Abstract
Many animal–bacteria cooperative associations occur in highly modified host organs that create a unique environment for housing and maintaining the symbionts. It has been assumed that these specialized organs develop through a program of symbiosis-specific or -enhanced gene expression in one or both partners, but a clear example of this process has been lacking. In this study, we provide evidence for the enhanced production of an enzyme in the symbiotic organ of the squid Euprymna scolopes, which harbors a culture of the luminous bacterium Vibrio fischeri. Our data show that this enzyme has a striking biochemical similarity to mammalian myeloperoxidase (MPO; EC 1.11.17), an antimicrobial dianisidine peroxidase that occurs in neutrophils. MPO and the squid peroxidase catalyze the same reaction, have similar apparent subunit molecular masses, and a polyclonal antibody to native human MPO specifically localized a peroxidase-like protein to the bacteria-containing regions of the symbiotic organ. We also provide evidence that a previously described squid cDNA encodes the protein (LO4) that is responsible for the observed dianisidine peroxidase activity. An antibody made against a fragment of LO4 immunoprecipitated dianisidine peroxidase activity from extracts of the symbiotic organ, and reacted against these extracts and human MPO in Western blot analysis. These data suggest that related biochemical mechanisms for the control of bacterial number and growth operate in associations that are as functionally diverse as pathogenesis and mutualism, and as phylogenetically distant as molluscs and mammals.
Cooperative associations between animals and prokaryotes are prevalent in virtually all habitats, yet relatively little is known about their complex cellular and molecular integration and regulation. In many of these partnerships, the animal host has evolved specific tissues or organs that house the bacterial symbionts and serve their needs by creating a highly modified environment (1). Significant, yet still unresolved, questions about the nature of this microhabitat include the following: (i) to form an association with bacteria, what host genes are newly evolved, or specific to the interaction, and what genes are co-opted from preexisting programs?; (ii) do cooperative and pathogenic associations employ a common “currency” of signal exchange between host and symbiont, or do they use alternate methods of signaling?; (iii) what role does the prokaryotic partner play in the direct induction of host tissue morphogenesis?; and (iv) in these stable associations, how is the number of symbionts controlled so that they persist yet do not overgrow the host? Whereas such questions remain largely unanswered for any cooperative animal–bacteria association, in both partners of the leguminous plant–Rhizobium sp. symbioses, dozens of genes have been identified whose expression is essential for the proper development of a mature partnership (2, 3). For example, some nodulins, the name given to proteins encoded by plant host genes that are expressed in the root nodule symbiosis, are not specific to the nodule, but are only expressed in the root portion of the plant in response to interactions with the specific symbiont (reviewed in ref. 4).
The symbiosis between the sepiolid squid Euprymna scolopes and the luminous bacterium Vibrio fischeri provides an opportunity to identify “nodulin analogs” in an animal host, because it shares features with the root nodule symbiosis that make it experimentally tractable; specifically, the Euprymna–Vibrio symbiosis is a partnership between a single bacterial species and its host, and both partners can be maintained independent of one another under laboratory conditions (5, 6). In the native symbiosis, the host squid cultures V. fischeri extracellularly in epithelia-lined crypts, which are surrounded by accessory structures (Fig. 1; ref. 7) that modify the emitted bacterial luminescence. In nature, the nocturnally active squid is always found harboring the symbionts and appears to require their luminescence in its behavior (9). Similar to animal gut symbioses, the Euprymna–Vibrio relationship begins anew each generation and persists throughout the life of the host. Within hours of hatching, the host organ is inoculated with V. fischeri, which is only one of a myriad of bacterial species in the surrounding seawater (10, 11).
Figure 1.
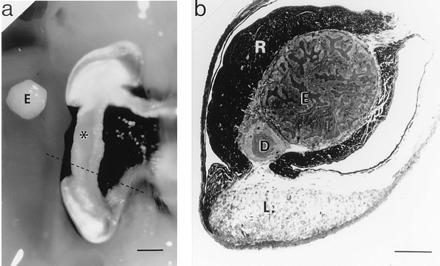
The symbiotic organ of E. scolopes. (a) Ventral view of the right lobe of the organ. Alongside of this lobe is placed the bacteria-containing epithelial tissue (E), which has been dissected out the left lobe of the organ. ∗, Position of this tissue in the right lobe. The dashed line indicates the location of the cross section shown in b. (Bar = 1 mm.) (b) Light micrograph of a cross section of an E. scolopes symbiotic organ. The section was stained with Richardson’s stain (8). E, bacteria-containing tissue comprised of epithelial cells surrounding crypts that house V. fischeri; D, ciliated duct; R, reflector tissue; L, light organ lens. (Bar = 450 μm.)
