Abstract
In the past two decades, scientists have elucidated the molecular mechanisms behind Drosophila sex determination and dosage compensation. These two processes are controlled essentially by two different sets of genes, which have in common a master regulatory gene, Sex-lethal (Sxl). Sxl encodes one of the best-characterized members of the family of RNA binding proteins. The analysis of different mechanisms involved in the regulation of the three identified Sxl target genes (Sex-lethal itself, transformer, and male specific lethal-2) has contributed to a better understanding of translation repression, as well as constitutive and alternative splicing. Studies using the Drosophila system have identified the features of the protein that contribute to its target specificity and regulatory functions. In this article, we review the existing data concerning Sxl protein, its biological functions, and the regulation of its target genes.
INTRODUCTION
A breakthrough in understanding the genetic basis underlying sex determination and dosage compensation in Drosophila came from a paper published by Tom Cline (22). He found that Sex-lethal (Sxl)—a gene whose expression depends on the X:A signal—is the key gene controlling both sex determination and dosage compensation processes. Sxl is activated in females (2X;2A) but not in males (X;2A) (reviewed in reference 25).
How can the X:A signal function as the primary input for sex determination and dosage compensation? Two possible alternatives can be visualized. One possibility is that the X/A signal may be needed continuously by the cells during development to stay in the chosen sexual pathway and to maintain the proper dosage compensation. Under this hypothesis, X0 clones induced at any time during development of XX flies would survive and differentiate male structures. As a second alternative, the cells could use the X:A signal at a certain time in their development to set up their sex and dosage compensation processes. Under this second hypothesis, X0 clones induced before that time would survive and differentiate male structures whereas X0 clones induced later would die because their dosage compensation process would be upset. A clonal analysis strategy was used to verify which hypothesis is correct. Genotypes were constructed in a way to allow the removal, by mitotic recombination induced by X irradiation, of one of the two X chromosomes from a XX cell at different times during development. The results demonstrated that the X0 clones induced at around the blastoderm stage survive and differentiate male structures while clones induced later in development are lethal. However, if the X0 clones carry a loss-of-function Sxl mutant allele, they survive and differentiate male structures independent of the period of development when they are induced. These results have indicated that the X:A signal irreversibly sets, in a cell autonomous manner, the state of activity of Sxl sometime around the blastoderm stage. Once this is achieved, the X:A signal is no longer needed and the activity of Sxl remains fixed (4, 106).
Part of the X:A signal is constituted by proteins of the basic helix-loop-helix family. They form heteromeric complexes, which can function as negative or positive regulators of the early Sxl promoter. The twofold difference [X:A = 1.0 (2X;2A) versus X:A = 0.5 (XY;2A)] is transduced into an all-or-none response of Sxl through cis sequences (basic helix-loop-helix proteins binding sites) with variable binding affinities, placed at different locations at the early promoter (132).
Following the blastoderm stage, Sxl expression in females is maintained by a positive autoregulatory loop such that Sxl positively controls its own expression (8, 23). A new class of Sxl mutations that affect the sex determination process without affecting dosage compensation has provided genetic evidence for this autoregulatory function. These Sxl mutant alleles do not need the maternal Daughterless product for their expression in the zygote. In addition, they had the capacity of acting in trans, causing the activation of a wild-type Sxl+ allele in the absence of the maternal Daughterless product (23). The subsequent cloning and molecular characterization of Sxl demonstrated that this positive autoregulation is due to the requirement of the Sxl protein in the female-specific splicing of its own primary transcript (8). Regulation of Sxl expression throughout development is discussed in more detail in a later section.
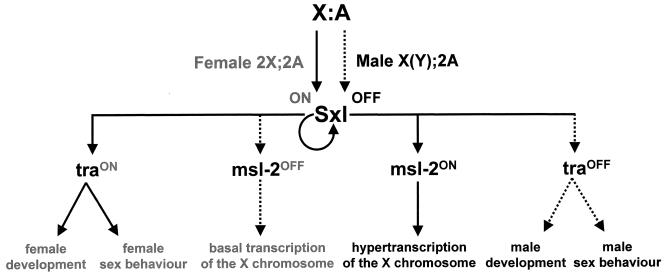
Sxl encodes one of the best-characterized members of the RNA binding family of proteins. Sxl controls the expression of two independent sets of regulatory genes (74). The sex determination genes form one set; mutations in these genes affect sex determination while having no effect on dosage compensation. The generically named male-specific lethal (msl) genes form the other set of genes; mutations in these genes affect dosage compensation while having no effect on sex determination (Fig. 1). In Drosophila, dosage compensation is achieved by hypertranscription of the male X chromosome.
FIG. 1.
The X:A signal and the control of sex determination, sexual behavior, and dosage compensation. The X:A signal targets the Sxl gene, controlling its expression. Autoregulation of Sxl is established only in embryos whose chromosomal constitution is 2X;2A (two X chromosomes; two sets of autosomes), but not in X(Y);2A (one X chromosome; two sets of autosomes) embryos. The autoregulatory feedback loop regulates Sxl expression throughout development and adult life. Sxl controls the expression of the tra and msl-2 genes, whose products are required for control of somatic sex determination/sexual behavior and dosage compensation, respectively. The dotted lines indicate that the expression of the gene is “off,” and the solid lines indicate that the expression of the gene is “on”.
This review is focused on the functions and properties of the Sxl protein and the control of its targets. The processes of sex determination and dosage compensation in D. melanogaster, as well as the X:A signal, have been reviewed extensively by others (3, 25, 27, 40, 86, 95, 107, 108, 112, 134).
CONTROL OF SOMATIC SEXUAL DETERMINATION BY Sxl
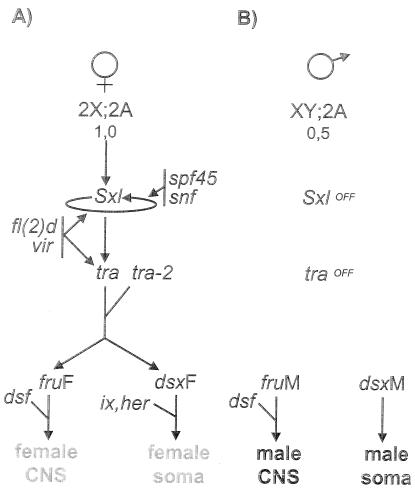
Control of the sex determination genes throughout development occurs by sex-specific splicing of their products. A hierarchical interaction exists among these genes: the product of a gene controls the sex-specific splicing of the pre-mRNA from the downstream gene in the genetic cascade (Fig. 2). Sxl is at the top of this cascade; its products control the splicing of Sxl itself and the downstream gene transformer (tra).
FIG. 2.
The somatic sex determination/sexual behavior cascade. A hierarchical interaction exists among the genes that form the backbone of the somatic sex determination/sexual behavior cascade. The product of a gene controls the sex-specific splicing of the pre-mRNA from the downstream gene in the genetic cascade. (A) The ratio between X chromosomes and autosomes (X:A signal) initiates the cascade by activating the expression of the Sxl gene. This activation occurs only in embryos that have the chromosomal constitution 2X;2A. Sxl regulates the splicing of its own pre-mRNA, a positive-feedback loop (8). The products of the fl(2)d, vir, snf, and spf45 genes are also necessary for this splicing regulation (42, 51, 52, 68, 101). The downstream target of Sxl is the tra gene; splicing control by Sxl allows the production of functional protein product (11). Tra forms a heterodimer with the Transformer-2 (Tra-2) protein (2) that modulates the splicing of two genes: double sex (dsx) (18, 56, 58) and fruitless (fru) (49, 60, 93). The generated sex-specific products control the expression of target genes necessary for female sex differentiation and behavior. (B) In X(Y);2A embryos, no Sxl protein is produced. As a consequence, tra RNA follows a different splicing pattern and no functional product is generated. fru and dsx produce male-specific transcripts. FruM and DsxM control the expression of target genes necessary for male sex differentiation and behavior. hermaphrodite (her) (91, 92) and intersex (20) are also required for proper sex differentiation. The dissatisfaction (dsf) gene is implicated in both male and female sexual behavior (31, 32). CNS, central nervous system.
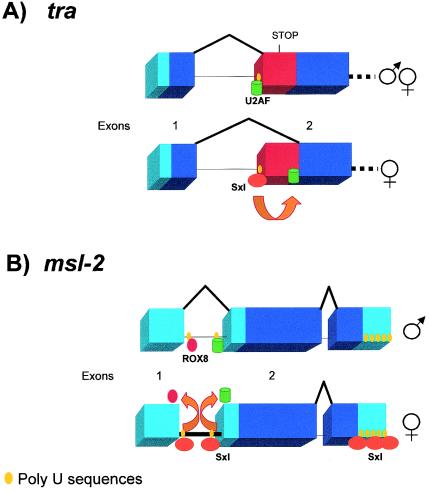
tra is transcribed in both sexes, but its RNA follows alternative splicing pathways. Intron 1 of tra has two alternative 3′ splice sites. A non-sex-specific transcript is generated when the proximal 3′ splice site is used. Use of this splice site introduces a stop codon in the open reading frame, leading to the production of a truncated, nonfunctional peptide. In females, approximately half of the tra pre-mRNA is spliced differently due to the intervention of the Sxl protein. In this case, the distal 3′ splice site is used. As a result, the stretch containing the termination codon is not included in the mature transcript and synthesis of full-length Tra protein occurs (11) (Fig. 3A).
FIG. 3.
Sxl and its target genes. Sxl controls the expression of tra (A) and msl-2 (B). Boxes represent exons, and horizontal lines represent introns. (A) A non-sex-specific transcript is generated when the proximal 3′ splice site is used. Use of this splice site introduces a stop codon in the open reading frame, leading to the production of a truncated and nonfunctional peptide. In females, about half of the tra pre-mRNA is spliced in a different manner due to the intervention of Sxl that competes with U2AF for binding to the poly(Y) tract associated with the proximal 3′ splice site: U2AF is then diverted to the distal poly(Y) tract, thus promoting the usage of the female-specific 3′ splice site. (B) Inhibition of msl-2 expression in females occurs in two steps. First, Sxl prevents the splicing of the first intron by competing with U2AF and Rox8 for binding to two poly(U) sequences located at the 5′ and 3′ ends of this intron. Later, binding of Sxl to these poly(U) sequences and to poly(U) stretches located at 3′ UTR will inhibit translation.
It has been determined by two independent groups that Sxl regulates tra RNA splicing by a blockage mechanism, not by enhancing the use of the female-specific 3′ splice site. Sxl binding sites are poly(U) sequences present at the polypyrimidine [poly(Y)] tract of the non-sex-specific 3′ splice site (57, 116). Actually, this poly(Y) tract contains two stretches of uridine within a highly conserved 24-nucleotide sequence (84). This region is also the binding site for the U2 auxiliary factor (U2AF), an essential splicing factor that is important for the recognition of the 3′ splice site. U2AF, but not Sxl, also binds to the poly(Y) tract associated with the female-specific 3′ splice site, but with 100-fold lower affinity (126). Chimeric proteins containing the effector domain of U2AF fused to the complete RNA binding domain of Sxl promote rather than inhibit splicing to the non-sex-specific 3′ splice site. This suggests that Sxl and U2AF compete for binding to the poly(Y) tract associated with the non-sex-specific 3′ splice site. Binding of Sxl to this sequence displaces U2AF, diverting it to the low-affinity distal poly(Y) tract and promoting the usage of the female-specific 3′ splice site (45, 126) (Fig. 3A).
CONTROL OF DOSAGE COMPENSATION BY Sxl
In Drosophila, dosage compensation takes place in males by hypertranscription of the single X chromosome and is mediated essentially by a group of genes known as male-specific lethal genes (msl1, msl2, msl3, and maleless [mle]). Three additional genes are also involved in dosage compensation: mof, roX1, and roX2. The products of all these genes form a heteromultimeric complex, known as Msl, that associates preferentially with many sites along the male X chromosome. The X chromosome acquires a chromatin structure, reflected by its pale bloated appearance, that allows easier access to the transcription machinery and, hence, its hypertranscription (reviewed in reference 121).
