Abstract
We used the species specificity and repetitious nature of subtelomeric kinetoplastida sequences to generate a duplex PCR assay for the simultaneous detection of Trypanosoma cruzi and Trypanosoma rangeli in experimentally and naturally infected triatomine (Reduviid) bugs and in infected human subjects. The assay was species specific and was capable of detecting 1/20th of T. cruzi and 1/4th of T. rangeli cell equivalents without complementary hybridization. In addition, the PCR-based assay was robust enough for direct application to difficult biological samples such as Reduviid feces or guts and was capable of recognizing all T. cruzi and T. rangeli strains and lineages. Because the assay primers amplify entirely different target sequences, no reaction interference was observed, facilitating future adaptation of this assay to an automated format.
Trypanosoma cruzi causes Chagas' disease, a fatal illness endemic to many Latin American countries. T. cruzi shares reservoirs and vectors with the related protozoan Trypanosoma rangeli, which is pathogenic to Reduviid bugs but apparently harmless to humans (1). These parasites share morphological similarities and antigenic determinants, making microscopic and serological diagnosis a challenge (13, 27). In recent years, we have studied an area where Chagas' disease is endemic and the two parasites coexist (3, 4). In this area, T. rangeli is more efficiently transmitted (23) and has the larger population exposure (3). Considering that inaccurate serological tests can lead to misdiagnosis, unnecessary chemotherapy (frequently accompanied by side effects) and psychological trauma to the patients and their families, we sought to develop a molecular test to be used in difficult cases and as a gold standard for our serology. The assay consists of a simple duplex PCR that allows the simultaneous detection of T. cruzi and T. rangeli DNA in complex biological samples.
MATERIALS AND METHODS
Parasites.
T. rangeli and T. cruzi strains and isolates (Table 1) were cultured in LIT (liver infusion tryptose) medium supplemented with 10% fetal bovine serum or in biphasic blood-agar/NNN (0.146% NaCl, 0.045% KCl, 0.05% CaCl2, 0.019% NaHCO3 [pH 7.2]) medium at 28°C. Leishmania promastigotes were cultured as described previously (10). For experimental infections and assays with parasitic cultures, we used T. cruzi isolate MHOM/VE/92/2-92-YBM and T. rangeli isolate MHOM/VE/2000/CH.
TABLE 1.
T. cruzi and T. rangeli strains, isolates, and clones used in this work
| Strain | Origin | Host | Source or referencea |
|---|---|---|---|
| T. rangeli | |||
| DOG-82 | Venezuela | Dog | Añez et al. (2) |
| Triatomino 1 | Venezuela | Rhodnius prolixus | N. Añez (ULA, Venezuela) |
| Triatomino 2 | Venezuela | Rhodnius prolixus | N. Añez (ULA, Venezuela) |
| San Agustin | Colombia | Human | Tibayrenc et al. (26) |
| Palma-2 | Venezuela | Rhodnius prolixus | Steindel et al. (25) |
| JRM | Venezuela | Human | N. Añez (ULA, Venezuela) |
| CH-00 | Venezuela | Human | N. Añez (ULA, Venezuela) |
| Mono | Venezuela | Macacus sp. | N. Añez (ULA, Venezuela) |
| T. cruzi | |||
| CL | Brazil | Triatoma infestans | Brener and Chiari (6) |
| CL Brener | Brazil | Clone from CL strain | Batista et al. (5) |
| G | Brazil | Didelphis marsupialis | Camargo (7) |
| Y | Brazil | Human | Silva and Nussenzweig (22) |
| JMP | Venezuela | Human | N. Añez (ULA, Venezuela) |
| YBM | Venezuela | Human | N. Añez (ULA, Venezuela) |
| Tulahuen | Chile | Triatoma infestans | Pizzi et al. (19) |
| Dm28c | Venezuela | Didelphis marsupialis | Contreras et al. (12) |
| Dm30c | Venezuela | Didelphis marsupialis | |
| Maracay | Venezuela | Human | Requena et al. (20) |
ULA, Universidad de Los Andes.
DNA extraction from parasites.
Genomic DNA from T. rangeli and T. cruzi epimastigotes and Leishmania and Crithidia promastigotes was extracted from log-phase cultures of 108 parasites as previously described (18). DNA samples from other trypanosomatids were kindly provided by Franco Da Silveira (Escola Paulista de Medicina, Universidad Federal de Saõ Paulo, Sao Paulo, Brasil) and Mendoza-Leon (Universidad Central de Venezuela, Caracas, Venezuela).
