Abstract
Polypeptide growth factors, such as platelet-derived growth factor (PDGF), promote the reinitiation of DNA synthesis and cell growth through multiple intracellular signaling pathways that converge in the nucleus to regulate the activity of transcription factors, thereby controlling the expression of growth-promoting genes. Among them, the AP-1 (activating protein-1) family of transcription factors, including c-Fos and c-Jun family members, plays a key role, as AP-1 activity is potently activated by PDGF and is required to stimulate cell proliferation. However, the nature of the pathways connecting PDGF receptors to AP-1 is still poorly defined. In this study, we show that PDGF regulates AP-1 by stimulating the expression and function of c-Fos through extracellular signal-regulated kinase (ERK). The latter involves the direct phosphorylation by ERK of multiple residues in the carboxyl-terminal transactivation domain of c-Fos, which results in its increased transcriptional activity. Interestingly, the phosphorylation of c-Fos by ERK was required for the ability of PDGF and serum to stimulate the activity of c-Fos as well as AP-1-dependent transcription. Furthermore, we provide evidence that the ERK-dependent activation of c-Fos is an integral component of the mitogenic pathway by which PDGF regulates normal and aberrant cell growth.
Exposure of quiescent cells to polypeptide growth factors initiates a cascade of biochemical events that results in key cell fate decisions, including cell proliferation, differentiation, survival, or death. Many of these biological responses are highly dependent on changes in the protein level and activity of an array of transcription factors, which in turn coordinate the expression of sets of genes that are involved in each specific cellular outcome. Among them, the transcription complex AP-1 (activating protein-1) is rapidly and transiently induced in response to a wide variety of external signals (4, 25). AP-1 is composed of Fos (c-Fos, Fos B, Fra-1, and Fra-2) and Jun (c-Jun, JunB, and JunD) family proteins. Whereas homo- and heterodimers of Jun proteins can bind DNA directly, Fos members require interaction with any of the Jun proteins to act as transcriptional activators (4, 38). Fos-Jun dimers activate transcription by binding to core TGAC/GTC/AA sequences, known as tetradecanoyl phorbol acetate-responsive elements or AP-1 sites (3, 4, 30). A wide array of genes have been found to contain AP-1 sites in their promoter-enhancer regions, including collagenase, stromelysin, metallothionein IIA, interleukin-2, transforming growth factor β, and cyclin D1, among others (4), and the timely activation of AP-1 has been implicated in the control of a number of key cellular processes (40, 41). Among them, the role of AP-1 in cell growth control is further supported by the early observations that deregulated expression of certain Jun and Fos family members can result in neoplastic transformation (4, 32) and that inhibition of AP-1 proteins by microinjection of blocking antibodies prevents cell growth in response to serum (27).
How the activity of AP-1 is regulated in response to growth factors has been the subject of intense investigation. In the case of c-Jun, recent work has revealed that its expression and activity are tightly regulated by members of the mitogen-activated protein kinase (MAPK) family, including c-Jun N-terminal kinases (JNKs), extracellular signal-regulated protein kinase 5 (ERK5), and p38 kinases, by acting on transcription factors that bind to the c-jun promoter (34). Similarly, c-Fos expression is also regulated at multiple steps. In fact, perhaps the most-studied aspect of c-Fos biology is the control of c-fos mRNA synthesis, as the activity of its promoter can be modulated by a myriad of extracellular signals acting through any of its several cis inducible elements, among which the serum response element (SRE) is believed to play a central regulatory role (41, 46, 47). This site confers to the c-fos promoter the ability to respond to growth factors, cytokines, cellular stress, and other stimuli that promote transcription from the SRE through a number of intracellular pathways, including the stimulation of ERK1 and -2 (ERKs), JNK, and p38 kinases (24, 45, 48).
Another component involved in the process of AP-1 activation is the posttranslational processing of preexisting or newly synthesized Fos and Jun proteins (25, 38). In particular, the reversible phosphorylation of Fos and Jun family members may result in the positive or negative modulation of their transactivating properties (21, 22), as well as their stability, translocation to the nucleus, and rate of binding to target DNA sequences. This mechanism of posttranslational control was extensively documented for c-Jun. In this case, JNK phosphorylation at Ser-63 and Ser-73 within the N-terminal transactivation domain (TAD) potentiates the ability of c-Jun to activate transcription either as a homodimer or a heterodimer with c-Fos (25). In contrast, the nature of the potential kinases regulating the transactivating activity of c-Fos is not yet clearly defined. In this regard, a still-unidentified kinase activity, termed Fos-related kinase (FRK), has been shown to cause a phosphorylation-dependent activation of c-Fos (13). Experimental evidence suggests that FRK is a Ras- and growth factor-responsive proline-directed threonine kinase and, thus, most probably related to MAPKs (13). c-Fos phosphorylation by ribosomal S6 kinase (RSK) and ERK has also been described to occur soon after serum stimulation (8, 9), but the significance of these events in the context of c-Fos activity and function remains unclear.
Of interest, the rapid accumulation of c-fos mRNA upon exposure of quiescent mouse fibroblasts to platelet-derived growth factor (PDGF) was one of the earliest observations linking the activity of this potent polypeptide mitogen to the nuclear expression of growth-promoting genes (11, 28). Since then, the contribution of AP-1 activity to cell proliferation (19, 51), expression of tissue-remodeling proteases (26, 39), and cytokine secretion (16) in response to PDGF has been extensively documented. However, how PDGF controls the activity of AP-1 is not yet fully understood. For example, in mouse fibroblasts PDGF provokes a robust activation of c-fos expression but a modest increase in the expression of c-jun (12). Furthermore, PDGF stimulates the activity of ERK1 and ERK2 potently, but in most cells it has a limited effect on the activity of JNK and p38 family members (12, 29), thus raising the possibility that PDGF may utilize a biochemical route distinct from that regulating c-Jun to stimulate AP-1 activity.
In the present study, we set out to investigate the nature of the intracellular signaling pathways connecting the activation of PDGF receptors to AP-1 stimulation. We found that PDGF regulates AP-1 primarily by stimulating the expression and function of c-Fos through ERK. The latter involves the direct phosphorylation of the C-terminal TAD of c-Fos by ERK, thereby enhancing its transcriptional activity. Furthermore, we provide evidence that the ERK-dependent activation of c-Fos is an integral component of the mitogenic pathway by which PDGF regulates normal and aberrant cell growth.
MATERIALS AND METHODS
Recombinant human PDGF (rHu PDGF-BB) was purchased from Intergen Company. The anti-Gal4 DNA binding domain (DBD) monoclonal antibody (RK5C1), the anti-N-terminal c-Fos polyclonal antibody (sc-52), the anti-phospho-ERK mouse monoclonal antibody (sc-7383), and the anti-total ERK2 rabbit polyclonal antibody (C-14) were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.). The anti-C-terminal c-Fos polyclonal antibody (ab2) was from Geneka Biotechnology Inc. (Montreal, Quebec, Canada). The anti-phospho threonine-proline monoclonal antibody was from Cell Signaling Technology (Beverly, Mass.). The anti-tag monoclonal antibodies (for hemagglutinin [HA], AU5, and six-His) were from Covance (Berkeley, Calif.). The MEK inhibitor U0126 was obtained from Promega Corp. (Madison, Wis.). The p38 and MEK inhibitors SB203580 hydrochloride and PD98059 were purchased from Calbiochem-Novabiochem Corp. (La Jolla, Calif.). The JNK inhibitor SP600125 [anthra(1,9-cd)pyrazol-6(2H)-one], was from Biomol (Plymouth Meeting, Pa.). Protein phosphatase-2A (PP2A) and recombinant ERK2 (active) were purchased from Upstate Biotechnology (Lake Placid, N.Y.). [32P]ATP was obtained from New England Nuclear (Chicago, Ill.). All other reagents were of analytical grade.
