vi) Cell envelope alterations can protect cells by changing architecture, composition, stability, and activity. (vii) The production of alkali by the ADI or urease system increases the internal pH of the cell. (viii) Metabolic properties can be altered.
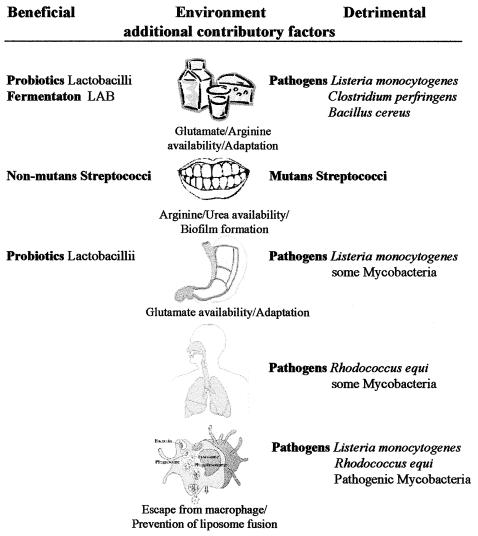
While gram-positive bacteria may encounter a wide range of low-pH environments, most studies have focused on those where bacterial growth and survival have an impact on human health and economics, i.e., survival and growth in food, effect on the oral cavity, gastric transit, and intracellular survival (Fig. 2). Acid has been used as a food preservative for millennia, primarily in the form of food fermentations. In each case, the fermentation process involves the oxidation of carbohydrates to generate a range of products including organic acids, alcohol, and carbon dioxide. Such products have a preservative effect through limiting the growth of spoilage and pathogenic members of the flora in the food product. Weak acids have potent antimicrobial activity because the undissociated form of weak acids pass freely through the cell membrane. Since the cytoplasmic pH is generally higher than that of the growth medium, the weak acid dissociates, releasing a proton and leading to acidification of the cytoplasm. Such is the effectiveness of weak organic acids in preventing bacterial growth that they are commonly added directly to foods, either alone or in combination with other preservatives. Such an addition is unnecessary in foods fermented with, for example, the homofermentative Lactococcus lactis, which converts 90% of metabolized sugar to lactic acid. An understanding of the acid resistance mechanisms used by lactic acid bacteria (LAB) to survive the by-products of their own metabolism and the responses available to spoilage and pathogenic organisms in low-pH foods is thus of great importance.
FIG. 2.
Acidic environments where the survival of bacteria has an impact on health or the economy. While contributory factors are listed, these do not include genes and acid resistance systems associated with individual bacteria.
Following the intake of dietary carbohydrate, acidogenic (acid-producing) and aciduric (acid-resistant) bacteria constituting the oral biofilm ferment metabolizable sugars, producing acid that can lower the plaque pH from 7.0 to 4.0 within 3 min. This is significant, given that tooth demineralization can occur at pH 5.2 and below (119, 122). The demineralization phases are followed by periods of alkalinization that promote remineralization. Dental caries occurs when the acidification phases prevail over the alkalinization phases, allowing the establishment of a more acidogenic flora, which in turn results in lower plaque pHs and enhanced and prolonged enamel demineralization. This process ultimately results in the formation of a carious lesion. Thus, the ability of bacteria to produce acid in combination with growth at low pH is regarded as a virulence factor. Dental caries is one of the most common causes of tooth loss in the developed world, constituting a substantial economic burden. The bacteria most closely associated with the initiation and progression of dental caries are mutans streptococci (Streptococcus mutans and S. sobrinus), lactobacilli, and possibly Actinomyces species, with acidogenicity and aciduricity being two of the most important virulence factors.
Further down the alimentary canal, ingested microbes enter the stomach. The stomach acts as a reservoir that controls the rate of entry of material into and, more importantly with respect to this review, the number of bacteria entering the small intestine. Humans secrete approximately 2.5 liters of gastric juice each day, generating a fasting gastric pH of 1.5, which increases to between pH 3.0 and 5.0 during feeding (116). The role of gastric acid is most apparent in patients who are achlorydic, i.e., unable to produce HCl, or have undergone a partial gastrectomy. In these circumstances, the number of aerobes, anaerobes, and coliforms increases so that there is overgrowth of bacteria in the stomach. It has also been observed that neutralization of acid by food or bicarbonate decreases the infectious dose; for example, an increase in the pH of the stomach results in gastric transit of greater numbers of Listeria (44, 63, 64, 198, 205, 258) while gastrectomy and chronic gastritis are risk factors for the development of intestinal tuberculosis (62). Interestingly, preadaptation under conditions similar to those encountered by Mycobacterium avium in its natural habitat allows the organisms to resist the acidic conditions of the stomach (20). For obvious reasons, it is also vital that probiotics be able to transit the stomach despite its low pH (118). Intrinsic resistance to gastric acid is a relatively rare probiotic property, with Lactobacillus acidophilus and bifidobacteria being among the most resistant (34). Gastric survival is also a highly valued property because it overcomes the limitations associated with needing to use food carriers with high buffering potential to aid gastric transit and ensure the delivery of viable cells into the small intestine.
Invasive pathogens also need to survive or avoid additional exposure to acid following phagocytosis by macrophages. Initially the phagosomal pH drops due to the activity of the vacuolar ATPase. Then, following phagosome-lysosome fusion, myleoperoxidase is released into the phagolysosome by the host and catalyzes the formation of potent hypochlorous acid (HOCl) from chloride ions and H2O2. The Listeria monocytogenes hemolysin protein responds to the initial drop in pH brought about by the vacuolar ATPases to permeabilize the phagosomal membrane and permit the bacterium to escape prior to phagosome-lysosome fusion (14). Pathogenic mycobacteria alter the phagosomal pH by excluding vacuolar ATPase from the phagosome membrane or inhibiting phagosome-lysosome fusion (197). The latter strategy would also appear to be used by Rhodococcus equi (260).
In this review the mechanisms of acid resistance that are most common among gram-positive bacteria are described. In addition, the consequences of survival of gram-positive bacteria at low pH with the aid of these and other, less common, systems are discussed. We also discuss how this knowledge might be adapted to enhance the survival of beneficial bacteria and reduce the survival of pathogenic bacteria in low-pH environments.
MECHANISMS OF RESISTANCE
Proton Pumps
F1F0-ATPase.
The multisubunit F1F0-ATPase links the production of ATP to the transmembrane proton motive force (PMF) and can either generate ATP at the expense of the PMF established by respiring cells or generate a PMF using ATP produced by fermentative substrate-level phosphorylation. The PMF can facilitate the extrusion of protons from the cell cytoplasm, resulting in a drop in intracellular pH. The membrane-embedded F0 complex, composed of subunits a, b, and c, has proton-translocating activity, although its channel activity is increased by coexpression of some F1 proteins, suggesting a possible role for an F1 protein in the assembly or gating of the channel. The peripherally bound F1 complex, consisting of subunits α, β, γ, δ, and ɛ, has ATPase activity when it is released from the membranes and catalyzes the coupled interconversion of proton translocation and ATP synthesis or hydrolysis when it is complexed to the F0 complex in sealed membranes (87).
(i) Enterococcus hirae.
Initial studies of the importance of the F1F0-ATPases in the regulation of cytoplasmic pH were made with Enterococcus hirae (formerly Streptococcus faecalis). This organism does not have a respiratory chain and thus is not capable of using the F1F0 complex for synthesis of ATP via oxidative phosphorylation. As a result, the sole function of this complex in E. hirae is the extrusion of hydrogen ions (H+) and consequently the establishment of pH homeostasis (111). ATPase activity in E. hirae is controlled primarily at the level of pH-dependent subunit assembly with a decrease in cytoplasmic pH (pH1), resulting in increased ATPase activity (6, 132, 133). Regulation of transcription or translation does not seem to play an important role. While translation is stimulated at low pH, it occurs at too low a level to account for the increase in the level of membrane-bound ATPase (6). A basal level of ATPase also exists at alkaline pH despite the lack of an obvious role in these conditions (133). It is speculated that it may be present to allow the rapid resumption of growth should the pH drop suddenly (124). In addition, the acid optimum of the complex contributes to pH homeostasis by preventing overalkalization of the cytoplasm (131). The importance of the F1F0-ATPase was demonstrated by the isolation of mutants that were sensitive to acid as a result of defects in the complex. Such sensitivity is generally typified by an inability to grow at pH 6, despite growth rates matching those of the parent when grown at pH 7.8 (218, 219).
(ii) F1F0-ATPase in other genera.
It was found that different acid tolerances in species of oral lactic acid bacteria corresponded to their relative permeabilities to protons. As well as the passive inflow, the permeability of a cell to protons is dictated by an active outflow through the proton-translocating membrane ATPase. Thus, the pHs at which the permeabilities of the relatively acid-tolerant Streptococcus mutans (pH 5) and S. salivarius (pH 6) and the relatively sensitive S. sanguis (pH 7) were lowest are reflected by the pH optima of the ATPase enzymes in these species of pH 6.0, 7.0, and 7.5, respectively. These figures correspond to the relative acid tolerances of these bacteria, indicating the importance of the role played by the ATPase in resistance to low-pH stress (17, 217). The S. sanguis and S. mutans ATPase operons have been compared, but it has not been possible to identify differences that might explain their contrasting activity at low pH (186). It is interesting that the pH profiles of these F1 subunits are similar, differing dramatically only following binding with F0 (217). The link between ATPase and acid tolerance has also been demonstrated for ruminal bacteria, where membranes of acid-sensitive bacteria such as Ruminococcus albus and Fibrobacter succinogenes contain much lower levels of ATPase than do those of the relatively tolerant Megasphaera elsdenii and Streptococcus bovis. While the variations in tolerance cannot be explained by the pH optima of the enzymes for these bacteria, since they are all similar, the activity profiles show that the ATPases from the tolerant bacteria are less sensitive to low pH (160), which is also the explanation for the acid resistance of the Leuconostoc oenos mutant LoV8413 (80). In accordance with the above observations, the acid sensitivities of the Lactococcus lactis subsp. lactis C2 mutant (5, 257) and the Lactobacillus helveticus mutant CPN4 (254) are explained by reduced ATPase activity at low pH, and thus the variations in acid tolerance in all of the above examples can be attributed to their relative ATPase activities at low pH.
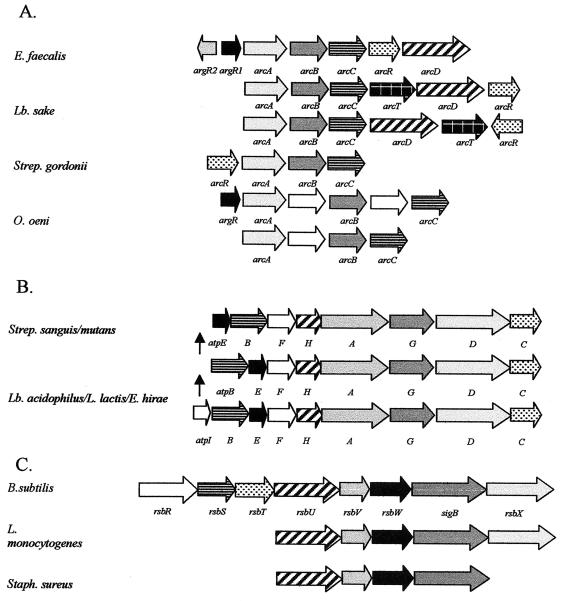
To date, all streptococci appear to possess an F0 gene order consisting of atpEBF, in contrast to atpBEF in most other bacteria (Fig. 3). However, the significance of this remains unclear since rearrangement of the genes within the Escherichia coli operon does not appear to influence the assembly and function of the enzyme (246). Interestingly, the S. mutans, S. sanguis, Lactobacillus acidophilus, and Lactococcus lactis operons all lack an atpI homologue upstream of the structural gene. The role of AtpI in the biology of bacterial ATPase is unknown, and although mutations have a slightly detrimental effect, ATPase assembly and function are not impaired. In S. sanguis (137) and S. mutans (213), a relatively large intergenic region with no known role occupies this position. The external pH has no effect on the transcriptional start sites for the ATPase operons in S. mutans (213) or S. sanguis (137), implying that the intergenic region is unlikely to be regulated.
FIG. 3.
(A) Alignment of ADI gene clusters. arcA, arginine deiminase; arcB, ornithine transcarbamylase; arcC, carbamate kinase; arcR, Crp/Fnr-type regulator; arcD, ornithine/arginine antiporter; arcT, noncharacterized transaminase; argR, arginine of the ArgR-AhrC family. The identities of a number of the genes indicated above remain putative (L. sakei [262, 263], E. faecalis [11], S. gordonii [78], S. pyogenes [74], S. suis [250], and O. oeni [228]). (B) Alignment of F1F0-ATPase operons (C) Alignment of of genes involved in regulation of sigB. (Panel B is based on data in reference 186; Panel C is based on data in reference 247.)
In contrast to the situation in Enterococcus hirae, the promoter of the S. mutans ATPase operon is up-regulated at pH 5 relative to pH 7. Optimal expression is dependent on the presence of upstream inverted repeats, although pH-dependent regulation is still observed in their absence (137, 186). A pH-dependent increase in ATPase transcription is also observed in S. pneumoniae. A point mutation in the −10 box of the pH-inducible promoter abolishes pH regulation (155). L. acidophilus atp mRNA production also increases on exposure to low pH, resulting in a twofold increase in the activity of the enzyme (138).
In addition to lactic acid bacteria, the F1F0-ATPase complex plays a major role in the acid resistance mechanisms of other gram-positive bacteria. The involvement of the Listeria monocytogenes F1F0-ATPase in resistance to acid stress was confirmed when it was found that cells treated with N,N′-dicyclohexylcarbodiimide (DCCD), an inhibitor of ATPase activity, were much more sensitive to exposure to pH 3.0 than were untreated cells. Treatment with DCCD resulted in a reduction in the level of survivors to 1% (stationary phase) or 0.001% (log phase) of that of untreated controls (67). F1F0-ATPase also plays a role in the induction of an acid tolerance response (ATR), i.e., the phenomenon whereby the tolerance of an organism to lethal pH increases as a consequence of a prior exposure to a sublethal pH (50). It was found that acid-adapted logarithmic-phase cells treated with DCCD are greatly sensitized to low pH stress. Two-dimensional gel electrophoresis has shown that the b subunit of the L. monocytogenes ATPase is induced by acid stress, and it may also be significant that the atpC (ɛ subunit) and atpD (β subunit) genes are preceded by boxes associated with PrfA, the positive regulator of virulence genes (97). In M. smegmatis, the delta pH at external pH of 5.0 was dissipated by protonophores and DCCD. These results demonstrate that proton extrusion by the F1F0-ATPase plays a key role in maintaining the internal pH near neutral (191).
In addition to the F1F0-ATPase, cation transport ATPases such as K+-ATPases can contribute to pH homeostasis through the exchange of K+ for H+. It was found that at pHo (pH outside the cell) 5.0 the pHi (pH inside the cell) of glucose-energized S. mutans is 5.5 in the absence of K+ but increases to 6.14 in the presence of 25 mmol of K+ per liter (66). Similar observations were reported for L. lactis (128) and E. hirae (9, 130).
Glutamate decarboxylase.
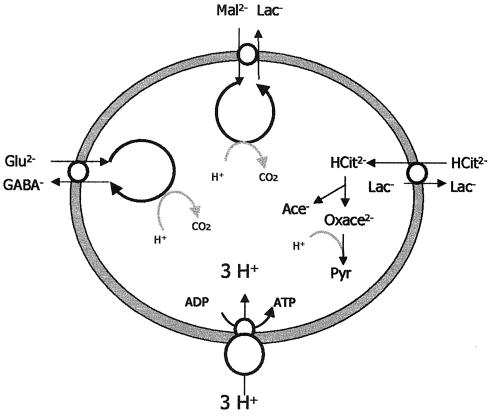
Almost 60 years ago, it was proposed that amino acid decarboxylases function to control the pH of the bacterial environment by consuming hydrogen ions as part of the decarboxylation reaction (93). Examples of this are the lysine, arginine, and glutamate decarboxylases (GAD), which operate by combining an internalized amino acid (lysine, arginine, or glutamate) with a proton and exchanging the resultant product (cadaverine, agmatine, or γ-aminobutyrate) for another amino acid substrate. Thus, an extracellular amino acid is converted to an extracellular product, but the consumption of an intracellular proton results in an increase in intracellular pH. Of the three systems mentioned, only the GAD system has been associated with pH control by gram-positive cells.
Among gram-positive bacteria, genetic characterization of GAD systems has been undertaken for L. lactis (202) and L. monocytogenes (51). However, biochemical studies have been used to analyze the corresponding systems in Clostridium (56, 90), some species of Mycobacterium (184), Bacteroides, Fusobacterium, Eubacterium (10), and Lactobacillus brevis (230). According to the most simple model (212), after uptake of glutamate by the specific transporter, cytoplasmic decarboxylation results in the consumption of an intracellular proton. The reaction product, γ-aminobutyrate (GABA), is exported from the cell via an antiporter. The net result is an increase in the pH of the cytoplasm due to the removal of hydrogen ions and a slight increase in the extracellular pH due to the exchange of extracellular glutamate for the more alkaline GABA. It has also been proposed (202), based on results presented by Higuchi et al. (115) showing that ATP could be generated by glutamate metabolism in lactobacilli, that the GAD system functions to provide metabolic energy by coupling electrogenic antiport and amino acid decarboxylation. It is thought that three successive decarboxylation-antiport cycles create a PMF sufficient for the synthesis of one molecule of ATP via the F0F1 ATPase (Fig. 4). The significance of the GAD system is linked to the presence of glutamate in foods. The use of glutamate for food flavor enhancement is a long-established process and can be traced back to the Orient, where seaweeds rich in glutamate were used for this purpose. In modern foods, the sodium salt monosodium l-glutamate monohydrate, usually termed MSG, is by far the most common source, although the potassium, ammonium, and calcium salts are also used (120). Glutamate is also present in a number of food ingredients, while the amino acid itself is used to adjust the acidity of foods. Protein-rich foods such as meat and milk, as well as many vegetables, also have a high content of bound glutamate, while their fermented derivatives have increased levels of free glutamate. If one also considers the antihypertensive effects of foods containing GABA, it is easy to appreciate the importance of glutamate decarboxylation.
FIG. 4.
Generation of ATP by transport and proton-consuming decarboxylation. The movement of glutamate (Glu2−) ions into the cell, glutamate decarboxylation, and extrusion of GABA ions (GABA−), the movement of malate (Mal2−) ions into the cell, malate decarboxylation, and extrusion of lactate (Lac−) ions (L. lactis), and the movement of citrate (HCit2−) ions into the cell, oxaloacetate decarboxylation, and extrusion of Lac ions (Lac−) (L. lactis) all create an electrogenic potential. These decarboxylation reactions and the consumption of a proton increase the alkalinity of the cytoplasm. Three cycles of decarboxylation and antiport create a PMF sufficient for the synthesis of ATP via the F1F0-ATPase. Ace−, acetate; Oxace2−, oxaloacetate; Pyr, pyruvate.
(i) Lactic acid bacteria.
The Lactococcus lactis gadCB operon was identified as a result of analysis of its chloride-dependent promoter. The predicted lactococcal GadC is homologous to GadC of Shigella flexneri (51% identity and 17% similarity [244]) and GadC of E. coli (114). On the basis of these homologies and the predicted presence of 12 potentially membrane-spanning hydrophobic domains, gadC is thought to encode an amino acid antiporter. The gadB product is most homologous to Synechocystis GAD enzymes (48% identity and 15% similarity [126]) and is highly similar to GadA and GadB of E. coli (43 and 44% identity, respectively [214]). GadB was named because its encoding genes in E. coli and S. flexneri are adjacent to a gadC gene (202). Southern blot analysis of a number of strains of L. lactis subsp. lactis has shown gadB to be present in a single copy (166). A constitutively expressed gene, gadR, was found upstream. GadR positively regulates chloride-induced expression of gadC and gadB.
