Abstract
We previously cloned an antigenic gene (named nfa1) from a cDNA library of Naegleria fowleri by immunoscreening. The nfa1 gene had a coding nucleotide sequence consisting of 357 bases and produced a recombinant 13.1-kDa protein (Nfa1). In this study, to get more information regarding the recombinant Nfa1 protein (rNfa1), we produced an anti-Nfa1 polyclonal antibody from mice immunized with rNfa1 and used a peroxidase staining method to carry out immunocytochemistry experiments. In addition, we observed the effect of the presence of an anti-Nfa1 antibody on the in vitro cytotoxicity of N. fowleri against Chinese hamster ovary (CHO) cells. Trophozoites of N. fowleri in cultivation reacted strongly with a peroxidase-labeled anti-Nfa1 antibody. In inflammatory and necrotic regions of brain tissue infected with N. fowleri, labeled trophozoites that were stained brown were also observed. When examined using a transmission electron microscope, the Nfa1 protein showed pseudopodium-specific immunolocalization on a trophozoite of N. fowleri. When examined using a light microscope, CHO cells grown in cocultures with N. fowleri trophozoites (group I) for 48 h showed morphologically severe destruction but CHO cells grown in cocultures with N. fowleri trophozoites and an anti-Nfa1 polyclonal antibody (group II) showed less destruction. The results of a lactate dehydrogenase release assay showed that group I CHO cells exhibited 81% cytotoxicity and group II CHO cells exhibited 13.8% cytotoxicity.
Naegleria fowleri, a free-living amoeba found in widespread areas in moist soil, water, and sediment, exists as a virulent pathogen causing fatal primary amoebic meningoencephalitis (PAME) in experimental animals and humans (2, 4, 8, 12). As evidence that PAME has been a factor in previous years, numerous cases have been diagnosed retrospectively from autopsy material (14).
No distinctive clinical features differentiate PAME from acute pyogenic or bacterial meningoencephalitis. The final diagnosis of PAME is made on the basis of the isolation and growth in culture of N. fowleri from spinal fluid or the demonstration of the presence of amoebic trophozoites in brain tissue from biopsies (10). Because the major location of infection is the central nervous system and because PAME progresses rapidly, diagnosis in the early stages is difficult (10). Previous studies of the detection of antibodies in infections with pathogenic free-living amoebae have been performed using various methods such as agglutination tests, immunofluorescence tests, and an enzyme-linked immunoelectrotransfer blot technique (5, 6, 15, 20). Monoclonal antibodies to N. fowleri have been produced and used as probes for the identification of N. fowleri in brain sections of PAME patients and to distinguish pathogenic N. fowleri from other Naegleria species (16, 21). The occurrence of antibodies against Naegleria spp. was investigated in wild mammals, humans, and laboratory animals (7, 8, 11, 13). Nevertheless, the search for antigenic molecules has not been very successful. The antigenicity of pathogenic N. fowleri was observed with immunoprecipitation and enzyme-linked immunoelectrotransfer blotting, and several protein bands seemed to be important in infection and immunization (11, 18). However, an attempt to search for an antigen-related gene at the molecular level has not been fully carried out (19).
Using the immunoscreening technique with infected and immune mouse sera as a first step toward the production of antigenic molecules, Shin et al. previously cloned an antigen-related gene from a cDNA expression library of N. fowleri and named it the nfa1 gene (17). The nfa1 gene consisted of 357 bases that translated into 119 amino acid residues which produced a 13.1-kDa recombinant protein. In Western blotting experiments, the recombinant protein (rNfa1) reacted strongly with infected and immune sera and the anti-Nfa1 antibody (17). As a second step, to get more information on the rNfa1 protein we carried out some immunological studies with an anti-Nfa1 polyclonal antibody. We produced an anti-Nfa1 polyclonal antibody from mice immunized with rNfa1 protein and carried out immunocytochemistry experiments using a peroxidase-staining method. In addition, we used light microscopy and a lactate dehydrogenase (LDH) release assay to observe the effect of an anti-Nfa1 antibody on the cytotoxicity of N. fowleri against CHO cells.