DNA constructs.
The full-length cDNA from the murine c-fos gene, containing amino acids (aa) 1 to 380, was PCR amplified and cloned as an EcoRI/NotI fragment into pCEFL-AU5, a modified pcDNAIII expression vector containing the elongation factor 1 (EF-1) promoter driving the expression of an in-frame N-terminal AU5 tag (34). The expression vectors for the constitutively active MAPK kinases (MAPKKs; pCEV29-MEKEE, pCEFL-MEKK1, and pCEFL-MEK3EE), the HA-tagged MAPKs (ERK2, JNK1, and p38α), c-Jun (pCEFL-c-Jun) and a dominant negative MEK1 with alanine replacements of Ser-218 and -222 (pcDNAIII-MEKAA) have been previously described (10, 33). The B-chain of the human PDGF gene (c-sis) (18) was cloned into the pcDNAIII expression vector. The pAP-1 luciferase reporter plasmid was obtained from Stratagene. This construct has been designed by inserting seven tandem repeats of the AP-1 response element from the polyoma enhancer (TGA C TAA) into the pA3 vector (17). To obtain the Gal4-c-FosTAD fusion protein, the C-terminal domain of mouse c-Fos (aa 209 to 380) (42) was PCR amplified from its full-length cDNA and cloned in a pcDNAIII vector encoding the DBD of the yeast transcription factor Gal4 (pcDNAIII-GBDEX). A similar construct was made by fusing the full-length c-Fos cDNA to the same expression vector (Gal4-c-FosFL). A Gal4-driven luciferase reporter, pGal4-Luc (34), was designed by inserting six tandem copies of Gal4-responsive elements and a minimum TATA initiator in place of the simian virus 40 (SV40) promoter in pGL3 (Promega).
Individual point mutations of pCEFL-AU5-c-Fos and Gal4-c-FosTAD on Thr-232, Thr-331, Thr-325, and Ser-374 were generated by site-directed mutagenesis (QuikChange site-directed mutagenesis kit; Stratagene). To obtain a pCEFL-AU5-c-Fos clone containing multiple alanine replacements on Thr-232, Thr-325, Thr-331, and Ser-374 (c-Fos-m), we used the QuikChange multisite-directed mutagenesis kit (Stratagene). The sequences of the primers used for mutagenesis will be provided upon request. Add-back mutants including a single potential phospho-acceptor site were generated by reintroducing each original residue on c-Fos-m, which was used as a template for mutagenesis. BamHI/NotI PCR fragments from c-FosTAD-m and the different mutants were also cloned in pcDNAIII-GBDEX to obtain expression plasmids for the corresponding Gal4-c-FosTAD fusion proteins.
To obtain the polyhistidine-tagged (six-His) c-Fos TAD constructs, the TADs from c-Fos wild type and the different c-Fos mutants were PCR amplified and cloned as BamHI/NotI fragments in a modified pRSET-A bacterial expression vector (Invitrogen Corp.) in which the HindIII recognition sequence at the polylinker was replaced by a NotI site.
Cell cultures.
NIH 3T3 mouse fibroblasts were routinely cultured in serum-supplemented medium composed of Dulbecco's modified Eagle's medium (DMEM; Life Technologies, Inc.), 10% calf serum, and penicillin-streptomycin-amphotericin B (Life Technologies, Inc.). HEK-293T cells were grown in DMEM containing 10% fetal bovine serum (FBS) and the above antimicrobial mixture.
Preparation of nuclear extracts.
Cells were collected after overnight starvation followed by induction with PDGF (20 ng/ml) or serum with or without previous treatment with MAPK inhibitors. The monolayers were washed with Tris-buffered saline and lysed with buffer containing 10 mM HEPES (pH 7.9), 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM dithiothreitol (DTT), and 0.5% NP-40. Cell lysates were centrifuged, and the resulting nuclei (pellet) were disrupted with extraction buffer (20 mM HEPES [pH 7.9], 0.5 M NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT). Cell debris were separated by low-speed centrifugation. Nuclei aliquots (2 to 5 μg of protein/μl) were stored at −70°C until use. All steps were performed at 4°C.
Transient transfections.
NIH 3T3 and HEK-293T cells were plated in complete medium and allowed to grow overnight to 70 to 80% confluence in six-well plates unless otherwise noted. The cells were transfected for 3 h in serum-free DMEM containing up to 2 μg of total plasmid DNA together with the Lipofectamine Plus reagent (Life Technologies, Inc.) according to the protocol suggested by the manufacturer.
Luciferase reporter assays.
Cells were transfected with different expression vectors together with 0.1 μg of each luciferase reporter and 0.01 μg of pRL-null (a plasmid encoding the luciferase gene from Renilla reniformis), which served as an internal control for transfection efficiency. The total amount of transfected DNA was normalized by adjusting it with pcDNAIII-β-gal, an expression vector for the enzyme β-galactosidase. Cells were lysed in passive lysis buffer (Promega) 24 h posttransfection. Cell lysates (50 μl/well) were transferred to a 96-well luminometer plate, and firefly and Renilla luciferase activities were assayed using the Dual-Luciferase reporter system (Promega). Light emission was quantified using a Microliter plate luminometer as specified by the manufacturer (DINEX Tech).
Focus-forming assays.
NIH 3T3 cells were grown up to 10 to 20% confluence in 10-cm plates and transfected following the calcium-phosphate precipitation technique. The day after transfection, cells were washed and incubated in DMEM supplemented with 5% calf serum. The cultures were maintained in the same medium, with medium changes every 3 days, until the appearance of foci from transformed cells was evident (usually 15 to 25 days after transfection).
Purification of GST and six-His fusion proteins.
Escherichia coli BL-21 Lys cells (Promega Corp.) were transformed with the vectors pGEX-4T3 and pRSET-A encoding glutathione acetyl transferase (GST) and polyhistidine (six-His) fusion proteins, respectively. Bacteria were grown in Luria-Bertani medium until the optical density reached 0.5. Protein synthesis was stimulated by the addition of 1 mM isopropyl-β-thiogalactopyranoside, and bacteria were allowed to grow for 3 h. The cells were collected by centrifugation (3,000 × g, 30 min) and resuspended in buffer containing phosphate-buffered saline (PBS), 1% Triton X-100, 1 mM EDTA, 2 μg of aprotinin/ml, 2 μg of leupeptin/ml, and 1 mM PMSF. The cell suspension was sonicated, and cellular debris was removed by centrifugation. The supernatant was incubated (1 h, 4°C) with 300 μl of glutathione-Sepharose 4B (Amersham-Pharmacia Biotech). The beads were pelleted by centrifugation, washed three times with PBS, 1% Triton X-100, 2 μg of aprotinin/ml, 2 μg of leupeptin/ml, 1 mM PMSF and then twice with PBS, 2 μg of aprotinin/ml, 2 μg of leupeptin/ml, 1 mM PMSF. GST fusion proteins were eluted with glutathione buffer (10 mM glutathione in 50 mM Tris [pH 8.0], 2 μg of aprotinin/ml, 2 μg of leupeptin/ml, 1 mM PMSF).
Six-His-tagged proteins were isolated using nickel-nitrilotriacetic acid magnetic agarose beads (Qiagen) following the manufacturer's protocol for bacterial expression and purification under native conditions.