Studies of the Euprymna–Vibrio partnership have begun to reveal the patterns of host control of symbiont growth and some of the mechanisms underlying these patterns. From a cDNA library of the E. scolopes symbiotic organ, we recently isolated a cDNA clone, pLO4, representing an mRNA that is 250 times more abundant in the symbiotic organ than in other nonsymbiotic host tissues (12). The derived amino acid sequence of the gene suggests that the protein is most similar to a specific halide peroxidase, myeloperoxidase (MPO; EC 1.11.17). In mammalian neutrophils, MPO participates in a complex and highly orchestrated antimicrobial response (for review, see 13 and 14). Briefly, upon the phagocytosis of pathogenic microbes neutrophils undergo a “respiratory burst,” which results in the production of toxic oxygen species, including hydrogen peroxide (H2O2). Within the phagosome, MPO catalyzes the conversion of H2O2 and a chloride ion to hypochlorous acid (HOCl), an even more potent microbicidal compound (14).
As a result of the identification of a squid gene that was highly expressed in the light organ, and that was most similar to one encoding mammalian MPO, we hypothesized that this gene encodes a protein whose activity could play a role in the control of symbiont specificity and growth. In this study, we provide biochemical, immunological, and ultrastructural evidence that this host squid gene encodes a protein that has striking functional similarities to mammalian MPO. These characterizations of the gene product suggest that not only vertebrates but also invertebrates employ similar molecular mechanisms that can function to either foster beneficial relationships or control pathogenesis.
MATERIALS AND METHODS
Specimens of E. scolopes were collected on Oahu, Hawaii, transported to 24°C recirculating aquaria at the University of Southern California in Los Angeles, and maintained as described (7). All reagents were purchased from Sigma unless otherwise noted.
Biochemical and Immunological Comparisons of the Squid Peroxidase with Human MPO.
To measure dianisidine peroxidase activity, the type of activity characteristic of halide peroxidases, adult E. scolopes were anesthetized by cooling, and symbiotic organs and other tissues were removed by dissection. Peroxidase activity in tissue extracts was detected spectrophotometrically using a method modified from Krawisz et al. (15), which measures the increase in absorbance at 460 nm due to the formation of the chromophore O-dianisidine and HOCl from O-dianisidine-HCl and H2O2. Preliminary experiments were conducted to confirm that assay conditions used for the measurement of mammalian halide peroxidase activity were optimal for squid peroxidases. Human neutrophil MPO (Calbiochem) and assay buffer without added tissue extracts were used as positive and negative controls, respectively. Peroxidase activity was also measured in the presence of 1 mM salicylhydroxamic acid (16), an inhibitor to dianisidine peroxidases.
The electrophoretic mobility of peroxidases from squid tissue extracts was compared with that of human MPO on native and denaturing gels. Following electrophoresis, native gels were incubated with 0.002% H2O2 and 5 μM O-dianisidine-HCl in 50 mM sodium phosphate buffer (pH 6.0) to reveal the position of proteins with halide peroxidase activity, which appeared as dark orange bands due to the generation of the chromophore O-dianisidine. To estimate the subunit molecular masses of the proteins, the stained bands in the lanes containing either human MPO or the extracts of bacteria-containing epithelial tissues of the light organ were excised from native gels, extracted in buffer, and subsequently run on a denaturing (SDS) polyacrylamide gel. Positions of the subunits were identified with Coomassie staining.
Symbiotic organs were prepared for immunocytochemistry as previously described (17), except that an IgG fraction of rabbit anti-human MPO (Calbiochem) was used as the primary antibody at a 1:150 dilution. A 1:50 dilution of goat anti-rabbit IgG complexed to 15 nm colloidal gold spheres (Ted Pella, Reading, CA) was used as the secondary antibody. Samples treated with the rabbit anti-human MPO were designated immune preparations. To control for nonspecific binding of the secondary antibody solution, a 1:150 dilution of nonimmune rabbit IgG (Calbiochem) was substituted for the primary antibody incubation. These samples were designated nonimmune preparations. Sections of symbiotic organs were analyzed on a JEOL 100CX transmission electron microscope, and the density of gold spheres was quantified.
Analyses with an Expressed Peptide of the pLO4 cDNA Clone.
To determine whether the squid cDNA clone in pLO4 could encode the peroxidase activity found in the squid tissues, a polyclonal antibody was produced against a fusion protein generated from the pLO4 cDNA as described below. The ability of the resulting antibody to recognize the squid peroxidase was then determined by using immunoprecipitation assays, Western blot analyses, and immunocytochemistry.
To express the product from the putative squid peroxidase cDNA, an EcoRI fragment of pLO4 (nucleotides 1–1274) was inserted into the expression vector pMAL-c2 (New England Biolabs). The resulting open reading frame, which contained the gene encoding the Escherichia coli maltose binding protein fused to the pLO4 fragment, was sequenced to confirm orientation. The encoded fusion protein (FPO) was then expressed and purified according to manufacturer’s instructions. A polyclonal antiserum to purified FPO was then produced by immunizing rabbits with 1 mg of the fusion protein. Immunizations and antiserum preparation were performed by Berkeley Antibody (Richmond, CA).
