Abstract
Candida albicans is the most common fungal pathogen of humans and has developed an extensive repertoire of putative virulence mechanisms that allows successful colonization and infection of the host under suitable predisposing conditions. Extracellular proteolytic activity plays a central role in Candida pathogenicity and is produced by a family of 10 secreted aspartyl proteinases (Sap proteins). Although the consequences of proteinase secretion during human infections is not precisely known, in vitro, animal, and human studies have implicated the proteinases in C. albicans virulence in one of the following seven ways: (i) correlation between Sap production in vitro and Candida virulence, (ii) degradation of human proteins and structural analysis in determining Sap substrate specificity, (iii) association of Sap production with other virulence processes of C. albicans, (iv) Sap protein production and Sap immune responses in animal and human infections, (v) SAP gene expression during Candida infections, (vi) modulation of C. albicans virulence by aspartyl proteinase inhibitors, and (vii) the use of SAP-disrupted mutants to analyze C. albicans virulence. Sap proteins fulfill a number of specialized functions during the infective process, which include the simple role of digesting molecules for nutrient acquisition, digesting or distorting host cell membranes to facilitate adhesion and tissue invasion, and digesting cells and molecules of the host immune system to avoid or resist antimicrobial attack by the host. We have critically discussed the data relevant to each of these seven criteria, with specific emphasis on how this proteinase family could contribute to Candida virulence and pathogenesis.
INTRODUCTION
Medical mycology is a relatively new field within the area of medical microbiology. Fungal diseases became recognized as being of clinical importance in the second half of the last century, mainly due to advances in medical technologies. However, within the last 20 years, the advent of the AIDS epidemic has opened up the clinical mycology field. The discovery that reduction of the CD4+ lymphocyte population of the cell-mediated immune system could predispose patients to a multitude of opportunistic fungal infections uncovered a whole new area of host susceptibility and disease. As a result, a notable increase in basic research on pathogenic fungi, predominantly Candida species, Cryptococcus neoformans, and Aspergillus fumigatus, has taken place (162). The outcome of this research has led to the unraveling of many fundamental biological processes that take place in the main fungal pathogens, particularly Candida albicans.
Candida infections are a problem of growing clinical importance. The incidence of infections has increased dramatically over the past two to three decades, and this trend will inevitably continue into the 21st century. C. albicans is the most common fungal pathogen of humans and has become the fourth leading cause of nosocomial infections (59, 167). At the most serious level, mortality rates from systemic candidiasis are high. However, the majority of patients, notably immunosuppressed individuals with human immunodeficiency virus (HIV) infection, experience some form of superficial mucosal candidiasis, most commonly thrush, and many suffer from recurrent infections. In addition, nearly three-quarters of all healthy women experience at least one vaginal yeast infection and about 5% endure recurrent bouts of disease (211, 212).
Candida species usually reside as commensal organisms as part of an individual's normal microflora and can be detected in approximately 50% of the population in this form. However, if the balance of the normal flora is disrupted or the immune defenses are compromised, Candida species often become pathogenic. Determining exactly how this transformation from commensal to pathogen takes place and how it can be prevented is a continuing challenge for the medical mycology field. Given the limited number of suitable and effective antifungal drugs, the continuing increase in the incidence of Candida infections, together with increasing drug resistance, highlights the need to discover new and better agents that target fundamental biological processes and/or pathogenic determinants of C. albicans.
PATHOGENESIS AND VIRULENCE OF CANDIDA INFECTIONS
The physiological status of the host is the primary factor governing the etiology of candidiasis. However, the observation that only slight alterations in the host can turn normally harmless commensal yeasts into agents able to inflict severely debilitating illness points to the pathogenic potential of Candida species. Indeed, it appears that the transition from harmless commensal to unrelenting pathogen is a fine line and one that is attributable to an extensive repertoire of virulence determinants selectively expressed under suitable predisposing conditions (232).
All pathogenic microorganisms have developed mechanisms that allow successful colonization or infection of the host (69). As a result, most pathogens, including Candida species, have developed an effective battery of putative virulence factors and specific strategies to assist in their ability to colonize host tissues, cause disease, and overcome host defenses. The virulence factors expressed or required by Candida species, and in particular C. albicans, to cause infections may well vary depending on the type of infection (i.e., mucosal or systemic), the site and stage of infection, and the nature of the host response. It seems apparent that a panel of virulence attributes are involved in the infective process, but no single factor accounts for Candida virulence and not all expressed virulence attributes may be necessary for a particular stage of infection (40, 161). Although many factors have been suggested to be virulence attributes for C. albicans, hyphal formation, surface recognition molecules, phenotypic switching, and extracellular hydrolytic enzyme production have been the most widely studied in recent years (24). The reader is guided to several excellent reviews on the topics of hyphal formation, surface recognition molecules, and phenotypic switching listed in Table 1.
TABLE 1.
Principal virulence attributes of C. albicans
| Virulence attribute | Putative virulence roles | References |
|---|---|---|
| Adhesins (e.g., Als family, Hwp1, Int1)a | Adhesion and colonization | 22, 23, 33, 93, 230, 231 |
| Hypha production | Adhesion, invasion, tissue damage | 18-20, 62, 63, 79, 127 |
| Extracellular hydrolytic enzymes (e.g., Sap, Plb, and Lip families)b | Nutrient acquisition, invasion, tissue damage, evasion of host response | 94, 95, 97, 98 |
| Phenotypic switching | Adhesion, evasion of host response | 214-216, 218 |
Als, agglutinin-like sequence; Hwp1, hyphal cell wall protein 1; Int1, integrin-like protein.
Sap, secreted aspartyl proteinases 1 to 10; Plb, phospholipase B1 and B2; Lip, Lipases 1 to 10.
The significance of these different putative virulence factors to C. albicans pathogenicity can possibly be ascertained by determining whether similar homologous attributes exist in other nonpathogenic or less pathogenic yeasts such as Saccharomyces cerevisiae (145). Sequencing of the Candida genome with 10.4 coverage has recently been completed (http://www-sequence.stanford.edu/group/candida), and a comparative genomic analysis between C. albicans and S. cerevisiae has been performed (239). Preliminary information on the Candida genome suggested that although approximately 90% of all S. cerevisiae genes have a counterpart in C. albicans, 6 to 7% of C. albicans genes are not found in S. cerevisiae (135). Interestingly, the genes that appear to have no equivalent in S. cerevisiae tend to be grouped in protein families, such as the agglutinin-like sequence (ALS) and secreted aspartyl proteinase (SAP) families, and are implicated in C. albicans virulence.
HYDROLYTIC ENZYMES
One factor that contributes to the process of virulence is hydrolytic enzyme production, which is known to play a central role in the pathogenicity of bacteria (69), protozoa (141), and pathogenic yeasts (163). Although many microorganisms possess a variety of hydrolytic enzymes, proteinases are by far the most commonly associated with virulence.
All proteinases catalyze the hydrolysis of peptide bonds (CO—NH) in proteins but can differ markedly in specificity and mechanism of action (7). Proteinases are classified on the basis of their catalytic mechanism and not according to their anatomical origin, substrate specificity, or physiological function. In 1978, Enzyme Nomenclature distinguished four classes of proteinases: serine, cysteine, and aspartyl proteinases and metalloproteinases. Examples of serine proteinases are the divergent trypsin, chymotrypsin, and subtilisin subfamilies; cysteine proteinases include streptococcal proteinase and papain; and metalloproteinases include collagenases and microvillus proteinases (6). Aspartyl proteinases are ubiquitous in nature and are involved in a myriad of biochemical processes (41). Well-known aspartyl proteinases include the HIV aspartyl proteinase, and pepsin and renin in humans.
Extracellular Proteinases of Pathogenic Fungi
Extracellular proteinases of saprophytic fungi such as Aspergillus niger or Neurospora crassa are secreted primarily to provide nutrients for the cells; however, pathogenic fungi appear to have adapted this biochemical property to fulfill a number of specialized functions during the infective process in addition to the simple role of digesting molecules for nutrient acquisition. These more direct virulence functions may include digesting or distorting host cell membranes to facilitate adhesion and tissue invasion, which has been demonstrated in plants (35, 155) and insects (209), or damaging cells and molecules of the host immune system to avoid or resist antimicrobial attack by the host (191).
Most studies investigating the role of extracellular hydrolytic enzymes in fungal pathogenicity have focused on human-pathogenic fungi, including the filamentous fungus Aspergillus fumigatus (104, 176, 208), the dermatophytes Trichophyton rubrum (5) and Trichophyton mentagrophytes (236), and the dimorphic yeasts Cryptococcus neoformans (21), Coccidioides immitis (253), and C. albicans (50, 75, 94, 97, 145). While little is known about the extracellular proteinases of most dimorphic human pathogenic fungi, the proteolytic system of C. albicans is well described.
SECRETED ASPARTYL PROTEINASES OF CANDIDA
The three most significant extracellular hydrolytic enzymes produced by C. albicans are the secreted aspartyl proteinases (Sap), phospholipase B enzymes, and lipases. Of these, the Sap proteins, encoded by a family of 10 SAP genes (66, 146, 147), have been the most comprehensively studied as key virulence determinants of C. albicans and are the subject of this review. For more information on phospholipases and lipases, the reader is guided to references 75 and 98.
C. albicans is not the only Candida species known to produce extracellular proteinases. Many of the pathogenic Candida species have been shown to posses SAP genes, including C. dubliniensis (76), C. tropicalis (147, 234, 254), and C. parapsilosis (53, 147), all of which produce active extracellular proteinases in vitro (76, 185). C. tropicalis is thought to possess four SAP genes (254), whereas C. parapsilosis possesses at least two SAP genes (53). Little published information is available with regard to the importance of Sap proteins in the virulence of C. dubliniensis. However, since C. dubliniensis probably possesses at least nine SAP genes (76) (J. R. Naglik, unpublished data), it is highly likely that proteinase production contributes to the virulence of this fungus. Less pathogenic or nonpathogenic Candida species do not appear to produce significant amounts of proteinase, even though they may possess aspartyl proteinase genes (see “Correlation between Sap production in vitro and Candida virulence” below). Finally, one should note that all secreted Candida secreted proteinases belong to the same class of enzyme: the aspartyl proteinases. Neither extracellular serine nor cysteine proteinases nor metalloproteinases have been identified in pathogenic Candida species.
Molecular and Biochemical Properties of the Candida SAP Family
Basic structure of the C. albicans SAP family.
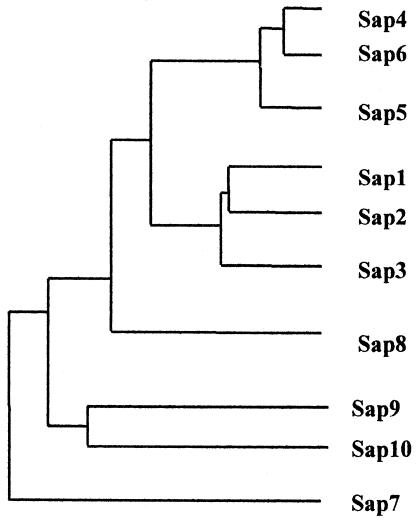
All 10 SAP genes of C. albicans encode preproenzymes approximately 60 amino acids longer than the mature enzyme (Table 2), which are processed when transported via the secretory pathway. The mature enzymes contain sequence motifs typical for all aspartyl proteinases, including the two conserved aspartate residues of the active site and conserved cysteine residues implicated in the maintenance of the three-dimensional structure. Most Sap proteins contain putative N-glycosylation sites (Table 2), but it remains to be determined which Sap proteins are glycosylated. Unlike Sap1 to Sap8, Sap9 and Sap10 both have C-terminal consensus sequences typical for glycosylphosphotidylinositol (GPI) proteins (66, 146). The dendrogram illustrated in Fig. 1 displays the relationship by sequence homology of the C. albicans Sap isoenzyme family.
TABLE 2.
C. albicans SAP genes and deduced proteinsa
| Gene | Prepropeptide size (aa)c | No. of KR and KK sitesb | ORF size (bp)c | Mature enzyme size (aa)c | No. of N-glycosylation (propeptide) sitesd | Chromosomed | Reference |
|---|---|---|---|---|---|---|---|
| SAP1 | 50 | 2 KR | 1,173 | 341 | 1 (0) | 6 | 101 |
| SAP2 | 56 | 2 KR | 1,194 | 342 | 0 (2) | R | 250 |
| SAP3 | 58 | 1 KR | 1,194 | 340 | 1 (1) | 3 | 247 |
| SAP4 | 75 | 4 KR | 1,254 | 342 | 0 (1) | 6 | 144 |
| SAP5 | 76 | 4 KR | 1,254 | 342 | 0 (0) | 6 | 147 |
| SAP6 | 76 | 4 KR | 1,254 | 342 | 0 (1) | 6 | 147 |
| SAP7 | 211 | 1 KK | 1,764 | 377 | 1 (4) | 1 | 147 |
| SAP8 | 73 | 2 KR | 1,215 | 405 | 1 (1) | 3 | 146 |
| SAP9 | 50 | 1 KR, 1 KK | 1,632 | 544 | 4 (0) | 3 | 146 |
| SAP10 | 38 | 1 KR | 1,326 | 403 | 8 (0) | 4 | 66 |
Reprinted from reference 98 with permission.
All Sap proteins contain Lys/Arg (KR)- or Lys/Lys (KK)-processing sites and four conserved cysteine residues.
The sizes of the prepropeptide (signal peptide and propeptide) and the mature enzyme are shown in amino acids (aa), and the size of the open reading frame (ORF) is shown in base pairs.
The number of potential N-glycosylation sites, including those located in the propeptide (in parentheses), and the chromosomal location of the gene are also shown. Sap9 and 10 have structural elements typical of GPI proteins (27).
FIG. 1.
Dendrogram displaying the relationship by sequence homology of the C. albicans Sap isoenzyme family. Three distinct groups are clustered within the family. Sap1 to Sap3 are up to 67% identical, and Sap4 to Sap6 are up to 89% identical, while Sap7 is only 20 to 27% identical to other Sap proteins. Sap9 and Sap10 both have C-terminal consensus sequences typical for GPI proteins and constitute the third distinct group. Similar Sap families exist in C. dubliniensis and C. tropicalis. Reprinted from reference 225a with permission.
Structural studies of the C. albicans proteinase family have concentrated on Sap2 (1, 39), which is the most abundant secreted protein in vitro when grown in the presence of protein as the sole source of nitrogen (96, 246). The overall structure of Sap2 conforms to the classical aspartic proteinase fold typified by pepsin. For more details relating to Sap2 structure and how it may affect subtrate specificty, see “Degradation of human proteins and structural analysis in determining Sap substrate specificity” below.
Processing, activation, and regulation of the C. albicans proteinases.
The pathway of proteinase synthesis starts in the nucleus, from where the newly synthesized mRNA is transferred to the cytoplasm and translated into the preproenzyme on the rough endoplasmic reticulum. The N-terminal signal peptide is removed in the rough endoplasmic reticulum by a signal peptidase (242), and the proenzyme transferred to the Golgi apparatus, where it is further processed after Lys-Arg sequences by a Kex2 proteinase (159, 235). Alternative but less efficient processing pathways for Saps are thought to exist (9, 115). Once activated, the enzyme is packaged into secretory vesicles and transported to the plasma membrane and either remains attached to the cell membrane, is incorporated into the cell wall via a GPI anchor (Sap9 and Sap10), or is released into the extracellular space.
Since the SAP gene family encode preproenzymes, the regulation of proteinase expression can be controlled either at the mRNA or protein level. Comparisons of Sap protein and mRNA levels at identical time points (246) and kinetic studies of proteinase secretion by protein labeling and immunoprecipitation (pulse chase experiments) (91) suggested that proteinase synthesis and secretion were tightly coupled, strongly implying that regulation of Sap activity occurred predominantly at the mRNA level. Studies using the proteinase inhibitor pepstatin A demonstrated that SAP2 expression in C. albicans was also regulated via a positive-feedback mechanism, since the proteolytic products of Sap2 and peptides of 8 amino acid residues or more induced this gene (96, 125).
Biochemical properties of the C. albicans proteinases.
The Sap proteinases have been purified and characterized either by direct purification from Candida culture supernatants (reviewed in reference 94) or by expression in recombinant Pichia pastoris (16, 254) or Escherichia coli (115). This has provided a more detailed insight into the characteristics of the Sap isoenzymes, whose general biochemical properties are described in Table 3.
TABLE 3.
Properties of purified C. albicans proteinases
| Proteinase | Mol mass (kDa) | pH range | pH for optimal activity | Isolectric point (pI) | Reference(s) |
|---|---|---|---|---|---|
| Sap1 | 38, 40, 40 | 2.5-5.5 | 3.2-4.5 | 4.0 | 16, 210, 247 |
| Sap2 | 40, 41, 43, 45, 48, 49 | 2.5-5.5 | 3.2-3.5 | 4.25, 4.4 | 16, 148, 177, 182, 210, 247 |
| Sap3 | 41, 42 | 2.0-5.0 | 3.2-3.5 | 5.7 | 16, 148, 210, 247 |
| Sap4 | 40a | 4.0-7.0 | 5.0 | ND | 16 |
| Sap5 | 37a | 3.0-7.0 | 5.0 | ND | 16 |
| Sap6 | 40a | 3.0-7.0 | 5.0 | ND | 16 |
| Sap7 | NDb | NDb,c | NDb | NDb | |
| Sap8 | 41 | ND | ND | ND | 148 |
| Sap9 | NDb | NDb | NDb | NDb | |
| Sap10 | NDb | NDb | NDb | NDb |
Molecular masses estimated from sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
No information regarding the biochemical properties are presently available.
ND, not determined.
The Sap1 to Sap10 proteins are between 35 to 50 kDa in size (66, 146, 147) and account for all of the extracellular proteolytic activity of C. albicans. Distinct difference in pH optima are evident between the heterologously expressed proteinases Sap1 to Sap6, with Sap1 to Sap3 having highest activity at lower pH values and Sap4 to Sap6 having highest activity at higher pH values, with a pH range of activity between 2.0 and 7.0 (Table 3). This versatile property may prove vital to the success of C. albicans as an opportunistic pathogen, by allowing the fungus to survive and cause infections on a variety of different tissues such as numerous mucosal surfaces, skin, and internal organs. In addition to different pH optima, a cleavage site specificity of Candida Sap proteins is suggested (70, 78). Koelsch et al. (115) investigated the functional aspects and substrate specificities of Sap1, Sap2, Sap3, and Sap6. All four cleaved peptide bonds between larger hydrophobic amino acids but had preferences at the P1 and P′1 sites. For example, Sap1, Sap2, and Sap6 preferred phenylalanine while Sap3 preferred leucine at the P1 site.
Based on present data (see the following sections), it is highly probable that the main roles of the C. albicans proteinases are to provide nutrition for the cells, to aid penetration and invasion, and to evade immune responses. However, the biochemical and proteolytic properties of the Sap7 to Sap10 enzymes are not presently known, and thus the full functional repertoire of the Sap family has yet to be elucidated.
For more details, the reader is guided to review articles on discovery and characterization of the SAP gene family; structure, processing, activation and regulation; purification, activity and enzymatic properties; and in vitro SAP gene expression in culture medium (50, 88, 94, 95, 97, 98, 145, 163, 185, 227). The present review is restricted to studies addressing the relationship between proteinase production and C. albicans pathogenesis.
SECRETED ASPARTYL PROTEINASES AND C. ALBICANS PATHOGENESIS
Over the past two decades, a plethora of studies have contributed to our understanding and knowledge of the SAP gene family. The existence of 10 SAP genes in C. albicans and their controlled expression and regulation raises a number of questions concerning the roles and functions of these proteinases during the infective process. The complexity of Sap involvement in C. albicans virulence has been highlighted by the fact that Sap production is associated with a number of other putative virulence attributes of C. albicans including hyphal formation, adhesion, and phenotypic switching. Although the consequences of proteinase secretion during human infections is not precisely known, the roles and functions of the Sap family can perhaps be deduced from in vitro and in vivo animal model data. As a result, nearly all the studies have implicated the proteinases in C. albicans virulence in one of the following seven ways, with perhaps the most definitive data obtained from the behavior of the various SAP-disrupted strains: (i) correlation between Sap production in vitro and Candida virulence, (ii) degradation of human proteins and structural analysis in determining Sap substrate specificity, (iii) association of Sap production with other virulence processes of C. albicans, (iv) Sap protein production and Sap immune responses in animal and human infections, (v) SAP gene expression during Candida infections, (vi) Modulation of C. albicans virulence by aspartyl proteinase inhibitors, and (vii) The use of SAP-disrupted mutants to analyze C. albicans virulence.
This review critically discusses the data relevant to each of these seven criteria, with specific emphasis on how this proteinase family could contribute to Candida virulence and pathogenesis in human infections. Each of the seven criteria is discussed in its own section.
Correlation between Sap Production In Vitro and Candida Virulence
Main focus points.
The main focus points are as follows.
(i) The virulence of C. albicans species appears to correlate with the level of Sap activity in vitro and may correlate with the number of SAP genes.
(ii) Infected patients (oral or vaginal) harbor C. albicans strains that are significantly more proteolytic than are isolates from asymptomatic carriers.
(iii) HIV infection appears to lead to the selection of C. albicans strains with heightened virulence attributes such as proteinase production.
Numerous studies have correlated extracellular proteolytic activity in vitro with the virulence of Candida species and have shown that only the most virulent species such as C. albicans, C. tropicalis and C. parapsilosis produce more proteinases in vitro than do less virulent species (185). Less common clinical isolates such as C. kefyr, C. glabrata, and C. guilliermondii appear to be nonproteolytic when tested in culture medium with bovine serum albumin (BSA) as the sole nitrogen source (131, 174, 189). The apparent clear-cut correlation between proteinase production and virulence may be due in part to the sensitivity of the assays used to determine proteolytic activity. For example, the BSA-agar method, which has been routinely used over the years (163), is a relatively insensitive hydrolysis assay that is unlikely to detect low levels of proteinase activity. This notion was supported when a sensitive, rapid fluorescence-based assay was developed that was able to detect aspartyl proteinase activity in all Candida species tested, in the order C. albicans > C. tropicalis > C. kefyr > C. lusitaniae > C. krusei (26). Since the proteolytic activity could be inhibited with pepstatin, this study demonstrated that some non-C. albicans species that were previously thought not to possess Sap activity are in fact proteolytic. It should be noted that although more virulent Candida species may in fact produce more detectable proteolytic activity in vitro, this does not necessarily imply that the production of Sap enzymes is the sole reason for virulence.
Many Candida species possess aspartyl proteinase genes. Monod et al. (147) used Southern analysis with SAP1 as a probe to demonstrate the presence of four bands with sequence similarity in EcoRI-digested genomic DNA of C. guilliermondii, although this yeast does not produce proteinase in vitro in BSA-containing medium. However, even though C. guilliermondii and possibly other Candida species may possess SAP-like genes, it is not known whether they are functional in vivo. Although these less pathogenic Candida species may be capable of producing secreted proteinases, in general the amount of Sap activity produced in vitro and possibly the number of SAP genes appear to be directly correlated to the virulence of the Candida species.
Sap production by clinical C. albicans isolates from humans.
C. albicans strains from different patient groups with various clinical infections have been isolated, and the level of Sap activity produced in vitro has been correlated with virulence (Table 4). Most of these studies have concentrated on C. albicans strains isolated from the vaginal lumen or the oral cavity and on the effect of HIV infection on proteinase production and C. albicans strain selection.
TABLE 4.