In vitro kinase assays.
HEK-293T cells were allowed to grow to up to 70 to 80% confluence in 6-cm plates and then transfected with expression vectors for HA-tagged kinases alone or in combination with the respective upstream activating molecules. The cells were incubated in serum-supplemented medium overnight after transfection (Lipofectamine Plus) and then serum starved for 6 h (ERK2) or 2 h (JNK and p38α) before lysis for kinase reaction. The plates were washed with iced-cold PBS and resuspended in lysis buffer (25 mM HEPES [pH 7.5], 0.3 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 20 mM β-glycerophosphate, 1 mM Na-vanadate, 1% NP-40, 1 mM PMSF, 20 μg of aprotinin/ml, and 20 μg of leupeptin/ml). Cleared lysates were subjected to immunoprecipitation using anti-HA monoclonal antibodies (1 h, 4°C) followed by incubation with protein G-Sepharose (Gamma-Bind G Sepharose; Amersham-Pharmacia Biotech). The beads were pelleted by centrifugation and washed three times with PBS, 1% NP-40, 2 mM Na-vanadate, followed by one wash with 100 mM Tris (pH 7.5), 0.5 M LiCl and one final wash with kinase reaction buffer (12.5 mM morpholinepropanesulfonic acid [MOPS] [pH 7.5], 12.5 mM glycerophosphate, 7.5 mM MgCl2, 0.5 mM EGTA, 0.5 mM sodium fluoride, 0.5 mM Na-vanadate). The reactions were initiated by the addition of 30 μl of reaction mixture (kinase buffer plus 10 μCi of [γ32P]ATP, 20 μM unlabeled ATP, 1 mM DTT, 0.5 to 1 μg of substrate) to the immunocomplexes. After 30 min at 30°C, the reactions were terminated by the addition of sodium dodecyl sulfate (SDS) sample buffer (400 mM Tris-HCl [pH 6.8], 10% SDS, 50% glycerol, 500 mM DTT, 2 μg of bromophenol blue/ml), followed by 5 min of boiling. ERK2 kinase activity was assayed using 0.5 μg of myelin basic protein (Sigma) as a positive control. JNK and p38α activities were assayed using 1 μg of purified GST-ATF2 (aa 1 to 96). Denatured samples were resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) on 12% polyacrylamide gels, and autoradiographs were taken from dried gels using X-Omat Kodak films. Cold kinase assays were performed identically except for the omission of radioactive ATP from the reaction mix. The samples were immunoblotted using anti-phospho-specific antibodies as described below.
Western blot analysis.
Cleared cell lysates were combined with SDS sample buffer, boiled for 5 min, and resolved by SDS-10% PAGE. Fractionated proteins were electrophoretically transferred to polyvinylidene fluoride membranes (Immobilon-P; Millipore). Nonspecific binding sites were blocked with 5% nonfat dried milk in PBS containing 0.05% Tween 20 (PBS-T) and incubated (1 h, room temperature) with the appropriate dilution of each primary antiserum or monoclonal antibody. The membranes were repeatedly washed with PBS-T prior to incubation with horseradish peroxidase-conjugated goat anti-mouse or anti-rabbit secondary antibodies (ICN-Cappel). Immunoreactive protein bands were visualized by enhanced chemiluminescence detection (ECL Plus System; Amersham Biosciences).
Northern blot analysis.
NIH 3T3 cells were grown to 70% confluence in 10-cm plates and serum starved overnight. Cells were left untreated or treated with 20 ng of PDGF/ml for different times, in the absence or presence of U0126. Cells were washed with cold PBS, and total RNA was extracted by homogenization in TRIzol (GIBCO BRL) according to the manufacturer's specifications. For Northern blotting, 20 μg of total RNA was fractionated in 2% formaldehyde-agarose gels, transferred to nylon membranes, and hybridized with a 32P-labeled cDNA probe from the murine c-Fos TAD, which was prepared using the Prime-a-Gene labeling system (Promega). Accuracy in gel loading and transfer was confirmed by fluorescence under UV light upon ethidium bromide staining of the gels.
Phosphatase treatment.
NIH 3T3 and HEK-293T cells overexpressing c-Fos proteins were rapidly washed with cold PBS, scraped, and collected by centrifugation. Cells were disrupted by freeze-thaw cycles and resuspended in a buffer containing 20 mM MOPS (pH 7.3), 150 mM NaCl, 1 mM MgCl2, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, and 20 μg of leupeptin/ml. Protein aliquots were incubated in the absence or presence of 0.5 U of PP2A (2 h, 30°C). The reactions were stopped by adding SDS sample buffer and processed for immunoblotting as described above.
RESULTS
Stimulation of AP-1 activity by PDGF depends on MAPKs.
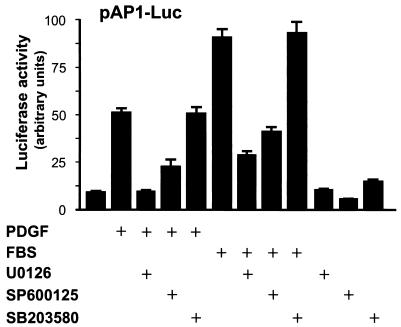
Stimulation of fibroblasts with PDGF, which acts on its cognate receptor tyrosine kinase, leads to the rapid activation of a number of intracellular signaling pathways, many of which converge in the nucleus to ultimately regulate the activity of AP-1-responsive promoters. Among these biochemical routes, cytoplasmic kinase cascades acting on members of the MAPK family have been shown to play a prominent role (48). To begin analyzing the mechanism whereby PDGF stimulates AP-1 activation, we used a reporter plasmid (pAP-1-Luc) that carries a luciferase gene under the control of seven tandem repeats of AP-1 response elements, which can be activated by c-Jun and c-Fos family members (17). As expected, PDGF and serum potently induced AP-1 transcription in NIH 3T3 cells (Fig. 1). As an initial approach to examine whether MAPK pathways connect PDGF signaling to AP-1, we made use of inhibitors with specificity for the ERK1/2 cascade, JNK 1/2/3, and two of the four p38 isoforms (14). As shown in Fig. 1, JNK-dependent pathways played a role in this response, as judged by the blocking effect displayed by SP600125, which inhibits JNK activity by competing for the ATP binding site in this kinase (7). This result is consistent with the ability of JNK to phosphorylate c-Jun and thereby regulate its transcriptional activity and mRNA expression (24). In contrast the inhibitor for p38α and p38β, SB203580 (44), had no apparent effect on AP-1 activity. Interestingly, U0126, which specifically inhibits ERK activation by preventing its phosphorylation by the upstream stimulating kinase MEK (15), nearly abolished the activation of AP-1 by PDGF and serum. Thus, this finding suggested also a key role for ERK in signaling from serum and growth factor receptors, including PDGF receptors, to the stimulation of AP-1-dependent transcription.
FIG. 1.
JNK and ERK pathways mediate AP-1 activation by PDGF and serum. NIH 3T3 cells were transfected with pAP-1-Luc and pRL-null, cultured under serum-free conditions, and pretreated for 30 min with inhibitors for each MAPK pathway (U0126, SP600125, and SB203580; 10 μM each) before PDGF (20 ng/ml) or FBS (10%) stimulation. Dual luciferase activities were determined 4 h after treatment, as described in Materials and Methods. The data represent the firefly luciferase activity normalized by Renilla luciferase activity present in each sample, expressed as absolute counts. Values are the average ± standard deviation of triplicate samples from a typical experiment. Nearly identical results were obtained in three additional experiments.
c-Fos expression and posttranslational modifications in response to PDGF depend on ERK activation.