For immunoprecipitation using anti-FPO, total soluble extracts of the bacteria-containing tissues of the host symbiotic organs were incubated 24 h at 4°C with protein A Sepharose beads to which anti-FPO had been bound. To control for nonspecific binding to the beads, extracts were also incubated in parallel with beads that had been exposed to preimmune serum. Peroxidase activity remaining after 24 h was compared with the activity in extracts that had been held at 4°C in the absence of beads. In all cases, no more than 40% of the original activity was lost during the 24-h incubation period. To further ensure specificity of the interaction between the peroxidase activity and the protein A beads with anti-FPO, we determined whether the fusion protein itself, which has no peroxidase activity, could competitively inhibit binding of the peroxidase.
The cross-reactivity of anti-FPO was determined in both immunoblots and immunocytochemistry. In each case, tissue preparation and labeling conditions were the same as those used for studies with anti-human MPO.
RESULTS AND DISCUSSION
Peroxidase Activity in Various Squid Tissues.
The discovery in the E. scolopes symbiotic organ of an abundant cDNA whose derived amino acid sequence has 30% sequence identity to a mammalian haloperoxidases (12) suggested that a correspondingly high level of activity of that type of peroxidase might be present in the organ. To determine whether such peroxidase activity could be detected in host tissues, dianisidine peroxidase activity was measured in both the symbiotic organ and in nonsymbiotic tissues. Activity in the symbiotic organ averaged 0.83 ± 0.49 mol of substrate·min−1·mg protein−1, a value that was over 800 times higher than that of mantle tissue and 1300 times higher than that of the digestive gland (Fig. 2). Purified human MPO activity was 23.5 mol·min−1·mg protein−1. MPO is reported to comprise ≈5% of the total soluble protein of the neutrophil (13). Thus, the specific activity of this protein from extracts of neutrophils would be expected to be ≈1.2 mol of substrate·min−1·mg protein−1, in a range similar to extracts of squid symbiotic tissue. Although the Euprymna–Vibrio symbiosis is the first invertebrate mutualism exhibiting the expression of an MPO-like gene, as well as the corresponding enzymatic activity, a similar enzyme activity has been described in the bay mussel, Mytilus edulis, where an antimicrobial function was suggested (20).
Figure 2.
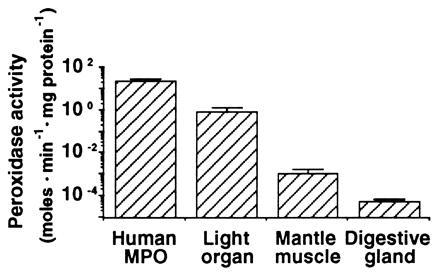
Dianisidine peroxidase activity in extracts of E. scolopes tissues. Squid light organ, mantle, and digestive gland were each homogenized over ice in ground glass tissue homogenizers containing 500 μl of buffer (50 mM sodium phosphate, pH 6.0). The homogenates were centrifuged at 8000 × g for 20 min at 4°C in a Sorvall RC-5B superspeed centrifuge, and the resulting supernatant fluids were kept on ice. For each assay, 17 μl of the supernatant fluid were added to 483 μl of an assay buffer containing 50 mM sodium phosphate, 0.002% H2O2, and 5 μM O-dianisidine·HCl (pH 6.0), and the absorbance of the solution was monitored at A460 for 1 min. Concentration of soluble protein was determined spectrophotometrically (18). Enzyme activity was expressed as mol O-dianisidine produced·min−1·mg soluble protein−1 as determined from Beer’s Law (ɛ = 11.3 cm2·millimol−1; ref. 19). Activity of commercially available human MPO was measured for comparison. Bars are averages ± standard deviation (n = 18).
In addition to the presence of relatively high levels of dianisidine peroxidase activity, 98% of the peroxidase activity was lost in the squid symbiotic tissue extracts when salicylhydroxamic acid, a dianisidine peroxidase inhibitor that binds directly to the heme cofactor (16), was included in the assay. The control human MPO was inhibited 92% under our experimental conditions. Taken together, these results demonstrate that squid light organ tissues contain substantial peroxidase activity similar to the type of activity exhibited by human MPO.
Biochemical and Immunological Relatedness of the Squid and Mammalian Peroxidases.