The msl, mof, and roX genes are transcribed in both males and females. However, a stable Msl complex is formed only if the products of all these genes are present. This occurs exclusively in males, since only males express the Msl-2 protein. In females, the production of this protein is prevented by the Sxl protein, which is exclusively expressed in this sex. In fact, ectopic expression of msl-2 in females is sufficient to assemble the Msl complex (6, 63, 64).
Splicing Regulation of the msl-2 Transcript
msl-2 expression is regulated in two steps in D. melanogaster. Splicing is the first step to achieve repression of msl-2 expression. The msl-2 RNA contains multiple putative Sxl binding sites [poly(U) sequences] at both 5′ and 3′ untranslated regions (UTR). Moreover, at the 5′ UTR, these binding sites are located close to the splice junctions of a small intron, which is alternatively spliced (Fig. 3B). In males, this intron is spliced out in the majority of msl-2 transcripts; while in females, the splicing of this intron is inhibited (5, 63, 135). These poly(U) sequences are necessary in a second regulatory step, which is described below. Splicing inhibition is the way to avoid their removal.
It has been demonstrated that Sxl binds to the msl-2 5′ UTR in vitro and inhibits the splicing of the first intron (36). Binding at both poly(U) stretches, located close to the 5′ and 3′ splice sites, is necessary for efficient splicing inhibition (35).
Two different processes occur at each end of the intron. At the 3′ end, a very long poly(U) stretch (16 residues) and the 3′ splice site AG are separated by a 13-nucleotides sequence. As in tra RNA, the U-rich stretch is also the poly(Y) tract. It was shown that the presence of these 13 nucleotides is critical for mediated intron retention. In these circumstances, binding of the U2 small nuclear ribonucleoprotein (snRNP) to the substrate is not very efficient and “splicing conditions” are not ideal, which facilitates Sxl inhibition. A deletion of the sequence that separates the poly(U) [poly(Y) tract] and the AG allows a better interaction between the small unit of the general splicing factor U2AF (U2AF35) and the AG. This interaction is responsible for stabilizing the binding of the large subunit of U2AF (U2AF65) to the poly(Y) tract, which improves the binding of U2 snRNP to the substrate. In the wild-type situation, the long distance between the poly(Y) tract and AG (13 nucleotides) disrupts the U2AF35-AG interaction. As a consequence, binding of U2AF65 to the poly(U) sequence is relatively unstable. In this scenario, Sxl has a better chance to compete with U2AF, bind to the poly(U) sequence, and inhibit splicing (76).
The 5′ splice site of the alternatively spliced intron of msl-2 has an unusual structure. At position +5, there is a U instead of the conserved G nucleotide, followed by an 11-nucleotide poly(U) stretch. In this particular situation, the proximity of the 5′ splice site and the poly(U) stretch is crucial for U1 snRNP binding and splice site activation. Full spliceosome assembly is disrupted, for instance, when a spacer of 12 nt is introduced between the poly(U) stretch and the 5′ splice site. In vitro experiments performed with HeLa nuclear extracts have demonstrated that the recognition of this weak 5′ splice site is enhanced by the presence of the RNA binding protein apoptosis-promoting factor TIA-1 (123). TIA-1 associates with the poly(U) sequence, facilitating U1 snRNP binding (34). When Sxl is present, the two proteins compete for binding to the msl-2 RNA 5′ splice site region. Binding of Sxl to the U11 stretch displaces TIA-1 and inhibits U1 snRNP recruitment to the 5′ splice site (Fig. 3B) (35). The Drosophila homologue of TIA-1 is a gene called Rox8 (16), for which very little information is found in the literature. In vivo experiments have to be performed to validate the authors' model and confirm the role of this gene in msl-2 splicing.
Sxl Controls msl-2 Translation and Stability
The presence of the alternatively spliced intron in the mature transcript does not affect msl-2 ORF, since it is located at the 5′ UTR. This suggests that for msl-2, in contrast to tra and Sxl, gene expression must be controlled by a different repression mechanism in addition to alternative splicing. Endogenous msl-2 RNA is not retained in the nuclei of wild-type females. Therefore, we can exclude nuclear retention as a mechanism for repression (6). Alternatively, regulation could occur at the level of translation. Two possible models of translational repression have been proposed: (i) the secondary structure of the intron could interfere with translation initiation, and (ii) Sxl could work not only as a splicing regulator but also as a translational inhibitior by its association with poly(U) sites present at the UTRs.
Mutations in both splice sites and retention of the intron did not affect the expression of Msl-2 protein in transgenic male flies (6, 64); this discards the first translation repression model. Constructs with mutated poly(U) sequences at the UTRs were used to test the role of Sxl as a translation inhibitor. Mutation of 5′ UTR poly(U) sequences interferes to some extent with translation inhibition by Sxl (6, 36, 64). However, high levels of repression are achieved only when Sxl binds simultaneously to the poly(U) elements located at the 5′ and 3′ ends (6, 64).
Very little is known about the mechanism by which Sxl represses msl-2 mRNA translation. The requirement of Sxl binding sites at the 5′ and 3′ UTR suggests that the binding of Sxl interferes with the synergism between the cap and the poly(A) tail [i.e., the interaction between elF4E and poly(A) binding protein (PABP)]. Nevertheless, the fact that RNAs lacking a poly(A) tail are as effectively repressed by Sxl in vitro as their counterparts that have 73-residue poly(A) tails excludes this possibility (37). msl-2, to our knowledge, is the only described example in the literature in which elements located at both the 5′ and 3′ UTRs are necessary at the same time for proper translation repression by an RNA binding protein.
Northern blot analyses have indicated a substantial difference between the levels of msl-2 mRNA in males and females (135). In addition, quantitative RNase protection assays have demonstrated that wild-type females contain only 20% of the msl-2 mRNA that males have (64). Furthermore, these authors have analyzed how the deletion of Sxl binding sites in transgenic flies affects the accumulation of msl-2 mRNA. The levels of msl-2 mRNA in females increase dramatically when Sxl binding sites are removed from the transgene. These data suggest that in addition to functioning as a splicing and translation repressor, Sxl might interfere with the stability of msl-2 mRNA. However, it cannot be ruled out that a different poly(U) RNA binding protein exerts the destabilizing effect on msl-2 mRNA.
msl-2 Regulation throughout Evolution
The Drosophila virilis msl-2 5′ UTR contains Sxl binding sites, but, in contrast to D. melanogater, they are not placed in an intronic region (6). Thus, D. virilis msl-2 expression appears to be controlled only at the level of mRNA translation whereas D. melanogaster msl-2 is regulated at the levels of splicing and translation. It will be interesting to see which mechanism of repression was favored by evolution: the D. melanogaster two regulatory steps or the D. virilis single step. To draw final conclusions about the evolution of msl-2 expression and the efficiency of the two mechanisms, we need to (i) have access to the sequences of msl-2 homologues in other drosophilids (and even in other Diptera) and (ii) learn more about msl-2 regulation and expression in the two species (D. melanogaster and D. virilis). We could also ask if the presence of an intron in the msl-2 pre-mRNA brings an advantage to male flies. Perhaps the presence of an intron at the 5′ UTR of D. melanogaster male msl-2 pre-mRNAs could be important for the production of large amounts of MSL-2 protein. It was shown that the presence of an intron and its splicing can enhance the level of gene expression (70, 73).
MODEL OF DOSAGE COMPENSATION IN FEMALES
Gergen (38) has reported that dosage compensation is established early in development and that the Daughterless and Sex-lethal proteins are involved in regulating X chromosome activity at the blastoderm stage. In addition, no obvious effect of msl mutations on dosage compensation of the X-linked gene runt has been observed. These results suggest the existence of a msl-independent dosage compensation mechanism and the involvement of Sxl in such a process. Nevertheless, it cannot be ruled out that the participation of the Sxl protein in this process is indirect, for instance, through the control of a not-yet-identified msl-like gene(s) that functions only at early stages of development.
Several years later, the finding that Sxl can also function as a translation repressor for msl-2 motivated Kelley et al. to propose that the Sxl protein was directly responsible for the msl-independent dosage compensation and to look for other genes whose expression could be modulated by Sxl at the translation level (63). They searched the 3′ UTRs of Drosophila genes containing three or more copies of Sxl binding sites. Surprisingly, 20 of the 22 identified genes are on the X chromosome (Table 1), among them is Sxl itself (see below). The authors have proposed a msl-independent dosage compensation model that would occur in females rather than in males. They have hypothesized that Sxl could directly regulate the expression level of several genes, causing a twofold decrease effect, by binding to the 3′ UTR and modulating translation and/or mRNA stability. The gene runt, for instance, expressed early in development, is dosage compensated by an Sxl-dependent mechanism (9). Another piece of data that is in agreement with the idea of a dosage compensation model in females is the fact that in embryonic nuclear extracts run on sucrose gradients, Sxl sediments in very large aggregates that include RNA. It is possible that these aggregates could include more transcripts than the small subset of known Sxl targets (104).
TABLE 1.
Drosophila genes identified by database searchesa
| Database designation | Gene or product | Chromosome | Location |
|---|---|---|---|
| 3′ UTR genesb | |||
| DROACS2 | Scute | X | 1B1-7 |
| DMZESTE | Zeste | X | 3A3 |
| DMIRCRGHA | Roughest | X | 3C5 |
| DROHELHELA | Helix-loop-helix protein | X | 4C3-4 |
| DROSXLMS11 | Sex lethal | X | 6F-7B |
| DMCUT | Cut | X | 7B1-2 |
| DROFSHB | Fs(1) homeotic | X | 7D |
| DROINTBETN | Integrin beta subunit | X | 7D |
| DMOTD | Orthodenticle-ocelliless | X | 8A1-2 |
| DMGS2 | Glutamine synthase 2 | X | 10B8-11 |
| DRODPTP10D | Protein tyrosine phosphatase | X | 10D |
| DROHSC3A | Heat shock 70 cognate 3 | X | 10E |
| DMDISCO | Disco | X | 14B3-4 |
| DMBJ6 | No-on transient A | X | 14C |
| DMCS14D | Anonymous cDNA | X | 14D |
| DROANTPS2 | Position-specific antigen 2 | X | 15A1-5 |
| DROSFP | Forked | X | 15F1-3 |
| DROBARH2 | Bar | X | 16A1-2 |
| DMRUNTR | Runt | X | 19E1-3 |
| DMU03717 | Folded gastrulation | X | 20A-B |
| NM_164536 | Msl-2 | 2 | 23F |
| DROMSL1X | Msl-1 | 2 | 36F-37A |
| 3′ EST genesc | |||
| AW940554 | 12E-14A | ||
| AW941340 | 9E7-10AE3 | ||
| BG638455 | 5C7-5D6 | ||
| AW940679 | 16B4-16D6 |
Adapted from reference 63 with permission from the publisher.
Drosophila genes identified by a database search for 3′ UTRs with three or more poly(U) stretches (AU7 or U8).
Drosophila 3′ EST, identified in the X chromosome by a database search for three or more poly(U) stretches (U8).
REGULATION OF Sxl AND ITS TRANSCRIPTS
Sxl expression is controlled early in development (around the blastoderm stage) and at the transcriptional level throughout the remainder of development and adult life at the splicing level (Fig. 4).
FIG. 4.
