Abstract
Liver receptor homolog 1 (LRH-1) and pancreatic-duodenal homeobox 1 (PDX-1) are coexpressed in the pancreas during mouse embryonic development. Analysis of the regulatory region of the human LRH-1 gene demonstrated the presence of three functional binding sites for PDX-1. Electrophoretic mobility shift assays and chromatin immunoprecipitation analysis showed that PDX-1 bound to the LRH-1 promoter, both in cultured cells in vitro and during pancreatic development in vivo. Retroviral expression of PDX-1 in pancreatic cells induced the transcription of LRH-1, whereas reduced PDX-1 levels by RNA interference attenuated its expression. Consistent with direct regulation of LRH-1 expression by PDX-1, PDX-1−/− mice expressed smaller amounts of LRH-1 mRNA in the embryonic pancreas. Taken together, our data indicate that PDX-1 controls LRH-1 expression and identify LRH-1 as a novel downstream target in the PDX-1 regulatory cascade governing pancreatic development, differentiation, and function.
Drosophila fushi tarazu factor 1 (FTZ-F1, NR5A3) is a member of the nuclear receptor superfamily that regulates the transcription of the homeobox fushi tarazu (ftz) gene during early development (30, 39). The best-characterized FTZ-F1 homologue is steroidogenic factor-1 (SF-1, NR5A1), a nuclear receptor expressed in particular regions of the steroidogenic organs, the hypothalamus, and the pituitary gonadotropic cells, where it controls critical developmental and physiological processes (48). Liver receptor homolog 1 (LRH-1, NR5A2) has been characterized as a paralogue of SF-1 in tissues of endodermal origin, such as liver, pancreas, and intestine. LRH-1 was first cloned in the mouse (GenBank accession number M81385), and several orthologues have subsequently been cloned in different species, including Xenopus laevis (FTZ-F1-related receptor xFF1) (15), chicken (orphan receptor 2.0) (29), rat (fetoprotein transcription factor) (19), zebrafish (FTZ-F1-related receptor zFF1) (35), frog (Rana rugosa FTZ-F1) (41), and human (pancreas homologue receptor 1) (4); human fetoprotein transcription factor (18); human B1-binding factor (34); and Cyp7a promoter binding factor (43).
In the adult animal, LRH-1 has a critical function in diverse pathways controlling cholesterol homeostasis, as evidenced by its role in the control of the expression of cholesterol 7α-hydroxylase (Cyp7A1), the rate-limiting enzyme of the bile acid biosynthesis pathway (37, 43), sterol 12α-hydroxylase (Cyp8B1), involved in cholic acid synthesis (9), multidrug resistance protein 3, implicated in the enterohepatic circulation of bile salts (24), and the cholesteryl ester transfer protein (38) and scavenger receptor class B type I (50), two key players in reverse cholesterol transport.
Besides its role in metabolism, LRH-1 also controls the expression of a number of developmental genes, such as α-fetoprotein, a marker of early liver development (19), and the transcription factors HNF-3β (49), HNF-4α, and HNF-1α (47), which coordinate hepatic developmental gene expression. Conversely, the expression of the mouse LRH-1 gene is under the control of the transcription factors GATA, Nkx, basic helix-loop-helix factors, and HNF-4α (47) whereas the human LRH-1 gene is regulated by HNF-3β and HNF-1 (65), all transcription factors involved in developmental control of gene expression. These findings establish a critical role for LRH-1 in hepatic development and homeostasis, yet the contribution of LRH-1 to the formation and function of the pancreas, a tissue in which LRH-1 is abundantly expressed, is still poorly understood.
Pancreatic-duodenal homeobox 1 (PDX-1; also called IUF-1, IPF-1, IDX-1, STF-1, and GSF) is a homeodomain transcription factor essential for pancreatic development (for review, see reference 13). Targeted disruption of the PDX-1 gene in mice as well as homozygous mutation of the human PDX-1 gene leads to pancreas agenesis (26, 44, 56). Moreover, heterozygous mutations in the human PDX-1 gene are linked to maturity-onset diabetes of the young type 4 and late-onset type 2 diabetes mellitus (23, 56).
PDX-1 is first detected in mouse embryos at embryonic day 8.5 (E8.5), and its expression is localized throughout the pancreas during embryonic development (45). During adulthood, PDX-1 is predominantly expressed in the β-cells of the islets, where it regulates directly or indirectly the expression of genes such as insulin (45), glucokinase (62), islet amyloid polypeptide (52), and the glucose transporter type 2 (61). This transcriptional regulation occurs via the binding of monomeric PDX-1 through a GTAATC consensus site. PDX-1 is also found in pancreatic ductal and acinar cells, where it controls the expression of elastase I as a heterodimeric complex with two homeodomain proteins, PBX-1b and MRG1 (58).
In this study, we characterized the developmental regulation of the LRH-1 gene. Most importantly, we show here that LRH-1 and PDX-1 are coexpressed during pancreatic development and that LRH-1 expression is regulated by PDX-1, both in vitro and in vivo. Altogether, our data suggest that LRH-1 is a major player in pancreatic development and homeostasis.
MATERIALS AND METHODS
Antibodies.
The anti-PDX-1 polyclonal rabbit antibody was raised against amino acids 269 to 284 (SPQPSSIAPLRPQEPR) of the murine PDX-1 protein. Validation of the antibody was performed with the immunogenic peptide as a competitor in electrophoretic mobility shift assays (EMSAs) and immunoprecipitation (data not shown). Anti-acetylated H3 and H4 antibodies were purchased from Euromedex (Souffelweyersheim, France). Anti-LRH-1 monoclonal antibody H2325 was produced with the baculovirus gp64 display system (Invitrogen, Carlsbad, Calif.). The human LRH-1 cDNA encoding amino acids 161 to 280 was amplified by PCR and ligated into the surface glycoprotein gp64 gene of Autographa californica multiple nuclear polyhedrosis virus to create a fusion protein expressed on the viral surface. The monoclonal antibody was produced exactly as described previously (59).
In situ hybridizations.
Wild-type embryos from stages E7.5 to E16.5 were directly embedded in Cryomatrix (Shandon, Pittsburgh, Pa.). In situ hybridizations were performed on 10-μm cryosections with 35S-labeled antisense RNA probes as described previously (10). Negative controls were performed in parallel with sense RNA probes for murine LRH-1 and murine PDX-1 (data not shown). PDX-1−/− mice were genotyped as described previously (26, 44, 56).
Cloning of human LRH-1 promoter and mutagenesis.
The human LRH-1 promoter was cloned with the Genome Walker kit (Clontech Laboratories Inc., Palo Alto, Calif.). PCR amplifications were performed according to the manufacturer's instructions with primers AP-1 (5′-GTAATACGACTCACTATAGGGC-3′) and JSA 55L (5′-GTGCAGCTTGTCAAATTTCGTGGCCTTGGG-3′) and primers AP-2 (nested to AP-1, 5′-ACTATAGGGCACGCGTGT-3′) and JSA 54L (nested to JSA 55L, 5′-CCCTGGACTCTGTACTTTTTCCAACATTAG-3′). Four major bands (600 bp, 1.2 kb, 1.5 kb, and 2.4 kb), differing from their 5′ ends, were purified on agarose gels with the Qiaquick gel extraction kit (Qiagen, Hilden, Germany) and subcloned into the T/A cloning vector pTAdv (Clontech). Positive clones were used to PCR amplify the promoter fragment with AP-1 (MluI site) and JSA 74L (BglII site, 5′-GAAGATCTTCCATGATGGTTTCTAATCAGA-3′), and PCR products were then purified on a 1% agarose gel and cloned into the pGL3-basic vector (Promega Life Science, Madison, Wis.) digested with MluI/BglII.
The different pGL3-human LRH-1 promoter constructs were sequenced and used in transient transfections. The promoter region was analyzed on the web site http://transfac.gbf.de/ with programs designed for promoter and transcription binding site predictions (Matinspector Transfac version 2.2). PDX-1 binding sites were identified and mutations were performed with the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) and the following forward primers: mutPdx2, 5′-CCCTAAATAAACCGGTAGACCGTAAATTC-3′; mutPdx3, 5′-CAACCTGCATTTACTTGGCCTAAAAGGAG-3′; and mutPdx5, 5′-TCACTTAAAGAGCGCATGTCTGCCAATGTTATC-3′ (italicized bases are mutated).
Cell culture and transient transfection assays.
Panc-1 (human pancreatic carcinoma), MiaPaca-2 (human pancreatic carcinoma), LTPA (murine pancreatic carcinoma), NIT-1 (mouse insulinoma), and 293gp and 293T (human embryonic kidney) cells were maintained according to the suppliers' instructions (American Type Culture Collection, Manassas, Va.). Cells were transfected according to the CaCl2 precipitation technique, and luciferase assays were carried out as described previously (50). Expression plasmid pBKCMV (Stratagene) containing mouse PDX-1 cDNA was described previously (36). Luciferase activity measurements were normalized for β-galactosidase activity to correct for differences in transfection efficiency, and graphed values represent the means of three independent experiments.
Electrophoretic mobility shift assays.