The high levels of dianisidine peroxidase activity, as well as the previously described gene and derived amino acid sequences (12), suggested that the squid peroxidase may share other biochemical and immunological properties with mammalian halide peroxidases, particularly MPO. To partially purify the squid protein for comparisons with mammalian MPO, we ran a native polyacrylamide gel of E. scolopes tissue extracts and human MPO, and stained the gel to reveal the positions of protein bands positive for dianisidine peroxidase activity (Fig. 3a). Peroxidase activity of human MPO was detected as a bright orange band that ran close to the top of the gel. In squid tissues, activity was detected as tight, orange bands running near the center of the gel in the soluble protein extracts of the bacteria-containing central core of the light organ (see Fig. 1a), although not in the isolated bacterial fraction itself (Fig. 3a, lane 7). Extracts of the gill and the ink sac, which included the ink gland, also exhibited activity. Activity in the gill could be predicted because the mollusc gill functions as a sight of pathogen accumulation and removal (22); thus, this protein may also function in defense against pathogens. Further, similar peroxidase activity was not surprising in the ink gland, because a cDNA similar to that in the squid light organ was recently cloned by Palumbo and Jackson (23) from the ink gland of the cuttlefish, Sepia officianalis. These authors suggested that the peroxidase in S. officianalis functions in ink production; however, ink of cephalopods is also reported to exhibit antimicrobial activity (24, 25). Barely detectable peroxidase activity was observed in the E. scolopes retina and was not detectable in extracts from the digestive gland.
Figure 3.
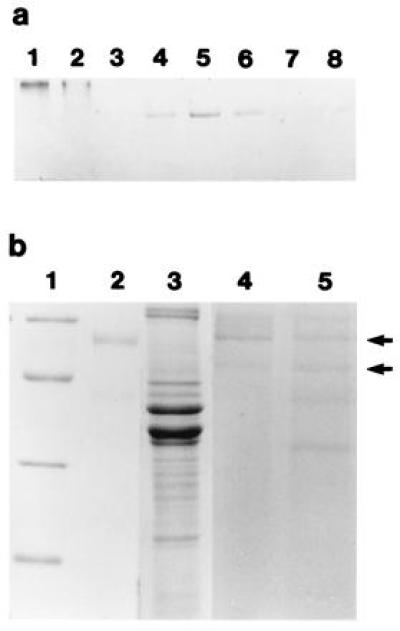
Characterization of the squid tissue extracts and human MPO using native and denaturing PAGE. PAGE in the absence of the detergent SDS was carried out on a 7.5% resolving gel at 14°C (techniques modified from ref. 21). Gels were stained in 50 mM sodium phosphate, 0.002% H2O2, and 5 μM O-dianisidine·HCl (pH 6.0). Orange bands were excised and homogenized in a 1-ml ground-glass tissue homogenizer in 50 μl of 330 mM Tris·HCl, 3.3% SDS, and 2 M 2-mercaptoethanol (pH 6.8). The proteins were extracted from the gel in this buffer for 24 h at room temperature before being separated by SDS/PAGE on a 12.5% resolving gel (21). (a) Native activity gel. Lanes: 1 and 2, purified human MPO, 0.5 and 0.25 μg, respectively; 3–8, total soluble proteins from squid tissue extracts, 15 μg of protein loaded in each lane; 3, squid digestive gland; 4, bacteria-containing central core of the light organ; 5, squid gill; 6, ink sac with ink gland; 7, soluble fraction of the sonicated pellet of the central core; and 8, squid retina. (b) SDS/PAGE stained with Coomassie blue. Lanes: 1, molecular mass markers, 66, 45, 30, and 22.5 kDa, from top to bottom; 2 and 3, purified human MPO (1 μg) and total soluble light organ extract (25 μg), respectively; 4 and 5, protein profiles from excised orange peroxidase bands from human MPO and symbiotic organ, respectively (protein concentrations not determined). Arrows point to subunits at 53 and 60 kDa which are present in both purified MPO (lane 4) and symbiotic organ (lane 5).
The excised, extracted bands derived from the native gels of the bacteria-containing epithelial tissue of the light organ revealed subunits of apparent molecular masses of 35, 44, 53, and 60 kDa, whereas two subunits of apparent molecular masses 53 and 60 kDa were present from human MPO native gel bands (Fig. 3b). Thus, although the symbiotic organ peroxidase and human MPO migrate differently as native proteins, the active bands shared subunit molecular masses of 53 and 60 kDa on denaturing gels. The 35- and 44-kDa bands derived from the squid extracts may represent contaminating proteins in the excised bands that comigrated with the peroxidase on the native gels. Alternatively, the 44-kDa band may also be a peroxidase subunit, since our SDS/PAGE experiments with commercially available human MPO often revealed a band at 44 kDa when the protein was loaded directly onto the denaturing gel. In addition, in these gels human MPO and squid tissue extracts shared bands that migrated at 44 and 60 kDa with similar immunological properties (see below), further suggesting that the 44-kDa band is a peroxidase subunit. Mammalian MPO is a heterotetramer consisting of two 57-kDa subunits, which bind the heme cofactor, and two 14-kDa subunits (13). The 57-kDa subunits often run anomalously high on denaturing gels, presumably due to glycosylation of the protein (13), which may explain the discrepancy between our subunit molecular mass determinations and the actual known molecular mass of the subunit. Using the described methods, we could not detect a 14-kDa peptide in the active, excised bands from either the purified human MPO or the squid protein.