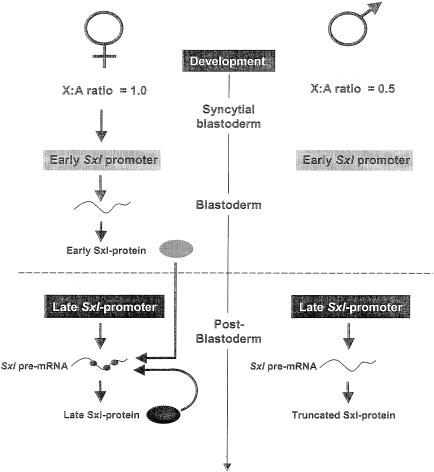
Regulation of Sxl expression. The primary genetic X:A signal acts on the early Sxl promoter and controls Sxl expression at the transcription level (67, 124). Due to a twofold difference in the number of X chromosomes (autosomes are the same in both sexes), Sxl transcription is either initiated in females but not in males or initiated in both sexes but much more efficiently in females than in males. As a result, early Sxl protein is abundantly produced in females whereas it remains undetectable in males. After the blastoderm stage, the late Sxl promoter starts functioning in both sexes, and production of the late Sxl transcripts persists throughout the remainder of the fly's life. In females, the abundant early Sxl protein imposes the female-specific splicing pathway on the late Sxl RNA, leading to the production of late Sxl protein, and the feedback loop is established. In male individuals in which no (or insufficient) early Sxl protein is produced, a different splicing pattern takes place and a truncated and nonfunctional version of Sxl is generated.
The Sxl gene produces two separate sets of transcripts, linked to the function of its two promoters, the so-called early and late promoters (Fig. 5). In females, the early promoter is activated around the blastoderm stage by the X:A signal, which controls Sxl at the transcriptional level (67, 124). Due to a twofold difference in the number of X chromosomes—autosomes are the same in both sexes—Sxl transcription is either initiated in females but not in males or initiated in both but much more efficiently in females than in males. As a result, early Sxl protein is abundantly produced in females whereas it remains undetectable in males (for further extensive discussion of the X:A signal, see references 25, 107, and 108). The late Sxl promoter is activated in both sexes after the blastoderm stage, and the production of the late transcripts persists throughout the remainder of development and adult life. Nothing is known about the regulation of the late Sxl promoter. The presence of the early Sxl protein in females directs the first copies of late Sxl RNAs into the female mode of splicing. This gives rise to the late set of Sxl proteins and, consequently, sets up the female mode of splicing. In contrast, in males, the first copies of late Sxl RNAs follow the male mode of splicing since few or no early Sxl proteins are available, resulting in the establishment of the male-splicing state of Sxl (Fig. 4). Therefore, the developmental meaning of the X:A signal is to “switch on” the early Sxl promoter at a specific time, providing females with the early Sxl proteins needed to establish female-specific control of Sxl once the late constitutive promoter of this gene starts to function.
FIG. 5.
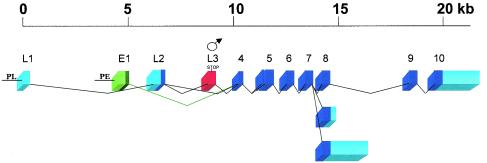
Molecular organization of the Sxl gene. The Sxl gene produces two separate sets of transcripts, linked to the function of its two promoters, the so-called early (PE) and late (PL) promoters. Alternative splicing and different polyadenylation sites give rise to several different transcripts. Boxes represent exons, and horizontal lines represent introns. The embryo-specific exon (E1) is represented in green (light green for the noncoding region, and dark green for the coding region). The male-specific exon is represented in red. Other exons are represented in blue (light blue for the noncoding region, and dark blue for the coding region).
The late sets of male and female Sxl mRNAs are similar, except for the presence in the male mRNAs of an additional exon (L3), which contains a translation stop codon. Consequently, the male late transcripts give rise to inactive truncated Sxl proteins (7). The elimination of the male-specific exon L3 in females requires the Sxl protein (Fig. 6A) and is discussed in detail in the next section.
FIG. 6.
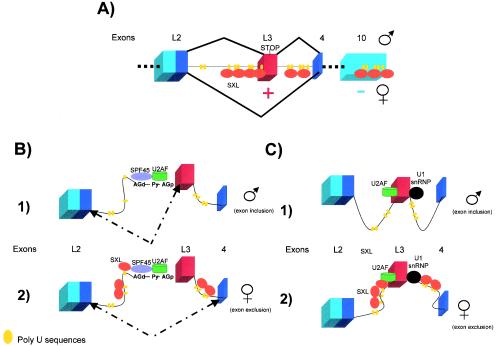
Sxl controls the alternative splicing of its own pre-mRNA. Boxes represent exons, and lines represent introns. The male-specific exon (Sxl) is represented in red. Other exons are represented in blue (light blue for the noncoding region, and dark blue for the coding region). (A) The Sxl protein binds to poly(U) sequences in introns 2 and 3 and excludes exon L3, which contains translation stop codons, from the mature transcript. This process establishes a positive autoregulatory loop. On the other hand, Sxl binds to poly(U) sequences located at the 3′ UTR, inhibiting the translation of its own mRNA. The balance between the negative and positive posttranscriptional controls keeps the concentration of Sxl constant at levels that are nontoxic for the cell and are sufficient to regulate Sxl splicing and its target genes. (B) Model for the control of the L3 3′ splice site by SPF45 and Sxl. The splicing pattern is indicated by dashed arrows. 1, In male flies, U2AF recognizes the distal AG (AGd) and the poly(Y) tract (Py) during the first catalytic splicing step. In the second step, SPF45 recognizes the proximal AG (pAG). This AG is then, used as the site of exon ligation. 2, In female flies, Sxl binds to poly(U) sequences located at the introns adjacent to exon L3 and interacts with SPF45, blocking both proximal and distal 3′ splice site AGs from being used. Splicing then occurs between exons 2 and 4. (C) Model of regulation of exon 3 splicing based in interactions between Sxl and U2AF and U1snRNP. 1, In males, a functional spliceosome is formed and exon 3 is included in the mature transcript. 2, In females, Sxl interacts with the splicing factors U2AF and U1snRNP, bound to the 3′ and 5′ splice sites, respectively, and inhibits the formation of a functional spliceosome. As a consequence, exon 3 is not included in the mature female transcript.
Differences found between the early and late Sxl transcripts are due predominantly to activation of different promoters and alternative splicing. The early Sxl transcripts follow a fixed splicing pattern in which exon L2 and the male-specific exon L3 are not included in the mature early transcripts and exon E1 is directly spliced to exon 4 (Fig. 5). Hence, early and late Sxl proteins have different amino-terminal ends. The splicing pattern of early Sxl transcripts was analyzed in transgenic flies that express a Sxl minigene containing the region between exon E1 and exon 4 under the control of a heat shock promoter (55). The analysis of the splicing pattern of the mRNAs produced by the transgene in a distinct genetic background has revealed a series of negative results, leading the authors to conclude that the exon E1-exon 4 splicing pattern is not affected by the X:A signaling, by the presence of the Sxl protein, or by genes required for proper function of the Sxl protein [fl(2)d, vir, and snf] (see below). Sequences important for the correct splicing of early Sxl transcripts have been searched by using a similar strategy (136). The results have suggest that the relative strength of the exon E1 and L2 5′ splice sites and sequences located in exon E1 and early intron 1 might play a role in the exclusion of exon L2 and L3 from the early Sxl mature transcripts. Nevertheless, a more detailed mutation-deletion analysis must be performed to map and identify elements required for the proper splicing of early Sxl transcripts.
ROLE OF Sxl IN THE SPLICING OF ITS OWN PRIMARY TRANSCRIPT
The mechanism by which Sxl precisely controls the skipping of the Sxl male exon (L3) is not totally understood. In contrast to most of the examples of exon-skipping events described in the literature, Sxl promotes a 100% switch between the two alternatively spliced forms, suggesting the existence of a complex mechanism of regulation with several checkpoints.
As in tra pre-mRNA, a long poly(U) sequence (a potential binding site for Sxl) is part of the poly(Y) polypyrimidine tract associated with one of the 3′ splice sites preceding the Sxl male-specific exon. Nevertheless, several lines of evidence suggest that competition between Sxl and U2AF for binding to the site is insufficient to explain Sxl-mediated exon skipping. Mutations within this poly(U) stretch do not abolish splicing regulation (53, 96), and multiple U-rich sequences placed in introns 2 and 3, and relatively distant from 5′ and 3′ splice sites (Fig. 6A), play an important role in the control of exon L3 skipping (53, 96). Multiple poly(U) sequences are also present in the adjacent introns of the male-specific exon of the Sxl genes of D. virilis (13) and D. subobscura (88). However, in both species the poly(Y) tract associated with the distal 3′ splice site, preceding the male-specific exon, does not contain a U-rich sequence. Finally, ectopic expression in male transgenic flies of a chimeric protein containing the effector domain of U2AF fused to the complete RNA binding domain of Sxl does not disrupt Sxl pre-mRNA splicing regulation, in contrast to what occurs with tra splicing (45). In combination, these data suggest that Sxl controls tra and Sxl alternative splicing by different mechanisms.
It has been proposed that the blockage of the exon 3 5′ splice site is the key regulatory step of Sxl RNA exon skipping (53). In this case, the relative strengths of the competing 5′ splice sites (exons L2 and L3) would play an important role. Although the splice sequences associated with exons L2 and L3 are conserved among D. melanogaster, D. virilis, and D. subobscura (the exon L3 5′ splice site is stronger than the exon L2 5′ splice site), mutations that change the strength of these splice sites do affect the default and regulated splicing (90) (the Sxl transcription unit is shown in Fig. 5).
Analysis of the two alternative 3′ splice sites preceding exon L3 has brought surprising results. Deletion or mutation of the distal 3′ splice site, including both the poly(Y) tract and the 3′ splice site AG, has resulted in the accumulation of transcripts in which exon L3 is skipped in the absence of Sxl, suggesting the importance of this splice site in exon definition (90). Mutations of the AG associated with the proximal 3′ splice site have not affected exon definition, but exon skipping in the presence of Sxl is strongly reduced. While the distal 3′ splice site plays an important role in exon definition, the majority of the actual splicing events take place using the proximal 3′ splice site (90). This dual recognition of the 3′ splice site could create an opportunity for Sxl repression. In vitro experiments with Sxl 3′ splice site constructs have been used to establish a mechanistic model. It has been proposed that Sxl blocks the splicing process at second catalytic step by interacting with and inhibiting the function of the splicing factor SPF45. The functions of this recently characterized splicing factor are related to the recognition of the proximal 3′ splice site AG, commitment of this site to splicing, and catalytic activation during the second step (68) (Fig. 6B).
Based on biochemical and genetic interactions, an alternative model has recently been suggested (78). It was observed that Sxl interacts with U2AF and U170K, a component of the U1 snRNP, in embryonic extracts. Besides, both U1-70K and U2AF mutants have shown Sxl splicing defects. It has been proposed, then, that Sxl interacts with U1 snRNP and U2AF, which are bound respectively to the 5′ and 3′ splice sites of male exon 3, producing a complex that prevents the formation of a functional spliceosome. In contrast to what has been suggested by the other authors (68), this process would occur after splice site recognition but before the catalysis process (78) (Fig. 6C). We do not think that either model excludes the other. As mentioned at the beginning of this section, Sxl exon-skipping regulation is very precise, in contrast from most examples found in the literature. It is possible that Sxl acts at different steps of the splicing process. A multiple-step regulation would permit pre-mRNAs that escaped regulation at the initial steps to be “rechecked” and directed to the female mode of splicing.
GENES REQUIRED FOR THE FEMALE-SPECIFIC SPLICING OF THE Sxl PRIMARY TRANSCRIPT
The female-specific splicing of Sxl pre-mRNA requires, in addition to the Sxl protein, the function of other genes, such as sans fille (snf), female-lethal-2-d [fl(2)d], and virilizer (vir).
snf
The snf gene is required for Sxl function in both somatic and germ cells. Females homozygous for snf loss-of-function mutations are sterile because the germ cells do not differentiate properly, but they continue to divide, giving rise to ovarian tumors (81, 100, 118). In these mutant germ cells, Sxl pre-mRNA is spliced as in males, so that truncated Sxl proteins are produced (13, 82). This indicates that snf plays a role in the process of sex determination in the germ cells. Its role in sex determination in the soma has been inferred from a female-lethal synergistic interaction between mutations at both the snf and Sxl genes (81, 100, 118). Molecular analyses of this lethal interaction have revealed that snf function is required to establish the female-specific splicing of Sxl primary RNA (1).