Double-stranded oligonucleotides containing the PDX-1 binding site described in the insulin promoter (12) (consensus: Ins, 5′-GACCTTAATGGGCCAAACAGCA-3′) or present in the human LRH-1 promoter were labeled with T4 polynucleotide kinase, and EMSA binding reactions were performed as described previously (50). For competition experiments, increasing amounts (from 50- to 200-fold molar excess) of unlabeled wild-type or mutated double-stranded oligonucleotide (Pdx 2, 5′-CTAAATAAATTAATAGACCGTA-3′; mutPdx 2, 5′-CTAAATAAACCGGTAGACCGTA-3′; Pdx 3, 5′-TGCATTTACTTAATTTAAAAGG-3′; mutPdx 3, 5′-TGCATTTACTTGGCCTAAAAGG-3′; Pdx 5, 5′-CTTAAAGAATTAATGTCTGCCA-3′; and mutPdx 5, 5′-CTTAAAGAGCGCATGTCTGCCA-3′) were included just before adding labeled wild-type PDX-1 oligonucleotide. Preimmune serum (negative control) or anti-PDX-1 serum was incubated for 30 min with in vitro-translated PDX-1 proteins or NIT-1 nuclear extracts before adding radioactive probes. DNA-protein complexes were separated by electrophoresis on a 4% polyacrylamide gel in 0.25× TBE (Tris-borate-EDTA) buffer at 4°C.
RNA extraction, Northern blotting, and RT-PCR.
RNA extraction and Northern blot analysis were performed as described previously (51). 32P-labeled full-length murine LRH-1 cDNA (accession number M81385) was used as a probe and hybridized to the mouse embryo multiple tissue Northern blot (Clontech). Reverse transcription of mRNA was performed at 37°C for 1 h with the Moloney murine leukemia virus reverse transcriptase (Life Technologies, Burlington, Canada) and random hexanucleotides, followed by a 15-min inactivation at 70°C.
Reverse transcription (RT)-PCR was performed with oligonucleotides specific to hPDX-1 cDNA (5′-GGCGCACCTTCACCACCACCTC-3′ and 5′-GCCGCCGCGCTTCTTGTCCT-3′), human LRH-1 cDNA (5′-TCAATGCCGCCCTGCTGGACTACACAATG-3′ and 5′-CTTCTTCCCCCTCCCCACTCCCCCAATCT-3′), murine PDX-1 cDNA (5′-CGACGACCCGGCTGGCGCTCACCTC-3′ and 5′-CTTCGCCCCCACCGCCCCCACTCG-3′), murine LRH-1 cDNA (5′-CTAGTTTGGATACTGGAGATTTTCA-3′ and 5′-ATAGGAGTAATTCACCATTTTAAAT-3′), human and murine acidic ribosomal phosphoprotein 36B4 (5′-ATGTGAAGTCACTGTGCCAG-3′ and 5′-GTGTAATCCGTCTCCACAGA-3′), and murine glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (5′-GCTCACTGGCATGGCCTTCCGTGT-3′ and 5′-TGGAAGAGTGGGAGTTGCTGTTGA-3′) under the following conditions: 94°C for 5 min, followed by 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s, and a final extension step at 72°C for 10 min. Amplification products were loaded on a 1% agarose gel.
ChIP.
For in vitro chromatin immunoprecipitation (ChIP), Panc-1, MiaPaca-2, and retrovirus-infected LTPA cells were used. For in vivo ChIP, gut/pancreas combined, pancreas, liver, and gastrointestinal tracts were microdissected from 20 e13.5, E16.5, and E17.5 embryos. Cells and organs were fixed for 10 min and 16 h, respectively, in phosphate-buffered saline containing 1% formaldehyde and protease inhibitor cocktail and subsequently rinsed five times in phosphate-buffered saline. Cells were collected and centrifuged at 4°C for 5 min at 2,000 × g and resuspended in lysis buffer (50 mM HEPES [pH 7.5], 140 mM NaCl, 1% Triton X-100, and protease inhibitor cocktail). After 30 min of lysis, three cycles of sonication (three pulses of 9 s, 20% amplitude for cells, 80% amplitude for organs) were performed to prepare DNA fragments ranging in size from 200 bp to 1,000 bp, followed by centrifugation for 10 min.
Supernatants were collected and cleared by incubation with protein A-Sepharose (2.5 mg), sonicated salmon sperm DNA (2 μg), and 10 μl of preimmune serum for 2 h at 4°C. Then 20 μl of supernatant was collected and used as the input. Immunoprecipitation was carried out overnight at 4°C with preimmune serum (negative control) or antibodies raised against PDX-1, acetylated histone H3 (dilution 1:200), or acetylated histone H4 (dilution 1:200). After centrifugations, washing, and elution, the cross-linking was reversed by heating the samples at 65°C overnight. DNA was then purified with the Qiagen PCR purification kit, and PCR was performed with primers 5′-GATTGCTTAAGTCCATGAGTCTGAGGTT-3′ and 5′-CAAAGTGCTGGAATTATAGGCGTGAG-3′, 5′-CAAGCAAGCTAATGCCTCTTCTAAC-3′ and 5′-CTGCAGCCCAGAGTGTGGAAAGTTG-3′, and 5′-AGTGCCAGCCTCGTCCCGTAGACAAAATG-3′ and 5′-AAGTGGGCCCCGGCCTTCTCCAT-3′ to amplify the human LRH-1, the murine LRH-1, and the murine GAPDH promoters, respectively. The ChIP assay was performed at least twice for each condition.
Retroviral infection.
Virus production and infection were performed as described previously (50). For retroviral infection specifically, the 293gp packaging cell line was transfected with Lipofectamine (Life Technologies), with 15 μg of empty control retroviral vector or retroviral vector containing the cDNA for murine PDX-1, and with 5 μg of the ecotropic vector SV-E-MLV-env, containing the Moloney murine leukemia virus envelope cDNA downstream of the simian virus 40 promoter-enhancer.
RNAi and immnuoblotting.
The RNA interference (RNAi) experiment was performed with the pSUPER RNAi system, following the manufacturer's instructions (DNAengine, Seattle, Wash.). Briefly, a double-stranded oligonucleotide targeting nucleotides 709 to 727 (5′-GATCCCCCCCGAGGAAAACAAGAGGATTCAAGAGATCCTCTTGTTTTCCTCGGGTTTTTGGAA-3′) of murine PDX-1 mRNA was cloned in the BglII and HindIII sites of the pSUPER vector. Transient transfection was carried out in 60-mm plates containing mock- and PDX-infected LTPA cells, with Lipofectamine (Life Technologies) and 10 μg of the empty pSUPER or PDX-1 RNAi vector. After 48 h, cells were harvested for whole-cell protein extracts, and Western blotting was performed as described previously (17). RNAi experiments were repeated three times. Intensities of the LRH-1 and PDX-1 protein signals were quantified by phosphorimager analysis, and the induction was calculated after normalization to β-actin protein levels.
RESULTS
LRH-1 and PDX-1 are coexpressed in the developing mouse pancreas.
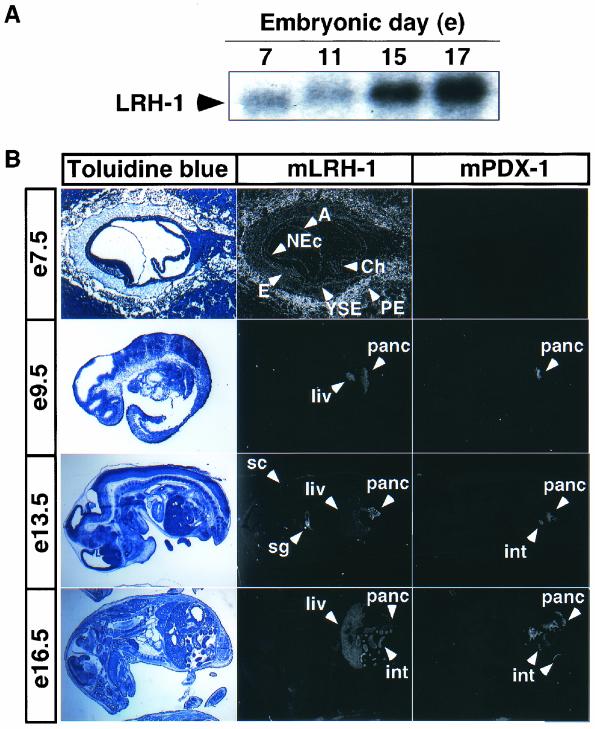
LRH-1 is expressed early in the developing pancreas (49). In view of the presence of homeobox response elements in the promoter region of the LRH-1 gene (47), we hypothesized that PDX-1, a homeobox transcription factor essential for pancreatic development, may control pancreatic LRH-1 expression. LRH-1 mRNA was already detected at E7 by Northern blot analysis and increased significantly from E7 to E17 (Fig. 1A). We then checked whether LRH-1 and PDX-1 were coexpressed with in situ hybridizations in embryos from stage E7.5 to E16.5. In situ hybridizations on E7.5 embryos showed that LRH-1 was present in the yolk sac endoderm, the embryonic endoderm, the neural ectoderm, the amnion, the chorion, and parietal endodermal cells attached to the Reichert's membrane, and it remained expressed at later stages in tissues of endodermal origin, such as liver, intestine, and pancreas (Fig. 1B). LRH-1 expression was also detected in the spinal cord and salivary gland. The in situ hybridization for PDX-1 was consistent with a previous report documenting PDX-1 expression in pancreas starting from E8.5 (45). PDX-1 was thereafter detected in pancreas and small intestine, as described previously (1, 14, 25, 44) (Fig. 1B).