Correlation between Sap production and activity with C. albicans virulence
| Main findings | Reference(s) |
|---|---|
| Oral candidiasis | |
| Increased Sap activity occurred in C. albicans strains isolated from HIV-positive patients with oral candidiasis compared with HIV-negative C. albicans carriers | 46, 252 |
| C. albicans from HIV-positive patients with oropharyngeal candidiasis produced more proteinase activity than did C. albicans from HIV-negative asymptomatic oral carriers or HIV-negative subjects with oral candidiasis. | 166 |
| Vaginal candidiasis | |
| C. albicans isolates from patients with vaginal candidiasis were significantly more proteolytic than isolates from asymptomatic vaginal carriers. | 4, 28, 43 |
| C. albicans isolates from HIV-positive women with vaginitis produced significantly higher levels of Sap than did C. albicans strains isolated from HIV-positive asymptomatic carriers or HIV-negative subjects with candidal vaginitis. | 49 |
| Animal models | |
| High-Sap-producing oral and vaginal C. albicans strains from HIV-positive patients were more pathogenic in the mouse and rat vaginitis models than were lower-Sap-producing C. albicans strains from HIV-negative patients. | 48, 49 |
| There was a correlation between C. albicans adherence to buccal epithelial cells, proteinase production, and lethality in mice. Higher-Sap-producing strains showed higher levels of tissue colonization in the liver, kidneys, and spleen | 2 |
| Other diseases | |
| There was no difference in Sap production from oral C. albicans isolated from patients with autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy and healthy controls | 87 |
| Production of Sap did not differ between patients with antibiotic-associated diarrhea and control subjects. | 118 |
| Higher levels of Sap were produced by C. albicans from children with acute diarrhea than those with chronic diarrhea, which may account for the more severe symptoms. | 136 |
Three reports showed that C. albicans isolates from symptomatic patients with vaginal candidiasis were significantly more proteolytic (1.5- to 2-fold) than isolates from asymptomatic vaginal carriers (4, 28, 43). A more recent study by the same group found that C. albicans strains isolated from 21 HIV-positive women with vaginitis produced significantly higher levels of Sap (fourfold) than did C. albicans strains isolated from either 7 HIV-positive asymptomatic carriers or 31 HIV-negative subjects with candidal vaginitis (49) (Table 4).
A similar approach has been applied to oral isolates of C. albicans, mainly from HIV-positive individuals. C. albicans isolates from 100 HIV-positive patients with oropharyngeal candidiasis produced significantly more proteinase activity than did isolates from 122 patients without HIV infection (50 with oral candidiasis and 72 asymptomatic Candida carriers) (166). The higher level of proteinase activity correlated with the increased level of cell surface-associated and secreted Sap, as revealed by cytofluorometry and Western blotting, respectively. A similar study, but using fewer patients, reported a comparable increase in Sap activity in C. albicans strains isolated from the oral cavities of 44 HIV-positive patients (advanced disease) with oral candidiasis compared with that in 30 HIV-negative C. albicans carriers (46). However, since a control group of HIV-negative subjects with oral candidiasis was not included, it is unclear whether the observed increase in Sap production resulted from the advanced HIV status of the individuals or from the Candida infection. Likewise, Wu et al. (252) found that oral C. albicans isolates from HIV-positive subjects (n = 18) produced significantly more proteinase than did isolates from HIV-negative individuals (n = 18) when they were investigated in a BSA agar plate assay. Finally, high Sap-producing oral (48) and vaginal (49) C. albicans strains isolated from HIV-positive patients were more pathogenic in the mouse and rat vaginitis models, respectively, than were lesser Sap-producing C. albicans strains from HIV-negative patients (Table 4).
In summary, these studies using oral and vaginal clinical isolates showed a positive correlation between the level of Sap production in vitro and the virulence of C. albicans. Whether these observations reflect an elevated “fitness” or a specific adapted response of C. albicans strains during infection is not clear, but the data tentatively support a role for the proteinases during the infective process in vivo.
C. albicans strain selection in HIV infection.
There is mounting evidence that Candida species colonizing the oral cavities of HIV-infected individuals are subject to selective pressures that may lead to the emergence of strains with altered genotypic and phenotypic characteristics and enhanced expression of known and putative virulence determinants. Studies have shown that the genotype of the infecting Candida cells in HIV infection is stable, and, as a result, HIV-infected patients tend to be colonized by a single endogenous strain of Candida that persists throughout recurrent bouts of oral candidiasis, even after antifungal therapy (34, 130, 143, 169, 175, 204, 245, 248). However, these and other studies also suggest that in the majority of AIDS patients the original commensal strains are replaced and that this replacement of genotypes occurs only once, early in the course of HIV infection, producing a genetically conserved population (140, 204).
Both Ollert et al. (166) and De Bernardis et al. (46) showed that the increase in Sap activity was observed only in patients with advanced HIV infection and not in those with earlier stages of HIV infection or HIV-negative subjects. This indicated that more virulent biotypes of C. albicans with heightened proteinase production might be selected in HIV-infected patients. However, it should be pointed out that this selection appeared to occur before patients developed AIDS and was independent of CD4+ counts.
One intriguing possibility for the observed differences in Sap production between HIV-positive and HIV-negative patients may be due to the direct binding of HIV proteins to Candida cells. Treatment of C. albicans with gp160, but not with gp120, led to an elevation of free and cell-bound aspartyl proteinase (82). In addition, culture supernatants obtained from C. albicans treated with gp160 or gp41, but not with gp120, showed a strong increase in proteinase activity. Why or how HIV gp160 or gp41, but not gp120, influences proteinase production and whether they modulate Sap secretion directly or indirectly through another mechanism remain to be elucidated. HIV infection might also promote C. albicans virulence in another way, since the HIV transactivating protein Tat binds RGD sequences present on the surface of C. albicans to induce hyphal production (81), a process known to be linked with virulence and the expression of the SAP4 to SAP6 subfamily (96, 246).
The pathobiological effects of HIV infection, including possible epithelial cell surface changes (170), reduced salivary flow rate (203), and alterations in the oral microflora (171), might also influence the candidal microenvironment. Together with impaired humoral or cell-mediated mucosal immunity and/or impaired nonspecific host defenses, these selective pressures in HIV infection are likely to contribute to the selection of Candida strains, some of which may possess altered or heightened virulence attributes such as proteinase production (232). However, the mechanism by which these selective pressures contribute to strain selection in HIV infection remains to be elucidated and may prove particularly challenging to resolve.
Degradation of Human Proteins and Structural Analysis in Determining Sap Substrate Specificity
Main focus points.
The main focus points are as follows.
(i) Sap2 has very broad substrate specificity and can degrade many human proteins.
(ii) The crystal structure of Sap2 indicates that the C. albicans proteinase family is unique among the aspartyl proteinases.
(iii) Computer modeling suggests that the electrostatic charge of the different Sap proteins may contribute to different substrate specificities and tissue targeting.
The first observation of proteolytic activity in C. albicans was demonstrated by Staib (222) when yeast cells were grown in media containing BSA as the sole source of protein. Three years later, Remold et al. (177) attributed this activity to the production of an extracellular proteinase. Since then and up to the early 1990s, a plethora of studies reported on the purification and biochemical properties of an extracellular proteinase from C. albicans and the effect of environmental factors such as pH and temperature on proteolytic activity (Table 3). The culture conditions used to induce proteinase activity in these early reports have subsequently been shown to favor SAP2 expression (96, 246). Therefore, any attempts to determine the substrate specificities and potential targets of the Sap family in vivo were based on the activity of Sap2 in vitro. At present, it is not clear whether the digestion of substrates by Sap2 in vivo is similar to that shown in vitro or whether the substrates for Sap2 are similar or different from those of the other proteinases in the Sap family. The full range of substrate specificities for all the secreted proteinases has not been adequately studied, but the in vitro proteolytic properties of Sap2 have been described in some detail.
Broad substrate specificity of Sap2.
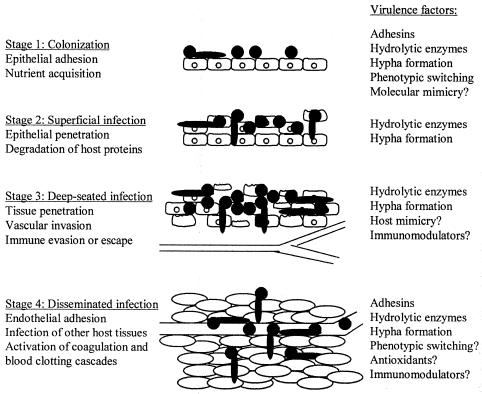
One of the most noticeable properties of Sap2 is the variety of proteins it can cleave. The contribution of this broad activity to Candida pathogenesis, along with other virulence attributes of C. albicans, is illustrated in Fig. 2. Sap2 is known to degrade many human proteins including molecules that protect mucosal surfaces such as mucin (36, 52) and secretory immunoglobulin A (IgA) (78, 184). Not only could this provide essential nitrogen for growth, but also it could enhance attachment, colonization, and penetration of host tissue by the removal of host barriers. Digestion of secretory IgA is particularly noteworthy because it is considerably more resistant to proteolysis than are monomeric or serum immunoglobulins, is able to neutralize many toxins and enzymes (109), and can inhibit C. albicans attachment to buccal epithelial cells (243). Supporting these data, Wu and Samaranayake (251) noted that reduction of total salivary protein concentration correlated with the degree of Sap expression, suggesting that Candida Sap proteins degrade salivary proteins in the oral cavity. Sap2 can also degrade molecules of the extracellular matrix such as keratin, collagen and vimentin (94, 163, 174). Morschhauser et al. (152) showed that induction of C. albicans proteinase caused digestion of soluble and immobilized extracellular matrix proteins produced by a human endothelial cell line, suggesting that Sap proteins may facilitate the dissemination of C. albicans via the circulatory system.
FIG. 2.
Schematic diagram illustrating the contribution of the various virulence attributes to C. albicans pathogenicity. C. albicans commonly colonizes the epithelial surface (stage 1) and causes superficial infections (stage 2), but under conditions when the host is compromised, the fungus establishes deep-seated infections (stage 3) by penetrating further into the epithelial tissue. Occasionally, C. albicans causes disseminated infections (stage 4), which allow the fungus to colonize and infect other host tissues and can be fatal. This infective process involves numerous virulence attributes including adhesins, hydrolytic enzyme production (Sap proteins, phospholipases, and lipases), hypha formation, and phenotypic switching. Sap2 (and possibly other Sap proteins) is known to degrade many human proteins, including mucin, extracellular matrix proteins, numerous immune system molecules, endothelial cell proteins, and coagulation and clotting factors. Therefore, the action of Sap proteins could be involved in all four stages of infection and probably greatly enhances the pathogenic ability of C. albicans. Modified from reference 160 with permission.
C. albicans proteinases may also evade host defenses by directly degrading molecules such as salivary lactoferrin, lactoperoxidase, cathepsin D (an intracellular lysosomal enzyme of leukocytes), and complement (72, 94, 106). In addition, Sap2 can degrade α2-macroglobulin, a natural proteinase inhibitor in human plasma (187), and cystatin A, a cysteine proteinase inhibitor found in human epidermal tissues and fluids (238). Furthermore, the proinflammatory cytokine interleukin-1β can be activated from its precursor by Sap2, suggesting a role for proteinases in the activation and maintenance of the inflammatory response at epithelial surfaces in vivo (8). Under certain conditions, Sap2 can also activate Hageman factor, a serine proteinase of the kallikrein-kinin system, which may cause increased vascular permeability in vivo (107). Similarly, Sap2 may also act on the blood clotting system by activating coagulation cofactor X (183), clotting factor XII, or prothrombin, which may in turn result in the generation of thrombin and hence blood clotting (105). The activation of such host proteolytic cascades may not appear to be advantageous to C. albicans; however, the resulting deleterious effects to the host may have some “downstream” beneficial affects that may assist or promote C. albicans infections.
Deducing proteinase specificity via three-dimensional structure and molecular modeling.
The substrate specificity of Sap2 is noticeably very broad, and some researchers thought that this broad specificity could be deduced from its three-dimensional structure. Accordingly, two reports on the crystal structure of Sap2 complexed with a potent inhibitor (A-70450 [see “Modulation of C. albicans virulence by aspartyl proteinase inhibitors” below]) were published (1, 39), which indicated that the Sap2 structure conforms to the classical aspartyl proteinase fold typified by pepsin. However, comparisons of Sap2 with pepsin have revealed a number of major differences that may contribute to the broad substrate specificity of Sap2 and which make the C. albicans proteinase family unique among the aspartyl proteinases (1, 39). Specifically, Sap2 has an enlarged and well-defined cavity for binding the third residue N-terminal to the cleaved bond in the substrate and two “flaps” overlying this cavity, the latter observation being a hallmark of the Sap family.
At present, the structure of Sap2 alone has been determined, and although other members of the Sap family are known to contribute to C. albicans virulence, very few data are available regarding their structures or substrate specificities. To address this, Stewart et al. (227) undertook a comparative structural study with the sequences of SAP1 SAP6 by molecular modeling. Although the structures of Sap1 to Sap6 and the electrostatic charge of their active sites were generally similar, sufficient differences existed to allow for different substrate specificities, with the difference between Sap1 to Sap3 and Sap4 to Sap6 being clearly evident. Furthermore, a potentially significant trend in the total electrostatic charge of the Sap1 to Sap6 enzymes was observed; the six enzymes had an overall net charge of −8, −21, −22, −5, +2, and +2, respectively. It is somewhat puzzling that the charge of the non-active-site regions of Sap1 to Sap6 varies so much, but it might contribute in part to our understanding of the different pH optima and range of pH activities of the Sap enzymes (ranging from pH 2.0 to 7.0). One more interesting observation resulting from the molecular modeling of Sap1 to Sap6 was the clear difference in the carboxy end-terminal extension between SAP1 to SAP3 and SAP4 to SAP6 at amino acid positions 323 to 324 and 335 (SAP1 to SAP3, NE and A, SAP4 to SAP6 = RK and Q, E) (227). While speculative, this carboxy-terminal extension appears to resemble an “attachment” anchor (C. Abad-Zapatero, personal communication). If true, this may support the hypothesis that the C. albicans proteinases may target specific cell proteins or tissue compartments during the infective process.
Discussion.
Although proteinases other than Sap2 (specifically Sap1 and Sap3 to Sap6) have recently been purified (16, 210, 246), it remains to be determined whether they have the same broad substrate specificities as Sap2. Furthermore, purified proteins of Sap7, Sap8, Sap9, and Sap10 have not been isolated or biochemically characterized, and thus the proteolytic properties of these proteinases remain totally unknown. On the one hand, it might seem unlikely that the different members of the Sap family have the same broad substrate specificities as Sap2, since it would seem unnecessary for C. albicans to possess a family of 10 proteins which are differentially expressed under a variety of environmental conditions and in different tissues (see “SAP gene expression during Candida infections” below) simply to digest the same substrates. On the other hand, C. albicans may require a family of extracellular proteolytic enzymes, each optimized to certain environmental conditions or different local pH values and/or particular tissues, to help the fungus colonize and infect multiple sites of the body. However, there are clearly differences in the substrate specificities of the Sap proteins; for example, sequence similarities of C. albicans Sap9 and Sap10 to the S. cerevisiae yapsins, including potential C-terminal consensus sequences for GPI anchors, suggest that Sap9 and Sap10 may have different specificities and functions from the other C. albicans Sap proteins (A. Albrecht, I. Pichova, M. Monod, and B. Hube, unpublished data).
In all likelihood, there is probably considerable overlap in the substrate specificities of many of the Sap members. Since the pH activity of the individual hydrolytic enzymes range between pH 2.0 and 7.0 (75, 182, 210, 247), this would allow for the concomitant expression of a number of similar SAP genes at environments with different pH values. In addition, there are differences in the promoter sequences of the different SAP genes, which indicates that their expression might be controlled by different SAP-specific transcriptional regulators and possibly suggests that the SAP genes might have evolved to possess distinct functions. Moreover, the coordinated regulation of the SAP genes with other virulence factors, including hyphal formation and phenotypic switching, would permit several proteinases to act in unison to carry out a series of tasks to not only digest a complex mixture of target proteins but also to provide C. albicans with a biological advantage to specifically enhance the pathogenic ability of the fungus (97). With these considerations in mind, it is entirely plausible that C. albicans has adapted to certain niche sites by expressing a combination of SAP genes (and other virulence genes), which are called upon as and when required.
Association of Sap Production with Other Virulence Processes of C. albicans
Main focus points.
The main focus points are as follows.
(i) Sap proteins facilitate C. albicans adherence to many host tissues and cell types.
(ii) Hypha formation and SAP4 to SAP6 expression are coordinately regulated, but the signaling pathways remain to be elucidated.
(iii) SAP1 appears to be regulated by phenotypic switching, but the contribution of switching to C. albicans virulence in vivo is not yet clear.
Many of the early proteinase studies focused on the influence of culture conditions on Sap expression and proteolytic activity in vitro (reviewed in reference 94). However, after the discovery of a SAP gene family, it became apparent that this enzyme family had a more significant and complex contribution to C. albicans pathogenicity. C. albicans is a polymorphic pathogen, which can exist in a yeast or a hyphal state and can undergo phenotypic switching (214). Therefore, it seemed logical to assume that due to the large number of proteinases present in C. albicans, the SAP gene family may be differentially expressed in the different morphological forms. As a result, the relationship between proteinase production and hyphal production, phenotypic switching, and other putative virulence attributes of C. albicans including adherence was investigated.
Sap production and C. albicans adherence.
Adhesion of Candida to host tissues allows the fungus to attain a foothold and to colonize a specific niche environment. Under suitable predisposing conditions when the host is compromised, this colonized site provides the base for candidal proliferation, invasion, and, in some instances, dissemination. Adherence of C. albicans to host cells is a complex, multifactorial process involving several types of candidal adhesins on a morphogenically changing cell surface (reviewed in the references in Table 1), and one mechanism through which Candida adherence might be promoted is via the production of proteinases.
One of the first early studies to link proteinase production to adherence in C. albicans showed that strongly proteolytic strains of C. albicans adhered significantly more strongly to human buccal epithelial cells in vitro than did strains producing less proteinase (74). A more recent report correlated proteinase production with increased adherence to buccal epithelial cells and death of mice; the higher-Sap-producing strains showed greater levels of tissue colonization in the liver, kidneys, and spleen (2). However, the majority of studies linking Sap production with C. albicans adherence have been performed using the proteinase inhibitor pepstatin, which inhibits Sap2 (and probably Sap1 and Sap3) very efficiently (168). Borg and Rüchel (12) demonstrated a marked reduction in C. albicans adhesion and invasion of human mucosa by pepstatin, and a similar reduction of C. albicans adherence using pepstatin was also shown with human epidermal cells (60, 165). Pepstatin could also inhibit the development of cavitations after yeast cells adhered to epidermal corneocytes (173). Some years earlier, Klotz et al. (114) observed that yeast cells formed cavitations and burrowed rapidly into vascular endothelium in vitro by a mechanism independent of germ tube formation. However, at that time the burrowing was not associated with proteinase production, but the results of the work by Ray and Payne (173) clearly implicated proteinases in the process.
The actual Sap proteins involved in adherence and cavitation (and possibly subsequent penetration) of host tissues were not studied, but a recent report by Kvaal et al. (121) indicated that Sap1 might be involved. Using a gene misexpression strategy in the switching strain WO-1, in which white-phase cells misexpressed the opaque-specific gene SAP1, the authors demonstrated in a cutaneous mouse model that SAP1 conferred two opaque-specific characteristics upon white cells: increased adhesion and the capacity to cavitate skin (237). Interestingly, the addition of pepstatin inhibited cavitation but not the enhanced adhesion (which confirmed the data of Ray and Payne [173]), suggesting that cavitation was the consequence of secreted Sap1 enzyme, while increased adhesion was the result of other cell-associated factors. Other, more recent studies using HIV aspartyl proteinase inhibitors have also implicated Sap1 to Sap3 in C. albicans adherence; however, these studies are explained in full in “Modulation of C. albicans virulence by aspartyl proteinase inhibitors” (below).
(i) How do Sap proteins contribute to adherence?
These pepstatin studies demonstrating the inhibition of C. albicans adherence clearly indicate that the Sap family plays some kind of role in C. albicans adherence. Although the precise mechanisms by which Sap proteins contribute to the adherence process are not clear, two hypotheses are currently favored. In the first, C. albicans proteinases could act as ligands to surface moieties on host cells, which does not necessarily require activity of the enzymes. In the second, C. albicans utilizes Sap proteins as active enzymes to modify target proteins or ligands on the fungal surface or on host cells (i.e., epithelial cells), which may alter surface hydrophobicity or lead to conformational changes, thus allowing better adhesion of the fungus (145). If the Sap proteins can indeed function directly as C. albicans adhesins, this will add to the growing number of virulence properties already possessed by the Sap family (i.e., tissue damage, invasion, and evasion of host defenses) and establish the proteinases as one of the most versatile and multifunctional virulence gene families possessed by C. albicans.
Sap production and yeast-to-hypha transition.
Research efforts by many investigators in different laboratories have concentrated on the study of C. albicans morphogenesis, as well as the identification and characterization of cell wall components that are growth phase (yeast and hypha) specific and associated with virulence. The foundation of these studies is based on two factors: (i) the common acceptance that the hyphal form is related to the invasive properties of C. albicans and (ii) the importance of morphogenesis as a biological phenomenon.
The ability of C. albicans to transform into hyphae may be considered a pathogenic determinant in the initial processes of superficial tissue invasion, whereby hyphae may promote the adherence and penetration of C. albicans to host tissues. In culture medium, the main proteinases associated with hyphal formation are the SAP4 to SAP6 subfamily (96, 246), and pH and hypha induction alone are sufficient for the induction of SAP4 to SAP6 (it should be noted that SAP4 transcripts were not detected in several experiments).
(i) Coordinate regulation of hypha-formation and SAP4 to SAP6 expression.
Although hypha formation and SAP4 to SAP6 expression were linked, more direct proof was required to determine whether the two phenomena were coordinately regulated. Sequence analysis of the promoter regions of SAP4 to SAP6 revealed the presence of consensus sequences [CATTC(A/C)] for the TEA/ATTS transcription factor Tec1 (206). C. albicans mutant strains lacking TEC1 failed to produce hyphal cells in vitro and were not able to express SAP4 to SAP6, suggesting the existence of joint or coordinated regulatory pathways for hypha production and proteinase expression. The concept of coordinated pathways was supported by the observation that SAP4 to SAP6 expression increased in a hyperfilamentous strain lacking CPP1 (a mitogen-activated protein kinase phosphatase) (205) and had a modified expression pattern in a strain lacking EFG1 (a key transcriptional regulator of dimorphism), which has a strongly reduced ability to form hyphae (228). Moreover, in a murine systemic intraperitoneal model, the EFG1-deficient mutant had a strongly reduced ability to produce hyphae, which was associated with reduced expression of SAP4 to SAP6 and an inability to invade or damage parenchymal organs including the liver and pancreas (65). Interestingly, a triple null C. albicans mutant lacking SAP4 to SAP6 showed strongly reduced invasiveness but still produced hyphal cells. Finally, SAP5 activation during in vivo infection was shown not to depend on growth of C. albicans in the hyphal form; however, the two major hyphal signaling pathways in C. albicans (defined by Cph1 and Efg1) were required for SAP5 expression (224). Together, these studies indicate that not only are hypha formation and proteinase production coordinately regulated but also C. albicans hyphal cells require the support of hydrolytic enzymes (specifically SAP4 to SAP6) in order to be fully invasive in vivo.
The observation that certain transcriptional factors which regulate the yeast-to-hypha transition also regulate proteinase expression has recently been addressed using C. albicans DNA microarrays. Transcriptional profiling of C. albicans mutants lacking factors that regulate the dimorphic transition has helped to elucidate signaling pathways and to clarify the coordinated regulation between morphology and proteinase production. The reader is guided to “Functional genomics and Candida” (below) for more details.
Sap production and phenotypic switching.
The selection of phenotypically altered strains may be enhanced in C. albicans by a phenomenon known as high-frequency phenotypic switching, whereby Candida cells randomly switch their phenotype, especially in response to stress (217, 218). While many prokaryotic and eukaryotic microorganisms can switch between alternate phenotypes under different environmental conditions (207), C. albicans appears to have an enhanced ability for chromosomal rearrangement and genetic reorganization. Unlike switching in other microbial pathogens, switching in C. albicans is pleiotropic, affecting several morphological and physiological parameters and a number of virulence traits (214), all of which may allow the fungus to adapt to different host environments during the course of an infection. Recent work suggests that phenotypic switching is based on heritable changes in chromatin structure and supports the notion that acetylation of histones plays a selective role in regulating the switching process (113, 220).