The snf gene encodes a 28-kDa nuclear protein that is homologous to the mammalian U1A and U2B′′ snRNP proteins (33, 47). The function of snf is not limited to sex determination, i.e., to the splicing regulation of Sxl RNA, since it has been found that a mutation abolishing snf function completely shows a non-sex-specific lethal phenotype (33). This shows that the Snf protein is a component of the constitutive splicing machinery. It has been proposed that Snf participates in Sxl RNA splicing by forming a nonproductive RNP complex with U1 snRNP and Sxl that blocks the 5′ splice site of the male-specific exon L3 of Sxl RNA, preventing incorporation of the L3 exon into the mature mRNA (101). The interaction of Sxl protein with Snf occurs mainly through the first RNA binding domain (RBD) (102). Although Sxl protein controls the female-specific splicing of tra and msl-2 pre-mRNAs, (see above), genetic analysis has ruled out a direct role of snf in tra and msl-2 pre-mRNA splicing (26).
fl(2)d
The fl(2)d gene has dual functions: a female-specific function involved in the splicing of Sxl and tra RNAs and a non-sex-specific function not related to Sxl (41, 42, 44). Mutations at fl(2)d mimic the behavior of loss-of-function mutations in Sxl, with respect to the processes of sex determination and dosage compensation in the soma (41). Moreover, it has been shown that fl(2)d mutant females express the Sxl transcripts characteristic of males, indicating the requirement of fl(2)d in the female-specific splicing of Sxl pre-mRNA (41). The fl(2)d gene is also required for the development of female germ cells. Loss-of-function mutations at either fl(2)d or Sxl are equivalent with regard to germ cell development, indicating that likely fl(2)d is also involved in the female-splicing regulation of Sxl RNA in these cells (42). A direct role of fl(2)d on tra pre-mRNA splicing has been reported (44).
The fl(2)d gene encodes a nuclear protein that contains two stretches of 5 and 6 histidines separated by prolines and two adjacent stretches of 10 glutamines followed by a glutamine-rich region, elements that suggest that protein-protein interactions may be important for its function (89). Since the female-specific function of fl(2)d is related to its requirement for proper splicing regulation of the Sxl and tra RNAs by the Sxl protein, the question arises of how Fl(2)d can affect the function of the Sxl protein.
Recently, data supporting the idea that Fl(2)d functions as a splicing regulator have been reported (85). Fl(2)d forms a complex with Sxl and Vir in vivo. Moreover, Fl(2)d has homology to a human protein (Wilms' tumor-associated protein [WTAP]) that interacts with the Wilms' tumor 1 protein (WT1) (72, 85). The WT1 gene is implicated in mammalian sex determination, and its mutation is associated with Wilms' tumor, a common type of pediatric kidney cancer (48, 69). One isoform of WT1 is associated and localized with splicing factors (65). In vitro splicing assays have been carried out with WTAP-depleted and mock depleted HeLa extracts to test the direct role of Fl(2)d/WTAP in splicing regulation. Depletion of WTAP has been shown to interfere with the capacity of Sxl in controlling tra alternative splicing. Addition of recombinant Fl(2)d protein to depleted extracts restores the ability of Sxl to inhibit tra splicing in vitro. The function of Fl(2)d as a splicing regulator is not restricted to the sex determination genes; it is also required for proper control of Ultrabithorax (Ubx) alternative splicing (17). More detailed biochemical analysis is necessary to determine the exact function of fl(2)d in splicing regulation. We could speculate that Fl(2)d might modulate the binding of Sxl to target sequences and/or form, together with Sxl and other proteins, a large complex necessary for splicing inhibition.
Fl(2)d also interacts with the product of the dim-7 gene. This gene is the Drosophila homologue of human RanBP7, a member of the importin β family of nuclear import receptors (39). This result opens the possibility that, besides its role in splicing, Fl(2)d can also play a role in the transport of Sxl and other proteins.
vir
The vir gene has dual functions: a female-specific function involving the splicing of Sxl and tra RNAs and a vital function not related to sex determination (51, 52). Mutations within vir mimic the behavior of loss-of-function mutations in Sxl with respect to the processes of sex determination and dosage compensation in the soma (51, 52). Moreover, it has been shown that females with vir mutations express Sxl transcripts characteristic of males, indicating the involvement of Vir in the female-specific splicing of Sxl pre-mRNA (52). vir is also required for female germ cell development, since it regulates Sxl function. The early production of Sxl protein in undifferentiated XX germ cells is independent of vir function, but this function is required later in oogenesis for Sxl expression (111).
vir encodes a nuclear protein of 210 kDa. It contains a putative transmembrane domain, a coiled-coil region, and PEST sequences (79). It has been speculated that Vir is located in the nuclear membrane and/or the nucleoplasm, where it may mediate the transport of a specific mRNA subset (79).
NEGATIVE AUTOREGULATION: Sxl AS ITS OWN TRANSLATION REPRESSOR
The presence of Sxl in the list of genes that contain Sxl binding sites at the 3′ UTR prompted Yanowitz et al. to test the possibility that this protein works as its own translational repressor (133). Several results clearly indicate that this is the case. First, Sxl mRNA can be detected by reverse transcription-PCR after immunoprecipitation of embryo extracts by anti-Sxl antibody, showing its association with the Sxl protein. Second, transgenic females carrying an Sxl cDNA with a reduced 3′ UTR express higher levels of Sxl protein than do their siblings that carry a complete cDNA including a long 3′ UTR that contains several Sxl binding sites. Finally, males carrying Sxl transgenes that contain a reduced 3′ UTR do not survive, in agreement with the presence of Sxl protein from these transgenes.
Sxl gives rise to a variety of transcripts, some of which are stage and tissue specific (67, 88, 99, 103). Figure 5 shows a schematic view the transcriptional unit of Sxl. Most of the differences among the transcripts are found at the 3′ UTR and are due to the usage of different poly(A) signals and/or alternative splicing of exons 8, 9, and 10. Transcripts having a short 3′ UTR are more abundant in the germ line and during the early stages of embryogenesis (5 to 8 h) (99). In later stages of development and during adult life the most abundant transcripts have a long 3′ UTR (up to 3,600 nucleotides) containing several Sxl binding sites (13 in the largest transcript) (103). It has been speculated that the presence of short transcripts at early stages of development allows a substantial amount of Sxl protein be produced and also permits a positive autoregulatory loop via splicing to be established. Once accumulation of the Sxl protein is no longer required, short mRNAs are substituted for larger transcripts whose expression can be repressed by Sxl. The balance between the negative and positive posttranscriptional controls keeps the concentration of Sxl at levels that do not interfere with other cellular functions and are sufficient to regulate Sxl splicing and its target genes (133). In this regard, it has been shown that high levels of the Diptera Musca domestica and Ceratitis capitata Sxl proteins ectopically expressed in D. melanogaster have deleterious consequences (75, 94). Although Sxl might play distinct functions in different Diptera species, this effect could be general and independent of the usual target genes of Sxl.
Several other examples of RNA binding proteins that regulate the translation of their own mRNAs are known, e.g., Drosophila Elav (105), Saccharomyces cerevisae ribosomal protein L32 (29), thymidylate synthase (21), Arabidopsis thaliana AtGRP7 (50), PABP (131), and the fragile X mental retardation protein (109). This type of autoregulation may be a common strategy among RNA binding proteins that modulate the splicing and/or translation of other genes as well as their own genes. Changes in the cellular levels of these proteins might interfere with their ability to regulate the expression of target genes. A negative autoregulatory loop could prevent overexpression, keeping the protein levels within an appropriate range.
Sxl AND DEVELOPMENT OF THE GERM LINE
The germ line, like the soma, also exhibits sexual dimorphism. Cells with the 2X;2A chromosomal constitution follow the oogenic pathway, and XY;2A cells develop as sperm. There is, however, a fundamental difference between the genetic control of sexual development in the soma and the germ line. The sexual development of somatic cells is determined solely by cell-autonomous signals, i.e., the chromosome constitution of the cells (the X:A ratio signal). In contrast, cell-autonomous (X:A ratio) and somatic inductive signals determine the sex of the germ line (80, 119; for extensive reviews, see references 25 and 83). Sxl is also required for oogenesis: 2X;2A germ cells lacking the Sxl protein do not enter oogenesis but follow an abortive spermatogenesis pathway characterized by the formation of multicellular cysts (80, 110, 119). The onset of Sxl expression occurs later in germ cells than in somatic cells. By the time this gene is activated in the somatic cells (around blastoderm stage), the pole cells (the precursors of the germ cells) still do not express Sxl (12). Expression of this gene in germ cells is first detected in 16- to 20-h-old embryos (54). A female germ line-specific Sxl transcript has been identified (99).
The Sxl protein is localized predominantly in the nucleus of somatic cells, whereas in germ cells it shows a more complex distribution pattern (13). In nondifferentiated germ cells, Sxl is accumulated in the cytoplasm, and it is translocated into the nucleus as these cells start to differentiate. The biological meaning of this protein shift remains unknown. It has been speculated that the cytoplasmic localization of the Sxl protein might be relevant for initiation of the germ cell differentiation pathway, maybe by regulating the translation or interfering with the stability of cytoplasmic mRNAs (12). In this context, it has been argued that the shift from cytoplasm to nuclei might represent a switch in the regulatory function of Sxl from being a translational regulator in nondifferentiated cells to a splicing regulator in differentiated germ cells (83).
Sxl proteins are abundant in ovaries but are not detectable in unfertilized eggs (13), although these eggs contain large amounts of Sxl mRNAs (99). Blockage of the translation of these mRNAs is necessary. After the blastoderm stage, the late Sxl promoter starts functioning in both sexes and produces the late Sxl transcripts. The presence of maternal Sxl protein in male embryos imposes the female-specific splicing pathway on the late Sxl RNA, leading to the production of late Sxl proteins, and the feedback loop is established. This causes male-specific death because the presence of Sxl protein prevents hypertranscription of the single X chromosome in males; that is, dosage compensation does not occur.
In the soma, Sxl is initially activated by the X:A signal (see above). How is Sxl activated in the germ line? Both germ line cell-autonomous (X:A signal) and somatic inductive signals from the gonadal soma act to regulate Sxl in germ cells (24, 43, 80, 130). So far, the somatic inductive signal has not been identified. With respect to the X:A signal, it has been shown that the genes that form this signal in the soma are not required to activate Sxl in the germ line, indicating that the X:A signal is formed by different genes in somatic and germ cells (43, 120). Moreover, the gene daughterless (da), whose maternal product is involved in the initial activation of Sxl in the soma (22), is not required for activation of Sxl in germ cells (28). It has been shown that increasing doses of snf+ cause feminization of 2X;3A germ cells (46) The authors have also reported that the same feminization effect of 2X;3A germ cells can be obtained with increasing doses of Sxl+. It has also been proposed that snf participates in a germ line X-chromosome counting signal. This signal, in conjunction with the somatic signals and eventually with other germ line X-chromosome dose signals, could determine the sexual fate of the germ cell. However, it can be also true that the observed feminization effect of 2X;3A germ cells due to increasing doses of snf+ is a consequence of the participation of Snf in the female-specific splicing of Sxl pre-mRNA. In this scenario, more Sxl protein is produced in the intersexual 2X;3A condition, causing a shift toward femaleness.