FIG. 1.
LRH-1 and PDX-1 are coexpressed in the developing pancreas. (A) LRH-1 hybridization of Northern blots containing polyadenylated mRNA from E7, E11, E13, and E17 mouse embryos. Murine LRH-1 mRNA expression was detected at stage E7. (B) In situ hybridizations on adjacent sections of E7.5, E9.5, E13.5, and E16.5 mouse embryos. Coexpression of LRH-1 and PDX-1 was observed in the pancreas (panc) and in the intestine (int). LRH-1 was also detected in the yolk sac endoderm (YSE), the embryonic endoderm (E), the neural ectoderm (NEc), the amnion (A), the chorion (Ch), the parietal endodermal cells attached to the Reichert's membrane (PE), the liver (liv), the salivary gland (sg), and the spinal cord (sc). Counterstaining with toluidine blue is depicted on the left panel.
Human LRH-1 promoter is transcriptionally regulated by PDX-1 in transfected cells.
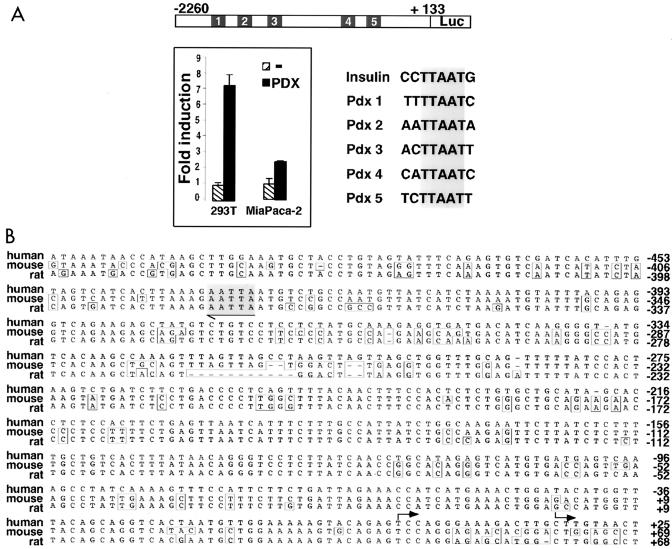
In view of the coexpression of LRH-1 and PDX-1 in the developing pancreas, we analyzed whether the promoter of the human LRH-1 gene was regulated by PDX-1. Interestingly, LRH-1 promoter activity was induced approximately 8-fold and 2.5-fold upon cotransfection of a PDX-1 expression vector in kidney 293T and pancreatic MiaPaca-2 cells, respectively (Fig. 2A).
FIG. 2.
Identification of PDX-1 binding sites in the human LRH-1 promoter. (A) Computational analysis of a ±2-kb fragment of the regulatory region of the human LRH-1 gene demonstrating the presence of five potential binding sites for PDX-1 (Pdx 1 to 5) homologous to the PDX-1 consensus binding site present in the rat insulin promoter. In the 293T and MiaPaca-2 cell lines, cotransfection of an expression vector encoding murine PDX-1 and a luciferase reporter containing ±2 kb of the human LRH-1 promoter induces human LRH-1 promoter activity compared to an empty expression vector. (B) Alignment of ±500 bp of the human, mouse, and rat LRH-1 promoters. The highly conserved site 5 is indicated in gray. Sequence differences are boxed. Transcription initiation sites are indicated by arrows.
Five potential binding sites homologous to the PDX-1 consensus site YHTTAATK (46), termed Pdx 1 to 5, were identified in the human LRH-1 promoter. One of these PDX-1 consensus sites, Pdx 5, is perfectly conserved between the mouse, rat, and human LRH-1 genes (Fig. 2B, gray box). Unfortunately, we were not able to find mouse and rat homologous sequences that we could align with the human sequence covering the Pdx 1 to 4 sites, since these murine and rat sequences are not yet available in the genomic database. In consequence, we cannot rule out the presence of genomic homology blocks in rodents corresponding to the human Pdx 1 to 4 sites.
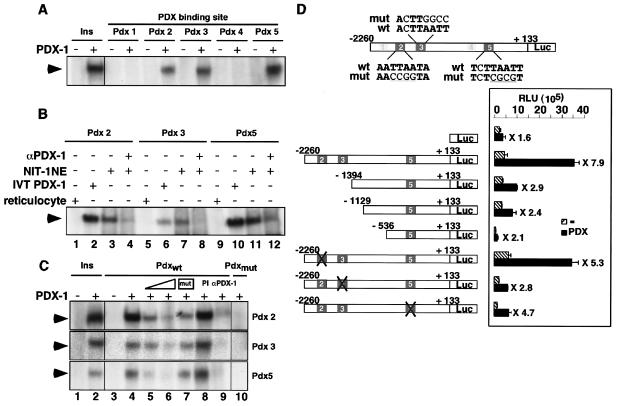
Next, all five potential PDX-1 binding sites in the human LRH-1 promoter were tested by EMSA for their ability to bind PDX-1 protein. In contrast to the putative binding sites Pdx 1 and Pdx 4, which were both unable to bind in vitro synthesized PDX-1, Pdx 2, 3, and 5 bound all PDX-1 protein to about the same extent as the binding observed between PDX-1 and the consensus PDX-1 binding site in the rat insulin promoter (Fig. 3A) (12).
FIG. 3.
Human LRH-1 promoter is responsive to PDX-1 in vitro. (A) EMSA showing PDX-1 binding to three out of the five potential PDX-1 binding sites, Pdx 2, 3, and 5. Binding of PDX-1 to the PDX-1 consensus site in the insulin promoter (Ins) is shown as a positive control. (B) EMSA showing binding of in vitro-synthesized PDX-1 (IVT PDX-1, lanes 2, 6, and 10) and NIT-1 insulinoma nuclear extracts (NIT-1 NE, lanes 3, 7, and 11) to the Pdx 2, 3, and 5 sites of the human LRH-1 promoter. Incubation of NIT-1 nuclear extracts with an anti-PDX-1 antibody decreased the intensity of the retarded band, demonstrating the specificity of the binding (lanes 4, 8, and 12). No binding could be observed with nonprogrammed reticulocyte lysate (lanes 1, 5, and 9). (C) EMSA with the Pdx 2, 3, and 5 sites of the human LRH-1 promoter as probes. Increasing amounts of unlabeled wild-type oligonucleotides competed for binding with the radiolabeled Pdx 2, 3, and 5 oligonucleotides (lanes 5 and 6), whereas competition with the respective mutated oligonucleotides (mutations in sequences are indicated in panel D) did not displace the protein-DNA complex (lane 7). Incubation with a PDX-1 antibody abrogated the formation of the retarded band (α-PDX-1, lane 9), whereas no modification in PDX-1 binding was observed with a preimmune serum (PI, lane 8). No binding of PDX-1 to radiolabeled Pdx 2, 3s and 5 mutated oligonucleotides was observed (Pdxmut, lane 10). (D) Activity generated from the pGL3-LRH-1-2260 reporter cotransfected in 293T cells with an empty vector or an expression vector encoding murine PDX-1. Various unilateral deletion mutations as well as site-directed mutations in the putative PDX-1 sites of the human LRH-1 promoter indicated that the DNA region located between −2260 and −1394 confers responsiveness to PDX-1.
We also investigated whether endogenous PDX-1 protein from insulinoma NIT-1 nuclear extracts was able to bind to the Pdx 2, 3, and 5 sites (Fig. 3B). A strong retarded band of the same size and roughly the same intensity as the one obtained with the in vitro synthetised PDX-1 was observed in EMSA (Fig. 3B, compare lanes 2, 6, and 10 to 3, 7, and 11). This band was specific to PDX-1, as demonstrated by its partial abrogation upon preincubation with an anti-PDX-1 antibody (Fig. 3B, lanes 4, 8, and 12). With in vitro synthesized PDX-1, binding to the Pdx 2, 3, and 5 sites could be competed away by adding increasing amounts of the respective unlabeled oligonucleotides (Fig. 3C, lanes 5 and 6), whereas the mutated Pdx oligonucleotides competed less efficiently (Fig. 3C, lane 7). Addition of an anti-PDX-1 antibody but not the preimmune serum in the EMSA resulted in a decrease in the intensity of binding of PDX-1, confirming that PDX-1 binds to these three sites (Fig. 3C, lanes 8 and 9). Moreover, radiolabeled double-stranded oligonucleotides containing the mutated PDX-1 binding sites 2, 3, and 5 were unable to bind PDX-1 protein (Fig. 3C, lane 10).
Several deletion constructs of the human LRH-1 promoter spanning the regions from −2260, −1394, −1129, and −536 up to +133 were next tested for their ability to respond to PDX-1 (Fig. 3D). Compared to that of the largest construct, the luciferase activity of the smaller LRH-1 promoter deletion constructs was considerably attenuated in the presence of PDX-1 (8-fold versus 2- to 3-fold), indicating that the two PDX-1 binding sites, Pdx 2 and Pdx 3, located in the sequence between bp −2260 and −1394 of the human LRH-1 gene were of major importance.