(i) Which SAP genes are regulated by phenotypic switching?
In a C. albicans strain named WO-1, the discovery and characterization of the white (W)-to-opaque (O) transition indicated that switching could affect a variety of cellular characteristics, including proteinase production (214). As a result, a correlation between Sap secretion and switching was subsequently described in C. albicans strains WO-1 (151) and 3153A (149). In both strains, transcripts of SAP1 were abundant in specific switching-regulated forms and Sap1 was primarily responsible for the higher extracellular proteolytic activity observed in these switching states. As a result, SAP1 was the first cloned switching-regulated gene detected in C. albicans (151), a year after the gene was first isolated by Hube et al. (101).
Sequence analysis of the 5′-untranslated regions of SAP1 from different C. albicans strains indicated that during switching, SAP1 expression was regulated by activation or deactivation of phase-specific trans-acting factors, which in turn were regulated by a “master switch” event (215). Since SAP1 was shown to be switching regulated, it was not surprising that the expression of this gene was not dependent on the presence of exogenous protein (96, 151, 246). This is in contrast to SAP2 (not switching regulated), which is expressed in both the white and opaque phenotypes of C. albicans strain WO-1 but only in the presence of exogenous protein (96, 246).
SAP3 expression may also be regulated by phenotypic switching (150, 247), but its regulation is different from that of SAP1 and SAP2 in that SAP3 is detected in C. albicans strains when SAP2 is expressed (96, 210, 246). Another SAP gene that is differentially expressed during switching is SAP8, since transcripts were detected in the opaque but not the white phenotype (99). However, since SAP8 was shown to be up-regulated at 25°C compared with 37°C (146) and since the opaque phenotype is stable only at 25°C (149, 151, 219), this suggested that SAP8 expression in opaque cells may be temperature regulated rather than switching regulated.
At present, in C. albicans strain WO-1, SAP1 is the only proteinase that is strictly regulated by phenotypic switching. However, phenotypic switching is a very complicated process, which is by no means fully understood. In fact, very little is known about this phenomenon outside of C. albicans strain WO-1. Other clinical or laboratory strains may have switching processes divergent from or even unrelated to that of WO-1, each affecting SAP gene expression and other virulence genes in distinctive ways. Therefore, it cannot yet be concluded which proteinases are regulated by switching in vivo or what contribution this phenomenon makes, in terms of SAP gene expression, to the virulence of C. albicans.
Discussion.
In summary, laboratory studies have indicated that the C. albicans SAP gene family is differentially expressed in the yeast, hyphal, and phenotypically switched states and may contribute to C. albicans adherence. At the most basic level, one could conclude that yeast cells predominantly express one set of SAP genes (SAP1 to SAP3), hyphae predominantly express another (SAP4 to SAP6), and phenotypically switched cells predominantly express yet another (SAP1 and SAP3). Although this might be attractive, it is almost certainly too simplistic, since these conclusions have usually been drawn from the use of one strain of C. albicans grown under laboratory-controlled conditions. In vivo, the environmental milieu and immune selective pressures may affect SAP gene expression and phenotypic switching in individual yeast and hyphal cells in a unique fashion, which cannot be tested or controlled for in the laboratory. Therefore, it is quite possible that the SAP genes expressed by C. albicans cells in the laboratory may not equate to the SAP genes expressed in vivo. Determination of exactly which SAP genes are expressed by the two morphological forms and during phenotypic switching at the single-cell level in vivo may provide a significant step forward in elucidating the complex interaction between the host environment and SAP gene regulation.
Sap Protein Production and Sap Immune Responses in Animal and Human Infections
Main focus points.
The main focus points are as follows.
(i) Sap proteins are produced in vivo during mucosal and systemic infections.
(ii) Proteinases are localized to the cell wall during C. albicans infections.
(iii) C. albicans proteinases are immunogenic and elicit mucosal and systemic antibody responses.
(iv) The inhibitory and protective effects of Sap antibodies against Candida infections remain unclear and the protective B- and T-cell epitopes of the Sap family are unknown.
Sap protein production during C. albicans infections.
Several studies have provided strong evidence demonstrating the production of Sap protein in vivo. Early work using murine models of disseminated candidiasis revealed the presence of Sap proteins on the surface of C. albicans cells in murine kidneys (116, 132). Using indirect-immunofluroescence microscopy, the presence of Sap proteins was also detected within the cell wall of yeast and hyphal cells in all organs of immunocompromised patients who had succumbed to systemic C. albicans infections, including the mucosa, central nervous system, lungs, heart, liver, pancreas, and kidneys (190). With regard to mucosal infections, elevated levels of Sap proteins were observed in vaginal fluids of candidiasis patients compared with Candida carrier subjects as determined by enzyme-linked immunosorbent assay and immunoblotting using Sap2 polyclonal antibodies (43), indicating a link between Sap production in vivo and infection.
The presence of Sap protein has also been demonstrated during phagocytosis of C. albicans by leukocytes. Macdonald and Odds (134) were the first to show that the resistance of C. albicans to phagocytosis was associated with Sap expression. Some years later, it was observed that C. albicans and C. tropicalis yeast cells that resisted phagocytic killing germinated intracellularly and expressed Sap on their surface (13). Recently, the Sap4 to Sap6 family have been implicated in the evasion of phagocytosis by C. albicans, since the expression of Sap4 to Sap6 but not Sap1 to Sap3 was upregulated on yeast and germ tubes after phagocytosis by murine peritoneal macrophages (16). In the same study, C. albicans mutants lacking SAP4 to SAP6 were significantly more susceptible to phagocytosis than were wild-type cells. These results strongly indicate that by preventing macrophage killing, Sap4 to Sap6 play at least one significant role in evading host immune defenses.
Sap localization to the cell wall.
More recent studies using immunogold-labeling techniques demonstrated that Sap proteins are localized to the cell wall during C. albicans infections. In a rat vaginitis model, Sap1 to Sap3 were present in the yeast cell wall during early stages of infection, a pattern that correlated with the in vitro localization of Sap (229). Three studies using polyclonal antibodies raised against Sap1 to Sap6 demonstrated the presence of Sap1 to Sap3 on the surface of both yeast and hyphal cells, while Sap4 to Sap6 antigens were found predominantly on hyphal cells (65, 119, 202). Also, in biopsy specimens of oral epithelial lesions collected from three HIV-infected patients with oropharyngeal candidiasis, most Sap was secreted at the locations where C. albicans directly adhered to epithelial cells or at sites in close contact between C. albicans and epithelial cells (199). However, it is important to recognize that the Sap1 to Sap3 and Sap4 to Sap6 antibodies used were not able to differentiate between the individual Sap proteins within each of the two subfamilies investigated. Therefore, the identities of the individual proteinases that localize to the cell wall or are detected in vivo during experimental infections could not be determined. Antibodies specific for individual Sap proteins do not yet exist, but the development of such antibodies would be a valuable addition to existing molecular techniques in determining the localization patterns of the individual proteinases as well as the expression patterns of the proteinases during different stages and types of C. albicans infections.
Antibodies against Sap induced by C. albicans infections.
(i) Antibodies produced during systemic infection.
Numerous studies have described the presence of Sap protein during C. albicans infections; however, few studies have described antibody responses to the C. albicans proteinases in human patients. Macdonald and Odds (133) were the first to detect proteinase-specific IgG antibodies in sera of patients with disseminated candidiasis at significantly higher levels than those found in healthy individuals. These findings were later confirmed by Ray and Payne (172) and Rüchel et al. (186, 187), further demonstrating the production of proteinases during systemic candidiasis. However, in the latter study, sera of a fifth of the patients suffering from candidiasis did not produce high titers of antibodies against purified Sap2, probably reflecting the inability of many high-risk patients to mount a normal immune response (185).
(ii) Antibodies produced during mucosal infection.
The above studies investigated anti-Sap IgG responses during systemic infections, but few studies have investigated the IgA response, and in particular secretory IgA, to Sap proteins during mucosal Candida infections, such as those in the oral cavity and vaginal lumen. This would clearly be more relevant than IgG responses, since IgA is the predominant antibody present at mucosal surfaces and is known to prevent the attachment of C. albicans to the mucosal epithelium (243).
Two reports have recently addressed this issue. Using a time-resolved immunofluorometric assay, total levels of IgA against Sap1, Sap2, and Sap6 were found to be higher in saliva from HIV-positive patients with oral and oropharyngeal candidiasis than from HIV-positive patients without oral candidiasis or HIV-negative healthy controls (57, 142). The authors concluded that during oral infection, HIV-positive patients have an increased mucosal antibody response specifically directed against C. albicans virulence antigens, in this case the proteinases (57). However, this study did not include an HIV-negative patient group with oral candidiasis, so the observed increase in the level of salivary IgA against the proteinases may be related to the HIV status of the individual as well as to candidiasis. Interestingly, over the 1-year period, variations in Candida colonization levels in the oral cavity and episodes of oropharyngeal candidiasis correlated with variations in salivary anti-Sap6 IgA antibody levels (142). This may indicate a direct relationship whereby as C. albicans numbers proliferate during mucosal infections, more Sap6 is produced, resulting in the induction of a corresponding mucosal IgA antibody response. However, since it is highly likely that the polyclonal antibodies induced by certain Sap proteins during human mucosal infections cross-react with other Sap proteins (especially within homologous subfamilies), more work is clearly needed before this hypothesis can be substantiated.
Functional anti-Sap antibodies.
Antibodies have many functions in many diseases, but the role of antibody immunity in protection against mucosal and systemic candidiasis is unclear. Indeed, the majority of patients with mucosal Candida infections have normal or even elevated levels of both serum and mucosal anti-Candida antibodies (37, 112, 137, 233). This indicates that although patients are able to produce high levels of antibodies in response to Candida infection, these high antibody titers are not able to clear candidal infection. However, secretory IgA antibodies are able to bind to Candida and reduce the adherence of Candida to epithelial cells, theoretically preventing or maintaining low levels of Candida colonization (126, 243).
(i) Systemic infections.
In patients with systemic candidiasis, there is some evidence for a role of anti-C. albicans antibodies in recovery from infection. Development of antibodies against C. albicans heat shock protein 90 (HSP90) appear to be protective against human and animal Candida infections; higher titers of antibodies against a 47-kDa breakdown product of C. albicans HSP90 were present in patients recovering from candidemia than in the early stages of infection (108, 139), and antibodies against this 47-kDa product were protective in a murine model of systemic candidiasis (138). In addition, Cutler and coworkers identified an IgM (84, 85) and IgG3 (86) antibody-recognizing phosphomannoprotein that is protective against systemic (84, 86) and vaginal (85, 86) candidiasis. These studies, at the very least, indicate that antibodies can be protective against Candida infections. However, very little information is available concerning a protective role of Sap antibodies against systemic Candida infections.
(ii) Mucosal infections.
More data are available with regard to the protective affects of mucosal Sap antibodies against C. albicans infections. De Bernardis et al. (45) published some promising data obtained with the rat vaginitis model, showing that immunization with Sap2 antigen, or administration of an anti-Sap2 monoclonal antibody or anti-Sap2 antibody-containing vaginal fluids, partially protected rats against candidal vaginitis. Protection by antibody was T-cell dependent and was conferred by the Sap2 antibodies, since preabsorption of the fluids with Sap2 antigen reduced the level of protection. However, it is likely that the protective anti-Sap2 antibodies cross-reacted with other proteinases such as Sap1 and Sap3, since these are thought to have similar epitopes (210, 246, 247). Although the mechanism of protection was not elucidated, this was the first demonstration that anti-Sap antibodies could afford protection against C. albicans infections in vivo and indicated that not only might Sap2 be the main proteinase contributing to rat vaginal infections but also that Sap2 could be the main target of the host immune response affording protection at mucosal sites.
(iii) Inhibitory antibodies.
While C. albicans proteinases are known to induce antibody responses in humans (57, 133, 142, 186), the inhibitory and protective effects of Sap antibodies produced by infected hosts remain unclear. While one study by Borg et al. (14) demonstrated that three monoclonal IgM antibodies raised against Sap2 did not inhibit enzyme activity, it is not generally known whether antibody responses to Sap proteins include inhibitory antibodies. The demonstration of direct inhibition of Sap activity by antibodies resulting in a protective effect in vivo would open up a new avenue of research and is an important area for further study.
(iv) Sap B-cell epitopes.
Only one study has endeavored to determine the B-cell epitopes of Candida Sap proteins for nondiagnostic purposes. Using sera from patients with oral and systemic candidiasis, Ghadjari et al. (73) delineated six Sap2 IgG and IgM epitopes. Human recombinant antibodies against two of these epitopes, though, were not protective in a mouse model of lethal systemic candidiasis. Nevertheless, this study provided some information regarding the Sap2 epitopes recognized by serum IgG and IgM antibodies. As yet, the mapping of epitopes relevant to mucosal immune protection by using saliva or vaginal secretions has not been performed for animals or humans. This would allow the identification of IgA epitopes, which would be particularly relevant to C. albicans infection at mucosal sites and would be more relevant for mucosal infections than would identifying serum IgG and IgM epitopes. Furthermore, B-cell and T-cell mapping of other members of the Sap family has not yet been undertaken, even though there is now solid evidence to indicate that they probably play significant but distinct roles during different C. albicans infections. Identification of such epitopes does not necessarily imply that they will be relevant to infection or host protection, but, given the crucial relationship between C. albicans pathogenesis and the immune status of the host, further studies in this area are clearly required.
SAP Gene Expression during Candida Infections
Main focus points.
The main focus points are as follows.
(i) SAP expression has been detected in all types of C. albicans infections by using various gene expression detection techniques.
(ii) In vitro reconstituted human epithelium (RHE) models appear to be good surrogates of human infections.
(iii) C. albicans expresses the SAP4 to SAP6 subfamily in all mucosal and systemic infections examined.
(iv) Future molecular studies will provide a more accurate representation of SAP gene expression during various types of C. albicans infections, particularly in humans.
The discovery that C. albicans possessed a multitude of proteinase genes that were differentially expressed under a variety of environmental conditions in vitro (96, 246) led to the attractive proposition that different members of the Sap family might also be differentially expressed in vivo and might contribute to different C. albicans infections. This concept, together with the knowledge that C. albicans inhabits a diverse number of host niches, was the driving force behind subsequent studies that investigated SAP gene expression in several models to ascertain which proteinases were expressed in which infections. This research involved human samples, animal models, and in vitro experimental human infections, and the findings are summarized in Table 5.
TABLE 5.
SAP gene expression in humans and in artificial and animal models
| Model | Infection | SAP genes | Assay | Main findings | Reference |
|---|---|---|---|---|---|
| Human | Oral | SAP1 to SAP7 | RT-PCR | SAP2 and SAP4 to SAP6 were the predominant proteinase genes expressed in the oral cavity of both patients with oral candidiasis and Candida carriers. SAP1 and SAP3 transcripts were observed only in patients. SAP7 mRNA expression (never previously demonstrated) was detected in carriers and patients. | 156 |
| Human | Oral and vaginal | SAP1 to SAP8 | RT-PCR | Certain SAP genes are preferentially expressed in the oral cavity and vaginal lumen. Individual SAP genes are more frequently expressed during active C. albicans infection than during carriage. | 157 |
| RHE | Oral | SAP1 to SAP6, SAP8 | RT-PCR | Progression of SAP expression occurred in the order SAP1 and SAP3 > SAP6 > SAP2 and SAP8. SAP6 expression was concomitant with germ tube formation and severe lesions. SAP4 and SAP5 transcripts were not detected. | 202 |
| RHE | Vaginal | SAP1 to SAP10 | RT-PCR | Progressive SAP expression occurred in the order SAP2, SAP9, and SAP10, followed by SAP2, SAP4, and SAP5, and finally SAP6 and SAP7. | 198 |
| RHE | Cutaneous | SAP1 to SAP6, SAP8 | RT-PCR | Progressive SAP expression occurred in the order SAP1 and SAP2 > SAP8 > SAP6 > SAP3 during the course of infection. Concomitant expression of SAP6 was found with germ tube formation and hyphal growth. | 201 |
| Mouse | Oropharyngeal (wild-type and transgenic mice expressing HIV-1) | SAP1 to SAP9 | RT-PCR | SAP7 and SAP8 were transiently expressed in both wild-type and transgenic mice. Sustained expression of other SAP gene occurred during the course of infection. SAP5 and SAP9 were strongly expressed throughout infection in transgenic mice. | 178 |
| Mouse | Oropharyngeal, intravenous, and intraperitoneal | SAP1 to SAP6 | RIVET | In esophageal candidiasis, SAP5 and SAP6 were strongly activated. In an intravenous model, SAP4 to SAP6 were the main SAP genes activated. In intraperitoneal infections, SAP5 was the main gene activated both in the initial stages and during invasion and dissemination. SAP1 to SAP3 were observed later in the disseminated models. SAP5 appeared to be the main gene activated mucosally and systemically. | 223 |
| Mouse | Gastrointestinal | SAP1 to SAP6 | RT-PCR | SAP4 and SAP6 were always detected, whereas SAP2, SAP3, and SAP5 were detected occasionally. SAP1 was not detected. | 119 |
| Mouse | Gastrointestinal | SAP1 to SAP6 | RIVET | There was a high percentage of SAP4 to SAP6 activation, which increased steadily during the course of infection. SAP1 to SAP3 activation was detected occasionally and at lower percentages than SAP4 to SAP6 activation. | 119 |
| Mouse | Intraperitoneal | SAP1 to SAP10 | RT-PCR | Within the first 72 h, SAP1, SAP2, SAP4, SAP5, SAP6, and SAP9 were most commonly expressed. SAP2, and SAP4 to SAP6 were detected in all samples. SAP3 was rarely detected, and SAP7 was never detected. | 65 |
| Rat | Vaginal | SAP1 and SAP2 | Northern blotting | SAP1 and SAP2 were expressed by two vaginopathic C. albicans strains and not by a nonvaginopathogenic strain, but only in the first week of infection. | 47 |
Human infections.
The precise roles and functions of the C. albicans proteinases during human infections are currently not clear (although information can be extrapolated from in vitro and in vivo animal model data [see below]). However, before these roles and functions can be investigated, the expression of the SAP gene family during different human infections needs to be assessed in order to determine which proteinases are produced in vivo. Naglik et al. (156) published the only detailed study of humans, in which SAP1 to SAP7 expression in the oral cavities of both patients with oral candidiasis (n = 10) and asymptomatic Candida carriers (n = 8) was analyzed (Table 5). SAP2 and SAP4 to SAP6 (SAP4 to SAP6 were detected together as a subfamily and not individually) were the predominant proteinase genes expressed in the oral cavities of infected patients and Candida carriers, while SAP1 and SAP3 transcripts were observed only in patients. SAP7 mRNA expression, which had never previously been demonstrated in vitro or in vivo, was readily detected in both carriers and patients. The same authors have since significantly extended their analysis to assess SAP gene expression in over 130 subjects with oral or vaginal C. albicans infection or asymptomatic carriage. The results indicate that not only are certain hydrolytic enzymes preferentially expressed in the oral cavity and vaginal lumen but also individual SAP and PLB genes are more frequently expressed during active C. albicans infection than during carriage (157). In addition, the data conclusively show that a spectrum of SAP genes can be expressed by different C. albicans strains in vivo during the course of the same disease (i.e., oral or vaginal) (Table 5).
Schaller et al. (202) also analyzed SAP1 to SAP6 and SAP8 expression in two patients with oral candidiasis: an HIV-negative female and an HIV-positive male, both suffering from pseudomembranous candidiasis. This analysis was undertaken to confirm the SAP expression data obtained using an in vitro model of oral candidiasis based on RHE and to determine whether the RHE models could be used as surrogates for human infections (see below). However, since only two patient samples were analyzed, no conclusions could be drawn regarding which SAP genes were associated with human oral candidiasis.
These are the only studies to have analyzed the expression of a C. albicans virulence gene family in any human infection, which highlights the distinct lack of human gene expression data available. Future experiments using patient samples should bring us one step closer to identifying the genes and proteins (SAP or otherwise) that are directly involved in Candida pathogenesis in humans.
In vitro artificial experimental infections based on RHE.
Most studies that analyze the virulence of Candida species use animal models (see below) rather than human subjects. However, recently a number of researchers have taken advantage of artificial RHE models and used them as an alternative to samples isolated directly from human patients. The RHE models (oral, vaginal, skin, and other cell types) can be obtained commercially and are prepared on an inert supporting membrane (190a). The models are highly reproducible, closely represent the human epithelium in vivo, and provide clear answers regarding gene expression that are not complicated by non-epithelial-cell factors. This, therefore, allows the direct analysis of the interaction between C. albicans and epithelial cells at the mucosal surface. Furthermore, it allows postulates to be proved experimentally.
Using an RHE model of oral candidiasis and the application of reverse transcription-PCR (RT-PCR), Schaller et al. (202) showed a distinct order of SAP gene expression (SAP1 and SAP3 > SAP6 > SAP2 and SAP8) during the course of infection. In this model, Sap1 to Sap3, but not Sap4 to Sap6, were associated with tissue damage (200). Interestingly, a different SAP expression pattern was observed using an RHE model of vaginal candidiasis: SAP2, SAP9, and SAP10 transcripts were observed initially, followed by SAP1, SAP4, and SAP5 (concomitant with lesions), with SAP6 and SAP7 being expressed during the later stages of infection (198). However, although the gene expression pattern was different from that in the oral RHE model, studies with mutants indicated that Sap1 and Sap2 were again the proteinases associated with tissue damage (198). Lastly, in an RHE model of cutaneous candidiasis, expression of SAP genes in the order SAP1 and SAP2 > SAP8 > SAP6 > SAP3 was demonstrated when Candida cells penetrated the corneal layer of the artificial skin (201). These studies with RHE models clearly suggest that Sap1 to Sap3 are the main C. albicans proteinases contributing to the early stages of mucocutaneous infections (Table 5).
Although the in vitro RHE models closely represent the human epithelium in vivo, it would be prudent to interpret the gene expression data with caution since the RHE models fall short of directly representing the in vivo situation in human patients. Parameters such as pH changes, nutrient limitation, competition with oral flora, and humoral, cell-mediated, and cytokine responses are likely not only to have a profound effect on the ability of C. albicans to grow unchallenged on the mucosal surface (as in the models) but also to provide strong local pressures that may influence the differential expression of the SAP gene family. However, despite this cautionary note, these RHE models have proved very effective for the direct observation of SAP gene expression in the initial stages of superficial infections and, at present, are the best in vitro surrogates for in vivo human Candida infections. The use of these artificial RHE models in the future should certainly lead to the discovery of new Candida genes that are expressed in human mucosal infections and should enlighten our understanding of host-Candida interactions.
Animal experimental models.
The development of animal models for specific diseases that precisely mimic their human equivalent is the “gold standard” of animal research. However, given the dissimilar nature of humans and since C. albicans is an opportunistic pathogen that adapts continuously to its environment, it is improbable that animal models can be developed that are fully representative of human Candida infections. Nevertheless, in truth, the use of animals is the best in vivo alternative to study mucosal and systemic candidiasis, with the mouse and rat models being the most commonly used. SAP gene expression in both mucosal and systemic C. albicans infections are described in Table 5.
(i) Mucosal models.
De Bernardis et al. (47) were the first to demonstrate SAP gene expression in vivo in an estrogen-dependent rat vaginitis model. Northern analysis indicated that SAP1 and SAP2 were expressed by two vaginopathic C. albicans strains but not by a nonvaginopathogenic strain, which supported a link between SAP expression and the virulence potential of certain C. albicans strains. However, now that 10 members of the SAP family have been identified, it would be interesting to examine the SAP1 to SAP10 expression pattern in this well-established rat model and compare the profiles with the RT-PCR and in vivo expression technology (IVET) mouse data described below. A comparison between humans, mice, and rats would provide an opportunity to determine how representative the rat or mouse models are to human Candida infections. The reader is guided to a detailed review recently published by Samaranayake and Samaranayake (192), which discusses the pros and cons of different animal models that have been used to study Candida infections.