To determine whether the squid peroxidase shared immunological properties with mammalian MPO, we performed various immunological analyses using an anti-human MPO antibody. The polyclonal antibody, made against native protein, did not successfully label immunoblots of denatured subunits of either squid symbiotic light organ extracts or purified human MPO (data not shown). However, using the anti-human MPO antibody, which would react with the protein in its native conformation, immunocytochemistry revealed a cross-reactive species in the bacteria-containing tissues of the symbiotic organ. Immune preparations of both the epithelium and the ciliated duct contained numbers of gold spheres that were statistically significantly above background numbers in control nonimmune preparations (Table 1). In these two tissues, which are immediately adjacent to the extracellular bacteria (see Introduction and Fig. 1), solitary and clustered gold spheres were concentrated in the apical ends of the epithelial cells of the crypts and ciliated duct (arrows in Fig. 4 b–d). Only background numbers of individual spheres were found in nonimmune preparations and in symbiotic light organ lens and reflector, two tissues that are not in direct contact with the symbionts. The clusters of spheres in the epithelial cells are similar in size and location to multiple black granules evident in the better preserved, conventionally fixed symbiotic organ epithelium (arrow in Fig. 4a). Although in the present study we did not confirm that the areas of immune cross-reactivity correspond to these electron dense areas, it should be noted that MPO is stored in granules in mammalian neutrophils (13). These comparisons of human MPO and the squid peroxidase demonstrate that they are not only similar biochemically, but also that an antibody to the human protein localizes a molecular species in the squid tissues that directly interact with the bacterial symbionts.
Table 1.
Immunolocalization of an MPO-like protein in the light organ of E. scolopes
| Squid tissue* | Antibody
cross-reactivity†
|
P value‡ | |
|---|---|---|---|
| Immune | Nonimmune | ||
| Epithelial (E)‡ | 27.8 ± 7.1 | 7.2 ± 3.4 | <0.005 |
| Ciliated duct (D) | 30.5 ± 9.6 | 11.2 ± 5.1 | <0.05 |
| Lens (L) | 10.5 ± 3.6 | 15.6 ± 4.3 | >0.1 |
| Reflector (R) | 2.3 ± 1.2 | 3.4 ± 1.6 | >0.1 |
Letters in parentheses corresponding to tissue types labeled in Fig. 1.
Number of immunogold spheres bound per 4 μm2 of tissue sections treated with either immune (containing anti-human MPO antibody) or nonimmune serum.
Results of two-tailed t tests comparing immune and nonimmune values.
Figure 4.
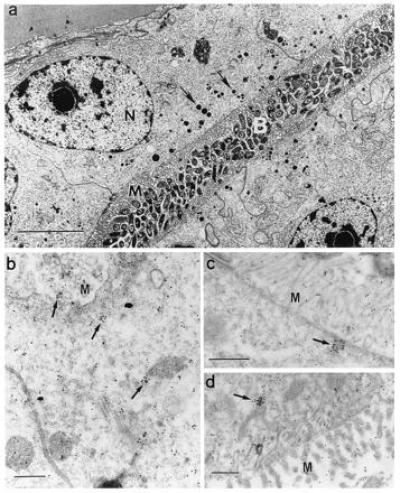
Cross-reactivity of squid tissues with polyclonal antibodies against native human MPO. (a) For reference and orientation, a low magnification micrograph of tissue fixed for conventional electron microscopy (7) illustrates the epithelial cells of the bacteria-containing region (nuclei, N), numerous microvilli (M) that extend into the bacteria-containing crypts (B), and vesicles (0.1–0.6 μm) along the apical surfaces of these cells (arrows). (Bar = 5 μm.) (b–d) Higher magnification micrographs of light organ tissues, fixed for immunocytochemistry, and reacted with antibodies against human MPO conjugated to gold spheres. (b and c) Apical surfaces of epithelial cells lining bacteria-containing crypts; numerous gold spheres occur singly and in clusters that average 1–2 μm (arrows). (d) Epithelia of the ciliated duct that connects the crypts with the exterior of the light organ, also with abundant gold spheres both alone and in clusters (arrow). M, epithelial microvilli. (Bars = 0.5 μm for b–d.)
Relationship of the Peroxidase Activity to the cDNA Insert in pLO4.