It was found that the lactococcal gadC and gadB genes were maximally expressed at low pH and at the onset of stationary phase in the presence of NaCl and glutamate and that insertional inactivation of gadB resulted in increased acid sensitvity. The induction by chloride is postulated as a mechanism for survival in the stomach, where the organism encounters a highly acidic, chloride-rich environment (202). It was also proposed that gadCB may be important for lactococcal survival during cheese production. A study of the production of GABA by cheese starters during cheese ripening (164) showed that of 31 colonies isolated from a variety of cheese starters, GABA production was observed in 17. The purified decarboxylase was found to be active in the pH range from 4.0 to 5.5 (optimum at pH 4.7) and to retain 90% activity between 30 and 70°C (166). Analysis of L. lactis subsp. cremoris strains showed a gadB homolog to be present but not to be active as a consequence of a frameshift mutation (165). This may, at least to some extent, explain why L. lactis subsp. lactis and cremoris can be differentiated on the basis of their abilities to respond to acid stress. It has been found that L. lactis subsp. lactis can induce an ATR while L. lactis subsp. cremoris cannot (129). In addition, the sublethal (minimum pH for growth) and lethal levels of acid stress were pH 4.5 and 2.5, respectively, for L. lactis subsp. lactis but higher, at pH 5.0 and 3.0, respectively, for L. lactis subsp. cremoris (129).
A number of strains of lactobacilli also catalyze the conversion of glutamate to GABA (115, 230). Of these, the Lactobacillus brevis GAD was purified and found to have optimal activity at 30°C and pH 4.2 and in the presence of 0.2 M pyridine-HCl, but not to be activated by NaCl (230). In addition, the ability of the GAD system to contribute to ATP synthesis was first demonstrated in Lactobacillus sp. strain E1 (115).
(ii) Listeria monocytogenes.
In L. monocytogenes LO28, two decarboxylase-encoding homologs, gadA and gadB, and an antiporter homolog, gadC, have been characterized (51). Indeed, a third decarboxylase and second antiporter homolog have been identified in the genome-sequenced strain EGDe (97). The system plays a role in the acid tolerance of Listeria during both the logarithmic and stationary phases of growth and is also required for the induction of an optimal acid tolerance response. A number of GAD mutants of strain LO28 have been generated. The acid resistance of these strains is in the following order: wild type > ΔgadA > ΔgadB = ΔgadC > ΔgadAB. Elimination of the GAD system results in sensitization to the low pH of the stomach, resulting in more rapid killing in this environment. This sensitivity manifests itself as a dramatic 6-log-unit reduction of the ΔgadAB mutant on exposure to porcine gastric fluid, whereas the parent survives relatively unscathed. It is notable that strains which display reduced GAD activity are more sensitive to gastric fluid. Indeed, the significant strain variation in GAD activity correlates significantly with the observed levels of acid tolerance, suggesting that GAD represents the key mechanism for maintenance of pH homeostasis in L. monocytogenes in this environment (51).
The ΔgadAB mutant was also used to determine the importance of the GAD system in the survival of L. monocytogenes in low-pH foods such as apple, orange, and tomato juices, yoghurts, salad creams, and mayonnaise (53). The greater acid sensitivity of the mutant was apparent even in foods with levels of free glutamate as low as 0.22 mM. It was also found that survival of the wild-type strain, in acidified reconstituted skim milk, diluted to reduce free glutamate levels, improves in response to supplementation with monosodium glutamate. With the exception of the dilute-milk model, no correlation was observed between glutamate concentrations and the levels of survival of the wild-type strain, nor the relative difference between the wild-type and ΔgadAB strains, suggesting that glutamate is present in excess in all instances (53). It would thus seem that at the mildly acidic pHs of the foods tested, the GAD system can function optimally in the presence of the amount of free glutamate naturally present in these foodstuffs.
(iii) Mycobacterium leprae.
Mycobacterium leprae GAD is also proposed to play a role in acid resistance, in this case in the intracellular environment in which the bacteria multiply, or to serve in the creation of energy (184). In addition, it was suggested that the presence of GAD activity in M. leprae is a reflection of the association of this organism with nerve tissue. Since glutamic acid is the most abundant amino acid in the nerve tissue and since GABA is an inhibitory neurotransmitter, glutamate decarboxylation may contribute to the impairment of sensation in lepromatous lesions (184).
(iv) Clostridium perfringens.
A GAD enzyme purified from Clostridum perfringens was shown to have a pH optimum of 4.7 (56). A rapid and continuous increase of GAD activity during the late logarithmic phase and early stationary phase was shown to correspond to a reduction in the pH of the culture medium to below 6 (55).
Electrogenic transport.
A number of other transporter-dependent metabolic pathways have been implicated in acid stress responses. Malolactic fermentation (MLF) is the conversion of the dicarboxylic malic acid to the monocarboxylic lactic acid. MLF plays a major role in the activity of the wine bacteria Oenococcus oeni (199) and has also been identified in Lactobacillus sakei (31), Lactobacillus plantarum (174), and Lactococcus lactis (192). The l-lactate produced is excreted via either a lactate-malate antiporter (L. lactis [182]) or an electrogenic uniport (O. oeni and L. planatum), both of which allow the synthesis of ATP at low pH (Fig. 4). This ATP production is probably the basis for the implication of MLF in O. oeni acid tolerance (229). Furthermore, due to the pK difference of the carboxylic groups of malate and lactate, the external replacement of malate by lactate results in alkalinization of the medium.
In L. lactis subsp. diacetylactis, bacterial growth at low pH is aided by induction of the citrate-lactate antiporter, CitP (95). It is likely that the generation of energy via electrogenic transport and the alkalinization of the medium are responsible for this stimulation of growth (156). The relief of lactate toxicity is thought to be of primary importance in this regard (151).
Protection or Repair of Macromolecules
A number of proteins that play a role in the protection or repair of macromolecules such as DNA and proteins are essential for optimal acid resistance. One such protein, RecA, acts as a mediator of homologous recombination and a regulator of the SOS response (243, 256). It participates in the housekeeping function of repairing and restarting stalled replication forks (54). RecA-deficient mutants of S. mutans are more susceptible to killing at pH 2.5. However, this deficiency can be almost completely overcome by acid adaptation. In addition, acid adaptation permits increased resistance to UV radiation and hydrogen peroxide. This suggests the existence of a RecA-independent, acid-induced DNA repair system (188). The consequences of intracellular acidification include loss of purines and pyrimidines from DNA at a greater rate than at neutral or alkaline pH (146). This involves protonation of the base followed by cleavage of the glycosyl bond (146). The residues left at sites of base loss are referred to as abasic sites or AP (apurinic-apyrimidinic) sites. The repair of AP sites is a multistep process and is initiated by AP endonucleases. It is thus unsurprising that the RecA-independent acid adaptation mechanism in S. mutans involves the induction of an AP endonuclease, designated Smn for (“S. mutans exonuclease”), whose DNA repair activities resemble those of the E. coli exonuclease III (106). Also in S. mutans, a uvrA-UV excinuclease gene homolog displayed increased expression at pH 5.0 compared to pH 7.5. The corresponding gene in B. subtilis is associated with the nucleotide excision repair pathway involved in DNA repair, which functions in locating and excising bulky DNA lesions (201). A uvrA mutant of S. mutans was more sensitive than the wild type to growth at pH 5.0, and acid-adapted mutants were unable to survive exposure to pH 3.0. Exposure to these conditions also resulted in more DNA damage being evident in the mutant than in the wild type (110). It has been suggested that the role of base excision repair involving the AP endonuclease could be to repair minor DNA damage whereas UvrA and the nucleotide excision repair pathway could be responsible for excising larger DNA lesions caused by acid and other DNA-damaging agents. The link between DNA damage repair and acid resistance has also been demonstrated in L. lactis. A moderate UV irradiation induces tolerance to several stresses, including acid shock (112), while four proteins upregulated during acid adaptation were also induced by DNA-damaging treatments (112).
Chaperonins intervene in numerous stresses for various tasks such as protein folding, renaturation, protection of denatured proteins, and evacuation of damaged proteins. Gene expression and protein levels of the DnaK chaperone are up-regulated in response to acid shock and sustained acidification in S. mutans (121). In E. coli, DnaK increases the stability of UvrA (261), suggesting that at least one of the reasons why the chaperonins play a role in the response to acid shock may be to ensure proper functioning of DNA repair mechanisms.
DnaK is a member of the class I heat shock genes in which regulation involves an inverted repeat (CIRCE element). Transcription of the other member of the CIRCE regulon, GroEL, also increases in response to acid shock. In S. mutans, both groESL and dnaK expression are negatively regulated by HrcA, also encoded within the dnaK operon. An hrcA mutant (SM11) was more sensitive to acid, not being able to lower the environmental pH as effectively as the parental phenotype, although it could mount an ATR. Even then, adapted SM11, cells demonstrated a lower F1F0-ATPase activity than did adapted wild-type cells (141). These phenotypes are likely to be due to the altered regulation of chaperone production.
A link between acid stress and chaperones has been demonstrated in a number of other gram-positive bacteria. Acid shock induces 33 proteins, including DnaK, GroEL, and UV-inducible proteins, in L. lactis subsp. lactis strain IL 403 (112). DnaK and GroEL are also induced following acid adaptation of Lactobacillus delbrueckii (145), while GroEL is induced in response to acid stress in Clostridium perfringens (238), S. mutans (249), and Listeria monocytogenes (178, 179).
In addition to the protective role of chaperonins, the HtrA protein is proposed to be involved in proteolysis of abnormal proteins synthesized under stressful conditions. An S. mutans HtrA mutant demonstrated a reduced ability to grow at low pH (76). The Clp ATPases also perform a similar role by targeting misfolded protein for degradation by the ClpP peptidase, which itself is not a member of the Clp ATPase family, in addition to protein reactivation and remodeling activities. The Clp proteins are members of the class III heat shock proteins that are regulated by CtsR. Strains lacking ClpP exhibited reduced growth yield at pH 5.0 (141), again demonstrating the need for removal of abnormal proteins.
The small heat-shock protein, Lo18, of O. oeni is induced after acid, heat, and ethanol stress (105). Upstream of hsp18, the gene encoding Lo18, are found two open reading frames, orfB and orfC, encoding a putative regulator of the LysR family and a protein with homologies to multidrug resistance systems. Heterologous expression of these genes in E. coli significantly enhanced viability under acidic conditions (162).
Cell Membrane Changes
In addition to a reliance on mechanisms directed to protect or repair DNA and protein damage in general, it has been shown that gram-positive bacteria are frequently sensitized to acid as a consequence of the mutation of genes involved in cell membrane biogenesis, assembly, and maintenance.
The importance of the role of the cell membrane is demonstrated by the changes in membrane fatty acid profiles in response to a pH drop. S. mutans strains grown at pH 5 had increased levels of monounsaturated fatty acids and longer-chain fatty acids than did those grown at pH 7. In contrast S. sobrinus 6715, which does not adapt well to pH stress, was less capable of changing its membrane composition (187). Acid-adapted I. monocytogenes was also more tolerant to nisin and other ionophores than was the nonadapted counterpart (67). It is suggested that the increase in the production of the straight-chain fatty acids C14:0 and C16:0 as well as the decreased C18:0 levels associated with acid adaptation may be responsible for this enhanced resistance to bacteriocins as well as, to some extent, the cross-protective effects of acid adaptation in general (235).
The dlt operon is involved in the synthesis of d-alanyl-lipoteichoic acid synthesis. Inactivation of dltC, encoding the d-alanyl carrier protein (Dcp), resulted in the generation of an acid-sensitive S. mutans strain. This mutant could not grow below pH 6.5, and unadapted cells were unable to survive a 3-h exposure in medium buffered at pH 3.5, while a pH of 3.0 was required to kill the wild type in the same period. Following acid adaptation, the level of mutant survival was 3 to 4 orders of magnitude lower than that of wild-type cells. Proton permeability assays revealed that the mutant was more permeable to protons than was the wild type (21). Another S. mutans mutant, with a deficiency in the DagK diacylglycerol kinase, demonstrated increased acid sensitivity. This enzyme is involved in phospholipid metabolism, and therefore membrane architecture and composition is again implicated (255).
A number of other genes have been implicated in contributing to membrane stability and activity at low pH. The acid-sensitive phenotype of a transposon mutant, S. mutans AS17, was found to be due to a partial polar effect on the ffh gene (104). Ffh, the 54-kDa subunit homologue of the eukaryotic signal recognition particle, has also been identified in B. subtilis and E. coli and is involved in protein translocation and membrane biogenesis (149). In S. mutans, ffh is located within the secretion and acid tolerance (sat) operon, ylxM, ffh, satC, satD, and satE, which has an acid-inducible promoter upstream of ylxM (134). This explains the induction of the ffh gene in the parent in response to acid shock (104), although it is the only gene of this operon that is involved in acid tolerance (134). The AS17 mutant is unable to induce an ATR (104), while a subsequently created nonpolar mutant was found to be unable to initiate growth at pH 5.0 (134). The finding that F1F0-ATPase levels in the mutant were twofold lower than in the parent strain may at least partially explain its acid sensitivity (104). Another protein, SGP (Streptococcus GTP-binding protein), is associated with the membrane when S. mutans is grown under a variety of stress-inducing conditions including low pH (8), while decreased growth is observed at low pH when sgp antisense RNA is expressed (204). SGP, like its E. coli homolog Era, exhibits properties associated with G-proteins, and while it is essential for normal growth, its function has yet to be determined. Era associates with the inner membrane of E. coli, and it has been suggested that it functions by acting as an interface between energy metabolism and macromolecular synthesis at the ribosome (123).
Production of Alkali
The capacity of oral biofilms to generate alkaline compounds, which can neutralize acids and prevent the emergence of a cariogenic microflora, is a critical factor in caries prophylaxis and in the inhibition of caries progression. The two most common ways in which oral bacteria generate alkali, or, more specifically, ammonia, involve the urease and arginine deiminase (ADI) pathways. The increased tolerance of cells results from the production of NH3, which combines with protons in the cytoplasm to produce NH4+, raising the internal pH. Urea diffusing from saliva into dental plaque is converted to ammonia and carbon dioxide by bacterial ureases, while ADI catabolizes arginine to ornithine, ammonia, and CO2. An extreme example of the role of urea involves patients with chronic renal failure, who have salivary urea levels 5- to 25-fold higher than healthy controls and who have a very low caries incidence despite having to consume a diet composed primarily of carbohydrates (177). Furthermore, the levels of free arginine in the parotid saliva of caries-free adults were significantly higher than in adults with a history of dental decay (236). This explains why caries-resistant subjects have greater ammonia concentrations and higher resting pH in their plaque (152).
Ammonia is also known to be produced by some mycobacteria and is thought to play a role in the increase in vacuolar pH brought about by phagocytosed mycobacteria (89). It has been proposed that this trait is responsible for the initial capacity of nonpathogenic mycobacteria to survive in macrophages (136).
Production of urease.
Urease is a nickel-containing oligomeric enzyme that catalyzes the hydrolysis of urea to two molecules of ammonia and one molecule of carbon dioxide (161). Levels of 3 to 10 mM urea are found in the saliva of healthy individuals (100), although this can range up to 30 mM in patients with renal disease (177). Urease is produced by a discrete subset of oral bacteria, including S. salivarius, Actinomyces naeslundii, and oral haemophili. Of the oral bacteria, S. salivarius produces the greatest quantities of urease (211). The enzyme has maximal activity at pH 7.0 and retains 80% activity between pH 5 and 8.5 but rapidly loses activity at more extreme acidic pHs, being essentially inactive at pH 4.3 (211). Regulation by the growth pH is the preeminent factor in determining the levels of urease produced. In addition, the glucose concentrations in the medium and the growth rate of the cells have an impact (36). At low pH values (pH 5.5), wild-type urease expression increased approximately 600-fold over that observed in cultures grown at pH 7.0 (38). This was supported by the observation that the level of urease-specific mRNA varied depending on the pH (36). The phosphoenolpyruvate-dependent phosphotransferase system also plays a significant role, as shown by the observation that carbon flow through the system could alleviate the repression of urease (38).
The significance of the urease system was best illustrated when a strain of S. mutans, which does not possess a urease system, was genetically engineered to express the urease genes of S. salivarius. Rats infected which this strain had a dramatically lower incidence and severity of all types of caries than did the controls, showing that alkali generation inhibits caries (42).
The S. salivarius urease operon consists of ureIABCEFGD, with ureC, ureB, and ureA encoding the α, β, and γ subunits that comprise the apoenzyme. The subunits encoded by ureDEFG function as accessory proteins and assist in the incorporation of nickel ions into the enzyme (37). Two promoters, PureI and PureA, were identified, with the former being regulated by acidic pH and carbohydrate levels (39).
The current model proposes the existence of a repressor that is phosphorylated in the absence of sugar and during growth at neutral pH values. When sugar becomes abundant, the phosphotransferase system would preferentially phosphorylate sugar, leaving the urease repressor unphosphorylated, thereby facilitating PureI-driven synthesis. Repression of PureI was also observed when it was cloned into the nonureolytic S. gordonii DL1, suggesting the involvement of a global regulatory circuit (35).
Arginine deiminase.
The ADI system has been identified in a variety of bacteria, including mycoplasmas, halobacteria, Pseudomonas sp., Bacillus spp., and a number of lactic acid and dental bacteria catabolizing arginine to ornithine, ammonia, and CO2 (59). The impact on humans of bacteria utilizing this system varies greatly. Its presence in oral bacteria is regarded as beneficial. Streptococcus gordonii, S. parasanguis, S. rattus, and S. sanguis (S. sanguinius), some lactobacilli, and a few other oral bacteria, including spirochetes, all utilize either the free form of arginine, which is secreted at concentrations averaging about 50 μM, or arginine in salivary peptides and proteins (236). Despite not seeming to play a role in the ATR, at least not in that of S. sanguis, the system does play a role in the acid resistance of a number of LAB (60). Its contribution to the acid resistance of lactic acid bacteria and, more specifically, to that of some of the less inherently acid resistant (i.e. non-mutans) oral lactic acid bacteria is of great importance. The presence of an ADI system in such bacteria is significant because it permits the survival of desirable, less acidogenic bacteria in plaque in the presence of their more aciduric and acidogenic neighbors (26). While ADI-positive bacteria can utilize arginine in the host diet (193), it has also been shown that in salivary levels of arginine, lysine, or peptides containing these amino acids that may be at least partially responsible for the low caries rates in caries-resistant individuals (236). Among the comparatively few ADI-positive organisms that colonize the mouth, S. gordonii is one of the most abundant on tooth biofilms. It is an early colonizer of teeth and forms a major portion of healthy human supragingival dental plaque. Its establishment may even inhibit subsequent colonization by the highly acidogenic, aciduric species responsible for dental caries (78). The system has three main enzymes: ADI, ornithine transcarbamylase and carbamate kinase, encoded by arcA, arcB, and arcC respectively. The three main enzymes involved in the system appear to be inherently acid tolerant, displaying activity at pH 3.1 and even lower in some species (30). The level to which this system can be induced may reflect its importance. The degree of induction is 48-fold in S. rattus, 1,467-fold in S. sanguis 10904, and over 300-fold in S. gordonii (61). One should note, however, that pH itself does not seem to be a major factor in ADI expression in oral streptococci.