Several groups have reported detailed SAP gene expression studies in murine mucosal models. In a mouse model of gastrointestinal infection, SAP4 and SAP6 were constitutively expressed whereas SAP2, SAP3, and SAP5 mRNA were only occasionally detected (119). Interestingly, SAP1 mRNA was not detected in this model, which is worthy of note because SAP1 expression has been observed in clinical C. albicans samples from infected human patients and carriers (156), a mouse oropharyngeal model (178), a rat vaginitis model (47), and oral (202) and vaginal RHE models (198) (Table 5), all of which are mucosal infections.
In a mouse model of oropharyngeal candidiasis, a sequential analysis of the temporal expression of SAP1 to SAP9 in normal and transgenic mice expressing HIV-1 showed that SAP1 to SAP6 and SAP9 transcripts were detected continuously throughout the course of infection, with SAP5 and SAP9 being most strongly expressed in transgenic mice (178). SAP7 and SAP8 were the only genes to be expressed transiently, supporting the current evidence that members of the SAP family are differentially expressed during C. albicans infections (Table 5).
The SAP expression data described above were obtained using RT-PCR, but IVET, a relatively new technique, has recently been developed to analyze gene activation during infective processes (241). One of these technologies, the recombination-based IVET technique (RIVET), uses site-specific recombination as a reporter of gene activation and is especially suited to detect the in vivo induction of genes that are only transiently expressed at a certain stage of infection (25), RIVET using FLP-recombinase has recently been adapted for use with C. albicans by Staib et al. (223) to analyze the expression of SAP1 to SAP6 in various mouse infection models. In a mouse model of esophageal candidiasis, SAP5 and SAP6 were strongly activated, indicating that these two SAP genes might be directly involved in the colonization and perhaps penetration of mucosal surfaces (Table 5).
(ii) Systemic models.
All SAP expression studies with animal systemic models have been performed with mice. In an intraperitoneal-infection model, SAP1, SAP2, SAP4, SAP5, SAP6, and SAP9 were discovered to be the most commonly expressed proteinase genes within the first 72 h (65). SAP2 and SAP4 to SAP6 transcripts were identified in all infected tissues at 4, 8, 24, and 72 h, SAP3 and SAP10 were less frequently expressed (in less than 50% of the organs), and SAP7 was never detected. The lack of SAP7 expression is noteworthy since this gene is expressed in human oral and vaginal infection and carriage (156, 157), in a mouse model of oropharyngeal candidiasis (178), and in the RHE vaginal model (198) (Table 5). It may be premature to conclude that this implicates SAP7 specifically in mucosal rather than systemic infections, but it tentatively supports some kind of role for this proteinase during human mucosal infections.
Using RIVET, SAP4 to SAP6 were the main SAP genes activated in an intravenous model of disseminated candidiasis and SAP5 was the main gene activated in disseminated intraperitoneal infections, both at the initial time of inoculation and during invasion and subsequent dissemination to the kidneys (223). However, SAP1 to SAP3 were also observed later in the infective process (223, 225). This pattern of SAP gene activation during systemic infections suggested a role for SAP4 to SAP6 during the initial phases of organ invasion and SAP1 to SAP3 in later phases, when C. albicans had already established infection.
RIVET was also used to assess SAP1 to SAP6 gene activation in a murine gastrointestinal model (119). SAP4 to SAP6 activation was detected in a high percentage of C. albicans cells, which increased steadily during the course of infection. In contrast, SAP1 to SAP3 activation was detected only occasionally and at lower percentages than SAP4 to SAP6. These results correspond well to the RT-PCR expression analysis in the same model, which demonstrated constitutive expression of SAP4 and SAP6 and only the occasional detection of SAP2 and SAP3, with SAP1 transcripts never being observed (119). The RT-PCR and RIVET studies thus appear to support some kind of role for SAP4 to SAP6 in both mucosal and systemic C. albicans infections.
Universal expression of the SAP4 to SAP6 subfamily at mucosal surfaces.
A common feature of all the studies described above is the ubiquitous expression of the SAP4 to SAP6 subfamily, regardless of the infection model (human or mouse, systemic or mucosal) or the assay technique used to detect SAP gene expression (RT-PCR or RIVET) (Table 5). Although a direct role for SAP4 to SAP6 in systemic Candida infections has been indicated from studies using SAP4- to SAP6-deficient strains (see “Use of SAP-disrupted mutants to analyze C. albicans virulence” below), the prevalent expression of SAP4 to SAP6 in mucosal infections but the lack of attenuation of virulence using SAP4 to SAP6-deficient strains poses numerous questions. (i) Are certain members of the SAP4 to SAP6 subfamily required by C. albicans for a housekeeping role or for some kind of essential or constant interaction with mucosal surfaces? (ii) Are SAP4 to SAP6 required simply for nutritional purposes in vivo, hence the necessity for their expression, or are they in fact essential for Candida virulence (i.e., invasion and tissue damage)? (iii) Alternatively, do SAP4 to SAP6 have nonenzymatic functions at mucosal surfaces? It is entirely feasible that although SAP4 to SAP6 are expressed by C. albicans in mucosal infections, they may not be required for virulence as they appear to be in systemic infections. At present, the roles and functions of the SAP4 to SAP6 subfamily during mucosal infections, and specifically during the infective process in humans, are not clear, but future functional studies might shed light on why SAP4 to SAP6 are so prevalent at mucosal surfaces.
Discussion and future directions for SAP expression studies.
While these SAP expression studies have been vital in determining which SAP genes are associated with which types of C. albicans infections, the true depth of their involvement in C. albicans pathogenicity has yet to be determined. This is confounded by the many differences in SAP gene expression profiles observed between the studies (Table 5). Although this may simply allude to the intricate and complex relationship between Sap production, Candida pathogenesis, and the host, a number of other explanations are possible. These include the technique used for SAP gene analysis (RT-PCR, IVET, or Northern blot analysis), the model used (human, mouse, rat, or RHE), or the site of infection studied (oral, oropharyngeal, vaginal, gastrointestinal, cutaneous, or systemic). In fact, there are no directly comparable studies by different authors using the same technique, model, or site of infection (Table 5).
In addition to these disparities, the environmental conditions alone at the different infection sites, including pH, substrate availability, and ionic content, can differ markedly and could partly account for the observed differences in SAP expression between the studies. With these considerations in mind, it is entirely probable that C. albicans has adapted to these niche sites in different hosts by expressing a different set of SAP genes. Furthermore, even within a single infection site, one would expect different gene expression patterns within single C. albicans cells, but all the studies to date have described the average SAP expression pattern of a population of cells.
Therefore, we require studies that use a single technique (RT-PCR or IVET) to analyze SAP gene expression in numerous infection models (human, mouse, rat, and RHE) or that use various gene expression techniques in a single infection model. Ideally, the former should reveal differences in SAP gene expression between infected sites and the latter should generate matching SAP expression data, since the infection is the same. The availability of more of these “multimodel” and/or “multitechnique” studies will add to our understanding of SAP gene expression in the host and its relevance to Candida pathogenesis.
Finally, the reader should note that the quantitative expression of the SAP genes or any other virulence gene family of Candida has not yet been assessed, and this is an important next step (particularly at the single-cell level) in addressing the up- and down-regulation of these virulence genes during various stages of different C. albicans infections. In addition, now that the C. albicans genome sequence is complete (http://www-sequence.stanford.edu/group/candida), the development of DNA microarrays specifically for Candida research should significantly expand our knowledge of which genes may be contributing not only to C. albicans infections but also specifically to different types of human infections.
Modulation of C. albicans Virulence by Aspartyl Proteinase Inhibitors
Main focus points.
The main focus points are as follows.
(i) Aspartyl proteinase inhibitors can inhibit C. albicans adherence and attenuate mucosal infections.
(ii) Proteinase inhibitors appear ineffective at preventing systemic C. albicans infections.
(iii) More conclusive studies using all main classes of proteinase inhibitors are required.
Drug-resistant Candida infections are likely to pose a serious therapeutic challenge over the next few decades. Consequently, discoveries of novel drug classes are crucial aims of antifungal research. One of the opportunities for the development of new compounds active against Candida species includes the development of drugs directed against the Candida proteinases. This is particularly pertinent now that the three-dimensional structure of one member of the Sap family (Sap2) has been determined (1, 39). Over the last two decades, and particularly within the last few years, two areas of interest—the role of Candida proteinases in pathogenesis and their potential as antifungal targets—have driven the use of aspartyl proteinase inhibitors (PIs) in Candida research. These studies using the classical aspartyl PI pepstatin, HIV PIs, and computer-assisted structure-based designed inhibitors have provided direct evidence demonstrating the contribution of Sap proteins to C. albicans virulence and have indicated that the proteinase family may be a possible target for antifungal research.
Pepstatin.
The contribution of proteinases to C. albicans adherence, invasion, and infection has long been advocated, but it was not until the use of the classical aspartyl PI pepstatin that many of these associations were confirmed. The studies that used pepstatin to demonstrate a link between Sap proteins and adherence are detailed in “Association of Sap production with other virulence processes of C. albicans” (above) and are not readdressed here, but the investigations using pepstatin to associate Sap proteins with Candida penetration and invasion of host tissues are described below.
(i) Use of pepstatin in vitro.
Colina et al. (36) showed that the digestion of mucin could be inhibited by pepstatin, indicating that Candida Sap proteins may degrade mucosal barrier proteins. This may allow C. albicans to gain access to the oral and gastrointestinal mucosa and may consequently indicate a role for Candida proteinases in dissemination from these colonized sites. Likewise, the addition of pepstatin inhibited the in vitro digestion of soluble and immobilized extracellular matrix proteins produced by a human endothelial cell line (152). Again, this suggests that Candida Sap proteins contribute to cell damage and invasion of the subendothelial extracellular matrix, which in turn could facilitate dissemination via the circulatory system. In an RHE model of oral candidiasis, pepstatin was used to demonstrate a role for Sap1 to Sap3 but not Sap4 to Sap6 in early tissue damage of oral epithelium (200). Pepstatin noticeably reduced the tissue damage caused by Sap1 to Sap3; however, even in the presence of pepstatin, epithelial tissue damage was observed in late stages of RHE infections. This suggested that certain members of the Sap family that might not be inhibited effectively by pepstatin or other hydrolytic enzymes of C. albicans such as phospholipases and lipases (which are not inhibited by pepstatin) might also contribute to the development of mucosal lesions.
(ii) Use of pepstatin in vivo.
While in vitro assays evaluating the effect of pepstatin on Candida infections are necessary and provide valuable information, the most compelling biological data for antifungal research come from in vivo studies of Candida infection. The first report of experiments using pepstatin in vivo was published in 1988 and described the use of a systemic model of murine candidiasis (58); however, no real protective effect was observed after intravenous challenge with C. albicans. A similar lack of protection was also reported by two subsequent studies, using an in vivo pepstatin concentration of up to 9 × 10−4 M (188, 258). However, a recent study of murine peritonitis demonstrated that the addition of pepstatin (1 mg/kg) significantly reduced the amount of liver and pancreatic tissue damage, as determined by decreased levels of alanine aminotransferase and α-amylase enzyme activities, respectively (120). Tissue damage appeared to be caused by Sap4 to Sap6, a conclusion obtained from using SAP-deficient strains (120). These findings indicated that pepstatin could attenuate C. albicans virulence in a systemic murine intraperitoneal model, probably by inhibiting tissue damage and invasion by Sap4 to Sap6.
While the evidence supporting a protective role for pepstatin in systemic animal models remain unconvincing, particularly after intravenous challenge with Candida, its potential use at mucosal surfaces is more convincing. Protective effects of pepstatin were demonstrated in a rat vaginitis model, where postinfective administration of pepstatin (1 mg/ml) greatly accelerated the clearance of C. albicans from the rat vagina (45, 49). Fallon et al. (64) showed in a mouse model that inhibition of Sap proteins by pepstatin (0.06 to 6 mg/kg) prevented the initial penetration of C. albicans through mucosal surfaces but not the dissemination of C. albicans once the cells had already reached the blood vessels. Thus, one conclusion may be that pepstatin can prevent disseminated infections by inhibiting Candida penetration through the mucosal route but cannot inhibit systemic infections when the Candida is administered via the intravenous route. In other words, the route of Candida challenge rather than the route of pepstatin administration might be more relevant to observe protective effects by pepstatin.
These studies indicate that proteinase inhibition by pepstatin can reduce the ability of C. albicans to colonize and invade host tissues and illustrate the potential benefits of pepstatin in vivo, chiefly in mucosal models. Unfortunately, however, pepstatin is not selective and is slightly toxic in animals, probably due to its inhibitory action on host aspartyl proteinases, including cathepsin D and renin (188). Moreover, pepstatin accumulates in the liver and not the kidneys, which is a major target organ of C. albicans in systemic infections, and is thus likely to be rapidly cleared from the blood in vivo. Given that the concentrations of pepstatin used in the above experiments are manyfold higher than those required to effectively inhibit Sap2 (inhibition constant [Ki] for Sap2 of 2.9 nM and 50% inhibitory concentrations [IC50s] of 27 nM [1, 226]), rapid clearance may partly explain the ineffectiveness of pepstatin in systemic models after intravenous administration of Candida. Therefore, while pepstatin is a potent inhibitor of the C. albicans aspartyl proteinases in vitro, its suitability as an antifungal agent in vivo is not compelling.
HIV PIs.
The interest in discovering new compounds that inhibit Sap activity was recently given a surprising and unexpected boost as a result of the evolving HIV epidemic. Since 1996, the treatment of HIV-positive patients with highly active antiretroviral therapy (HAART), which includes a cocktail of HIV reverse transcriptase and HIV PIs, has proved successful in delaying the onset of AIDS. Administration of HAART results in a significant improvement in the immune status of the HIV-positive individual, reflected by an increase in the CD4+ T-cell levels. These patients also tend to have a dramatically reduced incidence of oropharyngeal candidiasis (54, 92, 240). Since the Candida proteinases and the HIV proteinase are members of the same aspartyl proteinase family, these findings led to the hypothesis that HIV PIs may also act against Candida aspartyl proteinases in vivo and consequently prevent or reduce candidal infections directly (31, 83, 89). A spate of studies has followed to determine whether HIV PIs could inhibit C. albicans Sap proteins and whether they had any potential as therapeutic agents in the treatment of C. albicans infections. The main conclusions from these studies are that HIV PIs are able to inhibit Sap activity, C. albicans adherence, and tissue damage but have a limited effect on C. albicans viability (Table 6).
TABLE 6.
Effect of HIV PIs on Sap activity and C. albicans virulence
| HIV PI | Main findings | Reference(s) |
|---|---|---|
| In vitro | ||
| Ritonavir, indinavir, saquinavir | All three inhibited Sap2 activity. Only saquinavir was fungicidal, but only at high doses. | 80 |
| Indinavir | Indinavir weakly inhibited Sap2 and reduced the amount of cell-bound and secreted proteinase. C. albicans viability and growth were markedly reduced. | 83 |
| Ritonavir, indinavir, saquinavir, nelfinavir | Ritonavir and saquinavir inhibited Sap2 at micromolar concentrations, whereas indinavir and nelfinavir had no effect. Ritonavir and saquinavir also inhibited Sap proteins from C. tropicalis, C. parapsilosis, and C. lusitaniae. | 168 |
| Ritonavir, indinavir, saquinavir, nelfinavir | All four specifically inhibited Sap1 to Sap3, but not Sap4 to Sap6. Ritonavir and saquinavir inhibited C. albicans adherence to epithelial cells, whereas indinavir had no effect. No effect on C. albicans viability. | 15 |
| Saquinavir, indinavir | Both inhibited Sap2 activity. Saquinavir strongly attenuated tissue damage in an in vitro RHE model of oral candidiasis. | 117 |
| Indinavir, ritonavir | Neither had an antifungal effect. | 55, 56 |
| Ritonavir, indinavir, saquinavir | All three inhibited C. albicans adhesion to human epithelial cells; ritonavir was the most potent. None modulated phagocytosis of Candida. | 10 |
| In vivo models | ||
| Indinavir, ritonavir | Both inhibited Sap2 activity. Both exerted a therapeutic effect in an estrogen-dependent rat vaginitis model. | 29 |
| Indinavir, nelfinavir, ritonavir | The anti-Sap effect of PI-HAART was associated with clinical resolution of oral candidiasis in HIV-positive patients. | 30 |
(i) Inhibition of Candida proteinase activity by HIV PIs in vitro.
A total of four HIV PIs, namely ritonavir, saquinavir, indinavir, and nelfinavir, have been investigated for their ability to inhibit Candida proteinase activity (15, 29, 80, 83, 117, 168). Ritonavir was consistency found to be the most potent inhibitor of C. albicans Sap2 activity (15, 80, 168), with Ki of 0.34 μM (168), while saquinavir, indinavir, and nelfinavir inhibited Sap2 activity to differing extents. One study did not observe inhibition of Sap2 activity with indinavir or nelfinavir, which equated well with the poor Ki values of these compounds against Sap2 (Ki = >1,000 μM for both) (168). Of the four HIV PIs, only ritonavir and saquinavir were able to inhibit the activity of secreted proteinases purified from C. tropicalis, C. parapsilosis, and C. lusitaniae as well as those purified from C. albicans (168) (Table 6).
With regard to the inhibition of different members of the Sap family, Borg-von Zepelin et al. (15) discovered that all four HIV PIs specifically inhibited the C. albicans Sap1 to Sap3 but not Sap4 to Sap6 enzymes. Interestingly, ritonavir and saquinavir, but not indinavir, inhibited the adherence of C. albicans to Vero epithelial cell cultures in a dose-dependent manner, thus supporting a role for Sap1 to Sap3 in the adherence process (15). Similar results were also obtained by Bektic et al. (10) using HeLa S3 epithelial cell cultures; these investigators showed that ritonavir and saquinavir, as well as indinavir on this occasion, inhibited C. albicans adhesion, with ritonavir being the most potent. However, none of the HIV PIs modulated the phagocytosis of Candida by polymorphonuclear leukocytes (10). Since Sap4 to Sap6 are thought to assist C. albicans in evading phagocytosis by murine macrophages (16), these data support the notion that the HIV PIs might not inhibit Sap4 to Sap6 activity (15).
(ii) Effectiveness of HIV PIs against Candida infection in RHE and animal models.
The HIV PIs have been tested in artificial and animal models to determine their effectiveness at attenuating C. albicans infections (Table 6). Using an RHE model of oral candidiasis, saquinavir, but not indinavir, strongly attenuated the ability of C. albicans to cause tissue damage (117). Since there is good evidence that tissue damage in this model may result from the activity of the Sap1 to Sap3 subfamily (200), the data support the evidence that Sap1 to Sap3 contribute to the development of mucosal tissue damage. With hindsight, it is regrettable that ritonavir was not investigated for its ability to reduce tissue damage in this RHE model, since ritonavir appears to be the most potent inhibitor of Sap1 to Sap3 (15, 80, 168).
In a rat vaginitis model, ritonavir and indinavir exerted a therapeutic effect with an efficacy comparable to that of fluconazole (29). Since C. albicans infection appears to be dependent on Sap1 to Sap3 expression in this model (44, 45, 47) and since ritonavir and indinavir inhibit Sap1 to Sap3 but not Sap4 to Sap6 (15), the data indicate that these HIV PIs may indeed act against Sap1 to Sap3 during the infective process in vivo, resulting in reduced C. albicans infection.
(iii) Effectiveness of HIV PIs against Candida infection in humans.
Notwithstanding the basic scientific question of whether the HIV PIs can inhibit proteinase activity per se or C. albicans infection in experimental models, the underlying principle in performing the above studies is to determine whether the HIV PIs could be used therapeutically to treat Candida infections in humans. Only one report has addressed this issue to date, and this study claimed that the anti-Sap effect of the HIV PIs appeared to be associated with the clinical resolution of oral candidiasis (30). However, in vitro data indicate that the HIV PIs are very weak inhibitors of Sap2 compared with pepstatin (168) (Table 7) and do not affect C. albicans viability (15) (Table 6). In addition, very approximate estimates of the IC50s obtained for the HIV PIs in vivo (∼2 to 100 μM) (15, 226) were between two and four orders of magnitude higher than the IC50 of pepstatin (∼27 nM) (26, 226), which itself has unconvincing protective effects against systemic Candida infections and modest protective effects against mucosal infections (see “Pepstatin” above). Another issue that complicates this matter is that in vivo ritonavir and saquinavir are >97% bound to plasma protein, primarily α1-acid glycoprotein (3). As a consequence, very little drug remains to diffuse to mucosal surfaces where Candida resides during episodes of oropharyngeal candidiasis. Also, given that the ritonavir and saquinavir concentrations in saliva are approximately 100-fold lower than those found in human plasma (in the nanogram-per-milliliter range) (90), it appears unlikely that ritonavir and saquinavir (and the other HIV PIs) could inhibit Sap activity to such an extent as to be therapeutically active against mucosal Candida infections in humans.
TABLE 7.
Inhibition constants (Ki) and IC50 of inhibitors of C. albicans Sap2
| Inhibitor | Sap2 Ki (nM) | IC50 (nM) | In vivo effects |
|---|---|---|---|
| Pepstatin | 2.9a | 27a | Protective effect in murine and rat mucosal models (45, 49). Lack of protection in murine disseminated-infection models (58, 64, 188, 258). Possible protection in murine peritonitis (117). |
| Ritonavir | 300b | 2,000a | Therapeutic effect in a rat vaginitis model (29). |
| Saquinavir | 6,800b | 300,000a | Not tested in vivo. |
| Indinavir | >106b | 100,000a | Therapeutic effect in a rat vaginitis model (29). |
| A-70450 | 0.17a | 1.4a | No protection in a murine disseminated-infection model (1, 227). |
| A-79912 | NDc | 3.8a | No protection in a murine disseminated-infection model (1, 227). |
| Peptidomimetics | 0.6-14.4b | ND | Not tested in vivo. |
Based on the current data, there is insufficient information to support the potential role of HIV PIs as therapeutic agents for human Candida infections in HAART-treated HIV-positive patients. Further detailed work in this area is essential before any such proposal can be seriously recommended.
Computer-assisted structure-based designed inhibitors.
The pepstatin studies provided good evidence indicating a role for Sap proteins in adherence, invasion, and infection of host tissues (see above); however, the conclusion that pepstatin was not suitable as an antifungal agent led some laboratories to use computer-assisted design methods to develop new drugs to inhibit the C. albicans proteinases. The most advanced of these development programs was headed by Cele Abad-Zapatero at Abbott Laboratories, who noted that some renin inhibitors also inhibited C. albicans Sap2 activity. One inhibitor, A-70450, with a Ki for Sap2 of 0.17 nM and an IC50 of 1.4 nM, was a significant improvement on the inhibition properties of pepstatin (IC50 = 27 nM; Ki = 2.9 nM) (26, 226). Analogues of this new compound were synthesized, and one, named A-79912, retained potent activity against Sap2 (IC50 = 3.8 nM) and, favorably, had significantly reduced activity against key host aspartyl proteinases including renin (IC50 = 110 nM) and cathepsin D (IC50 = 22,000 nM) (1, 226).
Both A-70450 and A-79912 were extremely effective at inhibiting Sap2 activity in vitro, but, disappointingly, both were ineffective at attenuating C. albicans virulence in a mouse model of disseminated candidiasis under conditions that give a strong antifungal effect for the azole antifungal fluconazole (1, 227). This suggested that both these novel inhibitors may have similar inherent problems to pepstatin when used in vivo, in that the lack of protection may be due to the lack of inhibitor potency, low specificity, or the inability to completely inhibit all the Sap proteins or at least the members associated with C. albicans virulence in systemic infections. Be that as it may, A-70450 and A-79912 are the most potent PIs designed to date (Table 7). With the prospect of elucidating the three-dimensional structures of more members of the Sap family, structure-based inhibitor design methods may be the most practical way forward to generate inhibitors with the potency and selectivity needed for the inhibition of the Sap family in vivo and potentially for the subsequent treatment of Candida infections (226).