DNA samples for titration of duplex PCR.
For titration of the duplex amplification reactions (see Fig. 4), DNA was released from parasites by diluting 105 epimastigote cells in 100 μl of 10 mM Tris (pH 7.5)-150 mM NaCl and boiling the cell suspension for 10 min.
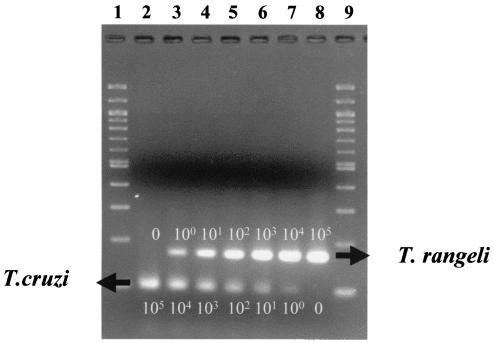
FIG. 4.
T. cruzi/T. rangeli duplex PCR assay titration experiment. Lanes 2 to 8, PCR products obtained from serial dilutions of lysed parasites in the following T. rangeli DOG-82/T. cruzi CL Brener ratios: lane 2, 0/105; lane 3, 100/104; lane 4, 101/103; lane 5, 102/102; lane 6, 103/101; lane 7, 104/100; lane 8, 105/0. Lanes 1 and 9 contain a 100-bp size ladder (Gibco-BRL).
Samples for duplex PCR.
Samples of parasite cultures, feces, or intestinal tracts of triatomines and blood samples from mice and human patients were collected on filter paper (Whatman 3MM), air dried, and stored at room temperature until used. Rhodnius prolixus nymphs were fed on mice infected with T. cruzi and/or T. rangeli. Infected triatomine feces and guts were collected 30 to 45 days postfeeding. Samples were dispersed in a drop of phosphate-buffered saline solution and examined microscopically to confirm the presence of flagellates. Infected insects were killed by ether anesthesia, and their intestinal tract was dissected, homogenized in 300 μl of phosphate-buffered saline, and examined microscopically. A single triatomine bug captured in a domicile located in an area where T. cruzi and T. rangeli are endemic was subjected to the same treatment.
Experimental mice were infected by an intradermic injection of 105 T. cruzi blood trypomastigotes. After 15 days, mice were sacrificed, and their hearts were removed and preserved at −20°C. To simulate infected mouse blood for additional experiments, 5 × 106 epimastigotes of T. cruzi, T. rangeli, or both were mixed with 1 ml of blood.
Preparation of samples for duplex PCR.
For blood samples collected on filter paper, 6-mm-diameter filter disks were cut and placed in Eppendorf microtubes with 100 μl of distilled sterile water. In the case of heart biopsies, approximately 2 mm3 of material was used in place of the filter disks. All samples were incubated with 5 μl of 20-mg/ml proteinase K (Promega) at 56°C for 30 min. After boiling for 10 min, Chelex-100 (Bio-Rad) was added to a final concentration of 10%, and the mixture was boiled again for 15 min. The samples were centrifuged at 12,000 rpm for 20 min, and the supernatant was transferred to a fresh tube, phenol-chloroform extracted, and precipitated with 99% ethanol plus 1 μl of 2% glycogen. Precipitated DNA was resuspended in 30 μl of H2O and stored at −20°C.
PCR conditions.
For T. cruzi detection, we used a modified version of a PCR assay targeted to the 189-bp telomeric junction (Tc189, GenBank accession number AF100651) (Fig. 1) (11). The primers were T189Fw2 (5′-CCAACGCTCCGGGAAAAC-3′) and Tc189Rv3 (5′-GCGTCTTCTCAGTATGGACTT-3′). For T. rangeli detection, we used an assay targeted to a conserved subtelomeric region (SubTr, GenBank accession number AF426020) (8) (Fig. 1). The primers were TrF3 (5′-CCCCATACAAAACACCCTT-3′) and TrR8 (5′-TGGAATGACGGTGCGGCGAC-3′). All oligonucleotides were purchased from Operon Technologies, Inc., Alameda, Calif. Single PCR amplifications were conducted in a final volume of 25 μl containing 0.2 mM deoxynucleoside triphosphate mixture, 1.5 mM MgCl2, 50 mM KCl, 10 mM Tris-HCl (pH 9.0), 0.1% Triton X-100, 0.4 μM each of the forward and reverse primers, 1.25 U of Taq DNA polymerase (Promega), and 1 to 10 ng of purified DNA template. An initial denaturation step at 94°C for 4 min was followed by 35 cycles of 30 s at 55°C, 40 s at 72°C, and 1 min at 94°C. This amplification was followed by incubations of 1 min at 55°C and 3 min at 72°C. Amplification was confirmed in a 2% agarose gel stained with ethidium bromide.