We next mutated each of the PDX-1 binding sites able to bind PDX-1 in EMSA to confirm that these DNA sequences mediate the effect of PDX-1 in transfection. Compared to the wild-type human LRH-1 promoter, individual mutation of sites 2, 3, and 5 led to a reduced capacity of PDX-1 to activate the human LRH-1 promoter, ranging from 30% (mutated site 3) to 60% (mutated site 2) (Fig. 3D). A further small reduction in promoter activity was observed when both sites 2 and 3 or 3 and 5 were mutated (data not shown).
In order to analyze the transcriptional effect of endogenous PDX-1 on the human LRH-1 promoter, we performed transient transfection studies with the site 3 mutant in MiaPaca-2 cells. In MiaPaca-2 cells as well, a decrease in basal LRH-1 promoter activity of the site 3 mutant was observed compared to the wild-type construct, defining this site as an important regulatory sequence for basal activity (data not shown). Together, these data identify site 3 as an important functional PDX-1 site in the human LRH-1 promoter but also suggest that sites 2 and 5 contribute to the regulation of the human LRH-1 gene.
PDX-1 binds to the LRH-1 promoter in Panc-1, MiaPaca-2, and LTPA cells.
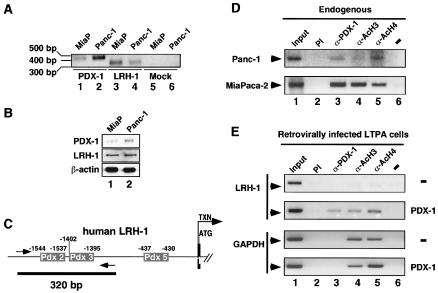
To establish the relevance of the binding of PDX-1 to the LRH-1 promoter, we performed chromatin immunoprecipitation (ChIP) assays on two human pancreatic cell lines, Panc-1 and MiaPaca-2. RT-PCR analysis and Western blotting demonstrated that both cell lines coexpressed LRH-1 and PDX-1 (Fig. 4A and B). Genomic DNAs derived from both pancreatic cell lines were cross-linked to proteins and sonicated. After immunoprecipitation with antibodies specific to PDX-1 or to acetylated histones H3 and H4, the recovered DNA was analyzed with primers spanning a 320-bp region comprising the Pdx 2 and Pdx 3 sites, schematically depicted in Fig. 4C.
FIG. 4.
PDX-1 binds to the LRH-1 promoter in pancreatic cell lines. (A) RT-PCR experiment showing coexpression of PDX-1 and LRH-1 in two human pancreatic cell lines, MiaPaca-2 (MiaP) and Panc-1. A mock PCR was performed with water as the template and is shown as a negative control for PCR amplification of PDX-1 (lane 5) and LRH-1 (lane 6). (B) Immunoblot demonstrating coexpression of PDX-1 and LRH-1 proteins in the MiaPaca-2 (lane 1) and Panc-1 (lane 2) pancreatic cell lines. (C) Schematic representation of the 5′ region of the human LRH-1 gene. The Pdx 2, 3, and 5 binding sites are indicated. Transcription (arrow, TXN) and translation (ATG) initiation sites are shown. Amplimers are highlighted by the arrows, whereas the black line depicts the 320-bp PCR product. (D) ChIP assay demonstrating binding of endogenous PDX-1 to the human LRH-1 promoter. Cross-linked chromatin from Panc-1 and MiaPaca-2 cells was incubated with antibodies against PDX-1 (lane 3), acetylated histone H3 (lane 4), acetylated histone H4 (lane 5), or preimmune serum (lane 2, PI). Immunoprecipitates were analyzed by PCR with primers specific for the human LRH-1 promoter, as shown in panel B. A positive (input chromatin, lane 1) and a negative (with water as a template for PCR, lane 6) control are shown. (E) ChIP assay demonstrating increased binding of the LRH-1 promoter by PDX-1 in the murine LTPA pancreatic cell line stably overexpressing PDX-1. Cross-linked chromatin from LTPA cells transduced with an empty (−) or PDX-1-expressing (PDX-1) retroviral vector were analyzed by ChIP as described for panel C. The murine LRH-1 promoter bound more PDX-1 (lane 3) and acetylated histones H3 (lane 4) and H4 (lane 5) in PDX-1-expressing LTPA cells relative to control cells.
Consistent with the previous EMSA data, the antibody specific to PDX-1 precipitated endogenous PDX-1 protein bound to the LRH-1 promoter under basal conditions in both MiaPaca-2 and Panc-1 cells (Fig. 4D, lane 3). No immunoprecipitation was observed when a preimmune serum was used for ChIP, demonstrating the specificity of the anti-PDX-1 antibody (Fig. 4D, lane 2). In addition, the same 320-bp fragment could also be amplified when anti-acetylated H3 or H4 antibodies were used to precipitate chromatin. The finding that binding of PDX-1 to the human LRH-1 promoter is associated with the presence of acetylated histones suggests that this promoter is transcriptionally active in these cells (Fig. 4D, lanes 4 and 5). Moreover, retroviral expression of PDX-1 in pancreatic LTPA cells resulted in an increased binding of PDX-1 to the LRH-1 promoter compared to the nonexpressing cell line (Fig. 4E, lane 3, compare top and bottom panel, LRH-1). The increased occupancy of the LRH-1 promoter by PDX-1 was associated with a robust increase in acetylated H3 or H4 on the promoter. Under similar conditions, no modification in the presence of acetylated histone H3 or H4 on the GAPDH promoter was observed (Fig. 4E, lanes 4 and 5, GAPDH). Although these data suggest that this increased presence of acetylated H3 and H4 is caused by overexpression of PDX-1, they do not prove a causal relationship.
PDX-1 binds to LRH-1 promoter during pancreatic development.
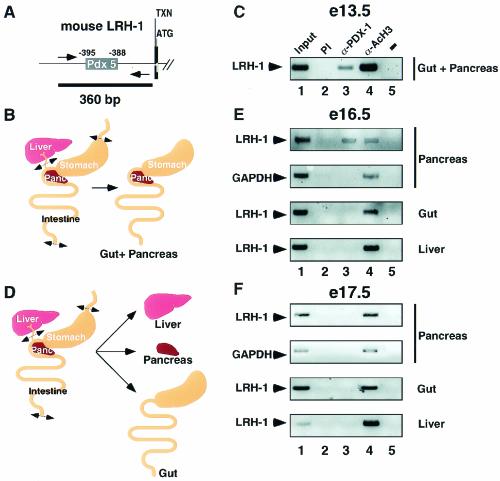
To further investigate the relevance of PDX-1 binding to the LRH-1 promoter during development, we next performed ChIP assays in mouse embryos. Since the Pdx 5 site is highly conserved between the mouse, rat, and human genes (Fig. 2B), we designed primers amplifying 360 bp of this region to analyze PDX-1 binding to the murine LRH-1 promoter in vivo (Fig. 5A). Because of the difficulties in isolating pancreas in E13.5 embryos, we initially microdissected the gut combined with the pancreas (Fig. 5B). At this stage, PDX-1 was present in the embryonic gut-pancreas mixture and bound to the LRH-1 promoter (Fig. 5C, lane 3). Moreover, the promoter of the LRH-1 gene was immunoprecipitated by anti-acetylated H3 antibody, indicating transcriptional activity of LRH-1 during this phase of endodermal development (Fig. 5C, lane 4).
FIG. 5.
PDX-1 binds to the LRH-1 promoter during pancreatic development. (A) Schematic representation of the 5′ region of the mouse LRH-1 gene. The Pdx 5 binding site is indicated. Transcription (arrow, TXN) and translation (ATG) initiation sites are shown. Amplimers are highlighted by the arrows, whereas the black line depicts the 360-bp PCR product. (B) Scheme depicting the microdissection of the gut and pancreas used for the in vivo ChIP on E13.5 mouse embryos. (C) In vivo ChIP assay demonstrating binding of endogenous PDX-1 to the murine LRH-1 promoter in the gut and pancreas from E13.5 embryos. Chromatin from gut and pancreas was incubated with antibodies against PDX-1 (lane 3), acetylated histone H3 (lane 4), or preimmune serum (lane 2). Immunoprecipitates were analyzed by PCR with primers specific for the mouse LRH-1 promoter. A negative control with water as the template for PCR is shown in lane 5. Input chromatin was used as a positive control in lane 1. (D) Schematic representation showing the microdissection of the isolated organs used for in vivo ChIP on E16.5 (E) and E17.5 (F) embryos. (E) In vivo ChIP assay demonstrating occupancy of the murine LRH-1 promoter by PDX-1 in pancreas but not gut or liver of E16.5 embryos. Cross-linked chromatin from pancreas, gut, and liver was prepared for the ChIP assay as described for panel B. Immunoprecipitates were analyzed by PCR with primers specific for the murine LRH-1 and murine GAPDH (positive control) promoters. (F) PDX-1 does not occupy the LRH-1 promoter in endodermal tissues dissected from E17.5 mouse embryos. Cross-linked chromatin from the pancreas, gut, and liver of E17.5 embryos was analyzed by ChIP as described for panel D.