As in the soma, Sxl expression in the germ line is regulated at the splicing level by the Sxl protein (13, 46). It has been found that the gene ovo and ovarian tumor (otu) are needed to promote female-specific splicing of the Sxl pre-mRNA in germ cells (13, 82, 87). Other genes are also involved in this regulation (see above). In addition to its role in D. melanogaster germ line sex determination, Sxl is necessary for homologous chromosomal recombination, a process that occurs only during female gametogenesis (15).
Target for Sxl in the Germ Line
In a very recent publication, it has been reported that the gene gutfeeling (guf)—the Drosophila homologue of the ornithine decarboxylase antizyme—is a target of Sxl in the germ line (127). This gene is supposedly involved in polyamine synthesis (127).
The authors have presented clear genetic evidence demonstrating an interaction between Sxl and guf. Nevertheless, a more detailed molecular analysis must be performed to determine at which level(s) Sxl controls guf expression. Sxl seems to bind both spliced and unspliced transcripts. A preliminary analysis has pointed out no differences between male and female guf transcripts. It is possible that sex-specific splicing occurs only in a stage- and/or cell specific fashion, as suggested by the authors, but more careful and quantitative assays should be done before any final conclusions are made. guf mRNA levels are inverted with respect to the amount of Sxl protein in the germ cells. Based on this observation, we could suggest that this constitutes another example of Sxl protein interfering with the stability of a target mRNA (see the discussion of msl-2 mRNA stability, above). However, at this point it cannot be ruled out that the observed effect is indirect.
guf itself apparently controls Sxl expression in the germ line. Moreover, guf participates in Sxl localization and its translocation from the cytoplasm into the nucleus (127). What is the biological significance of the Sxl-guf regulatory loop? Part of it could be the control of mitosis, as proposed by the authors. Levels of the cell cycle cyclin B gene are supposedly connected in a positive and negative fashion to Sxl and guf expression, respectively.
DOMAINS AND FUNCTIONS OF Sxl
RNA Binding Domains
Sxl has two segments in tandem, located in the central portion, with similarity to the RNA binding domain sequences (RBD) or RNA recognition motif (RRM) (Fig. 7). The described family members have about ninety amino acids and the same global fold: two α-helices packed against a four-stranded antiparallel β-sheet. For details about domain sequences and the RNA recognition properties, see reference 62.
FIG. 7.
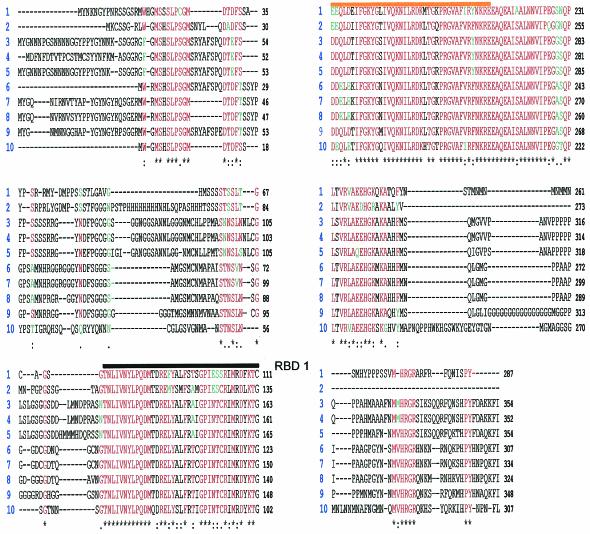
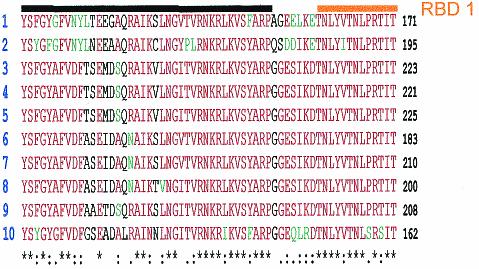
The Sxl protein in different Diptera species. The comparison was done using ClustalV software and then adjusted manually. Sequences are as follows: 1, Sciara ocelaris, accession number AA019468; 2, Anopheles gambiae, accession number EAA03881; 3, Drosophila melanogaster, accession number P19339; 4, Drosophila melanogaster embryonic form (emb.), accession number A4218; 5, Drosophila subobscura, accession number Q24668; 6, Chrysomya rufifacies, accession number O97018; 7, Lucilia cuprina, accession number Q9BKK4; 8, Musca domestica, accession number O17310; 9, Ceratitis capitata, accession number O61374; and 10, Megaselia scalaris, accession number O01671. A black line is drawn above RBD1, and an orange line is drawn above RBD2. Amino acids represented in red are highly conserved (identical in six species or more). Amino acids represented in green are the ones that have equivalent biochemical properties in relation to a highly conserved amino acid. Nonconserved amino acids are represented in black. An asterisk at the bottom of the sequences indicates that in this position, the referred amino acid is conserved in all analyzed species. A colon at the bottom of the sequences indicates that in this position a highly conserved amino acid and amino acid(s) with equivalent biochemical properties are present.
Deletions of the amino and the carboxyl termini did not interfere with the ability of Sxl RBDs to properly bind in vitro to their target sequences. Nevertheless, both RNA binding domains in cis are required for site-specific RNA binding (61, 98, 102, 128). The properties of several Sxl constructs were tested in vitro (102). Either RBD1 or RBD2 alone showed reduced RNA binding activities. Duplications of the RBDs (RBD1-RBD1 and RBD2-RBD2) did not affect the RNA binding capacity but interfered with RNA recognition properties. Proteins in which the order of the two RBDs was been reversed (RBD2-RBD1) bound very weakly to oligonucleotides that contained only a single Sxl binding site. Nevertheless, the binding was close to normal if an oligonucleotide containing two binding sites was used as a probe, reflecting possible reestablishment of protein-protein interactions.
Amino-Terminal Domain
The amino terminus of D. melanogaster Sxl is very rich in glycine, a characteristic that is evolutionarily conserved (Fig. 7). It was shown that this domain is implicated in protein-protein interaction (Sxl multimerization) and is absolutely required for proper control of Sxl RNA alternative splicing (68, 128, 129). A deletion of the first 38 amino acids disrupts Sxl RNA splicing regulation (128), and deletion of the first 94 amino acids interferes with the ability of Sxl to inhibit the second catalytic step of the splicing of Sxl intron 2 (68) (for details, see above). Moreover, a chimeric protein containing this region and β-galactosidase behaved like a dominant negative, altering Sxl RNA splicing, interfering with Sxl protein expression, and causing female death (30). This domain also mediates interactions with other RNA binding proteins that contain glycine-rich regions (129) and with SPF45 (68).
It is interesting that the first 25 amino acids are completely different between the early and late Sxl isoforms due to alternative splicing (see “Regulation of Sxl and its transcripts” above). Very little information is found in the literature about the early Sxl protein. A comparative study between the early and late isoforms should be performed to determine the differences between the mechanisms used by the two isoforms to control alternative splicing.
It has been observed that Sxl binds in a cooperative manner to RNAs containing two or more poly(U) sequences (61, 128). There is some conflict concerning the elements required for protein-protein interaction and, consequently, for cooperative binding. According to Samuels et al. (102), protein-protein interaction is mediated by the RBDs, not by the amino-terminal region, and can occur in the absence of added exogenous RNA. Sakashita and Sakamoto (98) also reached the same conclusion concerning the importance of RBDs for Sxl-Sxl interaction but, in contrast to Samuels et al. (102) and in agreement with Wang and Bell (128), have claimed that homodimerization of Sxl is RNA dependent.
There is also some controversy concerning the function of the amino-terminal region in tra RNA alternative splicing regulation. It has been proposed that this region is not necessary for tra regulation (45), while others have proposed the opposite (133). Differences in how the groups have performed the experiments, which include different constructs (40-amino-acid deletion of the amino-terminal region in one case and 94-amino-acid deletion in the other), use of different promoters, and use of different reverse transcription-PCR methods to detect the splice forms might explain the discrepancy. If the amino-terminal region is really implicated in tra alternative splicing control, the model of tra alternative splicing (126) should be revisited. Other regulatory steps besides the U2AF-Sxl competition for binding the non-sex-specific poly(Y) tract might exist.
The amino-terminal domain is not required for inhibiting msl-2 expression (37, 133). The two Sxl RBDs by themselves are able to control in vitro msl-2 mRNA translation (37).
Glycine-rich domains are present in other RNA binding proteins that regulate splicing. This is the case for the heterogeneous nuclear RNA (hnRNP) A1. A1 controls an exon-skipping event of its own mRNA by binding to two elements located in the introns that flank the alternatively spliced exon. Deletion of the glycine-rich domain does not affect RNA binding but does interfere with proper splicing regulation (10). In the proposed model, A1 binding to the intronic elements is followed by interaction among the bound A1 molecules. This gives rise to a loop structure that brings the more distant pair of splice sites into close proximity, thereby improving their rate of commitment. In this case, 5′ splice site selection would be an effect of conformational changes of the pre-mRNA (10). A similar mechanism may occur in female-specific Sxl RNA splicing.
Sxl BINDING SITE
Deletions and mutations of the intronic sequences in tra and Sxl have indicated that poly(U) sequences are the likely target of Sxl protein (53, 57, 96, 116). However, other nuclear RNA binding proteins like Elav, PTB, and U2AF also have affinity for poly(U) sequences (71, 115). It has been hypothesized that some additional sequences and/or secondary structure and/or differences in the length of the poly(U) tract are required for the binding specificity and regulation of target RNAs.
The specificity of Sxl was verified (97, 115, 129) by applying in vitro selection and amplification of ligand RNAs from a random pool (59, 125) and by performing gel shift titrations using Sxl fusion proteins and a number of RNAs generated from a splicing substrate vector (104). The majority of Sxl targets found by Singh et al. (115) consisted of long poly(U) stretches (17 to 20 uridines) interrupted by 2 to 4 guanosines, resulting in the consensus sequence UUUUUGUU(U/G)U(G/U)UUU(G/U)UU. In vivo, poly(U) tracts of this length are found only in the tra intron and in the Sxl 3′ UTR. SXL targets (consensus sequence AUnNnAGU) defined by Sakashita and Sakamoto (97), have a poly(U) tract that varies from 7 to 13 nucleotides, a value that is more consistent with the poly(U) stretches identified in the Sxl introns and 3′ UTR and in msl-2 UTRs; a similar result was obtained by Wang et al. (129). Samuels et al. (104) have tested only poly(U)-containing sequences that have 8 or fewer uridines. Sxl binding to poly(U) tracts containing 6 or fewer uridines was very inefficient. The presence of a single A immediately upstream of the poly(U) stretch increases the SXL affinity (97, 104), a feature shared by many putative SXL binding sites present in Sxl and msl-2.
The binding selectivity of Sxl in vitro is too low to explain why and how in vivo Sxl regulates the splicing of only a selected number of genes. For instance, Sxl binds in vitro to the fuji tarazu (ftz) poly(Y) tract, a non-Sxl target, with the same affinity with which it binds to the tra poly(Y) tract (61). As we discussed previously, there is a possibility that in vivo SXL activities, including binding to designated targets, are modulated by interacting proteins like female lethal 2 d [fl(2)d] (89).
Sxl OF OTHER DIPTERAN SPECIES
Sxl has been characterized in different Drosophila species. The structure and sequence organization of Sxl of D. virilis (14) and D. subobscura (88) are well conserved, and, as in D. melanogaster, Sxl regulation occurs by sex-specific alternative splicing: the Sxl transcripts in males have an additional exon containing stop translation codons. The Sxl of D. virilis, however, is unusual for the presence in males of an open reading frame, downstream of the last stop codon in the male-specific exon, that encodes a Sxl protein. This is identical to the female Sxl protein except for the first 25 amino acids of the amino-terminal region, which are encoded by differentially spliced exons. The male Sxl protein accumulates predominantly in the embryonic ectoderm, suggesting a putative role in the development of the central nervous system (14). The Sxl protein was also detected in males of other species (D. americana, D. flavomontana, and D. borealis) of the virilis radiation (14). As in D. melanogaster, the pole cells at the blastoderm stage of D. subobscura and D. virilis embryos do not express Sxl protein (14, 88).
