Abstract
When it was first proposed that the budding yeast Saccharomyces cerevisiae might serve as a model for human aging in 1959, the suggestion was met with considerable skepticism. Although yeast had proved a valuable model for understanding basic cellular processes in humans, it was difficult to accept that such a simple unicellular organism could provide information about human aging, one of the most complex of biological phenomena. While it is true that causes of aging are likely to be multifarious, there is a growing realization that all eukaryotes possess surprisingly conserved longevity pathways that govern the pace of aging. This realization has come, in part, from studies of S. cerevisiae, which has emerged as a highly informative and respected model for the study of life span regulation. Genomic instability has been identified as a major cause of aging, and over a dozen longevity genes have now been identified that suppress it. Here we present the key discoveries in the yeast-aging field, regarding both the replicative and chronological measures of life span in this organism. We discuss the implications of these findings not only for mammalian longevity but also for other key aspects of cell biology, including cell survival, the relationship between chromatin structure and genome stability, and the effect of internal and external environments on cellular defense pathways. We focus on the regulation of replicative life span, since recent findings have shed considerable light on the mechanisms controlling this process. We also present the specific methods used to study aging and longevity regulation in S. cerevisiae.
INTRODUCTION
When it was first proposed that the budding yeast Saccharomyces cerevisiae might serve as a model for human aging in 1959, the suggestion was met with considerable skepticism, and for seemingly good reasons. Although yeast had proved a valuable model for understanding basic cellular processes in humans, it was difficult to accept that such a simple unicellular organism could provide information about human aging, which is one of the most complex of biological phenomena. While it is true that causes of aging are likely to be multifarious, there is a growing realization that all eukaryotes possess surprisingly conserved longevity pathways that govern the pace of aging. This realization has come, in part, from studies of S. cerevisiae, which has emerged as a highly informative and respected model for the study of life span regulation. Genomic instability has been identified as a major cause of aging, and over a dozen longevity genes have now been identified that suppress it. Translating these findings to higher organisms remains a major challenge over the next decade. Here we present the key discoveries in the yeast-aging field, regarding both the replicative and chronological measures of life span in this organism. We discuss the implications of these findings not only for mammalian longevity but also for other key aspects of cell biology, including cell survival, the relationship between chromatin structure and genome stability, and the effect of internal and external environments on cellular defense pathways. We focus on the regulation of replicative life span, since recent findings have shed considerable light on the mechanisms controlling this process. We also present the specific methods used to study aging and longevity regulation in S. cerevisiae.
Yeast as a Model for Aging Research
The usual reasons for using yeast as a model system—its ease of genetic manipulation, well annotated genome, and short generation time—can be given for using this organism as a model for aging. But these do not suffice. The reason is that aging is unlike other basic biological processes because it is probably nonadaptive (i.e., it confers no benefit to the species) (100). In fact, it is even misleading to call aging a process. There are no “death genes” directing aging as they do, say, growth or development. In fact, aging is at most a by-product of natural selection, arising from a lack of selection for longer-lived organisms. So, if a process needs to be adaptive to be well conserved between species, how can yeast aging research have any relevance to humans? Is there an aspect of aging that is adaptive?
In the early days of aging research, few researchers suspected the existence of single genes that control aging. This was based in part on the valid assumption that aging was an incredibly complex process that was impacted by hundreds, if not thousands, of genes. Then, genetic studies with model organisms in the 1990s began to uncover numerous single-gene mutations that extend life span (75, 92, 95). Today there are dozens of mutations known to extend life span in model organisms (109) (Table 1). This leads to the question: what had researchers overlooked? The major oversight appears to have been the failure to foresee that organisms have evolved genes, not to promote aging but to promote survival and longevity during times of adversity. Longevity regulation, as it has come to be known, is now generally accepted as a highly adaptive biological trait, especially in a changing environment (94, 100).
TABLE 1.
S. cerevisae longevity genes
| Longevity gene | Relevant function of gene product | Overexpression or mutation increases life spana | Increases rDNA silencing/stability? | Reference |
|---|---|---|---|---|
| Replicative life span | ||||
| CDC25 | Glucose response signaling pathway | M | Yes | 114 |
| CYR1 | Glucose response signaling pathway | M | ? | 114 |
| FOB1 | Replication fork block; contributes to rDNA instability | M | Yes | 37 |
| GPR1 | Glucose response signaling pathway | M | ? | 114 |
| GPA2 | Glucose response signaling pathway | M | ? | 114 |
| HAP4 | Heme-activated transcription factor; induces respiration | O | Yes | 115 |
| LAG1 | Synthesis of ceramides and inositol phosphorylceramide | M | ? | 35 |
| LAG2 | Transmembrane domain containing protein | O | ? | 29 |
| NPT1 | NAD biosynthesis | O | Yes | 2 |
| NMA1/2 | NAD biosynthesis | O | Yes | 2 |
| PNC1 | NAD biosynthesis | O | Yes | 3 |
| RAS1 | Glucose response signaling pathway | M | ? | 194 |
| RAS2 | Glucose response signaling pathway | O | ? | 194 |
| RPD3 | Histone deacetylase, affects rDNA silencing | M | Yes | 79 |
| RTG3 | Transcription factor that transduces mitochondrial signals | M | ? | 78 |
| SIP2 | Repressor of the Snf1 serine/threonine kinase | O | Yes | 117 |
| SNF4 | Activator of Snf1 | M | Yes | 117 |
| SIR2 | NAD+-dependent HDAC; stabilizes rDNA | O | Yes | 88 |
| SIR4 | Associates with Sir2; Sir4-42 extends life span | M | ? | 92 |
| TPK1 | Glucose response pathway | M | ? | 114 |
| UTH1 | Unknown function; responds to oxidative stress | M | Yes | 92 |
| UTH4 | Localization of Sir complex | M | Yes | 54 |
| ZDS1 | Deletion increases silencing at rDNA locus | M | Yes | 163 |
| Chronological life span | ||||
| CYR1 | Glucose response signaling pathway | M | ? | 45 |
| RAS2 | Glucose response signaling pathway | M | ? | 44 |
| SCH9 | Protein kinase Akt/PKB homolog | M | ? | 45 |
| SOD1/SOD2 | Cytosolic/mitochondrial superoxide dismutases | O | ? | 125 |
O, overexpression; M, mutation.
The concept of longevity regulation fits well with current evolutionary hypotheses such as the “disposable soma” theory of Kirkwood and Holliday (100) and the “hormesis hypothesis” of Masoro (132). These ideas are based on the concept that each organism has limited resources and that these resources can be allocated to only a finite number of cellular activities, the two primary ones being reproduction and somatic maintenance (reviewed in reference 101). In summary, the theory proposes that organisms do not live forever because they cannot afford to devote all their energy to somatic maintenance, due to the competing demands of growth and reproduction. The key to this new paradigm is that these same evolutionary forces have led to the evolution of species with the ability to alter the amount of energy they devote to somatic maintenance within an individual's lifetime.
A second reason to expect some conservation between yeast and human aging, put simply, is that they have the same fundamental biology. If aging really is due to “wear and tear,” and most evidence suggests that it is, then not all biological systems will experience wear and tear at the same rate. Some systems, such as basic metabolic pathways, are relatively robust because they are inherently self-regulating and often contain inbuilt redundancy. The systems that fail first are those whose maintenance is energy demanding and for which the damage is irreversible. A good example of such a system is the genome. The energy required to constantly survey and repair the genome is immense, and the wear and tear often irreversible. Consistent with this idea, as we describe below, a major cause of replicative aging in S. cerevisiae has been shown to arise from the cell's inability to fully maintain the integrity of its genome at its most highly repetitive locus, the ribosomal DNA (rDNA) (180). Although rDNA stability may not play a role in human aging, it is suspected that the cumulative loss of coding or structural DNA (e.g., the repeats found at telomeres) may contribute to aging in humans (33). The important point is that the mechanisms of aging may be related between distant species (but by no means as highly conserved as longevity regulatory pathways) because eukaryotes have the same basic biology and some systems are inherently difficult to maintain over a lifetime.
Definition of Yeast Aging
For studies of aging, be they in yeast or in humans, it is critical to make a clear distinction between events that cause aging and those that cause sickness or disease. The ability to distinguish between these two phenomena, which both reduce life span in model organisms, is critical to the study of aging. According to the Merck Manual of Geriatrics, aging is a decline that occurs to the majority of the population whereas a sickness or disease occurs to a minority (14). Applying this criterion to S. cerevisiae, for a mechanism or gene to be considered relevant to aging, it must occur in most wild-type strains. Otherwise, it is merely a strain-specific sickness.
It is thus of particular concern that some yeast strains used routinely in the field have average life spans that are half that of most wild-type strains (Table 2). Any one of a number of mutations that shorten the life span could have been introduced into laboratory strains over the preceding decades, and researchers would not have detected a difference unless they examined the life span of individual cells. Even very short-lived strains appear to grow at normal rates. One way to ensure that one is studying true aging is to test one's findings with multiple wild-type strains that are not relatively short-lived. This would be an arduous task that most laboratories have yet to embrace.
TABLE 2.
Replicative life spansa of wild-type haploid yeast strains
| Strain | Life span (no. of divisions)
|
Reference | |
|---|---|---|---|
| Avg | Maximum | ||
| Bky1-14c (uth4-I4c) | 16 | 25 | 93 |
| SP1 | 12 | 22 | 79 |
| DBY747 | 22 | 32 | 92 |
| PSY316 | 22 | 34 | 16 |
| W303-1A | 23 | 43 | 180 |
| X2180-1A | 24 | 39 | 40 |
| X30 | 24 | 42 | 145 |
| S 288C | 28 | 47 | 147 |
| PSY142 | 29 | 55 | 92 |
Life spans determined on complete 2% glucose media.
Replicative versus chronological yeast life span.
Many researchers have proposed that aging in dividing or “mitotic” human cells may be fundamentally different from aging in those that remain in a postmitotic state. Similarly, for S. cerevisiae a distinction is made between the aging of mitotic cells and those that are quiescent. Yeast “replicative life span” is defined as the number of divisions an individual yeast cell undergoes before dying. One attractive feature of S. cerevisiae, as opposed to many other simple eukaryotes, is that the progenitor cell is easily distinguished from its descendants because cell division is asymmetric: a newly formed “daughter” cell is almost always smaller than the “mother” cell that gave rise to it. Yeast mother cells divide about 20 times before dying and, as described below, undergo characteristic structural and metabolic changes as they age. Table 2 lists the average and maximum replicative life spans of the primary strains used in yeast aging research.
The alternative measure, “chronological life span,” also referred to as “postdiauxic survival,” is the length of time a population of yeast cells remains viable in a nondividing state following nutrient deprivation (124). Yeast cells grown in a nutrient-rich medium multiply until all readily utilizable nutrients are exhausted. At this point, cells cease dividing and enter a postdiauxic, hypometabolic state known as stationary phase, where they can remain viable for weeks. In synthetic medium, cells deplete the medium and cease dividing yet retain relatively high metabolism (124). Such cells have a short chronological life span relative to cells in rich medium and are thought to more closely resemble postmitotic cells in multicellular organisms (125).
REPLICATIVE AGING
Background
In 1950, Andrew Barton monitored the fate of individual yeast cells by micromanipulation and discovered that mother cells are mortal (11). For the next 40 years, most yeast aging research remained descriptive. It was noted that as yeast cells grow older they accumulate bud scars, divide more slowly, and, finally, become sterile (reviewed in reference 178). One key observation was that daughter cells arising from old mothers inherit characteristics of old age and have a shorter life span (85, 91). This effect was not the result of mutation because the premature-aging phenotype could be diluted through successive generations, eventually restoring a normal life span in descendants. Based on these observations, Egilmez and Jazwinski proposed that yeast aging might be due to the stochastic appearance of a senescence factor that then accumulated exponentially until it killed cells (42). Such a factor was proposed to diffuse from old mothers into daughters, thus explaining how old age could be inherited and then diluted through successive generations.
A second important observation came from a genetic screen for long-lived yeast mutants (92). Kennedy, Guarente and colleagues first identified starvation-resistant mutants, having noted that in many other organisms there is a strong correlation between stress resistance and longevity. Rescreening of the stress-resistant mutants for increased longevity led to the isolation of four so-called “youth mutants”, uth1 to uth4. Interestingly, three of these have been found to affect the same cellular process: the formation of silent heterochromatin.
In yeast, transcriptional silencing occurs at telomeres, the two mating-type loci (HML and HMR), and rDNA locus RDN1 (127, 142). The establishment of heterochromatin at telomeres and mating-type loci requires the yeast Sir2/3/4 protein complex. Sir2, but not Sir3 or Sir4, also mediates silencing at the rDNA (24, 188). In many strains, the overexpression of Sir2 increases the extent of silencing at both telomeres and rDNA, implying that Sir2 is a limiting component of the silencing apparatus (88, 190).
The most informative allele isolated in the screen for longevity mutants was UTH2-1 (later renamed SIR4-42), which extended replicative life span by 45% (92). This semi-dominant mutation truncated the Sir4 protein, causing the Sir complex to relocalize to the nucleolus (93), thus increasing rDNA silencing (D. A. Sinclair, unpublished results). The other UTH genes were also interesting. The UTH1 gene encodes a SUN domain protein whose expression is greatly induced by oxidative stress and whose deletion results in a global increase in silencing and life span extension (L. Guarente, unpublished results). UTH4, which encodes a Drosophila Pumilio homolog (39), influences the distribution of the Sir complex within the nucleus (54, 93). Deletion of UTH4 reduces life span and decreases rDNA silencing, whereas overexpression of UTH4 has the opposite effect (93). Although the biochemical function of Uth1 and Uth4 remain to be determined, a clear trend emerged from these studies: increased heterochromatin at the yeast rDNA locus correlates with increased longevity.
Characteristics of Replicatively Aged Cells
Because many mutations can shorten life span, it is necessary to have a set of characteristics that distinguish accelerated aging from cell sickness. Recognizing aging in humans is so natural for us that we take it for granted. Even without molecular analyses, human “accelerated-aging” or progeroid diseases such as Werner's syndrome and Rothmund-Thompson syndrome are clearly not perfect mimics of normal aging (107). However, recognizing genuine aging in yeast is a much harder task. Although yeast cells go through a number of morphological and biochemical changes, as described below, none of these phenotypes is specific for aging. The sterility of old cells, for example, has been proposed as a reliable aging marker (187). As yeast cells age, the Sir2/3/4 protein complex that silences the mating-type genes at HM loci relocalizes to the nucleolus, resulting in a sterile state of pseudo-diploidy. Unfortunately, this phenotype does not appear to be specific to old yeast. In 1999, three separate papers showed that the Sir complex is relocalized from silent loci to DNA breaks, also resulting in sterility (108, 134, 140). Clearly, any mutation that results in frequent DNA breaks will cause sterility in a haploid yeast cell. One of the most promising approaches to identifying aged yeast cells involves microarray analysis. Transcriptome profiles of old yeast are now publicly available and should greatly facilitate our ability to distinguish between premature aging and mere sickness (116).
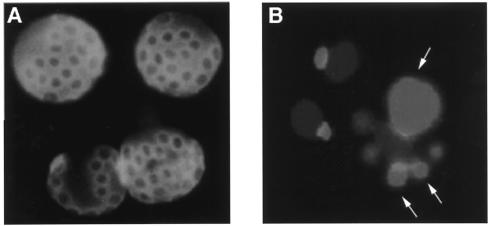
Bud scar accumulation.
When a daughter separates from a mother cell, the boundary between the two cells constricts, leaving behind on the mother cell's surface a circular chitin-containing remnant termed the bud scar (Fig. 1A). Bud scars remain permanently deposited on the surface of the mother cell. As a mother cell goes through successive rounds of cell division, bud scars accumulate on the cell surface, serving as a convenient marker for the number of divisions realized by a single cell. As a cell continues dividing, previous sites of budding are seldom reused. Although it has been hypothesized that the accumulation of bud scars may impose a theoretical upper limit on a cell's replicative potential (25), evidence indicates that it does not result in senescence of wild-type strains. First, most mutations that extend life span appear to have no relation to cell wall biosynthesis. Second, a bud scar typically occupies approximately 1% of the available cell surface, giving a theoretical upper limit of at least 100 divisions, but most laboratory strains have an average life span of 20 to 30 divisions (178) (Table 2). Third, increasing the cell size, and thus increasing the number of available budding sites, does not extend life span (147). Fourth, it is possible to artificially elevate the deposition of chitin by briefly incubating cells containing a temperature-sensitive allele of CDC24 at the nonpermissive temperature (185, 186). At this temperature, cdc24-ts cells arrest in an unbudded state and randomly accumulate cell wall chitin. Elevated chitin deposition does not adversely affect the life span (42), further demonstrating that bud scar accumulation is probably not a cause of yeast aging.
FIG. 1.
Yeast cell division and bud scar formation. (A) The budding of each daughter cell leaves a ring-shaped deposit, termed the bud scar, on the cell wall of the mother cell. These chitin-containing rings, formed at the neck of buds, can be stained with calcofluor, a fluorescent dye. The exact number of times an individual mother cell has undergone division can thus be determined by counting the number of bud scars present. (B) An aged yeast nucleus has an enlarged and fragmented nucleolus (arrows), unlike the nucleoli of two young cells in the upper left corner.
Increased cell size.
After three or four divisions, mother cells are easily distinguished from daughter cells due to their increased size. It has been proposed that one cause of aging might be a critical upper limit on size (145). Theoretically, increased cell volume could impose a replicative limit if the rate of nutrient diffusion to disparate parts of a large cell becomes limiting. However, the fact that life span can be extended over twofold by manipulating a variety of genes that are not apparently associated with cell size appears to counter this possibility. This model has also been tested directly (91). Cells were arrested in their cell cycle for various lengths of time during the G1 phase. Cells arrested at this phase remain metabolically active and continue to grow in size. After release from G1 arrest, cells that were greatly increased in size had an identical life span potential to those of untreated cells (91).
Loss of asymmetry.