We then repeated the in vivo ChIP on pancreas, gut, and liver isolated from E16.5 and E17.5 embryos (Fig. 5D). Interestingly, PDX-1 was only bound to the LRH-1 promoter in the pancreas at E16.5 (Fig. 5E, lane 3, LRH-1, pancreas), but not at E17.5 (Fig. 5F, lane 3, LRH-1, pancreas). Acetylated histone H3 occupied the LRH-1 promoter during all these stages of development. LRH-1 was reported to be expressed throughout the pancreas until E17, after which it becomes confined to the exocrine pancreas (49). Therefore, our results suggest that PDX-1 regulates LRH-1 expression during a well-defined time window when both factors are coexpressed in the pancreas (until E16.5 to 17). Later on during development, this regulation does not occur anymore, due to compartmentalization of LRH-1 in the exocrine and PDX-1 in the endocrine pancreas.
PDX-1 and LRH-1, however, were still coexpressed in the small intestine at E16.5 and E17.5, but no PDX-1 was detected on the LRH-1 promoter in this tissue (Fig. 5E and F, lane 3, LRH-1, gut), demonstrating that the binding of PDX-1 on the LRH-1 promoter is pancreas specific. Nevertheless, the murine LRH-1 promoter revealed an acetylation of histone H3 in pancreas, liver, and gastrointestinal tract, consistent with active LRH-1 expression during the development of these tissues (Fig. 5C, E, and F, lanes 4, LRH-1). The amplification of the GAPDH promoter with an anti-acetylated H3 antibody did not change during development.
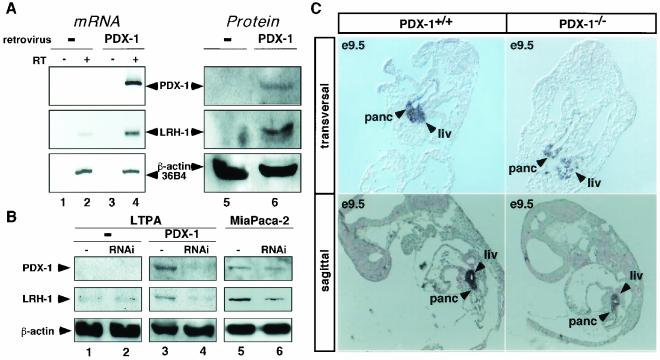
PDX-1 expression induces LRH-1 expression.
In order to confirm that PDX-1 regulates LRH-1 expression in vivo, we examined PDX-1 and LRH-1 expression in the pancreatic LTPA cell line infected with a control or a murine PDX-1-encoding retrovirus. Both mRNA and protein levels of PDX-1 were induced in the cell line infected with the PDX-1 retrovirus (Fig. 6A, lanes 4 and 6, >50-fold induction), whereas no expression of PDX-1 was observed in the cell line transduced with the empty retroviral vector (Fig. 6A, lanes 2 and 5). In addition, the expression of PDX-1 translated in a robust induction of LRH-1 expression at both the mRNA and protein levels (Fig. 6A, compare lanes 2 to 4 and 5 to 6, 16-fold induction), whereas no difference in the expression of 36B4 mRNA (Fig. 6A, compare lanes 2 and 4) or β-actin protein (Fig. 6A, compare lanes 5 and 6) was observed between control and PDX-1-infected cells. We speculate that this increased expression of LRH-1 in this cell line is caused by increased binding of PDX-1 to the LRH-1 promoter, as demonstrated by the ChIP experiments performed in these LTPA cells, shown in Fig. 4D.
FIG. 6.
PDX-1 regulates the expression of LRH-1 in vivo. (A) Expression of PDX-1 and LRH-1 analyzed by RT-PCR (mRNA) and immunoblotting (protein) in LTPA cells overexpressing PDX-1 (PDX-1) or not (−) by retroviral infection. Reverse transcription reactions were performed with (RT+) or without (RT−, negative control) reverse transcriptase. Subsequent PCR was performed with primers specific for PDX-1, LRH-1, and 36B4. Immunoblotting was performed with specific antibodies raised against PDX-1, LRH-1, and β-actin. The intensity of the signals was quantitated by phosphorimager analysis, and the induction was determined after normalization to β-actin signals. (B) Decreasing PDX-1 levels by RNAi reduces LRH-1 expression. Immunoblotting of protein isolated from control LTPA cells (−), LTPA cells retrovirally overexpressing PDX-1 (PDX-1), or MiaPaca-2 cells, all transfected with an empty RNAi vector (−, lanes 1, 3, and 5) or an RNAi vector targeting the PDX-1 mRNA (RNAi, lanes 2, 4, and 6). The induction was calculated as described for panel A. (C) Expression of LRH-1 in transversal (upper panel) and sagittal (lower panel) sections of PDX-1+/+ and PDX-1−/− mouse embryos from stage E9.5 analyzed by in situ hybridization. LRH-1 expression was significantly stronger in pancreas (panc) and liver (liv) of 2 PDX-1+/+ relative to PDX-1−/− tissues.
The results of the retroviral experiment above demonstrated clearly that ectopic expression of PDX-1 induces LRH-1. We next explored whether a knock-down of PDX-1, by use of RNAi technology, could result in a decrease of LRH-1 expression in these retrovirally infected cells. In the LTPA cell lines stably expressing a control or a PDX-1-encoding retrovirus, two different RNAi constructs, one corresponding to an empty vector (Fig. 5B, lanes 1 and 3) and one corresponding to sequences targeting nucleotides 709 to 727 of the murine PDX-1 mRNA (Fig. 6B, RNAi, lanes 2 and 4), were transiently transfected. When RNAi constructs were transfected in the cell line expressing PDX-1, the expression of the PDX-1 protein was significantly reduced compared to that in the cells transfected with the empty RNAi vector (70% reduction; Fig. 6B, PDX-1, compare lanes 3 and 4). The decrease in PDX-1 protein expression was associated with a robust decrease in LRH-1 protein levels (50% reduction; Fig. 6B, LRH-1, compare lanes 3 and 4). Moreover, in the LTPA cell line infected with the control vector and hence not expressing PDX-1, the RNAi vector for PDX-1 had no effect on the LRH-1 protein levels (Fig. 6B, compare lanes 1 and 2), demonstrating the specificity of this transcriptional regulation.
Furthermore, we also studied the expression of LRH-1 upon downregulation of endogenous PDX-1, with the RNAi constructs described above in transient transfection experiments in pancreatic MiaPaca-2 cells, known to express both LRH-1 and PDX-1. In these MiaPaca-2 cells, we observed a decrease of both LRH-1 (50% reduction) and PDX-1 (40% reduction) protein levels (Fig. 6B, RNAi, lanes 5 and 6). The decrease in PDX-1 and LRH-1 expression was somewhat less pronounced than the downregulation observed with the LTPA cell lines stably expressing a PDX-1-encoding retrovirus (Fig. 6B, RNAi, compare lanes 3 to 4 and 5 to 6).
We finally also studied PDX-1−/− mice to obtain in vivo evidence of the relevance of LRH-1 regulation by PDX-1. In three PDX-1−/− mice, LRH-1 expression was decreased in the developing pancreatic bud of E9.5 embryos compared to their wild-type littermates (Fig. 6C and data not shown). Interestingly, LRH-1 signal was also decreased in the embryonic liver of the mutant mice. Although we have no direct explanation, non-cell-autonomous effects can account for the observed effect at this stage of embryonic development. Since quantification of mRNA levels is difficult by either in situ hybridization analysis or quantitative PCR due to the small size of these developing organs, these conclusions need to be interpreted with caution.
DISCUSSION
Temporally and spatially controlled gene expression patterns are the basis of tissue development and differentiation. Since LRH-1 is expressed during early pancreatic development, this orphan nuclear receptor could constitute an important chain in the gene regulatory cascade leading to pancreatic development. To identify the players involved in the regulation of LRH-1, we cloned and characterized the regulatory region of the human LRH-1 gene. Computational analysis of the human LRH-1, murine LRH-1, and rat LRH-1 promoters revealed the presence of consensus sequences for several ubiquitous transcription factors, such as AP-1, GATA-1, USF, and Sp1, but also highlighted the existence of binding sites for tissue-specific transcription factors, such as HNF-3β, C/EBPβ, and PDX-1.
Since LRH-1 and PDX-1 are coexpressed throughout pancreatic development, we evaluated whether PDX-1 could be a transcriptional regulator of LRH-1. PDX-1 was shown to activate the human LRH-1 promoter in transient transfections and to bind to specific DNA sequences in the LRH-1 promoter, both in vitro and in vivo. Changes in PDX-1 levels in cells, by either retroviral overexpression or RNAi-mediated inhibition, induced concomitant changes in murine LRH-1 expression. In vivo ChIP analysis furthermore demonstrated that PDX-1 regulates pancreatic LRH-1 expression until E16.5 of mouse pancreatic development. Although LRH-1 and PDX-1 are also coexpressed in the small intestine, PDX-1 is not bound to the murine LRH-1 promoter in this tissue, demonstrating that this regulation occurs only in a temporally and tissue-restricted fashion, i.e., in a well-defined time window (E8.5 to E16.5) during pancreatic development, but not in the rest of the gut.