FIG. 1.
Schematic representation of T. cruzi and T. rangeli telomeric organization. Horizontal bars indicate the regions amplified by each set of primers. SubTr, T. rangeli conserved subtelomeric region. Blocks grouped with parentheses are sequences related to the T. cruzi trans-sialidase (TSA) superfamily.
Duplex PCR was performed in a final volume of 30 μl containing 2.5 mM MgCl2, 60 mM KCl, 12 mM Tris-HCl (pH 9.0), 0.12% Triton X-100, 0.24 mM deoxynucleoside triphosphate mix, 0.01% bovine serum albumin, 0.4 mM each Tc189Fw2 and Tc189Rv3 primers, 0.67 mM each TrF3 and TrR8 primers, 1.25 U of Taq DNA polymerase (Promega), and 4 μl of DNA preparation. Cycling was performed as above. PCR products were electrophoresed in a 3% agarose gel and stained with ethidium bromide. All PCR amplifications were performed in a GeneAmp PCR System 2400 (Perkin-Elmer).
For Southern blotting, gel-separated PCR products were transferred to nylon filters (Hybond-N; Amersham-Pharmacia-Biotech), UV cross-linked, dried, and stored until used.
Labeling and hybridizations with capture oligonucleotides.
For confirmatory hybridization experiments, we designed the following capture oligonucleotides: Tc189cap (5′-GCTCAAACACACTACACAAAG-3′) for the T. cruzi assay and Trcap (5′-CGGTAAAGCGAGTTTTGGT-3′) for the T. rangeli assay.
These oligonucleotides were labeled with dUTP-digoxigenin tails synthesized by terminal deoxynucleotidyl transferase as described previously (15). Hybridizations were carried out in 5× SSC (0.75 M NaCl, 0.075 M sodium citrate [pH 7.0], 0.1% sodium-N-laurylsarcosinate, 0.02% SDS, and 1% blocking reagent [Boehringer Mannheim]) at 55°C for 3 h. After hybridization, filters were washed at room temperature in 6× SSC-0.1% SDS for 5 min, then washed with 2× SSC-0.1% SDS at room temperature for 15 min, and finally washed at 58°C for 15 min with the same solution. Hybridized probes were detected by chemiluminescence with Lumi Phos 530, according to the manufacturer's instructions (Boehringer Mannheim).
RESULTS
Individual PCR assay specificity.
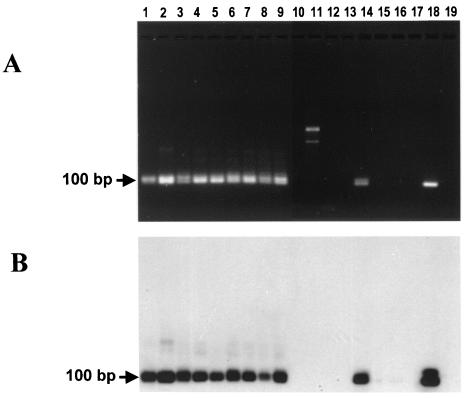
We first determined the optimal conditions and specificity of each assay. To adapt the original Tc189 PCR assay (11) to a duplex format, we designed a new set of primers that produced a lower-molecular-weight amplification band. When the modified Tc189 PCR assay was used in DNA samples from different members of the Trypanosomatidae family, a specific 100-bp amplification product was observed (Fig. 2A). This band was consistently amplified from samples of T. cruzi from different origins and of different “types” (Fig. 2A, lanes 1 to 9). A human sample originally thought to contain only T. rangeli also tested positive for T. cruzi in this PCR assay (Fig. 2A, lane 14; Fig. 3). The positive control containing DNA from a T. cruzi CL Brener telomeric recombinant (TcVA17; lane 18) (9) was positive for the 100-bp PCR fragment, and the negative controls, including DNA samples from Leishmania (lanes 15 and 16), Crithidia (lane 17), and T. rangeli (lanes 10 to 11) did not produce the 100-bp amplification product. In addition, there was a higher-molecular-weight PCR product (lane 11, see below) that confirmatory hybridization experiments with the capture digoxigenin-labeled Tc189cap oligonucleotide as probe proved to be unspecific (Fig. 2B).