The ADI system is also present in a number of lactic acid bacteria involved in different food fermentations, although its desirability varies. Lactobacillus sakei, used in meat fermentations, is unusual in that it is a facultative heterofermenter that possesses an ADI pathway. A mutant that lacked arginine deiminase activity was tested to determine the importance of the ADI system in L. sakei for meat fermentations. While no difference between the mutant and wild type was apparent, the authors point out that the levels of free arginine in such circumstances can vary greatly depending on the proteolytic activity (263). Of 70 strains of sourdough LAB screened for ADI, ornithine transcarbamylase, and carbamate kinase, only obligate heterofermenters possessed all three. One of these, Lactobacillus sanfranciscensis, was examined in greater detail. In addition to enhancing tolerance to acid stress, the system assisted cell growth, assisted cell survival during storage at 7°C, and favored the production of ornithine, an important precursor of crust aroma compounds (70). Arginine is also one of the major amino acids found in grape juice and wine. While the ADI system protects the LAB involved in MLF of wine (148), there is some concern about the creation of ethyl carbamate (urethane) precursors. Ethyl carbamate is an animal carcinogen that is formed in wine during the nonenzymatic reaction of alcohol with citrulline (228).
The ADI system acts as a virulence factor in Streptococcus pyogenes. S. pyogenes is a serious human pathogen and causes infections leading to acute disorders such as pharyngitis, erysipelas, otitis media, and impetigo and potentially to glomerulonephritis, acute rheumatic fever, and reactive arthritis. A protein originally designated streptococcal acid glycoprotein and originally characterized as being an inhibitor of stimulated human peripheral blood mononuclear cell proliferation (73, 74) was ultimately identified as an ADI. Its role in acid resistance is demonstrated by a massive 9-log-unit difference in survival between a wild-type strain and an isogenic streptococcal acid glycoprotein-negative mutant after 6 h at pH 4 in the presence of 10 mM arginine (72). This difference in acid sensitivity is likely to be responsible for the observation that this mutant demonstrates a reduced ability to enter and initially survive in epithelial cells. Streptococcus suis, which causes a number of diseases in young pigs and is also a cause of meningitis in humans, also possesses an equivalent system, although its role in acid resistance and virulence has yet to be established (250).
In addition to arcA, arcB, and arcC, a number of additional genes play a role in the ADI systems of a variety of organisms (Fig. 3). ArcD, an arginine-ornithine transporter, has been identified in Enterococcus faecalis, Lactobacillus sakei, and Streptococcus gordonii, while ArcT, found in L. sakei and S. gordonii, appears to be a dipeptidase. arcR encodes an activator of the ADI system and is a member of the Crp-Fnr family of regulators. Identification of acid-resistant mutants of Lactococcus lactis found that one such mutant resulted from disruption of a gene that demonstrated sequence similarity to ahrC, which encodes a regulator of arginine synthesis and catabolism in Bacillus subtilis (12). It is suggested that this mutation may have resulted in constitutive expression of arginine-metabolic enzymes in the ADI pathway (190). Similar regulators have been found in E. faecalis, where arginine induces expression of arcABCRD by means of two homologous ArgR/AhrC-type regulators (ArgR1/ArgR2). Transcription of argR1 and argR2 is influenced by glucose and arginine, and it is likely that induction of the system by arginine and repression by glucose is mediated by R1 and R2 (see next paragraph). There are R1- and R2-binding sites within the promoter regions of arcA, argR1, and argR2. These promoters also have ArcR- and CcpA-binding sites (11). Additional regulation in L. lactis subsp. cremoris MG1363 may be provided by the two-component signal transduction system, LlkinA-LlrA. While mutation of either of these components results in an ADI-negative and acid-sensitive phenotype, the precise role of this system has yet to be determined (169).
The ADI systems in LAB associated with cheese fermentations (57), L. sakei (262), oral LAB (78, 153), and E. faecalis (209) are subject to catabolite repression by glucose. In fact, the original observation that one of the major functions of the ADI system was to assist acid tolerance followed the observation that S. faecium and S. sanguis mutants which had defective catabolite repression systems and which could simultaneously degrade both glucose and arginine demonstrated greatly increased acid resistance in the presence of glucose (153). The derepression of these mutants can also be compared to the ability of the ADI system to protect cells that have been in stationary phase for a time when natural derepression occurs (28). In contrast, L. sanfranciscensis (70), O. oeni, and other wine LAB (147) catabolize glucose and arginine concurrently.
Regulators
Regulation of gene expression in response to the extracellular environment is an adaptive response that is required for bacterial replication and survival. Alternative sigma factors have been demonstrated to play an important role in the coordinate regulation of gene expression in bacteria undergoing major adaptive changes in physiology; it is therefore unsurprising that such sigma factors play a role in the response of bacteria to low pH.
Sigma factors.
σB is an alternative “stress” sigma factor that has been identified in L. monocytogenes, B. subtilis, and Staphylococcus aureus and whose function has been compared to those of RpoS/σS in gram-negative bacteria as a consequence of its regulation of a large stress-induced regulon in B. subtilis and S. aureus.
σB was initially identified in B. subtilis as controlling transcription of genes induced at the end of the exponential phase (107, 251). It exerts its influence on a regulon of over 100 csb (for “controlled by sigma B”) genes in response to environmental and energy stresses. The title of “stress sigma factor” is apt because B. subtilis σB mutants display a 50- to 100-fold reduced ability to survive heat (54°C), ethanol (9%), salt (10%), or acid (pH 4.3) shocks as well as freezing, desiccation, and exhaustion of glucose or phosphate (241). In fact, heat shock proteins regulated by σB constitute the class II heat shock regulon.
RsbW (for “regulator of sigma B”) functions as an anti-σ factor (19). RsbW thus prevents σB activity from occurring at inappropriate times, and its loss is deleterious to logarithmically growing cells (22). RsbV acts as an “anti-anti-σ factor” by preventing the formation of an RsbW-σB complex (81). RsbV is itself modulated by the phosphatases RsbP (237) and RsbU (240). The sensing of energy stress is mediated through the PAS domain of RsbP (237) and an α/β hydrolase, RsbQ (23), while environmental stress is sensed through the activation of RsbU by a regulatory cascade that includes the RsbS antagonist, the RsbT kinase, the RsbX phosphatase, the RsbR regulator, and four RsbR paralogues, YkoB, YojH, YqhA, and YtvA (1, 2, 127, 239).
The equivalent S. aureus genes were identified by analysis of a Tn 551 mutant that exhibited a dramatically reduced resistance to methicillin. The genes corresponding to rsbU, rsbV, rsbW, and sigB were found between orf136 and CTorf239, which did not show homology to any known proteins (253). No open reading frames corresponding to the B. subtilis rsbR, rsbS, rsbT, or rsbP genes have been identified (Fig. 3). Strain NCTC 8325, which demonstrates low methicillin resistance, has a truncated rsbU gene that seems to be caused by a frameshift. In the highly methicillin-resistant strain COL, a stretch of an additional 11 bp restores the reading frame. σB mutations, in strains with an intact rsbU gene, resulted in these strains being deficient in their ability to respond to acid (139) and impaired their ability to induce a stationary-phase ATR (32). It was confirmed that RsbU was indeed the major activator of the σB response to acid stress when it was shown that expression from the σB-dependent sarA P3 promoter was greatly reduced at pH 5.5 in the absence of RsbU (176).
The rsbU, rsbV, rsbW, sigB, and rsbX genes have also been identified in L. monocytogenes (15, 247). Again, a σB-dependent promoter has been identified upstream of rsbV, and this is induced by a drop in pH to 5.3. σB mutants were found to be extremely acid sensitive, with survival of mutant stationary-phase cells exposed to pH 2.5 being 3.6 log units lower than that of the isogenic parent after 2 h (247). Increased sigB transcription was seen following exposure of exponentially growing cells to acid (15). The percent survival of a stationary-phase culture of this mutant was also found to be 10,000-fold lower than that of the wild type after 3 h at pH 2.5. However, adaptation of this mutant at pH 4.5 improved the survival of the mutant by 1,000-fold, suggesting the existence of both σB-dependent and -independent pathways (86). The σB mutant also demonstrates reduced virulence potential, possibly because σB regulates one of the promoters immediately upstream of PrfA (163).
The Brevibacterium flavum sigB gene, a homolog of which has also been identified in B. lactofermentum (172), is significantly induced under several stress conditions, one of which is acid stress (108). While wild-type B. flavum demonstrates a reduced growth rate at pH 4.4, the growth of a sigB mutant was arrested at this pH (109).
The mycobacterial extracytoplasmic function sigma factors regulate gene expression in response to the extracellular environment. One of these, SigE, influences survival following stress in a number of mycobacteria, as demonstrated by the enhanced rate of killing of an M. smegmatis sigE mutant at pH 4 (252).
Two-component signal transduction systems.
Bacteria can also sense and respond to environmental changes through the use of two-component signal transduction systems (2CSs). 2CSs typically consist of a membrane-associated histidine kinase sensor and a cytoplasmic response regulator. 2CSs function in bacterial adaptation, survival, and virulence by sensing environmental parameters and giving the bacteria a mechanism through which they can respond.
A transposon mutant of L. monocytogenes has been identified which is more sensitive to low pH than the wild type during the logarithmic phase of growth and yet more acid resistant during stationary phase. The mutation responsible for this phenotype is located in a two-component system, lisRK (49). This system is also involved in the resistance of Listeria to agents that act at the cell envelope, such as ethanol, nisin, and cephalosporins (49, 52). On the basis of the roles of LisRK homologs in other bacteria (103, 157), as well as its regulation of a penicillin binding protein, it is likely that the growth phase variation in acid sensitivity of this mutant is linked to the regulation of cell envelope composition. The LisRK system is the only one of a number of 2CSs from Listeria that plays a role in acid stress resistance (125). In addition, of a number of PhoP-PhoS homologs identified in E. faecalis only one, EtaRS, a homolog of LisRK, had an impact on the acid resistance of the cell. Mutation of etaR resulted in a 100-fold decrease in the survival of logarithmic cells at pH 3.4 (227).
The role of two 2CSs from S. mutans, ComDE and HK11/RR11, which act in acid resistance and biofilm formation, is described in the next section.
Cell Density and Biofilms
The cell density of gram-positive bacteria affects their acid resistance, although this effect varies depending on whether the cells are planktonic or part of a biofilm. When attached to stainless steel, L. monocytogenes demonstrates increased resistance to acetic acid, and this resistance increases with the age of the culture (173). Cell density was found to modulate the acid adaptation of log-phase S. mutans BM71, since preadapted cells at a higher cell density from a dense biofilm displayed significantly higher resistance to the killing pH than did the cells at a lower density. The log-phase ATR could also be induced by a neutralized culture filtrate collected from a low-pH culture, suggesting that the culture filtrate contained an extracellular induction component(s) involved in acid adaptation in S. mutans. Mutants defective in the comC, comD, or comE genes, which encode a quorum-sensing system essential for cell density-dependent induction of genetic competence, had a diminished log-phase ATR. Addition of competence-stimulating protein to the comC mutant partially restored the ATR. These results show that cell density and biofilm growth modulate acid adaptation and suggest that optimal development of acid adaptation in this organism involves both low pH induction and cell-cell communication (143). This may explain why biofilm cells possess a number of novel proteins, of unknown function, that are not produced by planktonic cells. In addition to cell-cell communication, the formation of biofilms seems to physically protect cells from an exogenous source of protons because live cells are frequently observed in the deepest layers of the biofilm. It would seem, however, that biofilm formation does not protect bacteria from the internal low-pH shock that results from the acidic by-products of metabolism (221). Another 2CS, HK11-RR11, plays a role in the acid resistance of and biofilm formation by S. mutans. Deletion of hk11 resulted in slow growth at low pH, a diminished ATR, and defects in biofilm formation. HK11 may act in conjunction with one or more response regulators other than RR11 since mutation of this response regulator does not affect acid sensitivity (144).
In addition to the acid shocks produced by lactic acid or during sucrose consumption, starvation induces acid resistance of S. mutans in suspension and in biofilms, making it likely that during the “famine” between meals, the bacteria in oral biofilms may be more resistant to acid (259). The ability of S. mutans biofilms to cause dental caries is aided by a number of extracellular sucrose-metabolizing enzymes including three glucosyltransferases, encoded by gtfB, gtfC, and gftfD, and a fructosyltransferase, encoded by the ftf gene. Promoter reporter studies showed that in cells growing as part of a biofilm, the gtfBC genes were induced by sucrose, glucose, and a drop in pH (142).
The induction of acid tolerance at neutral pH has also been the subject of analysis to explain the growth phase acid sensitivity of Lactococcus lactis. “Growth phase acid sensitivity” is the term used to describe the observation that stationary-phase cells inoculated into fresh broth become gradually more acid sensitive, reaching their most sensitive state during the early logarithmic phase before tolerance increases to a maximum at the onset of the stationary phase. The growth phase acid tolerance of L. lactis does not result from changes in internal pH or F1F0-ATPase activities, and the increased sensitivity of early-log cells is not related to either glucose levels or lactate production. It was found that cells grown in partially spent TYG (tryptone, yeast extract, glucose) medium had increased relative acid tolerance, but that this enhanced tolerance is eliminated by the addition of tryptone or yeast extract. More specifically, a 1-kDa protease-sensitive putative active compound has been purified and is thought to be responsible for this phenotype (4).
Alteration of Metabolic Pathways
While studies of bacterial acid resistance have revealed mechanisms responsible for growth at mildly acidic pH, acid adaptation, and survival at more extreme pH, the systems that respond to autoacidification, which is of particular importance during fermentation by lactic acid bacteria, are less well understood. It has been established that as acidification progresses, ATP is rerouted to generate a proton gradient with a concomitant reduction in the energy available for biomass synthesis and catabolic flux until growth is eventually arrested. The metabolic perturbations that occur are brought about through altered transcription of genes involved in central metabolism, although, surprisingly, these alterations were often not evident during the period of sharp pH decline, in some cases possibly due to an increase in mRNA stability. In fact, an increase in the concentration of several glycolytic enzymes was observed postacidification, and this has the potential to facilitate rapid growth recovery if the pH stress is removed (84).
ACID RESISTANCE OF SELECTED GRAM-POSITIVE BACTERIA
Listeria monocytogenes
Listeria monocytogenes, the causative agent of listeriosis, may encounter low-pH environments at a number of stages during the course of its infectious cycle, including in low-pH foods, during gastric transit, following exposure to fatty acids in the intestine, and finally in the phagosome of macrophages, where Listeria not only survives acid stress but also requires a drop in pH in order to activate hemolysin, the toxin that permits its escape from the phagosome. Listeria must therefore have a number of mechanisms for responding to, and surviving in, acidic environments.
L. monocytogenes can induce a protein synthesis-dependent log-phase ATR when exposed to an induction pH (5.0 to 5.5), dramatically increasing the resistance of log-phase cells (69, 135, 170) to more extreme acidic conditions. One of the primary consequences of acid adaptation appears to be an increased intracellular pH (pHi), since it was found that at low external pH, acid-adapted and acid-resistant mutant cells maintained an elevated pHi relative to the wild type. This elevation in pHi is dependent on protein synthesis (171). The presence or absence of glucose has a large impact on pH. In the presence of glucose, the pHi drops below pH 7.0 only when the extracellular pH (pHo) drops below 4. However, in the absence of glucose, a pHo of 5.5 causes a drop in pHi. In both cases, a drop in pHi coincides with a net influx of protons, indicating that membrane transport processes are the main contributors to the process of pHi homeostasis in L. monocytogenes subjected to acid stress (206).
Two-dimensional analysis of the proteins induced during adaptation has identified 17, 30, and 21 proteins present at increased levels and 17, 8, and 9 present at reduced levels following adaptation with hydrochloric acid or lactic acid or in the nonadapted ATM56 mutant, respectively, in complex broth. In addition, six proteins were identified following HCl adaptation which were not visible on other gels (171). The proteins induced in response to acid induction and acid stress caused by the addition of HCl to minimal medium have also been examined (178, 179). It was found that 47 and 37 proteins were induced by pH 3.5 and 5.5, respectively, and that 23 of these were common (Table 1). These included ATPase-subunit B and GroEL, both of which have been discussed above.
TABLE 1.
Acid-induced proteins identified by using high-throughput screening strategies
| Organism (conditions) and gene | Predicted protein | Organism (conditions) and gene | Predicted protein | |
|---|---|---|---|---|
| S. oralis (pH 5.2, 2D-PAGE) (248) | 60-kDa chaperonin | |||
| Hsp33 protein (redox-regulated chaperone) | ||||
| Cell division protein DivIV A | ||||
| Superoxide dismutase | ||||
| Formate acetyitransferase 1 | ||||
| Pyruvate oxidase | ||||
| NADP-specific glutamate dehydrogenase | ||||
| Glyceraldehyde-3-phosphate dehydrogenase | ||||
| l-Lactate dehydrogenase | ||||
| Fructose bisphosphate aldolase | ||||
| Acetoin utilization AcuB protein | ||||
| Galactose-6-phosphate isomerase, LacB subunit | ||||
| ATP synthase alpha chain | ||||
| ATP synthase beta chain | ||||
| ATP synthase gamma chain | ||||
| ATP synthase delta chain | ||||
| ATP synthase epsilon chain | ||||
| Formate-tetrahydrofolate ligase | ||||
| Oxygen-insensitive NAD(P)H | ||||
| Nitroreductase/dihydropteridine reductase | ||||
| ABC transporter | ||||
| Hypothetical ABC transporter | ||||
| ATP-binding protein | ||||
| EIIA component of MTL phosphotransferase system | ||||
| Nicotinate phosphoribosyltransferase | ||||
| Adenylate kinase | ||||
| ReoA protein | ||||
| Polyribonucleotide nucleotidyltransferase | ||||
| Polypeptide deformylase | ||||
| Membrane protein precursor | ||||
| Succinyl-diaminopimelate desuccinylase | ||||
| Phosphoprotein phosphatase 2C | ||||
| M. tuberculosis (pH 5.5, microarray) (88) | ||||
| Rv3083 | Probable monooxygenase | |||
| Rv30084 (lipR) | Probable acetyl hydrolase | |||
| Rv3085 | Short-chain ADH | |||
| Rv3086 (adhD) | Zinc-containing ADH | |||
| Rv3087 | Conserved hypothetical protein | |||
| Rv3088 | Conserved hypothetical protein | |||
| Rv3089 (fadD13) | Acyl-CoA synthase | |||
| Rv0467 | Isocitrate lyase | |||
| Rv0468 (icl) | 3-Hydroxyacyl-CoA dehydrogenase | |||
| Rv3614c (fadB2) | Conserved hypothetical protein | |||
| Rv3615c | Conserved hypothetical protein | |||
| Rv3616c | Conserved hypothetical protein | |||
| Rv1130 | Conserved hypothetical protein | |||
| Rv1131 (gltA1) | Citrate synthase 3 | |||
| Rv2428 (ahpC) | Alkyl hydroperoxide reductase | |||
| Rv2429 (ahpD) | Member of AhpC family | |||
| Rv1245C | Putative dehydrogenase | |||
| Rv2007c (fdxA) | Ferredoxin | |||
| Rv1404 | Transcriptional regulator (MarR family) | |||
| MT0196 | Hypothetical protein | |||
| Rv1169c (PE) | PE family protein | |||
| Rv0582 | Hypothetical protein | |||
| Rv2450c (rpfE) | Conserved hypothetical protein (resuscitation-promoting factor) | |||
| L. monocytogenes (pH 3.5 and 5.5, 2D-PAGE) (179) | ATP synthase (subunit B) | |||
| Thoredoxin reductase homologue | ||||
| Chaperonin | ||||
| Sigma H factor | ||||
| Ferric uptake regulator | ||||
| Alcohol dehydrogenase | ||||
| YfiV, similar to transcriptional regulator | ||||
| Similar to transcriptional regulator | ||||
| Transcriptional regulator | ||||
| Similar to GroEL | ||||
| S. mutans (pH 5.2, 2D-PAGE) (249) | DnaK | |||
| GroEL | ||||
| Trigger factor, Ppiase | ||||
| Cell division FtsZ | ||||
| Cell division protein FtsA | ||||
| Superoxide dismutase (Mn) | ||||
| Neutral endopeptidase | ||||
| Glucan-1,6-α-glucosidase | ||||
| Phosphoglucomutase/ phosphomannomutase | ||||
| Enolase | ||||
| NADP-specific glutamate dehydrogenase | ||||
| l-Lactate dehydrogenase | ||||
| Enoyl-(acyl-carrier protein) reductase (NADH) | ||||
| Fructose bisphosphate aldolase | ||||
| Acetoin reductase | ||||
| Metal-dependent hydrolase | ||||
| Lactoylglutathione lyase | ||||
| Phosphoenolpyruvate:protein phosphotransferase | ||||
| Multiple sugar-binding transport protein ATP-binding transport protein MsmK | ||||
| EF-G | ||||
| EF-Tu | ||||
| Small-subunit ribosomal protein S1P | ||||
| EF-Ts | ||||
| DNA-directed RNA polymerase alpha chain | ||||
| Ribosome-recycling factor | ||||
| S. mutans (pH 5.5, differential-display PCR) (40) | ||||
| AP11 | Hippurate hydrolase | |||
| AP-712 | Acetolactate synthase, large subunit | |||
| AP-711 | Pyruvate carboxylase subunit B | |||
| GSP781 | Putative secreted protein (S. mutans) | |||
| AP-785 | Conserved hypothetical protein (Thermotoga maritima) | |||
| AP-185 | Branched-chain amino acid transferase | |||
| AR-186 | Aconitase hydratase | |||
| AR-731 | Isocitrate dehydrogenase | |||
| AR-732 | Citrate synthase |
The ability of L. monocytogenes to survive acid stress was found to be growth phase dependent, as indicated by the fact that 100% of stationary phase cells can survive exposure to pH 3.5 for 60 min. These cultures maintain high levels of acid resistance following inoculation into fresh broth but become more sensitive as the growth rate increases up to mid-log-phase, when they are at their most sensitive, resulting in a 2.5-log-unit reduction in cell numbers, before becoming gradually more resistant as the cells reenter the stationary phase (170). However, the phenomenon of cell density-dependent acid sensitivity in stationary-phase cultures observed in E. coli was found to be absent in Listeria and other gram-positive bacteria (68).