While the most direct method to demonstrate the correspondence between a gene and an enzyme activity is to obtain the amino acid sequence of the purified enzyme, in this study a limited quantity of tissue available and technical difficulties in sequencing the small amounts of purified protein obtained (N-terminal blockage and acid splitting of the peptide) precluded our obtaining these data. As a second approach to this question, we sought to investigate the relationship between the peroxidase activity and the putative peroxidase gene carried on pLO4 by studying the properties of the expressed gene product, which was generated from a fragment of LO4. First, immunoprecipitation using anti-fusion protein (FPO) was performed to determine whether the antibody could remove peroxidase activity from symbiotic light organ extracts. A symbiotic organ extract incubated with anti-FPO bound to Sepharose beads retained less than 35% of its peroxidase activity when compared with incubations with beads exposed to preimmune serum, which retained 82% of its activity relative to control incubations (Fig. 5; without fusion protein). However, when purified FPO was included in the incubations, activity remained high in both anti-FPO and anti-preimmune serum incubations (Fig. 5; with fusion protein). These results suggest that FPO competitively and specifically inhibited the immunoprecipitation, and that the decrease in activity observed in the anti-FPO incubation in the absence of the fusion protein was a direct result of the antiserum binding to a significant portion of the squid peroxidase.
Figure 5.
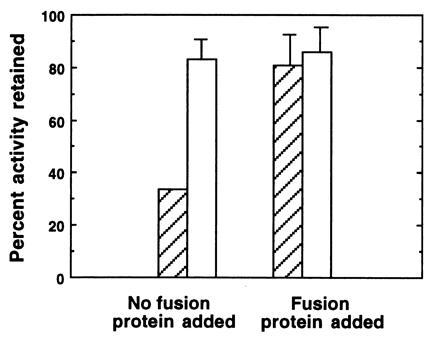
Immunoprecipitation of symbiotic organ extracts using anti-FPO. Tissues were dissected, homogenized and centrifuged as described in Fig. 2, except 0.05 M sodium phosphate with 0.1 M NaCl at pH 7.8 (PBS) was used as a buffer. In experimental trials (without fusion protein), 50 μl of the supernatant fluid were added to 10 μl of protein A Sepharose beads to which anti-FPO had been bound (hatched bars). To control for the potential effects of rabbit serum on enzyme activity, we also assayed samples in which 50 μl of supernatant fluid were added to 10 μl of Sepharose beads that had been preincubated with preimmune serum (open bars). In addition, in another set of controls, we determined whether the purified FPO could competitively inhibit binding of the putative peroxidase. In these experiments, 15 μl of a 2 mg/ml solution of purifed FPO in PBS were added to the supernatant fluid. To compensate for dilution effects, additional PBS was added to all samples for a final volume of 75 μl. In experimental and control samples, peroxidase activity was measured in all samples after a 24-h incubation period at 4°C. Bars are average percentages ± arcsine transformations (n = 5) of the initial peroxidase activity remaining in extracts after the 24-h incubation.
Immunoblot analyses using anti-FPO detected immune-reactive bands at 44 and 60 kDa in the symbiotic light organ extracts, but not in extracts of the digestive gland (Fig. 6). The fusion protein antibody also recognized protein subunits at similar molecular masses to those of the denatured human MPO at 44 and 60 kDa. The 14-kDa subunit of human MPO, while it did not appear in the lanes of denaturing gels following its elution from active bands on native gels, was detected when the protein was run directly on denaturing gels. This 14-kDa band of human MPO was not recognized by the antibody to the fusion protein (Fig. 6). Because the molecular mass of 44 kDa corresponds to neither of the subunits of human MPO (13), the 44-kDa band is assumed to be either a breakdown product of MPO, or an immune-reactive contaminant that occurs in the commercially purified MPO. The coincidence of positive bands at 44 kDa in both the purified human MPO and the squid, and the fact that we often see breakdown products of similar size on SDS gels, suggest that the 44-kDa band is indeed a breakdown product of MPO. In addition, using immunocytochemistry, anti-FPO antibody labeled tissues in the same areas as the anti-human MPO (data not shown), specifically at the apical surfaces of the epithelial cells containing bacterial symbionts.
Figure 6.
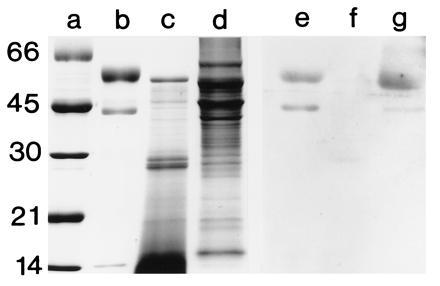
SDS/PAGE and immunoblot, using anti-FPO, of squid tissues and purified human MPO. For immunoblots, the soluble proteins were prepared as described in Fig. 3, except that they were homogenized in a 50 mM sodium phosphate buffer with 0.1 M NaCl (pH 7.2). SDS/PAGE was performed as described in Fig. 3. Proteins were electrophoretically transferred from unstained gels onto nitrocellulose membrane (modified from ref. 26). Immunoblots were performed with a chemiluminescence detection system (Renaissance Kit; DuPont/NEN). Membranes were first blocked for 12 h at 4°C in 50 mM Tris, 150 mM NaCl, 0.5% Tween 20 (pH 7.5) (TTBS) containing 3% powdered milk and a 1:100 dilution of goat serum. Following this, they were incubated in a 1:100 dilution of anti-FPO in TTBS for 12 h at 4°C and subsequently incubated in the secondary antibody, goat anti-rabbit IgG conjugated to horseradish peroxidase. Preimmune serum, obtained from the rabbit before immunization with FPO, was substituted for the 1° antiserum as a negative control. Lane: a, standards; b and e, purified human MPO (0.5 μg each lane); c and f, squid digestive gland (20 μg each lane); and d and g, bacteria-containing epithelial tissue of the symbiotic organ (20 μg each lane). The molecular masses of the standards are shown in kDa.