Sxl is present outside the genus Drosophila. Sxl has been characterized in Chrysomya rufifacies (77), Megaselia scalaris (113, 114), Musca domestica (75), and Ceratitis capitata (94) (Fig. 7 gives a comparison of the Sxl proteins of different species). Sxl of these species shows two main properties. First, Sxl does not present sex-specific regulation. Second, the Sxl proteins of these species and those of the drosophilids show a high degree of conservation in the two RBDs and the few amino acids that separate them—the linker region. However, a high degree of conservation is not found outside these two domains. The gene Sxl appears not to play the master regulatory role in sex determination in the non-drosophilids that it plays across the genus Drosophila. This suggests that during the evolution of the Drosophila lineage, Sxl was coopted to become the master regulatory gene in sex determination and dosage compensation.
CONCLUDING REMARKS AND PERSPECTIVES
Sxl constitutes a paradigm for the understanding of mechanisms of eukaryote gene expression. The very complex and unusual regulation of Sxl and its targets constitutes a unique model to study some aspects of posttranscriptional gene regulation. Although we know much about Sxl, there is still a lot to learn about this protein, which will lead us to a better understanding of the mechanisms of gene regulation in eukaryotes. For instance, the molecular mechanisms implicated in Sxl RNA alternative splicing and msl-2 and Sxl mRNA translation repression and the role of the interacting proteins Vir and Fl(2)d have not been completely defined. The biological meaning and the molecular mechanisms behind the regulatory loop involving Sxl and the newly described germ line target, guf, have to be elucidated. The role of the Sxl protein in recombination between homologous chromosomes that occurs in female gametogenesis is also not well understood. Recently, an intriguing study has been reported in which infection by Wolbachia restores fertility to D. melanogaster females that are mutant for germ line-specific Sxl alleles (117).
The list of genes whose expression is mediated by Sxl via alternative splicing or mRNA stability or at the translation level still needs to be completed; this includes testing the proposed hypothesis of a dosage compensation mechanism in females (63). This task is no longer a random fishing expedition because of the development of DNA microarray analysis and the Drosophila genome project databanks (www.fruitfly.org). Identification of the targets for RNA binding proteins has been expedited by the development of techniques that combine immunoprecipitation of mRNP and subsequent analysis on microarrays (122).
From a biological point of view, the characterization of Sxl, its target genes, and its interacting proteins is important for the understanding of the molecular strategy behind sex determination, sex behavior, dosage compensation, and development of the germ line. Sxl homologues seem not to be implicated in these processes in species outside the genus Drosophila, and their functions in these species remain unclear. The development of techniques that allow the transformation of other insects (19) and knockdown genes in flies by using RNA interference (66) can help scientists achieve this goal. The molecular characterization of Sxl in other insect species will help us to understand the molecular evolution of this gene. In fact, Sxl could be a paradigm for understanding of the molecular evolution of key regulatory genes in development.
Acknowledgments
We thank Amy Sims, Andre Pires da Silva, Kelly Gordon, Mike Burdick, Dale Beach, and Diego Zorio for comments on the manuscript and Mitzi Kuroda for authorizing the reproduction of published table.
This work was financed in part by grant PB98-0466 to L.S. by the D.G.I.C.Y.T.
REFERENCES
- 1.Albrecht, E. B., and H. K. Salz. 1993. The Drosophila sex determination gene snf is utilized for the establishment of the female-specific splicing pattern of Sex-lethal. Genetics 134:801-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amrein, H., M. Gorman, and R. Nothiger. 1988. The sex-determining gene tra-2 of Drosophila encodes a putative RNA binding protein. Cell 55:1025-1035. [DOI] [PubMed] [Google Scholar]
- 3.Amrein, H. 2000. Multiple RNA-protein interactions in Drosophila dosage compensation. Genome Biol. 1:1030.1-1030.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachiller, D., and L. Sánchez. 1991. Production of XO clones in XX females of Drosophila. Genet. Res. 57:23-28. [DOI] [PubMed] [Google Scholar]
- 5.Bashaw, G. J., and B. S. Baker. 1995. The msl-2 dosage compensation gene of Drosophila encodes a putative DNA-binding protein whose expression is sex specifically regulated by Sex-lethal. Development 121:3245-3258. [DOI] [PubMed] [Google Scholar]
- 6.Bashaw, G. J., and B. S. Baker. 1997. The regulation of the Drosophila msl-2 gene reveals a function for Sex-lethal in translational control. Cell 89:789-798. [DOI] [PubMed] [Google Scholar]
- 7.Bell, L. R., E. M. Maine, P. Schedl, and T. W. Cline. 1988. Sex-lethal, a Drosophila sex determination switch gene, exhibits sex-specific RNA splicing and sequence similar to RNA binding proteins. Cell 55:1037-1046. [DOI] [PubMed] [Google Scholar]
- 8.Bell, L. R., J. I. Horabin, P. Schedl, and T. W. Cline. 1991. Positive autoregulation of Sex-lethal, by alternative splicing maintains the female determined state in Drosophila. Cell 65:229-239. [DOI] [PubMed] [Google Scholar]
- 9.Bernstein, M., and T. W. Cline. 1994. Differential effects of Sex-lethal mutations on dosage compensation early in Drosophila development. Genetics 136:1051-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanchette, M., and B. Chabot. 1999. Modulation of exon skipping by high-affinity hnRNP A1-binding sites and by intron elements that repress splice site utilization. EMBO J. 18:1939-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boggs, R. T., P. Gregor, S. Idriss, J. M. Belote, and M. McKeown. 1987. Regulation of sexual differentiation in D. melanogaster via alternative splicing of RNA from the transformer gene. Cell 50:739-747. [DOI] [PubMed] [Google Scholar]
- 12.Bopp, D., L. R. Bell, T. W. Cline, and P. Schedl. 1991. Developmental distribution of female-specific Sex-lethal proteins in Drosophila melanogaster. Genes Dev. 5:403-425. [DOI] [PubMed] [Google Scholar]
- 13.Bopp, D., J. I. Horabin, R. A. Lersh, T. W. Cline, and P. Schedl. 1993. Expression of Sex-lethal gene is controlled at multiple levels during the Drosophila oogenesis. Development 118:797-812. [DOI] [PubMed] [Google Scholar]
- 14.Bopp, D., G. Calhoun, J. I. Horabin, M. Samuels, and P. Schedl. 1996. Sex-specific control of Sex-lethal is a conserved mechanism for sex determination in the genus Drosophila. Development 122:971-982. [DOI] [PubMed] [Google Scholar]
- 15.Bopp, D., C. Schutt, J. Puro, H. Huang, and R. Nothiger. 1999. Recombination and disjunction in female germ cells of Drosophila depend on the germline activity of the gene Sex-lethal. Development 126:5785-5794. [DOI] [PubMed] [Google Scholar]
- 16.Brand, S., and H. M. Bourbon. 1993. The developmentally-regulated Drosophila gene rox8 encodes an RRM-type RNA binding protein structurally related to human TIA-1-type nucleolysins. Nucleic Acids Res. 21:3699-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burnette, J. M., A. R. Hatton, and A. J. Lopez. 1999. Trans-acting factors required for inclusion of regulated exons in the Ultrabithorax mRNAs of Drosophila melanogaster. Genetics 151:1517-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burtis, K. C., and B. S. Baker. 1989. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell 56:997-1010. [DOI] [PubMed] [Google Scholar]
- 19.Catteruccia, F., T. Nolan, T. G. Loukeris, C. Blass, C. Savakis, F. C. Kafatos, and A. Crisanti. 2000. Stable germline transformation of the malaria mosquito Anopheles stephensi. Nature 405:959-962. [DOI] [PubMed] [Google Scholar]
- 20.Chase, B. A., and B. S. Baker. 1995. A genetic analysis of intersex, a gene regulating sexual differentiation in Drosophila melanogaster females. Genetics 139:1649-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chu, E., and C. J. Allegra. 1996. The role of thymidylate synthase as an RNA binding protein. Bioessays 18:191-198. [DOI] [PubMed] [Google Scholar]
- 22.Cline, T. W. 1978. Two closely-linked mutations in Drosophila melanogaster that are lethal to opposite sexes and interact with daughterless. Genetics 90:683-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cline, T. W. 1984. Autoregulatory functioning of a Drosophila gene product that establishes and maintains the sexually determined state. Genetics 107:231-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cline, T. W. 1986. A female specific lethal lesion in an X-linked positive regulator of the Drosophila sex determination gene Sex-lethal. Genetics 113:642-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cline, T. W. 1993. The Drosophila sex determination signal: how do flies count to two? Trends Genet. 9:385-390. [DOI] [PubMed] [Google Scholar]
- 26.Cline, T. W., D. Z. Rudner, D. A. Barbash, M. Bell, and R. Vutein. 1996. Functioning of the Drosophila integral U1/U2 protein Snf independent of U1 and U2 small nuclear ribonucleoprotein particles is revealed by snf+ gene dose effects. Proc. Natl. Acad. Sci. USA 96:14451-14458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cline, T. W., and B. J. Meyer. 1996. Vive la difference: males vs females in flies vs worms. Annu. Rev. Genet. 30:637-702. [DOI] [PubMed] [Google Scholar]
- 28.Cronmiller, C., and T. Cline. 1987. The Drosophila sex determination gene daughterless has different functions in the germ line versus the soma. Cell 48:479-487. [DOI] [PubMed] [Google Scholar]
- 29.Dabeva, M. D., and J. R. Warner. 1993. Ribosomal protein L32 of Saccharomyces cerevisiae regulates both splicing and translation of its own transcript. J. Biol. Chem. 268:19669-19674. [PubMed] [Google Scholar]
- 30.Deshpande, G., G. Calhoun, and P. D. Schedl. 1999. The N-terminal domain of Sxl protein disrupts Sxl autoregulation in females and promotes female-specific splicing of tra in males. Development 126:2842-2853. [DOI] [PubMed] [Google Scholar]
- 31.Finley, K. D., B. J. Taylor, M. Milstein, and M. McKeown. 1997. dissatisfaction, a gene involved in sex-specific behavior and neural development of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 94:913-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finley, K. D., P. T. Edeen, M. Foss, E. Gross, N. Ghbeish, R. H. Palmer, B. J. Taylor, and M. McKeown. 1998. dissatisfaction encodes a tailless-like nuclear receptor expressed in a subset of CNS neurons controlling Drosophila sexual behavior. Neuron 21:1363-1374. [DOI] [PubMed] [Google Scholar]
- 33.Flickinger, T. W., and H. K. Salz. 1994. The Drosophila sex determination gene snf encodes a nuclear protein with sequences and functional similarity to the mammalian U1A snRNP protein. Genes Dev. 8:914-925. [DOI] [PubMed] [Google Scholar]
- 34.Forch, P., O. Puig, N. Kedersha, C. Martinez, S. Granneman, B. Seraphin, P. Anderson, and J. Valcárcel. 2000. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell 6:1089-1098. [DOI] [PubMed] [Google Scholar]