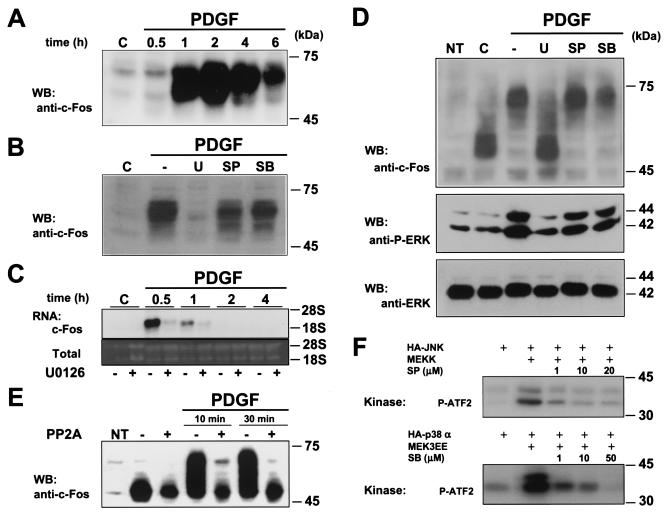
The phosphorylation of c-Jun by JNK is the best-known mechanism regulating the transcriptional activation of c-Jun-containing AP-1 complexes. However, ERK does not regulate the transcriptional activity of c-Jun (25). Furthermore, although ERK can activate the c-jun promoter in HeLa cells (20), it does not stimulate any of the cis-acting transcriptional elements controlling the expression of the c-jun promoter in NIH 3T3 cells (34). Instead, accumulated evidence indicates that activated ERK can promote the expression of c-fos by phosphorylating the ternary complex factor that binds the c-fos SRE (45, 48). Accordingly, exposure of quiescent NIH 3T3 cells to PDGF potently stimulated c-fos mRNA (Fig. 2C) and protein expression (Fig. 2A). c-Fos was detectable as early as 30 min after stimulation and remained highly expressed for up to 6 h. As evidenced from this time course, c-Fos migrates as multiple bands when resolved by SDS-PAGE, which suggests that c-Fos is covalently modified while being synthesized (6).
FIG. 2.
ERK signaling regulates c-Fos expression and posttranslational modifications. (A) PDGF induction of c-Fos expression in NIH 3T3 cells. Cells were grown overnight in the absence of serum and then stimulated with PDGF (20 ng/ml) for the indicated times. Nuclear fractions were collected, and c-Fos was detected by Western blotting. (B) ERK dependency of c-Fos expression. Cells were serum starved overnight, incubated for 30 min in the absence (c) or presence of U0126 (U), SP600125 (SP), or SB203580 (SB) (10 μM each), and stimulated with PDGF (20 ng/ml; 2 h), and c-Fos expression in nuclear fractions was determined as described for panel A. (C) ERK dependency of c-fos mRNA expression. Cells were serum starved overnight, incubated for 30 min in the absence or presence of U0126, and stimulated with PDGF (20 ng/ml) for the indicated times. c-fos transcript levels were detected by Northern blotting as described in Materials and Methods. (D) ERK dependency of c-Fos posttranslational modifications. Cells were transiently transfected with pCEFL-AU5-c-Fos (1 μg/well) and grown in serum-free medium overnight after transfection. Cells were stimulated with PDGF (20 ng/ml; 30 min), and total lysates were processed for Western blotting using anti-c-Fos antibodies (top panel). Pretreatment with MAPK inhibitors was performed as described above. Endogenous active (middle panel) and total (bottom panel) ERK expression from the same samples served as controls for PDGF and U0126 effects on ERK phosphorylation. (E) c-Fos phosphorylation induced by PDGF. NIH 3T3 cells were transfected with pCEFL-AU5-c-Fos (1 μg/well), cultured in serum-free medium overnight, and stimulated with PDGF for 10 or 30 min. Cell lysates were subjected to PP2A digestion followed by immunoblotting using anti-c-Fos antibodies (see Materials and Methods). (F) SP600125 (SP) and SB203580 (SB) inhibitory action on JNK and p38 activities. HEK-293T cells were transfected with HA-JNK1 or HA-p38α alone or in combination with their upstream activating kinases (MEKK and MEK3EE, respectively). In vitro kinase assays were performed as described in Materials and Methods in the absence or presence of the indicated final concentrations of the inhibitors. 32P-labeled substrate (P-ATF2) is indicated. NT, nontransfected cells.
To analyze the nature of the MAPKs that are involved in the induction of c-Fos by PDGF, we first examined whether the inhibitors of ERK-, JNK-, and p38-dependent pathways could affect the expression of c-Fos in response to PDGF. As shown in Fig. 2B, while the JNK and p38 inhibitors had a limited effect on c-Fos expression, the MEK inhibitor prevented the ability of PDGF to enhance c-Fos expression. In line with these results, the MEK inhibitor prevented the increase in c-fos mRNA levels after PDGF treatment (Fig. 2C). These observations suggested that the pathways leading to c-fos expression and the accumulation of its protein product in response to PDGF are strictly dependent on ERK, whereas JNK and p38 play a much more limited contribution.
Interestingly, the MEK inhibitor not only diminished the extent of c-Fos expression but also abolished the appearance of the slow-mobility forms of c-Fos induced by PDGF. These results indicated that the ERK pathway regulates the expression of c-fos (38, 47) as well as the posttranslational events occurring on the newly synthesized c-Fos molecules in response to PDGF. To analyze this possibility, we transiently overexpressed murine c-Fos from a constitutively active promoter, EF-1 (43), and examined the participation of different MAPKs in the posttranslational modification of c-Fos independently of those affecting c-fos message levels. As shown in Fig. 2D, the ectopically expressed c-Fos was readily detectable and experienced an electrophoretic mobility shift similar to that of the endogenous c-Fos when stimulated by PDGF. In agreement with our previous observation, the PDGF-induced mobility shift on c-Fos was strictly dependent on the activity of ERK kinases, as judged by the ability of the MEK inhibitor U0126 to abolish the appearance of the slow-migrating bands of c-Fos (Fig. 2D). Similar results were obtained with another broadly used MEK inhibitor, PD98059 (data not shown). In contrast, the JNK and p38 inhibitors were ineffective in altering the appearance of slow-migrating c-Fos species. These changes in gel migration were due to c-Fos phosphorylation, as accumulation of slow-migrating bands was reduced by PP2A treatment, a phosphatase that specifically hydrolyzes serine and/or threonine phosphoesters (Fig. 2E). Although we cannot rule out the possibility that PDGF may induce multiple modifications on c-Fos, the combined results from the phosphatase treatment and the blockade by U0126 suggest that either phosphorylation or a phosphorylation-dependent posttranslational event accounts for the c-Fos mobility shift induced by PDGF. As controls, the inhibitory actions of SP600125 on JNK1 and SB203580 on p38α were confirmed by in vitro kinase assays (Fig. 2F). Taken together, these results indicate that the ability of PDGF to regulate c-Fos involves at least two distinct ERK-dependent mechanisms, one regulating c-Fos expression and another affecting the status of c-Fos phosphorylation.
The transcriptional activation of c-Fos by PDGF requires ERK signaling: role of the c-Fos C-terminal TAD.