N. fowleri strain Carter NF69 (ATCC 30215) and N. gruberi strain Singh (ATCC 30960) were axenically grown at 37°C in Nelson's medium (22) and at 27°C in modified PYNFH (peptone-yeast-nucleic acid-folic acid-hemin) medium (American Type Culture Collection medium serial no. 1034), respectively. Trophozoites grown in cultures were pelleted at 800 × g for 10 min and washed twice in phosphate-buffered saline (PBS), pH 7.2. For the production of amoeba lysate, an aqueous extract was prepared from freeze-thawed trophozoites (18) and filtered using a 0.45-μm-pore-size filter. The protein concentration was determined by the Bradford assay method (1).
To obtain a recombinant Nfa1 fusion protein, expression and purification of the gene product were performed according to a method described in a previous paper (17). The purified DNA (5 μg/μl) obtained using PCR-T7/NT TOPO expression vector (Invitrogen, Groningen, The Netherlands) containing the nfa1 gene was subsequently used to transform the Escherichia coli strain BL21(DE3)pLysS by the heat shock method. Cells were grown at 37°C on a LAC (Luria-Bertani medium containing 100 μg of ampicillin/ml and 34 μg of chloramphenicol/ml) plate for selection. Selected transformed colonies were grown at 37°C in LAC broth until an absorbance value of 0.5 to 0.8 at 600 nm was reached, and then 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added to the medium. After incubation for 4 h, the cells were harvested by centrifugation (6,000 × g for 15 min).
Cell extracts were compared with those of nontransformed BL21(DE3)pLysS by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the presence of the expressed gene product was confirmed by Western blotting using both immune and infected sera and anti-His and Xpress antibody (Invitrogen). The expressed recombinant fusion protein was purified using the XpressTM system protein purification kit (Invitrogen). Harvested cells were resuspended in a native binding buffer (20 mM sodium phosphate, 500 mM sodium chloride, pH 7.8) in a tube. After the addition of egg white lysozyme (100 μg/ml), the tube was incubated for 15 min on ice. Cells were sonicated, frozen immediately in liquid nitrogen, and then quickly thawed at 37°C. Final concentrations of 5 μg of RNase/ml and 5 μg of DNase/ml in the tube were treated on ice for 15 min. Insoluble debris was removed by centrifugation at 5,000 × g for 15 min, and the lysate was cleared using a 0.8-μm-pore-size syringe filter. A recombinant gene product was eluted using Probond resin columns (Invitrogen) by increasing the imidazole concentration. The eluted protein was dialyzed and concentrated by freeze-drying. The fusion partner of a recombinant protein was digested with EnterokinaseMax, and a nontagging recombinant protein was purified with EK-Away resin columns (Invitrogen) and Probond resin columns (Invitrogen).
Immune and infected sera were prepared according to a method described in a previous paper (17). In addition, sectioned brain tissue for use in immunocytochemistry experiments was obtained from a mouse prepared for infected-serum collection. For the production of anti-Nfa1 polyclonal serum, purified recombinant protein (50 μg/mouse) was mixed with an equal volume of Freund's complete adjuvant (Sigma) and the mixture was injected intraperitoneally into 8-week-old female BALB/c mice (purchased from the Korea Institute of Science and Technology, Daejeon, Korea). The mouse received biweekly booster injections with Nfa1 protein (25 μg/mouse) containing an equal volume of incomplete Freund's adjuvant (Sigma) for another 4 weeks. After the third booster injection, purified Nfa1 protein (5 μg/mouse) without adjuvant was injected intravenously. At 4 days later, anti-Nfa1 polyclonal serum was collected from mouse blood by centrifugation at 2,500 × g and 4°C for 30 min. An enzyme-linked immunosorbent assay (ELISA) was performed with purified Nfa1 protein (5 μg/ml) and rabbit anti-mouse whole immunoglobulin (1:10,000 dilution) conjugated with alkaline phosphate (Sigma). Western blotting against recombinant Nfa1 protein was performed according to a method described in a previous paper (17).