- 35.Forch, P., L. Merendino, C. Martinez, and J. Valcárcel. 2001. Modulation of msl-2 5′ splice site recognition by Sex-lethal. RNA 7:1185-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gebauer, F., L. Merendino, M. W. Hentze, and J. Valcárcel. 1998. The Drosophila splicing regulator Sex-lethal directly inhibits translation of male-specific-lethal 2 mRNA. RNA 4:142-150. [PMC free article] [PubMed] [Google Scholar]
- 37.Gebauer, F., D. F. Corona, T. Preiss, P. B. Becker, and M. W. Hentze. 1999. Translational control of dosage compensation in Drosophila by Sex-lethal: cooperative silencing via the 5′ and 3′ UTRs of msl-2 mRNA is independent of the poly(A) tail. EMBO J. 18:6146-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gergen, J. P. 1987. Dosage compensation in Drosophila: evidence that Daughterless and Sex-lethal control X chromosome activity at the blastoderm stage of embryogenesis. Genetics 117:477-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gorlich, D., M. Dabrowski, F. R. Bischoff, J. Kutay, P. Bork, E. Hartmann, S. Prehn, and E. Izaurralde. 1997. A novel class of RanGTP binding proteins. J. Cell Biol. 138:65-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Graham, P., J. K. Penn, and P. Schedl. 2003. Masters change, slaves remain. Bioessays 25:1-4. [DOI] [PubMed] [Google Scholar]
- 41.Granadino, B., S. Campuzano, and L. Sánchez. 1990. The Drosophila melanogaster fl(2)d gene is needed for the female-specific splicing of Sex-lethal RNA. EMBO J. 9:2597-2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Granadino, B., A. B. San Juán, P. Santamaría, and L. Sánchez. 1992. Evidence of a dual function in fl(2)d a gene needed for Sex-lethal expression in Drosophila melanogaster. Genetics 130:597-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Granadino, B., P. Santamaria, and L. Sánchez. 1993. Sex determination in the germ line of Drosophila melanogaster: activation of the gene Sex-lethal. Development 118:813-816. [DOI] [PubMed] [Google Scholar]
- 44.Granadino, B., L. O. F. Penalva, and L. Sánchez. 1996. The gene fl(2)d is needed for the sex-specific splicing of transformer pre-mRNA but not for double-sex pre-mRNA in Drosophila melanogaster. Mol. Gen. Genet. 253:26-31. [DOI] [PubMed] [Google Scholar]
- 45.Granadino, B., L. O. F. Penalva, M. R. Green, J. Valcárcel, and L. Sánchez. 1997. Distinct mechanisms of splicing regulation in vivo by the Drosophila protein Sex-lethal. Proc. Natl. Acad. Sci. USA 94:7343-7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hager, J. H., and T. W. Cline. 1997. Induction of female Sex-lethal RNA splicing in male germ cells: implications for Drosophila germline sex determination. Development 124:5033-5048. [DOI] [PubMed] [Google Scholar]
- 47.Harper, D. S., L. D. Fresco, and J. D. Keene. 1992. RNA binding specificity of a Drosophila snRNP protein that shares sequence homology with mammalian U1-A and U2-B” proteins. Nucleic Acids Res. 20:3645-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hastie, N. D. 2001. Life, sex, and WT1 isoforms—three amino acids can make all the difference. Cell 106:391-394. [DOI] [PubMed] [Google Scholar]
- 49.Heinrichs, V., L. C. Ryner, and B. S. Baker. 1998. Regulation of sex-specific selection of fruitless 5′ splice sites by Transformer and Transformer-2. Mol. Cell. Biol. 18:450-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heintzen, C., M. Nater, K. Apel, and D. Staiger. 1997. AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94:8515-8520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hilfiker, A., and R. Nöthiger. 1991. The temperature-sensitive mutation vir (virilizer) identifies a new gene involved in sex determination of Drosophila. Roux's Arch. Dev. Biol. 200:240-248. [DOI] [PubMed] [Google Scholar]
- 52.Hilfiker, A., H. Amrein, A. Dübendorfer, and R. Nöthiger. 1995. The gene virilizer is required for female-specific splicing controlled by Sxl, the master gene for sexual development in Drosophila. Development 121:4017-4026. [DOI] [PubMed] [Google Scholar]
- 53.Horabin, J. I., and P. Schedl. 1993. Sex-lethal autoregulation requires multiple cis-acting elements upstream and downstream of the male exon and appears to depend largely on controlling the use of the male exon 5′ splice site. Mol. Cell. Biol. 13:7734-7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Horabin, J., D. Bopp, J. Waterbury, and P. Schedl. 1995. Selection and maintenance of sexual identity in the Drosophila germline. Genetics 142:1521-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Horabin, J. I., and P. Schedl. 1996. Splicing of the Drosophila Sex-lethal early transcripts involves exon skipping that is independent of Sex-lethal protein. RNA 2:1-10. [PMC free article] [PubMed] [Google Scholar]
- 56.Hoshijima, K., K. Inoue, I. Higuchi, H. Sakamoto, and Y. Shimura. 1991. Control of doublesex alternative splicing by transformer and transformer-2 in Drosophila. Science 252:833-836. [DOI] [PubMed] [Google Scholar]
- 57.Inoue, K., K. Hoshijima, H. Sakamoto, and Y. Shimura. 1990. Binding of the Drosophila Sex-lethal gene product to the alternative splice site of transformer primary transcript. Nature 344:461-463. [DOI] [PubMed] [Google Scholar]
- 58.Inoue, K., K. Hoshijima, I. Higuchi, H. Sakamoto, and Y. Shimura. 1992. Binding of the Drosophila transformer and transformer-2 proteins to the regulatory elements of doublesex primary transcript for sex-specific RNA processing. Proc. Natl. Acad. Sci. USA 89:8092-8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Irvine, D., C. Tuerk, and L. Gold. 1991. SELEXION. Systematic evolution of ligands by exponential enrichment with integrated optimization by non-linear analysis. J. Mol. Biol. 222:739-761. [DOI] [PubMed] [Google Scholar]
- 60.Ito, H., K. Fujitani, K. Usui, K. Shimizu-Nishikawa, S. Tanaka, and D. Yamamoto. 1996. Sexual orientation in Drosophila is altered by the satori mutation in the sex-determination gene fruitless that encodes a zinc finger protein with a BTB domain. Proc. Natl. Acad. Sci. USA 93:9687-9692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kanaar, R., A. L. Lee, D. Z. Rudner, D. E. Wemmer, and D. C. Rio. 1995. Interaction of the Sex-lethal RNA binding domains with RNA. EMBO J. 14:4530-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kenan, D. J., C. C. Query, and J. D. Keene. 1991. RNA recognition: towards identifying determinants of specificity. Trends Biochem. Sci. 16:214-220. [DOI] [PubMed] [Google Scholar]
- 63.Kelley, R. L., I. Solovyeva, L. M. Lyman, R. Richman, V. Solovyev, and M. I. Kuroda. 1995. Expression of msl-2 causes assembly of dosage compensation regulators on the X chromosomes and female lethality in Drosophila. Cell 81:867-877. [DOI] [PubMed] [Google Scholar]
- 64.Kelley, R. L., J. Wang, L. Bell, and M. I. Kuroda. 1997. Sex lethal controls dosage compensation in Drosophila by a non-splicing mechanism. Nature 387:195-199. [DOI] [PubMed] [Google Scholar]
- 65.Kennedy, D., T. Ramsdale, J. Mattick, and M. Little. 1996. An RNA recognition motif in Wilms' tumour protein (WT1) revealed by structural modelling. Nat. Genet. 12:329-331. [DOI] [PubMed] [Google Scholar]
- 66.Kennerdel, J. R., and R. W. Carthew. 1998. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 95:1017-1026. [DOI] [PubMed] [Google Scholar]
- 67.Keyes, L. N., T. W. Cline, and P. Schedl. 1992. The primary sex determination signal of Drosophila acts at the level of transcription. Cell 68:933-943. [DOI] [PubMed] [Google Scholar]
- 68.Lallena, M. J., K. J. Chalmers, S. Llamazares, A. I. Lamond, and J. Valcárcel. 2002. Splicing regulation at the second catalytic step by Sex-lethal involves 3′ splice site recognition by SPF45. Cell 3:285-296. [DOI] [PubMed] [Google Scholar]
- 69.Lee, S. B., and D. A. Haber. 2001. Wilms tumor and the WT1 gene. Exp. Cell Res. 264:74-99. [DOI] [PubMed] [Google Scholar]
- 70.Le Hir, H., A. Nott, and M. J. Moore. 2003. How introns influence and enhance eukaryotic gene expression. Trends Biochem. Sci. 28:215-220. [DOI] [PubMed] [Google Scholar]
- 71.Lisbin, M. J., J. Qiu, and K. White. 2001. The neuron-specific RNA-binding protein ELAV regulates neuroglian alternative splicing in neurons and binds directly to its pre-mRNA. Genes Dev. 15:2546-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Little, N. A., N. D. Hastie, and R. C. Davies. 2000. Identification of WTAP, a novel Wilms' tumour 1-associating protein. Hum. Mol. Genet. 9:2231-2239. [DOI] [PubMed] [Google Scholar]
- 73.Lu, S., and B. R. Cullen. 2003. Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA 9:618-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lucchesi, J. C., and T. Skripsky. 1981. The link between dosage compensation and sex differentiation in Drosophila melanogaster. Chromosoma 82:217-227. [DOI] [PubMed] [Google Scholar]
- 75.Meise, M., D. HilfikerKleiner, A. Dubendorfer, C. Brunner, R. Nothiger, and D. Bopp. 1998. Sex-lethal, the master sex-determining gene in Drosophila, is not sex-specifically regulated in Musca domestica. Development 125:1487-1494. [DOI] [PubMed] [Google Scholar]
- 76.Merendino, L., S. Guth, D. Bilbao, C. Martinez, and J. Valcarcel. 1999. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402:838-842. [DOI] [PubMed] [Google Scholar]
- 77.Müller-Holtkamp, F. 1995. The Sex-lethal gene in Chrysomya rufifacies is highly conserved in sequence and exon-intron organization. J. Mol. Evol. 42:467-477. [DOI] [PubMed] [Google Scholar]
- 78.Nagengast, A. A., S. M. Stitzinger, C. H. Tseng, S. M. Mount, and H. K. Salz. 2003. Sex-lethal splicing autoregulation in vivo: interactions between SEX-LETHAL, the U1 snRNP and U2AF underlie male exon skipping. Development 130:463-471. [DOI] [PubMed] [Google Scholar]
- 79.Niessen, M., R. Schneiter, and R. Nothiger. 2001. Molecular identification of virilizer, a gene required for the expression of the sex-determining gene Sex-lethal in Drosophila melanogaster. Genetics 157:679-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nöthiger, R., M. Jonglez, M. Leuthold, P. Meier-Gersch-Willer, and T. Weber. 1989. Sex determination in the germ line of Drosophila depends on genetic signals and inductive somatic factors. Development 107:505-518. [DOI] [PubMed] [Google Scholar]
- 81.Oliver, B., N. Perrimon, and A. P. Mahowald. 1988. Genetic evidence that the sans-fille locus is involved in Drosophila sex determination. Genetics 120:159-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Oliver, B., Y. J. Kim, and B. S. Baker. 1993. Sex-lethal, master and slave: A hierarchy of germ line sex determination in Drosophila. Development 119:897-908. [DOI] [PubMed] [Google Scholar]
- 83.Oliver, B. 2002. Genetic control of germline sexual dimorphism in Drosophila. Int. Rev. Cytol. 219:1-60. [DOI] [PubMed] [Google Scholar]
- 84.O'Neil, M. T., and J. M. Belote. 1992. Interspecific comparison of the transformer gene of Drosophila reveals an unusually high degree of evolutionary divergence. Genetics 131:113-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ortega, A., M. Niksic, A. Bachi, M. Wilm, L. Sanchez, N. Hastie, and J. Valcarcel. 2003. Biochemical function of female-lethal (2)D/Wilms' tumor suppressor-1-associated proteins in alternative pre-mRNA splicing. J. Biol. Chem. 278:3040-3047. [DOI] [PubMed] [Google Scholar]
- 86.Pannuti, A., and J. C. Lucchesi. 2000. Recycling to remodel: evolution of dosage-compensation complexes. Curr. Opin. Genet. Dev. 10:644-650. [DOI] [PubMed] [Google Scholar]
- 87.Pauli, D., B. Oliver, and A. Mahowald. 1993. The role of the ovarian tumor locus in Drosophila melanogaster germ line sex determination. Development 119:123-134. [DOI] [PubMed] [Google Scholar]