FIG. 2.
PCR amplification of the T. cruzi Tc189-specific sequence. (A) Agarose gel electrophoresis of amplified products obtained with primers Tc189 Fw2 and Tc189 Rv3. (B) Southern blot of gel A after hybridization with digoxigenin-labeled Tc189cap oligonucleotide. Lanes: 1, T. cruzi CL Brener; 2, T. cruzi CL; 3, T. cruzi G; 4, T. cruzi Y; 5, T. cruzi Tulahuen; 6, T. cruzi Dm 28c; 7, T. cruzi Dm 30c; 8, T. cruzi Maracay; 9, T. cruzi JMP; 10, T. rangeli DOG-82; 11, T. rangeli Triat 1; 12, T. rangeli S. Agustín; 13, T. rangeli Mono; 14, T. rangeli Alba; 15, L. chagasi; 16, L. braziliensis; 17, C. fasciculata; 18, recombinant TcVA17 (positive control) (9); 19, H2O. Arrow indicates the 100-bp specific product.
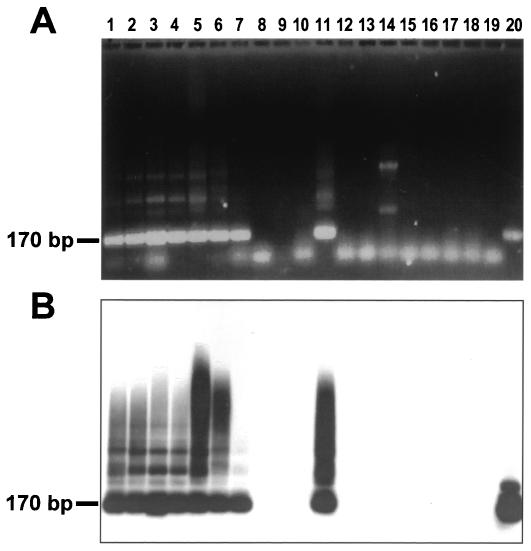
FIG. 3.
PCR amplification of T. rangeli SubTr-specific sequence. (A) Agarose gel electrophoresis of amplified products with TrF3 and TrRv6 primers. (B) Southern blot of gel A after hybridization with digoxigenin-labeled Trcap oligonucleotide. Lanes: 1, T. rangeli DOG-82; 2, T. rangeli triat 1; 3, T. rangeli triat 2; 4, T. rangeli S. Agustín; 5, T. rangeli Palma, 6, T. rangeli JRM; 7, T. rangeli Alba; 8, T. cruzi CL Brener; 9, T. cruzi G; 10, T. cruzi JMP; 11, T. rangeli Mono; 12, Trypanosoma sp. 13, Trypanosoma saimirii; 14, T. lewisi; 15, T. evansi; 16, L. amazonensis; 17, L. braziliensis; 18, L. chagasi; 19, C. fasciculata; 20, Recombinant TrVA5 (8) (positive control). Arrow indicates the 170-bp specific product.
To test the T. rangeli PCR assay (SubTr) (Fig. 3A), the above samples were examined with the addition of DNA from other species of the genus Trypanosoma, including T. saimirii, T. lewisi, and T. evansi. The main PCR product (Fig. 3A) had the expected size of 170 bp; there were also some minor specific amplification products of higher molecular weight. The assay was species specific. It recognized all T. rangeli isolates (lanes 1 to 7 and 11), and the positive control (lane 20), but not T. cruzi (lanes 8 to 10); an unidentified Trypanosoma sp. isolate obtained in Brazil from the two-clawed sloth Coleopus hofmanii, which is regarded as T. rangeli-like (lane 12) (30); T. saimirii (lane 13); T. lewisi (lane 14); T. evansi (lane 15); Leishmania amazonensis (lane 16); L. braziliensis (lane 17); L. chagasi (lane 18); or Crithidia fasciculata (lane 19). Hybridization experiments with the corresponding capture oligonucleotide (Trcap) (Fig. 3B) confirmed the specificity of both the 170-bp and the minor higher-molecular-weight products.
