Abstract
Skeletal myoblasts grown in vitro and induced to differentiate either form differentiated multinucleated myotubes or give rise to quiescent, undifferentiated “reserve cells” that share several characteristics with muscle satellite cells. The mechanism of determination of reserve cells is poorly understood. We find that the expression level of the metalloprotease disintegrin ADAM12 is much higher in proliferating C2C12 myoblasts and in reserve cells than in myotubes. Inhibition of ADAM12 expression in differentiating C2C12 cultures by small interfering RNA is accompanied by lower expression levels of both quiescence markers (retinoblastoma-related protein p130 and cell cycle inhibitor p27) and differentiation markers (myogenin and integrin α7A isoform). Overexpression of ADAM12 in C2C12 cells under conditions that promote cell cycle progression leads to upregulation of p130 and p27, cell cycle arrest, and downregulation of MyoD. Thus, enhanced expression of ADAM12 induces a quiescence-like phenotype and does not stimulate differentiation. We also show that the region extending from the disintegrin to the transmembrane domain of ADAM12 and containing cell adhesion activity as well as the cytoplasmic domain of ADAM12 are required for ADAM12-mediated cell cycle arrest, while the metalloprotease domain is not essential. Our results suggest that ADAM12-mediated adhesion and/or signaling may play a role in determination of the pool of reserve cells during myoblast differentiation.
Development and regeneration of skeletal muscle are tightly coupled to the cell cycle machinery (32, 62). During myogenic differentiation, proliferating myoblasts pause in G1, then permanently withdraw from the cell cycle, differentiate, and fuse into multinucleated myotubes (1, 45, 50, 68). In adult skeletal muscle, ≈5% of the nuclei belong to undifferentiated, G0-arrested cells (satellite cells) that play an essential role during muscle regeneration, muscle hypertrophy, and postnatal muscle growth (23, 54, 55). In response to injury, stretch, or exercise, satellite cells become activated, reenter the cell cycle, and start to proliferate. The progeny of activated satellite cells can either differentiate and fuse with adjacent muscle fibers, effectively replacing the damaged fibers (4, 21), or reenter G0, remain undifferentiated, and replenish the satellite cell compartment (55).
Mouse myoblastic cell line C2 and its subclone C2C12 have been used frequently as model systems of myogenic differentiation in vitro. Differentiation of myoblasts cultured in growth factor-rich medium (growth medium) is induced by transfer of cells to medium containing low concentration of mitogens (differentiation medium). Mitogen depletion leads to one of the following outcomes: a subpopulation of cells undergoes terminal differentiation and forms myotubes; another subpopulation called reserve cells remains undifferentiated and shares many characteristics with muscle satellite cells (3, 10, 18, 67); and a subset of cells undergoes apoptosis (62, 63). Reserve cells are arrested at the G0 phase of the cycle, have increased levels of the retinoblastoma-related protein p130, a quiescent cell marker (10), and decreased expression of myogenic transcription factor MyoD (31, 67). Interestingly, forced expression of p130 in myoblasts inhibits MyoD gene expression and its ability to transactivate muscle genes (10). Therefore, it has been proposed that induction of p130 is a part of a specific pathway that defines the pool of reserve cells during differentiation of myoblasts in vitro (10). The mechanisms involved in generation of reserve cells and upregulation of p130 in a fraction of myogenic cells upon mitogen deprivation remain, however, unknown.
ADAMs (proteins containing a disintegrin and metalloprotease domain) are cell surface receptors that are involved in cell-cell adhesion, surface proteolysis, and transmembrane signaling (5, 53, 60). A typical ADAM consists of a prodomain, metalloprotease, disintegrin-like and cysteine-rich domains, and, in most cases, epidermal growth factor-like, transmembrane, and cytoplasmic domains. ADAM12 has been implicated in skeletal muscle development and regeneration (6, 19, 20, 66), but its exact function in muscle is not clear. ADAM12 mRNA is detected predominantly in cardiac and smooth muscle (20), placenta (20), and neonatal skeletal muscle (66). In adult skeletal muscle, the expression level of ADAM12 is very low in both differentiated muscle fibers and quiescent satellite cells (6, 20, 66). The amount of ADAM12 protein increases dramatically in regenerating muscle (19), and its mRNA is readily detected in satellite cells following their activation (6).
During differentiation of C2C12 cells, ADAM12 mRNA and protein are present in undifferentiated cells, and during early stages of differentiation, and they both decrease to low levels as differentiation proceeds (8, 19, 66). Since differentiating C2C12 cultures are heterogeneous and contain both differentiated myotubes and undifferentiated reserve cells, we examined the distribution of ADAM12 between these two populations of cells to gain new insight into the function of ADAM12 during myoblast differentiation. We found that expression of ADAM12 is decreased in myotubes and preserved in reserve cells. Although inhibition of the expression of endogenous ADAM12 by small interfering RNA (siRNA) is accompanied by lower expression levels of both quiescence and differentiation markers, enhanced expression of ADAM12 induces a quiescent cell-like phenotype and does not stimulate differentiation. Our results suggest a novel role for ADAM12 in the establishment of the pool of reserve cells during myoblast differentiation.
MATERIALS AND METHODS
Antibodies.
Rabbit anti-ADAM12 antibody was raised against a peptide from mouse ADAM12 cytoplasmic domain (amino acids 774 to 791) and affinity purified (8). Rabbit anti-ADAM9 antibody was raised against bacterially expressed six-His-tagged cytoplasmic domain of mouse ADAM9 (8). Goat anti-p130 (C-20) polyclonal antibody was obtained from Santa Cruz Biotechnology (Santa Cruz, Calif.). Monoclonal antibodies were purchased from the following sources: anti-myoD1 (clone 5.8A) from Lab Vision (Fremont, Calif.), antimyogenin (clone F5D), anti-cyclin B1 (clone GNS1), and anti-p21 (clone F-5) from Santa Cruz Biotechnology, anti-retinoblastoma protein (pRb) (clone G3-245) and anti-p27 (clone G173-524) from BD Pharmingen (San Diego, Calif.), and antitubulin (clone DM 1A) from Sigma (St. Louis, Mo.). Rabbit anti-integrin α7 antibody recognizing the C terminus of the α7A isoform was a gift from S. J. Kaufman (University of Illinois, Urbana).
Expression constructs.
The following constructs were used in this study: the full-length mouse ADAM12 (amino acids 1 to 903); ADAM12(L73P), a mutant form of mouse ADAM12 in which Leu73 was replaced with proline; ADAM12(ΔPM), mouse ADAM12 lacking the prodomain and metalloprotease domain in which the N-terminal 424 amino acids were replaced with the Igκ secretion signal; ADAM9(ΔPM), mouse ADAM9 lacking the prodomain and metalloprotease domain in which the N-terminal 416 amino acids were replaced with the Igκ secretion signal; ADAM12(ΔPM)/9, containing the extracellular and transmembrane domains of ADAM12(ΔPM) (amino acids 425 to 727) and the cytoplasmic tail of ADAM9 (amino acids 719 to 845); and ADAM9(ΔPM)/12, containing the extracellular and transmembrane domains of ADAM9(ΔPM) (amino acids 417 to 718) and the cytoplasmic tail of ADAM12 (amino acids 728 to 903). The generation of all the constructs was described previously (8).
Cell culture, cell separation, transfections, and cell cycle analysis.
C2C12 myoblasts were cultured in growth medium (Dulbecco's modified Eagle's medium [DMEM] supplemented with 10% fetal bovine serum [FBS]) at 37°C in the presence of 5% CO2 under a humidified atmosphere. To induce differentiation, confluent cells were transferred to differentiation medium (DMEM containing 2% horse serum). In the experiments shown in Fig. 3, tumor necrosis factor alpha processing inhibitor (TAPI, also called IC-3, a hydroxamate-based inhibitor of metalloproteases; obtained from Biomol Research Laboratories, Plymouth Meeting, Pa.) was added to differentiation medium at the indicated concentrations; the medium was changed daily. After 4 days of incubation in differentiation medium, differentiated myotubes were detached by mild trypsinization (0.15% trypsin and 1 mM EDTA in Dulbecco's modified phosphate-buffered saline [DPBS], 1-min treatment). Undifferentiated reserve cells were then detached by 5 min of incubation with 0.25% trypsin and 1 mM EDTA in DPBS. Transient transfections were performed with Lipofectamine Plus (Life Technologies, Rockville, Md.) or Fugene 6 (Roche, Indianapolis, Ind.) according to the manufacturers' protocols. For cell cycle analysis, cells were detached with 0.25% trypsin and 1 mM EDTA in DPBS, washed with DPBS, and fixed with 80% ethanol at −20o. The cells were washed again and incubated for 20 min at 37°C in DPBS containing 500 μg of RNase A per ml and 10 μg of propidium iodide per ml, followed by flow cytometric analysis with a Becton Dickinson FACScan flow cytometer.
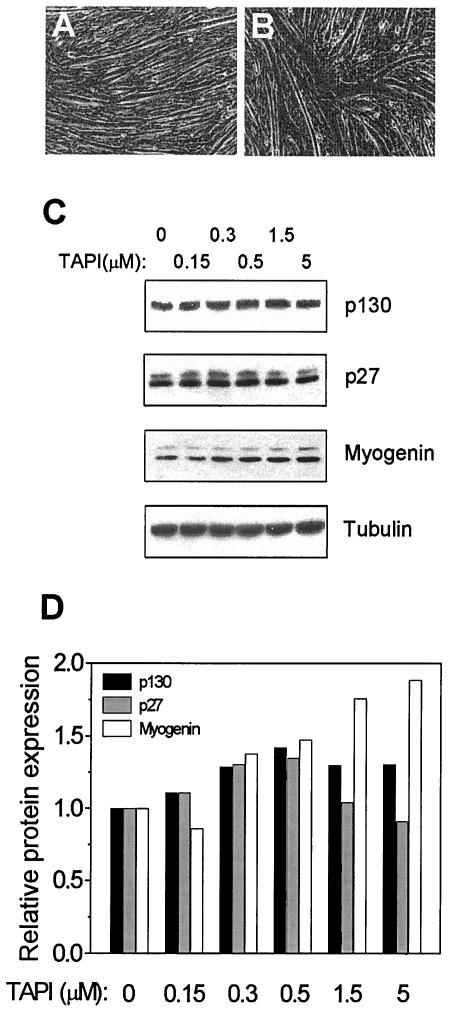
FIG. 3.
Effect of tumor necrosis factor alpha processing inhibitor on differentiation of C2C12 cells. Phase contrast images of cells incubated for 4 days in differentiation medium without (A) and with 5 μM TAPI (B) are shown. (C) Confluent cells were incubated for 4 days in differentiation medium containing the indicated concentrations of TAPI. The expression levels of p130, p27, and myogenin were analyzed by Western blotting. Tubulin was a loading control. (D) The intensities of the protein bands in panel C were quantified by densitometry with ScionImage software.
Northern blot analysis.