For the immunocytochemistry study of the Nfa1 protein, an InnoGenex Mouse-on-Mouse Iso-IHC kit (InnoGenex, San Ramon, Calif.) was used. Trophozoites of N. fowleri (105/well) were incubated in a 24-well culture plate at 37°C for 2 h. Three wells were used for normal serum, anti-Nfa1 polyclonal serum, and PBS control serum, respectively. Trophozoites were washed with PBS and fixed with a fixation solution (2.5% glutaraldehyde, 1% formalin, and 0.005% CaCl2 in cacodylate buffer) for 10 min. Fixed trophozoites were washed with PBS and permeabilized with a perforation solution (0.2% Triton X-100 and 1% bovine serum albumin in 0.1 M PBS) for 20 min. After the PBS washing, each well was blocked with 200 μl of peroxide reagent (0.3% H2O2 in methanol) for 20 min and washed with PBS. For blocking of nonspecific binding, normal mouse serum (1:200 dilution) and Power Block reagent were treated on each well for 12 h 10 min. After that, anti-Nfa1 polyclonal and normal sera were labeled by a 1/10 volume of biotin-labeling reagent and incubated for 30 min at room temperature. Labeled sera were mixed with a 1/10 volume of mouse blocking reagents and incubated for 3 min. Blocked labeled-serum mixture was placed into each well containing N. fowleri trophozoites, and each well was incubated for 30 min at room temperature and washed with PBS-0.1% Tween 20. Streptavidin-horseradish peroxidase conjugate (200 μl/well) was placed into each well, and each well was incubated for 20 min at room temperature and washed with PBS-0.1% Tween 20. A total of 4.4 μl of 0.5% diaminobenzidine was mixed with 200 μl of substrate buffer and placed into each well. Each well was incubated for 7 min at room temperature and washed with tap water. After the fixation solution was added, the samples were viewed and photographed using an inverted microscope. The mouse brain tissue infected with amoeba trophozoites was fixed with fixation solution, embedded into paraffin blocks, cut into 8-μm-thick tissues, and immunostained according to the procedures described above.
For preparation of transmission electron microscopy specimens for observation of the immunolocalization of the Nfa1 protein, amoeba trophozoites were treated according to the immunocytochemistry procedure described above. The prefixed samples were washed with 0.1 M cacodylate buffer (pH 7.2) and refixed with 1% osmium tetroxide (OsO4). The following process was performed according to standard methods. The blocks were sectioned using a Reichert-Jung Ultracut S apparatus and stained with Ultrostain 1H and Ultrostain 2 (Leica, Vienna, Austria). Specimens were observed and photographed with a Zeiss EM 902A transmission electron microscope (LEO, Oberkochen, Germany).
For experiments investigating the cytotoxicity of N. fowleri against CHO cells, N. fowleri trophozoites grown in Nelson's medium (22) were placed on each well in a 96-well plate containing CHO cells grown in cultures with complete Eagle's minimal essential medium (EMEM) (group I). For another experimental group (group II), an anti-Nfa1 polyclonal antibody (1:100 dilution) obtained from mice immunized with anti-Nfa1 protein was added to triplet wells. After incubation at 37°C for 48 h, the effect of the anti-Nfa1 polyclonal antibody was observed by light microscopy and an LDH release assay. The cytotoxicity revealed in the LDH release assay was observed with a CytoTox 96 nonradioactive