Duplex PCR assay.
Once the optimal conditions were established for the individual PCR assays, we combined them in a duplex assay and tested the sensitivity of this assay against a gradient of DNA concentrations from each parasite. As shown in Fig. 4, DNA was extracted from 105 cells of each parasite and sequentially diluted by factors of 10. DNA dilutions from the two parasites were mixed in inverted concentration ratios, so that the highest amount for one parasite, 105 cells per approximately 20 ng, matched the lowest dilution of the other (0, the negative control for reagents).
Duplex PCR yielded specific, appropriately sized amplification products from each reaction with no apparent primer interference (Fig. 4, lanes 2 to 8). Under these conditions, the assay detected DNA equivalent to a single cell.
Duplex PCR assay in other samples.
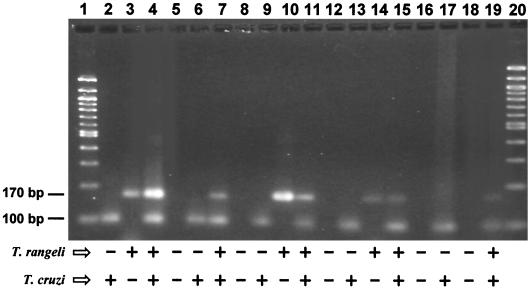
Next, we tested the ability of the duplex PCR assay to detect both parasites together within various samples (Fig. 5). The duplex assay was able to appropriately detect the presence of cultured T. cruzi (lane 2), T. rangeli (lane 3), and a mix of the two (lane 4). Experimentally infected triatomine feces were successfully identified as uninfected (lane 5), infected with T. cruzi (lane 6), or infected with both parasites (lane 7). Dissected intestinal tracts of triatomine bugs were PCR identified as coming from an uninfected bug (negative control, lane 8), from R. prolixus experimentally infected with T. cruzi (lane 9), and with both parasites (lane 11). In addition, a triatomine bug captured from the wild (Barinas State, Venezuela) tested positive for both parasites (lane 10). Further testing of the PCR assay's applicability showed that it was capable of distinguishing the presence and absence of the parasites in mouse blood in which infection of T. cruzi and/or T. rangeli was simulated (lanes 12 to 15), as well as in mouse heart samples from noninfected (negative control, lane 16) and T. cruzi-infected animals (15 days postinfection; lane 17). Finally, we tested a blood sample from a human subject living in an area where the disease is endemic, confirming the presence of both parasites in this patient, who was parasitologically and serologically positive for T. cruzi and T. rangeli (lane 19).
FIG. 5.
Duplex PCR in different samples visualized in ethidium bromide-stained 3% agarose gels. Cultured parasite cells, lanes: 2, T. cruzi; 3, T. rangeli; 4, T. cruzi + T. rangeli. R. prolixus feces, lanes: 5, noninfected; 6, infected with T. cruzi, 7, infected with T. cruzi plus T. rangeli. R. prolixus intestinal tract, lanes: 8, noninfected; 9, infected with T. cruzi; 10, T. rangeli; 11, T. cruzi plus T. rangeli. Simulated mice blood infections, lanes: 12, noninfected; 13, infected with T. cruzi; 14, infected with T. rangeli; 15, infected with T. cruzi plus T. rangeli. Mouse heart samples, lanes: 16, noninfected; 17, infected with T. cruzi. Lane 18, H2O. Lane 19, blood from a human case. Lanes 1 and 20 are the 100-bp size ladder (Gibco-BRL). Arrows indicate 100- and 170-bp specific amplification products for T. cruzi and T. rangeli, respectively.