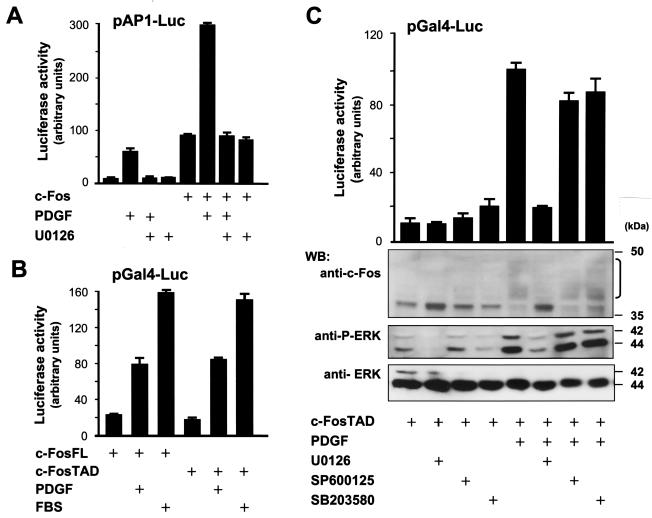
These observations prompted us to examine whether ERK-dependent mechanisms acting on c-Fos play a role in the regulation of AP-1 transcription by PDGF. As shown in Fig. 3A, the expression of c-Fos increased the activity of the AP-1 reporter plasmid by severalfold, and exposure to PDGF provoked a remarkable further increase in luciferase expression. Moreover, this effect of PDGF was dependent on ERK, as U0126 prevented the stimulation of AP-1 by this mitogen alone or in the presence of c-Fos, without affecting the basal activity of c-Fos as an AP-1 activator.
FIG. 3.
PDGF activation of c-Fos is ERK dependent. (A) ERK dependency of c-Fos activation of AP-1 by PDGF. NIH 3T3 cells were transfected with pAP-1Luc and pRL-null along with 0.2 μg of pCEFL-AU5-c-Fos or pcDNAIII-β-gal. Where indicated (+), cells were pretreated with U0126 (10 μM) for 30 min and left untreated or stimulated with PDGF (20 ng/ml) for 4 h before reading dual luciferase activities. (B) c-Fos transcriptional response to PDGF and serum. Cells were cotransfected with pGal4-c-FosFL or pGal4-c-FosTAD (2 ng/well each) along with pGal4-Luc and pRL-null. At 24 h after transfection, cells were stimulated with PDGF (20 ng/ml) or FBS (10%), as indicated, and dual luciferase activities were determined 4 h later. (C) ERK dependency of c-Fos TAD activation and posttranslational modifications by PDGF. Results shown in the bar graph are cells that were transiently transfected with pGal4-c-FosTAD (2 ng/well), pGal4-Luc, and pRL-null; luciferase assays were performed as described above. Where indicated, cells were pretreated with U0126, SP600125, or SB203580 (10 μM each) for 30 min before PDGF stimulation. PDGF-induced modified c-Fos species are denoted by the bracket. The Western blot shows results with cells that were transfected with pGal4-c-FosTAD (1 μg/well), grown in serum-free medium overnight, and stimulated with PDGF for 10 min. Total lysates were assayed by Western blotting using anti-c-Fos antibodies (top panel). Controls for phosphorylated and total ERKs are shown in parallel (middle and bottom blots). Reporter assay data represent the firefly luciferase activity normalized by Renilla luciferase activity present in each sample, expressed as absolute counts. All values are the average ± standard deviation of triplicate samples from a typical experiment. In each case, similar results were obtained in three additional experiments.
The C terminus of c-Fos harbors transactivating potential, and several motifs within it have been shown to control the activity of c-Fos as a transcription factor (42). Based on our previous results, we set out to investigate whether the ERK pathways had an effect on the transactivating function of c-Fos. For these studies, we examined c-Fos activation in a heterologous system, in which either the full-length c-Fos or its C-terminal TAD were expressed as fusion proteins in frame with the DBD of the yeast transcription factor Gal4, referred to as Gal4-c-FosFL and Gal4-c-FosTAD, respectively. The activity of these chimeras was assessed by their ability to stimulate transcription from a Gal4-regulated luciferase reporter plasmid (pGal4-Luc). As shown in Fig. 3B, both PDGF and serum induced the transcriptional activity of Gal4-c-FosFL, as well as that of Gal4-c-FosTAD. This response was strictly dependent on ERK, as U0126 (Fig. 3C) and PD98059 (data not shown) abolished the activation of the c-Fos TAD by PDGF without affecting its basal activity. Furthermore, when analyzed by Western blotting, the Gal4-c-FosTAD protein exhibited a retarded mobility upon PDGF treatment, which was also prevented by U0126 treatment, paralleling the ability of this drug to prevent ERK activation (Fig. 3C). In contrast, SP600125 and SB203580 did not display any effect. Similar results were obtained when analyzing the serum responsiveness of the Gal4-c-FosTAD fusions in reporter and immunoblotting experiments (data not shown).
ERK stimulates c-Fos and AP-1-dependent transcription.
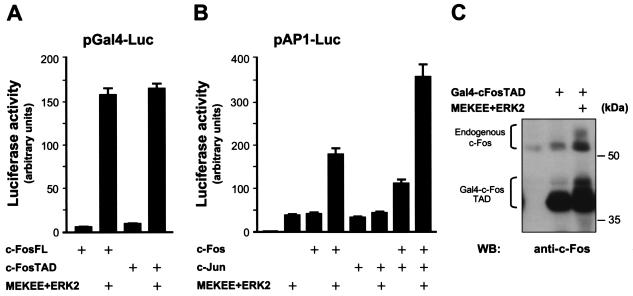
The fact that PDGF targets the c-Fos TAD through ERK-dependent pathways prompted us to test whether ERK directly stimulates the transcriptional activity of c-Fos. Indeed, Fig. 4A shows that both the Gal4-c-FosTAD and Gal4-c-FosFL chimeras were strongly activated by cotransfection of ERK2 and MEKEE, a constitutively active form of MEK1. In contrast, ERK2 did not affect the activity of Gal4-DBD alone (data not shown). Similarly, cotransfection of ERK2 and MEK EE stimulated the AP-1 reporter plasmid, and that was dramatically increased in the presence of cotransfected c-Fos (Fig. 4B). As c-Fos binds AP-1-responsive elements through Jun proteins (25), we further examined the possibility that c-Jun may mediate the effect of ERK2 on AP-1. As expected, overexpression of c-Jun alone also stimulated the AP-1 reporter (Fig. 4B). However, c-Jun did not enhance the AP-1 response to ERK2. In contrast, c-Jun and c-Fos when coexpressed potently increased the transcriptional response initiated by ERK2, together suggesting that ERK2 most likely signals to AP-1 through c-Fos as part of the AP-1 dimer. In line with these results, when overexpressed, activated ERK2 promoted the appearance of slower-migrating forms of the c-Fos TAD and endogenous c-Fos (Fig. 4C).
FIG. 4.
ERK2 activates c-Fos. (A) NIH 3T3 cells were transfected with p-Gal4c-FosFL or p-Gal4c-FosTAD (each 2 ng/well) along with pGal4-Luc, pRL-null, and MEKEE+ERK2 (0.5 μg each) or pcDNAIII-β-gal (controls). Cells were incubated in serum-deprived medium, lysed, and assayed for firefly and Renilla luciferase activities as for Fig. 3. (B) Effect of ERK2 on AP-1 activation by c-Fos. Cells were transfected with pAP-1Luc and pRL-null, alone or together with pCEFL-AU5-c-Fos (1 μg/well) pCEFL-c-Jun (1 μg/well), and/or MEKEE+ERK2 (0.5 μg each), and processed as above for AP-1-driven luciferase expression. (C) ERK2 induction of c-Fos posttranslational modifications. pGal4-c-FosTAD (1 μg/well) was expressed alone or in combination with ERK2+MEKEE (0.5 μg each) in HEK-293T cells. At 24 h posttransfection, c-Fos expression was detected by Western blotting using anti-c-Fos antibodies. The high-molecular-mass protein bands correspond to endogenous c-Fos proteins expressed in HEK-293T cells.