Total RNA was extracted from nondifferentiated C2C12 cells, reserve cells, or myotubes with Trizol reagent (Invitrogen, Carlsbad, Calif.). Equal amounts of total RNA (20 μg) were separated electrophoretically in a 1.5% agarose gel containing 2.2 M formaldehyde and transferred to a nylon membrane. The membrane was stained with methylene blue to visualize rRNA and to control for sample loading. The membrane was then hybridized at 65°C with a 2.7-kbp ADAM12 cDNA probe labeled with [α-32P]dCTP (3,000 Ci/mmol) by random priming (Prime a Gene labeling system; Promega, Madison, Wis.), washed at 68°C, and exposed for autoradiography at −70°C with an intensifying screen.
BrdU labeling.
5-Bromo-2′-deoxyuridine (BrdU) labeling was performed with the BrdU labeling and detection kit I (Roche, Indianapolis, Ind.), with several modifications. We plated 0.6 × 105 cells on glass coverslips (22 by 22 mm) placed in the wells of a six-well-plate. Twenty-four hours after plating, cells were transfected and incubated for an additional 24 h in growth medium. Unless indicated otherwise, cells were then transferred to DMEM containing 1% fetal bovine serum (FBS). Twenty-four hours later, 10 μM BrdU was added, and incubation was continued for 4 to 16 h. In some experiments, transfection was performed when cells were grown in 100-mm plates, and at the end of the 24-h period of incubation in 10% FBS, mitotic cells from one 100-mm plate were mechanically detached by repeated shaking and centrifuged at 1,000 rpm for 5 min. The cell pellet was resuspended in DMEM containing 1% FBS plus 10 μM BrdU and plated onto a coverslip placed in a six-well plate. At the indicated times, cells were fixed and permeabilized with 95% ethanol. To simultaneously visualize expression of the recombinant proteins and nuclear incorporation of BrdU, ethanol-fixed cells were first incubated with anti-ADAM12 or anti-ADAM9 primary antibodies and then with rhodamine-conjugated anti-rabbit immunoglobulin G secondary antibody, followed by incubation with 3.7% paraformaldehyde. Cells were then treated with 70% ethanol-50 mM glycine, pH 2.0, and stained with mouse anti-BrdU primary antibody and fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin G secondary antibody.
Small interfering RNA gene silencing assay.
The region of ADAM12 cDNA targeted by siRNA included nucleotides 3130 to 3148 (5′-TTTATGCAGAGTGTCTATT-3′); the sense siRNA was 5′-UUUAUGCAGAGUGUCUAUUdTdT-3′, and the antisense siRNA was 5′-AAUAGACACUCUGCAUAAAdTdT-3′. Initially, the siRNA duplex was prepared by a transcription-based method with the Silencer siRNA construction kit (Ambion, Austin, Tex.), according to the manufacturer's instructions. In later experiments, the ADAM12 siRNA duplex was obtained from Dharmacon Research (Lafayette, Colo.) and had a potency similar to that of the enzymatically synthesized siRNA. The green fluorescent protein duplex (Dharmacon) was used as a control to evaluate the potential presence of nonspecific effects of irrelevant siRNA on cell cycle progression and/or differentiation. C2C12 cells plated in a six-well plate were transfected at ≈70% confluency with ADAM12 or green fluorescent protein (GFP) siRNA with the TransIT-TKO transfection reagent (Mirus Corporation, Madison, Wis.), according to the manufacturer's instructions (50 nM siRNA and 10 μl of TransIT-TKO/well).
Immunoblotting.
Cellular proteins were extracted with 50 mM Tris-HCl (pH 7.4)-150 mM NaCl-1% (vol/vol) Triton X-100-1% sodium deoxycholate-0.1% sodium dodecyl sulfate (SDS)-1 mM 4-(2-aminoethyl)-benzene-sulfonylfluoride hydrochloride (AEBSF)-5 μg of aprotinin per ml-5 μg of leupeptin per ml-5 μg of pepstatin A per ml-5 mM EDTA-10 mM 1,10-phenanthroline, 2 ml/100-mm plate. In the experiments analyzing the phosphorylation status of pRb, phosphatase inhibitors (50 mM NaF, 2 mM Na3VO4, and 10 mM Na4P2O7) were included in extraction buffer. Samples were centrifuged for 30 min at 21,000 × g, and supernatants were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane.
To detect endogenous ADAM12, samples were enriched for glycoproteins prior to SDS-PAGE and immunoblotting. Cell extracts (2 ml) were incubated with concanavalin A-agarose (40-μl bed volume) for 2 h at 4°C, followed by washing the beads with extraction buffer and elution with 30 μl of SDS-PAGE sample buffer. Nitrocellulose membranes were blocked in DPBS containing 3% (wt/vol) dry milk and 0.3% (vol/vol) Tween 20 and then incubated with a primary antibody in blocking buffer, followed by incubation with a horseradish peroxidase-labeled secondary antibody and development with a chemiluminescence detection method (West Pico, Pierce, Rockford, Ill.). The following dilutions of primary antibodies were used: anti-ADAM12, 1:3,000; anti-myoD1, 1:500; antimyogenin, 1:50; anti-p130, 1:100; anti-pRb, 1:400; anti-cyclin B1, 1:100; anti-p21, 1:100; anti-p27, 1:1,000; and antitubulin, 1:40,000.
Immunostaining.
Cells on coverslips were fixed with 3.7% paraformaldehyde in DPBS for 15 min and permeabilized with 0.1% Triton X-100 in DPBS for 5 min. The coverslips were then incubated with anti-ADAM12 rabbit antibody (1:500 dilution) and one of the following mouse monoclonal antibodies: anti-myoD (1:10 dilution), antimyogenin (1:50 dilution), or anti-p27 (1:100 dilution), followed by incubation with rhodamine-conjugated anti-rabbit IgG antibody and fluorescein isothiocyanate-conjugated anti-mouse IgG antibody. In the experiments shown in Fig. 8, cells were stained with anti-ADAM12 antibody and goat anti-p130 antibody (1:40 dilution), followed by incubation with rhodamine-conjugated anti-rabbit IgG antibody and fluorescein isothiocyanate-conjugated anti-goat IgG antibody. After several washes in DPBS, cells were mounted on glass slides and examined by laser scanning confocal microscopy (Zeiss model LSM 410, equipped with an Axiovert 100 microscope).
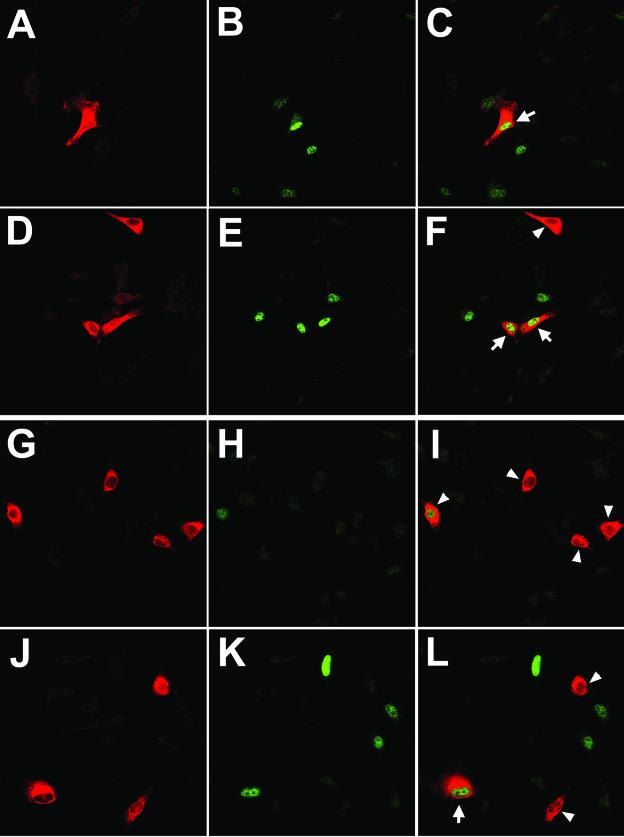
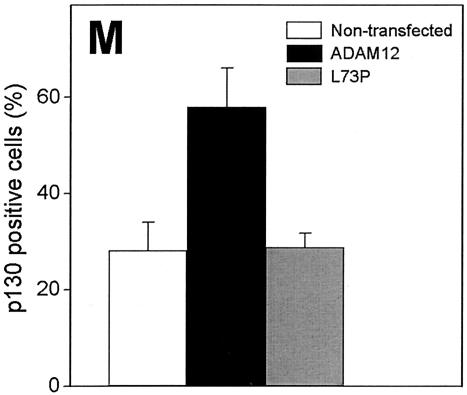
FIG. 8.
Overexpression of ADAM12 leads to induction of p130. Subconfluent C2C12 cells transfected with ADAM12 (A to F) or the L73P ADAM12 mutant (G to L) and incubated in medium containing 1% FBS were fixed and costained with anti-ADAM12 antibody (red; A, D, G, and J) and anti-p130 antibody (green; B, E, H, and K). Two representative microscopic fields are shown for ADAM12 and L73P transfectants. Panels C, F, I, and L are merged images of panels A and B, D and E, G and H, and J and K, respectively. Arrows indicate ADAM12- or L73P-overexpressing cells with induced expression of p130. Arrowheads mark ADAM12- or L73P-overexpressing cells that are p130 negative. (M) The percentage of p130-positive cells among ADAM12-overexpressing (solid bar), L73P-overexpressing (grey bar), and cells that were not stained with ADAM12 antibody (nontransfected cells, open bar) was quantified. Results shown are the averages from three experiments ± standard errors. At least 100 transfected cells were analyzed in each experiment.
RESULTS
To examine the relative abundance of ADAM12 protein in myotubes and reserve cells during C2C12 myoblast differentiation, the two subpopulations of cells were separated by brief and mild trypsinization (Fig. 1A). The reserve cell fraction contained smaller amounts of MyoD, myogenin, and integrin α7A than the myotube fraction and a significantly higher amount of the retinoblastoma-related protein p130 (Fig. 1A). Since downregulation of MyoD and upregulation of p130 expression are two hallmarks of reserve cells (10, 31, 67), and since myogenin and integrin α7A isoform (7) represent differentiation markers expressed in myotubes but not in reserve cells, this demonstrates that the myotube and the reserve cell fractions had been properly separated. Interestingly, p21 and p27, two cell cycle inhibitors that are strongly upregulated during myoblast differentiation (22, 49, 57, 69), showed different distribution pattern between myotubes and reserve cells: whereas p21 was more abundant in the myotube fraction, p27 was clearly more concentrated in reserve cells (Fig. 1A). Since p27 plays a unique role among the cell cycle inhibitors in the induction and maintenance of the quiescent state (42, 44, 48), higher expression of p27 in reserve cells than in myotubes is consistent with the postulated G0 arrest in reserve cells (10, 67).