PDX-1 null embryos (E9.5) were also examined in order to investigate the relevance of the role of PDX-1 in LRH-1 expression. Compared to wild-type embryos, PDX-1 null embryos show a decrease in LRH-1 expression in the pancreatic bud. Interestingly, we also observed lower levels of LRH-1 in the liver, a tissue where PDX-1 is not supposed to be expressed. Since the expression of PDX-1 begins at stage e8.5, it is possible that non-cell-autonomous effects might affect the expression of a number of genes critical in tissues that arise from the gut endoderm, such as the liver. The absence of PDX-1 could in fact alter the expression of a morphogen in the gut anlagen and affect the expression of a number of genes in the liver, as we observed here for LRH-1. Although the in vivo data go in the same direction as the in vitro data, they should be interpreted with caution in view of the complexity of the regulatory systems specifying development, the potential interference by other regulatory factors, and the difficulty in quantifying in situ hybridization experiments.
PDX-1 is a critical developmental transcriptional regulator in the pancreas and is itself controlled by a number of endodermal transcription factors. PDX-1 expression is under the control of Foxa2/HNF-3β, as shown by the characterization of HNF-3β−/− differentiated embryoid bodies and of β-cell-specific HNF-3β conditional knock-out mice (11, 20, 31, 54). Mice homozygous for a null mutation of HNF-3β have severe problems in gastrulation, and mutant embryos do not develop beyond E8.5, suggesting that HNF-3β is a key factor for development (3, 63). However, conditional deletion of HNF-3β in adult hepatocytes shows that this factor is dispensable for normal adult liver function, demonstrating different roles in embryos and adults (57).
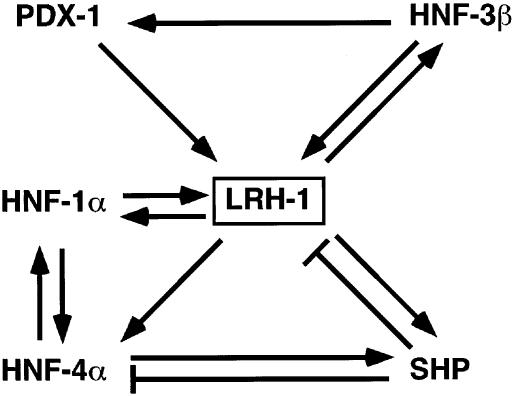
Interestingly, LRH-1, which we show to be a PDX-1 target gene, regulates HNF-3β in vitro (49), highlighting a potential autoregulatory loop involving HNF-3β, PDX-1, and LRH-1 (Fig. 7). Although the relevance of the regulation of HNF-3β by LRH-1 needs to be demonstrated in vivo, regulatory loops involving transcriptional cascades could play important roles in establishing control circuits that govern pancreatic development. More generally, these data also suggest a role for LRH-1 as an eventual more global regulator of endodermal development, given the fact that LRH-1 controls HNF-3β expression in vitro and LRH-1−/− embryos die at E7 (47; J.-S. Annicotte, K. Schoonjans, and J. Auwerx, unpublished data). It is also interesting that the basic helix-loop-helix transcription factor Ptf1a, which was first described as an exocrine-specific protein involved in exocrine pancreatic development and islet architecture (28), was recently shown to be crucial to orient undifferentiated foregut endoderm into the pancreatic fate (27). This raises the possibility that LRH-1, which, like Ptf1a, is expressed in an exocrine-specific manner in the adult, might participate in the early steps of the pancreatic differentiation program.
FIG. 7.
Position of LRH-1 as a central part of a transcriptional network in pancreatic development. Scheme depicting the transcription factors involved in pancreatic development acting up- and downstream of LRH-1. Arrows and stop symbols show positive and negative regulation, respectively.
The central position of LRH-1 in the transcriptional cascade controlling pancreatic development suggests that LRH-1 could also play an important role in homeostatic regulation during adulthood, as described previously for these other transcription factors that control endodermal development (reviewed in reference 55). Several reports demonstrate that PDX-1 is required not only for pancreas development but also for normal β-cell function and insulin secretion (2, 7). Also, the homeodomain transcription factor HNF-1α, which regulates expression of PDX-1, LRH-1, Beta2/NeuroD, and HNF-4α, has been reported to control insulin secretion (32). Mutations in the human HNF-1α gene are linked to maturity-onset diabetes of the young type 3 (8, 33). Moreover, in HNF-1α−/− mice, PDX-1 and Beta-2/NeuroD levels are downregulated in islets in newborns and adults, confirming the importance of these factors in insulin production (54). A recent study with compound heterozygous mutations for PDX-1+/−/HNF-3β+/− and PDX-1+/−/HNF-1α+/− further underscores the importance of PDX-1, HNF-1α, and HNF-3β in controlling glucose tolerance, glucose-stimulated insulin secretion, and islet architecture (53).
Another player in this regulatory network is HNF-4α, whose promoter is under the control of LRH-1, HNF-1α, and PDX-1 (5, 47, 60). Mutations in the human HNF-4α gene have been linked to maturity-onset diabetes of the young type 1 (64). Furthermore, a mutation of the PDX-1 binding site in the HNF-4α P2 alternative promoter was identified in a large family with maturity-onset diabetes of the young (60). This mutation cosegregates with diabetes and leads to decreased PDX-1 binding and transcriptional activation, suggesting that a transcriptional deregulation of PDX-1 target genes could be linked to maturity-onset diabetes of the young. The atypical nuclear receptor SHP has also been demonstrated to be a direct target gene of both LRH-1 and HNF-4α (22, 37, 54). Interestingly, mutations in the SHP gene were again linked to diabetes (42). Since all these transcription factors, which function either upstream or downstream of LRH-1, as highlighted in Fig. 7, affect glucose homeostasis, it is tempting to speculate that LRH-1 could also participate in this process and alludes to the existence of similar transcriptional regulatory cascades in developing and adult pancreas.
One problem in concluding that LRH-1 also plays a role in the endocrine pancreas is the fact that, unlike the other transcription factors, LRH-1 is not expressed in β-cells but is confined to the exocrine pancreas in the adult mouse (no data exist on LRH-1 expression in human islets). LRH-1 could, however, still take part in β-cell differentiation and homeostasis, a phenomenon linked to its expression in ductal epithelial cells in adult pancreas (49). Ductal cells are known to possess characteristics of pancreatic progenitor cells through their capacity to proliferate or to differentiate into endocrine β-cells (6, 16, 21, 40). Our observation might reinforce a potential link between LRH-1 in the exocrine pancreatic compartment and β-cell neogenesis. The exact role of LRH-1 in ductal cells hence warrants further investigation, and modulation of its activity might eventually open novel therapeutic strategies for the treatment of diabetes by in vitro differentiation of ductal cells into β-cells.
In conclusion, we demonstrate here that the expression of LRH-1 is controlled by the transcription factor PDX-1. LRH-1 might be a component of a transcriptional network involving PDX-1, HNF-1α, HNF-4α, and HNF-3β that determines pancreatic development and could also play a role in pancreas homeostasis in the adult. These data hence underscore the requirement for careful mechanistic and genetic studies to define whether LRH-1 contributes to diseases linked to pancreatic dysfunction, such as diabetes.
Acknowledgments
We thank Lluis Fajas for helpful discussions and Céline Haby, Thomas Ding, Estelle Heitz, Stéphanie Miard, and Maryam Rastegar for excellent technical assistance.
This work was supported by grants from the Centre Nationale de Recherche Scientifique (CNRS), the Institut National pour la Santé et la Recherche Médicale (INSERM), Hopitaux Universitaires de Strasbourg, European Union (QLG1-CT-1999-00674 and QLRT-2001-00930), and NIH (1-P01-DK59820-01). J.S.A. and E.F. are supported by fellowships from the Ligue Nationale Contre le Cancer and the Association pour la Recherche sur le Cancer, respectively.