Other inhibitors of Candida Sap proteins.
The identification of novel Candida Sap inhibitors, either through design by modification of existing inhibitors or by isolation of the inhibitors in their natural state from microbes, has been reported. Pichova et al. (168) designed a series of peptidomimetic inhibitors, derived from the structure of pepstatin A, and showed that most of the inhibitors were essentially equally active against four purified secreted proteinases isolated from C. albicans (Sap2), C. tropicalis (Sapt1), C. parapsilosis (Sapp1), and C. lusitaniae (Sap1). Unfortunately, these peptidomimetic inhibitors were not tested in animal models, but one might hypothesize that since they are based on pepstatin, similar ineffective protective properties may be observed during systemic Candida infections to found previously (58, 64, 188, 258).
Very recently, the first potent inhibitor of S. cerevisiae yapsin 1, termed Y1, with an apparent Ki of 64.5 nM, was reported (32). Y1 also inhibited Sap9 from C. albicans, which has sequence homology to the S. cerevisiae yapsins including potential GPI sites. Other groups have identified natural molecules isolated from microorganisms with inhibitory activity against the C. albicans proteinases. These include compounds from Streptomyces species (YF-0200R-A/B and YF-044P-D) (196, 197), numerous xanthones from Tovomita krokovii (256), the serratene triterpenes from Lycopodium cernuum (255), and phenolic compounds from Miconia myriantha. The in vivo efficacy of all these compounds was not investigated; however, the IC50s alone indicated that these compounds were not serious candidates as C. albicans Sap inhibitors.
Discussion and future directions for PI studies.
Studies using PIs have been instrumental in demonstrating the direct contribution of the Sap proteins to C. albicans virulence and have confirmed their status as true virulence factors of C. albicans. In addition, the data specifically implicates the Sap1 to Sap3 subfamily in causing mucosal tissue damage and disease pathology. Despite this, the in vivo benefits of the majority of these PIs has on the whole been rather disappointing. However, there are numerous shortfalls in the literature regarding PIs, and so there is still plenty of scope in using current and potentially new Sap inhibitors. For instance, the potency of Sap inhibition of the different classes of PIs has not been fully determined. Pepstatin inhibits Sap1 to Sap6 (16), but the strength of inhibition has been determined only for Sap2. Similarly, the HIV PIs inhibit only Sap1 to Sap3 and not Sap4 to Sap6 (15), and the potency of inhibition is weak compared with that of pepstatin (168, 226). The designed drugs A-70450 and A-79912 strongly inhibit Sap2, and although one assumes that they also strongly inhibit Sap1 and Sap3 (within the same subfamily as Sap2), their inhibitory activity against Sap4 to Sap6 has not been tested. In addition, the inhibition potency of all of these compounds against Sap7 to Sap10 are unknown.
This lack of information concerning the inhibition of activity of the different members of the C. albicans proteinase family has implications for the way in which data obtained from animal model experiments is interpreted. For example, since pepstatin inhibits Sap1 to Sap6 (16) and with the knowledge that Sap4 to Sap6 are readily expressed and contribute to systemic infections (see “SAP gene expression during Candida infections” above and “Use of SAP-disrupted mutants to analyze C. albicans virulence” below), one might predict that pepstatin would attenuate systemic as well as mucosal infections, which does not appear to be the case. Also, why do the HIV PIs appear to be so effective at inhibiting C. albicans adherence and attenuating experimental mucosal infections when they are weak inhibitors of Sap1 to Sap3 compared with pepstatin? The reader should be aware that the HIV PIs have not been tested in a systemic model of Candida infection, but since the HIV PIs do not appear to inhibit Sap4 to Sap6 activity (15), one might predict little or no attenuation of C. albicans virulence. Similarly, A-70450 and A-79912 have not been examined in a mucosal model of C. albicans infection, but only in systemic models, where, like pepstatin, they were shown to be ineffective. Since A-70450 and A-79912 strongly inhibit Sap2 (and in all probability Sap1 and Sap3) (26), it would be much more appropriate to test these two compounds in a mucosal model, where they might be predicted to be highly effective at inhibiting adhesion and tissue damage of the oral mucosa, similar to pepstatin (200).
More conclusive studies are required in which all three groups of inhibitors (pepstatin, HIV PIs, and the designed inhibitors) are tested for their ability to inhibit the activity of all members of the C. albicans proteinase family and to attenuate mucosal and systemic C. albicans infections in numerous experimental models. Only then will sufficient information be available to determine the efficacy of these agents at preventing C. albicans infections. Furthermore, with the completion of the Candida genome sequence (http://www-sequence.stanford.edu/group/candida) and with the potential of elucidating the three-dimensional structures of more members of the Sap family, the prospects of developing new and powerful inhibitors specific to the Candida proteinases seem excellent. Future studies will judge whether the Candida proteinases are genuine targets for treatment therapies of mucosal or systemic infections and will underline the therapeutic potential of drugs that are targeted against virulence traits rather than essential primary metabolic or biosynthetic processes.
Use of SAP-Disrupted Mutants To Analyze C. albicans Virulence
Main focus points.
The main focus points are as follows.
(i) Studies using C. albicans SAP-deficient strains have implicated Sap1 to Sap3 in mucosal but not systemic infections and Sap4 to Sap6 in systemic but not mucosal infections.
(ii) Disruption of SAP genes may be compensated by the up- or down-regulation of other SAP genes.
(iii) Functional analysis of SAP7 to SAP10 using disrupted mutants has not yet been investigated.
Genetic manipulation of C. albicans has always been complicated by the diploid nature of the fungus and the fact that it has an unproven naturally functional sexual stage. As a result, for a long time it was very difficult to create mutant strains for the analysis of virulence properties. In addition, C. albicans exhibits unusual codon usage, inserting the amino acid serine instead of leucine at the normally leucine-specific codon CTG (cytosine-thymine-guanine) by the use of a special tRNA (195). Another difficulty is that very few suitable dominant selectable markers are available for the selection of genetic transformants, and researchers are usually dependent on the use of auxotrophic strains for the genetic engineering of C. albicans.
Before the development of modern molecular biology tools, many of the early studies investigating the role of proteinases in C. albicans virulence used less proteolytic or nonproteolytic C. albicans mutants that were induced by chemical or UV mutagenesis. Indeed, some of the strongest initial evidence supporting a role of Sap proteins in C. albicans pathogenesis came from these studies. Macdonald and Odds (134) showed that a proteolysis-deficient mutant strain of C. albicans was less pathogenic in mice than was the parental strain. Likewise, Ross et al. (180) isolated a proteinase-deficient mutant, which was 1,000-fold less virulent on the basis of the 50% lethal dose (LD)50 in mice than was the wild-type strain, an observation confirmed for the rat vaginitis model by De Bernardis et al. (43). Also, an earlier report by Kwon-Chung et al. (122) demonstrated similar reduced virulence with another proteolysis-deficient mutant and showed that a spontaneous revertant, which reacquired half of its original proteolytic activity, was almost as virulent as the parental strain.
However, as mentioned above, a major drawback of all these studies was that the proteinase-deficient mutants were obtained by chemical or UV mutagenesis. Thus, the results had to be interpreted with extreme vigilance because most, if not all, of these mutants almost certainly contained nonspecific mutations at other gene loci that may have affected the growth and/or virulence of C. albicans. To overcome these rather crude genetic manipulations of C. albicans and because of the diploid nature of the fungus, a method for gene disruption that allowed repeated use of the same selection procedure was developed (known as the Ura-blaster). The Ura-blaster protocol (reviewed in references 11 and 221) has allowed the analysis of a variety of genes involved in C. albicans pathogenesis, including those encoding the Sap proteins and phospholipases (124) and genes associated with hypha formation (17, 71, 128, 129, 228).
The genes for SAP1 to SAP6 and SAP8 (and recently SAP9 and SAP10 [A. Felk, A. Albrecht, and B. Hube, unpublished data]) have been disrupted in C. albicans strain SC5314 and tested in numerous in vitro and animal models to determine their roles and functions during mucosal and systemic Candida infections. Perhaps the most definitive data regarding the contribution of the SAP family to Candida pathogenicity have been obtained from the behavior of the various SAP-disrupted strains, and the main findings of these studies are summarized in Table 8.
TABLE 8.
Use of SAP-disrupted mutants in mucosal and systemic C. albicans infectionsa
| Study | Main findings |
|---|---|
| Mucosal infections | |
| 244 | sap1, sap2, and sap3 null mutants were less strongly adherent to buccal epithelial cells than was the parental strain in vitro. The sap4 to sap6 triple mutant had significantly increased adherence. |
| 200 | sap1, sap2, and sap3 mutants caused less tissue damage than did the parental strain in an in vitro RHE model of oral candidiasis. A sap1-sap3 double mutant caused fewer lesions than did the two single sap1 or sap3 mutants. The sap4 to sap6 triple mutant showed tissue damage equal to that for the parent strain. |
| 44 | sap1, sap2, and sap3 mutants, but not the sap4 to sap6 triple mutant, were less virulent in a rat vaginitis model than was the parent strain. The sap2 mutant was almost avirulent. |
| 119 | No demonstrable differences between SAP1 to SAP6 knockout strains and control strains in gastrointestinal infections of mice. |
| Systemic infections | |
| 103 | No difference in adherence to endothelial cells in vitro between the sap1, sap2, and sap3 null mutants and the parental strain. The sap2 mutant caused less damage to endothelial cells. |
| 120 | The sap4 to sap6 triple mutant, but not the sap1, sap2, or sap3 mutants, caused less tissue damage and invasion in murine peritoneal infections than did the parental strain. |
| 65 | Analysis of single sap4, sap5, and sap6 and double sap4-sap6, sap5-sap6, and sap4-sap5 mutants in a murine intraperitoneal model indicated that only those lacking SAP6 showed significantly reduced tissue damage. |
| 100, 194 | sap1, sap2, sap3, and sap4 to sap6 mutants were less lethal in two different animal models of disseminated infections than was the parental strain. The sap4 to sap6 triple mutant displayed the greatest attenuation. |
| Evasion of host immune responses | |
| 16 | The sap4 to sap6 triple mutant was eliminated 53% more effectively after phagocytosis by macrophages in vitro than was the parent strain. |
Reprinted from reference 97 with permission.
Contribution of Sap proteins to mucosal infections using SAP-deficient strains.
To determine whether the C. albicans proteinases played a role in mucosal infections, the ability of the SAP-deficient mutants to cause tissue damage in RHE models of oral and vaginal candidiasis was investigated. The sap1, sap2, and sap3 mutants caused less tissue damage than did the parental strain in both the oral (200) and vaginal (198) models, with a sap1-sap3 double mutant causing for fewer lesions than did either of the two single sap1 or sap3 mutants in the oral model (200). In contrast, a sap4 to sap6 mutant showed an equal level of tissue damage to that caused by the parent strain in both RHE models (198, 200). These results confirmed their initial observations that Sap1 and Sap3 were associated with mucosal tissue damage (202) and provided good evidence for these two proteinases in establishing mucosal C. albicans infections.
De Bernardis et al. (44) showed that sap1, sap2, and sap3 mutants, but not the sap4 to sap6 mutant, were less virulent in a rat vaginitis model than was the parent strain, with the sap2 mutant being almost avirulent. Reintroduction of the SAP2 gene recovered the vaginopathic potential of the sap2 mutant, showing that Sap2 may play a pathogenic role in rat vaginitis. This supports the data obtained with the RHE model and also implicates the Sap1 to Sap3 subfamily in rat mucosal infections (Table 8).
The ability of the C. albicans SAP-deficient mutants to adhere to epithelial cells was also investigated (244). Although the sap1, sap2, and sap3 null mutants were moderately less adherent to buccal epithelial cells than was the parent strain, the sap4 to sap6 triple mutant had significantly increased adherence. This was unexpected since one might have predicted that the sap4 to sap6 mutant would have decreased adherence, given the association of SAP4 to SAP6 expression with the hyphal form (96, 246), which is known to be more adherent than the yeast form (110, 111, 193). To explain the results, it was suggested that one possible function of Sap4 to Sap6 might be to remove or degrade cell surface components on the yeast or epithelial cells, causing an inhibitory effect to the host-fungus recognition and adhesion mechanism. Although this is plausible, a more likely explanation would be that other proteinases or other unrelated adhesin genes might be up-regulated to compensate for the loss of Sap4 to Sap6 in the mutant strain. Since Sap1 to Sap3 may contribute to C. albicans adherence (15, 121), up-regulation of one or more of these genes alone in the SAP4- to SAP6-deficient strain may account for at least some of the increased adherence observed.
Studies using SAP-deficient strains have implicated the Sap1 to Sap3 subfamily, either individually or collectively, in the virulence of C. albicans infections at oral and vaginal sites (Table 8). This appears to be in contrast to murine gastrointestinal infections (also mucosal), where no demonstrable differences between SAP1- to SAP6-deficient strains and the parent strain could be observed in the ability to invade the stomach or to disseminate to the brain or in the number of fungi persisting in the feces (119). Although SAP gene and protein expression were readily detectable in this model, there was a notable absence of SAP1 expression. Since SAP1 contributes to oral (200) and vaginal (44) infections and might be involved in C. albicans adherence (15, 121), the absence of SAP1 expression in the gastrointestinal tract may go some way toward explaining why SAP1-deficient mutants were not attenuated in virulence compared with the parent strain (119). It is possible that the SAP genes used by C. albicans during gastrointestinal infections differ in part from the SAP genes required for oral or vaginal infections. This certainly appears to be the case, at least for systemic candidiasis, where Sap4 to Sap6 and not Sap1 to Sap3 appear to be more pertinent to infection (see below).
Contribution of Sap proteins to systemic infections using SAP-deficient strains.
The use of SAP-deficient strains has also implicated the Sap family in systemic C. albicans infections (Table 8). When guinea pig and murine models of disseminated candidiasis were infected with the sap1, sap2, sap3, and sap4 to sap6 mutants, all animals had increased survival rates compared with those infected by the parent strain (100, 194). The sap4 to sap6 mutant displayed the greatest attenuation not only in terms of lethality but also in terms of fungal burden in host organs such as the kidneys, liver, and skin. However, the authors concluded that although Sap4 to Sap6 appeared to contribute more to systemic infections in both animal models than did Sap1 to Sap3, none of the C. albicans proteinases was a single dominant factor during disseminated infections.
In a model of murine peritonitis, the invasive properties of sap1, sap2, or sap3 mutants were indistinguishable from those of wild-type cells, but the sap4 to sap6 triple mutant showed strongly reduced invasiveness (65, 120). In addition, the sap4 to sap6 mutant induced significantly reduced levels of alanine transferase and α-amylase activity (markers of liver and pancreas damage, respectively) compared with the parent strain. Interestingly, when single sap4, sap5, sap6, sap4-sap5, sap5-sap6, and sap4-sap6 mutants were compared with wild-type cells, only mutants that lacked a functional SAP6 gene showed significantly reduced liver and pancreas tissue damage (65). The data indicated that the Sap4 to Sap6 subfamily, and probably Sap6 specifically, contributes to tissue damage and invasion in peritoneal infections.
Ibrahim et al. (103) used an in vitro model of human umbilical vein endothelial cell injury to show that although Sap1 to Sap3 activity was not required for the adherence of C. albicans to endothelial cells, Sap2 activity was required for the initial stages of cell injury. Extrapolated to the in vivo situation, the possible degradation of the endothelial barrier by Sap2 may enable C. albicans to evade immune attack or to escape from blood vessels and cause disseminated infections. In contrast to the individual sap1 to sap3 mutants, the sap4 to sap6 mutant had increased adherence to endothelial cells but exhibited no differences in cell injury compared with the wild type (A. S. Ibrahim, S. G. Filler, D. Sanglard, J. E. Edwards, Jr., and B. Hube, Program Abstr. 37th Intersci. Conf. Antimicrob. Agents Chemother., abstr. B9, 1997). The increase in adherence is in line with the results of another study that showed enhanced adherence of the sap4 to sap6 mutant to epithelial cells (244). However, the lack of cell injury was perhaps surprising, considering that contact (68) and candidal germination (67, 102), which is associated with Sap4 to Sap6 expression (96, 246), are required for the induction of endothelial cell injury. Furthermore, if other SAP genes (e.g., SAP1 to SAP3) are up-regulated to compensate for the loss of Sap4 to Sap6 in this mutant, this raises the question whether certain SAP genes play dual roles; e.g., Sap1 to Sap3 may contribute to C. albicans adhesion and tissue damage.
Contribution of Sap proteins to evasion of host immune responses using SAP-deficient strains.
The sap mutants were also instrumental in demonstrating a role for Sap4 to Sap6 in evading host immune responses (Table 8). In macrophage-killing assays, the sap4 to sap6 triple mutant was eliminated 53% more effectively after phagocytosis by murine macrophages than was the parent strain (16). Under normal circumstances C. albicans is taken up for elimination into the phagolysosome, which has a pH of 4.7 to 4.8 (164). As a result of ingestion, C. albicans may germinate and secrete Sap4 to Sap6, which are optimally active at the same pH as that found in the phagolysosome (16). One should note that C. albicans germination in vitro is normally observed at higher pH values (7.0) and is limited at lower pH values (5.0, the approximate pH of the phagolysosome). Nevertheless, in vivo, the production of functional Sap4 to Sap6 proteinases within the phagolysosome may result in the digestion of enzymes associated with candidal killing and might render C. albicans more resistant to macrophage attack. The macrophage proteins targeted by Sap4 to Sap6 are not yet known, but Sap4 to Sap6 may act either directly on phagolysosomal enzymes involved in microbial killing (cathepsin D and/or antifungal peptides) or on key enzymes of macrophage metabolism that are essential for optimal microbial killing (145). The results of this study strongly indicated that Sap4 to Sap6 might function in a way that enables C. albicans to evade host innate defenses by resisting macrophage attack, a host evasion strategy also observed for other microorganisms, including the bacterium Listeria monocytogenes (51).
Conclusions from SAP-deficient mutant studies.
These in vitro and animal model studies using SAP-deficient C. albicans strains have been instrumental in confirming a role for Sap proteins in oral, vaginal, and systemic Candida infections and have begun to dissect some of the fundamental functions the proteinase family may possess in vivo. However, these results should be interpreted with a little caution, since the disruption of one SAP gene may be compensated for by the up-regulation or down-regulation of other SAP genes, which may account for some of the observations reported. In addition, SAP gene disruption may alter or modify the expression profiles of other hydrolytic enzymes of C. albicans such as phospholipases and lipases or, indeed, other genes that may be associated with virulence. These forced adaptations that C. albicans may need to undergo as a result of SAP gene disruption may mask or disguise some of the roles and functions of individual Sap enzymes in the in vivo situation.
Furthermore, the contributions of Sap7, Sap8, Sap9, and Sap10, nearly half the SAP gene family, during C. albicans infections are presently unknown, and the role of these proteinases in pathogenesis have not yet been tested in animal models using the respective SAP-deficient strains. Data from these mutants should add significantly to our knowledge of Sap function in C. albicans pathogenesis, particularly with respect to Sap9 and Sap10, which both have C-terminal consensus sequences typical for GPI-anchored proteins and may therefore not be secreted from the cell. Preliminary data suggest that Sap9 and Sap10 are regulatory proteinases that may play a role in the cell surface integrity of the cell, which is in contrast to the putative functions of the other Sap proteins (A. Albrecht, A. Felk, I. Pichova, M. Schaller, M. Monod, and B. Hube, unpublished data). It is clear that the use of SAP-deficient strains has significantly advanced our understanding of the possible roles and functions of the proteinases during Candida infections. Future experiments using SAP-deficient strains should lead to a better understanding of the complex interplay between C. albicans, the SAP gene family, and the host.
FUNCTIONAL GENOMICS AND CANDIDA
Our understanding of Candida pathogenesis in general has advanced considerably in the last 20 years, notably in the identification of genes and proteins involved in cell signaling, morphogenesis, and virulence. However, we are presently witnessing significant developments in molecular microbiology and proceeding toward a new era of fungal research, and this is primarily due to one reason: the sequencing of a large number of medically important fungal genomes. Complete genome sequences of S. cerevisiae and Schizosaccharomyces pombe have already been published (77, 249). The preliminary assembly for the C. albicans genome sequence has been released (http://www-sequence.stanford.edu/group/candida), and sequencing projects are currently under way for Candida glabrata, Cryptococcus neoformans, Aspergillus sp. and several other fungal species (http://www.tigr.org/tdb/mdb/mdbinprogress.html). The rapidity of these sequencing initiatives is impressive, and with the development of new functional genomic technologies including DNA microarrays, which has become an indispensable tool for high-throughput gene expression analysis, we will soon be able to analyze the whole-genome responses of many fungal pathogens. As a result, our view and appreciation of fungal biology and pathogenesis will undergo something little short of a revolution in the very near future.
Candida DNA Microarrays
The sequencing of the Candida genome has generated a mountain of information, but the next critical step is to accurately decipher the biochemical and physiological meaning of the information (257). Gene function is the primary information that most researchers would like to extract from the Candida genome sequence; at present, DNA microarray analysis is one of the most valuable technologies available to achieve this on a genome-wide scale. Microarrays allow researchers to compare the behavior of huge numbers of genes in different tissues and under different environmental conditions to identify virulence genes, gene pathways, and regulatory networks.
Due to the very recent date of the sequencing of the genome, only a handful of Candida DNA microarray studies have been published; these include analysis of the genome-wide expression profile of C. albicans when exposed to the azole antifungal drugs (38, 42, 179), during the yeast-to- hypha transition (123, 153, 154, 158, 213), during stress (61), and when exposed to human blood (69a). The microarray studies described above have already been valuable in confirming the suspected coordinated regulation between SAP gene expression and hypha formation. However, because the current arrays are not highly effective at distinguishing between different members of multigene families, one needs to be cautious in interpreting the results.
In conclusion, transcript profiling is rapidly becoming a standard tool in academic laboratories for the global analysis of gene expression. Although microarray technology is a dynamic and developing process, its development ranks as one of the pioneering breakthrough technologies of the last decade. In the Candida field, most microarray data have been based on experiments in vitro; the use of arrays to evaluate global gene responses during in vivo infections has not yet been reported. Given the complexity of Candida infections in vivo, including the influence of the local environment, the microflora, and the host immune response on gene function, transcript profiling data obtained from in vivo infections may well differ from the data observed in vitro. In addition, mammalian gene arrays have not yet been used to study host-pathogen interactions from the host's perspective, in order to determine host gene expression profiles induced by the presence of Candida. Furthermore, the simultaneous analysis of both the host and Candida genome response using DNA microarrays during in vivo infections is a goal well worth striving for. This should provide important information concerning which Candida and host factors interact during various stages of different infections and will clearly lead to valuable insights into the delicate and complex relationship between Candida and the host.
FUTURE DIRECTIONS IN SAP RESEARCH
The presence of a SAP gene family in C. albicans clearly provides the fungus with an efficient and flexible proteolytic system that may prove vital to its success as an opportunistic pathogen. Sap production is probably a well-regulated process that is activated at specific time points during colonization and infection to obtain maximum benefits for C. albicans. Nearly all studies implicating proteinases in C. albicans pathogenesis have done so by using one or more of the seven main criteria described in this review, which demonstrate the multiple functions of the SAP genes and have established the proteinases as a very versatile and multifunctional virulence gene family of C. albicans. However, although it appears that we know a great deal about the C. albicans SAP family, our knowledge is actually rather limited. In fact, a number of fundamental questions need to be addressed in future studies.
First, since SAP mRNA expression patterns in vivo appear to be different from those observed under in vitro conditions, what, exactly, are the environmental stimuli that influence SAP expression in vivo? Also, do the same transcriptional factors coordinately regulate the simultaneous expression of the SAP genes with a combination of other Candida virulence genes when triggered by changes in the local environment?
Second, what are the signal transduction pathways that regulate C. albicans proteinase expression? These pathways are currently unknown, and no receptor (if indeed a receptor is involved) has yet been implicated in the regulation of proteinase secretion.