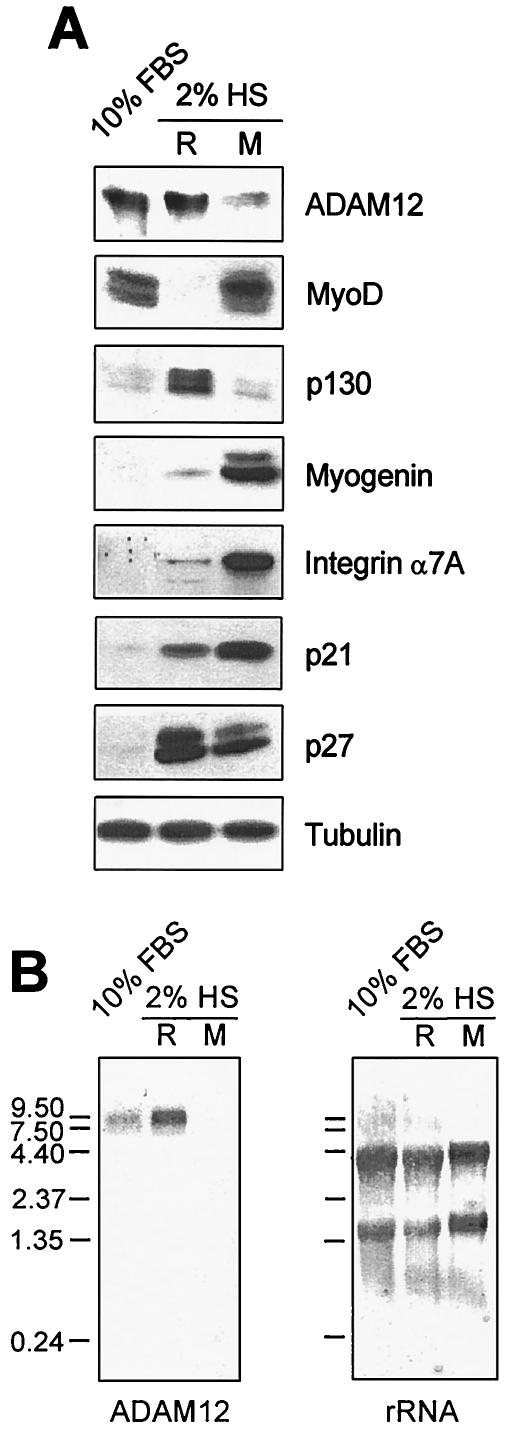
FIG. 1.
ADAM12 protein is more abundant in reserve cells than in myotubes during C2C12 cell differentiation. (A) Western blot analysis of ADAM12 expression in proliferating C2C12 cells grown in 10% FBS and in confluent C2C12 cells incubated for 4 days in differentiation medium containing 2% horse serum (HS). Cells incubated in 2% horse serum were separated by mild trypsinization into two fractions: nondifferentiated reserve cells (R), characterized by a lack of expression of myogenin, downregulation of MyoD, and upregulation of p130 expression; and differentiated myotubes (M) characterized by high expression of myogenin and integrin α7A, sustained expression of MyoD, and low expression of p130. Notice that while the cell cycle inhibitors p21 and p27 are both upregulated in cells switched to differentiation medium, p21 is more abundant in myotubes and p27 is more abundant in reserve cells. To detect endogenous ADAM12, cell extracts were enriched for glycoproteins on concanavalin A columns prior to Western blotting; the levels of all other proteins were analyzed by using total cell lysates. Since the mature 90-kDa cell surface form of ADAM12 (8) is degraded during the trypsinization used to separate reserve cells and myotubes, only the nascent 120-kDa intracellular form of ADAM12 is shown. Tubulin was a loading control. (B) The amount of ADAM12 mRNA in the same three populations of C2C12 cells as in panel A was analyzed by Northern blotting with ADAM12 cDNA as a probe (left). rRNA on the same membrane was stained with methylene blue to control for sample loading (right). The positions of RNA standards (in kilobases) are indicated.
The expression of ADAM12 in myotubes and in reserve cells was evaluated by Western and Northern blotting. As shown in Fig. 1A, the amount of ADAM12 protein was severalfold higher in reserve cells than in myotubes, and it was similar to the amount of ADAM12 in myoblasts before differentiation. Northern blot analysis further demonstrated the presence of a ≈9-kb band corresponding to ADAM12 mRNA (20, 66) in proliferating myoblasts and in reserve cells, but not in the myotube fraction (Fig. 1B). This suggests that ADAM12 may play a role in the induction and/or maintenance of the pool of reserve cells or the induction of differentiation.
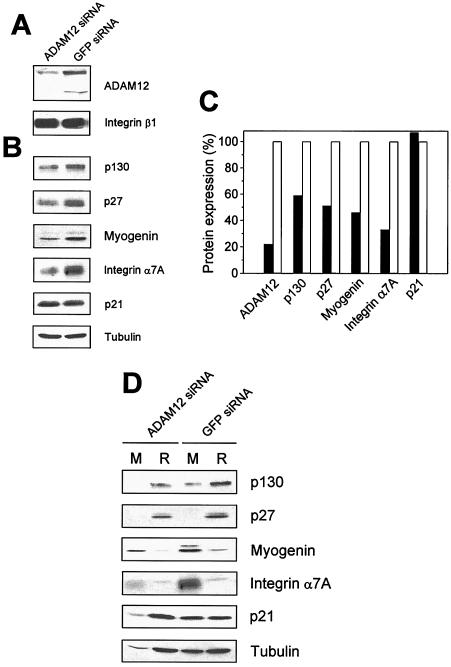
To examine whether ADAM12 is required for the formation of reserve cells or for differentiation, C2C12 cells were transfected with small interfering RNA (siRNA) designed to suppress ADAM12 expression. Two days after transfection, cells reached confluence and were transferred to differentiation medium containing 2% horse serum. After incubation for 2 days in differentiation medium, the level of ADAM12 in cells transfected with ADAM12 siRNA was decreased by ≈80% compared with the level in cells transfected with control siRNA against GFP (Fig. 2A). The expression levels of p130, p27, myogenin, and integrin α7A but not p21 in cells transfected with ADAM12 siRNA were significantly lower than they were in cells transfected with GFP siRNA (Fig. 2B and C). Consistent with the decreased expression of myogenin and integrin α7A, the amounts of differentiated myotubes that were formed in ADAM12 siRNA-transfected cells and separated by partial trypsinization were lower than the amounts of myotubes in GFP siRNA-transfected cells (Fig. 2D, see antitubulin immunoblot). p130 and p27, although diminished by ADAM12 siRNA, were still detected mainly in the population of nondifferentiated cells (Fig. 2D). Furthermore, p21, which was normally more abundant in myotubes than in reserve cells (see Fig. 1A) and was not downregulated by ADAM12 siRNA (Fig. 2B and C), was found in nondifferentiated cells after transfection with ADAM12 siRNA (Fig. 2D). Collectively, the results in Fig. 2 suggest that inhibition of ADAM12 expression in differentiating C2C12 cell cultures is accompanied by lower expression levels of both quiescence markers (p130 and p27) and differentiation markers (myogenin and integrin α7A).
FIG. 2.
Downregulation of ADAM12 by siRNA is correlated with decreased expression of p130, p27, myogenin, and integrin α7A, but not p21. C2C12 cells were transfected with small interfering RNA (siRNA) designed to suppress ADAM12 expression or with control siRNA against GFP. Two days after transfection, cells reached confluence and were transferred to differentiation medium containing 2% horse serum. After incubation for 2 additional days in differentiation medium, glycoprotein-enriched fractions obtained from C2C12 cells transfected with ADAM12 or GFP siRNA were subjected to Western blotting with anti-ADAM12 antibody (A). To control for equal protein loading, the same glycoprotein-enriched fractions were stained with anti-integrin β1 antibody. In cells transfected with GFP siRNA, the nascent (≈120 kDa) and the mature form (≈90 kDa) of ADAM12 are detected (8); in cells transfected with ADAM12 siRNA, the mature form is below the antibody detection limit. (B) The levels of expression of p130, p27, myogenin, integrin α7A, and p21 in cells transfected with ADAM12 or GFP siRNA were analyzed by Western blotting. Tubulin was a loading control. (C) The intensities of the bands in A and B were quantified by densitometry with ScionImage software. Solid and open bars represent cells transfected with ADAM12 and GFP siRNA, respectively. The experiment was repeated three times with similar results. Panels A to C show the results of a representative experiment. (D) Cells transfected with ADAM12 or GFP siRNA and incubated for 2 days in differentiation medium containing 2% horse serum were separated by mild trypsinization into myotube (M) and reserve cell (R) fractions, as described for Fig. 1. One fifth of the total myotube and reserve cell samples was then analyzed by Western blotting with the indicated antibodies. Notice that the amount of myotubes is lower in ADAM12 siRNA-transfected cells than in GFP siRNA-transfected cells, as documented by antitubulin blot.
We asked whether downregulation of p130 and p27 observed in cells transfected with ADAM12 siRNA can be explained by a decreased activity of ADAM12 metalloprotease in these cells. Recently, it was shown that hydroxamate-based inhibitors of metalloproteases (HIMPs) added at early stages of differentiation of C2C12 cells increase the size of myotubes, qualified as hypertrophy (25). The effective concentrations of the inhibitors required for this effect were in the micromolar range, which allowed the conclusion that the increased formation of myotubes was due to inhibition of ADAMs rather than matrix metalloproteases (12, 25).
As shown in Fig. 3, incubation of differentiating C2C12 cells for 4 days in the presence of TAPI, a member of the HIMP group of inhibitors, at concentrations ranging from 0 to 5 μM did not have a distinct effect on the expression level of p130 or p27 (Fig. 3C and D). In contrast, myogenin levels were increased by TAPI in a dose-dependent manner (Fig. 3C and D), which is a characteristic feature of a hypertrophic response in myoblasts (47). Microscopic examination of cells treated with TAPI confirmed the presence of large, branched myotubes, as reported earlier (Fig. 3A and B) (25). The response induced by general inhibition of metalloproteases is therefore very different from the effect observed after inhibition of ADAM12 expression by siRNA. Although the relative inhibition of ADAM12 and other ADAM metalloproteases by TAPI in C2C12 cells is not known, the results in Fig. 3 suggest that the metalloprotease domain of ADAM12 may not be essential for controlling the levels of p130, p27, myogenin, and integrin α7A.
Next, we examined the effect of enhanced expression of ADAM12 in C2C12 myoblasts on cell cycle progression. C2C12 cells transiently transfected with ADAM12 cDNA were incubated with BrdU, a marker of S-phase entry, followed by coimmunostaining of cells with anti-ADAM12 and anti-BrdU antibodies. Since anti-ADAM12 antibody used in this study poorly detects endogenous ADAM12 in C2C12 cells by immunofluorescence (8), the antibody-stained cells represented positively transfected cells with overexpression of ADAM12, whereas unstained cells corresponded to nontransfected cells with endogenous levels of ADAM12 (control cells).