DISCUSSION
Here we showed that subtelomeric sequences of T. cruzi and T. rangeli are appropriate for species-specific PCR detection of these parasites in complex biological samples. The high interspecies variability exhibited by Trypanosomatidae subtelomeric sequences is the basis for the high specificity achieved by PCRs targeted to these sequences. Previously, we used a similar approach to specifically detect Leishmania in visceral leishmaniasis (10). Here, we designed a species-specific duplex PCR assay to detect T. cruzi and T. rangeli. These reactions produced the expected PCR products when examined singly, with the T. cruzi primers yielding the expected 100-bp band and the T. rangeli primers yielding both the expected 170-bp band and minor high-molecular-weight bands. Confirmatory hybridization experiments proved that the assays did not cross-react with each other and that the high-molecular-weight bands observed in the T. rangeli assay were specific. Interestingly, the T. rangeli PCR assay did not react with DNA samples from isolates frequently found in squirrel monkeys, which are regarded by some authors as T. rangeli-like (30).
However, despite the minor bands present in the T. rangeli assay, we were able to duplex this reaction with the T. cruzi-specific assay and produce specific, correct amplification products. No amplification artifacts due to primer interference were observed even in samples with a large excess of DNA from one parasite over the other. The duplex assay was capable of detecting as little as one parasitic cell of each species in the samples tested. In additional experiments with parasite cell dilutions without confirmatory hybridization experiments (not shown), we detected an equivalent to 1/20th and 1/4th of T. cruzi and T. rangeli cells, respectively. This difference in detection level is likely due to a smaller number of telomeric targets in T. rangeli, as can be deduced from the smaller number of chromosomal bands exhibited by this organism in pulsed-field electrophoresis experiments (17). Importantly, the sensitivity reached by our duplex PCR in field samples is similar to that reported for single T. rangeli PCR typing assays with primers targeted to other nuclear targets (16, 21, 24, 29).
In the study of mixed infections, parasite culturing can cause the selection of one parasite over the other; hence, it is desirable to be able to analyze field samples directly. However, the presence of Taq DNA polymerase inhibitors in this type of sample poses a major problem for most PCR assays. A multiplex assay based on miniexon spacer sequences has been used to type T. cruzi and T. rangeli cells in culture (14). Another multiplex assay, targeted to the large-subunit rRNA gene, has been applied to detect these parasites in simulated infected vectors and in culture (24, 29). However, these assays have not been used previously to detect parasites in vertebrate blood. A related PCR assay based on kinetoplast DNA was highly sensitive for both trypanosomes, but the small size differences of the PCR products made it impractical to study mixed infections (28).
Therefore, to our knowledge this is the first report of a duplex PCR assay based on different DNA targets for the simultaneous detection of T. cruzi and T. rangeli. Our method ensures the simple, independent amplification of both targets and in addition is specific and robust enough to be used directly in the field and on complex samples. The duplex assay has the additional advantage of not requiring confirmatory hybridization experiments, and we are currently using it to examine patient blood and triatomine vector samples from an area in Barinas State, Venezuela, where T. rangeli infections greatly exceed those of T. cruzi (3). This method may allow the proper diagnosis of infections, allowing fewer false-positives and the attendant risk of chemotherapy.
Acknowledgments
This project received support from FONACIT grants G9900036 and G9900035, from CDCHT-UCLA grant 025E-ME-2002, from grant CDCHTULA/C101610007AA (to N.A.), and from grants RLA 0026 and FONACIT519800268 (to P.G.).