Third, what are the actual targets of the Sap family during human infections? Although the possible targets for Sap2 might be deduced from in vitro degradation studies, these need to be confirmed in the in vivo environment. Furthermore, the substrate specificities of Sap1, Sap3, and Sap4 to Sap10 are not yet known and may provide valuable insights into how these enzymes might contribute enzymatically and nonenzymatically to different infections at various bodily sites.
Fourth, given the recognized importance of the host-pathogen interaction in the development of candidiasis and the fine balance between commensialism and infection, our understanding of the immune response to the Sap family is distinctly unsatisfactory. The protective mechanisms of humoral and cell-mediated immune responses possibly elicited by different members of the Sap family need to be thoroughly investigated and may have the potential to open up a brand new avenue of research and lead to the exciting possibility that Sap proteins could be targeted for immunotherapeutic means.
Answers to these fundamental questions would significantly advance our understanding of the interplay between Candida proteinases and the host and bring us a good deal closer to discovering how important this protein family is to Candida pathogenicity. Future studies using DNA microarrays will allow us to identify new and no doubt surprising correlations between SAP gene expression and Candida biology and virulence. In time, this may lead to the development of new prophylactic and therapeutic strategies targeting specific virulence-associated genes, as well as signaling pathways or regulatory networks, which would be a valuable addition to the limited repertoire of antifungal agents currently available.
Acknowledgments
We gratefully acknowledge Cele Abad Zapatero for helpful discussions regarding Sap2 structure.
Our own work was supported by the Dunhill Medical Trust, the European commission (QLK2-2000-00795; “Galar Fungail consortium”), and the Deutsche Forschungsgemeinschaft (Hu 528/8).
REFERENCES
- 1.Abad-Zapatero, C., R. Goldman, S. W. Muchmore, C. Hutchins, K. Stewart, J. Navaza, C. D. Payne, and T. L. Ray. 1996. Structure of a secreted aspartic protease from C. albicans complexed with a potent inhibitor: implications for the design of antifungal agents. Protein Sci. 5:640-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abu-Elteen, K. H., A. Z. Elkarmi, and M. Hamad. 2001. Characterization of phenotype-based pathogenic determinants of various Candida albicans strains in Jordan. Jpn. J. Infect. Dis. 54:229-236. [PubMed] [Google Scholar]
- 3.Acosta, E. P., T. N. Kakuda, R. C. Brundage, P. L. Anderson, and C. V. Fletcher. 2000. Pharmacodynamics of human immunodeficiency virus type 1 protease inhibitors. Clin. Infect. Dis. 30(Suppl. 2):S151-S159. [DOI] [PubMed] [Google Scholar]
- 4.Agatensi, L., F. Franchi, F. Mondello, R. L. Bevilacqua, T. Ceddia, F. De Bernardis, and A. Cassone. 1991. Vaginopathic and proteolytic Candida species in outpatients attending a gynaecology clinic. J. Clin. Pathol. 44:826-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Apodaca, G., and J. H. McKerrow. 1990. Expression of proteolytic activity by cultures of Trichophyton rubrum. J. Med. Vet. Mycol. 28:159-171. [DOI] [PubMed] [Google Scholar]
- 6.Barrett, A. J. 1980. Introduction: the classification of proteinases, p. 1-13. In Symposium on protein degradation in health and disease. Excerpta Medica, Amsterdam, The Netherlands. [PubMed]
- 7.Barrett, A. J., and N. D. Rawlings. 1991. Types and families of endopeptidases. Biochem. Soc. Trans. 19:707-715. [DOI] [PubMed] [Google Scholar]
- 8.Beausejour, A., D. Grenier, J. P. Goulet, and N. Deslauriers. 1998. Proteolytic activation of the interleukin-1 beta precursor by Candida albicans. Infect. Immun. 66:676-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beggah, S., U. Lechenne, U. Reichard, S. Foundling, and M. Monod. 2000. Intra- and intermolecular events direct the propeptide-mediated maturation of the Candida albicans secreted aspartic proteinase Sap1p. Microbiology 146:2765-2773. [DOI] [PubMed] [Google Scholar]
- 10.Bektic, J., C. P. Lell, A. Fuchs, H. Stoiber, C. Speth, C. Lass-Florl, M. Borg-von Zepelin, M. P. Dierich, and R. Wurzner. 2001. HIV protease inhibitors attenuate adherence of Candida albicans to epithelial cells in vitro. FEMS Immunol. Med. Microbiol. 31:65-71. [DOI] [PubMed] [Google Scholar]
- 11.Berman, J., and P. E. Sudbery. 2002. Candida albicans: a molecular revolution built on lessons from budding yeast. Nat. Rev. Genet. 3:918-930. [DOI] [PubMed] [Google Scholar]
- 12.Borg, M., and R. Rüchel. 1988. Expression of extracellular acid proteinase by proteolytic Candida spp. during experimental infection of oral mucosa. Infect. Immun. 56:626-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borg, M., and R. Rüchel. 1990. Demonstration of fungal proteinase during phagocytosis of Candida albicans and Candida tropicalis. J. Med. Vet. Mycol. 28:3-14. [PubMed] [Google Scholar]
- 14.Borg, M., D. Watters, B. Reich, and R. Rüchel. 1988. Production and characterization of monoclonal antibodies against secretory proteinase of Candida albicans CBS 2730. Zentbl. Bakteriol. Microbiol. Hyg. Ser. A 268:62-73. [DOI] [PubMed] [Google Scholar]
- 15.Borg-von Zepelin, M., I. Meyer, R. Thomssen, R. Wurzner, D. Sanglard, A. Telenti, and M. Monod. 1999. HIV-Protease inhibitors reduce cell adherence of Candida albicans strains by inhibition of yeast secreted aspartic proteases. J. Investig. Dermatol. 113:747-751. [DOI] [PubMed] [Google Scholar]
- 16.Borg-von Zepelin, M., S. Beggah, K. Boggian, D. Sanglard, and M. Monod. 1998. The expression of the secreted aspartyl proteinases Sap4 to Sap6 from Candida albicans in murine macrophages. Mol. Microbiol. 28:543-554. [DOI] [PubMed] [Google Scholar]
- 17.Braun, B. R., and A. D. Johnson. 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277:105-109. [DOI] [PubMed] [Google Scholar]
- 18.Brown, A. J., and N. A. Gow. 1999. Regulatory networks controlling Candida albicans morphogenesis. Trends Microbiol. 7:333-338. [DOI] [PubMed] [Google Scholar]
- 19.Brown, A. J. P. 2002. Expression of growth-form specific factors during morphogenesis in Candida albicans, p. 87-93. In R. A. Calderone (ed.), Candida and candidiasis. ASM Press, Washington, D.C.
- 20.Brown, A. J. P. 2002. Morphogenetic signaling pathways in Candida albicans, p. 95-106. In R. A. Calderone (ed.), Candida and candidiasis. ASM Press, Washington, D.C.
- 21.Brueske, C. H. 1986. Proteolytic activity of a clinical isolate of Cryptococcus neoformans. J. Clin. Microbiol. 23:631-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calderone, R., and N. A. R. Gow. 2002. Host recognition by Candida species, p. 67-86. In R. A. Calderone (ed.), Candida and candidiasis. ASM Press, Washington, D.C.
- 23.Calderone, R. A., and P. C. Braun. 1991. Adherence and receptor relationships of Candida albicans. Microbiol. Rev. 55:1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calderone, R. A., and W. A. Fonzi. 2001. Virulence factors of Candida albicans. Trends Microbiol. 9:327-335. [DOI] [PubMed] [Google Scholar]
- 25.Camilli, A., D. T. Beattie, and J. J. Mekalanos. 1994. Use of genetic recombination as a reporter of gene expression. Proc. Natl. Acad. Sci. USA 91:2634-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Capobianco, J. O., C. G. Lerner, and R. C. Goldman. 1992. Application of a fluorogenic substrate in the assay of proteolytic activity and in the discovery of a potent inhibitor of Candida albicans aspartic proteinase. Anal. Biochem. 204:96-102. [DOI] [PubMed] [Google Scholar]
- 27.Caro, L. H., H. Tettelin, J. H. Vossen, A. F. Ram, H. van den Ende, and F. M. Klis. 1997. In silicio identification of glycosyl-phosphatidylinositol-anchored plasma-membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast 13:1477-1489. [DOI] [PubMed] [Google Scholar]
- 28.Cassone, A., F. De Bernardis, F. Mondello, T. Ceddia, and L. Agatensi. 1987. Evidence for a correlation between proteinase secretion and vulvovaginal candidosis. J. Infect. Dis. 156:777-783. [DOI] [PubMed] [Google Scholar]
- 29.Cassone, A., F. De Bernardis, A. Torosantucci, E. Tacconelli, M. Tumbarello, and R. Cauda. 1999. In vitro and in vivo anticandidal activity of human immunodeficiency virus protease inhibitors. J. Infect. Dis. 180:448-453. [DOI] [PubMed] [Google Scholar]
- 30.Cassone, A., E. Tacconelli, F. De Bernardis, M. Tumbarello, A. Torosantucci, P. Chiani, and R. Cauda. 2002. Antiretroviral therapy with protease inhibitors has an early, immune reconstitution-independent beneficial effect on Candida virulence and oral candidiasis in human immunodeficiency virus-infected subjects. J. Infect. Dis. 185:188-195. [DOI] [PubMed] [Google Scholar]
- 31.Cauda, R., E. Tacconelli, M. Tumbarello, G. Morace, De, F. Bernardis, A. Torosantucci, and A. Cassone. 1999. Role of protease inhibitors in preventing recurrent oral candidosis in patients with HIV infection: a prospective case-control study. J. Acquir. Immun. Defic. Syndr. 21:20-25. [DOI] [PubMed] [Google Scholar]
- 32.Cawley, N. X., M. Chino, A. Maldonado, Y. M. Rodriguez, Y. P. Loh, and J. A. Ellman. 2003. Synthesis and characterization of the first potent inhibitor of yapsin 1. Implications for the study of yapsin-like enzymes. J. Biol. Chem. 278:5523-5530. [DOI] [PubMed] [Google Scholar]
- 33.Chaffin, W. L., J. L. Lopez-Ribot, M. Casanova, D. Gozalbo, and J. P. Martinez. 1998. Cell wall and secreted proteins of Candida albicans: identification, function, and expression. Microbiol. Mol. Biol. Rev. 62:130-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Challacombe, S. J., J. Muir, S. A. Howell, and S. P. Sweet. 1995. Genetic variability of Candida albicans in HIV infection. Microb. Ecol. Health Dis. 8:63-70. [Google Scholar]
- 35.Choi, G. H., D. M. Pawlyk, B. Rae, R. Shapira, and D. L. Nuss. 1993. Molecular analysis and overexpression of the gene encoding endothiapepsin, an aspartic protease from Cryphonectria parasitica. Gene 125:135-141. [DOI] [PubMed] [Google Scholar]
- 36.Colina, A. R., F. Aumont, N. Deslauriers, P. Belhumeur, and L. de Repentigny. 1996. Evidence for degradation of gastrointestinal mucin by Candida albicans secretory aspartyl proteinase. Infect. Immun. 64:4514-4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coogan, M. M., S. P. Sweet, and S. J. Challacombe. 1994. Immunoglobulin A (IgA), IgA1, and IgA2 antibodies to Candida albicans in whole and parotid saliva in human immunodeficiency virus infection and AIDS. Infect. Immun. 62:892-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cowen, L. E., A. Nantel, M. S. Whiteway, D. Y. Thomas, D. C. Tessier, L. M. Kohn, and J. B. Anderson. 2002. Population genomics of drug resistance in Candida albicans. Proc. Natl. Acad. Sci. USA 99:9284-9289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cutfield, S. M., E. J. Dodson, B. F. Anderson, P. C. Moody, C. J. Marshall, P. A. Sullivan, and J. F. Cutfield. 1995. The crystal structure of a major secreted aspartic proteinase from Candida albicans in complexes with two inhibitors. Structure 3:1261-1271. [DOI] [PubMed] [Google Scholar]
- 40.Cutler, J. E. 1991. Putative virulence factors of Candida albicans. Annu. Rev. Microbiol. 45:187-218. [DOI] [PubMed] [Google Scholar]
- 41.Davies, D. R. 1990. The structure and function of aspartic proteinases. Annu. Rev. Biophys. Biophys. Chem. 19:189-215. [DOI] [PubMed] [Google Scholar]
- 42.De Backer, M. D., T. Hyina, X. J. Ma, S. Vandoninck, W. H. Luyten, and H. Vanden Bossche. 2001. Genomic profiling of the response of Candida albicans to itraconazole treatment using a DNA microarray. Antimicrob. Agents Chemother. 45:1660-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Bernardis, F., L. Agatensi, I. K. Ross, G. W. Emerson, R. Lorenzini, P. A. Sullivan, and A. Cassone. 1990. Evidence for a role for secreted aspartate proteinase of Candida albicans in vulvovaginal candidiasis. J. Infect. Dis. 161:1276-1283. [DOI] [PubMed] [Google Scholar]
- 44.De Bernardis, F., S. Arancia, L. Morelli, B. Hube, Sanglard, W. Schafer, and A. Cassone. 1999. Evidence that members of the secretory aspartyl proteinase gene family, in particular SAP2, are virulence factors for Candida vaginitis. J. Infect. Dis. 179:201-208. [DOI] [PubMed] [Google Scholar]
- 45.De Bernardis, F., M. Boccanera, D. Adriani, E. Spreghini, G. Santoni, and A. Cassone. 1997. Protective role of antimannan and anti-aspartyl proteinase antibodies in an experimental model of Candida albicans vaginitis in rats. Infect. Immun. 65:3399-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Bernardis, F., M. Boccanera, L. Rainaldi, C. E. Guerra, I. Quinti, and A. Cassone. 1992. The secretion of aspartyl proteinase, a virulence enzyme, by isolates of Candida albicans from the oral cavity of HIV-infected subjects. Eur. J. Epidemiol. 8:362-367. [DOI] [PubMed] [Google Scholar]
- 47.De Bernardis, F., A. Cassone, J. Sturtevant, and R. Calderone. 1995. Expression of Candida albicans SAP1 and SAP2 in experimental vaginitis. Infect. Immun. 63:1887-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Bernardis, F., P. Chiani, M. Ciccozzi, G. Pellegrini, T. Ceddia, G. D'Offizzi, I. Quinti, P. A. Sullivan, and A. Cassone. 1996. Elevated aspartic proteinase secretion and experimental pathogenicity of Candida albicans isolates from oral cavities of subjects infected with human immunodeficiency virus. Infect. Immun. 64:466-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Bernardis, F., F. Mondello, G. Scaravelli, A. Pachi, A. Girolamo, L. Agatensi, and A. Cassone. 1999. High aspartyl proteinase production and vaginitis in human immunodeficiency virus-infected women. J. Clin. Microbiol. 37:1376-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Bernardis, F., P. A. Sullivan, and A. Cassone. 2001. Aspartyl proteinases of Candida albicans and their role in pathogenicity. Med. Mycol. 39:303-313. [DOI] [PubMed] [Google Scholar]
- 51.de Chastellier, C., and P. Berche. 1994. Fate of Listeria monocytogenes in murine macrophages: evidence for simultaneous killing and survival of intracellular bacteria. Infect. Immun. 62:543-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Repentigny, L., F. Aumont, K. Bernard, and P. Belhumeur. 2000. Characterization of binding of Candida albicans to small intestinal mucin and its role in adherence to mucosal epithelial cells. Infect. Immun. 68:3172-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Viragh, P. A., D. Sanglard, G. Togni, R. Falchetto, and M. Monod. 1993. Cloning and sequencing of two Candida parapsilosis genes encoding acid proteases. J. Gen. Microbiol. 139:335-342. [DOI] [PubMed] [Google Scholar]
- 54.Diz Dios, P., A. Ocampo, C. Miralles, I. Otero, I. Iglesias, and N. Rayo. 1999. Frequency of oropharyngeal candidiasis in HIV-infected patients on protease inhibitor therapy. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodont. 87:437-441. [DOI] [PubMed] [Google Scholar]
- 55.Diz Dios, P., I. Otero, I. Iglesias, A. Ocampo, and C. Martinez. 1999. Failure of indinavir to inhibit Candida albicans in vitro. Eur. J. Clin. Microbiol. Infect. Dis. 18:755-756. [DOI] [PubMed] [Google Scholar]
- 56.Diz Dios, P., A. Ocampo, I. Iglesias, and I. Otero. 1999. Lack of ritonavir antifungal effect in vitro. Antimicrob. Agents Chemother. 43:997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Drobacheff, C., L. Millon, M. Monod, R. Piarroux, E. Robinet, R. Laurent, and D. Meillet. 2001. Increased serum and salivary immunoglobulins against Candida albicans in HIV-infected patients with oral candidiasis. Clin. Chem. Lab Med. 39:519-526. [DOI] [PubMed] [Google Scholar]
- 58.Edison, A. M., and M. Manning-Zweerink. 1988. Comparison of the extracellular proteinase activity produced by a low-virulence mutant of Candida albicans and its wild-type parent. Infect. Immun. 56:1388-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edmond, M. B., S. E. Wallace, D. K. McClish, M. A. Pfaller, R. N. Jones, and R. P. Wenzel. 1999. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin. Infect. Dis. 29:239-244. [DOI] [PubMed] [Google Scholar]
- 60.el-Maghrabi, E. A., D. M. Dixon, and J. W. Burnett. 1990. Characterization of Candida albicans epidermolytic proteases and their role in yeast-cell adherence to keratinocytes. Clin. Exp. Dermatol. 15:183-191. [DOI] [PubMed] [Google Scholar]
- 61.Enjalbert, B., A. Nantel, and M. Whiteway. 2003. Stress-induced gene expression in Candida albicans: absence of a general stress response. Mol. Biol. Cell 14:1460-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ernst, J. F. 2000. Regulation of dimorphism in Candida albicans. Contrib. Microbiol. 5:98-111. [DOI] [PubMed] [Google Scholar]
- 63.Ernst, J. F. 2000. Transcription factors in Candida albicans—environmental control of morphogenesis. Microbiology 146:1763-1774. [DOI] [PubMed] [Google Scholar]
- 64.Fallon, K., K. Bausch, J. Noonan, E. Huguenel, and P. Tamburini. 1997. Role of aspartic proteases in disseminated Candida albicans infection in mice. Infect. Immun. 65:551-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Felk, A., M. Kretschmar, A. Albrecht, M. Schaller, S. Beinhauer, T. Nichterlein, D. Sanglard, H. C. Korting, W. Schafer, and B. Hube. 2002. Candida albicans hyphal formation and the expression of the Efg1-regulated proteinases Sap4 to Sap6 are required for the invasion of parenchymal organs. Infect. Immun. 70:3689-3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Felk, A., W. Schafer, and B. Hube. 2000. Candida albicans secretory aspartic proteinase (SAP10) gene. Accession number AF146440.
- 67.Filler, S. G., A. S. Pfunder, B. J. Spellberg, J. P. Spellberg, and J. E. Edwards, Jr. 1996. Candida albicans stimulates cytokine production and leukocyte adhesion molecule expression by endothelial cells. Infect. Immun. 64:2609-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Filler, S. G., J. N. Swerdloff, C. Hobbs, and P. M. Luckett. 1995. Penetration and damage of endothelial cells by Candida albicans. Infect. Immun. 63:976-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Finlay, B. B., and S. Falkow. 1989. Common themes in microbial pathogenicity. Microbiol. Rev. 53:210-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69a.Fradin, C., M. Kretschmar, T. Nichterlein, C. Gaillardin, C. d’Enfert, and B. Hube. 2003. Stage-specific gene expression of Candida albicans in human blood. Mol. Microbiol. 47:1523-1543. [DOI] [PubMed] [Google Scholar]
- 70.Fusek, M., E. A. Smith, M. Monod, B. M. Dunn, and S. I. Foundling. 1994. Extracellular aspartic proteinases from Candida albicans, Candida tropicalis, and Candida parapsilosis yeasts differ substantially in their specificities. Biochemistry 33:9791-9799. [DOI] [PubMed] [Google Scholar]
- 71.Gale, C. A., C. M. Bendel, M. McClellan, M. Hauser, J. M. Becker, J. Berman, and M. K. Hostetter. 1998. Linkage of adhesion, filamentous growth, and virulence in Candida albicans to a single gene, INT1. Science 279:1355-1358. [DOI] [PubMed] [Google Scholar]
- 72.Germaine, G. R., and L. M. Tellefson. 1981. Effect of pH and human saliva on protease production by Candida albicans. Infect. Immun. 31:323-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ghadjari, A., R. C. Matthews, and J. P. Burnie. 1997. Epitope mapping Candida albicans proteinase (SAP 2). FEMS Immunol. Med. Microbiol. 19:115-123. [DOI] [PubMed] [Google Scholar]
- 74.Ghannoum, M., and K. Abu Elteen. 1986. Correlative relationship between proteinase production, adherence and pathogenicity of various strains of Candida albicans. J. Med. Vet. Mycol. 24:407-413. [DOI] [PubMed] [Google Scholar]
- 75.Ghannoum, M. A. 2000. Potential role of phospholipases in virulence and fungal pathogenesis. Clin. Microbiol. Rev. 13:122-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gilfillan, G. D., D. J. Sullivan, K. Haynes, T. Parkinson, D. C. Coleman, and N. A. R. Gow. 1998. Candida dubliniensis: phylogeny and putative virulence factors. Microbiology 144:829-838. [DOI] [PubMed] [Google Scholar]
- 77.Goffeau, A., B. G. Barrell, H. Bussey, R. W. Davis, B. Dujon, H. Feldmann, F. Galibert, J. D. Hoheisel, C. Jacq, M. Johnston, E. J. Louis, H. W. Mewes, Y. Murakami, P. Philippsen, H. Tettelin, and S. G. Oliver. 1996. Life with 6000 genes. Science. 274:546, 563-546, 567. [DOI] [PubMed] [Google Scholar]
- 78.Goldman, R. C., D. J. Frost, J. O. Capobianco, S. Kadam, R. R. Rasmussen, and C. Abad-Zapatero. 1995. Antifungal drug targets: Candida secreted aspartyl protease and fungal wall beta-glucan synthesis. Infect. Agents Dis. 4:228-247. [PubMed] [Google Scholar]
- 79.Gow, N. A. R., A. J. P. Brown, and F. C. Odds. 2002. Fungal morphogenesis and host invasion. Curr. Opin. Microbiol. 5:366-371. [DOI] [PubMed] [Google Scholar]
- 80.Gruber, A., J. Berlit, C. Speth, C. Lass-Florl, G. Kofler, M. Nagl, M. Borg von Zepelin, M. P. Dierich, and R. Wurzner. 1999. Dissimilar attenuation of Candida albicans virulence properties by human immunodeficiency virus type 1 protease inhibitors. Immunobiology 201:133-144. [DOI] [PubMed] [Google Scholar]
- 81.Gruber, A., C. P. Lell, C. Speth, H. Stoiber, C. Lass-Florl, A. Sonneborn, J. F. Ernst, M. P. Dierich, and R. Wurzner. 2001. Human immunodeficiency virus type 1 Tat binds to Candida albicans, inducing hyphae but augmenting phagocytosis in vitro. Immunology 104:455-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gruber, A., E. Lukasser-Vogl, M. Borg von Zepelin, M. P. Dierich, and R. Wurzner. 1998. Human immunodeficiency virus type 1 gp160 and gp41 binding to Candida albicans selectively enhances candidal virulence in vitro. J. Infect. Dis. 177:1057-1063. [DOI] [PubMed] [Google Scholar]
- 83.Gruber, A., C. Speth, E. Lukasser-Vogl, R. Zangerle, M. Borg von Zepelin, M. P. Dierich, and R. Wurzner. 1999. Human immunodeficiency virus type 1 protease inhibitor attenuates Candida albicans virulence properties in vitro. Immunopharmacology 41:227-234. [DOI] [PubMed] [Google Scholar]
- 84.Han, Y., and J. E. Cutler. 1995. Antibody response that protects against disseminated candidiasis. Infect. Immun. 63:2714-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Han, Y., R. P. Morrison, and J. E. Cutler. 1998. A vaccine and monoclonal antibodies that enhance mouse resistance to Candida albicans vaginal infection. Infect. Immun. 66:5771-5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Han, Y., M. H. Riesselman, and J. E. Cutler. 2000. Protection against candidiasis by an immunoglobulin G3 (IgG3) monoclonal antibody specific for the same mannotriose as an IgM protective antibody. Infect. Immun. 68:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hannula, J., M. Saarela, B. Dogan, J. Paatsama, P. Koukila-Kahkola, S. Pirinen, H. L. Alakomi, J. Perheentupa, and S. Asikainen. 2000. Comparison of virulence factors of oral Candida dubliniensis and Candida albicans isolates in healthy people and patients with chronic candidosis. Oral Microbiol. Immunol. 15:238-244. [DOI] [PubMed] [Google Scholar]
- 88.Hoegl, L., M. Ollert, and H. C. Korting. 1996. The role of Candida albicans secreted aspartic proteinase in the development of candidoses. J. Mol. Med. 74:135-142. [DOI] [PubMed] [Google Scholar]
- 89.Hoegl, L., E. Thoma-Greber, M. Rocken, and H. C. Korting. 1998. HIV protease inhibitors influence the prevalence of oral candidosis in HIV-infected patients: a 2-year study. Mycoses 41:321-325. [DOI] [PubMed] [Google Scholar]
- 90.Hoetelmans, R. M., M. van Essenberg, P. L. Meenhorst, J. W. Mulder, and J. H. Beijnen. 1997. Determination of saquinavir in human plasma, saliva, and cerebrospinal fluid by ion-pair high-performance liquid chromatography with ultraviolet detection. J. Chromatogr. Ser. B 698:235-241. [DOI] [PubMed] [Google Scholar]
- 91.Homma, M., H. Chibana, and K. Tanaka. 1993. Induction of extracellular proteinase in Candida albicans. J. Gen. Microbiol. 139:1187-1193. [DOI] [PubMed] [Google Scholar]
- 92.Hood, S., A. Bonington, J. Evans, and D. Denning. 1998. Reduction in oropharyngeal candidiasis following introduction of protease inhibitors. AIDS 12:447-448. [PubMed] [Google Scholar]
- 93.Hostetter, M. K. 1994. Adhesins and ligands involved in the interaction of Candida spp. with epithelial and endothelial surfaces. Clin. Microbiol. Rev. 7:29-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hube, B. 1996. Candida albicans secreted aspartyl proteinases. Curr. Top. Med. Mycolo. 7:55-69. [PubMed] [Google Scholar]
- 95.Hube, B. 2000. Extracellular proteinases of human pathogenic fungi, p. 126-137. In J. F. Ernst and A. Schmidt (ed.), Contributions to microbiology. Dimorphism in human pathogenic and apathogenic yeasts. S. Karger AG, Basel, Switzerland. [DOI] [PubMed]
- 96.Hube, B., M. Monod, D. A. Schofield, A. J. P. Brown, and N. A. R. Gow. 1994. Expression of seven members of the gene family encoding secretory aspartyl proteinases in Candida albicans. Mol. Microbiol. 14:87-99. [DOI] [PubMed] [Google Scholar]
- 97.Hube, B., and J. Naglik. 2001. Candida albicans proteinases: resolving the mystery of a gene family. Microbiology 147:1997-2005. [DOI] [PubMed] [Google Scholar]
- 98.Hube, B., and J. R. Naglik. 2002. Extracellular hydrolases, p. 107-122. In R. A. Calderone (ed.), Candida and candidiasis. ASM Press, Washington, D.C.