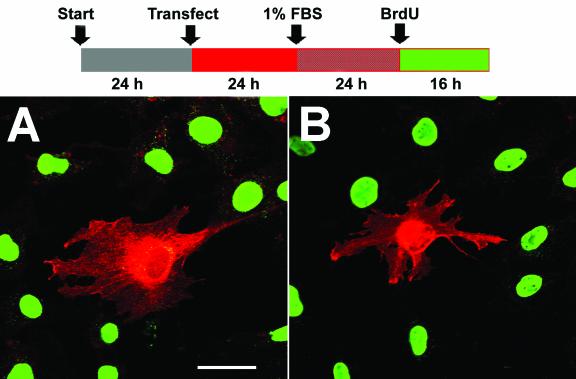
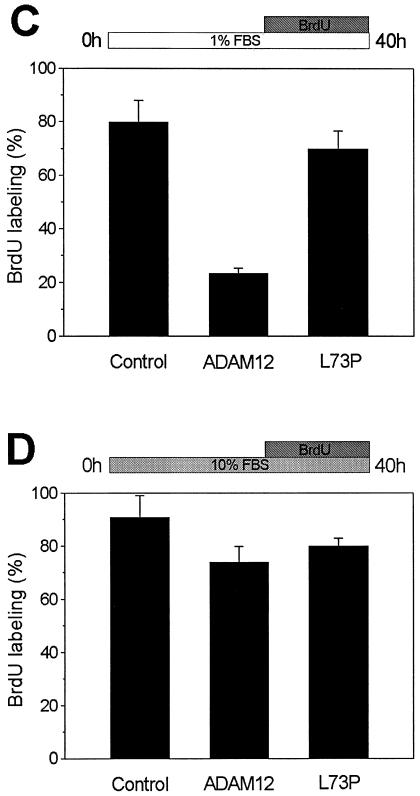
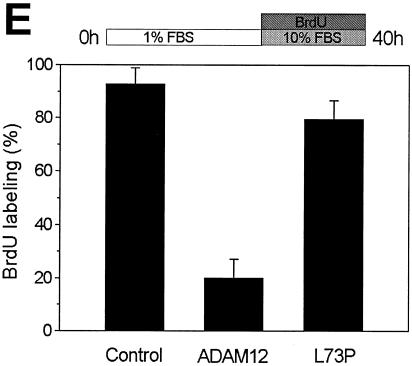
ADAM12 overexpression did not have a significant effect on BrdU labeling in cells incubated in the presence of 10% FBS (Fig. 4D). However, when cells were transferred to 1% FBS 24 h prior to BrdU labeling, only ≈25% of ADAM12-overexpressing cells and ≈80% of control cells showed nuclear incorporation of BrdU (Fig. 4A, B, and C). In contrast, overexpression of the L73P mutant form of ADAM12 that was trapped in the endoplasmic reticulum and was not transported to the cell surface (8) did not have a significant effect on BrdU labeling under these conditions. This suggests that inhibition of BrdU labeling by ADAM12 was not a mere consequence of protein overexpression, but rather a specific effect induced by the presence of a functional ADAM12 form at the cell surface. In addition, when cells were incubated first with 1% FBS and then transferred to 10% FBS during BrdU labeling, the inhibitory effect of ADAM12 on BrdU incorporation was evident (Fig. 4E).
FIG.4.
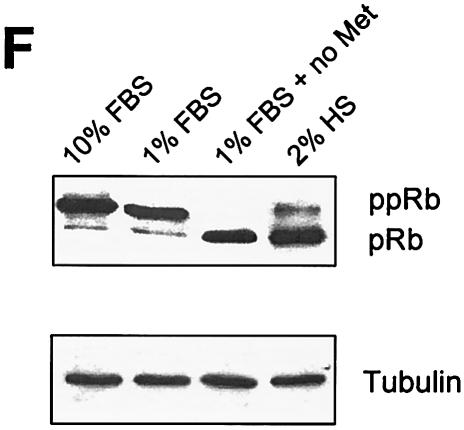
Enhanced expression of the wild type but not the L73P mutant of ADAM12 inhibits S-phase entry in C2C12 cells. (A and B) C2C12 cells were transfected with a vector encoding the wild-type ADAM12. Twenty-four hours after transfection, cells were transferred to medium containing 1% FBS. After an additional 24 h, cells were supplemented with 10 μM BrdU, and incubation was continued for 16 h. Cells were then fixed and coimmunostained with anti-ADAM12 (red) and anti-BrdU antibody (green); two representative microscopic fields are shown. Bar, 25 μm. (C) The percentage of BrdU-positive nuclei in the experiment shown in panels A and B was determined for cells that were not stained with ADAM12 antibody (control), for cells overexpressing ADAM12, and for cells overexpressing the L73P mutant form of ADAM12, which is not transported to the cell surface (8). (D) Transfected cells were incubated in the presence of 10% FBS, and BrdU labeling was determined as in panel C. (E) Following transfection, cells were incubated for 24 h with 1% FBS. BrdU labeling was then performed for 16 h in the presence of 10% FBS. In panels C, D, and E, at least 150 to 200 ADAM12-overexpressing cells in multiple fields were scored for BrdU staining. Results shown are the averages from five experiments ± standard errors. (F) Analysis of the phosphorylation status of the retinoblastoma protein in C2C12 cells incubated for 24 h in the presence of 10% FBS, 1% FBS, 1% FBS in the absence of methionine, or 2% horse serum. ppRb and pRb designate the hyperphosphorylated and hypophosphorylated forms of the retinoblastoma protein, respectively. Tubulin was a loading control.
The results in Fig. 4A to E indicate that ADAM12 overexpression inhibits entry into S phase in cells grown in 1% but not in 10% FBS. However, once the signaling events leading to the cell cycle arrest in ADAM12-overexpressing cells are initiated in 1% FBS, these events are irreversible and persist even in the presence of 10% FBS. To better understand the effect of FBS on ADAM12-mediated inhibition of the entry into S phase, we examined cell cycle progression in control, nontransfected C2C12 cells incubated in 1% or 10% FBS. We found that 1% FBS was almost as potent as 10% FBS in promoting the entry of C2C12 cells into S phase. First, cell cycle analysis by flow cytometry demonstrated that the number of cells in G0/G1 phase was only ≈10% higher when cells were incubated in 1% FBS compared to 10% FBS, and the number of cells in S phase was ≈10% lower (Table 1). Second, after 3 h of BrdU labeling, the percentages of BrdU-positive nuclei in cells incubated with 10% or 1% FBS were similar and equal to 40 and 39%, respectively (Table 1). After 16 h of BrdU labeling, 93% of cells incubated with 10% FBS and 78% of cells incubated with 1% FBS showed nuclear incorporation of BrdU (Table 1). Third, the extent of phosphorylation of the retinoblastoma protein (pRb) in cells incubated for 24 h in 1% and 10% FBS was similar. Western blotting with anti-pRb antibody demonstrated that under these two conditions, pRb was present predominantly in the hyperphosphorylated state (Fig. 4F). As expected, pRb became hypophosphorylated when cells were incubated for 4 days in differentiation medium containing 2% horse serum or when cells were forced into G0 by incubation in methionine-free medium with 1% FBS (Fig. 4F, see also Table 1) (10, 31). Taken together, these results indicate that the majority of C2C12 cells incubated for 16 h in 1% FBS are in a proliferative stage, in which the exit into quiescence or differentiation has not been initiated. Thus, inhibition of entry into S phase after overexpression of ADAM12 in these cells occurs under conditions that fully promote cell cycle progression. The inability of ADAM12 to induce similar inhibition in the presence of 10% FBS may be due simply to an excess of mitogens that override the inhibitory effect of ADAM12. To further investigate ADAM12-mediated inhibition of cell cycle progression, the experiments described in the rest of this study were performed in the presence of 1% FBS.
TABLE 1.
Effect of FBS concentration on C2C12 cell cycle progression
| FBS concn (%) | Mean % BrdU-labeled nucleia ± SE after:
|
% of cells in phase:
|
|||
|---|---|---|---|---|---|
| 3 h of labeling | 16 h of labeling | G1/G0 | S | G2/M | |
| 10 | 40 ± 7 | 93 ± 6 | 51.5 | 19.4 | 29.1 |
| 1 | 39 ± 5 | 78 ± 8 | 62.8 | 11.4 | 25.8 |
| 1 (no methionine) | <1 | ND | 97.0 | <2 | <2 |
ND, not determined.
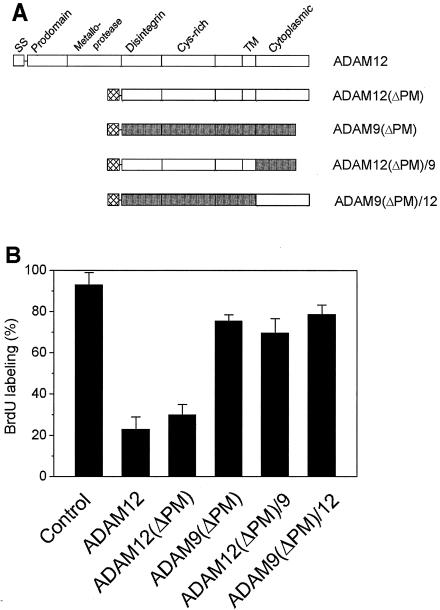
We explored the role of individual protein domains of ADAM12 in the inhibition of cell cycle progression. First, C2C12 cells were transfected to express ADAM12(ΔPM), an ADAM12 mutant in which the 424 N-terminal amino acids encoding the signal sequence, the prodomain, and the metalloprotease domain were replaced with an exogenous secretion signal. ADAM12(ΔPM) undergoes posttranslational modification similar to that of the full-length ADAM12 and is efficiently expressed at the cell surface (8). As shown in Fig. 5B, ADAM12(ΔPM) was almost as effective in inhibiting BrdU incorporation as the full-length ADAM12. This suggests that the prodomain and metalloprotease domain are not essential for inhibition of cell cycle progression by ADAM12.
FIG. 5.
Metalloprotease domain of ADAM12 is not essential for inhibition of cell cycle progression. (A) Expression constructs used to transfect C2C12 cells included mouse ADAM12, ADAM12(ΔPM) (a truncated form of ADAM12 lacking the prodomain and metalloprotease domain), ADAM9(ΔPM) (a truncated form of mouse ADAM9 lacking the prodomain and metalloprotease domain), a chimeric construct, ADAM12(ΔPM)/9, composed of the extracellular and transmembrane domains of ADAM12(ΔPM) and the cytoplasmic domain of ADAM9, and ADAM9(ΔPM)/12, containing the extracellular and transmembrane domains of ADAM9(ΔPM) and the cytoplasmic domain of ADAM12. (B) C2C12 cells were transfected with the expression constructs shown in panel A and incubated for 24 h in the presence of 1% FBS and then for 16 h with BrdU (as in Fig. 4A and B), fixed, and stained with mouse anti-BrdU antibody and either anti-ADAM12 antibody [ADAM12, ADAM12(ΔPM), and ADAM9(ΔPM)/12 transfectants] or anti-ADAM9 antibody [ADAM9(ΔPM) and ADAM12(ΔPM)/9 transfectants], as described in Materials and Methods. Cells were then viewed by confocal microscopy and scored for the expression of recombinant proteins and nuclear BrdU labeling. At least 150 to 200 positively transfected cells in multiple fields were analyzed in each experiment, and transfection with each construct was repeated at least three times.