As yeast cells age, they grow dramatically larger but continue to give rise to small daughter cells throughout most of their life span. However, very old mother cells tend to produce large, short-lived daughter cells (76, 85, 91). At or near the final division, daughter cells often do not separate from the mother until both cells are similar in size (76, 85). The loss of the asymmetry of age in old cells is intriguing because it provides a strong argument for the ERC model of senescence (discussed below). When old cells become packed with extrachromosomal rDNA circles (ERCs), these molecules “leak” into daughter cells, presumably causing the premature aging seen in these cells (180). The ERC model would also explain why old age is diluted by successive generations, so that the great granddaughter of an old mother, for example, has a normal life span.
Loss of fertility.
S. cerevisiae can exist in either a haploid or diploid state. When two fertile haploid cells of opposite mating type encounter one another, they mate to form a diploid zygote. To switch between the two mating types (MATa and MATα), a cell transposes the opposite silent information to the mating-type locus, where it is expressed. Young yeast cells are normally fertile since the two repositories of mating-type information, HMR and HML, remain in a transcriptionally silent state (127). As discussed above, the silent state at HM loci requires the Sir2-Sir3-Sir4 silencing complex. When yeast cells grow old, they become sterile (147), and this phenotype remains one of the most reliable external markers of yeast aging.
Sterility in old cells is caused by a loss of transcriptional silencing at the cryptic mating-type loci, HMRa and HMLα, resulting in simultaneous expression of both a and α information (187). To prove this, HMRa was deleted from a MATα strain. The resulting strain exhibited the same characteristic life span as the wild type but no longer became sterile. In addition to a loss of mating-type silencing, old cells lose silencing near telomeres (97) and the Sir complex relocalizes to the nucleolus in old cells (93).
Nucleolar fragmentation.
The nucleolus is a nuclear structure containing the rDNA genes and other components required for ribosome assembly (31, 137). Yeast rDNA, located on chromosome XII, contains 100 to 200 tandemly repeated copies of a 9.1-kb unit. In young yeast cells, the nucleolus forms a crescent-shaped structure retained near the nuclear periphery (168). In old cells, however, the nucleolus becomes enlarged and fragmented into multiple, rounded structures (181). (Fig. 1B). Fragmentation of the nucleolus and relocalization of Sir3 may be a response to the accumulation and aggregation of extrachromosomal rDNA circles (180). It is important to note though, that chromosomal rDNA remains intact in these fragmented nucleoli.
Metabolic changes.
The only study to specifically address the question of metabolism in aging yeast cells involved a combination of microanalytical biochemical assays and microarray analyses of mRNA levels (116). The major finding was that yeast cells, as they age, undergo a progressive shift away from glycolysis toward gluconeogenesis and energy storage. This shift is associated with the induction of genes involved in glycogen production, fatty acid degradation, gluconeogenesis, and the glyoxylate cycle. Biochemical profiling of enzymes and metabolic intermediates confirmed the shift to gluconeogenesis. Much of the up-regulation of genes involved in gluconeogenesis appears to be due to the translocation of the Mig1 transcriptional repressor from the nucleus to the cytoplasm as cells age (6) (see below). ATP levels do not decline with age, but there is a ∼30% drop in the NAD+ level between divisions 0 to 1 and 7 to 8. Interestingly, many of these changes are recapitulated in short-lived sip2 cells, which lack the putative repressor of Snf1, a kinase that regulates cellular responses to glucose deprivation. The role of the Snf1/Sip2 pathway in longevity is described in further detail below.
Mechanisms of Yeast Replicative Aging
Background
Over the past 40 years, numerous models have been proposed to explain the mortality of yeast cells. In the 1960s, researchers noted that yeast mother cells accumulate numerous bud scars, one for each daughter, and grow larger as they age. The first models for yeast aging suggested that there might be limits to the number of bud scars that can be accommodated by a cell or there might be an upper size limit. As described above, neither of these models withstood close scrutiny. Artificially increasing the size of yeast mothers by transiently blocking the cell cycle did not reduce life span, and the increased cell surface of dividing mother cells easily accomodates the new bud scars. In recent years, the demonstration that replicative life span can be significantly extended by specific mutations, with a concomitant increase in final cell size and bud scar number, showed that these attributes do not limit the life span of yeast cells.
Mortimer and Johnston first noted that daughter cells that budded from old mothers tend to have premature-aging characteristics, including a large size, a slow cell cycle, and a short life span (145). Based on this observation, Egilmez and Jazwinski proposed that yeast aging is caused by a (potentially cytoplasmic) senescence factor that accumulates exponentially as cells divide, reaching a level that eventually kills cells (42). This model is appealing for two reasons. First, it explains how daughters inherit old age from their mothers. When mothers become sufficiently old due to the accumulation of the senescence factor, some of this factor might leak into daughter cells, making them prematurely old themselves. Second, the senescence-factor model explains the dynamics of yeast mortality. All species that experience aging, from yeast to humans, have characteristic rates of mortality that can be described mathematically by the Gompertz-Makeham equation (53). Mortality curves derived by plotting percent survival against age have a characteristic “shoulder,” where most individuals remain viable early in life but then viability rapidly declines as the mortality rate increases exponentially. There is also a characteristic “tail,” where some lucky individuals seem to experience delayed mortality and remain viable longer than would be expected by extrapolating the curve. These kinetics imply that aging is the sum of two components (182): (i) a stochastic trigger that initiates a process and (ii) an aging process whose negative impact on life span increases exponentially with age. Egilmez and Jazwinski proposed that the triggering event might be the generation of a senescence factor and that aging would result from the exponential accumulation of this factor with each cell division (42). This idea is consistent with the ERC mechanism of yeast aging described below.
Over the past 5 years, researchers have come to the consensus that a primary cause of yeast replicative aging stems from changes within the nucleolus, the distinct nuclear region responsible for rRNA transcription and ribosome assembly. The first clue that nucleolar events were involved in aging came from the aforementioned genetic screen for mutations that increased yeast life span and the subsequent isolation of SIR4 (92). SIR4 encodes a component of the Sir2-Sir3-Sir4 silencing complex that catalyzes the formation of heterochromatin at the silent mating-type (HM) loci and telomeres. The mutation in SIR4 resulted in a C-terminal truncation of the protein, causing it to relocalize from telomeres and HM loci to the nucleolus (93). Although the mechanism by which the Sir4-42 protein increases life span was not immediately clear, the finding focused researchers attention on the nucleolus and the rDNA, eventually leading to the identification of a molecular cause of yeast aging.
rDNA stability.
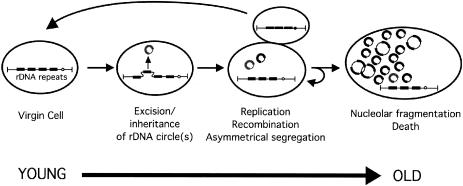
In 1997, Sinclair and Guarente proposed that yeast replicative aging stems from genomic instability at the rDNA locus, RDN1 (180). The yeast rDNA locus is inherently recombinagenic due to its repetitive nature and unidirectional mode of DNA replication. The locus, which comprises 10% of the total yeast genome, consists of 100 to 200 tandemly arrayed 9-kb repeats that encode the rRNAs. Each repeat contains a potential origin of DNA replication, with three ARS consensus sites within each origin. Roughly one of three origins within these repeats fires during each S phase (139). Homologous recombination between adjacent repeats is known to result in the excision of extrachromosomal circular forms of rDNA known as ERCs (Fig. 2). The important aspect of the aging mechanism is that ERCs replicate during S phase but are inefficiently segregated to daughter cells. Because ERCs can double in copy number every S phase, their abundance increases exponentially in mother cells at a rate timed by cell division (180).
FIG. 2.
Instability of repeated DNA as a cause of replicative aging in S. cerevisiae. The yeast rDNA locus is the most highly repetitive locus in the organism, consisting of ∼150 tandem 9.1-kb repeats. These repeats are stabilized, in part, by the NAD+-dependent HDAC Sir2. The initiating event is the generation of an ERC by homologous recombination between repeats within the rDNA array on chromosome XII. ERCs have a high probability of replicating and are segregated almost exclusively to the mother cell. They accumulate exponentially in mother cells, resulting in fragmented nucleoli, cessation of cell division, and cellular senescence. Stabilization of the rDNA locus or inhibition of ERC replication extends the life span. Daughters from very old mothers inherit ERCs due to the breakdown in the asymmetry of inheritance, explaining why daughters of old mothers are prematurely old. Ectopic release of an ERC in a young cell accelerates aging. Overexpression of SIR2 or deletion of FOB1 (encoding a replication fork block protein specific to the rDNA) reduces rDNA recombination and ERC formation, and the life span is extended by 30 to 50%. Reprinted from reference 180 with permission.
Experiments have shown that most virgin cells usually bud off mothers without inheriting an ERC, except when the mother cell is very old (180). This explains why the inheritance of aging is asymmetric and why the daughters of old mothers are prematurely aged. Southern blotting experiments have indicated that ERCs accumulate to more than 1,000 copies per old cell, which totals more DNA than the rest of the yeast genome. All of the ERCs in old cells are probably derived from a single initial recombination event (Fig. 2). The mechanism by which ERCs cause death is not known, but given their abundance, it is likely that they titrate away vital transcription and/or replication factors (182).
There is now convincing evidence that in a typical wild-type strain, ERCs are a major cause of aging. First, the ectopic release of an ERC into a virgin cell shortens the life span and causes premature aging, demonstrating that ERCs are sufficient to cause aging (180). Second, genetic manipulations that decrease the formation of ERCs greatly extend life span (2, 6, 36, 114, 117, 152, 180) whereas those that accelerate ERC formation have an opposite effect (88, 152).
Genetic manipulations that extend yeast life span by suppressing ERCs seem to fall into one of two categories. The most common suppress ERC formation by increasing the extent of heterochromatin at the rDNA. Examples of this class include SIR2 and PNC1 overexpression. These are discussed in detail below in the context of environmentally regulated chromatin. The other class includes those that directly suppress homologous recombination between rDNA repeats. A good example of a longevity gene in this class is FOB1 (35). FOB1 encodes a nucleolar protein that is required for a DNA replication fork block immediately downstream of the rDNA origins (86, 102). The fork block is thought to prevent transcription complexes from running head on into moving replication forks and to allow the rDNA locus to expand and contract via recombination. The Fob1-mediated fork block is thought to be responsible for the general instability of the rDNA locus, since strains lacking FOB1 have a 100-fold lower rate of rDNA recombination (118) and have manyfold fewer ERCs than aged-matched wild-type cells (36). As predicted by the ERC model, fob1Δ mutants live almost twice as long as wild-type controls. Weak mutations in FOB1 also extend the life span but not to the extent of the deletion. Furthermore, ERC levels in these strains correlate with the respective life span extensions. The argument that this life span extension is due to effects on rDNA silencing does not appear to be founded because rDNA silencing in fob1 cells is the same as or less than that in wild-type cells (M. Kaeberlein, personal communication).
Role of heterochromatin.
A major mechanism by which yeast cells suppress ERC formation is the packaging of DNA and histones into “silent” heterochromatin. Heterochromatin in yeast occurs at telomeres, HM loci, and the rDNA (142). The formation of heterochromatin at HM loci and telomeres is mediated by the silent information regulatory complex Sir2/3/4 (66, 192). Alternatively, heterochromatin at the rDNA locus is catalyzed by the RENT (for “regulator of nucleolar silencing and telophase exit”) complex, which includes Sir2, Net1, Cdc14, and Nan1 (51, 176). Of these proteins, Sir2 is the only factor that is indispensable for silencing at all three silent regions (24, 55, 188). To be precise, “rDNA silencing” is actually a misnomer. Although polymerase II-transcribed marker genes integrated at the rDNA are transcriptionally silenced, transcription of the native rDNA is seemingly unaffected by deletion of SIR2 (166). This suggests that the primary function of heterochromatin at the rDNA locus may be to suppress recombination rather than silence transcription. There have been several reports, though, which demonstrate “natural” silencing of polymerase II-transcribed genes within the rDNA (24, 209).
Other mechanisms of replicative aging.
It would be a mistake to assume that there is only one cause of aging for any species, and no doubt other causes of yeast aging will be found. As yet, apart from the ERC mechanism, there is no other known cause of replicative aging, although some strains have been found to age independently of ERC formation (96). One proposed cause of aging is dysregulation of rDNA transcription during the life span of a cell (74). This would also explain the strong correlation between increased rDNA silencing and life span. Another recent finding has uncovered a putative rDNA- and silencing-independent contribution to yeast aging (157). This mechanism appears to be governed by the Slt2 kinase via phosphorylation of Sir3, although the details of how this mechanism influences life span are not yet known.
CHRONOLOGICAL AGING
Background
As discussed above, one measure of yeast life span is the number of divisions a mother cell undergoes before senescing. Another distinct measure of life span in this organism is the ability of nondividing cells to maintain viability over time (122, 129). Under laboratory conditions, yeast cells grow rapidly until they exhaust the available nutrients. At this point, diploid yeast cells have the option of generating stress-resistant spores that can remain viable for years without nutrients. Haploid cells do not have this option. When pregrown in nutrient-rich medium, they enter a low metabolic state known as stationary phase, where they can survive for months. In synthetic defined medium, haploid cells maintain a high metabolic state even when nutrients are scarce, and they can survive for only about 1 to 2 weeks (124). It is under these conditions that chronological life span is usually measured because the high metabolic state is thought to more closely resemble the metabolic state of postmitotic cells in higher organisms (124).
Characteristics of Stationary-Phase Cells
A population of yeast cells that enters stationary phase undergoes a number of dramatic physiological and biochemical transformations. These primarily unbudded G0 cells accumulate glycogen and trehalose and develop thick cell walls (212). They are also significantly more thermotolerant and resistant to various forms of oxidative damage (191, 211). As may be expected, protein synthesis rates drop dramatically, although the number and identity of different proteins being synthesized is similar to those of exponentially growing cells (212). As Longo and Fabrizio point out, while the chronological life span of yeast may appear to be a starvation phase distinct from the aging of higher eukaryotic postmitotic cells, nondividing yeast cells are not starving (124). During these postdiauxic phases, yeast cells are breaking down glycogen and utilizing other stored nutrients, similarly to hibernating animals and the diapause state of other metazoans. Respiration is the primary source of energy in these cells, and the limited resources appear to be directed toward resisting cellular damage and stress (124).
Mechanisms
Free-radical theory of aging.
Among the first genes to be implicated in the chronological aging of yeast were SOD1 and SOD2, which encode cytoplasmic and mitochondrial superoxide dismutases, respectively (125). It was found that long-term survival in stationary phase required the expression of each of these genes, sparking speculation that chronological death is the result of extensive free-radical damage. Indeed, it has been further demonstrated that overexpression of both SOD1 and SOD2 can extend chronological survival by 30% and that loss of mitochondrial function occurs just before the death of both wild-type and sod2 mutant cells (45, 126). Analysis of the sod2 mutant led to the identification of aconitase as one of the primary mitochondrial targets of oxidative damage (126). Interestingly, aging fruit flies also show increased oxidation and inactivation of aconitase and in sod2 knockout mice, mitochondrial aconitase activity is 67% of the wild-type activity in some tissues (138, 217).
These results have given support to the free-radical theory of aging, first proposed a half a century ago by Harman (64). Subsequent identification of other yeast genes involved in chronological aging, though, has clearly shown that survival in stationary phase requires more than just antioxidant protection. For example, whereas overexpression of SOD1 and SOD2 extends life span by 30%, mutation of specific signaling proteins (discussed below) can increase longevity by as much as 300%. There is no question, though, that free radicals play an important role in this process (see below). At the very least, chronologically aging yeast has proven to be a valuable model for the study of oxidative damage in the postmitotic tissues of higher eukaryotes.
Interestingly, this research has provided novel insights into a process seemingly far removed from yeast biology, i.e., programmed cell death or apoptosis. Expression of the human antiapoptotic Bcl-2 protein in yeast partially reverses defects of sod mutants and increases the survival of wild-type cells (123). These results raised the possibility that components of the apoptotic machinery are present in yeast (reviewed in references 50 and 184). In support of this, the human proapoptotic Bax protein has been shown to promote cell death in yeast, and several putative apoptotic components (including a caspase-like protease) have been recently identified in yeast (130, 131, 133, 172). As suggested by Frohlich and Madeo, the study of old yeast cells that produce free radicals and that may die apoptotically will probably provide clues to similar events involved in mammalian aging (50).
SCH9 and CYR1.
The most notable of the above-mentioned discoveries came from a transposon-mediated mutagenesis screen performed by Fabrizio et al. (45). In a search for mutants that were both long-lived and stress resistant, the researchers isolated strains with transposon insertions in two genes, SCH9 and CYR1 (the transposon was in the promoter region of SCH9). Loss-of-function mutations in these genes had dramatic effects on chronological longevity. Mutant sch9 and cyr1 cells survived roughly three- and twofold longer than did the wild type, respectively. Interestingly, these two mutants not only had the greatest extension in chronological life span compared with wild-type cells but also were the only two genes isolated independently from both heat and oxidative stress selections. This supports a model whereby survival in stationary phase and extension of chronological life span depends on development of resistance to multiple forms of stress.