- 99.Hube, B., D. Sanglard, M. Monod, A. J. P. Brown, and N. A. R. Gow. 1997. Extracellular proteolytic activity of Candida species, p. 109-122. In H. Vanden Bossche, D. A. Stevens, and F. C. Odds (ed.), Host-fungus interplay. Proceedings of the Fifth Symposium on Topics in Mycology, 1995. National Foundation for Infectious Diseases, Bethesda, Md.
- 100.Hube, B., D. Sanglard, F. C. Odds, D. Hess, M. Monod, W. Schafer, A. J. Brown, and N. A. Gow. 1997. Disruption of each of the secreted aspartyl proteinase genes SAP1, SAP2, and SAP3 of Candida albicans attenuates virulence. Infect. Immun. 65:3529-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hube, B., C. J. Turver, F. C. Odds, H. Eiffert, G. J. Boulnois, H. Kochel, and R. Ruchel. 1991. Sequence of the Candida albicans gene encoding the secretory aspartate proteinase. J. Med. Vet. Mycol. 29:129-132. [PubMed] [Google Scholar]
- 102.Ibrahim, A. S., S. G. Filler, M. A. Ghannoum, and J. E. Edwards, Jr. 1993. Interferon-γ protects endothelial cells from damage by Candida albicans. J. Infect. Dis. 167:1467-1470. [DOI] [PubMed] [Google Scholar]
- 103.Ibrahim, A. S., S. G. Filler, D. Sanglard, J. E. J. Edwards, and B. Hube. 1998. Secreted aspartyl proteinases and interactions of Candida albicans with human endothelial cells. Infect. Immun. 66:3003-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jaton-Ogay, K., S. Paris, M. Huerre, M. Quadroni, R. Falchetto, G. Togni, J. P. Latge, and M. Monod. 1994. Cloning and disruption of the gene encoding an extracellular metalloprotease of Aspergillus fumigatus. Mol. Microbiol. 14:917-928. [DOI] [PubMed] [Google Scholar]
- 105.Kaminishi, H., H. Hamatake, T. Cho, T. Tamaki, N. Suenaga, T. Fujii, Y. Hagihara, H. Maeda, S. H. Miyasaki, T. C. White, and N. Agabian. 1994. Activation of blood clotting factors by microbial proteinases. FEMS Microbiol. Lett. 176:1702-1710. [DOI] [PubMed] [Google Scholar]
- 106.Kaminishi, H., H. Miyaguchi, T. Tamaki, N. Suenaga, M. Hisamatsu, I. Mihashi, H. Matsumoto, H. Maeda, and Y. Hagihara. 1995. Degradation of humoral host defense by Candida albicans proteinase. Infect. Immun. 63:984-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kaminishi, H., M. Tanaka, T. Cho, H. Maeda, and Y. Hagihara. 1990. Activation of the plasma kallikrein-kinin system by Candida albicans proteinase. Infect. Immun. 58:2139-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kaufmann, S. H. 1990. Heat shock proteins and the immune response. Immunol. Today 11:129-136. [DOI] [PubMed] [Google Scholar]
- 109.Kilian, M., J. Mestecky, and M. W. Russell. 1988. Defense mechanisms involving Fc- dependent functions of immunoglobulin A and their subversion by bacterial immunoglobulin A proteases. Microbiol. Rev. 52:296-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kimura, L. H., and N. N. Pearsall. 1978. Adherence of Candida albicans to human buccal epithelial cells. Infect. Immun. 21:64-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kimura, L. H., and N. N. Pearsall. 1980. Relationship between germination of Candida albicans and increased adherence to human buccal epithelial cells. Infect. Immun. 28:464-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kirkpatrick, C. H., R. R. Rich, and J. E. Bennett. 1971. Chronic mucocutaneous candidiasis: model-building in cellular immunity. Ann. Intern. Med. 74:955-978. [DOI] [PubMed] [Google Scholar]
- 113.Klar, A. J., T. Srikantha, and D. R. Soll. 2001. A histone deacetylation inhibitor and mutant promote colony-type switching of the human pathogen Candida albicans. Genetics 158:919-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Klotz, S. A., D. J. Drutz, J. L. Harrison, and M. Huppert. 1983. Adherence and penetration of vascular endothelium by Candida yeasts. Infect. Immun. 42:374-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Koelsch, G., J. Tang, J. A. Loy, M. Monod, K. Jackson, S. I. Foundling, and X. Lin. 2000. Enzymic characteristics of secreted aspartic proteases of Candida albicans. Biochim. Biophys. Acta 1480:117-131. [DOI] [PubMed] [Google Scholar]
- 116.Kondoh, Y., K. Shimizu, and K. Tanaka. 1987. Proteinase production and pathogenicity of Candida albicans. II. Virulence for mice of C. albicans strains of different proteinase activity. Microbiol. Immunol. 31:1061-1069. [DOI] [PubMed] [Google Scholar]
- 117.Korting, H. C., M. Schaller, G. Eder, G. Hamm, U. Bohmer, and B. Hube. 1999. Effects of the human immunodeficiency virus (HIV) proteinase inhibitors saquinavir and indinavir on in vitro activities of secreted aspartyl proteinases of Candida albicans isolates from HIV-infected patients. Antimicrob. Agents Chemother. 43:2038-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Krause, R., E. Schwab, D. Bachhiesl, F. Daxbock, C. Wenisch, G. J. Krejs, and E. C. Reisinger. 2001. Role of Candida in antibiotic-associated diarrhea. J. Infect. Dis. 184:1065-1069. [DOI] [PubMed] [Google Scholar]
- 119.Kretschmar, M., A. Felk, P. Staib, M. Schaller, D. Hess, M. Callapina, J. Morschhauser, W. Schafer, H. C. Korting, H. Hof, B. Hube, and T. Nichterlein. 2002. Individual acid aspartic proteinases (Saps) 1-6 of Candida albicans are not essential for invasion and colonization of the gastrointestinal tract in mice. Microb. Pathog. 32:61-70. [DOI] [PubMed] [Google Scholar]
- 120.Kretschmar, M., B. Hube, T. Bertsch, D. Sanglard, R. Merker, M. Schroder, H. Hof, and T. Nichterlein. 1999. Germ tubes and proteinase activity contribute to virulence of Candida albicans in murine peritonitis. Infect. Immun. 67:6637-6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kvaal, C., S. A. Lachke, T. Srikantha, K. Daniels, J. McCoy, and D. R. Soll. 1999. Misexpression of the opaque-phase-specific gene PEP1 (SAP1) in the white phase of Candida albicans confers increased virulence in a mouse model of cutaneous infection. Infect. Immun. 67:6652-6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kwon-Chung, K. J., D. Lehman, C. Good, and P. T. Magee. 1985. Genetic evidence for role of extracellular proteinase in virulence of Candida albicans. Infect. Immun. 49:571-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lane, S., C. Birse, S. Zhou, R. Matson, and H. Liu. 2001. DNA array studies demonstrate convergent regulation of virulence factors by Cph1, Cph2, and Efg1 in Candida albicans. J. Biol. Chem. 276:48988-48996. [DOI] [PubMed] [Google Scholar]
- 124.Leidich, S. D., A. S. Ibrahim, Y. Fu, A. Koul, C. Jessup, Vitullo, W. Fonzi, F. Mirbod, S. Nakashima, Y. Nozawa, and M. A. Ghannoum. 1998. Cloning and disruption of caPLB1, a phospholipase B gene involved in the pathogenicity of Candida albicans. J. Biol. Chem. 273:26078-26086. [DOI] [PubMed] [Google Scholar]
- 125.Lerner, C. G., and R. C. Goldman. 1993. Stimuli that induce production of Candida albicans extracellular aspartyl proteinase. J. Gen. Microbiol. 139:1643-1651. [DOI] [PubMed] [Google Scholar]
- 126.Liljemark, W. F., and R. J. Gibbons. 1973. Suppression of Candida albicans by human oral streptococci in gnotobiotic mice. Infect. Immun. 8:846-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liu, H. 2001. Transcriptional control of dimorphism in Candida albicans. Curr. Opin. Microbiol. 4:728-735. [DOI] [PubMed] [Google Scholar]
- 128.Liu, H., J. Kohler, and G. R. Fink. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723-1726. [DOI] [PubMed] [Google Scholar]
- 129.Lo, H. J., J. R. Kohler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939-949. [DOI] [PubMed] [Google Scholar]
- 130.Lupetti, A., G. Guzzi, A. Paladini, K. Swart, M. Campa, and S. Senesi. 1995. Molecular typing of Candida albicans in oral candidiasis: karyotype epidemiology with human immunodeficiency virus-seropositive patients in comparison with that with healthy carriers. J. Clin. Microbiol. 33:1238-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Macdonald, F. 1984. Secretion of inducible proteinase by pathogenic Candida species. Sabouraudia 22:79-82. [PubMed] [Google Scholar]
- 132.Macdonald, F., and F. C. Odds. 1980. Inducible proteinase of Candida albicans in diagnostic serology and in the pathogenesis of systemic candidosis. J. Med. Microbiol. 13:423-435. [DOI] [PubMed] [Google Scholar]
- 133.Macdonald, F., and F. C. Odds. 1980. Purified Candida albicans proteinase in the serological diagnosis of systemic candidosis. JAMA 243:2409-2411. [PubMed] [Google Scholar]
- 134.Macdonald, F., and F. C. Odds. 1983. Virulence for mice of a proteinase-secreting strain of Candida albicans and a proteinase-deficient mutant. J. Gen. Microbiol. 129:431-438. [DOI] [PubMed] [Google Scholar]
- 135.Magee, P. T., and S. Scherer. 1998. Genome mapping and gene discovery in Candida albicans. ASM News 64:505-511. [Google Scholar]
- 136.Mathaba, L. T., A. E. Paxman, P. B. Ward, D. A. Forbes, and J. R. Warmington. 2000. Genetically distinct strains of Candida albicans with elevated secretory proteinase production are associated with diarrhoea in hospitalized children. J. Gastroenterol. Hepatol. 15:53-60. [DOI] [PubMed] [Google Scholar]
- 137.Mathur, S., G. V. Koistinen, E. O. Horger, T. A. Mahvi, and H. H. Fudenberg. 1977. Humoral immunity in vaginal candidiasis. Infect. Immun. 15:287-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Matthews, R. C., J. P. Burnie, D. Howat, T. Rowland, and F. Walton. 1991. Autoantibody to heat-shock protein 90 can mediate protection against systemic candidosis. Immunology 74:20-24. [PMC free article] [PubMed] [Google Scholar]
- 139.Matthews, R. C., J. P. Burnie, and S. Tabaqchali. 1984. Immunoblot analysis of the serological response in systemic candidosis. Lancet ii:1415-1418. [DOI] [PubMed] [Google Scholar]
- 140.McCullough, M. J., B. C. Ross, B. D. Dwyer, and P. C. Reade. 1994. Genotype and phenotype of oral Candida albicans from patients infected with the human immunodeficiency virus. Microbiology 140:1195-1202. [DOI] [PubMed] [Google Scholar]
- 141.McKerrow, J. H., E. Sun, P. J. Rosenthal, and J. Bouvier. 1993. The proteases and pathogenicity of parasitic protozoa. Annu. Rev. Microbiol. 47:821-853. [DOI] [PubMed] [Google Scholar]
- 142.Millon, L., C. Drobacheff, R. Piarroux, M. Monod, G. Reboux, R. Laurent, and D. Meillet. 2001. Longitudinal study of anti-Candida albicans mucosal immunity against aspartic proteinases in HIV-infected patients. J. Acquir. Immune Defic. Syndr. 26:137-144. [DOI] [PubMed] [Google Scholar]
- 143.Miyasaki, S. H., J. B. Hicks, D. Greenspan, I. Polacheck, L. A. MacPhail, T. C. White, N. Agabian, and J. S. Greenspan. 1992. The identification and tracking of Candida albicans isolates from oral lesions in HIV-seropositive individuals. J. Acquir. Immune Defic. Syndr. 5:1039-1046. [PubMed] [Google Scholar]
- 144.Miyasaki, S. H., T. C. White, and N. Agabian. 1994. A fourth secreted aspartyl proteinase gene (SAP4) and a CARE2 repetitive element are located upstream of the SAP1 gene in Candida albicans. J. Bacteriol. 176:1702-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Monod, M., and M. Borg-von Zepelin. 2002. Secreted proteinases and other virulence mechanisms of Candida albicans. Chem. Immunol. 81:114-128. [DOI] [PubMed] [Google Scholar]
- 146.Monod, M., B. Hube, D. Hess, and D. Sanglard. 1998. Differential regulation of SAP8 and SAP9, which encode two new members of the secreted aspartic proteinase family in Candida albicans. Microbiology 144:2731-2737. [DOI] [PubMed] [Google Scholar]
- 147.Monod, M., G. Togni, B. Hube, and D. Sanglard. 1994. Multiplicity of genes encoding secreted aspartic proteinases in Candida species. Mol. Microbiol. 13:357-368. [DOI] [PubMed] [Google Scholar]
- 148.Morrison, C. J., S. F. Hurst, S. L. Bragg, R. J. Kuykendall, H. Diaz, D. W. McLaughlin, and E. Reiss. 1993. Purification and characterization of the extracellular aspartyl proteinase of Candida albicans: removal of extraneous proteins and cell wall mannoprotein and evidence for lack of glycosylation. J. Gen. Microbiol. 139:1177-1186. [DOI] [PubMed] [Google Scholar]
- 149.Morrow, B., H. Ramsey, and D. R. Soll. 1994. Regulation of phase-specific genes in the more general switching system of Candida albicans strain 3153A. J. Med. Vet. Mycol. 32:287-294. [DOI] [PubMed] [Google Scholar]
- 150.Morrow, B., T. Srikantha, J. Anderson, and D. R. Soll. 1993. Coordinate regulation of two opaque-phase-specific genes during white-opaque switching in Candida albicans. Infect. Immun. 61:1823-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Morrow, B., T. Srikantha, and D. R. Soll. 1992. Transcription of the gene for a pepsinogen, PEP1, is regulated by white-opaque switching in Candida albicans. Mol. Cell. Biol. 12:2997-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Morschhauser, J., R. Virkola, T. K. Korhonen, and J. Hacker. 1997. Degradation of human subendothelial extracellular matrix by proteinase-secreting Candida albicans. FEMS Microbiol. Lett. 153:349-355. [DOI] [PubMed] [Google Scholar]
- 153.Murad, A. M., C. d'Enfert, C. Gaillardin, H. Tournu, F. Tekaia, D. Talibi, D. Marechal, V. Marchais, J. Cottin, and A. J. Brown. 2001. Transcript profiling in Candida albicans reveals new cellular functions for the transcriptional repressors CaTup1, CaMig1 and CaNrg1. Mol. Microbiol. 42:981-993. [DOI] [PubMed] [Google Scholar]
- 154.Murad, A. M., P. Leng, M. Straffon, J. Wishart, S. Macaskill, D. MacCallum, N. Schnell, D. Talibi, D. Marechal, F. Tekaia, C. d'Enfert, C. Gaillardin, F. C. Odds, and A. J. Brown. 2001. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 20:4742-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Murphy, J. M., and J. D. Walton. 1996. Three extracellular proteases from Cochliobolus carbonum: cloning and targeted disruption of ALP1. Mol. Plant-Microbe Interact 9:290-297. [DOI] [PubMed] [Google Scholar]
- 156.Naglik, J. R., G. Newport, T. C. White, L. L. Fernandes-Naglik, J. S. Greenspan, D. Greenspan, S. P. Sweet, S. J. Challacombe, and N. Agabian. 1999. In vivo analysis of secreted aspartyl proteinase expression in human oral candidiasis. Infect. Immun. 67:2482-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Naglik, J. R., C. A. Rodgers, P. J. Shirlaw, J. L. Dobbie, L. L. Fernandes-Naglik, D. Greenspan, N. Agabian, and S. J. Challacombe. 2003. Differential expression of Candida albicans secreted aspartyl proteinase and phospholipase B genes in humans correlates with active oral and vaginal infections. J. Infect. Dis. 188:465-475. [DOI] [PubMed] [Google Scholar]
- 158.Nantel, A., D. Dignard, C. Bachewich, D. Harcus, A. Marcil, A. P. Bouin, C. W. Sensen, H. Hogues, H. M. Van Het, P. Gordon, T. Rigby, F. Benoit, D. C. Tessier, D. Y. Thomas, and M. Whiteway. 2002. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell 13:3452-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Newport, G., and N. Agabian. 1997. KEX2 influences Candida albicans proteinase secretion and hyphal formation. J. Biol. Chem. 272:28954-28961. [DOI] [PubMed] [Google Scholar]
- 160.Odds, F. C. 1994. Candida species and virulence. ASM News 60:313-318. [Google Scholar]
- 161.Odds, F. C. 1994. Pathogenesis of Candida infections. J. Am. Acad. Dermatol. 31:S2-S5. [DOI] [PubMed] [Google Scholar]
- 162.Odds, F. C. 2000. Pathogenic fungi in the 21st century. Trends Microbiol. 8:200-201. [DOI] [PubMed] [Google Scholar]
- 163.Ogrydziak, D. M. 1993. Yeast extracellular proteases. Crit. Rev. Biotechnol. 13:1-55. [DOI] [PubMed] [Google Scholar]
- 164.Ohkuma, S., and B. Poole. 1978. Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents. Proc. Natl. Acad. Sci. USA 75:3327-3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ollert, M. W., R. Sohnchen, H. C. Korting, U. Ollert, S. Brautigam, and W. Brautigam. 1993. Mechanisms of adherence of Candida albicans to cultured human epidermal keratinocytes. Infect. Immun. 61:4560-4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ollert, M. W., C. Wende, M. Gorlich, C. G. McMullan-Vogel, M. Borg-von Zepelin, C. W. Vogel, and H. C. Korting. 1995. Increased expression of Candida albicans secretory proteinase, a putative virulence factor, in isolates from human immunodeficiency virus-positive patients. J. Clin. Microbiol. 33:2543-2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Pfaller, M. A., R. N. Jones, S. A. Messer, M. B. Edmond, and R. P. Wenzel. 1998. National surveillance of nosocomial blood stream infection due to Candida albicans: frequency of occurrence and antifungal susceptibility in the SCOPE Program. Diagn. Microbiol. Infect. Dis. 31:327-332. [DOI] [PubMed] [Google Scholar]
- 168.Pichova, I., L. Pavlickova, J. Dostal, E. Dolejsi, O. Hruskova-Heidingsfeldova, J. Weber, T. Ruml, and M. Soucek. 2001. Secreted aspartic proteases of Candida albicans, Candida tropicalis, Candida parapsilosis and Candida lusitaniae. Inhibition with peptidomimetic inhibitors. Eur. J. Biochem. 268:2669-2677. [DOI] [PubMed] [Google Scholar]
- 169.Powderly, W. G. 1992. Mucosal candidiasis caused by non-albicans species of Candida in HIV-positive patients. AIDS 6:604-605. [PubMed] [Google Scholar]
- 170.Qureshi, M. N., C. E. Barr, T. Seshamma, J. Reidy, R. J. Pomerantz, and O. Bagasra. 1995. Infection of oral mucosal cells by human immunodeficiency virus type 1 in seropositive persons. J. Infect. Dis. 171:190-193. [DOI] [PubMed] [Google Scholar]
- 171.Rams, T. E., M. J. Andriolo, D. Feik, S. N. Abel, T. M. McGivern, and J. Slots. 1991. Microbiological study of HIV-related periodontitis. J. Periodontol. 62:74-81. [DOI] [PubMed] [Google Scholar]
- 172.Ray, T. L., and C. D. Payne. 1987. Detection of Candida acid proteinase (CAP) antibodies in systemic candidiasis by enzyme immunoassay. Clin. Res. 35:711. [Google Scholar]
- 173.Ray, T. L., and C. D. Payne. 1988. Scanning electron microscopy of epidermal adherence and cavitation in murine candidiasis: a role for Candida acid proteinase. Infect. Immun. 56:1942-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ray, T. L., C. D. Payne, R. Ruchel, B. Ritter, and M. Schaffrinski. 1990. Comparative production and rapid purification of Candida acid proteinase from protein-supplemented cultures. Infect. Immun. 273:391-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Redding, S., J. Smith, G. Farinacci, M. Rinaldi, Fothergill, J. Rhine-Chalberg, and M. Pfaller. 1994. Resistance of Candida albicans to fluconazole during treatment of oropharyngeal candidiasis in a patient with AIDS: documentation by in vitro susceptibility testing and DNA subtype analysis. Clin. Infect. Dis. 18:240-242. [DOI] [PubMed] [Google Scholar]
- 176.Reichard, U., M. Monod, F. Odds, and R. Ruchel. 1997. Virulence of an aspergillopepsin-deficient mutant of Aspergillus fumigatus and evidence for another aspartic proteinase linked to the fungal cell wall. J. Med. Vet. Mycol. 35:189-196. [DOI] [PubMed] [Google Scholar]
- 177.Remold, H., H. Fasold, and F. Staib. 1968. Purification and characterization of a proteolytic enzyme from Candida albicans. Biochim. Biophys. Acta 167:399-406. [DOI] [PubMed] [Google Scholar]
- 178.Ripeau, J. S., M. Fiorillo, F. Aumont, P. Belhumeur, and L. de Repentigny. 2002. Evidence for differential expression of Candida albicans virulence genes during oral infection in intact and human immunodeficiency virus type 1-transgenic mice. J. Infect. Dis. 185:1094-1102. [DOI] [PubMed] [Google Scholar]
- 179.Rogers, P. D., and K. S. Barker. 2002. Evaluation of differential gene expression in fluconazole-susceptible and -resistant isolates of Candida albicans by cDNA microarray analysis. Antimicrob. Agents Chemother. 46:3412-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ross, I. K., F. De Bernardis, G. W. Emerson, A. Cassone, and P. A. Sullivan. 1990. The secreted aspartate proteinase of Candida albicans: physiology of secretion and virulence of a proteinase-deficient mutant. J. Gen. Microbiol. 136:687-694. [DOI] [PubMed] [Google Scholar]
- 181.Reference deleted.