We next asked whether the disintegrin, cysteine-rich, transmembrane, or cytoplasmic domain of ADAM12 can be replaced with the corresponding domains from ADAM9. Unlike ADAM12, ADAM9 is ubiquitously expressed in many tissues and cell types. We evaluated nuclear BrdU incorporation in C2C12 cells expressing ADAM9(ΔPM), a deletion mutant of ADAM9 containing an N-terminal truncation equivalent to that in ADAM12(ΔPM), a chimeric ADAM9(ΔPM)/12 protein containing the extracellular and transmembrane domains of ADAM9(ΔPM) and the cytoplasmic domain of ADAM12, and the ADAM12(ΔPM)/9 chimera, containing the extracellular and transmembrane domains of ADAM12(ΔPM) and the cytoplasmic domain of ADAM9 (see Fig. 5A). While all three of these recombinant proteins were intact and efficiently expressed at the cell surface (8), they did not have a significant effect on C2C12 cell entry into S phase. After 16 h of incubation in the presence of BrdU and 1% FBS, ≈80% of control cells and ≈75% of ADAM9(ΔPM)-, ≈73% of ADAM9(ΔPM)/12-, and ≈68% of ADAM12(ΔPM)/9-expressing cells showed clear BrdU staining (Fig. 5B). This suggests that the inhibition of cell cycle progression requires the presence of the region extending from the disintegrin to the transmembrane domain of ADAM12 (or a shorter, unidentified fragment from that region), as well as the cytoplasmic domain of ADAM12, and the effect of ADAM12 cannot be replaced by using the corresponding domains of ADAM9.
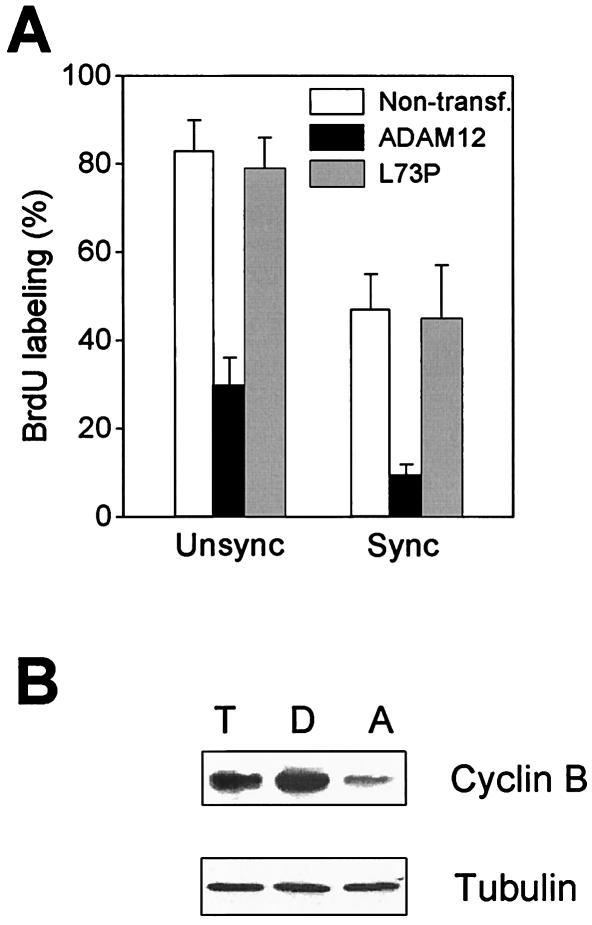
Next, we examined whether ADAM12-induced arrest occurred between the M and S phases of the cell cycle. Transfected, unsynchronized cells were subjected to repeated shake-off to dislodge the population of cells in mitosis (31, 38). Detached cells were then allowed to reattach in the presence of 1% FBS and BrdU and to progress into G1 and then into S phase. Western blotting with antibody against cyclin B, a marker of M phase, demonstrated that the level of cyclin B was much higher in detached cells than in cells that remained attached on the plate after shake-off (Fig. 6B), confirming that the majority of the detached cells were in M phase. As shown in Fig. 6A, while 40 to 45% of the M-phase-synchronized control cells or L73P mutant-transfected cells became BrdU positive 8 h after mitotic shake-off, only ≈10% of ADAM12-transfected cells showed nuclear incorporation of BrdU during the same period. This suggests that enhanced expression of ADAM12 arrests C2C12 cells at the G0 or G1 phase of the cycle.
FIG. 6.
Overexpression of ADAM12 arrests C2C12 cells between M and S phases of the cycle. (A) Cells were transfected with the full-length ADAM12 or the L73P ADAM12 mutant and incubated for 24 h in the presence of 1% FBS and then for 16 h with BrdU (as in Fig. 4A and B) (unsynchronized cells [unsync]). Alternatively, transfected cells were incubated for 24 h in the presence of 10% FBS, mitotic cells were dislodged by shake-off, plated on a coverslip, and incubated for 8 h in the presence of 1% FBS (M-synchronized cells [sync]). Cells were then fixed and stained with anti-ADAM12 and anti-BrdU antibodies. The percentage of BrdU labeling among cells that were not stained with anti-ADAM12 antibody (open bars), among ADAM12-overexpressing cells (solid bars), and among L73P-overexpressing cells (grey bars) was determined. At least 150 to 200 transfected cells in multiple fields were analyzed in the experiment with unsynchronized cells, and 50 to 100 transfected cells were scored in the experiments with M-synchronized cells. Results shown are the averages from four experiments ± standard errors. (B) Total cells before the shake-off procedure (T), dislodged cells (D), and cells that remained attached after shake-off (A) were analyzed by Western blotting with anti-cyclin B to verify the effective synchronization of the dislodged cells in M phase. Tubulin was a loading control.
G0/G1 arrest after overexpression of ADAM12 indicates that cells might have either exited into differentiation (G1 arrest) or entered quiescence (G0 arrest). To distinguish between these two possibilities, we evaluated the effect of enhanced expression of ADAM12 on the levels of MyoD as well as differentiation markers (myogenin and p21) and quiescence markers (p130 and p27). The level of MyoD oscillates during the myoblast cell cycle and is highest at the end of M phase and in early to mid-G1 at the time when the exit into differentiation occurs (31). Conversely, upon entry into G0, MyoD drops to an undetectable level, which is one of the hallmarks of the quiescent state in myoblastic cell lines in vitro (10, 31, 67) and in satellite cells in vivo (14, 64).
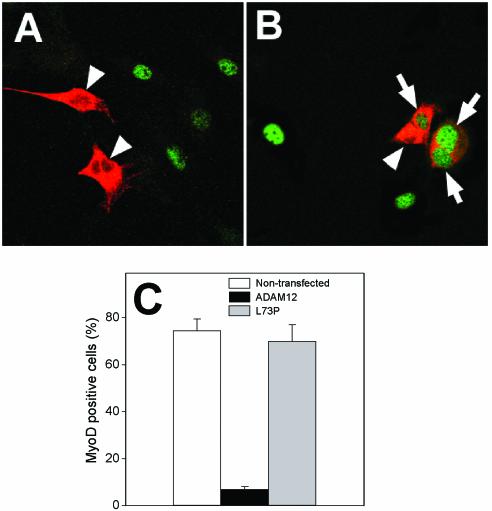
When C2C12 cells were transfected with ADAM12 cDNA, synchronized at M phase by mitotic shake-off, and then allowed to attach, ≈75% of ADAM12-unstained cells and ≈70% of L73P mutant-transfected cells were MyoD positive 4 h after shake-off (Fig. 7B and C). In contrast, less than 10% of ADAM12-overexpressing cells showed nuclear MyoD staining at that time point (Fig. 7A and C) or after an additional 4 h of incubation (result not shown). Consistently, we did not detect any induction of myogenin, a transcriptional target of MyoD and early marker of differentiation, or p21 in ADAM12-overexpressing cells (result not shown). Since induction of p21 is a prerequisite for the irreversible cell cycle withdrawal that proceeds differentiation (22, 49, 57, 62, 63), this suggests that ADAM12-induced cell cycle arrest is not compatible with myogenic differentiation.
FIG. 7.
Overexpression of ADAM12 reduces MyoD expression. C2C12 cells transfected with ADAM12 (A) or the L73P ADAM12 mutant (B) were synchronized by mitotic shake-off, plated on coverslips, incubated for 4 h in the presence of 1% FBS, fixed, and stained with anti-ADAM12 (red) or anti-myoD antibody (green). Arrows indicate L73P-transfected cells with nuclear MyoD staining. Arrowheads mark ADAM12- or L73P-overexpressing cells, which are MyoD negative. (C) The percentage of MyoD-positive cells among nontransfected, ADAM12-overexpressing, and L73P-overexpressing cells was quantified 4 h after shake-off. The results shown are the averages from three experiments ± standard errors; 50 to 100 transfected cells were analyzed in each experiment.
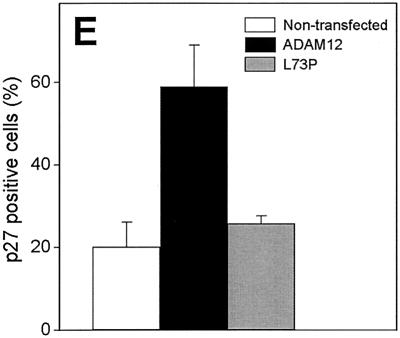
We examined whether enhanced expression of ADAM12 leads to induction of p130 and p27. p130 is a critical determinant of G0 arrest (33, 41, 58) and appears to be required for the induction and/or maintenance of quiescence in C2C12 reserve cells (10). p27, in addition to being upregulated in terminally differentiated myoblasts (13, 17, 69), is also expressed in satellite cells that, when activated, lose p27 expression and reenter the cell cycle (11, 59). When ADAM12-transfected, subconfluent C2C12 cells were incubated for 24 h in 1% FBS, p130 was detected in 25 to 30% of the cells that were not stained with ADAM12 antibody or that were transfected with the L73P ADAM12 mutant (Fig. 8G to L). In contrast, p130 was detected in ≈60% of ADAM12-overexpressing cells (Fig. 8A to F). Similarly, while only 20 to 25% of nontransfected or L73P-transfected cells were stained with p27 antibody (Fig. 9C and D), the percentage of p27-positive cells among ADAM12-transfected cells reached ≈60% (Fig. 9A and B). These results indicate that enhanced expression of ADAM12 is sufficient for upregulation of p130 and p27 expression in C2C12 cells.