REFERENCES
- 1.Añez, N. 1984. Studies on Trypanosoma rangeli/Tejera, 1920. VII. Its effect on the survival of infected triatomine bugs. Mem. Inst. Oswaldo Cruz 79:249-255. [DOI] [PubMed] [Google Scholar]
- 2.Añez, N., E. Nieves, and D. Cazorla. 1987. Studies on Trypanosoma rangeli Tejera, 1920. IX. Course of infection in different stages of Rhodnius prolixus. Mem. Inst. Oswaldo Cruz 82:1-6. [DOI] [PubMed] [Google Scholar]
- 3.Añez, N., G. Crisante, A. Rojas, H. Carrasco, H. Parada, Y. Yépez, R. Borges, P. Guevara, and J. L. Ramírez. 2001. Detection and significance of inapparent infection in Chagas disease in western Venezuela. Am. J. Trop. Med. Hyg. 65:227-232. [DOI] [PubMed] [Google Scholar]
- 4.Añez, N., H. Carrasco, H. Parada, G. Crisante, A. Rojas, N. Gonzalez, J. L. Ramírez, P. Guevara, C. Rivero, R. Borges, and J. V. Scorza. 1999. Acute Chagas' disease in western Venezuela: a clinical, seroparasitologic, and epidemiologic study. Am. J. Trop. Med. Hyg. 60:215-222. [DOI] [PubMed] [Google Scholar]
- 5.Batista, J. A., S. M., Texeira, J. E. Donelson, L. V. Kirchhoff, and C. M. de Sa. 1994. Characterization of a Trypanosoma cruzi poly(A)-binding protein and its genes. Mol. Biochem. Parasitol. 67:301-312. [DOI] [PubMed] [Google Scholar]
- 6.Brener, Z., and E. Chiari. 1963. Variações morfológias observadas amostras de Trypanosoma cruzi. Rev. Inst. Med. Trop. São Paulo 5:220-224. [PubMed] [Google Scholar]
- 7.Camargo, E. P. 1964. Growth and differentiation in Trypanosoma cruzi: Origin of metacyclic trypomastigotes in liquid media. Rev. Inst. Med. Trop. São Paulo 6:93-100. [PubMed] [Google Scholar]
- 8.Chiurillo, M. A., A. Peralta, and J. L. Ramírez. 2002. Comparative study of Trypanosoma rangeli and Trypanosoma cruzi telomeres. Mol. Biochem. Parasitol. 120:305-308. [DOI] [PubMed] [Google Scholar]
- 9.Chiurillo, M. A., I. Cano, J. Franco Da Silveira, and J. L. Ramírez. 1999. Organization of telomeric and subtelomeric regions of chromosomes from the protozoan parasite Trypanosoma cruzi. Mol. Biochem. Parasitol. 100:173-183. [DOI] [PubMed] [Google Scholar]
- 10.Chiurillo, M. A., M. Sachdeva, V. S. Dole, Y. Yepes, E. Miliani, L. Vazquez, A. Rojas, G. Crisante, P. Guevara, N. Añez, R. Madhubala, and J. L. Ramírez. 2001. Detection of Leishmania causing visceral leishmaniasis in the Old and New World by a polymerase chain reaction assay based on telomeric sequences. Am. J. Trop. Med. Hyg. 65:573-582. [DOI] [PubMed] [Google Scholar]
- 11.Chiurillo, M., M. Santos, J. Franco Da Silveira, and J. L. Ramirez. 2002. An improved general approach for cloning and characterizing telomeres: the protozoan parasite Trypanosoma cruzi as model organism. Gene 294:197-204. [DOI] [PubMed] [Google Scholar]
- 12.Contreras, V. T., J. M. Salles, N. Thomas, C. M. Morel, and S. Goldenberg. 1985. In vitro differentiation of Trypanosoma cruzi under chemically defined conditions. Mol. Biochem. Parasitol. 16:315-327. [DOI] [PubMed] [Google Scholar]
- 13.D'Alessandro, A., and N. G. Saravia. 1992. Trypanosoma rangeli, p. 1-54. In J. P. Kreier and J. R. Baker (ed.), Parasitic protozoa, vol. 2. Academic Press, New York, N.Y.