- 88.Penalva, L. O. F., H. Sakamoto, A. NavarroSabate, E. Sakashita, B. A. Granadino, C. Segarra, and L. Sánchez. 1996. Regulation of the gene Sex-lethal: A comparative analysis of Drosophila melanogaster and Drosophila subobscura. Genetics 144:1653-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Penalva, L. O. F., M. F. Ruiz, A. Ortega, B. Granadino, L. Vicente, C. Segarra, J. Valcárcel, and L. Sanchez. 2000. The Drosophila fl(2)d gene, required for female-specific splicing of Sxl and tra pre-mRNAs, encodes a novel nuclear protein with a HQ-rich domain. Genetics 155:129-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Penalva, L. O. F., M. J. Lallena, and J. Valcárcel. 2001. Switch in 3′ splice site recognition between exon definition and splicing catalysis is important for Sex-lethal autoregulation. Mol. Cell. Biol. 21:1986-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pultz, M. A., G. S. Carson, and B. S. Baker. 1994. A genetic analysis of hermaphrodite, a pleiotropic sex determination gene in Drosophila melanogaster. Genetics 136:195-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pultz, M., and B. Baker. 1995. The dual role of hermaphrodite in the Drosophila sex determination regulatory hierarchy. Development 121:99-111. [DOI] [PubMed] [Google Scholar]
- 93.Ryner, L. C., S. F. Goodwin, D. H. Castrillon, A. Anand, A. Villella, B. S. Baker, J. C. Hall, B. J. Taylor, and S. A. Wasserman. 1996. Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell 87:1079-1089. [DOI] [PubMed] [Google Scholar]
- 94.Saccone, G., I. Peluso, D. Artiaco, E. Giordano, D. Bopp, and L. C. Polito. 1998. The Ceratitis capitata homologue of the Drosophila sex-determining gene Sex lethal is structurally conserved, but not sex-specifically regulated. Development 125:1495-1500. [DOI] [PubMed] [Google Scholar]
- 95.Saccone, G., A. Pane, and L. C. Polito. 2002. Sex determination in flies, fruitflies and butterflies. Genetica 116:15-23. [DOI] [PubMed] [Google Scholar]
- 96.Sakamoto, H., K. Inoue, I. Higuchi, Y. Ono, and Y. Shimura. 1992. Control of Drosophila Sex-lethal pre-mRNA splicing by its own female-specific product. Nucleic Acids Res. 20:5533-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sakashita, E., and H. Sakamoto. 1994. Characterization of RNA binding specificity of the Drosophila Sex-lethal protein by in vitro ligand selection. Nucleic Acids Res. 22:4082-4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sakashita, E., and H. Sakamoto. 1996. Protein-RNA and protein-protein interactions of the Drosophila Sex-lethal mediated by its RNA-binding domains. J. Biochem. 120:1028-1033. [DOI] [PubMed] [Google Scholar]
- 99.Salz, H. K., E. M. Maine, L. N. Keyes, M. E. Samuels, T. W. Cline, and P. Schedl. 1989. The Drosophila female-specific sex-determination gene, Sex-lethal, has stage-, tissue-, and sex-specific RNAs suggesting multiple modes of regulation. Genes Dev. 3:708-719. [DOI] [PubMed] [Google Scholar]
- 100.Salz, H. K. 1992. The genetic analysis of snf: a Drosophila sex determination gene requires for activation of Sex-lethal in both the germline and the soma. Genetics 130:547-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Salz, H. K., and T. W. Flickinger. 1996. Both loss-of-function and gain-of-function mutations in snf define a role for snRNP proteins in regulating Sex-lethal pre-mRNA splicing in Drosophila development. Genetics 144:95-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Samuels, M., G. Deshpande, and P. Schedl. 1998. Activities of the Sex-lethal protein in RNA binding and protein:protein interactions. Nucleic Acids Res. 26:2625-2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Samuels, M. E., P. Schedl, and T. W. Cline. 1991. The complex set of late transcripts from the Drosophila sex determination gene Sex-lethal encodes multiple related polypeptides. Mol. Cell. Biol. 11:3584-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Samuels, M. E., D. Bopp, R. A. Colvin, R. F. Roscigno, M. A. Garcia-Blanco, and P. Schedl. 1994. RNA binding by Sxl proteins in vitro and in vivo. Mol. Cell. Biol. 14:4975-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Samson, M. L. 1998. Evidence for 3′ untranslated region-dependent autoregulation of the Drosophila gene encoding the neuronal nuclear RNA-binding protein ELAV. Genetics 150:723-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sánchez, L., and R. Nöthiger. 1983. Sex determination and dosage compensation in Drosophila melanogaster. production of male clones in XX females. EMBO J. 2:485-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sánchez, L., B. Granadino, and M. Torres. 1994. Sex determination in Drosophila melanogaster: X-linked genes involved in the initial step of Sex-lethal activation. Dev. Genet. 15:251-264. [DOI] [PubMed] [Google Scholar]
- 108.Sánchez, L., P. López, and B. Granadino. 1998. Early events associated with sex determination in Drosophila melanogaster, p. 98-119. In R. Chatterjee and L. Sánchez (ed.), Genome analysis in eukaryotes: developmental and evolutionary aspects. Narosa Publishing House-Springer Verlag, New Delhi, India.
- 109.Schaeffer, C., B. Bardoni, J. L. Mandel, B. Ehresmann, C. Ehresmann, and H. Moine. 2001. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 20:4803-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schüpbach, T. 1985. Normal female germ cell differentiation requires the female X chromosome to autosome ratio and expression of Sex-lethal in Drosophila melanogaster. Genetics 109:529-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schütt, C., A. Hilfiker, and R. Nothiger. 1998. Virilizer regulates Sex-lethal in the germline of Drosophila melanogaster. Development 125:1501-1507. [DOI] [PubMed] [Google Scholar]
- 112.Schütt, C., and R. Nothiger. 2000. Structure, function and evolution of sex-determining systems in Dipteran insects. Development. 127:667-677. [DOI] [PubMed] [Google Scholar]
- 113.Sievert, V., S. Kuhn, and W. Traut. 1997. Expression of the sex Determination cascade genes Sex-lethal and doublesex in the phorid fly Megaselia scalaris. Genome 40:211-214. [DOI] [PubMed] [Google Scholar]
- 114.Sievert, V., S. Kuhn, A. Paululat, and W. Traut. 2000. Sequence conservation and expression of the Sex-lethal homologue in the fly Megaselia scalaris. Genome 43:382-390. [DOI] [PubMed] [Google Scholar]
- 115.Singh, R., J. Valcárcel, and M. R. Green. 1995. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 268:1173-1176. [DOI] [PubMed] [Google Scholar]
- 116.Sosnowski, B. A., J. M. Belote, and M. McKeown. 1989. Sex-specific alternative splicing of RNA from the transformer gene results from sequence-dependent splice site blockage. Cell 58:449-459. [DOI] [PubMed] [Google Scholar]
- 117.Starr, D. J., and T. W. Cline. 2002. A host parasite interaction rescues Drosophila oogenesis defects. Nature 428:76-79. [DOI] [PubMed] [Google Scholar]
- 118.Steinmann-Zwicky, M. 1988. Sex determination in Drosophila: the X chromosomal gene liz is required for Sxl activity. EMBO J. 7:3889-3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Steinmann-Zwicky, M., H. Schmid, and R. Nöthiger. 1989. Cell-autonomous and inductive signals can determine the sex of the germ line of Drosophila by regulating the gene Sxl. Cell 57:157-166. [DOI] [PubMed] [Google Scholar]
- 120.Steinmann-Zwicky, M. 1993. Sex determination in Drosophila: sis-b, a major numerator element of the X:A signal ratio in the soma, does not contribute to the X:A ratio in the germ line. Development 117:763-767. [DOI] [PubMed] [Google Scholar]
- 121.Stuckenholz, C., Y. Kageyama, and M. I. Kuroda. 1999. Guilt by association: non-coding RNAs, chromosome-specific proteins and dosage compensation in Drosophila. Trends Genet. 15:454-458. [DOI] [PubMed] [Google Scholar]
- 122.Tenenbaum, S. A., C. C. Carson, P. J. Lager, and J. D. Keene. 2000. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc. Natl. Acad. Sci. USA 97:14085-14090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tian, Q., M. Streuli, H. Saito, S. F. Schlossman, and P. Anderson. 1991. A polyadenylate binding protein localized to the granules of cytolytic lymphocytes induces DNA fragmentation in target cells. Cell 67:629-639. [DOI] [PubMed] [Google Scholar]
- 124.Torres, M., and L. Sánchez. 1991. The sisterless-b function of the Drosophila gene scute is restricted to the state when the X:A ratio signal determines the activity of Sex-lethal. Development 113:715-722. [DOI] [PubMed] [Google Scholar]
- 125.Tsai, D. E., D. S. Harper, and J. D. Keene. 1991. U1-snRNP-A protein selects a ten nucleotides consensus sequence from a degenerate RNA pool presented in various structural contexts. Nucleic Acids Res. 19:4931-4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Valcárcel, J., R. Singh, P. D. Zamore, and M. R. Green. 1993. The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature 362:171-175. [DOI] [PubMed] [Google Scholar]
- 127.Vied, C., N. Halachmi, A. Salzberg, and J. I. Horabin. 2003. Antizyme is a target of Sex-lethal in the Drosophila germline and appears to act downstream of Hedgehog to regulate Sex-lethal and Cyclin B. Dev. Biol. 253:214-229. [DOI] [PubMed] [Google Scholar]
- 128.Wang, J., and L. R. Bell. 1994. The Sex-lethal amino terminus mediates cooperative interactions in RNA binding and is essential for splicing regulation. Genes Dev. 8:2072-2085. [DOI] [PubMed] [Google Scholar]
- 129.Wang, J., Z. Dong, and L. R. Bell. 1997. Sex-lethal interactions with protein and RNA. Roles of glycine-rich and RNA binding domains J. Biol. Chem. 272:22227-22235. [DOI] [PubMed] [Google Scholar]
- 130.Waterbury, J. A., J. I. Horabin, D. Bopp, and P. Schedl. 2000. Sex determination in the Drosophila germline is dictated by the sexual identity of the surrounding soma. Genetics 155:1742-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wu, J., and J. Bag. 1998. Negative control of the poly(A)-binding protein mRNA translation is mediated by the adenine-rich region of its 5′-untranslated region. J. Biol. Chem. 273:34535-34542. [DOI] [PubMed] [Google Scholar]
- 132.Yang, D., H. Lu, Y. Hong, T. Jinks, P. Estes, and J. Erickson. 2001. Interpretation of X chromosome dose at Sex-lethal requires non-E-box sites for the basic helix-loop-helix proteins SisB and Daughterless. Mol. Cell. Biol. 21:1581-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yanowitz, J. L., G. Deshpande, G. Calhoun, and P. D. Schedl. 1999. An N-terminal truncation uncouples the sex-transforming and dosage compensation functions of Sex-lethal. Mol. Cell. Biol. 19:3018-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zarkower, D. 2001. Establishing sexual dimorphism: conservation amidst diversity? Nat. Rev. Genet. 2:175-185. [DOI] [PubMed] [Google Scholar]
- 135.Zhou, S., Y. Yang, M. J. Scott, A. Pannuti, K. C. Fehr, A. Eisen, E. V. Koonin, D. L. Fouts, R. Wrightsman, J. E. Manning, et al. 1995. Male-specific lethal 2, a dosage compensation gene of Drosophila, undergoes sex-specific regulation and encodes a protein with a RING finger and a metallothionein-like cysteine cluster. EMBO J. 14:2884-2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhu, C., J. Urano, and L. R. Bell. 1997. The Sex-lethal early splicing pattern uses a default mechanism dependent on the alternative 5′ splice sites. Mol. Cell. Biol. 17:1674-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]