REFERENCES
- 1.Ahlgren, U., J. Jonsson, and H. Edlund. 1996. The morphogenesis of the pancreatic mesenchyme is uncoupled from that of the pancreatic epithelium in IPF1/PDX1-deficient mice. Development 122:1409-1416. [DOI] [PubMed] [Google Scholar]
- 2.Ahlgren, U., J. Jonsson, L. Jonsson, K. Simu, and H. Edlund. 1998. beta-cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the beta-cell phenotype and maturity onset diabetes. Genes Dev. 12:1763-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ang, S. L., and J. Rossant. 1994. HNF-3 beta is essential for node and notochord formation in mouse development. Cell 78:561-574. [DOI] [PubMed] [Google Scholar]
- 4.Becker-André, M., E. André, and J. F. DeLamarter 1993. Identification of nuclear receptor mRNAs by RT-PCR amplification of conserved zinc-finger motif sequences. Biochem. Biophys. Res. Commun. 194:1371-1379. [DOI] [PubMed] [Google Scholar]
- 5.Boj, S. F., M. Parrizas, M. A. Maestro, and J. Ferrer. 2001. A transcription factor regulatory circuit in differentiated pancreatic cells. Proc. Natl. Acad. Sci. USA 98:14481-14486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouwens, L., and D. G. Pipeleers. 1998. Extra-insular beta cells associated with ductules are frequent in adult human pancreas. Diabetologia 41:629-633. [DOI] [PubMed] [Google Scholar]
- 7.Brissova, M., M. Shiota, W. E. Nicholson, M. Gannon, S. M. Knobel, D. W. Piston, C. V. Wright, and A. C. Powers. 2002. Reduction in pancreatic transcription factor PDX-1 impairs glucose-stimulated insulin secretion. J. Biol. Chem. 277:11225-11232. [DOI] [PubMed] [Google Scholar]
- 8.Byrne, M. M., J. Sturis, S. Menzel, K. Yamagata, S. S. Fajans, M. J. Dronsfield, S. C. Bain, A. T. Hattersley, G. Velho, P. Froguel, G. I. Bell, and K. S. Polonsky. 1996. Altered insulin secretory responses to glucose in diabetic and nondiabetic subjects with mutations in the diabetes susceptibility gene MODY3 on chromosome 12. Diabetes 45:1503-1510. [DOI] [PubMed] [Google Scholar]
- 9.del Castillo-Olivares, A., and G. Gil. 2001. Suppression of sterol 12alpha-hydroxylase transcription by the short heterodimer partner: insights into the repression mechanism. Nucleic Acids Res. 29:4035-4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dolle, P., and D. Duboule. 1989. Two gene members of the murine HOX-5 complex show regional and cell-type specific expression in developing limbs and gonads. EMBO J. 8:1507-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duncan, S. A., M. A. Navas, D. Dufort, J. Rossant, and M. Stoffel. 1998. Regulation of a transcription factor network required for differentiation and metabolism. Science 281:692-695. [DOI] [PubMed] [Google Scholar]
- 12.Dusing, M. R., E. A. Florence, and D. A. Wiginton. 2001. Pdx-1 is required for activation in vivo from a duodenum-specific enhancer. J. Biol. Chem. 276:14434-14442. [DOI] [PubMed] [Google Scholar]
- 13.Edlund, H. 2002. Pancreatic organogenesis-developmental mechanisms and implications for therapy. Nat. Rev. Genet. 3:524-532. [DOI] [PubMed] [Google Scholar]
- 14.Edlund, H. 1998. Transcribing pancreas. Diabetes 47:1817-1823. [DOI] [PubMed] [Google Scholar]
- 15.Ellinger-Ziegelbauer, H., A. K. Hihi, V. Laudet, H. Keller, W. Wahli, and C. Dreyer. 1994. Ftz-F1-related orphan receptors in Xenopus laevis: transcriptional regulators differentially expressed during early embryogenesis. Mol. Cell. Biol. 14:2786-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elsasser, H. P., A. Biederbick, and H. Kern. 1994. Growth of rat pancreatic acinar cells quantitated with a monoclonal antibody against the proliferating cell nuclear antigen. Cell Tissue Res. 276:603-609. [DOI] [PubMed] [Google Scholar]
- 17.Fajas, L., R. L. Landsberg, Y. Huss-Garcia, C. Sardet, J. A. Lees, and J. Auwerx. 2002. E2Fs regulate adipocyte differentiation. Dev. Cell 3:39-49. [DOI] [PubMed] [Google Scholar]
- 18.Galarneau, L., R. Drouin, and L. Bèlanger. 1998. Assignment of the fetoprotein transcription factor gene (FTF) to human chromosome band 1q32.11 by in situ hybridization. Cytogenet. Cell Genet. 82:269-270. [DOI] [PubMed] [Google Scholar]
- 19.Galarneau, L., J.-F. Paré, D. Allard, D. Hamel, L. Lévesque, J. D. Tugwood, S. Green, and L. Bélanger. 1996. The α1-fetoprotein locus is activated by a nuclear receptor of the drosphila Ftz-F1 family. Mol. Cell. Biol. 16:3853-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerrish, K., M. Gannon, D. Shih, E. Henderson, M. Stoffel, C. V. Wright, and R. Stein. 2000. Pancreatic beta cell-specific transcription of the pdx-1 gene. The role of conserved upstream control regions and their hepatic nuclear factor 3beta sites. J. Biol. Chem. 275:3485-3492. [DOI] [PubMed] [Google Scholar]
- 21.Githens, S. 1988. The pancreatic duct cell: proliferative capabilities, specific characteristics, metaplasia, isolation, and culture. J. Pediatr. Gastroenterol. Nutr. 7:486-506. [PubMed] [Google Scholar]
- 22.Goodwin, B., S. A. Jones, R. R. Price, M. A. Watson, D. D. McKee, L. B. Moore, C. Galardi, J. G. Wilson, M. C. Lewis, M. E. Roth, P. R. Maloney, T. M. Willson, and S. A. Kliewer. 2000. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell 6:517-526. [DOI] [PubMed] [Google Scholar]
- 23.Hani, E. H., D. A. Stoffers, J. C. Chevre, E. Durand, V. Stanojevic, C. Dina, J. F. Habener, and P. Froguel. 1999. Defective mutations in the insulin promoter factor-1 (IPF-1) gene in late-onset type 2 diabetes mellitus. J. Clin. Investig. 104:R41-R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inokuchi, A., E. Hinoshita, Y. Iwamoto, K. Kohno, M. Kuwano, and T. Uchiumi. 2001. Enhanced expression of the human multidrug resistance protein 3 by bile salt in human enterocytes. A transcriptional control of a plausible bile acid transporter. J. Biol. Chem. 276:46822-46829. [DOI] [PubMed] [Google Scholar]
- 25.Jonsson, J., U. Ahlgren, T. Edlund, and H. Edlund. 1995. IPF1, a homeodomain protein with a dual function in pancreas development. Int. J. Dev. Biol. 39:789-798. [PubMed] [Google Scholar]
- 26.Jonsson, J., L. Carlsson, T. Edlund, and H. Edlund. 1994. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature 371:606-609. [DOI] [PubMed] [Google Scholar]
- 27.Kawaguchi, Y., B. Cooper, M. Gannon, M. Ray, R. J. MacDonald, and C. V. Wright. 2002. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat. Genet. 32:128-134. [DOI] [PubMed] [Google Scholar]
- 28.Krapp, A., M. Knofler, B. Ledermann, K. Burki, C. Berney, N. Zoerkler, O. Hagenbuchle, and P. K. Wellauer. 1998. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev. 12:3752-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kudo, T., and S. Sutou. 1997. Molecular cloning of chicken FTZ-F1-related orphan receptors. Gene 2197:261-268. [DOI] [PubMed] [Google Scholar]
- 30.Lavorgna, G., H. Ueda, J. Clos, and C. Wu. 1991. FTZ-F1, a steroid hormone receptor-like protein implicated in the activation of fushi tarazu. Science 252:848-851. [DOI] [PubMed] [Google Scholar]
- 31.Lee, C. S., N. J. Sund, M. Z. Vatamaniuk, F. M. Matschinsky, D. A. Stoffers, and K. H. Kaestner. 2002. Foxa2 controls Pdx1 gene expression in pancreatic beta-cells in vivo. Diabetes 51:2546-2551. [DOI] [PubMed] [Google Scholar]
- 32.Lee, Y. H., B. Sauer, and F. J. Gonzalez. 1998. Laron dwarfism and non-insulin-dependent diabetes mellitus in the Hnf-1alpha knockout mouse. Mol. Cell. Biol. 18:3059-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lehto, M., T. Tuomi, M. M. Mahtani, E. Widen, C. Forsblom, L. Sarelin, M. Gullstrom, B. Isomaa, M. Lehtovirta, A. Hyrkko, T. Kanninen, M. Orho, S. Manley, R. C. Turner, T. Brettin, A. Kirby, J. Thomas, G. Duyk, E. Lander, M. R. Taskinen, and L. Groop. 1997. Characterization of the MODY3 phenotype. Early onset diabetes caused by an insulin secretion defect. J. Clin. Investig. 99:582-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, M., Y.-H. Xie, Y.-Y. Kong, X. Wu, L. Zhu, and Y. Wang. 1998. Cloning and characterization of a novel human hepatocyte transcription factor, hB1F, which binds and activates enhancer II of hepatitis B virus. J. Biol. Chem. 273:29022-29031. [DOI] [PubMed] [Google Scholar]
- 35.Liu, D., Y. Le Drean, M. Ekker, F. Xiong, and C. L. Hew. 1997. Teleost FTZ-F1 homolog and its splicing variant determine the expression of the salmon gonadotropin IIbeta subunit gene. Mol. Endocrinol. 11:877-890. [DOI] [PubMed] [Google Scholar]