- 14.Fernandes, O., S. S. Santos, E. Cupolillo, B. Mendoca, R. Derre, A. C. V. Junqueira, L. C. Santos, N. R. Sturm, R. D. Naiff, T. V. Barret, D. A. Campbell, and J. R. Coura. 2001. A mini-exon multiplex polymerase chain reaction to distinguish the major groups of Trypanosoma cruzi and T. rangeli in the Brazilian Amazon. Trans. R. Soc. Trop. Med. Hyg. 95:97-99. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez, N., I. Galindo, P. Guevara, E. Novak, J. V. Scorza, N. Añez, J. Franco Da Silveira, and J. L. Ramirez. 1994. Identification and detection of Trypanosoma cruzi by with a DNA amplification fingerprint obtained from the ribosomal intergenic spacer. J. Clin. Microbiol. 32:153-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grisard, E., D. A. Campbell, and J. Romanha. 1999. Mini-exon gene sequence polymorphism among Trypanosoma rangeli strains isolated from distinct geographical regions. Parasitology 118:375-382. [DOI] [PubMed] [Google Scholar]
- 17.Henriksson, J., A. Solari, M. Rydåker, O. E. Sousa, and U. Pettersson. 1996. Karyotype variability in Trypanosoma rangeli. Parasitology 112:385-391. [DOI] [PubMed] [Google Scholar]
- 18.Medina-Acosta, E., and G. A. Cross. 1993. Rapid isolation of DNA from trypanosomatid protozoa with a simple ′mini-prep' procedure. Mol. Biochem. Parasitol. 59:327-329. [DOI] [PubMed] [Google Scholar]
- 19.Pizzi, T. P., M. D. Rubio, R. Prager, and R. C. Silva. 1952. Accion de la cortisona en la infeccion experimental por Trypanosoma cruzi. Bol. Chil. Parasitol. 7:22-24. [PubMed] [Google Scholar]
- 20.Requena, J. M., A. Jimenez-Ruiz, M. Soto, M. C. Lopez, and C. Alonso. 1992. Characterization of a highly repeated interspersed DNA sequence of Trypanosoma cruzi: its potential use in diagnosis and strain classification. Mol. Biochem. Parasitol. 51:271-280. [DOI] [PubMed] [Google Scholar]
- 21.Silber, A. M., J. Búa, B. M. Porcel, E. L. Segura, and A. M. Ruiz. 1997. Trypanosoma cruzi: Specific Detection of Parasites by PCR in infected humans and vectors with a set of primers (BP1/BP2) targeted to a nuclear DNA sequence. Exp. Parasitol. 85:141-152. [DOI] [PubMed] [Google Scholar]
- 22.Silva, L. H. P., and V. Nussenzweig. 1953. Sobre uma cepa de Trypanosoma cruzi altamente virulenta para o camundongo branco. Folia Clin. Biol. 20:191-203. [Google Scholar]
- 23.Sousa, O., and C. M. Johnson. 1973. Prevalence of Trypanosoma cruzi and Trypanosoma rangeli in triatomines (Hemiptera, reduviidae) collected in the Republic of Panama. Am. J. Trop. Med. Hyg. 22:18-23. [DOI] [PubMed] [Google Scholar]
- 24.Souto, R. P., N. Vargas, and B. Zingales. 1999. Trypanosoma rangeli: discrimination from Trypanosoma cruzi based on a variable domain from large subunit ribosomal RNA gene. Exp. Parasitol. 91:306-314. [DOI] [PubMed] [Google Scholar]
- 25.Steindel, M., E. Dias-Neto, R. Ribeiro-Rodrigues, C. J. Carvalho-Pinto, E. C. Grisard, C. L. P. Menezes, S. M. F. Murta, A. J. G. Simpson, and A. J. Romanha. 1994. Randomly amplified polymorphic DNA (RAPD) and isoenzyme analysis of Trypanosoma rangeli strains. J. Euk. Microbiol. 41:261-267. [DOI] [PubMed] [Google Scholar]
- 26.Tibayrenc, M., K. Neubauer, C. Barnabe, F. Guerrini, D. Skarecky, and F. J. Ayala. 1993. Genetic characterization of six parasitic protozoa: parity between random-primer DNA typing and multilocus enzyme electrophoresis. Proc. Natl. Acad. Sci. USA 90:1335-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Urdaneta-Morales, S., and F. Tejero. 1992. Trypanosoma rangeli (Tejera, 1920): observations upon pleomorphism. Mem. Inst. Oswaldo Cruz 87:511-516. [DOI] [PubMed] [Google Scholar]
- 28.Vallejo, G. A., F. Guhl, E. Chiari, and A. M. Macedo. 1999. Species-specific detection of Trypanosoma cruzi and Trypanosoma rangeli in vector and mammalian host by polymerase chain reaction amplification of kinetoplast minicircle DNA. Acta Trop. 72:203-212. [DOI] [PubMed] [Google Scholar]
- 29.Vargas, N., R. P. Souto, J. C. Carranza, G. A. Vallejo, and B. Zingales. 2000. Amplification of a specific repetitive DNA sequence for Trypanosoma rangeli identification and its potential application in epidemiological investigations. Exp. Parasitol. 96:147-159. [DOI] [PubMed] [Google Scholar]
- 30.Ziccardi, M., and R. Lourenco-De-Oliveira. 1997. The infection rates of trypanosomes in squirrel monkeys at two sites in the Brazilian Amazon. Mem. Inst. Oswaldo Cruz 92:465-470. [DOI] [PubMed] [Google Scholar]