The acid resistance of Listeria influences its survival in foods. In addition to the role of the GAD system described above, the phenomenon of acid adaptation aids the survival of this food pathogen in acidic foods such as cottage cheese, yoghurt, whole-fat cheddar cheese, orange juice, and salad dressing and thus must be taken into consideration when using low pH as a preservative (92). In addition to the pH, the natural flora of the food can have a significant impact. It was found that the presence of the natural microbial flora that may be established in a meat-processing plant in nonacidic (water) washings had a detrimental effect on the ability of L. monocytogenes to subsequently induce an ATR, thus sensitizing it to subsequent exposure to acid. This may be due to L. monocytogenes having to adapt and survive under the conditions created in the water washings by the gram-negative spoilage flora. Since bacterial pathogens seem to be acid sensitized under the stressful conditions prevailing in nonacidic meat washings, it may be possible to use water, steam, or other nonacid treatments with or without low-pH meat decontamination technologies to manipulate the natural flora in a way that may be advantageous to meat safety (200).
In addition to acidic foods, Listeria encounters low-pH stress in vivo. In agreement with results showing the importance of the role of the GAD acid resistance system in the survival of L. monocytogenes in gastric juice, the increased gastric transit of Listeria following an increase in the pH of the stomach demonstrates the role of stomach acid as a primary barrier against listeriosis (44, 63, 205, 258). A neutropenic-mouse model was used to demonstrate that oral administration of sodium bicarbonate shortly before intragastric inoculation significantly increased the severity of the resulting systemic infection by L. monocytogenes strain EGD (64). When the murine gastric pH was lowered through the use of acidified water, it was observed that acid-adapted cells enjoyed increased resistance relative to their unadapted counterparts (198). These results suggest that control of gastric pH through dietary practices or use of inhibitors of gastric acid secretion may be potential aggravating risk factors for food-borne listeriosis.
The induction of an ATR does not improve the survival of L. monocytogenes cells inoculated into the intraperitoneal cavity of mice. However, it has been speculated that this may be a consequence of cells naturally developing acid tolerance following uptake by macrophages, thus eliminating any advantage that would be conferred by prior adaptation (91). Acid-adapted cells do, however, enjoy an advantage over nonadapted cells in cell culture assays, as is evidenced by a ninefold increase in the invasion efficiency of the enterocyte-like cell line Caco-2. This advantage also extends to growth in lipopolysaccharide-treated J774.A1 macrophages. In these cells, four- to fivefold-greater numbers of acid-adapted Listeria were present 4 h postinfection, relative to the number in untreated cells. At 8 h postinfection, the majority of acid-adapted cells were free in the cytoplasm whereas a large number of the nonadapted cells remained in intact phagosomes (46). Both adapted and nonadapted bacteria were phagocytosed and multiplied efficiently in nonactivated THP-1 macrophages. Following activation of the macrophages, the rate of invasion of acid-adapted and nonadapted cells remained equal. However, the active macrophages became nonpermissive to growth of the nonadapted cells while acid-adapted cells demonstrated a greatly enhanced intracellular growth rate (47).
The enhanced survival of acid-adapted L. monocytogenes in activated human macrophages is associated with an altered pattern of expression of genes involved in acid resistance and cell invasion. Transcription of plcA, which is involved in vacuolar escape and cell-to-cell spread, is decreased by 12% after acid adaptation. In contrast, increased transcription of sodC (310%) and the combined gad genes (3000%) was observed after acid adaptation (47). It is interesting that while Hly and PlcA are both expressed in the phagosome and contribute to vacuolar escape, the pH ranges for their enzymatic activities are different (96, 99). Hly expression requires a more acidic pH (4.5 to 6.5) than does PlcA (pH 5.5 to 7.5) and is triggered by the immediate drop in the pH of the macrophage phagosome following phagocytosis (14), with optimal activity in vivo observed at pH 5.94. The key to the survival of the bacteria appears to be their escape before phagosome-lysosome fusion (71). It was found that inhibitors of vacuolar ATPases such as the lipophilic weak base ammonium chloride bafilomycin A1 or the carboxylic ionophore monensin completely prevent perforation of the vacuole, suggesting that a drop in pH is a prerequisite for phagosomal escape (14, 48). On the basis of these observations, it is speculated that after cell invasion, the acidic environment of the phagosome could prevent plcA expression while enabling Hly to be accumulated in an enzymatically active state. Pore formation by Hly would then raise the phagosomal pH to neutral values, thereby increasing PlcA expression (47). Such tight regulation is essential, since many virulence factors, including listerial hemolysin and phosphatidylinositol-specific phospholipase C (208), are highly antigenic for the host immune response. Thus, both premature and delayed expression are counterproductive to bacterial survival. This was demonstrated when a leucine-to-threonine conversion was generated at position 461 of hemolysin and resulted in a 10-fold increase in activity at neutral pH. As a consequence of premature permeabilization of the host membrane, resulting in host cell death, there was a 100-fold virulence defect in the mouse listeriosis model (98).
To further appreciate the consequences of the emergence of acid-resistant strains, spontaneous acid-tolerant mutants were isolated which were almost as resistant, without acid induction, as adapted wild-type cells and which could be further induced to display even greater levels of survival (46, 170). Compared with wild-type cells, these mutants displayed enhanced acid tolerance during all stages of growth, enjoyed improved survival in acidic foods (92), displayed enhanced invasiveness and growth in enterocyte-like and macrophage-like cell lines, respectively (46), and caused higher rates of mortality in mice (170). Further evidence for the role of the ATR in virulence has also been obtained by the identification of a mutant, ATR-1, that is unable to induce acid tolerance and displays diminished virulence in mice (154). In this case, it was subsequently found that any ATR-1 survivors from this virulence study had all reverted to being able to induce an ATR (unpublished results).
Acid resistance is subject to strain variation. Of a selection of 15 strains of L. monocytogenes of clinical origin, none displayed significant sensitivity after exposure to pH 2.5 for 2 h, yet 2 of a selection of 15 strains of meat origin were acid sensitive (82). It was thought to be significant that no acid-sensitive strains were selected from among the clinical isolates, again suggesting the importance of acid resistance in the infectious process (82).
Rhodococcus equi
Rhodococcus equi is a facultative intracellular pathogen causing suppurative bronchopneumonia, lymphadenitis, and/or enteritis in foals younger than 3 months (185). There has also been an increase in the number of cases of infections in patients with AIDS or some other form of immunodeficiency (79), with the route of infection suspected to be via inhalation of dust. A large (85- to 90-kb) plasmid is associated with virulent strains (225) that appear to proliferate in phagosomes prior to fusion with lysosomes (260).
During its stay in alveolar macrophages, R. equi is thought to encounter low pH and oxidative stress. R. equi seems to be particularly acid resistant, since pH 4.0 (HCl) is bacteriostatic and more severe challenges (pH 3.0 and 2.5) are necessary to inactivate the bacteria, while 20% of a population of mid-exponential-phase cells survived after 30 min at pH 2 (18). Virulent and avirulent strains are equally capable of inducing an ATR, with maximal protection provided after adaptation at pH 5 for 90 min. Two-dimensional gel electrophoresis indicated that six membrane proteins were overexpressed and six were repressed at pH 5. Of the cytoplasmic proteins, seven were overexpressed and five were repressed at the same pH. These results were common to both virulent and avirulent strains. However, in another strategy involving a comparison of cDNA generated from plasmid mRNA with and without acid induction, it was found that vapA (for “virulence-associated protein”) exhibited induction by a factor of 29.5 at pH 5 while Western blot analysis suggested 5-fold enhancement (18). These results were in line with a previous study showing VapA induction in cultures grown at acidic pHs (223). In addition, vapD, which seems to be cotranscribed with another open reading frame (named orf3), was induced. While the roles of vapA and vapD have yet to be elucidated, it is known that VapA is surface exposed (224), which is consistent with the high immunogenic response to this protein (225, 226). It is speculated that VapD is also surface exposed due to its high percent homology to VapA.
Mycobacterium spp.
Although many mycobacteria are free-living saprophytes, it is unsurprising that studies of the acid resistance of mycobacteria have concentrated to a greater extent on the pathogenic species: (i) Mycobacterium tuberculosis, which, as the causative agent of tuberculosis in humans, is responsible for around 3 million deaths per year; (ii) Mycobacterium avium, which can survive in macrophages and causes one of the most common opportunistic infections in AIDS patients (83); and (iii) Mycobacterium avium subsp. paratuberculosis, the causative agent of paratuberculosis (Johne's disease), a chronic granulomatous enteritis in ruminants. M. avium subsp. paratuberculosis has also been detected in the intestinal tissue of human patients with Crohn's disease, a chronic enteritis of unknown etiology with pathological and clinical similarities to paratuberculosis (41), suggesting a possible zoonotic relevance.
In vivo, mycobacteria encounter an acidic pH in the lungs, stomach, and macrophage phagosome. Evidence that M. tuberculosis encounters an acidic environment in the lungs has been established by the activity of the antimycobacterial agent pyrazinamide, which has no activity against M. tuberculosis in vitro except at an acidic pH. Pyrazinamide rapidly reduces the level of extracellular M. tuberculosis in the lungs of tuberculous patients (159). The acidic pH of the stomach has long been assumed to be a mechanism of host defense against M. tuberculosis (62). Gastrectomy and chronic gastritis (decrease or absence of the acid barrier) are risk factors for the development of intestinal tuberculosis (62). The large majority of M. avium infections in AIDS patients are acquired via the gastrointestinal tract (65, 194).
Of the mycobacteria studied to date, the growth of M. tuberculosis was most restricted at pH 6 (requiring elevated levels of Mg2+ [see below]). M. kansasii and M. smegmatis also grew very poorly in acidic media with limiting Mg2+. However, M. fortuitum, M. marinum, M. scrofulaceum, M. avium, and M. chelonae grew at pH 6 in an unrestricted manner (180). In fact, it has been shown that M. avium, independent of the growth phase, is able to resist exposure to pH 2.2 for 2 h. Prolonging exposure for up to 24 h decreased survival in a growth-phase-dependent manner, with cells grown to the stationary phase being more tolerant. Acid resistance of M. avium was observed without preadaptation to acid, suggesting that the bacterial cell wall may resist acid without the de novo synthesis of proteins, lipid, or carbohydrate or that the use of presynthesized components such as proton pumps or amino acid decarboxylases may confer protection. However, the increased resistance of M. avium to acid following preincubation under conditions similar to those found in the environment suggests that a natural resistance to low pH may exist under such conditions (20). In contrast to the high constitutive level of acid resistance exhibited by M. avium, the relatively acid-sensitive M. smegmatis exhibits a strong ATR; i.e., adapted cells enjoy a two- to threefold increase in percent viability after a 4-h exposure to a lethal pH (168).
Regardless of the initial site of infection of the pathogenic mycobacteria, persistence in macrophages is a crucial step. M. tuberculosis, M. leprae, and M. avium use a combination of two strategies to survive in macrophages. The phagosomal pH can be altered, with the exclusion of the vacuolar ATPase from the phagosome membrane being a major factor (58, 216), or phagosome-lysosome fusion can be inhibited (181). Ammonia is produced by some mycobacteria and may also contribute to the increase in vacuolar pH (89). This ability may explain an observed initial capacity of the nonpathogenic mycobacteria to survive in phagosomes.
Dual-label spectrofluorometry was used to show that live M. avium cells were exposed to a pH of ca. 5.7 at 6 h postphagocytosis while similarly labeled Salmonella enterica serovar Typhimurium or heat-killed M. avium cells were at a pH of less than 4.8 (216). Similar values were seen with phagosomes containing the nonphagocytic mycobacteria M. gordonae (pH 5.8) or M. smegmatis (pH 5.2) after 5 h (136). The less acidic phagosomal pH encountered by M. avium approximated closely the optimal pH for survival and growth of M. avium in simple broths and is similar to the optimal pH reported for M. tuberculosis and M. scrofulaceum (pH 6 to 6.5) (33), suggesting that the less acidic environment encountered by M. avium within macrophages favors its survival (101). A Mycobacterium-Coxiella burnetii coinfection model was used to test whether M. avium or M. tuberculosis could survive after transfer to an acidified phagolysosome-like vacuole. It was found that while colocalization did not affect M. avium growth (since the doubling time in fact decreased from 1.9 to 1.5 days), the growth of M. tuberculosis was impaired (the doubling time increased from 2.2 to 7.1 days). This correlates with the enhanced acid sensitivity of M. tuberculosis relative to M. avium (33). It is suggested that the sensitivity of M. tuberculosis to low pH may contribute to the control of the infection in humans that is thought to depend on macrophage activation by an emerging immune response. If activation is delayed and the number of organisms increases before acidification of the vacuole occurs, extensive growth of the mycobacteria may take place and clinical disease eventually results (180). Although M. tuberculosis grows within the phagocytic vacuoles of macrophages, it has to cope with both nutrient limitation (94) and a moderately acidic pH (6.1 to 6.5). Since the stress brought about by low pH is more severe in a nutrient-limited environment, the response of mycobacteria to pH stress in a defined (nutrient-poor) medium was studied (77). In such a medium, the growth of M. tuberculosis was very restricted by acidic pH. Higher levels of Mg2+ were required for growth in mildly acidic media (pH 6 to 6.5) compared to media at pH 7. Ca2+, Zn2+, or Mn2+ could not supplant Mg2+ at pH 6.25, but Ca2+ could partially substitute at pH 7. Thus, M. tuberculosis must acquire a sufficient level of Mg2+ to grow in mildly acidic environments such as the macrophage phagosome, for which it has been estimated that the level of Mg2+ is in the low micromolar range (94). It has been shown that an M. tuberculosis mutant with a mutation in the mgtC gene, encoding a magnesium transporter, exhibits attenuated growth in media with reduced Mg2+ levels but only when the media are mildly acidic (25). This mutant is also attenuated for growth in human macrophages and in vivo in the lungs and spleens of mice. While the role of Mg2+ is unknown, it may be participating in the maintenance of a neutral pH, since the Mg2+-dependent proton-translocating ATP synthase is reported to play a role in acid tolerance in bacteria (13). Homologues of the ATP synthase are present in the M. tuberculosis genome (45), while the role of the M. smegmatis F1F0-ATPase in response to low pH has been described in a previous section. Alternatively, the effect may be indirect; for example, increased levels of Mg2+ in the environment may be required due to a less active mycobacterial Mg2+ transport system at low pH or as a consequence of a cellular demand for increased levels of Mg2+ for other functions under acidic conditions.
Clostridium perfringens
Perfringens food poisoning is the term used to describe the common food-borne illness caused by C. perfringens. A more serious, but rare, illness termed pig-bel may be caused by ingesting food contaminated with type C strains. It is likely that the acid resistance of the organism contributes to its survival in foods and in vivo, since C. perfringens is capable of escaping from the macrophage phagosome (167).
In addition to the roles of GAD and GroEL to the acid resistance of clostridia, C. perfringens induces a number of acid shock proteins in response to adaptation at pH 4.5 for 20 min, resulting in a 15-fold increase in acid resistance (238).
Staphylococcus aureus
Staphylococcus aureus is able to invade tissues only via broken skin or mucous membranes; hence, intact skin is an excellent human defense. Since abscesses are generally the initial foci of infection, nonprofessional phagoctes are responsible for most of the initial killing of S. aureus. The ability to avoid or survive attack by neutrophils and other phagocytes is thus critical for the pathogen. S. aureus is killed rapidly at pH 2 but develops resistance if adapted first via a sigB-mediated pathway (32). This leads to induction of sodA. A sodA mutant demonstrates increased acid sensitivity, suggesting that acid stress in turn leads to oxidative stress (43). This is significant when one considers the combined roles of low pH and oxidative stress in the macrophage phagolysosome. Of 10 S. aureus mutants defective in the starvation-induced stationary phase of growth, 6 were more sensitive than the wild type strain at pH 2. Two of these had mutations in genes encoding homologs of hypoxanthine-guanine phosphoribosyltransferase and heme A synthase, respectively, which seemed to result in decreased viability regardless of the stress. The other mutants lacked SodA, which has been discussed above; RpoE, which seems to play a regulatory role in stationary phase; a protein of unknown function; and UmuC. UmuC plays a role in the SOS response in B. subtilis, i.e., the response induced in response to DNA damage (242), again demonstrating the importance of DNA repair systems in the response to low pH (245).
Bacillus cereus
B. cereus food poisoning is a general description, although two recognized types of illness are caused by two distinct metabolites. The diarrhoeal syndrome is caused by a high-molecular-weight protein or protein complex, while the vomiting (emetic) syndrome is caused by a low-molecular-weight, circular, heat-stable peptide. The presence of large numbers of B. cereus cells, greater than 106 organisms/g, in foods is indicative of active growth and proliferation of the organism and is consistent with a potential hazard to health.
Low pH is again likely to be one of the stresses encountered by this pathogen in food. B. cereus undergoes acid adaptation at pH 6.3, resulting in enhanced protection when subsequently exposed to pH 4.6, heat, ethanol, salt, or hydrogenperoxide. Acid adaptation was associated with induction of SodA, and acid-adapted cells maintained a higher internal pH. An acid-sensitive mutant appeared to suffer from membrane damage that appeared to affect ability of the strain to control its internal pH (24). This mutant underexpresses a number of proteins.
Oral Streptococci
The acid resistance mechanisms used by oral streptococci, especially S. mutans, have been described in great detail in previous sections. However, while S. mutans is one of the most acidogenic of the species found in dental plaque, being capable of producing acid from fermentable carbohydrate at a higher rate and over a much greater pH range than most other streptococci (75), the role of other bacteria in the progression of the disease was not investigated until recently, when a number of studies highlighted the pathogenic potential of non-mutans streptococci. S. oralis forms a significant proportion of the aciduric microflora of plaque (3, 16). It has also been demonstrated that non-mutans streptococci, including S. oralis, exhibit acid-induced tolerance of low pH and acidogenicity (202, 222, 233, 234). As a consequence, it has been suggested that the contribution of non-mutans streptococci to dental caries may need to be reevaluated.