Potential ERK sites on the c-Fos TAD are essential for transactivation in response to PDGF.
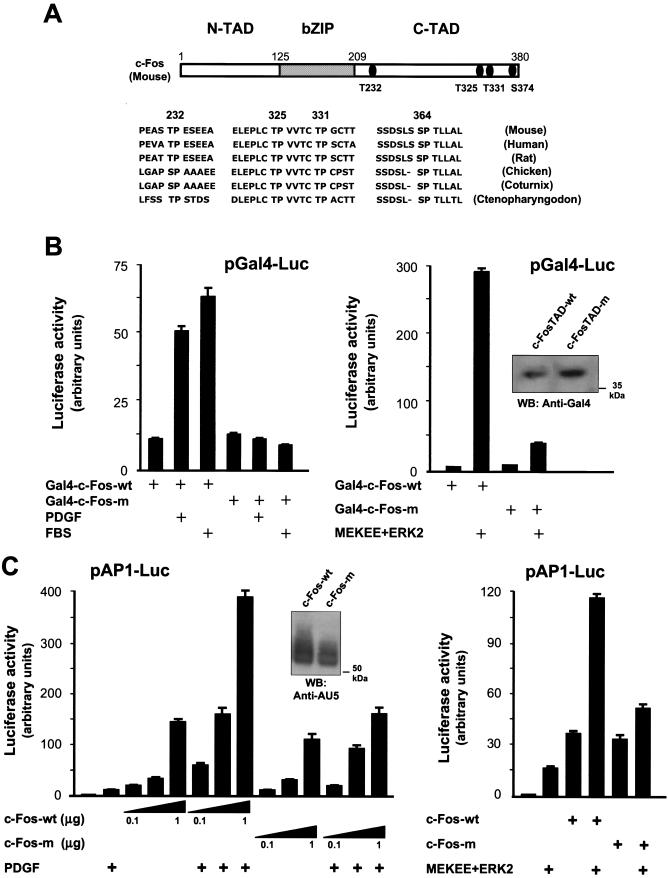
All known MAPKs catalyze the phosphorylation of serine or threonine residues adjacent to a proline in the +1 position (T/S+P). Searching for this pattern in the primary sequence of the mouse c-Fos TAD, we identified four candidate MAPK sites (Thr-232, -325, and -331 and Ser-374). Figure 5A represents the structural domains of c-Fos depicting the location of these four residues. Furthermore, sequence alignment revealed that these particular sites are highly conserved among c-Fos proteins from a variety of distantly related species, from mammals to fish (Fig. 5A). We therefore investigated whether these potential phosphorylation sites were required for the ability of c-Fos to be activated by ERK2 and PDGF.
FIG.5.
Candidate MAPK phosphorylation sites confer PDGF responsiveness to the c-Fos TAD. (A) Schematic representation of c-Fos primary structure with emphasis on the major functional domains and the relative location of the four conserved sites that follow the consensus motif for MAPK phosphorylation in the C-terminal TAD (above). bZIP, region that comprises the leucine zipper and DBD; N- and C-TAD, N-terminal and C-terminal TADs, respectively. Details are shown for known c-Fos sequences from diverse species, showing the conservation of all four potential MAPK phosphorylation sites (below). Note that Thr-325, Thr-331, and Ser-374 lie within sequence motifs that are also highly conserved. (B) Effect of mutations in potential MAPK phosphorylation sites on the basal activity and inducibility by PDGF of the c-Fos TAD. Where indicated, cells were transfected with pGal4-FosTAD or pGal4-c-FosTAD-m (2 ng/well each) along with pGal4-luc, pRL-null, and MEKEE+ERK2 (0.5 μg each) (right panel). Cells were serum starved and stimulated with PDGF (20 ng/ml) or FBS (10%) (left panel) for 4 h before reading firefly and Renilla luciferase activities. Total lysates from Gal4c-FosTAD- or Gal4-c-FosTAD-m-overexpressing cells were analyzed by Western blotting using anti-Gal4 DBD antibodies (inset). (C) Effect of mutations in MAPK sites on the c-Fos activation of AP-1 transcription. NIH 3T3 fibroblasts were transfected with pAP-1-Luc, pRL-null, and 1 μg (right panel) or increasing concentrations (left panel) of pCEFL-AU5-c-Fos or pCEFL-AU5-c-Fos-m. Cells were incubated overnight in the absence of serum and then stimulated with PDGF for 4 h before measuring dual luciferase activities. Total lysates of c-Fos- or c-Fos-m-overexpressing cells were analyzed by Western blotting using anti-AU5 antibodies (left panel, inset). MEKEE and ERK2 were cotransfected at 0.5 μg each (right panel). Reporter assay data represent the firefly luciferase activity normalized by Renilla luciferase activity present in each sample, expressed as absolute counts. All values are the average ± standard deviation of triplicate samples from a typical experiment. Similar results were obtained in three additional experiments.
As an approach, we replaced each of these serine and threonine residues with nonphosphorylatable alanine residues by site-directed mutagenesis. Initially, we analyzed the effect of individual point mutations on the activity of the Gal4-c-FosTAD chimera. Mutated constructs were transfected into NIH 3T3 cells, and their responsiveness to PDGF and activated ERK2 was compared to that of the wild-type Gal4c-FosTAD. Surprisingly, the transcriptional activity of all four Gal4-c-FosTAD point mutants was induced to an extent comparable to that of the wild-type protein. Similarly, full-length c-Fos proteins carrying the same point mutations did not exhibit an impaired ability to activate AP-1 transcription or to be induced by PDGF and ERK2 (data not shown). However, when all the candidate MAPK sites were replaced simultaneously by alanines, the transcriptional activation of the c-Fos TAD (Gal4-c-FosTAD-m) in response to PDGF and serum was abolished, and the activation by ERK2 was dramatically reduced when compared to that of the wild-type c-Fos TAD, despite the fact that both constructs were expressed at comparable levels (Fig. 5B).
We also introduced these four alanine replacements in the full-length c-Fos protein and examined the ability of the resulting c-Fos mutant, c-Fos-m, to promote AP-1-dependent transcription. As shown in Fig. 5C, c-Fos-wt and c-Fos-m were expressed at comparable levels, and their basal transcriptional activities were nearly identical. However, c-Fos-m no longer responded efficiently to signals that emanated from PDGF receptors or when induced by MEKEE/ERK2 expression. These findings supported the relevance of the four potential ERK phospho-acceptor sites on the c-Fos TAD in the ability of the full-length c-Fos to transduce signals resulting in AP-1 activation.
Potential ERK phosphorylation sites are required for c-Fos phosphorylation by PDGF.