FIG. 9.
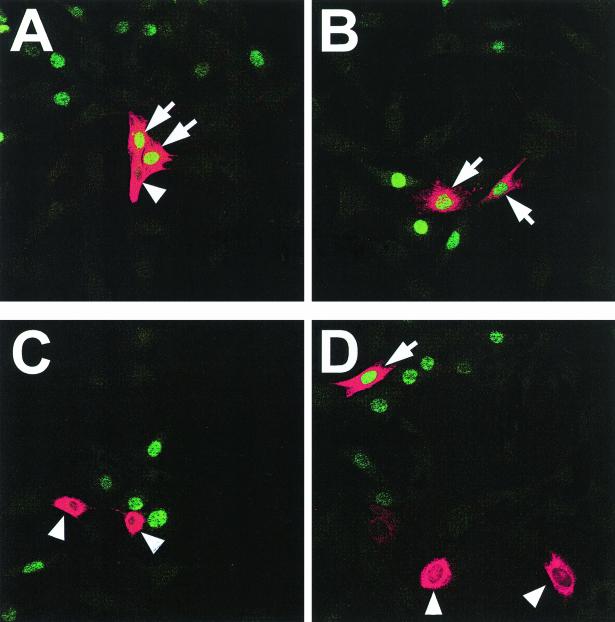
Overexpression of ADAM12 induces p27 expression. Subconfluent C2C12 cells transfected with ADAM12 (A and B) or the L73P ADAM12 mutant (C and D) and incubated in medium containing 1% FBS were fixed and costained with anti-ADAM12 antibody (red) and anti-p27 antibody (green). Two representative microscopic fields are shown for ADAM12 and L73P transfections. Arrows indicate ADAM12- and L73P-overexpressing cells with induced expression of p27. Arrowheads mark ADAM12- and L73P-overexpressing cells that are p27 negative. (E) The percentage of p27-positive cells among ADAM12-overexpressing (solid bar), L73P-overexpressing (grey bar), and cells that were not stained with ADAM12 antibody (nontransfected cells, open bar) was quantified. Results shown are the averages from three experiments ± standard errors. At least 100 transfected cells were analyzed in each experiment.
DISCUSSION
The generation of reserve cells has been best described and characterized for the C2 and C2C12 cell lines (3, 10, 18, 67), which are frequently used as models of muscle differentiation. Although these are transformed, tumorigenic cells and many aspects of their differentiation program may differ from the differentiation program present in normal myoblasts, the presence of reserve cells was also observed during differentiation of primary myoblasts (31). Therefore, induction of reserve cells may represent a more general phenomenon which is not restricted solely to C2C12 cells. The mechanism by which a population of myoblasts cultured in vitro and induced to differentiate do not form myotubes and become quiescent reserve cells is, however, not known. Since reserve cells can resume proliferation when replated at a low density in growth medium and can give rise to myotubes and a new generation of reserve cells when transferred to differentiation medium, they do not seem to have any “defects” in the pathways controlling the differentiation program. Moreover, upregulation of p130 and p27 and downregulation of MyoD indicate that generation of a pool of reserve cells is an active process that resembles satellite cell renewal. Our results suggest that ADAM12, a membrane protein mediating cell adhesion and cell-cell communication, may be involved in the generation of reserve cells during differentiation of C2C12 myoblasts.
Despite recent studies aimed at the biochemical characterization of ADAM12, the role of ADAM12 in development and/or regeneration of skeletal muscle is not clear. ADAM12 is as an active metalloprotease that cleaves insulin-like growth factor binding proteins 3 and 5 (40, 56) and heparin binding-epidermal growth factor (2). The disintegrin and cysteine-rich domains of ADAM12 support cell adhesion by binding to integrin α9β1 and syndecan-4, respectively (15, 16, 26, 27). The ADAM12 cytoplasmic domain interacts with protein tyrosine kinase Src (28), phosphatidylinositol 3-kinase (29), and cytoskeletal proteins α-actinin-2 (19) and α-actinin-1 (9).
According to an early report, C2C12 clones with constitutive expression of a truncated, 67-kDa version of ADAM12 lacking the N-terminal prodomain and metalloprotease domain showed increased cell-cell fusion (66). Intriguingly, stable transfection of C2C12 cells with the 120-kDa full-length ADAM12 inhibited cell fusion (66). Therefore, it was postulated that the active form of ADAM12 is the one that lacks the pro- and metalloprotease domains and that overexpression of the full-length protein exerts a dominant-negative effect on the function of the endogenous ADAM12 (66). However, while the cleavage of ADAM12 between the pro- and metalloprotease domains and generation of the 90-kDa form have been observed frequently (8, 24, 40, 66) (and are also typical of most other ADAM proteins), intracellular processing of ADAM12 leading to the removal of the metalloprotease domain has not been well documented, and therefore the participation of ADAM12 in cell-cell fusion is not clear. Finally, recent expression of ADAM12 in transgenic mice under control of the muscle creatine kinase promoter (which drives ADAM12 overexpression in differentiated myofibers but not in satellite cells) results in pronounced accumulation of adipocytes in skeletal muscle (30). Interestingly, overexpression of ADAM12 in myofibers of dystrophin-deficient mdx mice alleviates the muscle pathology in these animals (34).
To gain new insight into the function of ADAM12 during myoblast differentiation, we examined the distribution of ADAM12 between different subpopulations of differentiating C2C12 cultures. We found that expression of ADAM12 is higher in quiescent reserve cells than in myotubes. This contradicts a previous report, in which immunofluorescence analysis of differentiating C2C12 cell cultures indicated staining in myotubes but not in mononucleated myoblasts (66). Our results agree, on the other hand, with reports of ADAM12 being present in undifferentiated cells, being elevated during the first 1 to 2 days after transfer of cells to differentiation medium (when few myotubes are formed) and decreasing to almost undetectable levels as differentiation proceeds and the number of myotubes increases (19, 66).
In this study, we were not able to detect endogenous ADAM12 by immunocytochemistry or by Western blotting of total cell lysates. However, ADAM12 was readily observed in immunoblots of glycoprotein-enriched fractions obtained from myoblasts or reserve cells. Positive identification of the ≈120- and ≈90-kDa protein bands as the mature and the processed forms of ADAM12, respectively, was confirmed by using ADAM12-transfected cells as positive controls and ADAM12 antisense clones as negative controls (8). Importantly, Northern blot analysis of ADAM12 mRNA expression (Fig. 1B) was in agreement with the results obtained by Western blotting (Fig. 1A) and further suggested that ADAM12 is expressed predominantly in nondifferentiated C2C12 myoblasts and in reserve cells and to a much lesser extent in differentiated myotubes. Interestingly, while ADAM12 was not detected in myofibers of normal muscle, a strong anti-ADAM12 antibody staining was observed in regenerating, newly formed myofibers in dystrophic mdx muscle (19). Thus, the mechanisms regulating ADAM12 expression in normal and dystrophic myofibers may be different.
To examine whether ADAM12 is required for the formation of reserve cells or for differentiation, we used the siRNA approach. We showed that C2C12 cells in which the expression of ADAM12 has been attenuated by siRNA have a decreased potential both to differentiate and to form quiescent reserve cells. Reduced differentiation of C2C12 cells after inhibition of ADAM12 expression is in agreement with a previous report in which C2C12 clones stably transfected with an ADAM12 antisense mRNA construct showed decreased formation of myotubes (66). However, while this result was interpreted as a direct impairment of cell-cell fusion in ADAM12-deficient myoblasts (66), our results rather indicate that decreased expression of ADAM12 leads to inhibition of an early step of differentiation that involves expression of myogenin. Interestingly, p21 is upregulated normally in ADAM12 siRNA-transfected cells, which suggests that p21-mediated cell cycle exit does not require the presence of ADAM12.
The decreased potential of ADAM12 siRNA-transfected C2C12 cells to form reserve cells is a novel finding that has not been observed before. We show that decreased expression of ADAM12 in confluent C2C12 cells that are transferred to differentiation medium results in diminished expression of p27 and p130, two quiescent cell markers. Furthermore, enhanced expression of ADAM12 leads to cell cycle arrest, upregulation of p27 and p130 expression, and downregulation of MyoD (a quiescence-like phenotype). Remarkably, overexpression of ADAM12 does not induce p21 or myogenin (markers of differentiation). Based on these results, we conclude that while ADAM12 function appears both necessary and sufficient for generation of reserve C2C12 cells, it is required but not sufficient for differentiation.
Induction of the quiescence-like phenotype in C2C12 cells overexpressing ADAM12 may help to explain previous observations in which stable transfection of C2C12 cells with the full-length ADAM12 led to inhibition of myoblast fusion (66). As quiescence and differentiation represent two alternative outcomes for cells after transfer to differentiation medium and they are mutually exclusive, the increased propensity of ADAM12-overexpressing cells to enter G0 should clearly be manifested as a decreased ability to form myotubes.
Inhibition of differentiation in ADAM12-deficient myoblasts reported here and observed previously by others (66) is not supported by a recent report describing generation of mice with targeted disruption of the ADAM12 gene (35). It has to be stressed, however that although most of the muscles in ADAM12−/− mice develop normally, the extent of compensation of the lack of ADAM12 by other ADAM family members is currently not known (35). The lack of a clear muscle phenotype in ADAM12 knockout mice, on the other hand, is not inconsistent with the postulated role of ADAM12 in determination of quiescent reserve cells in vitro or self-renewal of satellite cells in vivo. If ADAM12 is involved in induction of quiescence in myoblasts, then the genetic manipulations to uncover this role of ADAM12 should rather consist of transgenic expression of ADAM12 in myoblasts prior to differentiation and testing muscle regenerative potential of such ADAM12 transgenic animals.
This situation is somewhat a reverse of the situation involving MyoD expression, as the induction of myoblast quiescence is associated with downregulation of MyoD. Indeed, while MyoD−/− mice develop normally due a compensatory role of Myf-5 (51), MyoD−/− mice interbred with mdx dystrophic mice lacking dystrophin display severe defects in muscle regeneration due to increased propensity of satellite cell for self-renewal (i.e., G0 entry) rather than progression through the myogenic program (43, 52, 65). A similar susceptibility for self-renewal of the myoblasts in ADAM12-overexpressing mdx mice should therefore yield a phenotype similar to that observed in mdx/MyoD−/− animals. Mice with deficient expression of ADAM12 (ADAM12−/− or mdx/ADAM12−/− genotype) or mdx mice with overexpression of ADAM12 targeted selectively to differentiated myofibers with muscle creatine kinase promoter (34) may not need to develop symptoms observed in mdx/MyoD−/− animals.