However, until such a reevaluation occurs, S. mutans will continue to receive greatest attention. Even then, care needs to be taken when studying laboratory strains. A study involving two fresh isolates and two laboratory strains found that all strains could induce a mid-log-phase ATR but that only the fresh isolates exhibited a stationary-phase ATR. These also differed from the laboratory strains by exhibiting enhanced acid tolerance as a consequence of glucose starvation, although this was diminished in a defined medium with a low concentration of amino acids. It was found that one of the fresh isolates, H7, produced at least 56 extracellular proteins whereas a laboratory strain, LT11, produced only 19. H7 was also found to have two- to fourfold-higher levels of intracellular glycogen in the stationary phase than strain LT11 did. It has been suggested that S. mutans H7 possesses the required endogenous metabolism to support amino acid and peptide uptake in the early stationary phase, which resulted in the formation of basic end products that, in turn, contributed to enhanced intracellular pH homeostasis (220).
Lactic Acid Bacteria
The three main systems involved in lactococcal pH homeostasis, i.e., the ADI system, the H+-ATPase proton pump, and the GAD system, have been described in greater detail in previous sections.
LABs are also capable of inducing an ATR in response to mild-acid treatment (112, 175, 190). The systems induced by this response include pH homeostatic, protection, and repair mechanisms. Acid adaptation of Lactococcus lactis subsp. cremoris strains MG1363 and 712 both require de novo protein synthesis (190), as does Lactobacillus casei (all oral LABs) (150). In contrast, acid adaptation in L. lactis subsp. lactis strain IL 403 is reported to be independent of protein synthesis (112), although induction of UV-inducible proteins and heat shock proteins is observed (112). Heat shock proteins are also induced following acid adaptation of Lactobacillus delbrueckii (145).
Genes and proteins involved in pH homeostasis and cell protection or repair play a role in acid adaptation, but this role can also extend to more general acid tolerance mechanisms. Rallu et al. isolated 21 acid-tolerant insertional mutants of L. lactis MG1363 that resulted in the identification of 18 corresponding loci (189, 190) (Table 2). In all, four classes of mutants were defined: (i) class 1 mutants are more resistant than the wild type during the exponential phase only; (ii) class 2 mutants are more resistant than the wild type during the early stationary phase; (iii) class 3 mutants are more resistant than the wild type during all stages of growth but do not exhibit enhanced long-term (2-day) survival; and (iv) class 4 mutants are more resistant than the wild type during all stages of growth and also exhibit enhanced long-term (2-day) survival.
TABLE 2.
Acid-tolerant mutants of L. lactis MG1363 identified by Rallu et al. (189)
| Mutant | Insert location | Putative functiona | Class | Other informationb |
|---|---|---|---|---|
| arl12 | glnH | Amino acid ABC transporter (binding) | 2 | |
| arl13 | glnP | Amino acid ABC transporter (permease) | 2 | |
| arl11 | Putative glnP-glnQ intergenic region | 2 | ||
| arl14 | glnQ | Amino acid ABC transporter (ATP binding) | 1 | |
| arl15 | yzmB (pstS) ribosome-binding site | Phosphate ABC transporter (binding) | 1 | Ox+++ |
| arl16 | yzmE (pstB) | Phosphate ABC transporter (ATP binding) | 3 | |
| arl17 | hpt | Purine salvage | 4 | Ox+/Heat+/CS+ |
| arl18 | relA | (p)ppGpp synthetase | 4 | Ox+/Heat+/CS+ |
| arl19, arl20 | guaA | Purine biosynthesis | 4 | Ox+/Heat+/CS+ |
| arl21, arl22 | deoB promoter | Purine and pyrimidine salvage | 3 | Heat+ |
| arl23 | recN | DNA metabolism | 3 | Heat+ |
| arl8, arl10 | yybT | No homology | 4 | |
| arl1 | No homology | 3 | Ox+++ | |
| arl2 | No homology | 3 | Ox+ | |
| arl3 | No homology | 2 | ||
| arl4 | Upstream of putative glnU promoter | No homology | 3 | Ox+ |
| arl5 | No homology | 4 | Heat+/CS+ | |
| arl7 | No homology (purine metabolism?) | 4 | Heat+/CS+ |
ABC, ATP, binding cassette.
Ox+, increased survival in the presence of oxidative stress; Ox+++, greatly increased survival in the presence of oxidative stress; Heat+, increased survival in the presence of heat stress; CS+, increased resistance to carbon starvation.
The authors propose that the class 1 and 2 mutants are altered in their abilities to maintain pH homeostatis, and thus acid stress specifically, while class 3 and 4 mutants are involved primarily in protection of cell components, with the majority playing a role in general stress responses.
Classes 1 and 2 consist of mutants affected in glutamate or phosphate transport. Increased acid resistance may result from decreased intracellular levels of these metabolites acting to trigger a stress response or may be a consequence of reduced proton uptake. The high acid tolerance of L. lactis mutants with defects in the high-affinity phosphate transporter is possibly due to the induction of a regulon that includes stress response genes, as is the case in B. subtilis (85). This finding also correlates with the observation that low pH decreases the activity of the phosphate transporter (183).
The enhanced acid and general stress resistance of the class 3 and 4 mutants is thought to be a result of reduced intracellular concentrations of purine, phosphate, or guanine acting as signals for the induction of the stringent response. A model has been proposed (232) in which ribosomes may act as stress sensors (231), with RelA binding to the ribosomes and modulating its (p)ppGpp synthetase activity according to their activity (29) while SGP senses, and possibly modulates, the GTP/GDP ratio (8).
INDUCTION AT LOW pH
More recently, a variety of techniques have become available which permit the identification, on a large scale, of genes and proteins that are induced in response to a low-pH environment (Table 1). Improvements in two-dimensional gel electrophoresis as well as differential-display PCR, subtractive hybridization, and gene arrays have provided us, and will continue to provide us, with a wealth of information. However, care must be taken when interpreting these results, because not all of these genes and proteins will necessarily play a direct role in the pH stress response. This will require the use of more classical techniques such as the creation of mutants and determination of the consequences of these mutations on an individual basis.
It is interesting to note instances, however, where a number of genes associated with the same pathway or role are induced. For instance, many enzymes involved in glycolysis were up-regulated following growth of S. oralis and S. mutans at low pH (248, 249). Subsequent analysis of metabolic flux in L. lactis following autoacidification (discussed previously [84]) showed that increased production of a number of glycolytic enzymes was a recurrent phenomenon following exposure of lactic acid bacteria to low pH. It may be that this increase aids the production of ATP at low pH and consequently aids increased proton extrusion via the F1F0-ATPase. In fact, induction of ATPase subunits in S. oralis, although not in S. mutans, was observed at low pH (Table 1).
Another trend is observed in M. tuberculosis cells incubated at pH 5.5. Under such circumstances, the induced genes included those encoding isocitrate lyase and four other genes also potentially involved in the metabolism of fatty acids, fadB2, gltA1, Rv1130, and Rv1169c (88). Isocitrate lyase is a key enzyme in the glyoxylate pathway used to metabolize fatty acids to glucose through an acetyl coenzyme A (CoA) intermediate. The icl genes from both M. tuberculosis and M. avium were previously shown to be induced in macrophages (102, 117), and recent evidence demonstrated the requirement of these genes for survival in macrophages and for persistence in mice (158). The fact that icl is induced by acid suggests that M. tuberculosis may sense low pH as a signal that the bacteria are entering the phagosome and should express genes, such as those involved in fatty acid metabolism, needed for long-term survival in that environment.
While a number of genes associated with the citrate pathway of glutamate biosynthesis were repressed at low pH, the involvement, if any, of this pathway in responding to acidic environments has yet to be elucidated. These results are representative of the potential benefits of proteomics and transcipteomics and indicate how the use of such strategies in combination with other techniques has the potential to greatly enhance our understanding of the global impact of low-pH stress (40).
CONCLUSION
The mechanisms utilized by gram-positive bacteria to survive low-pH stress operate in a number of different ways. Although not available to all gram-positive bacteria, the most successful means of surviving low-pH stress is the complete avoidance of extremely acidic environments. This is the case when pathogenic bacteria avoid exposure to mature phagosomes by preventing acidification or escaping into the cytoplasm. However, such strategies are carefully regulated by a number of environmental factors, none more important than the sensing of mild acidification, to prevent the potentially lethal consequences of inappropriate production of potentially antigenic proteins. However, even if escape from similarly extreme environments is not a realistic option, bacteria that are forewarned by mild acidification can prepare through the induction of a wide range of protective measures. These are described at length above and encompass systems that alter cell membrane composition, extrude protons, protect macromolecules, alter metabolic pathways, and generate alkalis. In situations where bacteria are “caught unprepared” by a low-pH stress, they are forced into a rearguard action which involves a heavy reliance on proton pumps, most notably the F1F0-ATPase, in an effort to keep the internal pH stable for a sufficient length of time to permit the induction of additional systems.
As is apparent from this review, while our understanding of the mechanisms used by gram-positive bacteria to grow or survive in low-pH environments has increased, much remains to be determined. Sequencing of whole genomes will undoubtedly result in the identification of genes and systems homologous to those described in this review as playing a role in the acid stress response. This will make it possible to speculate about how the newly sequenced organism may respond to different low-pH environments. Other relatively new technologies such as differential display and subtractive hybridization, the use of gene arrays, and two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) combined with mass spectroscopy have resulted in a recent surge in information pertaining to pH-inducible genes. These genes fall into three categories: (i) those which are known to play a role in stress resistance, (ii) those for which such a role could be envisaged, and (iii) those apparently unconnected to stress resistance or of unknown function. It is necessary to determine, through the use of classical techniques, the role, if any, of individual genes in the last two categories in pH stress resistance. This area represents a major challenge for scientists in this field but should lead to an exciting future when a clearer understanding of the global response of bacteria to low pH will emerge.
The anticipation of such an understanding does not prevent the application of information that has already been gleaned. An understanding of the systems involved may ultimately result in the reduction of numbers of pathogenic bacteria in low-pH foods, the mouth, the stomach and the macrophages (where relevant) while possibly suggesting ways in which the gastric transit of probiotic bacteria, survival of noncariogenic bacteria in the mouth, or enhanced acid production by a starter culture might be improved. If the theory put forward by Rowbury (195) is correct, i.e., that bacteria that are removed from foods by extreme heat treatments may leave residual extracellular induction factors with the potential to aid the survival of other bacteria contaminating another food ingested during the same meal, then it may be possible that responses induced by cells in, for example, the stomach may benefit other cells present in the small intestine.
The link between foods and low-pH stress exists at a number of levels. The age-old practice of fermenting with acid-producing starter cultures to improve food quality and limit the growth of spoilage and pathogenic bacteria represents a fine example of the manipulation of acid-resistant bacteria. However, future applications may involve the enhancement of such techniques. With respect to food quality, it has been speculated that the presence of the ADI system may benefit Lactobacillus sakei during certain meat fermentations (263). Since the levels of free arginine in such circumstances can vary greatly depending on proteolytic activity, it may be of benefit to determine the consequences of artificially adding arginine. In sourdough LAB, this system, in addition to enhancing tolerance to acid stress, assists cell growth and cell survival during storage at 7°C and favors the production of ornithine, an important precursor of crust aroma compounds (70). Whether this system improves or reduces wine quality requires further analysis. While it protects the LAB involved in malolactic fermentation of wine (148), there is some concern about the creation of ethyl carbamate (urethane) precursors.
With a view to improving food safety, the observation that a number of starter cultures possess GAD activity could be significant because the selection, isolation, or creation of such cultures with heightened activity would minimize the amount of residual glutamate available to undesirable bacteria that depend on this amino acid for prolonged survival at low pH. Alternatively, if it were possible to generate safe glutamate analogues that would specifically inhibit or compete for bacterial decarboxylases, one could use starters that are not dependent on GAD activity to make safer foods. The use of mildly acidic conditions to limit the growth of undesirable bacteria in foods can have counterproductive consequences. While growth may be limited, the induction of an ATR frequently results in enhanced resistance to more extreme stresses (pH or otherwise) encountered downstream. Our ever-improving understanding of the signals and responses involved in this process will help to minimize adaptive responses.
The significance of the urease system in the oral cavity was best illustrated when a strain of S. mutans, which lacks urease, was genetically engineered to express the urease genes of S. salivarius. Rats infected which this strain had a dramatically lower incidence and severity of all types of caries compared with controls, showing that alkali generation inhibits the development of caries (42). Increased production of saliva, containing 3 to 10 mM urea in healthy individuals, is the aim behind the use of sugar-free chewing gum. Utilization of salivary urea by non-mutans streptococci and other oral bacteria brings about an increase in pH and reduces the risk of enamel demineralization. This strategy has been taken a step further through the use of urea-containing chewing gum. Rinsing with urea also results in a rapid rise in plaque pH (210), while a number of mouthwashes contain urea in the form of the “whitening” agent urea peroxide. It may also be possible to benefit from the neutralizing effects of the ADI system in a similar way. It is also thought that early colonization by ADI+ strains results in cariogenic bacteria being outcompeted (78). Our ability to apply this research may become more important if fluoride-resistant mutants were to to emerge. Two fluoride-resistant mutants of S. sobrinus, another mutans Streptococcus species, were shown to exhibit significantly greater acid production than their parent strains in the presence of 0.5 and 5.0 mM fluoride. The fluoride-resistant strains were also more aciduric than the parent strains in the presence of fluoride but not in its absence (207).
In addition to processing foods to minimize bacterial survival, the opposite can be desirable. Foods, especially those with a good buffering capacity, are frequently used as carriers for probiotic bacteria to ensure survival and passage through the acidic stomach. Using the same example as outlined above, probiotic bacteria that possess an active GAD system would greatly benefit from glutamate supplementation. Furthermore, induction of some, or all, of the systems that play a role in acid adaptation could greatly aid the survival of these cultures not only in the stomach but also against other stresses subsequently encountered in the gastrointestinal tract. This theory was taken a step further when it was shown that heterologous expression of the O. oeni gene hsp18 in E. coli resulted in an enhanced acid-resistant phenotype. While such an application would have implications for the food-grade status of the manipulated organism, it does demonstrate one potential application that could emerge from the study of resistance mechanisms of gram-positive bacteria.
The few examples given above merely represent the most obvious applications. However, given that so many gram-positive bacteria depend on their acid resistance mechanisms, as illustrated in this review, the development of techniques to improve or reduce the survival of bacteria at low pH will reap rich dividends.
REFERENCES
- 1.Akbar, S., T. A. Gaidenko, C. M. Kang, M. O'Reilly, K. M. Devine, and C. W. Price. 2001. New family of regulators in the environmental signaling pathway which activates the general stress transcription factor sigma(B) of Bacillus subtilis. J. Bacteriol. 183:1329-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akbar, S., C. M. Kang, T. A. Gaidenko, and C. W. Price. 1997. Modulator protein RsbR regulates environmental signalling in the general stress pathway of Bacillus subtilis. Mol. Microbiol. 24:567-578. [DOI] [PubMed] [Google Scholar]
- 3.Alam, S., S. R. Brailsford, S. Adams, C. Allison, E. Sheehy, L. Zoitopoulos, E. A. Kidd, and D. Beighton. 2000. Genotypic heterogeneity of Streptococcus oralis and distinct aciduric subpopulations in human dental plaque. Appl. Environ. Microbiol. 66:3330-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alemayehu, D., E. O'Sullivan, and S. Condon. 2000. Changes in acid tolerance of Lactococcus lactis during growth at constant pH. Int. J. Food Microbiol. 55:215-221. [DOI] [PubMed] [Google Scholar]
- 5.Amachi, S., K. Ishikawa, S. Toyoda, Y. Kagawa, A. Yokota, and F. Tomita. 1998. Characterization of a mutant of Lactococcus lactis with reduced membrane-bound ATPase activity under acidic conditions. Biosci. Biotechnol. Biochem. 62:1574-1580. [DOI] [PubMed] [Google Scholar]
- 6.Arikado, E., H. Ishihara, T. Ehara, C. Shibata, H. Saito, T. Kakegawa, K. Igarashi, and H. Kobayashi. 1999. Enzyme level of enterococcal F1Fo-ATPase is regulated by pH at the step of assembly. Eur. J. Biochem. 259:262-268. [DOI] [PubMed] [Google Scholar]
- 7.Audia, J. P., C. C. Webb, and J. W. Foster. 2001. Breaking through the acid barrier: an orchestrated response to proton stress by enteric bacteria. Int. J. Med. Microbiol. 291:97-106. [DOI] [PubMed] [Google Scholar]
- 8.Baev, D., R. England, and H. K. Kuramitsu. 1999. Stress-induced membrane association of the Streptococcus mutans GTP-binding protein, an essential G protein, and investigation of its physiological role by utilizing an antisense RNA strategy. Infect. Immun. 67:4510-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bakker, E. P., and F. M. Harold. 1980. Energy coupling to potassium transport in Streptococcus faecalis. Interplay of ATP and the protonmotive force. J. Biol. Chem. 255:433-440. [PubMed] [Google Scholar]
- 10.Banks, E. R., S. D. Allen, J. A. Siders, and N. A. O'Bryan. 1989. Characterization of anaerobic bacteria by using a commercially available rapid tube test for glutamic acid decarboxylase. J. Clin. Microbiol. 27:361-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barcelona-Andres, B., A. Marina, and V. Rubio. 2002. Gene structure, organization, expression, and potential regulatory mechanisms of arginine catabolism in Enterococcus faecalis. J. Bacteriol. 184:6289-6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baumberg, S., and U. Klingel. 1993. Biosynthesis of arginine, proline, and related compounds, p. 299-306. In A. L. Sononshein, R. Losick, and J. A. Hochs (ed.), Bacillus subtilis and other gram-positive bacteria. ASM Press, Washington, D.C.