cytotoxicity assay kit (Promega, Madison, Wis.). About 104 CHO cells were prepared with 100 μl of EMEM in a round-bottomed 96-well plate. The amoeba trophozoites (105) and/or an anti-Nfa1 polyclonal antibody (1 mg/ml) was added to each well, and five sets of triplicate wells for the controls were prepared. A cast plate was incubated in a humidified chamber at 37°C and then in a 5% CO2 incubator for 45 min and was centrifuged at 250 × g for 4 min. The supernatant (50 μl) from each well was transferred to a fresh flat-bottomed 96-well plate. Equal volumes of the reconstituted substrate mix were added to each well. The plate was incubated at room temperature for 30 min. A total of 50 μl of stop solution was added, and after 1 h, the absorbance was recorded at 490 nm. Percentages of cytotoxicity were calculated using the following formula:
 |
(1) |
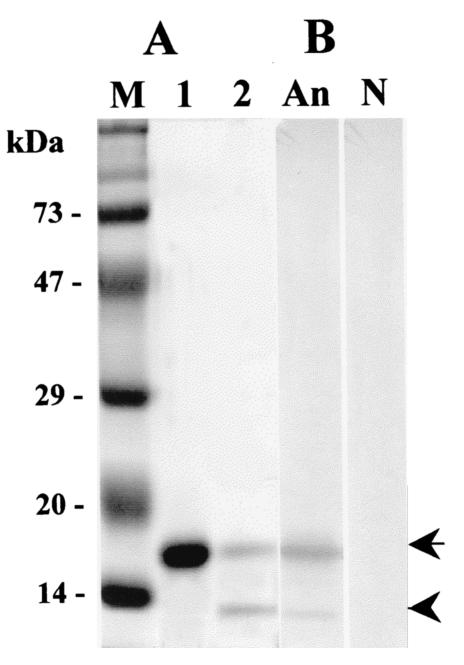
To obtain a purified recombinant Nfa1 protein, 17-kDa fusion protein expressed from E. coli strain BL21 containing the nfa1 gene was purified using a nickel resin column and digested with enzyme (Fig. 1A). Finally, a purified protein eluted from gels had a molecular mass of 13.1 kDa, which corresponded well to the size assumed from the amino acid length.
FIG. 1.
(A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis band patterns of a recombinant fusion protein (lane 1) and an enterokinase-digested Nfa1 protein (lane 2) expressed from the nfa1 gene. (B) Results of Western blotting with an anti-Nfa1 polyclonal antibody (lane An) and normal mouse serum (lane N). Arrow, uncut 17-kDa fusion protein; arrowhead, cut 13.1-kDa Nfa1 protein; lane M, molecular size markers.
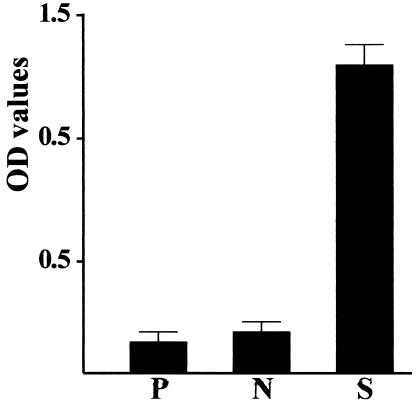
An anti-Nfa1 polyclonal antibody was collected from mice immunized with recombinant Nfa1 protein, and its specific reactivity against the Nfa1 protein was observed using ELISA and Western blotting. A405 values determined by ELISA for anti-Nfa1 polyclonal antibodies ranged from 0.231 ± 0.025 (mean ± standard deviation; n = 5) (1:10,000 dilution) to 1.339 ± 0.044 (1:200 dilution), which contrasts with the values for normal sera and the PBS control sera of 0.122 ± 0.005 (1:200 dilution) and 0.116 ± 0.005, respectively (Fig. 2). The anti-Nfa1 polyclonal antibody was also reactive to recombinant Nfa1 protein in Western blotting, showing a 13.1-kDa protein band (Fig. 1B).
FIG. 2.
Antibody titers of anti-Nfa1 polyclonal sera (1:200 dilution) obtained from Nfa1-immunized mice. The recombinant Nfa1 protein (5 μg/ml) was used as the antigen. A405 values for each serum sample (n = 5) were determined by an ELISA reader. OD, optical density; P, PBS control; S, anti-Nfa1 polyclonal sera; N, normal mouse sera (1:200 dilution).
As the results of immunocytochemistry experiments showed, trophozoites of N. fowleri in cultivation reacted strongly with a peroxidase-labeled anti-Nfa1 antibody, appearing brown by light microscopy (Fig. 3A). When normal serum was used, the trophozoites did not show any color (Fig. 3B). In addition, N. gruberi trophozoites did not react with a peroxidase-labeled anti-Nfa1 antibody (Fig. 3C). In the inflammatory and necrotic regions of brain tissue infected with N. fowleri, amoeba trophozoites treated with an anti-Nfa1 antibody were also observed to be brown (Fig. 4A and C), whereas trophozoites treated with normal serum were not stained with peroxidase (Fig. 4B and D).