Function of MPO and Implications for the Role of the Squid Dianisidine Peroxidase in Symbiosis.
The shared characteristics of MPO and the peroxidase in the symbiotic tissues of E. scolopes present a seeming paradox; i.e., how does this host squid enzyme, which is related to a mammalian protein associated with defense against microorganisms, function in a cooperative symbiosis? The occurrence of this type of enzyme in both pathogenic and mutualistic symbioses suggests the possibility that the biochemical “language” of interaction between animals and their bacterial symbionts is similar, if not identical, in these two different types of associations. In pathogenesis, this enzyme clearly functions to curtail an unwanted association, whereas in mutualisms, perhaps it functions either to ensure both specificity and control symbiont number, or to prevent invasion of host tissues by the microbial partner. Further, evidence is accumulating that the biochemical “dialogue” in both animal and plant, pathogenic and mutualistic relationships, may involve not only this type of peroxidase, but also the other components of the respiratory burst pathway. Recently, a respiratory burst-mediated antibacterial response has been reported in plant pathogen interactions (27); and, the role of reactive oxygen species in the relationship between legumes and their rhizobial partners is an area of active research and considerable controversy (28).
Similar investigations have begun to explore the role of reactive oxygen species in the E. scolopes–V. fischeri association (29, 30). Studies of the symbiont V. fischeri are providing evidence that these bacteria may have specific responses to, and/or defenses against, the activity of this enzyme. Cultured V. fischeri have an inducible protective response to both hydrogen peroxide and hypochlorous acid, and bacteria isolated from the light organs exhibit high catalase activity (29). Mutants in the ability to respond to oxidative stress are presently being generated to determine whether they show a deficiency in symbiotic competence. In addition, recently an ADP ribosyltransferase activity has been described in V. fischeri that is similar to the cholera toxin-associated activity characteristic of the related species V. cholerae (30). Among the many effects of this activity in the V. cholerae pathogenesis is an inhibition of host cell respiratory burst. While the role of the ADP ribosyltransferase activity has yet to be studied in the intact symbiosis between E. scolopes and V. fischeri, if this enzyme is active in the establishment and maintenance of this association, it may serve to modulate expression of the squid peroxidase to levels that promote a stable symbiosis.
SUMMARY
This study characterizes a peroxidase in symbiotic light organs of the squid E. scolopes. Our results provide evidence that this dianisidine peroxidase, whose activity was significantly higher in the light organ than in other nonsymbiotic squid tissues, shares several biochemical and immunological features with the human antimicrobial enzyme MPO: (i) the squid peroxidase and human MPO shared similar subunit molecular masses; (ii) an antibody to human MPO localized immune reactive areas to the interface between the cells of the light organ and the bacterial symbionts; and (iii) an antibody made to the product of an E. scolopes peroxidase-like cDNA cross-reacted in Western blot analyses with peptide subunits of similar molecular mass in both symbiotic organ homogenates and human MPO; and, this antibody localized similar sites in the symbiotic organ as anti-human MPO.
One of the key questions in the study of cooperative animal–bacterial symbioses is: what new genes have evolved for the control of these beneficial relationships, and what already existing genes have been co-opted to participate in the interaction? The presence of an MPO-like peroxidase in the Euprymna–Vibrio symbiosis suggests that at least one aspect of the dynamic dialogue between these partners may rely on the modulation of a preexisting antibacterial program. Alternatively, genes such as that encoding MPO in the host and ADP ribosyltransferase activity in the bacteria may have evolved for the precise modulation of many animal–bacterial interactions, and it is the “fine tuning” of expression of such genes that determines whether the nature of the symbiosis is cooperative or pathogenic.
Acknowledgments
We thank E. G. Ruby, K. Visick, and J. Graf for comments on the manuscript. This work was supported by National Science Foundation Grant IBN 9220492 and Office of Naval Research Grant N00017-91-1347 to M.J.M.-N.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. §1734 solely to indicate this fact.
Abbreviation: MPO, myeloperoxidase.