- 13.Bearson, S., B. Bearson, and J. W. Foster. 1997. Acid stress responses in enterobacteria. FEMS Microbiol. Lett. 147:173-180. [DOI] [PubMed] [Google Scholar]
- 14.Beauregard, K. E., K. D. Lee, R. J. Collier, and J. A. Swanson. 1997. pH-dependent perforation of macrophage phagosomes by listeriolysin O from Listeria monocytogenes. J. Exp. Med. 186:1159-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Becker, L. A., M. S. Cetin, R. W. Hutkins, and A. K. Benson. 1998. Identification of the gene encoding the alternative sigma factor sigmaB from Listeria monocytogenes and its role in osmotolerance. J. Bacteriol. 180:4547-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beighton, D., S. R. Brailsford, E. Lynch, H. Y. Chen, and D. T. Clark. 1999. The influence of specific foods and oral hygiene on the microflora of fissures and smooth surfaces of molar teeth: a 5-day study. Caries Res. 33:349-356. [DOI] [PubMed] [Google Scholar]
- 17.Bender, G. R., S. V. Sutton, and R. E. Marquis. 1986. Acid tolerance, proton permeabilities, and membrane ATPases of oral streptococci. Infect. Immun. 53:331-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benoit, S., S. Taouji, A. Benachour, and A. Hartke. 2000. Resistance of Rhodococcus equi to acid pH. Int. J. Food Microbiol. 55:295-298. [DOI] [PubMed] [Google Scholar]
- 19.Benson, A. K., and W. G. Haldenwang. 1993. Bacillus subtilis sigma B is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc. Natl. Acad. Sci. USA 90:2330-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bodmer, T., E. Miltner, and L. E. Bermudez. 2000. Mycobacterium avium resists exposure to the acidic conditions of the stomach. FEMS Microbiol. Lett. 182:45-49. [DOI] [PubMed] [Google Scholar]
- 21.Boyd, D. A., D. G. Cvitkovitch, A. S. Bleiweis, M. Y. Kiriukhin, D. V. Debabov, F. C. Neuhaus, and I. R. Hamilton. 2000. Defects in d-alanyl-lipoteichoic acid synthesis in Streptococcus mutans results in acid sensitivity. J. Bacteriol. 182:6055-6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boylan, S. A., A. Rutherford, S. M. Thomas, and C. W. Price. 1992. Activation of Bacillus subtilis transcription factor sigma B by a regulatory pathway responsive to stationary-phase signals. J. Bacteriol. 174:3695-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brody, M. S., K. Vijay, and C. W. Price. 2001. Catalytic function of an alpha/beta hydrolase is required for energy stress activation of the sigma(B) transcription factor in Bacillus subtilis. J. Bacteriol. 183:6422-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Browne, N., and B. C. Dowds. 2002. Acid stress in the food pathogen Bacillus cereus. J. Appl. Microbiol. 92:404-414. [DOI] [PubMed] [Google Scholar]
- 25.Buchmeier, N., A. Blanc-Potard, S. Ehrt, D. Piddington, L. Riley, and E. A. Groisman. 2000. A parallel intraphagosomal survival strategy shared by mycobacterium tuberculosis and Salmonella enterica. Mol. Microbiol. 35:1375-1382. [DOI] [PubMed] [Google Scholar]
- 26.Burne, R. A. 1998. Oral streptococci—products of their environment. J. Dent. Res. 77:445-452. [DOI] [PubMed] [Google Scholar]
- 27.Burne, R. A., R. G. Quivey, Jr., and R. E. Marquis. 1999. Physiologic homeostasis and stress responses in oral biofilms. Methods Enzymol. 310:441-460. [DOI] [PubMed] [Google Scholar]
- 28.Campbell, J., III, G. R. Bender, and R. E. Marquis. 1985. Barotolerant variant of Streptococcus faecalis with reduced sensitivity to glucose catabolite repression. Can. J. Microbiol. 31:644-650. [DOI] [PubMed] [Google Scholar]
- 29.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 30.Casiano-Colon, A., and R. E. Marquis. 1988. Role of the arginine deiminase system in protecting oral bacteria and an enzymatic basis for acid tolerance. Appl. Environ. Microbiol. 54:1318-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Champomier-Verges, M. C., S. Chaillou, M. Comet, and M. Zagorec. 2001. Lactobacillus sakei: recent developments and future prospects. [Research in Microbiology 152 (2001) 839]. Res. Microbiol. 152:839. (Erratum, 153:15-123, 2002.) [DOI] [PubMed] [Google Scholar]
- 32.Chan, P. F., S. J. Foster, E. Ingham, and M. O. Clements. 1998. The Staphylococcus aureus alternative sigma factor sigmaB controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. J. Bacteriol. 180:6082-6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chapman, J. S., and J. S. Bernard. 1962. The tolerances of unclassified mycobacteria. Am. Rev. Respir. Dis. 86:582-583. [DOI] [PubMed] [Google Scholar]
- 34.Charteris, W. P., P. M. Kelly, L. Morelli, and J. K. Collins. 1998. Development and application of an in vitro methodology to determine the transit tolerance of potentially probiotic Lactobacillus and Bifidobacterium species in the upper human gastrointestinal tract. J. Appl. Microbiol. 84:759-768. [DOI] [PubMed] [Google Scholar]
- 35.Chen, Y. Y., M. J. Betzenhauser, and R. A. Burne. 2002. cis-Acting elements that regulate the low-pH-inducible urease operon of Streptococcus salivarius. Microbiology 148:3599-3608. [DOI] [PubMed] [Google Scholar]
- 36.Chen, Y. Y. and R. A. Burne. 1996. Analysis of Streptococcus salivarius urease expression using continuous chemostat culture. FEMS Microbiol. Lett. 135:223-229. [DOI] [PubMed] [Google Scholar]
- 37.Chen, Y. Y., K. A. Clancy, and R. A. Burne. 1996. Streptococcus salivarius urease: genetic and biochemical characterization and expression in a dental plaque streptococcus. Infect. Immun. 64:585-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen, Y. Y., T. H. Hall, and R. A. Burne. 1998. Streptococcus salivarius urease expression: involvement of the phosphoenolpyruvate:sugar phosphotransferase system. FEMS Microbiol. Lett. 165:117-122. [DOI] [PubMed] [Google Scholar]
- 39.Chen, Y. Y., C. A. Weaver, D. R. Mendelsohn, and R. A. Burne. 1998. Transcriptional regulation of the Streptococcus salivarius 57.I urease operon. J. Bacteriol. 180:5769-5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chia, J. S., Y. Y. Lee, P. T. Huang, and J. Y. Chen. 2001. Identification of stress-responsive genes in Streptococcus mutans by differential display reverse transcription-PCR. Infect. Immun. 69:2493-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiodini, R. J. 1989. Crohn's disease and the mycobacterioses: a review and comparison of two disease entities. Clin. Microbiol. Rev. 2:90-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clancy, K. A., S. Pearson, W. H. Bowen, and R. A. Burne. 2000. Characterization of recombinant, ureolytic Streptococcus mutans demonstrates an inverse relationship between dental plaque ureolytic capacity and cariogenicity. Infect. Immun. 68:2621-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clements, M. O., and S. J. Foster. 1999. Stress resistance in Staphylococcus aureus. Trends Microbiol. 7:458-462. [DOI] [PubMed] [Google Scholar]
- 44.Cobb, C. A., G. D. Curtis, D. S. Bansi, E. Slade, W. Mehal, R. G. Mitchell, and R. W. Chapman. 1996. Increased prevalence of Listeria monocytogenes in the faeces of patients receiving long-term H2-antagonists. Eur. J. Gastroenterol. Hepatol. 8:1071-1074. [DOI] [PubMed] [Google Scholar]
- 45.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry, III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M.-A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 46.Conte, M. P., G. Petrone, A. M. Di Biase, M. G. Ammendolia, F. Superti, and L. Seganti. 2000. Acid tolerance in Listeria monocytogenes influences invasiveness of enterocyte-like cells and macrophage-like cells. Microb. Pathog. 29:137-144. [DOI] [PubMed] [Google Scholar]
- 47.Conte, M. P., G. Petrone, A. M. Di Biase, C. Longhi, M. Penta, A. Tinari, F. Superti, G. Fabozzi, P. Visca, and L. Seganti. 2002. Effect of acid adaptation on the fate of Listeria monocytogenes in THP-1 human macrophages activated by gamma interferon. Infect. Immun. 70:4369-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Conte, M. P., G. Petrone, C. Longhi, P. Valenti, R. Morelli, F. Superti, and L. Seganti. 1996. The effects of inhibitors of vacuolar acidification on the release of Listeria monocytogenes from phagosomes of Caco-2 cells. J. Med. Microbiol. 44:418-424. [DOI] [PubMed] [Google Scholar]
- 49.Cotter, P. D., N. Emerson, C. G. Gahan, and C. Hill. 1999. Identification and disruption of lisRK, a genetic locus encoding a two-component signal transduction system involved in stress tolerance and virulence in Listeria monocytogenes. J. Bacteriol. 181:6840-6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cotter, P. D., C. G. Gahan, and C. Hill. 2000. Analysis of the role of the Listeria monocytogenes F0F1-ATPase operon in the acid tolerance response. Int. J. Food Microbiol. 60:137-146. [DOI] [PubMed] [Google Scholar]
- 51.Cotter, P. D., C. G. Gahan, and C. Hill. 2001. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 40:465-475. [DOI] [PubMed] [Google Scholar]
- 52.Cotter, P. D., C. M. Guinane, and C. Hill. 2002. The LisRK signal transduction system determines the sensitivity of Listeria monocytogenes to nisin and cephalosporins. Antimicrob. Agents Chemother. 46:2784-2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cotter, P. D., K. O'Reilly, and C. Hill. 2001. Role of the glutamate decarboxylase acid resistance system in the survival of Listeria monocytogenes LO28 in low pH foods. J. Food Prot. 64:1362-1368. [DOI] [PubMed] [Google Scholar]
- 54.Cox, M. M., M. F. Goodman, K. N. Kreuzer, D. J. Sherratt, S. J. Sandler, and K. J. Marians. 2000. The importance of repairing stalled replication forks. Nature 404:37-41. [DOI] [PubMed] [Google Scholar]
- 55.Cozzani, I., R. Barsacchi, G. Dibenedetto, L. Saracchi, and G. Falcone. 1975. Regulation of breakdown and synthesis of l-glutamate decarboxylase in Clostridium perfringens. J. Bacteriol. 123:1115-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cozzani, I., A. Misuri, and C. Santoni. 1970. Purification and general properties of glutamate decarboxylase from Clostridium perfringens. Biochem. J. 118:135-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Crow, V. L., and T. D. Thomas. 1982. Arginine metabolism in lactic streptococci. J. Bacteriol. 150:1024-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Crowle, A. J., R. Dahl, E. Ross, and M. H. May. 1991. Evidence that vesicles containing living, virulent Mycobacterium tuberculosis or Mycobacterium avium in cultured human macrophages are not acidic. Infect. Immun. 59:1823-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cunin, R., N. Glansdorff, A. Pierard, and V. Stalon. 1986. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Rev. 50:314-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Curran, T. M., J. Lieou, and R. E. Marquis. 1995. Arginine deiminase system and acid adaptation of oral streptococci. Appl. Environ. Microbiol. 61:4494-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Curran, T. M., Y. Ma, G. C. Rutherford, and R. E. Marquis. 1998. Turning on and turning off the arginine deiminase system in oral streptococci. Can. J. Microbiol. 44:1078-1085. [DOI] [PubMed] [Google Scholar]
- 62.Curtis, K. J., and M. H. Sleisenger. 1978. Infectious and parasitic diseases, p. 193-224. In M. H. Sleisenger and J. S. Fordtran (ed.), Gastrointestinal disease. The W. B. Saunders Co., Philadelphia, Pa.
- 63.Czuprynski, C. J., and E. Balish. 1981. Killing of Listeria monocytogenes by conventional and germfree rat sera. Infect. Immun. 33:348-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Czuprynski, C. J., and N. G. Faith. 2002. Sodium bicarbonate enhances the severity of infection in neutropenic mice orally inoculated with Listeria monocytogenes EGD. Clin. Diagn. Lab. Immunol. 9:477-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Damsker, B., and E. J. Bottone. 1985. Mycobacterium avium-Mycobacterium intracellulare from the intestinal tracts of patients with the acquired immunodeficiency syndrome: concepts regarding acquisition and pathogenesis. J. Infect. Dis. 151:179-181. [DOI] [PubMed] [Google Scholar]
- 66.Dashper, S. G., and E. C. Reynolds. 1992. pH regulation by Streptococcus mutans. J. Dent. Res. 71:1159-1165. [DOI] [PubMed] [Google Scholar]
- 67.Datta, A. R., and M. M. Benjamin. 1997. Factors controlling acid tolerance of Listeria monocytogenes: effects of nisin and other ionophores. Appl. Environ. Microbiol. 63:4123-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Datta, A. R., and M. M. Benjamin. 1999. Cell density dependent acid sensitivity in stationary phase cultures of enterohemorrhagic Escherichia coli O157:H7. FEMS Microbiol. Lett. 181:289-295. [DOI] [PubMed] [Google Scholar]
- 69.Davis, M. J., P. J. Coote, and C. P. O'Byrne. 1996. Acid tolerance in Listeria monocytogenes: the adaptive acid tolerance response (ATR) and growth-phase-dependent acid resistance. Microbiology 142:2975-2982. [DOI] [PubMed] [Google Scholar]
- 70.De Angelis, M., L. Mariotti, J. Rossi, M. Servili, P. F. Fox, G. Rollan, and M. Gobbetti. 2002. Arginine catabolism by sourdough lactic acid bacteria: purification and characterization of the arginine deiminase pathway enzymes from Lactobacillus sanfranciscensis CB1. Appl. Environ. Microbiol. 68:6193-6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.de Chastellier, C., and P. Berche. 1994. Fate of Listeria monocytogenes in murine macrophages: evidence for simultaneous killing and survival of intracellular bacteria. Infect. Immun. 62:543-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Degnan, B. A., M. C. Fontaine, A. H. Doebereiner, J. J. Lee, P. Mastroeni, G. Dougan, J. A. Goodacre, and M. A. Kehoe. 2000. Characterization of an isogenic mutant of Streptococcus pyogenes Manfredo lacking the ability to make streptococcal acid glycoprotein. Infect. Immun. 68:2441-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Degnan, B. A., M. A. Kehoe, and J. A. Goodacre. 1997. Analysis of human T cell responses to group A streptococci using fractionated Streptococcus pyogenes proteins. FEMS Immunol. Med. Microbiol. 17:161-170. [DOI] [PubMed] [Google Scholar]
- 74.Degnan, B. A., J. M. Palmer, T. Robson, C. E. Jones, M. Fischer, M. Glanville, G. D. Mellor, A. G. Diamond, M. A. Kehoe, and J. A. Goodacre. 1998. Inhibition of human peripheral blood mononuclear cell proliferation by Streptococcus pyogenes cell extract is associated with arginine deiminase activity. Infect. Immun. 66:3050-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.de Soet, J. J., B. Nyvad, and M. Kilian. 2000. Strain-related acid production by oral streptococci. Caries Res. 34:486-490. [DOI] [PubMed] [Google Scholar]
- 76.Diaz-Torres, M. L., and R. R. Russell. 2001. HtrA protease and processing of extracellular proteins of Streptococcus mutans. FEMS Microbiol. Lett. 204:23-28. [DOI] [PubMed] [Google Scholar]
- 77.Dilworth, M. J., and A. R. Glenn. 1999. Problems of adverse pH and bacterial strategies to combat it. Novartis Found. Symp. 221:4-14. [DOI] [PubMed] [Google Scholar]
- 78.Dong, Y., Y. Y. Chen, J. A. Snyder, and R. A. Burne. 2002. Isolation and molecular analysis of the gene cluster for the arginine deiminase system from Streptococcus gordonii DL1. Appl. Environ. Microbiol. 68:5549-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Donisi, A., M. G. Suardi, S. Casari, M. Longo, G. P. Cadeo, and G. Carosi. 1996. Rhodococcus equi infection in HIV-infected patients. AIDS 10:359-362. [DOI] [PubMed] [Google Scholar]
- 80.Drici-Cachon, Z., J. Guzzo, J. F. Cavin, and C. Divies. 1996. Acid tolerance in Leuconostoc oenos. Isolation and characterization of an acid-resistant mutant. Appl. Microbiol. Biotechnol. 44:785-789. [Google Scholar]
- 81.Dufour, A., and W. G. Haldenwang. 1994. Interactions between a Bacillus subtilis anti-sigma factor (RsbW) and its antagonist (RsbV). J. Bacteriol. 176:1813-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dykes, G. A., and S. M. Moorhead. 2000. Survival of osmotic and acid stress by Listeria monocytogenes strains of clinical or meat origin. Int. J. Food Microbiol. 56:161-166. [DOI] [PubMed] [Google Scholar]
- 83.Ellner, J. J., M. J. Goldberger, and D. M. Parenti. 1991. Mycobacterium avium infection and AIDS: a therapeutic dilemma in rapid evolution. J. Infect. Dis. 163:1326-1335. [DOI] [PubMed] [Google Scholar]
- 84.Even, S., N. D. Lindley, P. Loubiere, and M. Cocaign-Bousquet. 2002. Dynamic response of catabolic pathways to autoacidification in Lactococcus lactis: transcript profiling and stability in relation to metabolic and energetic constraints. Mol. Microbiol. 45:1143-1152. [DOI] [PubMed] [Google Scholar]
- 85.Eymann, C., H. Mach, C. R. Harwood, and M. Hecker. 1996. Phosphate-starvation-inducible proteins in Bacillus subtilis: a two- dimensional gel electrophoresis study. Microbiology 142:3163-3170. [DOI] [PubMed] [Google Scholar]
- 86.Ferreira, A., C. P. O'Byme, and K. J. Boor. 2001. Role of sigma(B) in heat, ethanol, acid, and oxidative stress resistance and during carbon starvation in Listeria monocytogenes. Appl. Environ. Microbiol. 67:4454-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fillingame, R. H., and S. Divall. 1999. Proton ATPases in bacteria: comparison to Escherichia coli F1F0 as the prototype. Novartis Found. Symp. 221:218-229. [DOI] [PubMed] [Google Scholar]
- 88.Fisher, M. A., B. B. Plikaytis, and T. M. Shinnick. 2002. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J. Bacteriol. 184:4025-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Flynn, J. L., and J. Chan. 2001. Immunology of tuberculosis. Annu. Rev. Immunol. 19:93-129. [DOI] [PubMed] [Google Scholar]
- 90.Fonda, M. L. 1985. l-Glutamate decarboxylase from bacteria. Methods Enzymol. 113:11-16. [DOI] [PubMed] [Google Scholar]
- 91.Gahan, C. G., and C. Hill. 1999. The relationship between acid stress responses and virulence in Salmonella typhimurium and Listeria monocytogenes. Int. J. Food Microbiol. 50:93-100. [DOI] [PubMed] [Google Scholar]
- 92.Gahan, C. G., B. O'Driscoll, and C. Hill. 1996. Acid adaptation of Listeria monocytogenes can enhance survival in acidic foods and during milk fermentation. Appl. Environ. Microbiol. 62:3128-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gale, E. F. 1946. The bacterial amino acid decarboxylases. Adv. Enzymol. 6:1-32. [Google Scholar]
- 94.Garcia-del Portillo, F., J. W. Foster, M. E. Maguire, and B. B. Finlay. 1992. Characterization of the micro-environment of Salmonella typhimurium-containing vacuoles within MDCK epithelial cells. Mol. Microbiol. 6:3289-3297. [DOI] [PubMed] [Google Scholar]
- 95.Garcia-Quintans, N., C. Magni, D. de Mendoza, and P. Lopez. 1998. The citrate transport system of Lactococcus lactis subsp. lactis biovar diacetylactis is induced by acid stress. Appl. Environ. Microbiol. 64:850-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Geoffroy, C., J. L. Gaillard, J. E. Alouf, and P. Berche. 1987. Purification, characterization, and toxicity of the sulfhydryl-activated hemolysin listeriolysin O from Listeria monocytogenes. Infect. Immun. 55:1641-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. G. Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 98.Glomski, I. J., M. M. Gedde, A. W. Tsang, J. A. Swanson, and D. A. Portnoy. 2002. The Listeria monocytogenes hemolysin has an acidic pH optimum to compartmentalize activity and prevent damage to infected host cells. J. Cell Biol. 156:1029-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Goldfine, H., and C. Knob. 1992. Purification and characterization of Listeria monocytogenes phosphatidylinositol-specific phospholipase C. Infect. Immun. 60:4059-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Golub, L. M., S. M. Borden, and I. Kleinberg. 1971. Urea content of gingival crevicular fluid and its relation to periodontal diseases in humans. J. Periodontal Res. 6:243-251. [DOI] [PubMed] [Google Scholar]
- 101.Gomes, M. S., S. Paul, A. L. Moreira, R. Appelberg, M. Rabinovitch, and G. Kaplan. 1999. Survival of Mycobacterium avium and Mycobacterium tuberculosis in acidified vacuoles of murine macrophages. Infect. Immun. 67:3199-3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Graham, J. E., and J. E. Clark-Curtiss. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. USA 96:11554-11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Graham, M. R., L. M. Smoot, C. A. Migliaccio, K. Virtaneva, D. E. Sturdevant, S. F. Porcella, M. J. Federle, G. J. Adams, J. R. Scott, and J. M. Musser. 2002. Virulence control in group A Streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc. Natl. Acad. Sci. USA 99:13855-13860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gutierrez, J. A., P. J. Crowley, D. G. Cvitkovitch, L. J. Brady, I. R. Hamilton, J. D. Hillman, and A. S. Bleiweis. 1999. Streptococcus mutans ffh, a gene encoding a homologue of the 54 kDa subunit of the signal recognition particle, is involved in resistance to acid stress. Microbiology 145:357-366. [DOI] [PubMed] [Google Scholar]
- 105.Guzzo, J., F. Delmas, F. Pierre, M. P. Jobin, B. Samyn, J. Van Beeumen, J. F. Cavin, and C. Divies. 1997. A small heat shock protein from Leuconostoc oenos induced by multiple stresses and during stationary growth phase. Lett. Appl. Microbiol. 24:393-396. [DOI] [PubMed] [Google Scholar]
- 106.Hahn, K., R. C. Faustoferri, and R. G. Quivey, Jr. 1999. Induction of an AP endonuclease activity in Streptococcus mutans during growth at low pH. Mol. Microbiol. 31:1489-1498. [DOI] [PubMed] [Google Scholar]
- 107.Haldenwang, W. G., and R. Losick. 1980. Novel RNA polymerase sigma factor from Bacillus subtilis. Proc. Natl. Acad. Sci. USA 77:7000-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Halgasova, N., G. Bukovska, J. Timko, and J. Kormanec. 2001. Cloning and transcriptional characterization of two sigma factor genes, sigA and sigB, from Brevibacterium flavum. Curr. Microbiol. 43:249-254. [DOI] [PubMed] [Google Scholar]
- 109.Halgasova, N., G. Bukovska, J. Ugorcakova, J. Timko, and J. Kormanec. 2002. The Brevibacterium flavum sigma factor SigB has a role in the environmental stress response. FEMS Microbiol. Lett. 216:77-84. [DOI] [PubMed] [Google Scholar]
- 110.Hanna, M. N., R. J. Ferguson, Y. H. Li, and D. G. Cvitkovitch. 2001. uvrA is an acid-inducible gene involved in the adaptive response to low pH in Streptococcus mutans. J. Bacteriol. 183:5964-5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Harold, F. M., E. Pavlasova, and J. R. Baarda. 1970. A transmembrane pH gradient in Streptococcus faecalis: origin, and dissipation by proton conductors and N,N′-dicyclohexylcarbodimide. Biochim. Biophys. Acta 196:235-244. [DOI] [PubMed] [Google Scholar]
- 112.Hartke, A., S. Bouche, J. C. Giard, A. Benachour, P. Boutibonnes, and Y. Auffray. 1996. The lactic acid stress response of Lactococcus lactis subsp. lactis. Curr. Microbiol. 33:194-199. [DOI] [PubMed] [Google Scholar]
- 113.Henriksson, A., A. K. D. Khaled, and P. L. Conway. 1999. Lactobacillus colonization of the gastrointestinal tract of mice after removal of the non-secreting stomach region. Microb. Ecol. Health Dis. 11:96-99. [Google Scholar]
- 114.Hersh, B. M., F. T. Farooq, D. N. Barstad, D. L. Blankenhorn, and J. L. Slonczewski. 1996. A glutamate-dependent acid resistance gene in Escherichia coli. J. Bacteriol. 178:3978-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Higuchi, T., H. Hayashi, and K. Abe. 1997. Exchange of glutamate and gamma-aminobutyrate in a Lactobacillus strain. J. Bacteriol. 179:3362-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hill, M. J. 2002. Factors controlling the microflora of the healthy upper gastointestinal tract, p. 57-85. In M. J. Hill and P. D. Marsh (ed.), Human microbial ecology. CRC Press, Inc., Boca Raton, Fla.