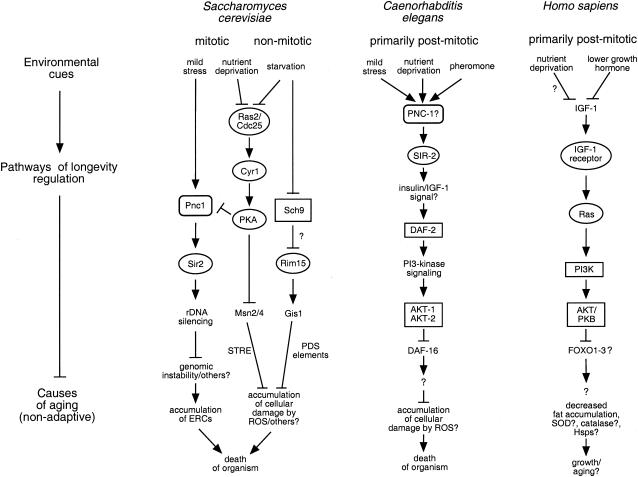
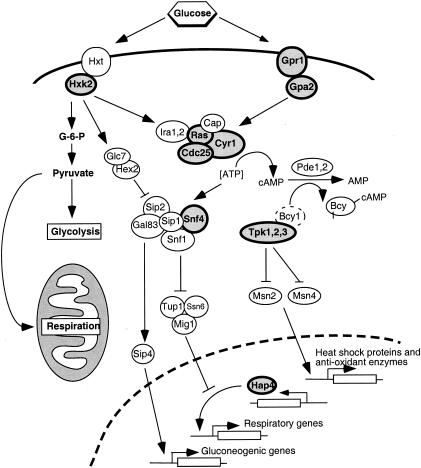
SCH9 encodes a serine/threonine protein kinase, whereas CYR1 encodes adenylate cyclase, required for stimulation of cyclic AMP (cAMP)-dependent protein kinase (PKA) activity (Fig. 3). These proteins appear to function in separate yet parallel signaling pathways, both of which mediate glucose/nutrient signaling, stimulate growth and glycolysis, and function to down-regulate stress resistance, glycogen accumulation, and gluconeogenesis (113, 124). Consistent with a role of cAMP signaling in regulation of chronological aging, deletion of the gene encoding the GTP-binding protein Ras2, an upstream regulator of Cyr1, doubles chronological life span (44) (Fig. 3). Similar to the above mutants, long-lived ras2 cells also display multiple stress resistances, since they are thermotolerant and more resistant to oxidative damage. Furthermore, the life span extension of both cyr1 and ras2 strains requires the stress response transcription factors Msn2 and Msn4. The life span extension in sch9 strains does not require these factors and may act via the protein kinase Rim15 (45). Rim15 in turn functions via the stress response transcription factor Gis1 (153), which binds postdiauxic shift elements found in the promoters of such genes as HSP26, HSP12, and SOD2 (Fig. 3). The above results again highlight the close association between stress response pathways and chronological longevity. Interestingly, mutation of the PKA pathway extends the replicative life span of yeast cells (114), arguing for some degree of overlap between these two distinct measures of aging (see below). It is worth noting that replicative life span extension is independent of Msn2 and Msn4 (114), so that while there may be some commonality in the pathways involved, the downstream mechanisms regulating these two processes are almost certainly distinct (Fig. 3).
FIG. 3.
Conserved longevity-regulatory pathways in S. cerevisiae, C. elegans, and possibly humans. There are two ways to study longevity in S. cerevisiae: replicative (mitotic) life span and chronological (nonmitotic) life span. Both these approaches have identified yeast genes with functional homologues in a conserved insulin-like signaling longevity pathway. These include SIR2, which encodes an NAD+-dependent deacetylase, RAS, which encodes a GTP-binding protein, and SCH9, which encodes a serine/threonine kinase. These findings are consistent with the idea that longevity regulation is highly adaptive and that components of a primordial pathway have been conserved in a diverse range of species.
In accordance with previous observations, it was also shown that the age-dependent inactivation of aconitase was significantly lower in both sch9 and cyr1 mutants (45). This suggests that increased survival of the strains is due, at least in part, to increased protection from oxidative damage. This was further supported by the recent confirmation that Sod2 functions downstream of Sch9 (44). Deletion of SOD2 abolishes the life span extension of an sch9 mutant; furthermore, expression of endogenous SOD2 was shown to be elevated in sch9 cells. As mentioned above, the limited survival obtained by overexpression of SOD1 and/or SOD2 indicates that these pathways probably regulate many downstream genes. Identification of these targets will be a major step toward understanding the relationship between stress resistance and chronological aging in this organism.
Conservation in Higher Eukaryotes
The kinase domain of Sch9 is 47 and 49% identical to that of Caenorhabditis elegans AKT-2 and AKT-1, respectively. AKT-1 and AKT-2 function in a longevity and diapause regulatory pathway downstream of the insulin receptor homolog DAF-2 (62, 94) (Fig. 3). Loss-of-function mutations in this insulin-insulin like growth factor type 1 (IGF-1) signaling pathway causes the worms to enter a state of diapause called dauer, a state normally initiated in response to nutrient limitation or crowding. Weak mutations in this pathway, though, can extend the life span of the adult worm by as much as twofold (84, 95). Similar to the yeast Ras-Cyr1-PKA and Sch9 pathways, the insulin-IGF-1 pathway in worms also acts to down-regulate stress resistance and the storage of reserve nutrients (95, 98, 144). Thus, not only is there conservation between specific factors involved, but also a similar strategy for the regulation of longevity and stress resistance may be conserved.
These results have been extended to Drosophila, where it has been shown that mutation of components of the fly insulin-IGF-1 pathway can extend life span by 85% (30, 200). These mutants also up-regulate nutrient storage and superoxide dismutase expression, similar to yeast and worms. Yeast Sch9 is also 49% identical to human Akt-1-Akt-2-PKB, involved in insulin and glucose signaling, apoptosis, and cellular proliferation (89) (Fig. 3). Whereas down-regulation of the yeast pathway leads to storage of glycogen, down-regulation of the human insulin-IGF-1 pathway stimulates the storage of fat, each of which is the primary carbon source during starvation for the respective organism (113, 124). The similarity in strategies, pathways, and factors involved in such distantly related organisms has led to the proposal that this common longevity regulatory system arose early in evolution as a way to delay reproduction and increase the chances of survival during times of nutrient limitation (94). This degree of conservation ensures that the study of chronological aging in yeast will continue to shed light on the regulation of, and mechanisms underlying, numerous critical processes in higher eukaryotes.
REGULATION OF REPLICATIVE AGING
Role of Sir2
Multiple lines of evidence point to Sir2 as being a key longevity protein that extends life span by suppressing rDNA recombination (60). Sir2 is a limiting component of yeast longevity. A single extra copy of the SIR2 gene suppresses rDNA recombination and extends life span by 40% (2, 88, 114). Conversely, deletion of SIR2 increases the frequency of rDNA recombination 10-fold, leading to accelerated aging. It has recently been shown that SIR2 is essential for the increased longevity provided by calorie restriction (114). In dispute of this, Jazwinski and colleagues have observed an extension of lifespan in sir2Δ mutants grown on 0.1% glucose (79). Recent experiments in our laboratory, though, confirm that sir2Δ strains grown on either 0.5 or 0.1% glucose show no life span extension compared to strains grown on medium containing 2% glucose (3). This disparity can probably be explained by strain-specific differences or sickness, since the average lifespan of the wild-type strain used by Jazwinski and colleagues, (SP1) is roughly half that of our wild-type strain (PSY316) (Table 2).
The role of Sir2 in longevity regulation appears to be conserved. Sir2 homologues can be found in a wide array of organisms, ranging from bacteria to humans, and it has been shown that increased dosage of the Sir2 homologue, sir2.1, can extend the life span of the nematode C. elegans (202). In addition, the nearest human homologue, SIRT1, inhibits apoptosis through deacetylation and negative regulation of p53 (128, 206). These two findings suggest that Sir2 and its homologues play a conserved role in the regulation of survival at both the cellular and organismal levels (62). A recent study with yeast has also implicated Sir2 in the asymmetric inheritance of oxidatively damaged proteins during cell division (1). It was found that carbonylated proteins, which accumulate during aging, are retained in the mother cell during cytokinesis in a Sir2-dependent manner. Although the authors do not claim this process to be a determinant of replicative life span, it probably contributes to the ability of cells to resist oxidative damage and to the overall fitness of daughter cells.
Overview of Sir2 enzymology.
Yeast Sir2 is the founding member of class III histone deacetylases (HDACs). Unlike class I or II HDACs, Sir2-like deacetylases are not inhibited by trichostatin A and have the unique characteristic of being NAD+ dependent (72, 105, 189, 196). Yeast Sir2 is known to have specificity for lysine 16 of histone H4 and lysines 9 and 14 of histone H3 (72, 105, 189). While many Sir2-like enzymes readily deacetylate histone substrates in vitro, at least two Sir2 homologues, yeast Hst2 and human SIRT2, are localized to the cytoplasm (20, 72, 196, 198). Studies have also demonstrated that human SIRT3 is localized to mitochondria (149, 170), and human SIRT1, a nuclear protein, has recently been shown to target p53 for deacetylation (128, 206). In addition, eubacterial species such as Salmonella lack histones entirely, yet their genomes still encode Sir2 homologues (203). The above results suggest that the Sir2 family of deacetylases may act on a broad range of substrates and that only a subset of these enzymes are likely to target histones for deacetylation in vivo.
Sir2-catalyzed deacetylation is considered an atypical reaction from both thermodynamic and mechanistic standpoints. Although trichostatin A-sensitive HDACs catalyze deacetylation without the need for a cofactor, Sir2 requires NAD+, even though deacetylation is a simple and energetically favorable hydrolysis reaction. Furthermore, NAD+ is not required catalytically for this reaction but is actually consumed by it. Hydrolysis of the glycosidic bond between the ribose and nicotinamide moieties of NAD+ liberates roughly 8.2 kcal of free energy per mol (143). This leads to the question of why the cell would couple cleavage of a high-energy bond in a metabolically valuable molecule to an already exothermic reaction. One possible explanation for these findings is that the NAD+ dependence of Sir2 may allow for regulation of its activity through changes in the availability of this cosubstrate (167). In other words, this may allow Sir2-like enzymes to “sense” the energetic and redox states of the cell and adjust their activity accordingly. An additional possibility is that the products formed from the unique Sir2 reaction may initiate a signal transduction pathway or may themselves allow for regulated control of the enzyme (16, 143). These possibilities are discussed in more detail below.
Evidence from crystal structures.
Sir2 enzymes couple NAD+ hydrolysis to the deacetylation of the ɛ amine of lysine residues with a 1:1 stoichiometry. The overall reaction actually consists of two hydrolysis steps, which are thought to be coupled (141). The first step is the cleavage of the high-energy glycosidic bond that joins the ADP-ribose moiety of NAD+ to nicotinamide. This is followed by cleavage of the C—N bond between the acetyl group and lysine. Upon cleavage, Sir2 then catalyzes the transfer of the acetyl group to ADP-ribose (189, 196, 198).
Crystal structures of two archaeal Sir2 homologues (Sir2-Af1 and Sir2-Af2), as well as the catalytic core of human SIRT2, have been solved and have begun to shed light on the specifics of the reaction mechanism (9, 27, 46, 141) (Fig. 4). The archaeal structures have been particularly revealing. Both the wild-type and several catalytically deficient varieties of Sir2-Af1 have been crystallized with an (albeit incomplete) NAD+ molecule (27, 141). In addition, Avalos et al. have published a crystal structure of Sir2-Af2 bound to an acetylated p53 peptide substrate (9). Conservation previously observed only at the sequence level is immediately apparent when the structures of these distantly related homologues are compared. The conserved core domain of these enzymes forms two regions which can bind an NAD+ molecule within a deep pocket between them (Fig. 4A). The larger of these two domains is reminiscent of the Rossmann fold, a motif found in many NAD(H)-NADP(H)-binding proteins (162), whereas the smaller domain coordinates a structural zinc atom.
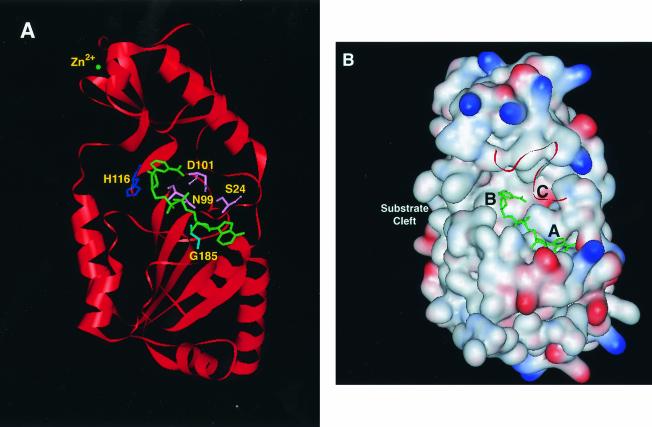
FIG. 4.
Crystal structure of the Sir2 deacetylase. (A) Ribbon diagram of Sir2-Af1 complexed with NAD+ (PDB 1ICI based on the structure of Min et al. [141]). The NAD+ molecule and zinc atom are in green. Putative catalytic site B residue His116 is shown as stick model in dark blue. Conserved site C residues Ser24, Asn99, and Asp101 are shown in pink. Gly185, which is located in site A and which is perfectly conserved in all Sir2 family members, is shown in light blue. (B) Surface representation of Sir2-Af1. Blue and red patches show surface electrostatic potential distribution for positively and negatively charged residues, respectively. Amino acid residues 30 to 47 are shown as a ribbon diagram for better visualization of the NAD+-binding pocket. This pocket is spatially divided into three regions, termed sites A, B, and C, which contact different portions of the NAD+ molecule (141). An acetyllysine substrate is proposed to come in close proximity to site B by inserting into a tunnel within the indicated substrate-binding cleft (9). The inhibitor nicotinamide is proposed to bind in the C site (16). Models were generated using Web Lab Viewer Lite software.
(i) NAD+-binding site.
The NAD+-binding pocket itself is divided into three distinct regions, termed sites A, B, and C (Fig. 4B). Site A appears to bind the adenine-ribose moiety of NAD+, while site B contacts the nicotinamide-ribose (141). Many of the mutations that diminish or abolish the activity of Sir2-like enzymes map to site B (5, 27, 141, 197), and it is therefore thought to be directly involved in catalysis. Site C forms a deep core within the pocket and does not appear to make direct contact with NAD+ in any of the solved structures. However, mutation of conserved residues within this pocket (specifically, residues equivalent to Ser24, Asn99, and Asp101 of Sir2-Af1) can severely affect or abolish catalytic activity (27, 141) (Fig. 4A).
Min et al. have suggested that because these residues are on the surface of the NAD+-binding pocket, they probably do not contribute to protein stability but may instead be somehow involved in catalysis. They proposed that the first step in Sir2-catalyzed deacetylation may be reminiscent of a serine protease reaction. A conformational change in NAD+, due to a rotation around the ester bond joining the adenine-ribose to the pyrophosphate and/or the phosphodiester bonds within the pyrophosphate moiety, would position the nicotinamide in proximity to site C. In this conformation, Ser24 may act as a catalytic base in the cleavage of the glycosidic linkage between the nicotinamide and the ribose (141) (Fig. 4A). Arguing against this model, though, Chang et al. found that mutation of Ser24 in Sir2-Af1 decreases activity by only sixfold, suggesting that this residue is not essential for activity. From an examination of the structures of these mutants, they instead conclude that site C residues play a role in NAD+ binding and positioning rather than in catalysis (27). In either case, hydrolysis of the acetyl group from N-acetyllysine in the second step of the reaction seems to require conserved residues in site B. Specifically, His116 (His118 in Sir2-Af2) is proposed to be directly involved in catalyzing this step (27) (Fig. 4A).
(ii) Substrate-binding site.
A cleft between the Rossmann fold domain and zinc-binding domain serves as a protein substrate-binding site (9) (Fig 4B). The acetyllysine side chain appears to insert into a tunnel within this cleft, bringing it close to site B of the NAD+-binding region. In particular, the structure of Sir2-Af2 bound to a p53 peptide shows the acetyllysine in close proximity to the putative catalytic His118 (His116 in Fig. 4A). This is proposed to make the acetyl group of the acetyllysine a better nucleophile during deacetylation (9). Based on the available data, a current model for the mechanism of deacetylation proposes a nucleophilic attack by the carbonyl oxygen of the N-acetyl group of the substrate on the C-1′ of the nicotinamide ribose (167, 199). Since several models detailing the exact chemistry of the two hydrolysis steps have been proposed (9, 27, 141), further investigation is needed to elucidate the precise mechanism in its entirety.
(iii) Substrate specificity.
Close analysis of the Sir2-Af2 peptide structure may reveal insights into the substrate specificity of Sir2-like proteins. It appears from this structure that all of the contacts that anchor the substrate to its binding site on the enzyme are simple hydrogen bonds between the two peptide backbones (9). According to the authors, these interactions form an enzyme-substrate β-sheet which they refer to as a “β staple,” since the substrate appears to link two distinct regions of Sir2-Af2. As Tanny and Moazed point out, this means that any peptide with a stretch of amino acids containing an acetylated lysine and which are flexible enough to form a β staple can be a putative substrate for Sir2-like enzymes (199). This idea is consistent with the broad specificity displayed by many of these enzymes in vitro. Indeed, Avalos et al. tested the ability of archaeal Sir2-Af1, Sir2-Af2, human SIRT2, and yeast Sir2 to act on both acetylated histones and two monoacetylated p53 peptides. They found that all four enzymes were able to deacetylate each of the four substrates (9). While mutation of several nonconserved residues in this region did alter the specificity of Sir2-Af2, it is likely that in vivo substrate specificity is determined primarily through protein-protein interactions outside the catalytic domain of the enzyme (199).
Sir2 reaction products.
(i) Nicotinamide.
While a complete understanding of the chemistry underlying the reaction requires further investigation, the products of Sir2-catalyzed deacetylation have been determined with precision. It has been demonstrated that this reaction leads to production of a deacetylated lysine and two additional products in a 1:1:1 molar ratio (104, 196, 198). Cleavage of the glycosidic bond joining the nicotinamide moiety of NAD+ to ADP-ribose results in the release of free nicotinamide. Bitterman et al. have shown that nicotinamide is a strong inhibitor of Sir2 and have proposed that it blocks catalysis by occupying site C of the NAD+ binding pocket (16). This important regulatory molecule, which is a precursor of nicotinic acid in the cell and a form of vitamin B3, is discussed in detail below.
(ii) O-Acetyl ADP-ribose.