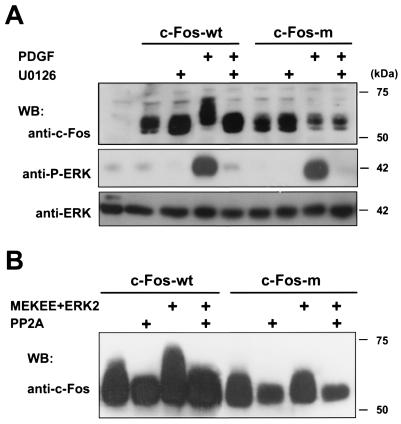
The availability of a mutant form of c-Fos that is impaired in its transcriptional response to PDGF and ERK provided an opportunity to examine in more detail the requirement of c-Fos phosphorylation in the c-Fos response to PDGF. We first analyzed the electrophoretic behavior of c-Fos-m upon PDGF stimulation. In contrast to the wild-type c-Fos, the electrophoretic mobility of c-Fos-m was not shifted by PDGF treatment of NIH 3T3 cells (Fig. 6A). Furthermore, the appearance of PP2A-sensitive slow-migrating c-Fos bands provoked by activated ERK2 was also greatly diminished in c-Fos-m (Fig. 6B), together suggesting that the four residues mutated in c-Fos-m are required for the phosphorylation of c-Fos by ERK in vivo, resulting in its shift in electrophoretic mobility.
FIG. 6.
(A) MAPK sites are required for c-Fos phosphorylation induced by PDGF. As indicated, NIH 3T3 cells were transfected with pCEFL-AU5-c-Fos or pCEFL--AU5-c-Fos-m (2 μg/well), pretreated with U0126 (10 μM) or vehicle for 30 min, and stimulated with PDGF for an additional 30 min. Cells were lysed, resolved by SDS-PAGE, and blotted using anti-c-Fos antibodies (top panel). Phospho-ERK and total ERK immunoreactivities from the same samples are shown below (middle and bottom panels). (B) MAPK sites are required for c-Fos phosphorylation by ERK2. Cell lysates from c-Fos- or c-Fos-m-overexpressing cells in the absence or presence of MEKEE+ERK2, as indicated, were subjected to PP2A digestion as described in Materials and Methods. c-Fos expression was detected by Western blotting using anti-c-Fos antibodies.
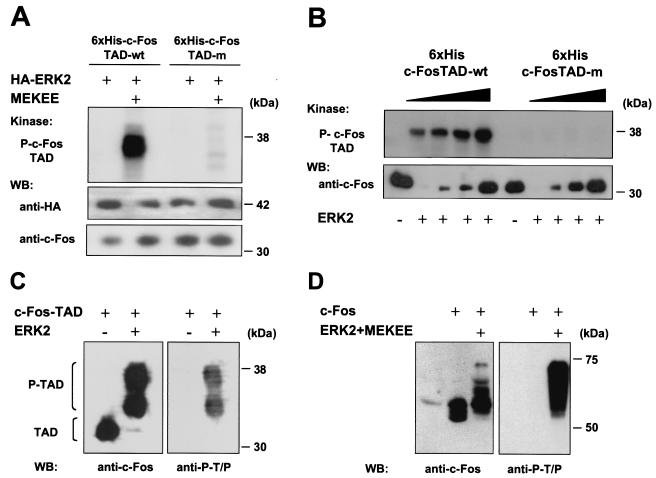
To investigate whether ERK2 was able to phosphorylate c-Fos directly, we then performed in vitro kinase assays using bacterially expressed six-His-tagged forms of the c-Fos TAD as a potential substrate for either an immunoprecipitated (Fig. 7A) or a recombinant purified preparation of ERK2 (Fig. 7B). The results, shown in Fig. 7A and B, indicate that ERK2 can phosphorylate c-Fos effectively. Furthermore, no detectable phosphorylation was observed when c-FosTAD-m was used as an ERK2 substrate in vitro. This observation confirmed the specificity of the kinase reaction and demonstrated that the mutated residues were strictly required for the phosphorylation of the c-Fos TAD by ERK2. As a complementary approach, we confirmed the phosphorylation of the c-Fos TAD by ERK2 by a cold kinase assay. In this case, using six-His-c-FosTAD as a substrate and unlabeled ATP, we observed that activated ERK2 directly catalyzed the phosphorylation of threonine residues adjacent to prolines on the c-Fos TAD (Fig. 7C). In contrast, c-FosTAD-m was not phosphorylated in parallel reactions (data not shown). Aligned with these observations, the phosphorylation of T+P sites on the c-Fos TAD was also seen in vivo when the full-length c-Fos was coexpressed in the presence of ERK2 (Fig. 7D).
FIG. 7.
ERK2 phosphorylates c-Fos in vitro and in vivo. (A and B) Bacterially expressed c-Fos TADs, six-His c-FosTAD-wt, and six-His c-FosTAD-m were purified and assayed as substrates for an immunoprecipitated (A) or recombinant purified (b) preparation of ERK2. In vitro kinase assays were performed as described in Materials and Methods. The purified six-His substrates (A and B, lower panels) and the HA-tagged ERK2 (A, middle panel) used in the kinase reactions were detected by Western blotting with the indicated antibodies. (C) In vitro cold kinase assay using purified six-His c-Fos TAD as a substrate for recombinant ERK2. Reaction products were analyzed by Western blotting using anti-c-Fos antibodies or anti-phospho-T/P antibodies. (D) HEK-293T cells were transfected with pCEFLc-Fos (1 μg/well) alone or in combination with ERK2+MEKEE (0.5 μg each). Total lysates were analyzed by Western blotting using anti pCEFL-AU5-c-Fos antibodies (left panel) or immunoprecipitated with anti-c-Fos and immunoblotted using anti-phospho-T/P antibodies (right panel).
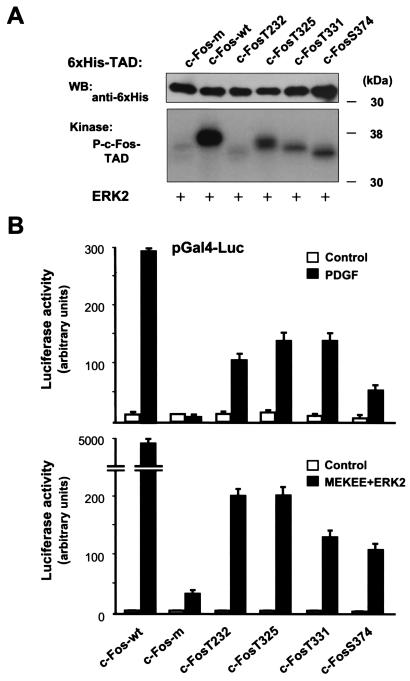
To study the contribution of each potential MAPK phosphorylation site to the phosphorylation of the c-Fos TAD, we then chose to use an add-back approach which consisted of reintroducing each of the original serine or threonine residues back into the backbone of c-FosTAD-m. As shown in Fig. 8A, when equal amounts of the purified c-Fos TAD proteins were used as substrates for ERK2, c-Fos-wt was effectively phosphorylated whereas c-Fos-m was not. When Thr-232 was the only potential ERK phospho-acceptor site, the corresponding protein was only poorly phosphorylated by ERK2. In contrast, ERK2 phosphorylated c-Fos TADs that included Thr- 325, Thr-331, or Ser-374 as unique phospho-acceptor sites, thus indicating that these residues can serve as in vitro targets for the enzymatic activity of ERK2.
FIG. 8.