- 117.Honer Zu Bentrup, K., A. Miczak, D. L. Swenson, and D. G. Russell. 1999. Characterization of activity and expression of isocitrate lyase in Mycobacterium avium and Mycobacterium tuberculosis. J. Bacteriol. 181:7161-7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hood, S. K., and E. A. Zottola. 1988. Effect of low pH on the ability of Lactobacillus acidophilus to survive and adhere to human intestinal cells. J. Food Sci. 53:1514-1516. [Google Scholar]
- 119.Imfeld, T., and F. Lutz. 1980. Intraplaque acid formation assessed in vivo in children and young adults. Pediatr. Dent. 2:87-93. [Google Scholar]
- 120.Institute of Food Technologists Expert Panel on Food Safety and Nutrition. 1987. Monosodium glutamate. Food Technol. 41:143-154. [Google Scholar]
- 121.Jayaraman, G. C., J. E. Penders, and R. A. Burne. 1997. Transcriptional analysis of the Streptococcus mutans hrcA, grpE and dnaK genes and regulation of expression in response to heat shock and environmental acidification. Mol. Microbiol. 25:329-341. [DOI] [PubMed] [Google Scholar]
- 122.Jensen, M. E., P. J. Polansky, and C. F. Schachtele. 1982. Plaque sampling and telemetry for monitoring acid production on human buccal tooth surfaces. Arch. Oral Biol. 27:21-31. [DOI] [PubMed] [Google Scholar]
- 123.Johnstone, B. H., A. A. Handler, D. K. Chao, V. Nguyen, M. Smith, S. Y. Ryu, E. L. Simons, P. E. Anderson, and R. W. Simons. 1999. The widely conserved Era G-protein contains an RNA-binding domain required for Era function in vivo. Mol. Microbiol. 33:1118-1131. [DOI] [PubMed] [Google Scholar]
- 124.Kakinuma, Y. 1998. Inorganic cation transport and energy transduction in Enterococcus hirae and other streptococci. Microbiol. Mol. Biol. Rev. 62:1021-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kallipolitis, B. H. and H. Ingmer. 2001. Listeria monocytogenes response regulators important for stress tolerance and pathogenesis. FEMS Microbiol. Lett. 204:111-115. [DOI] [PubMed] [Google Scholar]
- 126.Kaneko, T., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, Y. Nakamura, N. Miyajima, M. Hirosawa, M. Sugiura, S. Sasamoto, T. Kimura, T. Hosouchi, A. Matsuno, A. Muraki, N. Nakazaki, K. Naruo, S. Okumura, S. Shimpo, C. Takeuchi, T. Wada, A. Watanabe, M. Yamada, M. Yasuda, and S. Tabata. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3:109-36. [DOI] [PubMed] [Google Scholar]
- 127.Kang, C. M., M. S. Brody, S. Akbar, X. Yang, and C. W. Price. 1996. Homologous pairs of regulatory proteins control activity of Bacillus subtilis transcription factor sigma(b) in response to environmental stress. J. Bacteriol. 178:3846-3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kashket, E. R., and S. L. Barker. 1977. Effects of potassium ions on the electrical and pH gradients across the membrane of Streptococcus lactis cells. J. Bacteriol. 130:1017-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kim, W. S., J. Ren, and N. W. Dunn. 1999. Differentiation of Lactococcus lactis subspecies lactis and subspecies cremoris strains by their adaptive response to stresses. FEMS Microbiol. Lett. 171:57-65. [DOI] [PubMed] [Google Scholar]
- 130.Kobayashi, H. 1982. Second system for potassium transport in Streptococcus faecalis. J. Bacteriol. 150:506-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kobayashi, H. 1987. Regulation of cytoplasmic pH in streptococci, p. 255-269. In J. Reizer and A. Peterofsky (ed.), Sugar transport and metabolism in gram-positive bacteria. Ellis Horwood Ltd., Chichester, England.
- 132.Kobayashi, H., T. Suzuki, N. Kinoshita, and T. Unemoto. 1984. Amplification of the Streptococcus faecalis proton-translocating ATPase by a decrease in cytoplasmic pH. J. Bacteriol. 158:1157-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kobayashi, H., T. Suzuki, and T. Unemoto. 1986. Streptococcal cytoplasmic pH is regulated by changes in amount and activity of a proton-translocating ATPase. J. Biol. Chem. 261:627-630. [PubMed] [Google Scholar]
- 134.Kremer, B. H., K. M. van der, P. J. Crowley, I. R. Hamilton, L. J. Brady, and A. S. Bleiweis. 2001. Characterization of the sat operon in Streptococcus mutans: evidence for a role of Ffh in acid tolerance. J. Bacteriol. 183:2543-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kroll, R. G., and R. A. Patchett. 1992. Induced acid tolerance in Listeria monocytogenes. Lett. Appl. Microbiol. 14:224-227. [Google Scholar]
- 136.Kuehnel, M. P., R. Goethe, A. Habermann, E. Mueller, M. Rohde, G. Griffiths, and P. Valentin-Weigand. 2001. Characterization of the intracellular survival of Mycobacterium avium ssp. paratuberculosis: phagosomal pH and fusogenicity in J774 macrophages compared with other mycobacteria. Cell Microbiol. 3:551-566. [DOI] [PubMed] [Google Scholar]
- 137.Kuhnert, W. L. 1999. Ph.D. thesis. University of Rochester, Rochester, N.Y.
- 138.Kullen, M. J., and T. R. Klaenhammer. 1999. Identification of the pH-inducible, proton-translocating F1F0-ATPase (atpBEFHAGDC) operon of Lactobacillus acidophilus by differential display: gene structure, cloning and characterization. Mol. Microbiol. 33:1152-1161. [DOI] [PubMed] [Google Scholar]
- 139.Kullik, I., P. Giachino, and T. Fuchs. 1998. Deletion of the alternative sigma factor sigmaB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 180:4814-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lee, Y. K., and S. Selminen. 1995. The coming age of probiotics. Trends Food Sci. Technol. 6:241-245. [Google Scholar]
- 141.Lemos, J. A., Y. Y. Chen, and R. A. Burne. 2001. Genetic and physiologic analysis of the groE operon and role of the HrcA repressor in stress gene regulation and acid tolerance in Streptococcus mutans. J. Bacteriol. 183:6074-6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Li, Y., and R. A. Burne. 2001. Regulation of the gtfBC and ftf genes of Streptococcus mutans in biofilms in response to pH and carbohydrate. Microbiology 147:2841-2848. [DOI] [PubMed] [Google Scholar]
- 143.Li, Y. H., M. N. Hanna, G. Svensater, R. P. Ellen, and D. G. Cvitkovitch. 2001. Cell density modulates acid adaptation in Streptococcus mutans: implications for survival in biofilms. J. Bacteriol. 183:6875-6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li, Y. H., P. C. Lau, N. Tang, G. Svensater, R. P. Ellen, and D. G. Cvitkovitch. 2002. Novel two-component regulatory system involved in biofilm formation and acid resistance in Streptococcus mutans. J. Bacteriol. 184:6333-6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lim, E. M., S. D. Ehrlich, and E. Maguin. 2000. Identification of stress-inducible proteins in Lactobacillus delbrueckii subsp. bulgaricus. Electrophoresis 21:2557-2561. [DOI] [PubMed] [Google Scholar]
- 146.Lindahl, T., and B. Nyberg. 1972. Rate of depurination of native deoxyribonucleic acid. Biochemistry 11:3610-3618. [DOI] [PubMed] [Google Scholar]
- 147.Liu, S.-Q., G. C. Pritchard, M. J. Hardman, and G. J. Pilone. 1996. Arginine catabolism in wine lactic acid bacteria: is it via the arginine deiminase pathway or the arginine-urease pathway? J. Appl. Microbiol. 81:486-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Liu, S.-Q., and G. J. Pilone. 1998. A review: arginine metabolism in wine lactic acid bacteria and its practical application. J. Appl. Microbiol. 84:315-327. [Google Scholar]
- 149.Luirink, J., and B. Dobberstein. 1994. Mammalian and Escherichia coli signal recognition particles. Mol. Microbiol. 11:9-13. [DOI] [PubMed] [Google Scholar]
- 150.Ma, Y., T. M. Curran, and R. E. Marquis. 1997. Rapid procedure for acid adaptation of oral lactic-acid bacteria and further characterization of the response. Can. J. Microbiol. 43:143-148. [DOI] [PubMed] [Google Scholar]
- 151.Magni, C., D. de Mendoza, W. N. Konings, and J. S. Lolkema. 1999. Mechanism of citrate metabolism in Lactococcus lactis: resistance against lactate toxicity at low pH. J. Bacteriol. 181:1451-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Margolis, H. C., J. H. Duckworth, and E. C. Moreno. 1988. Composition of pooled resting plaque fluid from caries-free and caries- susceptible individuals. J. Dent. Res. 67:1468-1475. [DOI] [PubMed] [Google Scholar]
- 153.Marquis, R. E., G. R. Bender, D. R. Murray, and A. Wong. 1987. Arginine deiminase system and bacterial adaptation to acid environments. Appl. Environ. Microbiol. 53:198-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Marron, L., N. Emerson, C. G. Gahan, and C. Hill. 1997. A mutant of Listeria monocytogenes LO28 unable to induce an acid tolerance response displays diminished virulence in a murine model. Appl. Environ. Microbiol. 63:4945-4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Martin-Galiano, A. J., M. J. Ferrandiz, and A. G. de la Campa. 2001. The promoter of the operon encoding the F0F1 ATPase of Streptococcus pneumoniae is inducible by pH. Mol. Microbiol. 41:1327-1338. [DOI] [PubMed] [Google Scholar]
- 156.Marty-Teysset, C., C. Posthuma, J. S. Lolkema, P. Schmitt, C. Divies, and W. N. Konings. 1996. Proton motive force generation by citrolactic fermentation in Leuconostoc mesenteroides. J. Bacteriol. 178:2178-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mascher, T., D. Zahner, M. Merai, N. Balmelle, A. B. De Saizieu, and R. Hakenbeck. 2003. The Streptococcus pneumoniae cia regulon: CiaR target sites and transcription profile analysis. J. Bacteriol. 185:60-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.McKinney, J. D., B. K. Honer Zu, E. J. Munoz-Elias, A. Miczak, B. Chen, W. T. Chan, D. Swenson, J. C. Sacchettini, W. R. Jacobs, Jr., and D. G. Russell. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735-738. [DOI] [PubMed] [Google Scholar]
- 159.Mitchison, D. A. 1992. The Garrod Lecture. Understanding the chemotherapy of tuberculosis—current problems. J. Antimicrob. Chemother. 29:477-493. [DOI] [PubMed] [Google Scholar]
- 160.Miwa, T., H. Esaki, J. Umemori, and T. Hino. 1997. Activity of H+-ATPase in ruminal bacteria with special reference to acid tolerance. Appl. Environ. Microbiol. 63:2155-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mobley, H. L., M. D. Island, and R. P. Hausinger. 1995. Molecular biology of microbial ureases. Microbiol. Rev. 59:451-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Morel, F., F. Delmas, M. P. Jobin, C. Divies, and J. Guzzo. 2001. Improved acid tolerance of a recombinant strain of Escherichia coli expressing genes from the acidophilic bacterium Oenococcus oeni. Lett. Appl. Microbiol. 33:126-130. [DOI] [PubMed] [Google Scholar]
- 163.Nadon, C. A., B. M. Bowen, M. Wiedmann, and K. J. Boor. 2002. Sigma B contributes to PrfA-mediated virulence in Listeria monocytogenes. Infect. Immun. 70:3948-3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Nomura, M., H. Kimoto, Y. Someya, S. Furukawa, and I. Suzuki. 1998. Production of gamma-aminobutyric acid by cheese starters during cheese ripening. J. Dairy Sci. 81:1486-1491. [DOI] [PubMed] [Google Scholar]
- 165.Nomura, M., M. Kobayashi, S. Ohmomo, and T. Okamoto. 2000. Inactivation of the glutamate decarboxylase gene in Lactococcus lactis subsp. cremoris. Appl. Environ. Microbiol. 66:2235-2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Nomura, M., I. Nakajima, Y. Fujita, M. Kobayashi, H. Kimoto, I. Suzuki, and H. Aso. 1999. Lactococcus lactis contains only one glutamate decarboxylase gene. Microbiology 145:1375-1380. [DOI] [PubMed] [Google Scholar]
- 167.O'Brien, D. K., and S. B. Melville. 2000. The anaerobic pathogen Clostridium perfringens can escape the phagosome of macrophages under aerobic conditions. Cell Microbiol. 2:505-519. [DOI] [PubMed] [Google Scholar]
- 168.O'Brien, L. M., S. V. Gordon, I. S. Roberts, and P. W. Andrew. 1996. Response of Mycobacterium smegmatis to acid stress. FEMS Microbiol. Lett. 139:11-17. [DOI] [PubMed] [Google Scholar]
- 169.O'Connell-Motherway, M., D. van Sinderen, F. Morel-Deville, G. F. Fitzgerald, S. D. Ehrlich, and P. Morel. 2000. Six putative two-component regulatory systems isolated from Lactococcus lactis subsp. cremoris MG1363. Microbiology 146:935-947. [DOI] [PubMed] [Google Scholar]
- 170.O'Driscoll, B., C. G. Gahan, and C. Hill. 1996. Adaptive acid tolerance response in Listeria monocytogenes: isolation of an acid-tolerant mutant which demonstrates increased virulence. Appl. Environ. Microbiol. 62:1693-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.O'Driscoll, B., C. G. M. Gahan, and C. Hill. 1997. Two-dimensional polyacrylamide gel electrophoresis analysis of the acid tolerance response in Listeria monocytogenes LO28. Appl. Environ. Microbiol. 63:2679-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Oguiza, J. A., A. T. Marcos, M. Malumbres, and J. F. Martin. 1996. Multiple sigma factor genes in Brevibacterium lactofermentum: characterization of sigA and sigB. J. Bacteriol. 178:550-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Oh, D. H., and D. L. Marshall. 1996. Monolaurin and acetic acid inactivation of Listeria monocytogenes attached to stainless steel. J. Food Prot. 59:249-252. [DOI] [PubMed] [Google Scholar]
- 174.Olsen, E. B., J. B. Russell, and T. Henick-Kling. 1991. Electrogenic l-malate transport by Lactobacillus plantarum: a basis for energy derivation from malolactic fermentation. J. Bacteriol. 173:6199-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.O'Sullivan, E., and S. Condon. 1997. Intracellular pH is a major factor in the induction of tolerance to acid and other stresses in Lactococcus lactis. Appl. Environ. Microbiol. 63:4210-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Palma, M., and A. L. Cheung. 2001. sigma(B) activity in Staphylococcus aureus is controlled by RsbU and an additional factor(s) during bacterial growth. Infect. Immun. 69:7858-7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Peterson, S., J. Woodhead, and J. Crall. 1985. Caries resistance in children with chronic renal failure: plaque pH, salivary pH, and salivary composition. Pediatr. Res. 19:796-799. [DOI] [PubMed] [Google Scholar]
- 178.Phan-Thanh, L., and F. Mahouin. 1999. A proteomic approach to study the acid response in Listeria monocytogenes. Electrophoresis 20:2214-2224. [DOI] [PubMed] [Google Scholar]
- 179.Phan-Thanh, L., F. Mahouin, and S. Alige. 2000. Acid responses of Listeria monocytogenes. Int. J. Food Microbiol. 55:121-126. [DOI] [PubMed] [Google Scholar]
- 180.Piddington, D. L., A. Kashkouli, and N. A. Buchmeier. 2000. Growth of Mycobacterium tuberculosis in a defined medium is very restricted by acid pH and Mg2+ levels. Infect. Immun. 68:4518-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Pieters, J., and J. Gatfield. 2002. Hijacking the host: survival of pathogenic mycobacteria inside macrophages. Trends Microbiol. 10:142-146. [DOI] [PubMed] [Google Scholar]
- 182.Poolman, B., D. Molenaar, E. J. Smid, T. Ubbink, T. Abee, P. P. Renault, and W. N. Konings. 1991. Malolactic fermentation: electrogenic malate uptake and malate/lactate antiport generate metabolic energy. J. Bacteriol. 173:6030-6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Poolman, B., R. M. Nijssen, and W. N. Konings. 1987. Dependence of Streptococcus lactis phosphate transport on internal phosphate concentration and internal pH. J. Bacteriol. 169:5373-5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Prabhakaran, K., E. B. Harris, and W. F. Kirchhcimer. 1983. Glutamic acid decarboxylase in Mycobacterium leprae. Arch. Microbiol. 134:320-323. [DOI] [PubMed] [Google Scholar]
- 185.Prescott, J. F. 1991. Rhodococcus equi: an animal and human pathogen. Clin. Microbiol. Rev. 4:20-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Quivey, R. G., W. L. Kuhnert, and K. Hahn. 2001. Genetics of acid adaptation in oral streptococci. Crit Rev. Oral Biol. Med. 12:301-314. [DOI] [PubMed] [Google Scholar]
- 187.Quivey, R. G., Jr., R. Faustoferri, K. Monahan, and R. Marquis. 2000. Shifts in membrane fatty acid profiles associated with acid adaptation of Streptococcus mutans. FEMS Microbiol. Lett. 189:89-92. [DOI] [PubMed] [Google Scholar]
- 188.Quivey, R. G., Jr., R. C. Faustoferri, K. A. Clancy, and R. E. Marquis. 1995. Acid adaptation in Streptococcus mutans UA159 alleviates sensitization to environmental stress due to RecA deficiency. FEMS Microbiol. Lett. 126:257-261. [DOI] [PubMed] [Google Scholar]
- 189.Rallu, F., A. Gruss, S. D. Ehrlich, and E. Maguin. 2000. Acid- and multistress-resistant mutants of Lactococcus lactis: identification of intracellular stress signals. Mol. Microbiol. 35:517-528. [DOI] [PubMed] [Google Scholar]
- 190.Rallu, F., A. Gruss, and E. Maguin. 1996. Lactococcus lactis and stress. Antonie Leeuwenhoek 70:243-251. [DOI] [PubMed] [Google Scholar]
- 191.Rao, M., T. L. Streur, F. E. Aldwell, and G. M. Cook. 2001. Intracellular pH regulation by Mycobacterium smegmatis and Mycobacterium bovis BCG. Microbiology 147:1017-1024. [DOI] [PubMed] [Google Scholar]
- 192.Renault, P., C. Gaillardin, and H. Heslot. 1988. Role of malolactic fermentation in lactic acid bacteria. Biochimie 70:375-379. [DOI] [PubMed] [Google Scholar]