FIG. 3.
Immunoreactivity of the Nfa1 protein with a peroxidase-labeled anti-Nfa1 antibody. Trophozoites of N. fowleri in cultivation were treated with an anti-Nfa1 antibody (A) and normal mouse serum (B). Trophozoites of N. fowleri were stained brown. Trophozoites of N. gruberi (C) did not react with an anti-Nfa1 antibody and did not stain brown. Magnification, ×200.
FIG. 4.
Mouse brain tissue infected with N. fowleri trophozoites was treated with an anti-Nfa1 antibody (A and C) and unimmunized control serum (B and D). Trophozoites (arrows) were stained brown. (A and B) The inflammatory region of the brain tissue; (C and D) the necrotic region of the brain tissue. Magnification, ×200.
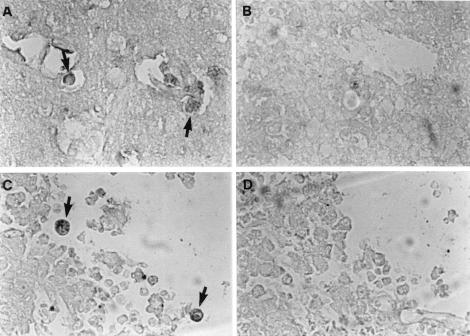
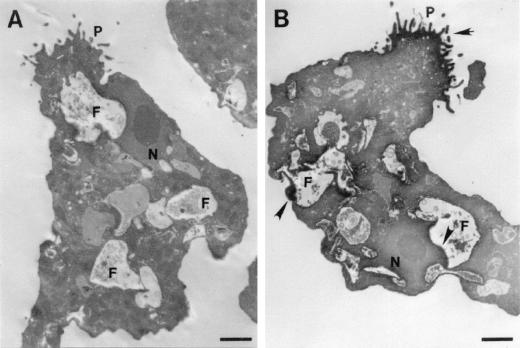
As observed by electron microscopy, the Nfa1 protein immunostained with a peroxidase-labeled anti-Nfa1 antibody was apparently localized on vigorous pseudopodia and sometimes around food vacuoles of a N. fowleri trophozoite (Fig. 5A). No immunolocalization signal was found for normal serum (Fig. 5B).
FIG. 5.
Cellular localization of the Nfa1 protein in an N. fowleri trophozoite. N. fowleri trophozoites were immunostained with normal serum (A) and anti-Nfa1 antibody (B) by using peroxidase and diaminobenzidine. Positive findings (arrowheads) were observed on pseudopodia (P) and around food vacuoles (F) of a trophozoite. N, nucleus. Bar, 2.5 μm.