- 182.Rüchel, R. 1981. Properties of a purified proteinase from the yeast Candida albicans. Biochim. Biophys. Acta 659:99-113. [DOI] [PubMed] [Google Scholar]
- 183.Rüchel, R. 1983. On the renin-like activity of Candida proteinases and activation of blood coagulation in vitro. Zentbl. Bakteriol. Mikrobiol. Hyg. Orig. A 255:368-379. [PubMed] [Google Scholar]
- 184.Rüchel, R. 1986. Cleavage of immunoglobulins by pathogenic yeasts of the genus Candida. Microbiol. Sci. 3:316-319. [PubMed] [Google Scholar]
- 185.Rüchel, R. 1992. Proteinase, p. 17-31. In J. E. Bennett, R. J. Hay, and P. K. Peterson (ed.), New strategies in fungal disease. Churchill Livingstone, Edinburgh, United Kingdom.
- 186.Rüchel, R., B. Boning-Stutzer, and A. Mari. 1988. A synoptical approach to the diagnosis of candidosis, relying on serological antigen and antibody tests, on culture, and on evaluation of clinical data. Mycoses 31:87-106. [PubMed] [Google Scholar]
- 187.Rüchel, R. and B. Boning. 1983. Detection of Candida proteinase by enzyme immunoassay and interaction of the enzyme with alpha-2-macroglobulin. J. Immunol. Methods 61:107-116. [DOI] [PubMed] [Google Scholar]
- 188.Rüchel, R., B. Ritter, and M. Schaffrinski. 1990. Modulation of experimental systemic murine candidosis by intravenous pepstatin. Zentbl. Bakteriol. 273:391-403. [DOI] [PubMed] [Google Scholar]
- 189.Rüchel, R., K. Uhlemann, and B. Boning. 1983. Secretion of acid proteinases by different species of the genus Candida. Zentbl. Bakteriol. Mikrobiol. Hyg. Orig. A 255:537-548. [PubMed] [Google Scholar]
- 190.Rüchel, R., F. Zimmermann, B. Boning-Stutzer, and U. Helmchen. 1991. Candidiasis visualised by proteinase-directed immunofluorescence. Virchows Arch. Ser. A 419:199-202. [DOI] [PubMed] [Google Scholar]
- 190a.Rupniak, H. T., C. Rowlatt, E. B. Lane, J. G. Steele, Trejdosiewicz, LK, B. Laskiewicz, S. Povey, and B. T. Hill. 1985. Characteristics of four new human cell lines derived from squamous cell carcinomas of the head and neck. J. Natl. Cancer Inst. 75:621-635. [PubMed] [Google Scholar]
- 191.Salyers, A., and D. Witt. 1994. Virulence factors that promote colonization, p. 30-46. In A. Salyers and D. Witt (ed.), Bacterial pathogenesis: a molecular approach. ASM Press, Washington, D.C.
- 192.Samaranayake, Y. H., and L. P. Samaranayake. 2001. Experimental oral candidiasis in animal models. Clin. Microbiol. Rev. 14:398-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sandin, R. L., A. L. Rogers, R. J. Patterson, and E. S. Beneke. 1982. Evidence for mannose-mediated adherence of Candida albicans to human buccal cells in vitro. Infect. Immun. 35:79-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sanglard, D., B. Hube, M. Monod, F. C. Odds, and N. A. Gow. 1997. A triple deletion of the secreted aspartyl proteinase genes SAP4, SAP5, and SAP6 of Candida albicans causes attenuated virulence. Infect. Immun. 65:3539-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Santos, M. A., and M. F. Tuite. 1995. The CUG codon is decoded in vivo as serine and not leucine in Candida albicans. Nucleic Acids. Res. 23:1481-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Sato, T., K. Nagai, M. Shibazaki, K. Abe, Y. Takebayashi, B. Lumanau, and R. M. Rantiatmodjo. 1994. Novel aspartyl protease inhibitors, YF-0200R-A and B. J. Antibiot. (Tokyo) 47:566-570. [DOI] [PubMed] [Google Scholar]
- 197.Sato, T., M. Shibazaki, H. Yamaguchi, K. Abe, H. Matsumoto, and M. Shimizu. 1994. Novel Candida albicans aspartyl protease inhibitor. II. A new pepstatin-ahpatinin group inhibitor, YF-044P-D. J. Antibiot. 47:588-590. [DOI] [PubMed] [Google Scholar]
- 198.Schaller, M., M. Bein, H. C. Korting, S. Baur, G. Hamm, M. Monod, S. Beinhauer, and B. Hube. 2003. The secreted aspartyl proteinases Sap1 and Sap2 cause tissue damage in an in vitro model of vaginal candidiasis based on reconstituted human vaginal epithelium. Infect. Immun. 71:3227-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Schaller, M., B. Hube, M. W. Ollert, W. Schafer, v. Borg, M. Zepelin, E. Thoma-Greber, and H. C. Korting. 1999. In vivo expression and localization of Candida albicans secreted aspartyl proteinases during oral candidiasis in HIV-infected patients. J. Investig. Dermatol. 112:383-386. [DOI] [PubMed] [Google Scholar]
- 200.Schaller, M., H. C. Korting, W. Schafer, J. Bastert, W. Chen, and B. Hube. 1999. Secreted aspartic proteinase (Sap) activity contributes to tissue damage in a model of human oral candidosis. Mol. Microbiol. 34:169-180. [DOI] [PubMed] [Google Scholar]
- 201.Schaller, M., C. Schackert, H. C. Korting, E. Januschke, and B. Hube. 2000. Invasion of Candida albicans correlates with expression of secreted aspartic proteinases during experimental infection of human epidermis. J. Investig. Dermatol. 114:712-717. [DOI] [PubMed] [Google Scholar]
- 202.Schaller, M., W. Schäfer, H. C. Korting, and B. Hube. 1998. Differential expression of secreted aspartyl proteinases in a model of human oral candidosis and in patient samples from the oral cavity. Mol. Microbiol. 29:605-615. [DOI] [PubMed] [Google Scholar]
- 203.Schiodt, M. 1992. HIV-associated salivary gland disease: a review. Oral Surg. Oral Med. Oral Pathol. 73:164-167. [DOI] [PubMed] [Google Scholar]
- 204.Schmid, J., F. C. Odds, M. J. Wiselka, K. G. Nicholson, and D. R. Soll. 1992. Genetic similarity and maintenance of Candida albicans strains from a group of AIDS patients, demonstrated by DNA fingerprinting. J. Clin. Microbiol. 30:935-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Schroppel, K., K. Sprosser, M. Whiteway, Y. Thomas, M. Rollinghoff, and C. Csank. 2000. Repression of hyphal proteinase expression by the mitogen-activated protein (MAP) kinase phosphatase cpp1p of Candida albicans is independent of the MAP kinase cek1p. Infect. Immun. 68:7159-7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Schweizer, A., S. Rupp, B. N. Taylor, M. Rollinghoff, and K. Schroppel. 2000. The TEA/ATTS transcription factor CaTec1p regulates hyphal development and virulence in Candida albicans. Mol. Microbiol. 38:435-445. [DOI] [PubMed] [Google Scholar]
- 207.Shapiro, J. 1983. Mobile genetic elements. Academic Press, Inc., Orlando, Fla.
- 208.Smith, J. M., C. M. Tang, S. Van Noorden, and D. W. Holden. 1994. Virulence of Aspergillus fumigatus double mutants lacking restriction and an alkaline protease in a low-dose model of invasive pulmonary aspergillosis. Infect. Immun. 62:5247-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Smithson, S. L., I. C. Paterson, A. M. Bailey, S. E. Screen, B. A. Hunt, B. D. Cobb, R. M. Cooper, A. K. Charnley; and J. M. Clarkson. 1995. Cloning and characterisation of a gene encoding a cuticle-degrading protease from the insect pathogenic fungus Metarhizium anisopliae. Gene 166:161-165. [DOI] [PubMed] [Google Scholar]
- 210.Smolenski, G., P. A. Sullivan, S. M. Cutfield, and J. F. Cutfield. 1997. Analysis of secreted aspartic proteinases from Candida albicans: purification and characterization of individual Sap1, Sap2 and Sap3 isoenzymes. Microbiology 143:349-356. [DOI] [PubMed] [Google Scholar]
- 211.Sobel, J. D. 1988. Pathogenesis and epidemiology of vulvovaginal candidiasis. Ann. N. Y. Acad. Sci. 544:547-557. [DOI] [PubMed] [Google Scholar]
- 212.Sobel, J. D. 1992. Pathogenesis and treatment of recurrent vulvovaginal candidiasis. Clin. Infect. Dis. 14(Suppl. 1):S148-S153. [DOI] [PubMed] [Google Scholar]
- 213.Sohn, K., C. Urban, H. Brunner, and S. Rupp. 2003. EFG1 is a major regulator of cell wall dynamics in Candida albicans as revealed by DNA microarrays. Mol. Microbiol. 47:89-102. [DOI] [PubMed] [Google Scholar]
- 214.Soll, D. R. 1992. High-frequency switching in Candida albicans. Clin. Microbiol. Rev. 5:183-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Soll, D. R. 1997. Gene regulation during high-frequency switching in Candida albicans. Microbiology 143:279-288. [DOI] [PubMed] [Google Scholar]
- 216.Soll, D. R. 2002. Phenotypic switching, p. 123-142. In R. A. Calderone (ed.), Candida and candidiasis. ASM Press, Washington, D.C.
- 217.Soll, D. R., B. Morrow, and T. Srikantha. 1993. High-frequency phenotypic switching in Candida albicans. Trends Genet. 9:61-65. [DOI] [PubMed] [Google Scholar]
- 218.Soll, D. R., B. Morrow, T. Srikantha, K. Vargas, and P. Wertz. 1994. Developmental and molecular biology of switching in Candida albicans. Oral Surg. Oral Med. Oral Pathol. 78:194-201. [DOI] [PubMed] [Google Scholar]
- 219.Srikantha, T., and D. R. Soll. 1993. A white-specific gene in the white-opaque switching system of Candida albicans. Gene 131:53-60. [DOI] [PubMed] [Google Scholar]
- 220.Srikantha, T., L. Tsai, K. Daniels, A. J. Klar, and D. R. Soll. 2001. The histone deacetylase genes HDA1 and RPD3 play distinct roles in regulation of high-frequency phenotypic switching in Candida albicans. J. Bacteriol. 183:4614-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Staab, J. F., and P. Sundstrom. 2003. URA3 as a selectable marker for disruption and virulence assessment of Candida albicans genes. Trends Microbiol. 11:69-73. [DOI] [PubMed] [Google Scholar]
- 222.Staib, F. 1965. Serum-proteins as nitrogen source for yeastlike fungi. Sabouraudia 4:187-193. [DOI] [PubMed] [Google Scholar]
- 223.Staib, P., M. Kretschmar, T. Nichterlein, H. Hof, and J. Morschhauser. 2000. Differential activation of a Candida albicans virulence gene family during infection. Proc. Natl. Acad. Sci. USA 97:6102-6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Staib, P., M. Kretschmar, T. Nichterlein, H. Hof, and J. Morschhauser. 2002. Transcriptional regulators Cph1p and Efg1p mediate activation of the Candida albicans virulence gene SAP5 during infection. Infect. Immun. 70:921-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Staib, P., M. Kretschmar, T. Nichterlein, G. Kohler, S. Michel, H. Hof, J. Hacker, and J. Morschhauser. 1999. Host-induced, stage-specific virulence gene activation in Candida albicans during infection. Mol. Microbiol. 32:533-546. [DOI] [PubMed] [Google Scholar]
- 225a.Stehr, F., A. Felk, M. Kretschmar, M. Schaller, W. Schafer, and B. Hube. 2000. Extracellular hydrolytic enzymes and their relevance during Candida albicans infections. Mycoses 43(Suppl. 2):17-21. [PubMed] [Google Scholar]
- 226.Stewart, K., and C. Abad-Zapatero. 2001. Candida proteases and their inhibition: prospects for antifungal therapy. Curr. Med. Chem. 8:941-948. [DOI] [PubMed] [Google Scholar]
- 227.Stewart, K. W., R. C. Goldman, and C. Abad-Zapatero. 1999. The secreted proteinases from Candida: challenges for structure-aided drug design, p. 117-138. In B. M. Dunn (ed.), Proteases of infectious agents. Academic Press, Inc., San Diego, Calif.
- 228.Stoldt, V. R., A. Sonneborn, C. E. Leuker, and J. F. Ernst. 1997. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 16:1982-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Stringaro, A., P. Crateri, G. Pellegrini, G. Arancia, A. Cassone, and F. De Bernardis. 1997. Ultrastructural localization of the secretory aspartyl proteinase in Candida albicans cell wall in vitro and in experimentally infected rat vagina. Mycopathologia 137:95-105. [DOI] [PubMed] [Google Scholar]
- 230.Sundstrom, P. 1999. Adhesins in Candida albicans. Curr. Opin. Microbiol. 2:353-357. [DOI] [PubMed] [Google Scholar]
- 231.Sundstrom, P. 2002. Adhesion in Candida spp. Cell Microbiol. 4:461-469. [DOI] [PubMed] [Google Scholar]
- 232.Sweet, S. P. 1997. Selection and pathogenicity of Candida albicans in HIV infection. Oral Dis. 3:S88-S95. [DOI] [PubMed] [Google Scholar]
- 233.Sweet, S. P., S. J. Challacombe, and J. R. Naglik. 1995. Whole and parotid saliva IgA and IgA-subclass responses to Candida albicans in HIV infection. Adv. Exp. Med. Biol. 371B:1031-1034. [PubMed] [Google Scholar]
- 234.Togni, G., D. Sanglard, R. Falchetto, and M. Monod. 1991. Isolation and nucleotide sequence of the extracellular acid protease gene (ACP) from the yeast Candida tropicalis. FEBS Lett. 286:181-185. [DOI] [PubMed] [Google Scholar]
- 235.Togni, G., D. Sanglard, M. Quadroni, S. I. Foundling, and M. Monod. 1996. Acid proteinase secreted by Candida tropicalis: functional analysis of preproregion cleavages in C. tropicalis and Saccharomyces cerevisiae. Microbiology 142:493-503. [DOI] [PubMed] [Google Scholar]
- 236.Tsuboi, R., I. Ko, K. Takamori, and H. Ogawa. 1989. Isolation of a keratinolytic proteinase from Trichophyton mentagrophytes with enzymatic activity at acidic pH. Infect. Immun. 57:3479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Tsuboi, R., H. Ogawa, K. Bramono, M. D. Richardson, G. S. Shankland, W. J. Crozier, Y. Sei, J. Ninomiya, A. Nakabayashi, and I. Takaiuchi. 1994. Pathogenesis of superficial mycoses. J. Med. Vet. Mycol. 32(Suppl. 1):91-104. [DOI] [PubMed] [Google Scholar]
- 238.Tsushima, H., H. Mine, Y. Kawakami, F. Hyodoh, and A. Ueki. 1994. Candida albicans aspartic proteinase cleaves and inactivates human epidermal cysteine proteinase inhibitor, cystatin A. Microbiology 140:167-171. [DOI] [PubMed] [Google Scholar]
- 239.Tzung, K. W., R. M. Williams, S. Scherer, N. Federspiel, T. Jones, N. Hansen, V. Bivolarevic, L. Huizar, C. Komp, R. Surzycki, R. Tamse, R. W. Davis, and N. Agabian. 2001. Genomic evidence for a complete sexual cycle in Candida albicans. Proc. Natl. Acad. Sci. USA 98:3249-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Valdez, H., B. M. Gripshover, R. A. Salata, and M. M. Lederman. 1998. Resolution of azole-resistant oropharyngeal candidiasis after initiation of potent combination antiretroviral therapy. AIDS 12:538. [PubMed] [Google Scholar]
- 241.Valdivia, R. H., and S. Falkow. 1997. Probing bacterial gene expression within host cells. Trends Microbiol. 5:360-363. [DOI] [PubMed] [Google Scholar]
- 242.von Heijne, G. 1985. Signal sequences. The limits of variation. J. Mol. Biol. 184:99-105. [DOI] [PubMed] [Google Scholar]
- 243.Vudhichamnong, K., D. M. Walker, and H. C. Ryley. 1982. The effect of secretory immunoglobulin A on the in-vitro adherence of the yeast Candida albicans to human oral epithelial cells. Arch. Oral Biol. 27:617-621. [DOI] [PubMed] [Google Scholar]
- 244.Watts, H. J., F. S. Cheah, B. Hube, D. Sanglard, and N. A. Gow. 1998. Altered adherence in strains of Candida albicans harbouring null mutations in secreted aspartic proteinase genes. FEMS Microbiol. Lett. 159:129-135. [DOI] [PubMed] [Google Scholar]
- 245.Whelan, W. L., D. R. Kirsch, K. J. Kwon-Chung, S. M. Wahl, and P. D. Smith. 1990. Candida albicans in patients with the acquired immunodeficiency syndrome: absence of a novel of hypervirulent strain. J. Infect. Dis. 162:513-518. [DOI] [PubMed] [Google Scholar]
- 246.White, T. C., and N. Agabian. 1995. Candida albicans secreted aspartyl proteinases: isoenzyme pattern is determined by cell type, and levels are determined by environmental factors. J. Bacteriol. 177:5215-5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.White, T. C., S. H. Miyasaki, and N. Agabian. 1993. Three distinct secreted aspartyl proteinases in Candida albicans. J. Bacteriol. 175:2997-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.White, T. C., M. A. Pfaller, M. G. Rinaldi, J. Smith, and S. W. Redding. 1997. Stable azole drug resistance associated with a substrain of Candida albicans from an HIV-infected patient. Oral Dis. 3(Suppl 1):S102-S109. [DOI] [PubMed] [Google Scholar]
- 249.Wood, V., R. Gwilliam, M. A. Rajandream, M. Lyne, R. Lyne, A. Stewart, J. Sgouros, N. Peat, J. Hayles, S. Baker, D. Basham, S. Bowman, K. Brooks, D. Brown, S. Brown, T. Chillingworth, C. Churcher, M. Collins, R. Connor, A. Cronin, P. Davis, T. Feltwell, A. Fraser, S. Gentles, A. Goble, N. Hamlin, D. Harris, J. Hidalgo, G. Hodgson, S. Holroyd, T. Hornsby, S. Howarth, E. J. Huckle, S. Hunt, K. Jagels, K. James, L. Jones, M. Jones, S. Leather, S. McDonald, J. McLean, P. Mooney, S. Moule, K. Mungall, L. Murphy, D. Niblett, C. Odell, K. Oliver, S. O'Neil, D. Pearson, M. A. Quail, E. Rabbinowitsch, K. Rutherford, S. Rutter, D. Saunders, K. Seeger, S. Sharp, J. Skelton, M. Simmonds, R. Squares, S. Squares, K. Stevens, K. Taylor, R. G. Taylor, A. Tivey, S. Walsh, T. Warren, S. Whitehead, J. Woodward, G. Volckaert, R. Aert, J. Robben, B. Grymonprez, I. Weltjens, E. Vanstreels, M. Rieger, M. Schafer, S. Muller-Auer, C. Gabel, M. Fuchs, C. Fritze, E. Holzer, D. Moestl, H. Hilbert, K. Borzym, I. Langer, A. Beck, H. Lehrach, R. Reinhardt, T. M. Pohl, P. Eger, W. Zimmermann, H. Wedler, R. Wambuttt, B. Purnelle, A. Goffeau, E. Cadieu, S. Dreano, S. Gloux, V. Lelaure, S. Mottier, F. Galibert, S. J. Aves, Z. Xiang, C. Hunt, K. Moore, S. M. Hurst, M. Lucas, M. Rochet, C. Gaillardin, V. A. Tallada, A. Garzon, G. Thode, R. R. Daga, L. Cruzado, J. Jimenez, M. Sanchez, F. del Rey, J. Benito, A. Dominguez, J. L. Revuelta, S. Moreno, J. Armstrong, S. L. Forsburg, L. Cerrutti, T. Lowe, W. R. McCombie, I. Paulsen, J. Potashkin, G. V. Shpakovski, D. Ussery, B. G. Barrell, and P. Nurse. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415:871-880. [DOI] [PubMed] [Google Scholar]
- 250.Wright, R. J., A. Carne, A. D. Hieber, I. L. Lamont, G. W. Emerson, and P. A. Sullivan. 1992. A second gene for a secreted aspartate proteinase in Candida albicans. J. Bacteriol. 174:7848-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Wu, T., and L. P. Samaranayake. 1999. The expression of secreted aspartyl proteinases of Candida species in human whole saliva. J. Med. Microbiol. 48:711-720. [DOI] [PubMed] [Google Scholar]
- 252.Wu, T., L. P. Samaranayake, B. Y. Cao, and J. Wang. 1996. In-vitro proteinase production by oral Candida albicans isolates from individuals with and without HIV infection and its attenuation by antimycotic agents. J. Med. Microbiol. 44:311-316. [DOI] [PubMed] [Google Scholar]
- 253.Yuan, L., and G. T. Cole. 1987. Isolation and characterization of an extracellular proteinase of Coccidioides immitis. Infect. Immun. 55:1970-1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Zaugg, C., M. Borg-von Zepelin, D. Reichard, D. Sanglard, and M. Monod. 2001. Secreted aspartic proteinase family of Candida tropicalis. Infect. Immun. 69:405-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Zhang, Z., H. N. El Sohly, M. R. Jacob, D. S. Pasco, L. A. Walker, and A. M. Clark. 2002. Natural products inhibiting Candida albicans secreted aspartic proteases from Lycopodium cernuum. J. Nat. Prod. 65:979-985. [DOI] [PubMed] [Google Scholar]
- 256.Zhang, Z., H. N. El Sohly, M. R. Jacob, D. S. Pasco, L. A. Walker, and A. M. Clark. 2002. Natural products inhibiting Candida albicans secreted aspartic proteases from Tovomita krukovii. Planta Med. 68:49-54. [DOI] [PubMed] [Google Scholar]
- 257.Zhou, J., and J. H. Miller. 2002. Microbial genomics—challenges and opportunities: the 9th International Conference on Microbial Genomes. J. Bacteriol. 184:4327-4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Zotter, C., U. F. Haustein, C. Schonborn, H. D. Grimmecke, and H. Wand. 1990. Effect of pepstatin A on Candida albicans infection in the mouse. Dermatol. Monatsschrift 176:189-198. [PubMed] [Google Scholar]