The other reaction product, which results from cleavage of the C—N bond between the acetyl group and lysine followed by transfer of the acetyl to ADP-ribose, has been the focus of considerable speculation. This follows from the fact that this unique reaction leads to the generation of two novel metabolites, namely, a regioisomer of 2′- and 3′-O-acetyl ADP-ribose (73, 167). The initial product of the transfer reaction appears to be the 2′ form of this molecule, which quickly converts to 3′-O-acetyl ADP-ribose, eventually forming an equilibrium between the two (27). It has been proposed that Sir2 may initiate a signal transduction cascade through generation of these compounds, since the metabolic instability of these molecules is reminiscent of the initiators of other signaling pathways (167). This is cited as one possible rationale for the consumption of NAD+ during deacetylation. In support of this idea, injection of O-acetyl ADP-ribose into living cells causes a delay or block in the cell cycle and maturation of Xenopus oocytes (19). Furthermore, it has recently been demonstrated that ADP-ribose hydrolases may potentially metabolize O-acetyl ADP-ribose in Xenopus cell extracts. Interestingly, though, the predominant activities observed in these cell extracts appear to be those of novel, unidentified enzymes (156). Elucidation of the fate of these small molecules may uncover novel downstream effectors of Sir2-like proteins and shed light on the signaling pathways involved in longevity regulation and other cellular processes.
Metabolism, Chromatin, and Longevity
NAD+ synthesis and Sir2 activity.
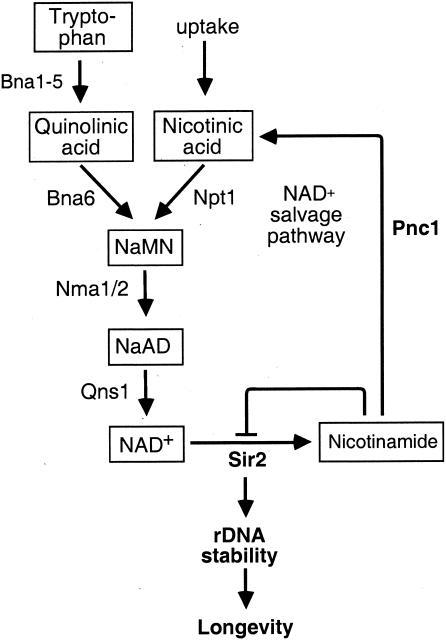
While the production of O-acetyl ADP-ribose provides an attractive hypothesis to explain the seemingly wasteful consumption of NAD+ in this reaction, others have been posited as well. As mentioned above, it has been proposed that the strict NAD+ dependence of Sir2-catalyzed deacetylation may allow for regulation of enzymatic activity through availability of the cosubstrate itself. This may permit the Sir2 to respond to changes in the energy status or redox state of the cell. In eukaryotes, there are two pathways of NAD+ biosynthesis: NAD+ may be synthesized de novo from tryptophan or recycled from nicotinamide via the NAD+ salvage pathway (48) (Fig. 5).
FIG. 5.
Pathways for NAD+ and nicotinamide metabolism in S. cerevisiae. In yeast, NAD+ can be recycled from nicotinamide via the NAD+ salvage pathway or synthesized de novo from tryptophan. Nicotinamide generated by Sir2 is converted into nicotinic acid by the nicotinamidase Pnc1 and subsequently into NaMN by Npt1. Nicotinic acid may also enter the pathway exogenously. The formation of desamido-NAD+ (NaAD) is catalyzed by one of two adenylyltransferases encoded by NMA1 and NMA2, and the subsequent formation of NAD+ by the NAD+ synthetase Qns1. Trptophan taken up from the medium is converted into quinolinic acid by Bna1 to Bna5. A quinolinic acid phosphoribosyltransferase, encoded by BNA6/QPT1, catalyzes the subsequent conversion to NaMN, which feeds into the salvage pathway.
(i) Kynurenine pathway.
De novo NAD+ synthesis, also known as the kynurenine pathway, is catalyzed by the BNA (for “biosynthesis of nicotinic acid”) genes, which catabolize tryptophan to quinolinic acid and subsequently to nicotinic acid mononucleotide (NaMN) (Fig. 5). Tryptophan taken up from the medium or from an intracellular source is converted to kynurenine in two steps by Bna2 and Bna3. Bna4 and Bna5 catalyze the ensuing conversion to 3-hydroxykynurenine and 3-hydroxyanthanilic acid, respectively. An oxidoreductase encoded by BNA1 then catalyzes the formation of quinolinic acid, which is converted into NaMN by the BNA6 gene product (also known as QPT1) (56, 165). It is interesting that the de novo pathway for NAD+ synthesis does not appear to be essential for either Sir2-dependent silencing or life span extension by calorie restriction. Deletion of either BNA1 or BNA6 does not result in an rDNA-silencing defect, unless the cells are grown on medium without nicotinic acid, compromising their salvage synthesis of NAD+ (see below) (165). In addition, a cdc25-10 strain, which is defective in glucose signaling and is considered a genetic mimic of calorie restriction (see below), is long-lived even in the absence of BNA6 (114).
(ii) Salvage synthesis of NAD+.
In yeast, the salvage pathway for NAD+ synthesis consists of four steps (Fig. 5). Nicotinamide produced from NAD+ cleavage is first converted to nicotinic acid by the nicotinamidase Pnc1 (3, 52). The specific role of this key enzyme in Sir2 regulation is discussed in detail below. Nicotinic acid may also enter the pathway exogenously, and a high-affinity transporter for this compound, Tna1, has recently been identified (120, 165). In either case, nicotinic acid is subsequently converted into NaMN by a phosphoribosyltransferase encoded by NPT1 (2). At this point, the NAD+ salvage and the de novo NAD+ synthesis pathways converge and NaMN is converted to desamido-NAD+ by a nicotinate mononucleotide adenylyltransferase (NaMNAT). In S. cerevisiae, there are two open reading frames (ORFs) with homology to bacterial NaMNAT genes (43, 189), named NMA1 and NMA2 (2). In Salmonella, the final step in the regeneration of NAD+ is catalyzed by an NAD+ synthetase (71). An as yet uncharacterized yeast ORF, YHR074w/QNS1, is predicted to encode an NAD+ synthetase.
The NAD+ salvage pathway has been a recent focus of attention because, unlike the de novo pathway, it is critical for Sir2 activity. Cells lacking NPT1 or PNC1, for example, show a loss of silencing at telomeres and the rDNA, reminiscent of a sir2 mutant (165). Furthermore, the replicative life span of a cdc25-10 strain (the genetic mimic of calorie restriction) is no longer extended if NPT1 is absent (114). Conversely, increased dosage of NAD+ salvage pathway genes increases silencing, and a single extra copy of the NPT1 gene extends the life span by 60% (2). These results suggest a model whereby up-regulation of NAD+ synthesis leads to an increase in cellular NAD+ levels and a subsequent stimulation of Sir2 activity. In support of this, NAD+ levels are significantly reduced in cells lacking NPT1 but not in those lacking BNA1 or BNA6, consistent with the observed silencing phenotypes of these mutants (165). Interestingly, although overexpression of NPT1 increases silencing and extends life span, NAD+ levels are unaltered in these strains (2).
Calorie restriction, respiration, and NAD+.
As mentioned above, controlled fluctuations in NAD+ levels provide an attractive rationale for the cofactor dependence of the Sir2 reaction. But is this the mechanism by which calorie restriction affects Sir2 activity? And if so, how would limiting calorie intake affect a change in cellular NAD+ levels? Guarente and colleagues have proposed that this is indeed the mechanism by which Sir2 is regulated, and a recent paper by Lin et al. (115) offers a possible explanation. S. cerevisiae, a facultative anaerobe, can generate energy through either fermentation or the more energy-efficient respiration. When glucose levels are high, cells preferentially utilize fermentation, since energy is in excess. When glucose becomes limiting, though, respiration is preferred because 16 ATP molecules can be produced from a single glucose, compared to only 2 molecules from fermentation (10). Thus, carbon is shunted toward the mitochondrial tricarboxylic acid cycle, increasing electron transport and respiration (155).
By measuring oxygen consumption, Lin et al. showed that yeast grown on low glucose respire at a threefold higher rate. Furthermore, calorie-restricted yeast that cannot respire due to a mutation in their electron transport chain do not live longer than unrestricted cells. In contrast, artificially inducing genes involved in respiration through overexpression of the HAP4 transcription factor causes a Sir2-dependent extension of life span, even under high-glucose conditions (115). The authors propose a model whereby increased respiration leads to increased oxidation of NADH to NAD+ in the mitochondria. This change in the NAD+/NADH ratio would be transmitted outside the mitochondria, where it would effectively stimulate Sir2 activity.
While this is an attractive hypothesis, an overall increase in the cellular NAD+ concentration or in the NAD+/NADH ratio has yet to be observed under these conditions. In fact, we and others have been unable to detect increases in NAD+ levels in calorie-restricted cells (116) or in genetic mimics of calorie restriction (2), even when the unbound NAD+ level was measured (R. M. Anderson, K. J. Bitterman, J. G. Wood, and D. A. Sinclair, unpublished results). Furthermore, and perhaps most striking, is the fact that although cells lacking PNC1 are compromised for Sir2-dependent silencing, their NAD+ levels are unaltered (165). It is formally possible that Sir2 is regulated either by nucleus-specific changes in NAD+ availability, flux through the salvage pathway, or a different mechanism altogether.
Regulation by nicotinamide.
(i) Inhibition of Sir2 by nicotinamide.
One intriguing possibility, which may resolve these apparent conflicts, concerns the other product of the Sir2-catalyzed reaction, nicotinamide, produced from the cleavage of the glycosidic bond of NAD+. Nicotinamide was recently demonstrated to be a strong inhibitor of yeast Sir2 and human SIRT1 activity, both in vitro and in vivo (16, 128) (Fig. 5). We found that cells grown in the presence of this compound show a nearly complete loss of Sir2-dependent silencing and have a replicative life span indistinguishable from that of a sir2 mutant. In vitro, the 50% inhibitory concentration of nicotinamide for SIRT1 was found to be ∼50 μM, leading us to speculate that this small molecule may be a physiologically relevant regulator of Sir2-like enzymes (16). Based on these findings, we proposed that fluctuations in cellular nicotinamide levels may directly control the activity of Sir2 proteins in vivo and that these fluctuations may in turn be regulated by enzymes involved in nicotinamide metabolism.
(ii) Regulation of nicotinamide levels by Pnc1.
PNC1 encodes a nicotinamidase that is situated in a key position to regulate NAD+-dependent deacetylases. This enzyme converts nicotinamide into nicotinic acid as part of the NAD+ salvage pathway. Thus, Pnc1 may reduce levels of this inhibitor and simultaneously increase the availability of NAD+ to Sir2. Importantly, we have shown that the product of this reaction, the structurally similar nicotinic acid, has no effect on Sir2 activity either in vitro or in vivo (16). This raised the possibility that high levels of Pnc1 induce Sir2 activity by removing the inhibitory effects of nicotinamide. In support of this model, we have found that overexpression of PNC1 extends the replicative life span by 70% in a Sir2-dependent manner and that the life span of these cells is not further increased by growth on low glucose. In addition, the life span of cells lacking PNC1 is not augmented by calorie restriction (3). These results demonstrate that PNC1 is necessary for life span extension by calorie restriction and that additional PNC1 is sufficient to mimic these stimuli.
The most striking aspect of this model is the fact that endogenous Pnc1 protein levels are greatly augmented in response to all stimuli that extend yeast life span. These include low glucose concentration, low amino acid concentrations, heat stress, and osmotic stress (3, 173, 195). Consistent with the involvement of the PKA pathway in regulating life span, Pnc1 levels are increased in a cdc25 mutant strain as well (3). Furthermore, in support of the notion that nicotinamide is the primarily regulator of Sir2, it was shown that manipulation of nicotinamide metabolism by proteins outside the NAD+ salvage pathway can also affect Sir2 activity. These genes were the human nicotinamide N-methyltransferase gene (NNMT) and its putative yeast homologue encoded by the YLR285w ORF. These results identify PNC1 as the first longevity gene that is responsive to all stimuli that are known to extend replicative life span and show that nicotinamide depletion is sufficient to activate Sir2. These findings support a model whereby life span extension via calorie restriction is the consequence of an active cellular response to low-intensity stress (Fig. 6).
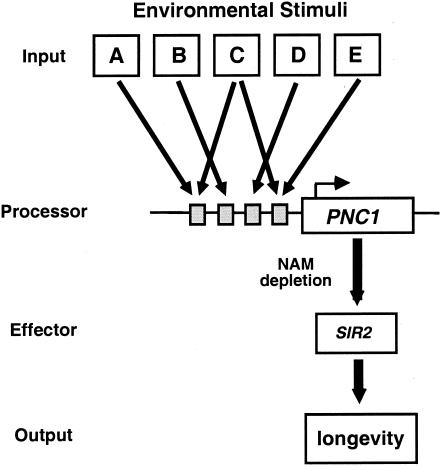
FIG. 6.
Model for the regulation of Sir2 activity and life span by nicotinamide. Disparate environmental stimuli including calorie restriction, heat, and osmotic stress serve as inputs to a common pathway of longevity. Cells coordinate a response to these inputs by inducing the transcription of PNC1, which encodes an enzyme that converts nicotinamide to nicotinic acid, thereby alleviating the inhibition of Sir2 and promoting longevity.
Why would an organism utilize a single gene and a small ubiquitous molecule to govern such a critical pathway as life span extension by calorie restriction? We speculate that this system may allow the organism to process inputs from multiple stimuli and facilitate a coordinated defense response. A more intriguing possibility, however, is that this design permits the rapid evolution of strategies to suit a changing environment via changes in the Pnc1 promoter. The Sir2 pathway is ancient, and perhaps early life-forms used nicotinamide directly as an indicator of nutrient availability. Its role as a signaling molecule regulating longevity and survival may have been conserved to the present day.
(iii) Nicotinamide and higher organisms.
It will be interesting to determine whether nicotinamide levels do indeed decrease under conditions of calorie restriction or mild stress and, if so, whether this mechanism is conserved in higher organisms. The possibility that nicotinamide is the major regulator of Sir2-like proteins raises many questions regarding the role of this small molecule and its targets in such organisms. There is already evidence for a link between nicotinamide metabolism and stress resistance in mammals. Poly(adenosine diphosphate-ribose) polymerase 1 (PARP) is a nuclear enzyme that cleaves NAD+ to covalently attach poly(ADP-ribose) to acceptor proteins (reviewed in reference 146). This two-step reaction generates nicotinamide, which exerts an inhibitory effect on PARP-1, allowing for autoregulation. PARP enzymes have been implicated in numerous cellular functions including DNA break repair, telomere length regulation, histone modification, and the transcriptional regulation of key proteins including intercellular cell adhesion molecule 1 and nitric oxide synthase (207). PARP enzymes are perhaps regulated by nicotinamide metabolism as part of a general stress response.
As mentioned above, nicotinamide also inhibits human SIRT1 both in vitro and in vivo (16, 128). SIRT1 negatively controls p53 activity, indicating that nicotinamide levels may also regulate apoptosis and DNA repair (128, 206). Consistent with this, increased expression of NNMT correlates with tumorigenesis (103) and decreased expression correlates with radiosensitivity (90). There is no obvious homologue of PNC1 in model organisms more complex than Drosophila, which probably reflects differences in nicotinamide metabolism. It will be important to determine whether nicotinamide regulates multiple Sir2-like proteins and, if so, which metabolic pathways in turn regulate nicotinamide levels. Clearly there is still much to be learned about this family of proteins and their potential roles in cell and organismal survival.
Rpd3
Another HDAC that has been shown to influence life span in multiple organisms is Rpd3. RPD3 encodes a class I HDAC which seems to have specificity for lysines 5 and 12 of histone H4 (164). The function of this enzyme in yeast has recently been shown to be required for the proper timing of replication origin firing (208) and for regulating the number of “open” rDNA repeats expressed during stationary phase (166). In addition, Rpd3 is known to repress the transcription of a number of critical genes including HO, TRK2, STE6, PHO5, and SPO13 (59, 193).
Mutation of RPD3 leads to histone hyperacetylation, similar to deletion of SIR2 (28). Surprisingly, though, loss of Rpd3 function has an opposite effect on silencing compared to a sir2 mutant. Whereas deletion of SIR2 abrogates silencing at mating loci, telomeres, and the rDNA, deletion of RPD3 increases silencing at all three of these loci (96, 164, 205). This disparity extends to the life span of the organism as well, since rpd3 mutants live significantly longer than wild-type cells (96). These findings have recently been extended to Drosophila melanogaster. While complete loss of Rpd3 function is lethal in this organism, Rogina et al. showed that a partial loss-of-function mutation extended life span in both male and female flies (159). In addition, it was demonstrated that Rpd3 appears to function in the calorie restriction pathway in this organism. This is consistent with the fact that levels of this protein are reduced under nutrient-limiting conditions (154, 159). A similar result has also been found for yeast RPD3 (79).
Because Rpd3 and Sir2 act on different lysine residues on histones H3 and H4 and because they have opposing effects on silencing and life span, it is clear that the overall pattern of core histone acetylation governs the degree of silencing at these loci. It has been proposed that in addition to changes in the state of silent heterochromatin, overall changes in gene expression patterns resulting from manipulation of these genes may regulate yeast life span (15, 28). Additional work is required to elucidate the precise mechanisms by which low levels of Rpd3 extend replicative life span in yeast and Drosophila.
Glucose Sensing and Calorie Restriction
Background.
The link between diet and aging has been known for over 50 years, ever since McCay et al. discovered that restricting calorie intake in rats significantly extends life span (135). Since then, this dietary regime, known as calorie restriction, has been shown to extend life span in every species tested thus far (132). In 2000, two groups laid the groundwork for elucidating the genetic pathways of calorie restriction in S. cerevisiae (78, 114). It was found that yeast grown on low glucose or with limiting amounts of nonessential amino acids showed a significantly extended replicative life span. Here we provide a brief overview of glucose-signaling pathways in yeast and what is known about their role in the calorie restriction pathway of life span regulation.
HXT and HXK genes.