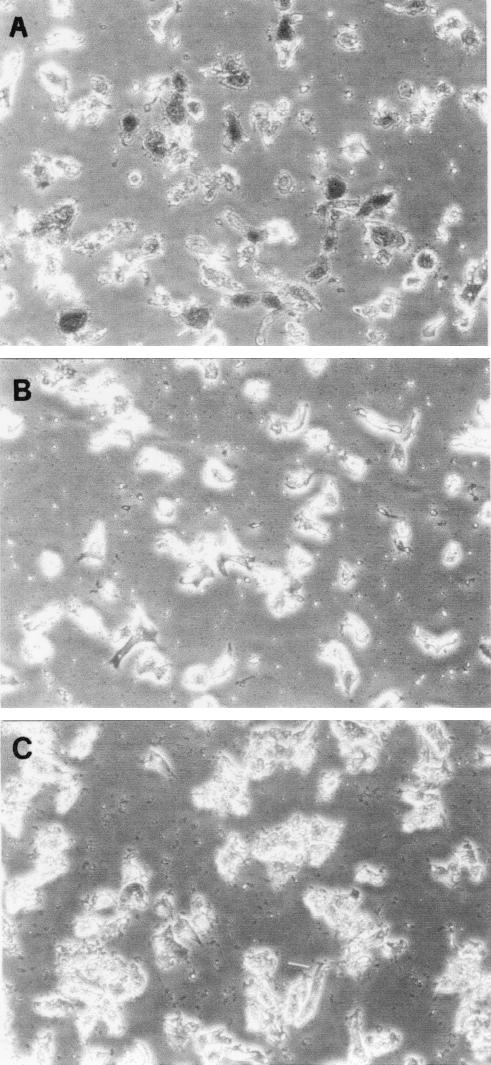
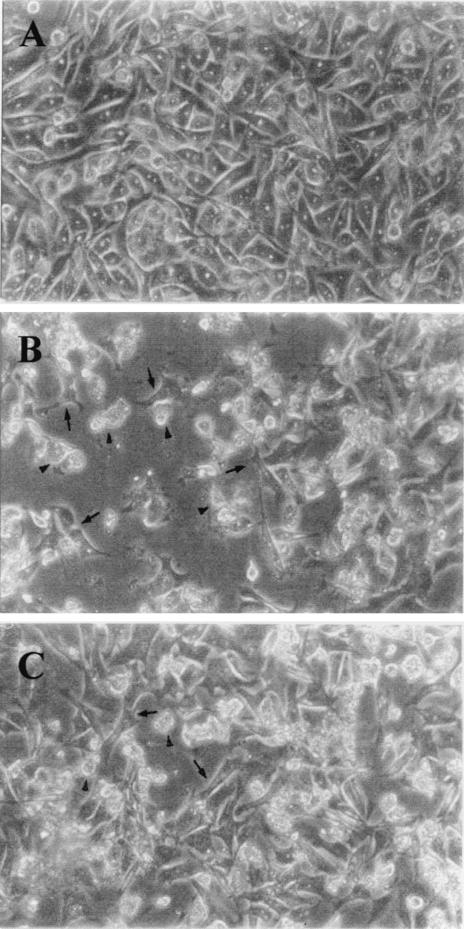
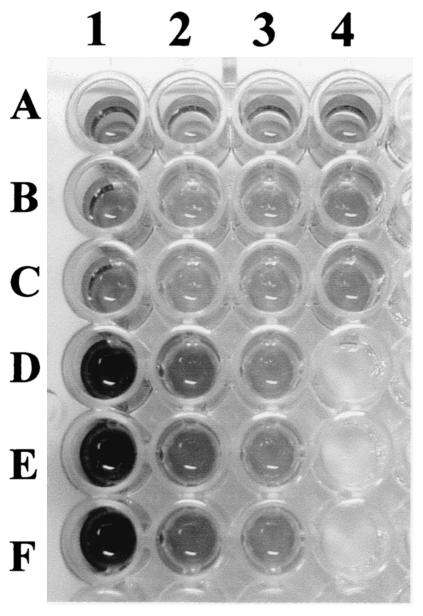
Regarding the effect of an anti-Nfa1 antibody on the cytotoxicity of N. fowleri, CHO cells grown in cocultures with N. fowleri trophozoites (group I) for 48 h showed morphologically severe destruction (Fig. 6). In contrast, CHO cells grown in cocultures with N. fowleri trophozoites and anti-Nfa1 polyclonal antibody (group II) showed less destruction than those of group I (Fig. 6). The results of an LDH release assay (Fig. 7) showed that group I CHO cells exhibited 80.7% cytotoxicity and group II CHO cells exhibited 13.8% cytotoxicity. Thus, the presence of an anti-Nfa1 polyclonal antibody led to a decrease in the cytotoxicity of N. fowleri.
FIG. 6.
Microscopy findings for CHO cells (arrows) grown in cultures in EMEM (A) and grown in cocultures with N. fowleri trophozoites (arrowheads) (B) and with N. fowleri and an anti-Nfa1 polyclonal antibody (C) at 48 h. Magnification, ×200.
FIG. 7.