- 36.Liu, Y., R. J. MacDonald, and G. H. Swift. 2001. DNA binding and transcriptional activation by a PDX1. PBX1b. MEIS2b trimer and cooperation with a pancreas-specific basic helix-loop-helix complex. J. Biol. Chem. 276:17985-17993. [DOI] [PubMed] [Google Scholar]
- 37.Lu, T. T., M. Makishima, J. J. Repa, K. Schoonjans, T. A. Kerr, J. Auwerx, and D. J. Mangelsdorf. 2000. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell 6:507-515. [DOI] [PubMed] [Google Scholar]
- 38.Luo, Y., C. P. Liang, and A. R. Tall. 2001. The orphan nuclear receptor LRH-1 potentiates the sterol-mediated induction of the human CETP gene by liver X receptor. J. Biol. Chem. 276:24767-24773. [DOI] [PubMed] [Google Scholar]
- 39.Mangelsdorf, D. J., C. Thummel, M. Beato, P. Herrlich, G. Schutz, K. Umesono, B. Blumberg, P. Kastner, E. Mark, P. Chambon, and R. M. Evans. 1995. The nuclear receptor superfamily: the second decade. Cell 83:835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muller, R., R. Laucke, B. Trimper, L. Cossel, and D. D. R. Frauenklinik Stendal. 1990. Pancreatic cell proliferation in normal rats studied by in vivo autoradiography with 3H-thymidine. Virchows Arch. B Cell Pathol. Incl. Mol. Pathol. 59:133-136. [DOI] [PubMed] [Google Scholar]
- 41.Nakajima, T., M. Takase, I. Miura, and M. Nakamura. 2000. Two isoforms of FTZ-F1 messenger RNA: molecular cloning and their expression in the frog testis. Gene 248:203-212. [DOI] [PubMed] [Google Scholar]
- 42.Nishigori, H., H. Tomura, N. Tonooka, M. Kanamori, S. Yamada, K. Sho, I. Inoue, N. Kikuchi, K. Onigata, I. Kojima, T. Kohama, K. Yamagata, Q. Yang, Y. Matsuzawa, T. Miki, S. Seino, M. Y. Kim, H. S. Choi, Y. K. Lee, D. D. Moore, and J. Takeda. 2001. Mutations in the small heterodimer partner gene are associated with mild obesity in Japanese subjects. Proc. Natl. Acad. Sci. USA 98:575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nitta, M., S. Ku, C. Brown, A. Y. Okamoto, and B. Shan. 1999. CPF: an orphan nuclear receptor that regulates liver-specific expression of the human cholesterol 7alpha-hydroxylase gene. Proc. Natl. Acad. Sci. USA 96:6660-6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Offield, M. F., T. L. Jetton, P. A. Labosky, M. Ray, R. W. Stein, M. A. Magnuson, B. L. Hogan, and C. V. Wright. 1996. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development 122:983-995. [DOI] [PubMed] [Google Scholar]
- 45.Ohlsson, H., K. Karlsson, and T. Edlund. 1993. IPF1, a homeodomain-containing transactivator of the insulin gene. EMBO J. 12:4251-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohlsson, H., S. Thor, and T. Edlund. 1991. Novel insulin promoter- and enhancer-binding proteins that discriminate between pancreatic alpha- and beta-cells. Mol. Endocrinol. 5:897-904. [DOI] [PubMed] [Google Scholar]
- 47.Paré, J. F., S. Roy, L. Galarneau, and L. Belanger. 2001. The mouse fetoprotein transcription factor (ftf) gene promoter is regulated by three gata elements with tandem e box and nkx motifs, and ftf in turn activates the hnf3beta, hnf4alpha, and hnf1alpha gene promoters. J. Biol. Chem. 276:13136-13144. [DOI] [PubMed] [Google Scholar]
- 48.Parker, K. L. 1998. The roles of steroidogenic factor 1 in endocrine development and function. Mol. Cell Endocrinol. 140:59-63. [DOI] [PubMed] [Google Scholar]
- 49.Rausa, F. M., L. Galarneau, L. Bélanger, and R. H. Costa. 1999. The nuclear receptor fetoprotein transcription factor is coexpressed with its target gene HNF-3β in the developing murine liver, intestine and pancreas. Mech. Dev. 89:185-188. [DOI] [PubMed] [Google Scholar]
- 50.Schoonjans, K., J. S. Annicotte, T. Huby, O. A. Botrugno, E. Fayard, Y. Ueda, J. Chapman, and J. Auwerx. 2002. Liver receptor homolog 1 controls the expression of the scavenger receptor class B type I. EMBO Rep. 3:1181-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schoonjans, K., J. Peinado-Onsurbe, A. M. Lefebvre, R. Heyman, M. Briggs, S. Deeb, B. Staels, and J. Auwerx. 1996. PPARα and PPARγ activators direct a tissue-specific transcriptional response via a PPRE in the lipoprotein lipase gene. EMBO J. 15:5336-5348. [PMC free article] [PubMed] [Google Scholar]
- 52.Serup, P., J. Jensen, F. G. Andersen, M. C. Jorgensen, N. Blume, J. J. Holst, and O. D. Madsen. 1996. Induction of insulin and islet amyloid polypeptide production in pancreatic islet glucagonoma cells by insulin promoter factor 1. Proc. Natl. Acad. Sci. USA 93:9015-9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shih, D. Q., M. Heimesaat, S. Kuwajima, R. Stein, C. V. Wright, and M. Stoffel. 2002. Profound defects in pancreatic beta-cell function in mice with combined heterozygous mutations in Pdx-1, Hnf-1alpha, and Hnf-3beta. Proc. Natl. Acad. Sci. USA 99:3818-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shih, D. Q., S. Screenan, K. N. Munoz, L. Philipson, M. Pontoglio, M. Yaniv, K. S. Polonsky, and M. Stoffel. 2001. Loss of HNF-1alpha function in mice leads to abnormal expression of genes involved in pancreatic islet development and metabolism. Diabetes 50:2472-2480. [DOI] [PubMed] [Google Scholar]
- 55.Shih, D. Q., and M. Stoffel. 2001. Dissecting the transcriptional network of pancreatic islets during development and differentiation. Proc. Natl. Acad. Sci. USA 98:14189-14191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stoffers, D. A., J. Ferrer, W. L. Clarke, and J. F. Habener. 1997. Early-onset type-II diabetes mellitus (MODY4) linked to IPF1. Nat. Genet. 17:138-139. [DOI] [PubMed] [Google Scholar]
- 57.Sund, N. J., S. L. Ang, S. D. Sackett, W. Shen, N. Daigle, M. A. Magnuson, and K. H. Kaestner. 2000. Hepatocyte nuclear factor 3β (Foxa2) is dispensable for maintaining the differentiated state of the adult hepatocyte. Mol. Cell. Biol. 20:5175-5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swift, G. H., Y. Liu, S. D. Rose, L. J. Bischof, S. Steelman, A. M. Buchberg, C. V. Wright, and R. J. MacDonald. 1998. An endocrine-exocrine switch in the activity of the pancreatic homeodomain protein PDX1 through formation of a trimeric complex with PBX1b and MRG1 (MEIS2). Mol. Cell. Biol. 18:5109-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tanaka, T., T. Takeno, Y. Watanabe, Y. Uchiyama, T. Murakami, H. Yamashita, A. Suzuki, R. Aoi, H. Iwanari, S. Jiang, M. Naito, K. Tachibana, T. Doi, A. I. Shulman, D. J. Mangelsdorf, R. Reiter, J. Auwerx, T. Hamakubo, and T. Kodama. 2002. The generation of monoclonal antibodies against human peroxisome proliferator-activated receptors (PPARs). J. Atheroscler. Thromb. 9:233-242. [DOI] [PubMed] [Google Scholar]
- 60.Thomas, H., K. Jaschkowitz, M. Bulman, T. M. Frayling, S. M. Mitchell, S. Roosen, A. Lingott-Frieg, C. J. Tack, S. Ellard, G. U. Ryffel, and A. T. Hattersley. 2001. A distant upstream promoter of the HNF-4alpha gene connects the transcription factors involved in maturity-onset diabetes of the young. Hum. Mol. Genet. 10:2089-2097. [DOI] [PubMed] [Google Scholar]
- 61.Waeber, G., N. Thompson, P. Nicod, and C. Bonny. 1996. Transcriptional activation of the GLUT2 gene by the IPF-1/STF-1/IDX-1 homeobox factor. Mol. Endocrinol. 10:1327-1334. [DOI] [PubMed] [Google Scholar]
- 62.Watada, H., Y. Kajimoto, J. Miyagawa, T. Hanafusa, K. Hamaguchi, T. Matsuoka, K. Yamamoto, Y. Matsuzawa, R. Kawamori, and Y. Yamasaki. 1996. PDX-1 induces insulin and glucokinase gene expressions in alphaTC1 clone 6 cells in the presence of betacellulin. Diabetes 45:1826-1831. [DOI] [PubMed] [Google Scholar]
- 63.Weinstein, D. C., A. Ruiz i Altaba, W. S. Chen, P. Hoodless, V. R. Prezioso, T. M. Jessell, and J. E. Darnell, Jr. 1994. The winged-helix transcription factor HNF-3 beta is required for notochord development in the mouse embryo. Cell 78:575-588. [DOI] [PubMed] [Google Scholar]
- 64.Yamagata, K., H. Furuta, N. Oda, P. J. Kaisaki, S. Menzel, N. J. Cox, S. S. Fajans, S. Signorini, M. Stoffel, and G. I. Bell. 1996. Mutations in the hepatocyte nuclear factor-4alpha gene in maturity-onset diabetes of the young (MODY1). Nature 384:458-460. [DOI] [PubMed] [Google Scholar]
- 65.Zhang, C. K., W. Lin, Y. N. Cai, P. L. Xu, H. Dong, M. Li, Y. Y. Kong, G. Fu, Y. H. Xie, G. M. Huang, and Y. Wang. 2001. Characterization of the genomic structure and tissue-specific promoter of the human nuclear receptor NR5A2 (hB1F) gene. Gene 273:239-249. [DOI] [PubMed] [Google Scholar]