In S. cerevisiae, glucose serves not only as the preferred carbon source but also acts as a signaling molecule (reviewed in reference 161). Yeast sense and transport glucose via the Hxt family of transporters (Fig. 7). External glucose levels regulate transcription of the HXT genes, which encode several isoforms of transporters, each possessing different affinities for hexoses (reviewed in reference 151). This finely tuned mechanism ensures the expression of low-affinity receptors in response to conditions of high glucose availability and intermediate-affinity receptors for situations in which the supply of glucose is limiting. On transport, glucose is phosphorylated to glucose-6-phosphate (G-6-P), a high-energy intermediate that is a key substrate for glycolysis. This reaction is catalyzed by the three kinases Hxk2, Glk1, and Hxk1. Notably, mutation of HXK2 leads to a significant increase in yeast replicative life span (114). Although the simplest interpretation of this result is that the mutation mimics low glucose, this is not necessarily the case because Hxk2 has an additional function: the down-regulation of genes involved in respiration, gluconeogenesis, and the glyoxylate cycle during growth at high glucose concentrations in a process called glucose repression.
FIG. 7.
Glucose-sensing and signaling pathways of S. cerevisiae. Proteins indicated by bold type have been shown via genetic analyses to extend the replicative life span when either deleted or mutated. Glucose in the environment is first sensed by either of two complementary receptor families, the Hxt hexose transporters or the G-protein-coupled receptor, Gpr1. Glucose-mediated signals arising from these receptors cause Cdc25 to catalyze the formation of GTP-bound Ras1-Ras2, which then binds to and regulates the adenylate cyclase complex. Cyr1 (Cdc35) within the complex then generates cAMP, which in turn activates PKA, consisting of subunits Tpk1, Tpk2, and Tpk3. PKA inhibits Msn2 and Msn4, two factors required for the expression of heat shock proteins and superoxide dismutases, which are important for regulating the chronological life span. Further downstream, both pathways also converge on the Snf1 serine/threonine kinase complex, which is responsible for derepressing genes that are turned off during growth at relatively high glucose concentrations. The activity of the Snf1 complex is modified by the Glc7-Hex2 complex and also perhaps by a proposed kinase(s) that may be responsive to intracellular ATP and AMP levels. The Snf1 complex inhibits the Mig1-Ssn6-Tup1 repressor complex, required for shutting off the transcription of genes needed for respiration, and activates Sip4, which turns on the transcription of genes required for gluconeogenesis under conditions of low glucose concentrations.
Snf1 complex.
Central to the derepression of glucose-repressed genes is the Snf1 kinase complex, consisting of the serine/threonine kinase Snf1, along with an activating subunit, Snf4 (26), and one of three cofactors, Sip1, Sip2, and Gal83 (81), that confer substrate specificity (Fig. 7). Low glucose levels stimulate the activity of the Snf1 complex (80), leading to the expression of genes involved in gluconeogenesis and respiration. The deletion of SIP2, encoding an inhibitory N-myristoylprotein subunit of Snf1, results in an accelerated-aging phenotype, characterized by the accumulation of ERCs and shortening of the replicative life span (6). Conversely, deletion of SNF4, an activating subunit of Snf1, increases the replicative life span by approximately 20%, relative to wild-type cells (6). Interestingly, LAG1, a gene required for ceramide synthesis and the transport of glycosylphosphatidylinositol-anchored proteins (12, 63), was shown to result in an increase replicative life span when deleted (38). This is suggestive of a general mechanism in which lipid-modified proteins may be involved in transducing longevity-determining signals, as in the case of Sip2.
Mig1, a component of the Tup1-Ssn6-Mig1 repressor complex, is downstream of the Snf1 complex (Fig. 7). The Mig1 repressor serves to repress genes involved in respiration, gluconeogenesis, and alternative carbon source utilization. Low glucose levels lead to the phosphorylation of Mig1 by the Snf1 complex, followed by its inactivation and export to the cytoplasm (37). Interestingly, deletion of MIG1 does not affect the replicative life span (6). Thus, although Snf1 function and respiratory activity are linked to life span, they must act via a mechanism that is independent of the Mig1 repressor pathway.
Given that low glucose levels and the corresponding increase in respiration correlate with increased life span (115), why would loss-of-function mutations in the Snf1 complex, which presumably mimic this metabolic state, lead to accelerated aging? A plausible explanation is that the effect of the Snf1 complex on longevity is independent of its role in glucose repression. This is supported by the recent finding that the Snf1 complex can phosphorylate histone H3 in vitro on Ser10 (117, 121), which is suggested to result in desilencing and hyperrecombination at the rDNA locus. Indeed, Snf1 kinase activity has also been shown to increase in aged cells (117). Thus, even if Snf1 were to increase the replicative life span by promoting respiration, this would probably be offset by the corresponding increase in recombination at the rDNA locus. Although this is highly suggestive of a pleiotropic role for Snf1, it still needs to be determined whether the increased kinase activity directly affects the rDNA locus in vivo.
cAMP signaling and replicative life span.
Glucose repression is dependent not only on signaling initiated by the Hxt transporters but also on the activity of a G-protein-coupled receptor complex, Gpr1-Gpa2, that activates cAMP synthesis (160) (Fig. 7). The Hxk and Gpr1-Gpa2 glucose-signaling pathways are both required for the appropriate repression of genes encoding proteins for respiration and gluconeogenesis. The yeast PKA pathway was first implicated in the regulation of longevity by Lin et al., who showed that mutations in GPR1, GPA2, CDC25 (encoding a GTP-GDP exchange factor), CYR1-CDC35 (encoding adenylate cyclase), or TPK1-3 (encoding the PKA catalytic subunits) each significantly extended the replicative life span (114). Interestingly, life span extension in a cdc25-10 strain requires Sir2. Although the mechanism by which cAMP regulates Sir2 is not known, it is interesting to speculate about whether Sir2 activity might be modulated by PKA via regulation of Pnc1 (Fig. 3). Another possibility is that intracellular AMP levels regulate Snf1 activity and thereby regulate longevity. Although the mammalian homologue of Snf1, AMP-activated protein kinase (AMPK), is allosterically activated by intracellular concentrations of AMP, this is not the case with Snf1 (70). Instead, Snf1 is proposed to be activated by an upstream kinase, which in turn may be directly activated by AMP levels. Although Snf1 can be activated in vitro using mammalian kinases such as AMPKK and inactivated by phosphatases (213), the putative yeast kinase that phosphorylates Snf1 is still unknown. Furthermore, it remains to be seen whether kinases from cAMP-mediated signaling and glucose signaling ultimately converge on a single downstream substrate or whether there are multiple substrates that may each affect longevity.
Retrograde Signaling And Life Span
Mitochondrial function is known to influence the transcription of nuclear genes. This phenomenon, termed retrograde signaling, can influence the replicative life span (99). In S. cerevisiae, three genes, RTG1, RTG2, and RTG3, are required for signaling between the mitochondria and nucleus (77, 111). Deletion of RTG3 significantly increases the replicative life span, in a manner independent of the calorie restriction pathway (78). RTG1 and RTG3 encode a pair of basic helix-loop-helix leucine zipper transcription factors that bind as heterodimers to “GGTCAC” consensus sequences. These sites, referred to as R-box elements, are found within the nuclear genome. Rtg3 localizes within the cytoplasm in a hyperphosphorylated state. Dephosphorylation of Rtg3 induces nuclear relocalization and the activation of tricarboxylic acid cycle genes (119, 171). It is still not known how the phosphorylation status of Rtg3 is controlled, although there is some evidence that the signal is transduced at least partly by Rtg2. The nuclear gene encoding peroxisomal citrate synthase, Cit2, is highly up-regulated as a consequence of retrograde signaling (112) and has thus been used as a convenient marker for the activation of this pathway.
Several different strains of yeast that lack mitochondria, known as “petites,” have increased replicative life spans relative to “grande” parental strains that contain functional mitochondria, although some standard laboratory strains such as W303 do not show this effect (99). It is also worth noting that these results appear to contradict those of Guarente and colleagues, who found that mitochondrial respiration is both necessary and sufficient for replicative life span extension (115). Long-lived petite strains have significant up-regulation of CIT2, indicative of an activation of the retrograde response. Thus, it is possible that strain differences within the retrograde response pathways may account for the discrepancies in life span observed when comparing grandes and petites. Although CIT2 is up-regulated as a result of mitochondrial defects, it is not required for the extension of life span observed in petites. Instead, the G protein Ras2, required for the proper functioning of mitochondria, is necessary for this life span extension (49, 201). Overexpression of RAS2 or deletion of RAS1 significantly extends the replicative life span (194). This is an interesting finding, given that these genes are interchangeable for signaling within the PKA pathway. One possibility is that Ras2 but not Ras1 is required to transduce retrograde signals between mitochondria and the nucleus, but this remains speculative.
The similarities between the Snf1 pathway and the retrograde response are intriguing. Both Snf1 signaling and the Rtg response are influenced by the PKA pathway, with retrograde signaling being dependent on the activity of Ras2 while Snf1 is responsive to intracellular AMP/ATP ratios. This suggests that yeast longevity is regulated by at least two pathways, one of which is responsive to the external environment and one responsive to the internal cellular environment. The elucidation of these pathways may prove to be difficult because there is probably considerable cross talk between them.
TELOMERE MAINTENANCE AND PREMATURE-AGING SYNDROMES
Background
Since the discovery that primary cells in tissue culture divide a finite number of times, a phenomenon known as the Hayflick limit (65), it has been proposed that this phenomenon may play a role in human aging, at least for actively dividing tissues (169). This limit on cell division is due primarily to the progressive shortening of telomeres as cells replicate. Loss of telomeric DNA is the consequence of the inability of lagging-strand synthesis to finish replicating the ends of linear chromosomes, originally termed the end replication problem (148, 210). To counter this problem, telomerase, a specialized reverse transcriptase that can synthesize telomeric DNA de novo (57, 58), is expressed in many immortal cell types including many tumorigenic cells. Unlike human somatic cells, wild-type yeast cells constitutively express telomerase, and telomeric DNA does not shorten with replicative age (93).
Yeast as a Model For Replicative Senescence
Nevertheless, S. cerevisiae has been used to study replicative senescence. By deleting genes encoding components of telomerase (e.g., EST2 or TLC1, encoding the catalytic protein subunit and RNA template of telomerase, respectively), yeast cells can be forced to undergo replicative senescence (110, 183). These genetic manipulations lead to progressive telomere shortening with each subsequent cell division, akin to what occurs in actively dividing human primary cells. The population of yeast cells in culture gradually ceases to divide when telomeres reach critically short lengths. The cessation of division is probably due to shortened telomeres being recognized as double-stranded breaks (DSBs) by DNA damage-sensing mechanisms. Indeed, telomeres are normally found associated with a protective “cap” of proteins, containing factors that are also required for repairing DNA DSBs (reviewed in reference 17). These factors include the Ku70-Ku80 heterodimer (204), which is involved in nonhomologous end joining and the ATM kinase orthologs, Mec1 and Tel1 (158), required for signaling the presence of DSBs.
In yeast, telomeres serve as repositories for silencing factors such as Sir2/3/4, of which Sir2 is present within the cell in limiting quantities (190). These factors probably serve to minimize recombination between telomeric repeats. Unlike the case for human cells, telomere shortening increases yeast replicative life span (8), presumably because the telomere-bound pool of Sir2 relocalizes to the rDNA, where it promotes DNA stability. Recently it has been shown that telomeric silencing also occurs in human cells (13). The integration of a luciferase reporter gene proximal to telomeres demonstrated that silencing of this transgene was increased relative to an internal genomic control. Microarray analysis has shown that senescing mammalian cells have significantly different expression levels of a wide variety of genes relative to nonsenescing controls (175). It has been hypothesized that this may be due, in part, to a redistribution of silencing factors away from telomeres and to more distal regions of the genome, thereby affecting gene expression (216). Such epigenetic effects may account for the differential gene expression observed in senescent cells (215).
Telomeres and Premature-Aging Syndromes
A link between premature aging and telomeres was recently uncovered in studies using yeast as a model. The premature-aging disease, Werner syndrome, is characterized by symptoms that resemble premature aging, including cataracts, osteoporosis, atherosclerosis, grey hair, and an increased incidence of certain cancers. Werner syndrome is due to loss-of-function mutations in the WRN gene, encoding a RecQ-like DNA helicase implicated in DNA repair during replication (reviewed in reference 150). The rate of spontaneous immortalization of senescent WRN−/− cells is extremely low, suggesting that they lack some ability to reestablish telomere length and hence bypass senescence.
S. cerevisiae possesses a single homologue of WRN, the slow-growth suppressor, SGS1. Interestingly, mutation of SGS1 in strains lacking telomerase causes them to undergo accelerated senescence (32), similar to the phenotype of WRN−/− cells. Moreover, sgs1 cells do not recover from senescence as rapidly as do SGS1 controls, also reminiscent of the human Werner phenotype (32, 69, 82). Examination of telomeric DNA in these cells indicated that sgs1 cells lose telomeric DNA more rapidly than do SGS1 cells (32) and that they cannot rebuild their shortened telomeres via the main (type II) recombination pathway that normally leads to extended telomeric terminal repeats (32, 69, 82). These results imply that the WRN helicase may be involved in maintaining the integrity of telomeric DNA in normal cells and that its function might also be important for the recovery of cells from replicative senescence, which is a key step in tumorigenesis.
A related RecQ-like helicase gene found mutated in Bloom's syndrome patients, BLM, leads to a number of defects, including an increased predisposition to certain cancers and genomic instability. Unlike WRN, BLM was found to fully suppress the increased rDNA recombination phenotype of sgs1 cells (68). Although sgs1 cells have a shorter replicative life span, they should not be regarded as true mimics of aging because the resultant life span is a composite of normal aging processes and mitotic arrest due to increased intrachromosomal recombination (136). Similarly, the relevance of RecQ helicases to normal aging in humans is still the subject of conjecture.
METHODS
Replicative Life Span Analysis
Currently, the most accurate way to determine yeast replicative life span is to micromanipulate away and count every daughter cell that emerges from a mother. The method is tedious, time-consuming, and often cited as the main barrier to entry into the field. Many groups have searched for a less labor-intensive method, so far without success. The entire assay takes about 2 weeks and demands that the researchers spend the majority of their time at the microscope during this period. The life span assay begins by twice passaging the strain to be assayed on the appropriate medium. At least 40 cells are then arrayed down the life span plate, using a 25- or 50-μm dissection needle. Life span plates have at least 2% agar and are poured thicker than usual. After 1 hour, virgin daughters that emerge are then used for the assay and the mothers are moved away and discarded. At 30°C, young cells divide every 1 to 2 h. Daughter cells are picked off mothers and moved at least one field of view away so that they do not compete with the mothers for nutrients. In the initial stages, it may be difficult to discern the daughter from its mother, so it is useful to mark the orientation of the mother and her bud and not let the mother divide more than once. Mothers are usually moved in the opposite direction to daughters each dissection. Cells may be placed at 4°C overnight to slow cell growth, but keeping cells at this temperature for more than 2 days is not advised.
Toward the end of a cell's life span, its division time will become considerably longer, to more than 2 h, and the size difference between mother and daughter will be obvious. A cell is considered dead when it ceases to divide after ∼6 h. Often, late-emerging daughters remain attached to the mother, and it is imperative that daughters from these cells not be scored. Potential pitfalls include the use of overcooked medium, use of a dissection needle with a rough surface, working with a wild-type strain that is short-lived or whose daughters do not dissociate from their mothers easily, and working in a dry atmosphere where cells and plates can become desiccated. We have found it helpful to place a humidifier in the vicinity of the microscope and to always place dissection plates in a plastic bag when they are incubating.
Chronological Life Span Analysis
Chronological life span in S. cerevisiae is determined by maintaining cells in either complete or defined medium. In these different media, cells cease dividing when they exhaust nutrients, but they maintain different metabolic states. In complete medium or in cells switched to water from defined medium, cells maintain low metabolic rates. In contrast, cells maintained in defined medium maintain relatively high metabolic rates. Defined medium is used more frequently than complete medium because the metabolic status of cells is thought to more closely resemble that of postmitotic human cells. Moreover, the average life span of cells in this medium is a few weeks rather than months, so that experiments proceed at a faster pace.
The standard chronological life span assay requires that cells be diluted to the same density in synthetic complete medium (2% [wt/vol] glucose) and incubated at 30°C. Cells cease dividing after the first 48 h, and most strains have a mean survival of 15 to 29 days (W303, PSY316, S288C, and BY4700). Some strains only live an average of 6 to 12 days (DBY746 and SP1). Every 2 or 3 days, an aliquot of the cultures is plated to complete YPD medium to score viability (expressed as CFU). The CFU on day 3 are considered to denote 100% survival. The assay is performed until less than 1% of the culture is viable. A “live-dead” fluorescence assay from Molecular Probes Inc. can also be used to determine viability.
Isolation of Old Cells
The study of the physical and biochemical changes that accompany yeast aging requires the separation of large numbers of old individuals from young ones. This prospect is complicated in single-celled, exponentially growing organisms because the fraction of old cells in the population is exceedingly small. Consider a yeast strain with an average life span of 20 divisions. Because the population doubles with each cell division, 50% of the population consists of virgin cells, 25% are one division old, 12.5% are 2 divisions old, and only 1 in four million cells has exceeded 20 divisions. The techniques for accomplishing this separation are reviewed here.
Fluorescence-activated cell sorting.
When yeast cells divide, they synthesize a bud that eventually becomes the daughter cell. The cell surface of the bud is derived de novo, with no contribution from previously synthesized mother cell components. This process allows mother cells to be “tagged” and then distinguished from daughter cells, even when they are greatly outnumbered in culture. Similar to magnetic sorting (see below), proteins on the cell surface are conjugated to biotin and then the cells are cultured in broth for up to 15 generations. By adding fluorochrome-conjugated avidin to the final population of cells, old cells become specifically labeled and may be separated using fluorescence-activated cell sorting (187). This technique allows the rapid purification of an old-cell population with greater than 99% purity, although yield is compromised for the sake of purity. Typically, this method allows procurement of 104 old cells.
Magnetic sorting.
The most efficient and widely used method for isolating old yeast cells relies on magnetic sorting of mother cells from the unbiotinylated daughter cells (187). Cells are biotinylated and allowed to divide as in fluorescence-activated cell sorting. Avidin-coated paramagnetic iron beads are then added to the final population. The beads coat the old cells, which are then separated from the bulk population by placing the culture near a magnet. Several rounds of magnetic sorting and resuspension allow the high-fidelity separation of old cells. Magnetic sorting is the most useful of the methods described here because it combines the purity of biotin-dependent sorting with the ability to rapidly sort large numbers of cells. Magnetic sorting can easily yield 108 old cells and has been used extensively to demonstrate a variety of age-associated molecular events (36, 114, 180).