Visualization (using an LDH release assay) of the cytotoxicity of N. fowleri trophozoites against CHO cells. CHO cells were grown in cultures in EMEM medium alone (lane 1, rows A, B, and C), in cocultures with N. fowleri trophozoites (lane 1, rows D, E, and F), and in cocultures with N. fowleri trophozoites and an anti-Nfa1 antibody (lane 2, rows D, E, and F). Other wells represent control groups.
N. fowleri has a worldwide distribution, and more than 145 cases of PAME had been reported worldwide by 1996 (8, 9, 12). Unfortunately, most cases have been diagnosed by finding trophozoites through either biopsy or autopsy. Even when an autopsy has been performed, it is not easy to distinguish amoebae from brain cells. So it is thought that the incidence of actual PAME cases is higher than that observed (10, 14).
In spite of many problems encountered in immunological diagnosis that require well-timed procedures during the period between the start of an infection and the formation of an antibody and an antigen with appropriate specificity, the importance of immunological diagnosis cannot be overemphasized. Thus, as the first step of developing a method for immunological diagnosis, the identification and production of specific antigens against N. fowleri are very important. By using immune and infected sera in a previous study, Shin et al. cloned an antigenic gene (nfa1) producing a recombinant 13.1-kDa protein (17). This recombinant 13.1-kDa protein had a strong immunoreactivity against both immune and infected sera (17). In this study, an anti-Nfa1 polyclonal antibody obtained by immunization with a recombinant 13.1-kDa protein was not cross-reactive with nonpathogenic N. gruberi and lysates of some Acanthamoeba species, although not all amoeba species were compared (data not shown). According to the results of immunohistochemistry experiments with the Nfa1 protein in vitro and in vivo, amoeba trophozoites were well immunostained. This staining signal may be the result of antigen presentation. In addition, the immunolocalization study of the Nfa1 protein revealed that this protein was abundant in pseudopodia of N. fowleri trophozoites. As most of the Nfa1 protein was located on the cell surface, it may be a useful antigen. In the studies that follow, the specificity of the Nfa1 antigen as a diagnostic agent should be observed in comparisons using various free-living amoebae.
On the other hand, the pseudopodium-specific localization observed suggests that the Nfa1 protein is required for amoeba movement. It was reported that highly virulent amoebae exhibited faster movement than weakly virulent amoebae (3). On the basis of the idea that the function of the pseudopodium-specific Nfa1 protein may involve a pathogen of N. fowleri, we estimated the cytotoxicity of N. fowleri trophozoites against CHO cells. CHO cells grown in cocultures with N. fowleri trophozoites and an anti-Nfa1 polyclonal antibody showed less destruction than those grown in cultivation with N. fowleri alone. As the results of the LDH release assay revealed, the presence of an anti-Nfa1 polyclonal antibody decreased the in vitro cytotoxicity of N. fowleri. In future studies, investigations of the biological roles of the Nfa1 protein have to be expanded, especially concerning whether the Nfa1 protein is related to the pathogenicity of N. fowleri.
Acknowledgments
This work was supported by a grant (R01-2000-000-00077-0) from the Basic Research Program of the Korea Science & Engineering Foundation and in part by a grant from the Ajou University School of Medicine in 2000.