To determine whether decreased expression of quiescent cell markers in ADAM12-deficient cells can be explained by the lack of ADAM12 metalloprotease activity, C2C12 cells were treated with TAPI, a member of the hydroxamate-based inhibitors of metalloproteases (HIMPs) (12, 25). As shown previously (25) and confirmed in this report, TAPI added to myoblasts at early stages of differentiation induces myotube hypertrophy. The mechanism of HIMP-induced hypertrophy does not involve IGF-1, calcineurin, or tumor necrosis factor alpha (25), but rather myostatin, a member of the transforming growth factor beta family and a negative regulator of skeletal muscle growth (37). When added ectopically to C2C12 cells, myostatin inhibits both cell proliferation (61) and differentiation (36). It has to be stressed, however, that myostatin-treated, differentiation-inhibited myoblasts are different from quiescent reserve cells, and myostatin does not function to increase the reserve cell population (36).
Myostatin is synthesized as a precursor protein and needs to be proteolytically processed to generate a biologically active form (37). Although it is believed that myostatin, like other members of the transforming growth factor beta superfamily, is processed intracellularly by furins, addition of HIMPs to C2C12 cells clearly results in the reduction of the processing of the endogenous myostatin produced by these cells (25). The reduced amounts of active myostatin in HIMP-treated cells have therefore been attributed to the hypertrophic growth of myotubes.
Since inhibition of ADAM12 expression by siRNA leads to inhibition of both differentiation and formation of reserve cells, a response that is very different from the effect induced by general inhibition of ADAMs by TAPI, we conclude that ADAM12 is presumably not involved in myostatin processing. Thus, myotube hypertrophy most likely results from inhibition of other ADAM metalloproteases by TAPI. Furthermore, the lack of the effect of TAPI on the expression levels of p27 and p130 (Fig. 3) and the opposing effects of TAPI and ADAM12 siRNA on the expression of myogenin and integrin α7A (Fig. 2 and 3) lead us to the conclusion that the metalloprotease domain of ADAM12 is not essential for controlling the levels of p130, p27, myogenin, and integrin α7A. It has to be stressed, however, that the relative inhibition of ADAM12 and other ADAMs by TAPI in C2C12 cells is not known, and it is possible that TAPI does not induce a response similar to the response generated by ADAM12 siRNA because of an ineffective inhibition of ADAM12 metalloprotease activity in TAPI-treated cells.
In summary, our results suggest that ADAM12 plays an important role in the induction of the quiescent state during myogenic differentiation in vitro. Although the mechanism by which ADAM12 induces exit into quiescence is not clear, our results point to a potential role of ADAM12-mediated cell-cell interactions in determination of the pool of reserve cells and, possibly, the renewal of the satellite cell compartment.
Acknowledgments
This work was supported by NIH COBRE Award P20 RR017708 and matching support from the State of Kansas and by AHA Award 0160386Z.
Footnotes
This is contribution 03-260-J from the Kansas Agricultural Experiment Station.
REFERENCES
- 1.Arnold, H. H., and B. Winter. 1998. Muscle differentiation: more complexity to the network of myogenic regulators. Curr. Opin. Genet. Dev. 8:539-544. [DOI] [PubMed] [Google Scholar]
- 2.Asakura, M., M. Kitakaze, S. Takashima, Y. Liao, F. Ishikura, T. Yoshinaka, H. Ohmoto, K. Node, K. Yoshino, H. Ishiguro, H. Asanuma, S. Sanada, Y. Matsumura, H. Takeda, S. Beppu, M. Tada, M. Hori, and S. Higashiyama. 2002. Cardiac hypertrophy is inhibited by antagonism of ADAM12 processing of HB-EGF: metalloproteinase inhibitors as a new therapy. Nat. Med. 8:35-40. [DOI] [PubMed] [Google Scholar]
- 3.Beauchamp, J. R., L. Heslop, D. S. Yu, S. Tajbakhsh, R. G. Kelly, A. Wernig, M. E. Buckingham, T. A. Partridge, and P. S. Zammit. 2000. Expression of CD34 and Myf5 defines the majority of quiescent adult skeletal muscle satellite cells. J. Cell Biol. 151:1221-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bischoff, R. 1994. The satellite cell and muscle regeneration, p. 97-118. In A. G. Engel and C. Franzini-Armstrong (ed.), Mycology, vol. 1. McGraw-Hill, New York, N.Y.
- 5.Black, R. A., and J. M. White. 1998. ADAMs: focus on the protease domain. Curr. Opin. Cell Biol. 10:654-659. [DOI] [PubMed] [Google Scholar]
- 6.Bornemann, A., R. Kuschel, and A. Fujisawa-Sehara. 2000. Analysis for transcript expression of meltrin alpha in normal, regenerating, and denervated rat muscle. J. Muscle Res. Cell Motil. 21:475-480. [DOI] [PubMed] [Google Scholar]
- 7.Burkin, D. J., and S. J. Kaufman. 1999. The α7β1 integrin in muscle development and disease. Cell Tissue Res. 296:183-190. [DOI] [PubMed] [Google Scholar]
- 8.Cao, Y., Q. Kang, Z. Zhao, and A. Zolkiewska. 2002. Intracellular processing of metalloprotease disintegrin ADAM12. J. Biol. Chem. 277:26403-26411. [DOI] [PubMed] [Google Scholar]
- 9.Cao, Y., Q. Kang, and A. Zolkiewska. 2001. Metalloprotease-disintegrin ADAM12 interacts with α-actinin-1. Biochem. J. 357:353-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carnac, G., L. Fajas, A. l'Honore, C. Sardet, N. J. C. Lamb, and A. Fernandez. 2000. The retinoblastoma-like protein p130 is involved in the determination of reserve cells in differentiating myoblasts. Curr. Biol. 10:543-546. [DOI] [PubMed] [Google Scholar]
- 11.Chakravarthy, M. V., M. L. Fiorotto, R. J. Schwartz, and F. W. Booth. 2001. Long-term insulin-like growth factor-I expression in skeletal muscles attenuates the enhanced in vitro proliferation ability of the resident satellite cells in transgenic mice. Mech. Ageing Dev. 122:1303-1320. [DOI] [PubMed] [Google Scholar]
- 12.Cherney, R. J., L. Wang, D. T. Meyer, C. B. Xue, E. C. Arner, R. A. Copeland, M. B. Covington, K. D. Hardman, Z. R. Wasserman, B. D. Jaffee, and C. P. Decicco. 1999. Macrocyclic hydroxamate inhibitors of matrix metalloproteinases and TNF-α production. Bioorg. Med. Chem. Lett. 9:1279-1284. [DOI] [PubMed] [Google Scholar]
- 13.Chu, C. Y., and R. W. Lim. 2000. Involvement of p27(kip1) and cyclin D3 in the regulation of cdk2 activity during skeletal muscle differentiation. Biochim. Biophys. Acta 1497:175-185. [DOI] [PubMed] [Google Scholar]
- 14.Cornelison, D. D., and B. J. Wold. 1997. Single-cell analysis of regulatory gene expression in quiescent and activated mouse skeletal muscle satellite cells. Dev. Biol. 191:270-283. [DOI] [PubMed] [Google Scholar]
- 15.Eto, K., C. Huet, T. Tarui, S. Kupriyanov, H. Z. Liu, W. Puzon-McLaughlin, X. P. Zhang, D. Sheppard, E. Engvall, and Y. Takada. 2002. Functional classification of ADAMs based on a conserved motif for binding to integrin α9β1: implications for sperm-egg binding and other cell interactions. J. Biol. Chem. 277:17804-17810. [DOI] [PubMed] [Google Scholar]
- 16.Eto, K., W. Puzon-McLaughlin, D. Sheppard, A. Sehara-Fujisawa, X. P. Zhang, and Y. Takada. 2000. RGD-independent binding of integrin α9β1 to the ADAM-12 and -15 disintegrin domains mediates cell-cell interaction. J. Biol. Chem. 275:34922-34930. [DOI] [PubMed] [Google Scholar]
- 17.Franklin, D. S., and Y. Xiong. 1996. Induction of p18INK4c and its predominant association with CDK4 and CDK6 during myogenic differentiation. Mol. Biol. Cell 7:1587-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friday, B. B., G. K. Pavlath. 2001. A calcineurin- and NFAT-dependent pathway regulates Myf5 gene expression in skeletal muscle reserve cells. J. Cell Sci. 114:303-310. [DOI] [PubMed] [Google Scholar]
- 19.Galliano, M. F., C. Huet, J. Frygelius, A. Polgren, U. M. Wewer, and E. Engvall. 2000. Binding of ADAM12, a marker of skeletal muscle regeneration, to the muscle-specific actin-binding protein, α-actinin-2, is required for myoblast fusion. J. Biol. Chem. 275:13933-13939. [DOI] [PubMed] [Google Scholar]
- 20.Gilpin, B. J., F. Loechel, M. G. Mattei, E. Engvall, R. Albrechtsen, and U. M. Wewer. 1998. A novel, secreted form of human ADAM12 (meltrin alpha) provokes myogenesis in vivo. J. Biol. Chem. 273:157-166. [DOI] [PubMed] [Google Scholar]
- 21.Grounds, M. D. 1999. Muscle regeneration: molecular aspects and therapeutic implications. Curr. Opin. Neurol. 12:535-543. [DOI] [PubMed] [Google Scholar]
- 22.Halevy, O., B. G. Novitch, D. B. Spicer, S. X. Skapek, J. Rhee, G. J. Hannon, D. Beach, and A. B. Lassar. 1995. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science 267:1018-1021. [DOI] [PubMed] [Google Scholar]
- 23.Hawke, T. J., and D. J. Garry. 2001. Myogenic satellite cells: physiology to molecular biology. J. Appl. Physiol. 91:534-551. [DOI] [PubMed] [Google Scholar]
- 24.Hougaard, S., F. Loechel, X. Xu, R. Tajima, R. Albrechtsen, and U. M. Wewer. 2000. Trafficking of human ADAM12-L: retention in the trans-Golgi network. Biochem. Biophys. Res. Commun. 275:261-267. [DOI] [PubMed] [Google Scholar]
- 25.Huet, C., Z. F. Li, H. Z. Liu, R. A. Black, M. F. Galliano, and E. Engvall. 2001. Skeletal muscle cell hypertrophy induced by inhibitors of metalloproteases: myostatin as a potential mediator. Am. J. Physiol. Cell Physiol. 281:C1624-C1634. [DOI] [PubMed] [Google Scholar]