Sucrose gradient centrifugation.
After only a few divisions, mother cells become substantially larger than the daughter cells they generate. To generate an age-matched population of cells, a saturated yeast culture can be separated on a 10 to 30% sucrose gradient, generating two distinct bands of cells, one of which is composed mostly of virgin cells (40). Virgin cells are synchronized in the cell cycle by exposure to mating pheromone and then allowed to proceed through as many as three divisions. Daughter cells are separated from mother cells by repeating the sucrose gradient step, except that aged rather than virgin cells are collected. By repeating cycles of synchronization, growth, and separation, it is possible to isolate a population of cells that are age matched up to 20 generations and are contaminated by less than 10% young cells. One drawback of this technique is that many rounds of manipulation are required to obtain old cells. However, it is advantageous because it allows large-scale isolation of a relatively pure population of cells of advanced age. Using this technique, researchers have been able to prepare old cells and use them to characterize various biomarkers of yeast aging (41).
Centrifugal elutriation.
A variation on the cell size-dependent separation scheme relies on centrifugal elutriation to continuously separate daughter cells from mother cells (214). This method allows the rapid, manipulation-free purification of age-matched, middle-aged yeast cells. Cells are grown in the chamber of an elutriation rotor and eluted under centrifugation. Daughter cells are collected and subjected to a second round of elutriation. During the second elutriation, daughter cells are discarded and mother cells are retained. Continuous growth and elution over 15 divisions can produce a population 10 generations or older with 70% purity. Like most techniques for isolating old cells, the purity of the final product is compromised at large numbers of divisions.
Membrane attachment.
One technique for separating mother cells from daughter cells has been described as a “baby machine” (67). Cells are attached to a membrane and allowed to divide. Mother cells remain attached to the membrane, but daughter cells are continuously eluted and collected by washing the membrane. Although this technique efficiently produces virgin cells, it is not used routinely due to the difficulty in obtaining and processing large numbers of old cells.
Isolation and Analysis of ERCs
Although ERCs are so abundant in old cells that they can be visualized on an EtBr-stained gel, they are typically quantified by Southern blotting (6, 7, 36, 88, 114, 117, 152). At least 5 × 106 cells isolated by magnetic sorting are used as starting material (180). To isolate ERCs, the cell wall is digested with lyticase and cells are lysed in dilute sodium dodecyl sulfate buffer. Lysis must be performed gently to avoid excessive shearing of linear genomic DNA. The DNA is cleaned by phenol extraction, precipitated, electrophoresed as uncut DNA, and probed by Southern blotting using an rDNA-specific probe. Two-dimensional chloroquine gel electrophoresis may be performed to determine the degree of ERC supercoiling (180).
CONCLUSIONS AND PERSPECTIVES
Although the use of S. cerevisiae as a model for aging research was first proposed nearly half a century ago, only in the last decade have the validity and power of this model been truly established. With the elucidation of the ERC mechanism of replicative aging, the identification of nearly two dozen longevity genes (Table 1), and the ever-emerging conservation of yeast longevity pathways in higher eukaryotes, S. cerevisiae has emerged as a highly informative and respected model for the study of metazoan longevity. Perhaps the primary reason that such conclusions remained dubious for so long was the inability to distinguish between aging, a nonadaptive by-product of natural selection, and longevity, a highly adaptive trait. In retrospect, it is no surprise that disparate organisms have retained common mechanisms that allow them to allocate limited resources in response to environmental stimuli and thus control the rate of aging.
Several areas of mammalian research have highlighted this conservation. Prop-1 and Pit-1 dwarf mice, which are deficient in serum growth hormone (GH), show decreased levels of IGF-1 and live up to 65% longer than do wild-type mice (21, 47). In support of the notion that this phenotype is the result of attenuated IGF-1 signaling is the fact that dwarf mice with high GH levels but only 10% of the normal IGF-1 levels also show a life span extension (34). Most recently, mice with a fat-specific insulin receptor knockout (FIRKO) also showed a significant life span extension compared to wild-type mice (18). Not surprisingly, the links between these signaling pathways and stress resistance appear to be conserved as well. Mice hepatocytes which are exposed to either GH or IGF-1 show a decrease in both superoxide dismutase and catalase activity as well as in the expression of heat shock proteins (22, 23, 174). Because of the many similarities between the yeast glucose and mammalian IGF-1 signaling pathways, it is highly likely that elucidation of the downstream targets and mechanisms which mediate life span extension in yeast will shed light on a number of critical processes in humans. Interestingly, high levels of IGF-1 have already been implicated in osteoarthritis and numerous forms of cancer including breast, lung, colorectal, and prostate cancers (177, 218).
In addition to the highly homologous RAS and serine/threonine kinase genes (SCH9 and Akt/PKB) found in the yeast glucose and mammalian IGF-1 signaling pathways, the functional conservation of Sir2 further emphasizes the critical importance of yeast as a model organism. While the initial identification of SIR2 as a yeast longevity gene may have seemed to be of little relevance to human aging, the subsequent implication of this gene in calorie restriction and the conservation of its effect on life span in higher organisms have brought Sir2 and its homologues to the forefront of aging research. Indeed, the discovery that human SIRT1 negatively modulates the activity of the p53 tumor suppressor not only underscores the importance of these enzymes but also highlights a conserved role for this family of enzymes as “survival genes.” In addition to its involvement in apoptosis and DNA repair, it is expected that SIRT1 and its homologues SIRT2 to SIRT7 play key regulatory roles in numerous other cellular processes. Finding the targets of the SIRT1 family and understanding how these enzymes are regulated in vivo are two of the many challenges facing the field as we explore the relevance of the yeast findings to mammalian biology. It will also be essential to determine the influence of NAD+ metabolism and the NAD+/NADH ratio on the regulation of all Sir2-like deacetylases.
Among other questions which need to be addressed in the near future, are those concerning the extent of overlap between yeast chronological and replicative life spans. In addition to the fact that mutations in the PKA pathway extend both types of life span, results from several other studies have revealed additional degrees of overlap. For example, the longer the period that yeast cells are maintained in stationary phase, the greater the decrease in their replicative life span once they are released (7). Furthermore, deletion of both SOD1 and SOD2 reduces both chronological and replicative life spans (106, 125). Elucidating the extent of commonality between these two aging paradigms will clearly be a significant asset to the field. Other specific questions include the mechanism by which nicotinamide inhibits Sir2, as well as the identification of other putative targets of nicotinamide regulation. Identification of genes involved in the retrograde response and elucidation of the mechanisms which tie this process to replicative life span will also be important. In the coming years, these and other questions will provide the driving force for researchers in the yeast aging field as the impact of this model organism continues to grow exponentially.
Acknowledgments
We thank members of the Sinclair laboratory for helpful discussions and manuscript preparation.
REFERENCES
- 1.Aguilaniu, H., L. Gustafsson, M. Rigoulet, and T. Nystrom. 2003. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science 299:1751-1753. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, R. M., K. J. Bitterman, J. G. Wood, O. Medvedik, H. Cohen, S. S. Lin, J. K. Manchester, J. I. Gordon, and D. A. Sinclair. 2002. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J. Biol. Chem. 277:18881-18890. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, R. M., K. J. Bitterman, J. G. Wood, O. Medvedik, and D. A. Sinclair. 2003. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature 423:181-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reference deleted.
- 5.Armstrong, C. M., M. Kaeberlein, S. I. Imai, and L. Guarente. 2002. Mutations in Saccharomyces cerevisiae gene SIR2 can have differential effects on in vivo silencing phenotypes and in vitro histone deacetylation activity. Mol. Biol. Cell 13:1427-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashrafi, K., S. S. Lin, J. K. Manchester, and J. I. Gordon. 2000. Sip2p and its partner snflp kinase affect aging in S. cerevisiae. Genes Dev. 14:1872-1885. [PMC free article] [PubMed] [Google Scholar]
- 7.Ashrafi, K., D. Sinclair, J. I. Gordon, and L. Guarente. 1999. Passage through stationary phase advances replicative aging in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 96:9100-9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Austriaco, N. R., Jr., and L. P. Guarente. 1997. Changes of telomere length cause reciprocal changes in the lifespan of mother cells in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:9768-9772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avalos, J. L., I. Celic, S. Muhammad, M. S. Cosgrove, J. D. Boeke, and C. Wolberger. 2002. Structure of a Sir2 enzyme bound to an acetylated p53 peptide. Mol. Cell 10:523-535. [DOI] [PubMed] [Google Scholar]
- 10.Bakker, B. M., K. M. Overkamp, A. J. van Maris, P. Kotter, M. A. Luttik, J. P. van Dijken, and J. T. Pronk. 2001. Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25:15-37. [DOI] [PubMed] [Google Scholar]
- 11.Barton, A. 1950. Some aspects of cell division in Saccharomyces cerevisiae. J. Gen. Microbiol. 4:84-86. [DOI] [PubMed] [Google Scholar]
- 12.Barz, W. P., and P. Walter. 1999. Two endoplasmic reticulum (ER) membrane proteins that facilitate ER-to-Golgi transport of glycosylphosphatidylinositol-anchored proteins. Mol. Biol. Cell 10:1043-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baur, J. A., Y. Zou, J. W. Shay, and W. E. Wright. 2001. Telomere position effect in human cells. Science 292:2075-2077. [DOI] [PubMed] [Google Scholar]
- 14.Beers, M., and R. Berkow. 2000. The Merck manual of geriatrics, 3rd ed. Merck Research Laboratories, Whitehouse Station, N.J.
- 15.Bernstein, B. E., J. K. Tong, and S. L. Schreiber. 2000. Genomewide studies of histone deacetylase function in yeast. Proc. Natl. Acad. Sci. USA 97:13708-13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bitterman, K. J., R. M. Anderson, H. Y. Cohen, M. Latorre-Esteves, and D. A. Sinclair. 2002. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J. Biol. Chem. 277:45099-45107. [DOI] [PubMed] [Google Scholar]
- 17.Blackburn, E. H. 2001. Switching and signaling at the telomere. Cell 106:661-673. [DOI] [PubMed] [Google Scholar]
- 18.Bluher, M., B. B. Kahn, and C. R. Kahn. 2003. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science 299:572-574. [DOI] [PubMed] [Google Scholar]
- 19.Borra, M. T., F. J. O'Neill, M. D. Jackson, B. Marshall, E. Verdin, K. R. Foltz, and J. M. Denu. 2002. Conserved enzymatic production and biological effect of O-acetyl-ADP-ribose by silent information regulator 2-like NAD+-dependent deacetylases. J. Biol. Chem. 277:12632-12641. [DOI] [PubMed] [Google Scholar]
- 20.Brachmann, C. B., J. M. Sherman, S. E. Devine, E. E. Cameron, L. Pillus, and J. D. Boeke. 1995. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 9:2888-2902. [DOI] [PubMed] [Google Scholar]
- 21.Brown-Borg, H. M., K. E. Borg, C. J. Meliska, and A. Bartke. 1996. Dwarf mice and the ageing process. Nature 384:33. [DOI] [PubMed] [Google Scholar]
- 22.Brown-Borg, H. M., and S. G. Rakoczy. 2000. Catalase expression in delayed and premature aging mouse models. Exp. Gerontol. 35:199-212. [DOI] [PubMed] [Google Scholar]
- 23.Brown-Borg, H. M., S. G. Rakoczy, M. A. Romanick, and M. A. Kennedy. 2002. Effects of growth hormone and insulin-like growth factor-1 on hepatocyte antioxidative enzymes. Exp. Biol. Med. 227:94-104. [DOI] [PubMed] [Google Scholar]
- 24.Bryk, M., M. Banerjee, M. Murphy, K. E. Knudsen, D. J. Garfinkel, and M. J. Curcio. 1997. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 11:255-269. [DOI] [PubMed] [Google Scholar]
- 25.Cabib, E., R. Ulane, and B. Bowers. 1974. A molecular model for morphogenesis: the primary septum of yeast. Curr. Top. Cell Regul. 8:1-32. [DOI] [PubMed] [Google Scholar]
- 26.Celenza, J. L., F. J. Eng, and M. Carlson. 1989. Molecular analysis of the SNF4 gene of Saccharomyces cerevisiae: evidence for physical association of the SNF4 protein with the SNF1 protein kinase. Mol. Cell. Biol. 9:5045-5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang, J. H., H. C. Kim, K. Y. Hwang, J. W. Lee, S. P. Jackson, S. D. Bell, and Y. Cho. 2002. Structural basis for the NAD-dependent deacetylase mechanism of Sir2. J. Biol. Chem. 277:34489-34498. [DOI] [PubMed] [Google Scholar]
- 28.Chang, K. T., and K. T. Min. 2002. Regulation of lifespan by histone deacetylase. Ageing Res. Rev. 1:313-326. [DOI] [PubMed] [Google Scholar]
- 29.Childress, A. M., D. S. Franklin, C. Pinswasdi, and S. Kale. 1996. LAG2, a gene that determines yeast longevity. Microbiology 142:2289-2297. [DOI] [PubMed] [Google Scholar]
- 30.Clancy, D. J., D. Gems, L. G. Harshman, S. Oldham, H. Stocker, E. Hafen, S. J. Leevers, and L. Partridge. 2001. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science 292:104-106. [DOI] [PubMed] [Google Scholar]
- 31.Cockell, M. M., and S. M. Gasser. 1999. The nucleolus: nucleolar space for RENT. Curr. Biol. 9:R575-R576. [DOI] [PubMed] [Google Scholar]
- 32.Cohen, H., and D. A. Sinclair. 2001. Recombination-mediated lengthening of terminal telomeric repeats requires the Sgs1 DNA helicase. Proc Natl Acad Sci USA 98:3174-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cong, Y. S., W. E. Wright, and J. W. Shay. 2002. Human telomerase and its regulation. Microbiol. Mol. Biol. Rev. 66:407-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coschigano, K. T., D. Clemmons, L. L. Bellush, and J. J. Kopchick. 2000. Assessment of growth parameters and life span of GHR/BP gene-disrupted mice. Endocrinology 141:2608-2613. [DOI] [PubMed] [Google Scholar]
- 35.Defossez, P. A., P. U. Park, and L. Guarente. 1998. Vicious circles: a mechanism for yeast aging. Curr. Opin. Microbiol. 1:707-711. [DOI] [PubMed] [Google Scholar]
- 36.Defossez, P. A., R. Prusty, M. Kaeberlein, S. J. Lin, P. Ferrigno, P. A. Silver, R. L. Keil, and L. Guarente. 1999. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol. Cell 3:447-455. [DOI] [PubMed] [Google Scholar]
- 37.DeVit, M. J., and M. Johnston. 1999. The nuclear exportin Msn5 is required for nuclear export of the Mig1 glucose repressor of Saccharomyces cerevisiae. Curr. Biol. 9:1231-1241. [DOI] [PubMed] [Google Scholar]
- 38.D'Mello N, P., A. M. Childress, D. S. Franklin, S. P. Kale, C. Pinswasdi, and S. M. Jazwinski. 1994. Cloning and characterization of LAG1, a longevity-assurance gene in yeast. J. Biol. Chem. 269:15451-15459. [PubMed] [Google Scholar]
- 39.Edwards, T. A., J. Trincao, C. R. Escalante, R. P. Wharton, and A. K. Aggarwal. 2000. Crystallization and characterization of Pumilo: a novel RNA binding protein. J. Struct. Biol. 132:251-254. [DOI] [PubMed] [Google Scholar]
- 40.Egilmez, N. K., J. B. Chen, and S. M. Jazwinski. 1990. Preparation and partial characterization of old yeast cells. J. Gerontol. 45:B9-B17. [DOI] [PubMed] [Google Scholar]
- 41.Egilmez, N. K., J. B. Chen, and S. M. Jazwinski. 1989. Specific alterations in transcript prevalence during the yeast life span. J. Biol. Chem. 264:14312-14317. [PubMed] [Google Scholar]
- 42.Egilmez, N. K., and S. M. Jazwinski. 1989. Evidence for the involvement of a cytoplasmic factor in the aging of the yeast Saccharomyces cerevisiae. J. Bacteriol. 171:37-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emanuelli, M., F. Carnevali, M. Lorenzi, N. Raffaelli, A. Amici, S. Ruggieri, and G. Magni. 1999. Identification and characterization of YLR328W, the Saccharomyces cerevisiae structural gene encoding NMN adenylyltransferase. Expression and characterization of the recombinant enzyme. FEBS Lett. 455:13-17. [DOI] [PubMed] [Google Scholar]
- 44.Fabrizio, P., L. L. Liou, V. N. Moy, A. Diaspro, J. SelverstoneValentine, E. B. Gralla, and V. D. Longo. 2003. SOD2 functions downstream of Sch9 to extend longevity in yeast. Genetics 163:35-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fabrizio, P., F. Pozza, S. D. Pletcher, C. M. Gendron, and V. D. Longo. 2001. Regulation of longevity and stress resistance by Sch9 in yeast. Science 292:288-290. [DOI] [PubMed] [Google Scholar]
- 46.Finnin, M. S., J. R. Donigian, and N. P. Pavletich. 2001. Structure of the histone deacetylase SIRT2. Nat. Struct. Biol. 8:621-625. [DOI] [PubMed] [Google Scholar]
- 47.Flurkey, K., J. Papaconstantinou, and D. E. Harrison. 2002. The Snell dwarf mutation Pit1(dw) can increase life span in mice. Mech. Ageing Dev. 123:121-130. [DOI] [PubMed] [Google Scholar]
- 48.Foster, J. W., Y. K. Park, T. Penfound, T. Fenger, and M. P. Spector. 1990. Regulation of NAD metabolism in Salmonella typhimurium: molecular sequence analysis of the bifunctional nadR regulator and the nadA-pnuC operon. J. Bacteriol. 172:4187-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fraenkel, D. G. 1985. On ras gene function in yeast. Proc. Natl. Acad. Sci. USA 82:4740-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frohlich, K. U., and F. Madeo. 2001. Apoptosis in yeast: a new model for aging research. Exp. Gerontol. 37:27-31. [DOI] [PubMed] [Google Scholar]
- 51.Ghidelli, S., D. Donze; N. Dhillon, and R. T. Kamakaka. 2001. Sir2p exists in two nucleosome-binding complexes with distinct deacetylase activities. EMBO J. 20:4522-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ghislain, M., E. Talla, and J. M. Francois. 2002. Identification and functional analysis of the Saccharomyces cerevisiae nicotinamidase gene, PNC1. Yeast 19:215-224. [DOI] [PubMed] [Google Scholar]
- 53.Gompertz, B. 1825. On the nature of the function expressive of the law of human mortality, and on a new mode of determining life contingencies. Philos. Trans. R. Soc. London Ser. B 115:513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gotta, M., S. Strahl-Bolsinger, H. Renauld, T. Laroche, B. K. Kennedy, M. Grunstein, and S. M. Gasser. 1997. Localization of Sir2p: the nucleolus as a compartment for silent information regulators. EMBO J. 16:3243-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gottschling, D. E., O. M. Aparicio, B. L. Billington, and V. A. Zakian. 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63:751-762. [DOI] [PubMed] [Google Scholar]
- 56.Grant, R. S., R. Passey, G. Matanovic, G. Smythe, and V. Kapoor. 1999. Evidence for increased de novo synthesis of NAD in immune-activated RAW264.7 macrophages: a self-protective mechanism? Arch. Biochem. Biophys. 372:1-7. [DOI] [PubMed] [Google Scholar]
- 57.Greider, C. W., and E. H. Blackburn. 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43:405-413. [DOI] [PubMed] [Google Scholar]
- 58.Greider, C. W., and E. H. Blackburn. 1987. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell 51:887-898. [DOI] [PubMed] [Google Scholar]
- 59.Grunstein, M. 1997. Histone acetylation in chromatin structure and transcription. Nature 389:349-352. [DOI] [PubMed] [Google Scholar]
- 60.Guarente, L. 2000. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 14:1021-1026. [PubMed] [Google Scholar]
- 61.Reference deleted.