REFERENCES
- 1.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 2.Carter, R. F. 1970. Description of a Naegleria species isolated from two cases of primary amoebic meningoencephalitis, and of the experimental pathological changes induced by it. J. Pathol. 100:217-244. [DOI] [PubMed] [Google Scholar]
- 3.Cline, M., R. Catchman, and F. Marciano-Cabral. 1986. Movement of Naegleria fowleri stimulated by mammalian cells in vitro. J. Protozool. 33:10-13. [DOI] [PubMed] [Google Scholar]
- 4.Culbertson, C. G. 1971. The pathogenicity of soil amoebas. Annu. Rev. Microbiol. 25:231-254. [DOI] [PubMed] [Google Scholar]
- 5.Cursons, R. T. M., T. S. Brown, E. A. Keys, K. M. Moriarty, and D. Till. 1980. Immunity to pathogenic free-living amoebae: role of humoral antibody. Infect. Immun. 29:401-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubray, B. L., E. W. Walter, and B. R. Jennings. 1997. Serology of Naegleria fowleri and Naegleria lovaniensis in a hospital survey. J. Protozool. 34:322-327. [DOI] [PubMed] [Google Scholar]
- 7.John, D. T., and S. L. Nussbaum. 1993. Naegleria fowleri infection acquired by mice through swimming in amebae-contaminated water. J. Parasitol. 69:871-874. [PubMed] [Google Scholar]
- 8.Kollars, T. M., Jr., and W. E. Wilhelm. 1996. The occurrence of antibodies to Naegleria species in wild mammals. J. Parasitol. 82:73-77. [PubMed] [Google Scholar]
- 9.Lares-Villa, F., J. F. De Jonckheere, H. De Moura, A. Rechi-Iruretagoyena, E. Ferreira-Guerrero, G. Fernandez-Quintanilla, C. Ruiz-Matus, and G. S. Visvesvara. 1993. Five cases of primary amebic meningoencephalitis in Mexicali, Mexico: study of the isolates. J. Clin. Microbiol. 31:685-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma, P., G. S. Visvesvara, A. J. Martinez, F. H. Theodore, P. M. Daggett, and T. K. Sawyer. 1990. Naegleria and Acanthamoeba infections: review. Rev. Infect. Dis. 12:490-513. [DOI] [PubMed] [Google Scholar]
- 11.Malik, A., S. Kumar, and S. R. Das. 1984. Antigenic analysis of pathogenic and nonpathogenic Acanthamoeba spp. using gel diffusion precipitation and immunoelectrophoresis tests. Int. J. Parasitol. 8:83-88. [Google Scholar]
- 12.Marciano-Cabral, F. 1988. Biology of Naegleria spp. Microbiol. Rev. 52:144-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marciano-Cabral, F., M. C. Cline, and S. G. Bradley. 1987. Specificity of antibodies from human sera for Naegleria species. J. Clin. Microbiol. 25:692-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez, A. J., and G. S. Visvesvara. 1997. Free-living amphizoic and opportunistic amoebas. Brain Pathol. 7:583-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reilly, M. F., F. Marciano-Cabral, D. W. Bradley, and S. G. Bradley. 1983. Agglutination of Naegleria fowleri and Naegleria gruberi by antibodies in human serum. J. Clin. Microbiol. 17:576-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryu, J. S., and K. I. Im. 1992. The production and characterization of anti-Naegleria fowleri monoclonal antibodies. Korean J. Parasitol. 30:33-41. [DOI] [PubMed] [Google Scholar]
- 17.Shin, H. J., M. S. Cho, S. Y. Jung, H. I. Kim, S. Park, H. J. Kim, and K. I. Im. 2001. Molecular cloning and characterization of a gene encoding a 13.1 kDa antigenic protein of Naegleria fowleri. J. Eukaryot. Microbiol. 48:713-714. [DOI] [PubMed] [Google Scholar]
- 18.Shin, H. J., and K. I. Im. 1992. Analysis of antigenic specificities of Naegleria fowleri using EITB. Yonsei Rep. Trop. Med. 23:9-13. [Google Scholar]
- 19.Toney, D. M., and F. Marciano-Cabral. 1992. Alterations in protein expression and complement resistance of pathogenic Naegleria amoebae. Infect. Immun. 60:2784-2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsang, V. C. W., J. M. Peralta, and A. R. Simons. 1983. Enzyme-linked immunoelectrotransfer blot techniques (EITB) for studying the specificities of antigens and antibodies separated by gel electrophoresis. Methods Enzymol. 92:377-391. [DOI] [PubMed] [Google Scholar]
- 21.Visvesvara, G. S., M. J. Peralta, F. H. Brandt, M. Wilson, C. Aloisio, and E. Franko. 1987. Production of monoclonal antibodies to Naegleria fowleri, agent of primary amoebic meningoencephalitis. J. Clin. Microbiol. 25:1629-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willaert, E. 1971. Isolement et culture in vitro des amibes de genre Naegleria. Ann. Soc. Belg. Med. Trop. 51:701-708. [PubMed] [Google Scholar]