- 193.Rogers, A. H., P. S. Zilm, N. J. Gully, and A. L. Pfennig. 1988. Response of a Streptococcus sanguis strain to arginine-containing peptides. Infect. Immun. 56:687-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Roth, R. I., R. L. Owen, D. F. Keren, and P. A. Volberding. 1985. Intestinal infection with Mycobacterium avium in acquired immune deficiency syndrome (AIDS). Histological and clinical comparison with Whipple's disease. Dig. Dis. Sci. 30:497-504. [DOI] [PubMed] [Google Scholar]
- 195.Rowbury, R. J. 2000. Killed cultures of Escherichia coli can protect living organisms from acid stress. Microbiology 146:1759-1760. [DOI] [PubMed] [Google Scholar]
- 196.Rowbury, R. J. 2001. Extracellular sensing components and extracellular induction component alarmones give early warning against stress in Escherichia coli. Adv. Microb. Physiol. 44:215-257. [DOI] [PubMed] [Google Scholar]
- 197.Russell, D. G. 2001. Mycobacterium tuberculosis: here today, and here tomorrow. Nat. Rev. Mol. Cell Biol. 2:569-577. [DOI] [PubMed] [Google Scholar]
- 198.Saklani-Jusforgues, H., E. Fontan, and P. L. Goossens. 2000. Effect of acid-adaptation on Listeria monocytogenes survival and translocation in a murine intragastric infection model. FEMS Microbiol. Lett. 193:155-159. [DOI] [PubMed] [Google Scholar]
- 199.Salema, M., J. S. Lolkema, M. V. San Romao, and M. C. Lourero Dias. 1996. The proton motive force generated in Leuconostoc oenos by L-malate fermentation. J. Bacteriol. 178:3127-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Samelis, J., J. N. Sofos, P. A. Kendall, and G. C. Smith. 2001. Influence of the natural microbial flora on the acid tolerance response of Listeria monocytogenes in a model system of fresh meat decontamination fluids. Appl. Environ. Microbiol. 67:2410-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Sancar, A. 1996. DNA excision repair. Annu. Rev. Biochem. 65:43-81. [DOI] [PubMed] [Google Scholar]
- 202.Sanders, J. W., K. Leenhouts, J. Burghoorn, J. R. Brands, G. Venema, and J. Kok. 1998. A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol. Microbiol. 27:299-310. [DOI] [PubMed] [Google Scholar]
- 203.Sansone, C., J. Van Houte, K. Joshipura, R. Kent, and H. C. Margolis. 1993. The association of mutans streptococci and non-mutans streptococci capable of acidogenesis at a low pH with dental caries on enamel and root surfaces. J. Dent. Res. 72:508-516. [DOI] [PubMed] [Google Scholar]
- 204.Sato, T., J. Wu, and H. Kuramitsu. 1998. The sgp gene modulates stress responses of Streptococcus mutans: utilization of an antisense RNA strategy to investigate essential gene functions. FEMS Microbiol. Lett. 159:241-245. [DOI] [PubMed] [Google Scholar]
- 205.Schlech, W. F., III, D. P. Chase, and A. Badley. 1993. A model of food-borne Listeria monocytogenes infection in the Sprague- Dawley rat using gastric inoculation: development and effect of gastric acidity on infective dose. Int. J. Food Microbiol. 18:15-24. [DOI] [PubMed] [Google Scholar]
- 206.Shabala, L., B. Budde, T. Ross, H. Siegumfeldt, M. Jakobsen, and T. McMeekin. 2002. Responses of Listeria monocytogenes to acid stress and glucose availability revealed by a novel combination of fluorescence microscopy and microelectrode ion-selective techniques. Appl. Environ. Microbiol. 68:1794-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Sheng, J. Y., and Z. Liu. 2000. Acidogenicity and acidurance of fluoride-resistant Streptococcus sobrinus in vitro. Chin. J. Dent. Res. 3:7-14. [PubMed] [Google Scholar]
- 208.Sibelius, U., E. C. Schulz, F. Rose, K. Hattar, T. Jacobs, S. Weiss, T. Chakraborty, W. Seeger, and F. Grimminger. 1999. Role of Listeria monocytogenes exotoxins listeriolysin and phosphatidylinositol-specific phospholipase C in activation of human neutrophils. Infect. Immun. 67:1125-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Simon, J. P., B. Wargnies, and V. Stalon. 1982. Control of enzyme synthesis in the arginine deiminase pathway of Streptococcus faecalis. J. Bacteriol. 150:1085-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Singer, D. L., and I. Kleinberg. 1983. Quantitative assessment of urea, glucose and ammonia changes in human dental plaque and saliva following rinsing with urea and glucose. Arch. Oral Biol. 28:923-929. [DOI] [PubMed] [Google Scholar]
- 211.Sissons, C. H., H. E. Perinpanayagam, E. M. Hancock, and T. W. Cutress. 1990. pH regulation of urease levels in Streptococcus salivarius. J. Dent. Res. 69:1131-1137. [DOI] [PubMed] [Google Scholar]
- 212.Small, P. L., and S. R. Waterman. 1998. Acid stress, anaerobiosis and gadCB: lessons from Lactococcus lactis and Escherichia coli. Trends Microbiol. 6:214-216. [DOI] [PubMed] [Google Scholar]
- 213.Smith, A. J., R. G. Quivey, Jr., and R. C. Faustoferri. 1996. Cloning and nucleotide sequence analysis of the Streptococcus mutans membrane-bound, proton-translocating ATPase operon. Gene 183:87-96. [DOI] [PubMed] [Google Scholar]
- 214.Smith, D. K., T. Kassam, B. Singh, and J. F. Elliott. 1992. Escherichia coli has two homologous glutamate decarboxylase genes that map to distinct loci. J. Bacteriol. 174:5820-5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Stingl, K., K. Altendorf, and E. P. Bakker. 2002. Acid survival of Helicobacter pylori: how does urease activity trigger cytoplasmic pH homeostasis? Trends Microbiol. 10:70-74. [DOI] [PubMed] [Google Scholar]
- 216.Sturgill-Koszycki, S., P. H. Schlesinger, P. Chakraborty, P. L. Haddix, H. L. Collins, A. K. Fok, R. D. Allen, S. L. Gluck, J. Heuser, and D. G. Russell. 1994. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science 263:678-681. [DOI] [PubMed] [Google Scholar]
- 217.Sturr, M. G., and R. E. Marquis. 1992. Comparative acid tolerances and inhibitor sensitivities of isolated F-ATPases of oral lactic acid bacteria. Appl. Environ. Microbiol. 58:2287-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Suzuki, T., C. Shibata, A. Yamaguchi, K. Igarashi, and H. Kobayashi. 1993. Complementation of an Enterococcus hirae (Streptococcus faecalis) mutant in the alpha subunit of the H+-ATPase by cloned genes from the same and different species. Mol. Microbiol. 9:111-118. [DOI] [PubMed] [Google Scholar]
- 219.Suzuki, T., T. Unemoto, and H. Kobayashi. 1988. Novel streptococcal mutants defective in the regulation of H+-ATPase biosynthesis and in F0 complex. J. Biol. Chem. 263:11840-11843. [PubMed] [Google Scholar]
- 220.Svensater, G., O. Bjornsson, and I. R. Hamilton. 2001. Effect of carbon starvation and proteolytic activity on stationary-phase acid tolerance of Streptococcus mutans. Microbiology 147:2971-2979. [DOI] [PubMed] [Google Scholar]
- 221.Svensater, G., J. Welin, J. C. Wilkins, D. Beighton, and L. R. Hamilton. 2001. Protein expression by planktonic and biofilm cells of Streptococcus mutans. FEMS Microbiol. Lett. 205:139-146. [DOI] [PubMed] [Google Scholar]
- 222.Takahashi, N., and T. Yamada. 1999. Acid-induced acid tolerance and acidogenicity of non-mutans streptococci. Oral Microbiol. Immunol. 14:43-48. [DOI] [PubMed] [Google Scholar]
- 223.Takai, S., N. Fukunaga, K. Kamisawa, Y. Imai, Y. Sasaki, and S. Tsubaki. 1996. Expression of virulence-associated antigens of Rhodococcus equi is regulated by temperature and pH. Microbiol. Immunol. 40:591-594. [DOI] [PubMed] [Google Scholar]
- 224.Takai, S., M. Iie, Y. Watanabe, S. Tsubaki, and T. Sekizaki. 1992. Virulence-associated 15- to 17-kilodalton antigens in Rhodococcus equi: temperature-dependent expression and location of the antigens. Infect. Immun. 60:2995-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Takai, S., K. Koike, S. Ohbushi, C. Izumi, and S. Tsubaki. 1991. Identification of 15- to 17-kilodalton antigens associated with virulent Rhodococcus equi. J. Clin. Microbiol. 29:439-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Tan, C., J. F. Prescott, M. C. Patterson, and V. M. Nicholson. 1995. Molecular characterization of a lipid-modified virulence-associated protein of Rhodococcus equi and its potential in protective immunity. Can. J. Vet. Res. 59:51-59. [PMC free article] [PubMed] [Google Scholar]
- 227.Teng, F., L. Wang, K. V. Singh, B. E. Murray, and G. M. Weinstock. 2002. Involvement of PhoP-PhoS homologs in Enterococcus faecalis virulence. Infect. Immun. 70:1991-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Tonon, T., J. P. Bourdineaud, and A. Lonvaud-Funel. 2001. The arcABC gene cluster encoding the arginine deiminase pathway of Oenococcus oeni, and arginine induction of a CRP-like gene. Res. Microbiol. 152:653-661. [DOI] [PubMed] [Google Scholar]
- 229.Tourdot-Marechal, R., L. C. Fortier, J. Guzzo, B. Lee, and C. Divies. 1999. Acid sensitivity of neomycin-resistant mutants of Oenococcus oeni: a relationship between reduction of ATPase activity and lack of malolactic activity. FEMS Microbiol. Lett. 178:319-326. [DOI] [PubMed] [Google Scholar]
- 230.Ueno, Y., K. Hayakawa, S. Takahashi, and K. Oda. 1997. Purification and characterization of glutamate decarboxylase from Lactobacillus brevis IFO 12005. Biosci. Biotechnol. Biochem. 61:1168-1171. [DOI] [PubMed] [Google Scholar]
- 231.VanBogelen, R. A., and F. C. Neidhardt. 1990. Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc. Natl. Acad. Sci. USA. 87:5589-5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.van de Guchte, M., P. Serror, C. Chervaux, T. Smokvina, S. D. Ehrlich, and E. Maguin. 2002. Stress responses in lactic acid bacteria. Antonic Leeuwenhoek. 82:187-216. [PubMed] [Google Scholar]
- 233.Van Houte, J., J. Lopman, and R. Kent. 1996. The final pH of bacteria comprising the predominant flora on sound and carious human root and enamel surfaces. J. Dent. Res. 75:1008-1014. [DOI] [PubMed] [Google Scholar]
- 234.Van Houte, J., C. Sansone, K. Joshipura, and R. Kent. 1991. Mutans streptococci and non-mutans streptococci acidogenic at low pH, and in vitro acidogenic potential of dental plaque in two different areas of the human dentition. J. Dent. Res. 70:1503-1507. [DOI] [PubMed] [Google Scholar]
- 235.van Schaik, W., C. G. Gahan, and C. Hill. 1999. Acid-adapted Listeria monocytogenes displays enhanced tolerance against the lantibiotics nisin and lacticin 3147. J. Food Prot. 62:536-539. [DOI] [PubMed] [Google Scholar]
- 236.Van Wuyckhuyse, B. C., H. E. Perinpanayagam, D. Bevacqua, R. F. Raubertas, R. J. Billings, W. H. Bowen, and L. A. Tabak. 1995. Association of free arginine and lysine concentrations in human parotid saliva with caries experience. J. Dent. Res. 74:686-690. [DOI] [PubMed] [Google Scholar]
- 237.Vijay, K., M. S. Brody, E. Fredlund, and C. W. Price. 2000. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the sigmaB transcription factor of Bacillus subtilis. Mol. Microbiol. 35:180-188. [DOI] [PubMed] [Google Scholar]
- 238.Villarreal, L., N. L. Heredia, and S. Garcia. 2000. Changes in protein synthesis and acid tolerance in Clostridium perfringens type A in response to acid shock. Int. Microbiol. 3:113-116. [PubMed] [Google Scholar]
- 239.Voelker, U., T. Luo, N. Smirnova, and W. Haldenwang. 1997. Stress activation of Bacillus subtilis sigma B can occur in the absence of the sigma B negative regulator RsbX. J. Bacteriol. 179:1980-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Voelker, U., A. Voelker, B. Maul, M. Hecker, A. Dufour, and W. G. Haldenwang. 1995. Separate mechanisms activate sigma B of Bacillus subtilis in response to environmental and metabolic stresses. J. Bacteriol. 177:3771-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Voelker, U., B. Maul, and M. Hecker. 1999. Expression of the sigmaB-dependent general stress regulon confers multiple stress resistance in Bacillus subtilis. J. Bacteriol. 181:3942-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Walker, G. C. 1995. SOS-regulated proteins in translesion DNA synthesis and mutagenesis. Trends Biochem. Sci. 20:416-420. [DOI] [PubMed] [Google Scholar]
- 243.Walker, G. C. 1996. The SOS response of Escherichia coli, p. 529-537. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 244.Waterman, S. R., and P. L. Small. 1996. Identification of sigma S-dependent genes associated with the stationary-phase acid-resistance phenotype of Shigella flexneri. Mol. Microbiol. 21:925-940. [DOI] [PubMed] [Google Scholar]
- 245.Watson, S. P., M. Antonio, and S. J. Foster. 1998. Isolation and characterization of Staphylococcus aureus starvation- induced, stationary-phase mutants defective in survival or recovery. Microbiology 144:3159-3169. [DOI] [PubMed] [Google Scholar]
- 246.Weber, J., and A. E. Senior. 1997. Catalytic mechanism of F1-ATPase. Biochim. Biophys. Acta 1319:19-58. [DOI] [PubMed] [Google Scholar]
- 247.Wiedmann, M., T. J. Arvik, R. J. Hurley, and K. J. Boor. 1998. General stress transcription factor sigmaB and its role in acid tolerance and virulence of Listeria monocytogenes. J. Bacteriol. 180:3650-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Wilkins, J. C., K. A. Homer, and D. Beighton. 2001. Altered protein expression of Streptococcus oralis cultured at low pH revealed by two-dimensional gel electrophoresis. Appl. Environ. Microbiol. 67:3396-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Wilkins, J. C., K. A. Homer, and D. Beighton. 2002. Analysis of Streptococcus mutans proteins modulated by culture under acidic conditions. Appl. Environ. Microbiol. 68:2382-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Winterhoff, N., R. Goethe, P. Gruening, M. Rohde, H. Kalisz, H. E. Smith, and P. Valentin-Weigand. 2002. Identification and characterization of two temperature-induced surface-associated proteins of Streptococcus suis with high homologies to members of the arginine deiminase system of Streptococcus pyogenes. J. Bacteriol. 184:6768-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Wong, S. L., C. W. Price, D. S. Goldfarb, and R. H. Doi. 1984. The subtilisin E gene of Bacillus subtilis is transcribed from a sigma 37 promoter in vivo. Proc. Natl. Acad. Sci. USA. 81:1184-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Wu, Q. L., D. Kong, K. Lam, and R. N. Husson. 1997. A mycobacterial extracytoplasmic function sigma factor involved in survival following stress. J. Bacteriol. 179:2922-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Wu, S., H. de Lencastre, and A. Tomasz. 1996. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J. Bacteriol. 178:6036-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Yamamoto, N., Y. Masujima, and T. Takano. 1996. Reduction of membrane-bound ATPase activity in a Lactobacillus helveticus strain with slower growth at low pH. FEMS Microbiol. Lett. 138:179-184. [Google Scholar]
- 255.Yamashita, Y., T. Takehara, and H. K. Kuramitsu. 1993. Molecular characterization of a Streptococcus mutans mutant altered in environmental stress responses. J. Bacteriol. 175:6220-6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Yasbin, R. E., D. Cheo, and D. Bol. 1993. DNA repair systems, p. 529-537. In R. E. Yasbin, D. Cheo, and D. Bol (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. ASM Press, Washington, D.C.
- 257.Yokota, A., S. Amachi, S. Ishii, and F. Tomita. 1995. Acid sensitivity of a mutant of Lactococcus lactis subsp. lactis C2 with reduced membrane-bound activity. Biosci. Biotechnol. Biochem. 59:2004-2007. [DOI] [PubMed] [Google Scholar]
- 258.Zachar, Z., and D. C. Savage. 1979. Microbial interference and colonization of the murine gastrointestinal tract by Listeria monocytogenes. Infect. Immun. 23:168-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Zhu, M., S. Takenaka, M. Sato, and E. Hoshino. 2001. Influence of starvation and biofilm formation on acid resistance of Streptococcus mutans. Oral Microbiol. Immunol. 16:24-27. [DOI] [PubMed] [Google Scholar]
- 260.Zink, M. C., J. A. Yager, J. F. Prescott, and M. A. Fernando. 1987. Electron microscopic investigation of intracellular events after ingestion of Rhodococcus equi by foal alveolar macrophages. Vet. Microbiol. 14:295-305. [DOI] [PubMed] [Google Scholar]
- 261.Zou, Y., D. J. Crowley, and B. Van Houten. 1998. Involvement of molecular chaperonins in nucleotide excision repair. DNaK leads to increased thermal stability of UvrA, catalytic UvrB loading, enhanced repair, and increased UV resistance. J. Biol. Chem. 273:12887-12892. [DOI] [PubMed] [Google Scholar]
- 262.Zuniga, M., M. Champomier-Verges, M. Zagorec, and G. Perez-Martinez. 1998. Structural and functional analysis of the gene cluster encoding the enzymes of the arginine deiminase pathway of Lactobacillus sake. J. Bacteriol. 180:4154-4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Zuniga, M., M. C. Miralles, and G. Perez-Martinez. 2002. The product of arcR, the sixth gene of the arc operon of Lactobacillus sakei, is essential for expression of the arginine deiminase pathway. Appl. Environ. Microbiol. 68:6051-6058. [DOI] [PMC free article] [PubMed] [Google Scholar]