References
- 1.Smith D C, Douglas A E. The Biology of Symbiosis. London: Edward Arnold; 1987. [Google Scholar]
- 2.Fisher R F, Long S R. Nature (London) 1992;357:655–660. doi: 10.1038/357655a0. [DOI] [PubMed] [Google Scholar]
- 3.Hirsch A M. New Phytol. 1992;122:211–237. doi: 10.1111/j.1469-8137.1992.tb04227.x. [DOI] [PubMed] [Google Scholar]
- 4.Verma D, Hu C-A, Zhang M. Physiol Plant. 1992;85:253–265. [Google Scholar]
- 5.McFall-Ngai M J, Ruby E G. Science. 1991;254:1491–1494. doi: 10.1126/science.1962208. [DOI] [PubMed] [Google Scholar]
- 6.Ruby E G, McFall-Ngai M J. J Bacteriol. 1992;14:4865–4870. doi: 10.1128/jb.174.15.4865-4870.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McFall-Ngai M J, Montgomery M K. Biol Bull. 1990;179:332–339. doi: 10.2307/1542325. [DOI] [PubMed] [Google Scholar]
- 8.Richardson K C, Jarrett L, Finke E H. Stain Technol. 1960;35:313–323. doi: 10.3109/10520296009114754. [DOI] [PubMed] [Google Scholar]
- 9.Moynihan M. Behaviour. 1983;85:25–41. [Google Scholar]
- 10.Wei S, Young R E. Mar Biol (Berlin) 1989;103:541–546. [Google Scholar]
- 11.Montgomery M K, McFall-Ngai M J. Development (Cambridge, UK) 1994;120:1719–1729. doi: 10.1242/dev.120.7.1719. [DOI] [PubMed] [Google Scholar]
- 12.Tomarev S I, Zinovieva R D, Weis V M, Chepelinsky A B, Piatigorsky J, McFall-Ngai M J. Gene. 1993;132:219–226. doi: 10.1016/0378-1119(93)90199-d. [DOI] [PubMed] [Google Scholar]
- 13.Johnson K R, Nauseef W M. In: Peroxidases in Chemistry and Biology. Everse J, Everse K E, Grisham M B, editors. Vol. 1. Boca Raton, FL: CRC; 1991. pp. 63–81. [Google Scholar]
- 14.Klebanoff S. In: Peroxidases in Chemistry and Biology. Everse J, Everse K E, Grisham M B, editors. Vol. 1. Boca Raton, FL: CRC; 1991. pp. 1–35. [Google Scholar]
- 15.Krawisz J E, Sharon P, Stenson W F. Gastroenterology. 1984;87:1344–1350. [PubMed] [Google Scholar]
- 16.Ikeda-Saito M, Shelley D A, Lu L, Booth K S, Caughey W S, Kimura S. J Biol Chem. 1991;266:3611–3616. [PubMed] [Google Scholar]
- 17.Weis V M, Montgomery M K, McFall-Ngai M J. Biol Bull. 1993;184:309–321. doi: 10.2307/1542449. [DOI] [PubMed] [Google Scholar]
- 18.Whitaker J, Granum P. Anal Biochem. 1980;109:156–159. doi: 10.1016/0003-2697(80)90024-x. [DOI] [PubMed] [Google Scholar]
- 19.Hurst J K, Albrich J M, Greens T R, Rosen H, Klebanoff S J. J Biol Chem. 1984;259:4812–4821. [PubMed] [Google Scholar]
- 20.Schlenck D, Martinez P G, Livingstone D R. Comp Biochem Physiol C. 1991;99:63–68. [Google Scholar]
- 21.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Bayne C. Malacol Rev. 1973;6:13–17. [Google Scholar]
- 23.Palumbo A, Jackson I J. Biochim Biophys Acta. 1995;1247:173–178. doi: 10.1016/0167-4838(94)00221-2. [DOI] [PubMed] [Google Scholar]
- 24.Sheu T, Chou C. J Chin Agric Soc. 1990;28:59–68. [Google Scholar]
- 25.Takai M, Kawai Y, Inoue N, Shinano H. Bull Jpn Soc Sci Fish. 1992;58:2373–2378. [Google Scholar]
- 26.Towbin H, Staehelin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levine A, Tenhaken R, Dixon R, Lamb C. Cell. 1994;79:583–593. doi: 10.1016/0092-8674(94)90544-4. [DOI] [PubMed] [Google Scholar]
- 28.McKhann H I, Hirsch A M. In: Bacterial Pathogenesis of Plants and Animals. Dangl J L, editor. Berlin: Springer; 1994. pp. 139–162. [Google Scholar]
- 29.Boettcher, K. J. (1994) Dissertation (Univ. of Southern California, Los Angeles).
- 30.Reich K, Schoolnik G. J Bacteriol. 1996;178:209–215. doi: 10.1128/jb.178.1.209-215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]