- 26.Iba, K., R. Albrechtsen, B. Gilpin, C. Fröhlich, F. Loechel, A. Zolkiewska, K. Ishihuro, T. Kojima, W. Liu, J. K. Langford, R. D. Sanderson, C. Brakebusch, R. Fässler, and U. M. Wewer. 2000. The cysteine-rich domain of human ADAM12 supports cell adhesion through syndecans and triggers signaling events that lead to β1 integrin-dependent cell spreading. J. Cell Biol. 149:1143-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iba, K., R. Albrechtsen, B. J. Gilpin, F. Loechel, and U. M. Wewer. 1999. Cysteine-rich domain of human ADAM12 (meltrin α) supports tumor cell adhesion. Am. J. Pathol. 154:1489-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang, Q., Y. Cao, and A. Zolkiewska. 2000. Metalloprotease-disintegrin ADAM12 binds to the SH3 domain of Src and activates Src tyrosine kinase in C2C12 cells. Biochem. J. 352:883-892. [PMC free article] [PubMed] [Google Scholar]
- 29.Kang, Q., Y. Cao, and A. Zolkiewska. 2001. Direct interaction between the cytoplasmic tail of ADAM12 and the SH3 domain of p85α activates phosphatidylinositol 3-kinase in C2C12 cells. J. Biol. Chem. 276:24466-24472. [DOI] [PubMed] [Google Scholar]
- 30.Kawaguchi, N., X. Xu, R. Tajima, P. Kronqvist, C. Sundberg, F. Loechel, R. Albrechtsen, and U. M. Wewer. 2002. ADAM12 protease induces adipogenesis in transgenic mice. Am. J. Pathol. 160:1895-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitzmann, M., G. Carnac, M. Vandromme, M. Primig, N. J. C. Lamb, and A. Fernandez. 1998. The muscle regulatory factors MyoD and myf-5 undergo distinct cell cycle-specific expression in muscle cells. J. Cell Biol. 142:1447-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitzmann, M., and A. Fernandez. 2001. Crosstalk between cell cycle regulators and the myogenic factor MyoD in skeletal myoblasts. Cell. Mol. Life Sci. 58:571-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kops, G. J., R. H. Medema, J. Glassford, M. A. Essers, P. F. Dijkers, P. J. Coffer, E. W. Lam, and B. M. Burgering. 2002. Control of cell cycle exit and entry by protein kinase B-regulated forkhead transcription factors. Mol. Cell. Biol. 22:2025-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kronqvist, P., N. Kawaguchi, R. Albrechtsen, X. Xu, H. D. Schroder, B. Moghadaszadeh, F. C. Nielsen, C. Frohlich, E. Engvall, and U. M. Wewer. 2002. ADAM12 alleviates the skeletal muscle pathology in mdx dystrophic mice. Am. J. Pathol. 161:1535-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurisaki, T., A. Masuda, K. Sudo, J. Sakagami, S. Higashiyama, Y. Matsuda, A. Nagabukuro, A. Tsuji, Y. Nabeshima, M. Asano, Y. Iwakura, and A. Sehara-Fujisawa. 2003. Phenotypic analysis of meltrin α (ADAM12)-deficient mice: involvement of meltrin α in adipogenesis and myogenesis. Mol. Cell. Biol. 23:55-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langley, B., M. Thomas, A. Bishop, M. Sharma, S. Gilmour, and R. Kambadur. 2002. Myostatin inhibits myoblast differentiation by downregulating MyoD expression. J. Biol. Chem. 277:49831-49840. [DOI] [PubMed] [Google Scholar]
- 37.Lee, S. J., and A. C. McPherron. 2001. Regulation of myostatin activity and muscle growth. Proc. Natl. Acad. Sci. USA 98:9306-9311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindon, C., D. Montarras, and C. Pinset. 1998. Cell cycle-regulated expression of the muscle determination factor Myf5 in proliferating myoblasts. J. Cell Biol. 140:111-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loechel, F., J. W. Fox, G. Murphy, R. Albrechtsen, and U. M. Wewer. 2000. ADAM12-S cleaves IGFBP-3 and IGFBP-5 and is inhibited by TIMP-3. Biochem. Biophys. Res. Commun. 278:511-515. [DOI] [PubMed] [Google Scholar]
- 40.Loechel, F., B. J. Gilpin, E. Engvall, R. Albrechtsen, and U. M. Wewer. 1998. Hum. ADAM12 (meltrin alpha) is an active metalloprotease. J. Biol. Chem. 273:16993-16997. [DOI] [PubMed] [Google Scholar]
- 41.Mayol, X., and X. Grana. 1998. The p130 pocket protein: keeping order at cell cycle exit/re-entrance transitions. Front. Biosci. 3:D11-D24. [DOI] [PubMed] [Google Scholar]
- 42.Medema, R. H., G. J. Kops, J. L. Bos, and B. M. Burgering. 2000. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature 404:782-787. [DOI] [PubMed] [Google Scholar]
- 43.Megeney, L. A., B. Kablar, K. Garrett, J. E. Anderson, and M. A. Rudnicki. 1996. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 10:1173-1183. [DOI] [PubMed] [Google Scholar]
- 44.Miskimins, W. K., G. Wang, M. Hawkinson, and R. Miskimins. 2001. Control of cyclin-dependent kinase inhibitor p27 expression by cap-independent translation. Mol. Cell. Biol. 21:4960-4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Molkentin, J. D., and E. N. Olson. 1996. Defining the regulatory networks for muscle development. Curr. Opin. Genet. Dev. 6:445-453. [DOI] [PubMed] [Google Scholar]
- 46.Montgomery, R. A., and H. C. Dietz. 1997. Inhibition of fibrillin 1 expression with U1 snRNA as a vehicle for the presentation of antisense targeting sequence. Hum. Mol. Genet. 6:519-525. [DOI] [PubMed] [Google Scholar]
- 47.Musaro, A., and N. Rosenthal. 1999. Maturation of the myogenic program is induced by postmitotic expression of insulin-like growth factor I. Mol. Cell. Biol. 19:3115-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olashaw, N., and W. Pledger. 2002. Paradigms of growth control: relation to Cdk activation. Science's STKE. (Online, http://stke.sciencemag.org/cgi/content/full/OC_sigtrans;2002/134/re7.) [DOI] [PubMed]
- 49.Parker, S. B., G. Eichele, P. Zhang, A. Rawls, A. T. Sands, A. Bradley, E. N. Olson, J. W. Harper, and S. J. Elledge. 1995. p53-independent expression of p21Cip1 in muscle and other terminally differentiating cells. Science 267:1024-1027. [DOI] [PubMed] [Google Scholar]
- 50.Perry, R. L. S., and M. A. Rudnicki. 2000. Molecular mechanisms regulating myogenic determination and differentiation. Front. Biosci. 5:750-767. [DOI] [PubMed] [Google Scholar]
- 51.Rudnicki, M. A., T. Braun, S. Hinuma, and R. Jaenisch. 1992. Inactivation of MyoD in mice leads to upregulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell 71:383-390. [DOI] [PubMed] [Google Scholar]
- 52.Sabourin, L. A., A. Girgis-Gabardo, P. Seale, A. Asakura, and M. A. Rudnicki. 1999. Reduced differentiation potential of primary MyoD-/- myogenic cells derived from adult skeletal muscle. J. Cell Biol. 144:631-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schlöndorff, J., and C. P. Blobel. 1999. Metalloprotease-disintegrins: modular proteins capable of promoting cell-cell interactions and triggering signals by protein-ectodomain shedding. J. Cell Sci. 112:3603-3617. [DOI] [PubMed] [Google Scholar]
- 54.Schultz, E., and K. M. McCormick. 1994. Skeletal muscle satellite cells. Rev. Physiol. Biochem. Pharmacol. 123:213-257. [DOI] [PubMed] [Google Scholar]
- 55.Seale, P., and M. A. Rudnicki. 2000. A new look at the origin, function, and “stem-cell” status of muscle satellite cells. Dev. Biol. 218:115-124. [DOI] [PubMed] [Google Scholar]
- 56.Shi, Z., W. Xu, F. Loechel, U. M. Wewer, and L. J. Murphy. 2000. ADAM12, a disintegrin metalloprotease, interacts with insulin-like growth factor-binding protein-3. J. Biol. Chem. 275:18574-18580. [DOI] [PubMed] [Google Scholar]
- 57.Skapek, S. X., J. Rhee, D. B. Spicer, A. B. Lassar. 1995. Inhibition of myogenic differentiation in proliferating myoblasts by cyclin D1-dependent kinase. Science 267:1022-1024. [DOI] [PubMed] [Google Scholar]
- 58.Smith, E. J., G. Leone, J. DeGregori, L. Jakoi, and J. R. Nevins. 1996. The accumulation of an E2F-p130 transcriptional repressor distinguishes a G0 cell state from a G1 cell state. Mol. Cell. Biol. 16:6965-6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spangenburg, E. E., M. V. Chakravarthy, and F. W. Booth. 2002. p27Kip1: a key regulator of skeletal muscle satellite cell proliferation. Clin. Orthop. 403:S221-S227. [DOI] [PubMed] [Google Scholar]
- 60.Stone, A. L., M. Kroeger, and Q. X. A. Sang. 1999. Structure-function analysis of the ADAM family of disintegrin-like and metalloproteinase-containing proteins. J. Protein Chem. 18:447-465. [DOI] [PubMed] [Google Scholar]
- 61.Thomas, M., B. Langley, C. Berry, M. Sharma, S. Kirk, J. Bass, and R. Kambadur. 2000. Myostatin, a negative regulator of muscle growth, functions by inhibiting myoblast proliferation. J. Biol. Chem. 275:40235-40243. [DOI] [PubMed] [Google Scholar]
- 62.Walsh, K., and H. Perlman. 1997. Cell cycle exit upon myogenic differentiation. Curr. Opin. Genet. Dev. 7:597-602. [DOI] [PubMed] [Google Scholar]
- 63.Wang, J., and K. Walsh. 1996. Resistance to apoptosis conferred by Cdk inhibitors during myocyte differentiation. Science 273:359-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yablonka-Reuveni, Z., and A. J. Rivera. 1994. Temporal expression of regulatory and structural muscle proteins during myogenesis of satellite cells on isolated adult rat fibers. Dev. Biol. 164:588-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yablonka-Reuveni, Z., M. A. Rudnicki, A. J. Rivera, M. Primig, J. E. Anderson, and P. Natanson. 1999. The transition from proliferation to differentiation is delayed in satellite cells from mice lacking MyoD. Dev. Biol. 210:440-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yagami-Hiromasa, T., T. Sato, T. Kurisaki, K. Kamijo, Y. Nabeshima, and A. Fujisawa-Sehara. 1995. A metalloprotease-disintegrin participating in myoblast fusion. Nature 377:652-656. [DOI] [PubMed] [Google Scholar]
- 67.Yoshida, N., S. Yoshida, K. Koishi, K. Masuda, and Y. Nabeshima. 1998. Cell heterogeneity upon myogenic differentiation: downregulation of MyoD and Myf-5 generates "reserve cells'. J. Cell Sci. 111:769-779. [DOI] [PubMed] [Google Scholar]
- 68.Yun, K., and B. Wold. 1996. Skeletal muscle determination and differentiation: story of a core regulatory network and its context. Curr. Opin. Cell Biol. 8:877-889. [DOI] [PubMed] [Google Scholar]
- 69.Zabludoff, S. D., M. Csete, R. Wagner, X. Yu, and B. J. Wold. 1998. p27Kip1 is expressed transiently in developing myotomes and enhances myogenesis. Cell Growth Differ. 9:1-11. [PubMed] [Google Scholar]