- 62.Guarente, L., and C. Kenyon. 2000. Genetic pathways that regulate ageing in model organisms. Nature 408:255-262. [DOI] [PubMed] [Google Scholar]
- 63.Guillas, I., P. A. Kirchman, R. Chuard, M. Pfefferli, J. C. Jiang, S. M. Jazwinski, and A. Conzelmann. 2001. C26-CoA-dependent ceramide synthesis of Saccharomyces cerevisiae is operated by Lag1p and Lac1p. EMBO J. 20:2655-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harman, D. 1956. Atheory based on free radical and radiation chemistry. J. Gerontol. 11:298-304. [DOI] [PubMed] [Google Scholar]
- 65.Hayflick, L., and P. S. Mooshead. 1961. The limited in vitro lifetime of human diploid fibroblasts. Exp. Cell Res. 25:585-621. [Google Scholar]
- 66.Hecht, A., S. Strahl Bolsingers, and M. Grunstein. 1996. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature 383:92-96. [DOI] [PubMed] [Google Scholar]
- 67.Helmstetter, C. E. 1991. Description of a baby machine for Saccharomyces cerevisiae. New Biol 3:1089-1096. [PubMed] [Google Scholar]
- 68.Heo, S. J., K. Tatebayashi, I. Ohsugi, A. Shimamoto, Y. Furuichi, and H. Ikeda. 1999. Bloom's syndrome gene suppresses premature ageing caused by Sgs1 deficiency in yeast. Genes Cells 4:619-625. [DOI] [PubMed] [Google Scholar]
- 69.Huang, P., F. E. Pryde, D. Lester, R. L. Maddison, R. H. Borts, I. D. Hickson, and E. J. Louis. 2001. SGS1 is required for telomere elongation in the absence of telomerase. Curr. Biol. 11:125-129. [DOI] [PubMed] [Google Scholar]
- 70.Hubbard, E. J., X. L. Yang, and M. Carlson. 1992. Relationship of the cAMP-dependent protein kinase pathway to the SNF1 protein kinase and invertase expression in Saccharomyces cerevisiae. Genetics 130:71-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hughes, K. T., B. M. Olivera, and J. R. Roth. 1988. Structural gene for NAD synthetase in Salmonella typhimurium. J. Bacteriol. 170:2113-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Imai, S., C. M. Armstrong, M. Kaeberlein, and L. Guarente. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795-800. [DOI] [PubMed] [Google Scholar]
- 73.Jackson, M. D., and J. M. Denu. 2002. Structural Identification of 2′- and 3′-O-acetyl-ADP-ribose as novel metabolites derived from the Sir2 family of beta-NAD+-dependent histone/protein deacetylases. J. Biol. Chem. 277:18535-18544. [DOI] [PubMed] [Google Scholar]
- 74.Jazwinski, S. M. 2000. Metabolic control and gene dysregulation in yeast aging. Ann. N. Y. Acad. Sci. 908:21-30. [DOI] [PubMed] [Google Scholar]
- 75.Jazwinski, S. M., J. B. Chen, and J. Sun. 1993. A single gene change can extend yeast life span: the role of Ras in cellular senescence. Adv. Exp. Med. Biol. 330:45-53. [DOI] [PubMed] [Google Scholar]
- 76.Jazwinski, S. M., N. K. Egilmez, and J. B. Chen. 1989. Replication control and cellular life span. Exp. Gerontol. 24:423-436. [DOI] [PubMed] [Google Scholar]
- 77.Jia, Y., B. Rothermel, J. Thornton, and R. A. Butow. 1997. A basic helix-loop-helix leucine zipper transcription complex in yeast functions in a signaling pathway from mitochondria to the nucleus. Mol. Cell. Biol. 17:1110-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jiang, J. C., E. Jaruga, M. V. Repnevskaya, and S. M. Jazwinski. 2000. An intervention resembling caloric restriction prolongs life span and retards aging in yeast. Faseb J. 14:2135-2135. [DOI] [PubMed] [Google Scholar]
- 79.Jiang, J. C., J. Wawryn, H. M. Shantha Kumara, and S. M. Jazwinski. 2002. Distinct roles of processes modulated by histone deacetylases Rpd3p, Hdalp, and Sir2p in life extension by caloric restriction in yeast. Exp. Gerontol. 37:1023-1030. [DOI] [PubMed] [Google Scholar]
- 80.Jiang, R., and M. Carlson. 1996. Glucose regulates protein interactions within the yeast SNF1 protein kinase complex. Genes Dev. 10:3105-3115. [DOI] [PubMed] [Google Scholar]
- 81.Jiang, R., and M. Carlson. 1997. The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol. Cell. Biol. 17:2099-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Johnson, F. B., R. A. Marciniak, M. McVey, S. A. Stewart, W. C. Hahn, and L. Guarente. 2001. The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J. 20:905-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reference deleted.
- 84.Johnson, T. E. 1990. Increased life-span of age-1 mutants in Caenorhabditis elegans and lower Gompertz rate of aging. Science 249:908-912. [DOI] [PubMed] [Google Scholar]
- 85.Johnston, J. R. 1966. Reproductive capacity and mode of death of yeast cells. Antonie Leeuwenhoek 32:94-98. [DOI] [PubMed] [Google Scholar]
- 86.Johzuka, K., and T. Horiuchi. 2002. Replication fork block protein, Fob1, acts as an rDNA region specific recombinator in S. cerevisiae. Genes Cells 7:99-113. [DOI] [PubMed] [Google Scholar]
- 87.Reference deleted.
- 88.Kaeberlein, M., M. McVey, and L. Guarente. 1999. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13:2570-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kandel, E. S., and N. Hay. 1999. The regulation and activities of the multifunctional serine/threonine kinase Akt/PKB. Exp. Cell Res. 253:210-229. [DOI] [PubMed] [Google Scholar]
- 90.Kassem, H., V. Sangar, R. Cowan, N. Clarke, and G. P. Margison. 2002. A potential role of heat shock proteins and nicotinamide N-methyl transferase in predicting response to radiation in bladder cancer. Int. J. Cancer 101:454-460. [DOI] [PubMed] [Google Scholar]
- 91.Kennedy, B. K., N. R. Austriaco, Jr., and L. Guarente. 1994. Daughter cells of Saccharomyces cerevisiae from old mothers display a reduced life span. J. Cell Biol. 127:1985-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kennedy, B. K., N. R. Austriaco, Jr., J. Zhang, and L. Guarente. 1995. Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell 80:485-496. [DOI] [PubMed] [Google Scholar]
- 93.Kennedy, B. K., M. Gotta, D. A. Sinclair, K. Mills, D. S. McNabb, M. Murthy, S. M. Pak, T. Laroche, S. M. Gasser, and L. Guarente. 1997. Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. Cell 89:381-391. [DOI] [PubMed] [Google Scholar]
- 94.Kenyon, C. 2001. A conserved regulatory mechanism for aging. Cell 105:165-168. [DOI] [PubMed] [Google Scholar]
- 95.Kenyon, C., J. Chang, E. Gensch, A. Rudner, and R. Tabtiang. 1993. A C. elegans mutant that lives twice as long as wild type. Nature 366:461-464. [DOI] [PubMed] [Google Scholar]
- 96.Kim, S., A. Benguria, C. Y. Lai, and S. M. Jazwinski. 1999. Modulation of life-span by histone deacetylase genes in Saccharomyces cerevisiae. Mol. Biol. Cell 10:3125-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kim, S., B. Villeponteau, and S. M. Jazwinski. 1996. Effect of replicative age on transcriptional silencing near telomeres in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 219:370-376. [DOI] [PubMed] [Google Scholar]
- 98.Kimura, K. D., H. A. Tissenbaum, Y. Liu, and G. Ruvkun. 1997. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277:942-946. [DOI] [PubMed] [Google Scholar]
- 99.Kirchman, P. A., S. Kim, C. Y. Lai, and S. M. Jazwinski. 1999. Interorganelle signaling is a determinant of longevity in Saccharomyces cerevisiae. Genetics 152:179-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kirkwood, T. B., and R. Holliday. 1979. The evolution of ageing and longevity. Proc. R. Soc. London B Ser. 205:531-546. [DOI] [PubMed] [Google Scholar]
- 101.Kirkwood, T. L., P. Kapahi, and D. P. Shanley. 2000. Evolution, stress, and longevity. J. Anat. 197:587-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kobayashi, T., D. J. Heck, M. Nomura, and T. Horiuchi. 1998. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 12:3821-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lal, A., A. E. Lash, S. F. Altschul, V. Velculescu, L. Zhang, R. E. McLendon, M. A. Marra, C. Prange, P. J. Morin, K. Polyak, N. Papadopoulos, B. Vogelstein, K. W. Kinzler, R. L. Strausberg, and G. J. Riggins. 1999. A public database for gene expression in human cancers. Cancer Res. 59:5403-5407. [PubMed] [Google Scholar]
- 104.Landry, J., J. T. Slama, and R. Sternglanz. 2000. Role of NAD(+) in the deacetylase activity of the SIR2-like proteins. Biochem. Biophys. Res. Commun. 278:685-690. [DOI] [PubMed] [Google Scholar]
- 105.Landry, J., A. Sutton, S. T. Tafrov, R. C. Heller, J. Stebbins, L. Pillus, and R. Sternglanz. 2000. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. USA 97:5807-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Laun, P., A. Pichova, F. Madeo, J. Fuchs, A. Ellinger, S. Kohlwein, I. Dawes, K. U. Frohlich, and M. Breitenbach. 2001. Aged mother cells of Saccharomyces cerevisiae show markers of oxidative stress and apoptosis. Mol. Microbiol. 39:1166-1173. [PubMed] [Google Scholar]
- 107.Lebel, M. 2001. Werner syndrome: genetic and molecular basis of a premature aging disorder. Cell. Mol. Life Sci. 58:857-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lee, S. E., F. Paques, J. Sylvan, and J. E. Haber. 1999. Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr. Biol. 9:767-770. [DOI] [PubMed] [Google Scholar]
- 109.Lee, S. S., R. Y. Lee, A. G. Fraser, R. S. Kamath, J. Ahringer, and G. Ruvkun. 2003. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat. Genet. 33:40-48. [DOI] [PubMed] [Google Scholar]
- 110.Lendvay, T. S., D. K. Morris, J. Sah, B. Balasubramanian, and V. Lundblad. 1996. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 144:1399-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liao, X., and R. A. Butow. 1993. RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell 72:61-71. [DOI] [PubMed] [Google Scholar]
- 112.Liao, X. S., W. C. Small, P. A. Srere, and R. A. Butow. 1991. Intramitochondrial functions regulate nonmitochondrial citrate synthase (CIT2) expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:38-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lillie, S. H., and J. R. Pringle. 1980. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J. Bacteriol. 143:1384-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lin, S. J., P. A. Defossez, and L. Guarente. 2000. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289:2126-2128. [DOI] [PubMed] [Google Scholar]
- 115.Lin, S. J., M. Kaeberlein, A. A. Andalis, L. A. Sturtz, P. A. Defossez, V. C. Culotta, G. R. Fink, and L. Guarente. 2002. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature 418:344-348. [DOI] [PubMed] [Google Scholar]
- 116.Lin, S. S., J. K. Manchester, and J. I. Gordon. 2001. Enhanced gluconeogenesis and increased energy storage as hallmarks of aging in Saccharomyces cerevisiae. J. Biol. Chem. 276:36000-36007. [DOI] [PubMed] [Google Scholar]
- 117.Lin, S. S., J. K. Manchester, and J. I. Gordon. 2003. Sip2, an N-myristoylated beta subunit of Snf1 kinase, regulates aging in Saccharomyces cerevisiae by affecting cellular histone kinase activity, recombination at rDNA loci, and silencing. J. Biol. Chem. 278:13390-13397. [DOI] [PubMed] [Google Scholar]
- 118.Lin, Y. H., and R. L. Keil. 1991. Mutations affecting RNA polymerase I-stimulated exchange and rDNA recombination in yeast. Genetics 127:31-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liu, Z., and R. A. Butow. 1999. A transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in response to a reduction or loss of respiratory function. Mol. Cell. Biol. 19:6720-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Llorente, B., and B. Dujon. 2000. Transcriptional regulation of the Saccharomyces cerevisiae DAL5 gene family and identification of the high affinity nicotinic acid permease TNA1 (YGR260w). FEBS Lett. 475:237-241. [DOI] [PubMed] [Google Scholar]
- 121.Lo, W. S., L. Duggan, N. C. Tolga, Emre, R. Belotserkovskya, W. S. Lane, R. Shiekhattar, and S. L. Berger. 2001. Snf1—a histone kinase that works in concert with the histone acetyltransferase Gen5 to regulate transcription. Science 293:1142-1146. [DOI] [PubMed] [Google Scholar]
- 122.Longo, V. D. 1999. Mutations in signal transduction proteins increase stress resistance and longevity in yeast, nematodes, fruit flies, and mammalian neuronal cells. Neurobiol. Aging 20:479-486. [DOI] [PubMed] [Google Scholar]
- 123.Longo, V. D., L. M. Ellerby, D. E. Bredesen, J. S. Valentine, and E. B. Gralla. 1997. Human Bcl-2 reverses survival defects in yeast lacking superoxide dismutase and delays death of wild-type yeast. J. Cell Biol. 137:1581-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Longo, V. D., and P. Fabrizio. 2002. Regulation of longevity and stress resistance: a molecular strategy conserved from yeast to humans? Cell. Mol. Life Sci. 59:903-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Longo, V. D., E. B. Gralla, and J. S. Valentine. 1996. Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae. Mitochondrial production of toxic oxygen species in vivo. J. Biol. Chem. 271:12275-12280. [DOI] [PubMed] [Google Scholar]
- 126.Longo, V. D., L. L. Liou, J. S. Valentine, and E. B. Gralla. 1999. Mitochondrial superoxide decreases yeast survival in stationary phase. Arch. Biochem. Biophys. 365:131-142. [DOI] [PubMed] [Google Scholar]
- 127.Loo, S., and J. Rine. 1995. Silencing and heritable domains of gene expression. Annu. Rev. Cell Dev. Biol. 11:519-548. [DOI] [PubMed] [Google Scholar]
- 128.Luo, J., A. Y. Nikolaev, S. Imai, D. Chen, F. Su, A. Shiloh, L. Guarente, and W. Gu. 2001. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 107:137-148. [DOI] [PubMed] [Google Scholar]
- 129.MacLean, M., N. Harris, and P. W. Piper. 2001. Chronological lifespan of stationary phase yeast cells; a model for investigating the factors that might influence the ageing of postmitotic tissues in higher organisms. Yeast 18:499-509. [DOI] [PubMed] [Google Scholar]
- 130.Madeo, F., E. Frohlich, M. Ligr, M. Grey, S. J. Sigrist, D. H. Wolf, and K. U. Frohlich. 1999. Oxygen stress: a regulator of apoptosis in yeast. J. Cell Biol. 145:757-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Madeo, F., E. Herker, C. Maldener, S. Wissing, S. Lachelt, M. Herlan, M. Fehr, K. Lauber, S. J. Sigrist, S. Wesselborg, and K. U. Frohlich. 2002. A caspase-related protease regulates apoptosis in yeast. Mol. Cell 9:911-917. [DOI] [PubMed] [Google Scholar]
- 132.Masoro, E. J. 2000. Caloric restriction and aging: an update. Exp. Gerontol. 35:299-305. [DOI] [PubMed] [Google Scholar]
- 133.Matsuyama, S., Q. Xu, J. Velours, and J. C. Reed. 1998. The mitochondrial F0F1-ATPase proton pump is required for function of the proapoptotic protein Bax in yeast and mammalian cells. Mol. Cell 1:327-336. [DOI] [PubMed] [Google Scholar]
- 134.McAinsh, A. D., S. Scott-Drew, J. A. Murray, and S. P. Jackson. 1999. DNA damage triggers disruption of telomeric silencing and Mec1p-dependent relocation of Sir3p. Curr. Biol. 9:963-966. [DOI] [PubMed] [Google Scholar]
- 135.McCay, C. M., L. A. Maynard, G. Sperling, and L. L. Barnes. 1975. Retarded growth, life span, ultimate body size and age changes in the albino rat after feeding diets restricted in calories. Nutr. Rev. 33:241-243. [Reprint of J. Nutr. 18:1-13, 1939.] [DOI] [PubMed] [Google Scholar]
- 136.McVey, M., M. Kaeberlein, H. A. Tissenbaum, and L. Guarente. 2001. The short life span of Saccharomyces cerevisiae sgs1 and srs2 mutants is a composite of normal aging processes and mitotic arrest due to defective recombination. Genetics 157:1531-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Melese, T., and Z. Xue. 1995. The nucleolus: an organelle formed by the act of building a ribosome. Curr. Opin. Cell Biol. 7:319-324. [DOI] [PubMed] [Google Scholar]
- 138.Melov, S., J. A. Schneider, B. J. Day, D. Hinerfeld, P. Coskun, S. S. Mirra, J. D. Crapo, and D. C. Wallace. 1998. A novel neurological phenotype in mice lacking mitochondrial manganese superoxide dismutase. Nat. Genet. 18:159-163. [DOI] [PubMed] [Google Scholar]
- 139.Miller, C. A., and D. Kowalski. 1993. cis-acting components in the replication origin from ribosomal DNA of Saccharomyces cerevisiae. Mol. Cell. Biol. 13:5360-5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mills, K. D., D. A. Sinclair, and L. Guarente. 1999. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell 97:609-620. [DOI] [PubMed] [Google Scholar]
- 141.Min, J., J. Landry, R. Sternglanz, and R. M. Xu. 2001. Crystal structure of a SIR2 homolog-NAD complex. Cell 105:269-279. [DOI] [PubMed] [Google Scholar]
- 142.Moazed, D. 2001. Common themes in mechanisms of gene silencing. Mol. Cell 8:489-498. [DOI] [PubMed] [Google Scholar]
- 143.Moazed, D. 2001. Enzymatic activities of Sir2 and chromatin silencing. Curr. Opin. Cell Biol. 13:232-238. [DOI] [PubMed] [Google Scholar]
- 144.Morris, J. Z., H. A. Tissenbaum, and G. Ruvkun. 1996. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature 382:536-539. [DOI] [PubMed] [Google Scholar]
- 145.Mortimer, R. K., and J. R. Johnston. 1959. Life span of individual yeast cells. Nature 183:1751-1752. [DOI] [PubMed] [Google Scholar]
- 146.Muiras, M. L. 2003. Mammalian longevity under the protection of PARP-1's multi-facets. Ageing Res. Rev. 2:129-148. [DOI] [PubMed] [Google Scholar]
- 147.Muller, I. 1985. Parental age and the life-span of zygotes of Saccharomyces cerevisiae. Antonie Leeuwenhoek 51:1-10. [DOI] [PubMed] [Google Scholar]
- 148.Olovnikov, A. M. 1971. Principle of marginotomy in template synthesis of polynucleotides. Dokl. Akad. Nauk. SSSR 201:1496-1499. (In Russian.) [PubMed] [Google Scholar]
- 149.Onyango, P., I. Celic, J. M. McCaffery, J. D. Boeke, and A. P. Feinberg. 2002. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc. Natl. Acad. Sci. USA 99:13653-13658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Oshima, J. 2000. The Werner syndrome protein: an update. Bioessays 22:894-901. [DOI] [PubMed] [Google Scholar]
- 151.Ozcan, S., and M. Johnston. 1999. Function and regulation of yeast hexose transporters. Microbiol. Mol. Biol. Rev. 63:554-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Park, P. U., P. A. Defossez, and L. Guarente. 1999. Effects of mutations in DNA repair genes on formation of ribosomal DNA circles and life span in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:3848-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Pedruzzi, I., N. Burckert, P. Egger, and C. De Virgilio. 2000. Saccharomyces cerevisiae Ras/cAMP pathway controls post-diauxic shift element-dependent transcription through the zinc finger protein Gis1. EMBO J. 19:2569-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pletcher, S. D., S. J. Macdonald, R. Marguerie, U. Certa, S. C. Stearns, D. B. Goldstein, and L. Partridge. 2002. Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Curr. Biol. 12:712-723. [DOI] [PubMed] [Google Scholar]
- 155.Pronk, J. T., H. Yde Steensma, and J. P. Van Dijken. 1996. Pyruvate metabolism in Saccharomyces cerevisiae. Yeast 12:1607-1633. [DOI] [PubMed] [Google Scholar]
- 156.Rafty, L. A., M. T. Schmidt, A. L. Perraud, A. M. Scharenberg, and J. M. Denu. 2002. Analysis of O-acetyl-ADP-ribose as a target for nudix ADP-ribose hydrolases. J. Biol. Chem. 277:47114-47122. [DOI] [PubMed] [Google Scholar]
- 157.Ray, A., R. E. Hector, N. Roy, J. H. Song, K. L. Berkner, and K. W. Runge. 2003. Sir3p phosphorylation by the Slt2p pathway effects redistribution of silencing function and shortened lifespan. Nat. Genet. 33:522-526. [DOI] [PubMed] [Google Scholar]
- 158.Ritchie, K. B., J. C. Mallory, and T. D. Petes. 1999. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 19:6065-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rogina, B., S. L. Helfand, and S. Frankel. 2002. Longevity regulation by Drosophila Rpd3 deacetylase and caloric restriction. Science 298:1745. [DOI] [PubMed] [Google Scholar]
- 160.Rolland, F., J. H. De Winde, K. Lemaire, E. Boles, J. M. Thevelein, and J. Winderickx. 2000. Glucose-induced cAMP signalling in yeast requires both a G-protein coupled receptor system for extracellular glucose detection and a separable hexose kinase-dependent sensing process. Mol. Microbiol. 38:348-358. [DOI] [PubMed] [Google Scholar]
- 161.Rolland, F., J. Winderickx, and J. M. Thevelein. 2001. Glucose-sensing mechanisms in eukaryotic cells. Trends Biochem. Sci. 26:310-317. [DOI] [PubMed] [Google Scholar]
- 162.Rossmann, M. G., and P. Argos. 1978. The taxonomy of binding sites in proteins. Mol. Cell. Biochem. 21:161-182. [DOI] [PubMed] [Google Scholar]
- 163.Roy, N., and K. W. Runge. 2000. Two paralogs involved in transcriptional silencing that antagonistically control yeast life span. Curr. Biol. 10:111-114. [DOI] [PubMed] [Google Scholar]
- 164.Rundlett, S. E., A. A. Carmen, R. Kobayashi, S. Bavykin, B. M. Turner, and M. Grunstein. 1996. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. USA 93:14503-14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sandmeier, J. J., I. Celic, J. D. Boeke, and J. S. Smith. 2002. Telomeric and rDNA silencing in Saccharomyces cerevisiae are dependent on a nuclear NAD(+) salvage pathway. Genetics 160:877-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sandmeier, J. J., S. French, Y. Osheim, W. L. Cheung, C. M. Gallo, A. L. Beyer, and J. S. Smith. 2002. RPD3 is required for the inactivation of yeast ribosomal DNA genes in stationary phase. EMBO J. 21:4959-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sauve, A. A., I. Celic, J. Avalos, H. Deng, J. D. Boeke, and V. L. Schramm. 2001. Chemistry of gene silencing: the mechanism of NAD+-dependent deacetylation reactions. Biochemistry 40:15456-15463. [DOI] [PubMed] [Google Scholar]
- 168.Schimmang, T., D. Tollervey, H. Kern, R. Frank, and E. C. Hurt. 1989. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J. 8:4015-4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Schneider, E. L., and Y. Mitsui. 1976. The relationship between in vitro cellular aging and in vivo human age. Proc. Natl. Acad. Sci. USA 73:3584-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Schwer, B., B. J. North, R. A. Frye, M. Ott, and E. Verdin. 2002. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J. Cell Biol. 158:647-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sekito, T., J. Thornton, and R. A. Butow. 2000. Mitochondria-to-nuclear signaling is regulated by the subcellular localization of the transcription factors Rtg1p and Rtg3p. Mol. Biol. Cell 11:2103-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Shaham, S., M. A. Shuman, and I. Herskowitz. 1998. Death-defying yeast identify novel apoptosis genes. Cell 92:425-427. [DOI] [PubMed] [Google Scholar]
- 173.Shama, S., C. Y. Lai, J. M. Antoniazzi, J. C. Jiang, and S. M. Jazwinski. 1998. Heat stress-induced life span extension in yeast. Exp. Cell Res. 245:379-388. [DOI] [PubMed] [Google Scholar]
- 174.Sharma, H. S., F. Nyberg, T. Gordh, P. Alm, and J. Westman. 2000. Neurotrophic factors influence upregulation of constitutive isoform of heme oxygenase and cellular stress response in the spinal cord following trauma. An experimental study using immunohistochemistry in the rat. Amino Acids 19:351-361. [PubMed] [Google Scholar]
- 175.Shelton, D. N., E. Chang, P. S. Whittier, D. Choi, and W. D. Funk. 1999. Microarray analysis of replicative senescence. Curr. Biol. 9:939-945. [DOI] [PubMed] [Google Scholar]
- 176.Shou, W., K. M. Sakamoto, J. Keener, K. W. Morimoto, E. E. Traverso, R. Azzam, G. J. Hoppe, R. M. R. Feldman, J. DeModena, D. Moazed, H. Charbonneaux, M. Nomura, and R. J. Deshaies. 2001. Net1 stimulates RNA polymerase I transcription and regulates nucleolar structure independently of controlling mitotic exit. Mol. Cell 8:45-55. [DOI] [PubMed] [Google Scholar]
- 177.Silberberg, R. 1972. Articular aging and osteoarthrosis in dwarf mice. Pathol. Microbiol. (Basel) 38:417-430. [DOI] [PubMed] [Google Scholar]
- 178.Sinclair, D., K. Mills, and L. Guarente. 1998. Aging in Saccharomyces cerevisiae. Annu. Rev. Microbiol. 52:533-560. [DOI] [PubMed] [Google Scholar]
- 179.Reference deleted.
- 180.Sinclair, D. A., and L. Guarente. 1997. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell 91:1033-1042. [DOI] [PubMed] [Google Scholar]
- 181.Sinclair, D. A., K. Mills, and L. Guarente. 1997. Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science 277:1313-1316. [DOI] [PubMed] [Google Scholar]
- 182.Sinclair, D. A., K. Mills, and L. Guarente. 1998. Molecular mechanisms of yeast aging. Trends Biochem. Sci. 23:131-134. [DOI] [PubMed] [Google Scholar]
- 183.Singer, M. S., and D. E. Gottschling. 1994. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266:404-409. [DOI] [PubMed] [Google Scholar]
- 184.Skulachev, V. P. 2002. Programmed death in yeast as adaptation? FEBS Lett. 528:23-26. [DOI] [PubMed] [Google Scholar]
- 185.Sloat, B. F., A. Adams, and J. R. Pringle. 1981. Roles of the CDC24 gene product in cellular morphogenesis during the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 89:395-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Sloat, B. F., and J. R. Pringle. 1978. A mutant of yeast defective in cellular morphogenesis. Science 200:1171-1173. [DOI] [PubMed] [Google Scholar]
- 187.Smeal, T., J. Claus, B. Kennedy, F. Cole, and L. Guarente. 1996. Loss of transcriptional silencing causes sterility in old mother cells of S. cerevisiae. Cell 84:633-642. [DOI] [PubMed] [Google Scholar]
- 188.Smith, J. S., and J. D. Boeke. 1997. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 11:241-254. [DOI] [PubMed] [Google Scholar]
- 189.Smith, J. S., C. B. Brachmann, I. Celic, M. A. Kenna, S. Muhammad, V. J. Starai, J. L. Avalos, J. C. Escalante-Semerena, C. Grubmeyer, C. Wolberger, and J. D. Boeke. 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. USA 97:6658-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Smith, J. S., C. B. Brachmann, L. Pillus, and J. D. Boeke. 1998. Distribution of a limited Sir2 protein pool regulates the strength of yeast rDNA silencing and is modulated by Sir4p. Genetics 149:1205-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Stephen, D. W., S. L. Rivers, and D. J. Jamieson. 1995. The role of the YAP1 and YAP2 genes in the regulation of the adaptive oxidative stress responses of Saccharomyces cerevisiae. Mol. Microbiol. 16:415-423. [DOI] [PubMed] [Google Scholar]
- 192.Strahl-Bolsinger, S., A. Hecht, K. Luo, and M. Grunstein. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11:83-93. [DOI] [PubMed] [Google Scholar]
- 193.Struhl, K. 1998. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12:599-606. [DOI] [PubMed] [Google Scholar]
- 194.Sun, J., S. P. Kale, A. M. Childress, C. Pinswasdi, and S. M. Jazwinski. 1994. Divergent roles of RAS1 and RAS2 in yeast longevity. J. Biol. Chem. 269:18638-18645. [PubMed] [Google Scholar]
- 195.Swiecilo, A., Z. Krawiec, J. Wawryn, G. Bartosz, and T. Bilinski. 2000. Effect of stress on the life span of the yeast Saccharomyces cerevisiae. Acta Biochim. Pol. 47:355-364. [PubMed] [Google Scholar]
- 196.Tanner, K. G., J. Landry, R. Sternglanz, and J. M. Denu. 2000. Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc. Natl. Acad. Sci. USA 97:14178-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Tanny, J. C., G. J. Dowd, J. Huang, H. Hilz, and D. Moazed. 1999. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell 99:735-745. [DOI] [PubMed] [Google Scholar]
- 198.Tanny, J. C., and D. Moazed. 2001. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: evidence for acetyl transfer from substrate to an NAD breakdown product. Proc. Natl. Acad. Sci. USA 98:415-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Tanny, J. C., and D. Moazed. 2002. Recognition of acetylated proteins: lessons from an ancient family of enzymes. Structure 10:1290-1292. [DOI] [PubMed] [Google Scholar]
- 200.Tatar, M., A. Kopelman, D. Epstein, M. P. Tu, C. M. Yin, and R. S. Garofalo. 2001. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science 292:107-110. [DOI] [PubMed] [Google Scholar]
- 201.Tatchell, K., L. C. Robinson, and M. Breitenbach. 1985. RAS2 of Saccharomyces cerevisiae is required for gluconeogenic growth and proper response to nutrient limitation. Proc. Natl. Acad. Sci. USA 82:3785-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Tissenbaum, H. A., and L. Guarente. 2001. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410:227-230. [DOI] [PubMed] [Google Scholar]
- 203.Tsang, A. W., and J. C. Escalante-Semerena. 1998. CobB, a new member of the SIR2 family of eucaryotic regulatory proteins, is required to compensate for the lack of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase activity in cobT mutants during cobalamin biosynthesis in Salmonella typhimurium LT2. J. Biol. Chem. 273:31788-31794. [DOI] [PubMed] [Google Scholar]
- 204.Tsukamoto, Y., J. Kato, and H. Ikeda. 1997. Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature 388:900-903. [DOI] [PubMed] [Google Scholar]
- 205.Vannier, D., D. Balderes, and D. Shore. 1996. Evidence that the transcriptional regulators SIN3 and RPD3, and a novel gene (SDS3) with similar functions, are involved in transcriptional silencing in S. cerevisiae. Genetics 144:1343-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Vaziri, H., S. K. Dessain, E. N. Eaton, S. I. Imai, R. A. Frye, T. K. Pandita, L. Guarente, and R. A. Weinberg. 2001. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107:149-159. [DOI] [PubMed] [Google Scholar]
- 207.Virag, L., and C. Szabo. 2002. The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol. Rev. 54:375-429. [DOI] [PubMed] [Google Scholar]
- 208.Vogelauer, M., L. Rubbi, I. Lucas, B. J. Brewer, and M. Grunstein. 2002. Histone acetylation regulates the time of replication origin firing. Mol. Cell 10:1223-1233. [DOI] [PubMed] [Google Scholar]
- 209.Vu, L., I. Siddiqi, B. S. Lee, C. A. Josaitis, and M. Nomura. 1999. RNA polymerase switch in transcription of yeast rDNA: role of transcription factor UAF (upstream activation factor) in silencing rDNA transcription by RNA polymerase II. Proc. Natl. Acad. Sci. USA 96:4390-4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Watson, J. D. 1972. Origin of concatemeric T7 DNA. Nat. New Biol. 239:197-201. [DOI] [PubMed] [Google Scholar]
- 211.Werner-Washburne, M., E. Braun, G. C. Johnston, and R. A. Singer. 1993. Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol. Rev. 57:383-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Werner-Washburne, M., E. L. Braun, M. E. Crawford, and V. M. Peck. 1996. Stationary phase in Saccharomyces cerevisiae. Mol. Microbiol. 19:1159-1166. [DOI] [PubMed] [Google Scholar]
- 213.Wilson, W. A., S. A. Hawley, and D. G. Hardie. 1996. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr. Biol. 6:1426-1434. [DOI] [PubMed] [Google Scholar]
- 214.Woldringh, C. L., K. Fluiter, and P. G. Huls. 1995. Production of senescent cells of Saccharomyces cerevisiae by centrifugal elutriation. Yeast 11:361-369. [DOI] [PubMed] [Google Scholar]
- 215.Wood, J. G., and D. A. Sinclair. 2002. TPE or not TPE? It's no longer a question. Trends Pharmacol. Sci. 23:1-4. [DOI] [PubMed] [Google Scholar]
- 216.Wright, W. E., and J. W. Shay. 1992. Telomere positional effects and the regulation of cellular senescence. Trends Genet. 8:193-197. [DOI] [PubMed] [Google Scholar]
- 217.Yan, L. J., R. L. Levine, and R. S. Sohal. 1997. Oxidative damage during aging targets mitochondrial aconitase. Proc. Natl. Acad. Sci. USA 94:11168-11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Yu, H., and T. Rohan. 2000. Role of the insulin-like growth factor family in cancer development and progression. J. Natl. Cancer Inst. 92:1472-1489. [DOI] [PubMed] [Google Scholar]