ERK2 phosphorylation sites in the c-Fos TAD. (A) Mapping of c-Fos TAD phosphorylated sites by ERK2. The indicated purified six-His c-Fos TAD wild-type (wt) or mutant proteins bearing none (m) or a single potential ERK phosphorylation site were used as substrates for recombinant ERK2 in in vitro kinase assays. Equal amounts of purified proteins in each reaction mixture were confirmed by Western blotting with anti-six-His antibody. Phosphorylated products (P-c-FosTAD) were resolved by SDS-PAGE. (B) Evaluation of the role of ERK phospho-acceptor sites in the c-Fos TAD in the transcriptional response of the c-Fos TAD to PDGF and ERK2 activation. Cells were transiently transfected with pGal4-c-FosTAD wild-type (wt) or mutant vectors bearing none (m) or single potential ERK phosphorylation sites (2 ng/well), together with pGal4-Luc and pRL-null, and luciferase assays were performed. The top graph shows results with cells that were left untreated (control) or treated with PDGF for 5 h (20 ng/ml). The bottom graph shows results with cells that were transfected with pCDNAIII- β-gal (control) or MEKEE+ERK2 (0.5 μg each). All Gal4 constructs were expressed at comparable levels (data not shown). Reporter assay data represent the firefly luciferase activity normalized by the Renilla luciferase activity present in each sample. All values are the average ± standard deviation of triplicate samples from a typical experiment. In each case, similar results were obtained in at least three additional experiments.
To explore the participation of these individual phosphorylation sites in the transcriptional activity of c-Fos, we next generated Gal4-c-FosTAD mutants containing single potential MAPK phospho-acceptor sites. As shown in Fig. 8B, the sole presence of any of the candidate sites was sufficient to confer to the c-Fos TAD a limited transcriptional responsiveness to PDGF (upper panel). However, none of these Gal4-c-FosTAD mutants was as potent as the wild-type TAD. This was even more dramatic when activated ERK2 was used to stimulate the transcriptional activity of the c-Fos TAD (lower panel). These results supported the emerging notion that each of these potential ERK phosphorylation sites can play a role in the transcriptional activation of c-Fos but that multiple phosphorylation events may be required to achieve the maximal transcriptional response of c-Fos.
c-fos potentiates the transforming potential of c-sis in NIH 3T3 cells.
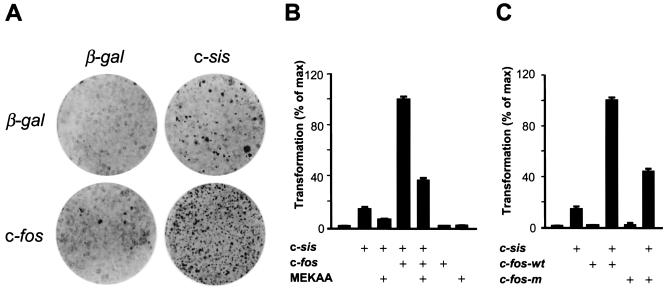
As we observed that the c-Fos TAD represents a target for PDGF receptor signaling, we next asked whether the transactivating effect of PDGF on c-Fos through ERK may have biological consequences, taking advantage of the high transforming potential of c-sis, which encodes the B-chain of PDGF (18), when expressed in NIH 3T3 cells. Indeed, as shown in Fig. 9A, even though c-Fos does not transform NIH 3T3 cells alone (37), it increased remarkably the number of foci induced by c-sis. Moreover, we also observed that foci initiation was dramatically accelerated, as foci induced by c-sis were visible 8 to 10 days after transfection whereas c-sis foci became evident as soon as 5 days of culture when c-fos and c-sis were expressed together. As expected from the autocrine action of PDGF resulting from c-sis expression, cells derived from c-sis foci expressed c-Fos under a hyperphosphorylated state (data not shown). This synergistic effect of c-sis and c-fos in promoting cell transformation suggested a functional role for c-Fos in the control of cell growth by PDGF.
FIG. 9.
c-fos potentiates the focus-forming activity of c-sis: requirement of ERK phosphorylation sites in c-Fos. (A) NIH 3T3 fibroblasts were transfected with 1 μg of c-fos expression vector (pCEFL-AU5-c-Fos) or pCDNAIII-β-galactosidase (β-gal) alone or in combination with 0.5 μg of c-sis (pcDNAIII-c-sis), as indicated. Cells were maintained in 5% calf serum and fixed and stained 25 days after transfection. Plates from a representative experiment are shown. (B) Cells were transfected with pcDNAIII-c-sis (0.5 μg/plate) alone or together with pCEFL-AU5-c-Fos and/or MEKAA (1 μg each). Transforming efficiency was estimated by counting the number of foci per dish after 25 days of transfection and expressed as the percentage with respect to that observed in c-sis and c-fos transfections, which was taken as 100%. Results from a representative experiment out of three independent ones are shown. (C) Cells were transfected with pcDNAIII-c-sis (0.5 μg) with or without pCEFL-AU5-c-Fos or pCEFL-AU5-c-Fos-m (1 μg each). Foci were counted 25 days after transfection and are represented as described for panel B.
Cooperation between c-fos and c-sis in cell transformation requires ERK phospho-acceptor sites in the c-Fos TAD.
As ERK mediates c-Fos activation by PDGF, we decided to examine whether the ERK cascade was involved in c-fos- and c-sis-induced transformation by using a dominant negative form of MEK1, MEK1AA. The specificity of this dominant interfering molecule has been previously documented (33). As shown in Fig. 9B, MEK1AA diminished the number of c-sis-induced foci, as well as the number resulting from c-sis and c-fos coexpression. These data suggested that c-fos cooperates with c-sis in transformation in an ERK-dependent manner. Nevertheless, other mechanisms might be also involved, as foci formation was not totally inhibited by cotransfecting MEK dominant negative molecules. Thus, to address more directly whether c-Fos phosphorylation on the TAD was required for the ability of c-sis to cooperate with c-fos in transformation, we examined the influence of c-Fos individual mutations on Thr-232, Thr-325, Thr-331, and Ser-374 on c-sis-induced focus formation. We observed that these single c-Fos point mutants increased the transforming efficiency of c-sis similarly to the c-fos wild type (data not shown). In contrast, the mutant c-fos carrying mutations in all ERK target sites, c-fos-m, was much less efficient in cooperating with c-sis than the wild-type c-fos (Fig. 9C). Of interest, when examined under identical experimental conditions, add-back mutants of c-Fos bearing single ERK phosphorylation sites behaved nearly identical to c-Fos-m (data not shown). These results are consistent with the idea that c-Fos acts downstream from PDGF receptors in a signaling route leading to normal and/or unregulated cell growth and that the ERK signaling pathway mediates the activation of c-Fos through the concerted phosphorylation of multiple sites on the c-Fos TAD.
DISCUSSION
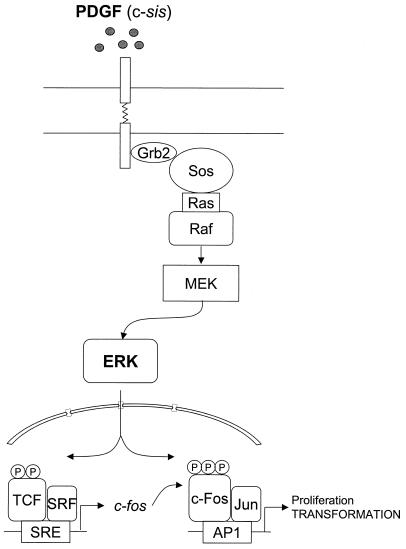
The activation of members of the AP-1 family of transcription factors, including c-Fos and c-Jun family members, is one of the earliest nuclear events provoked by stimulation of fibroblasts with serum or PDGF (11), and AP-1 activity is required for cell proliferation, as microinjection of antibodies blocking components of the AP-1 complex, including c-Fos, impairs the ability of serum-stimulated cells to reenter the S phase of the cell cycle (27). However, the precise nature of the pathways connecting PDGF to AP-1 is still not fully understood. In this study, we provide evidence that PDGF regulates AP-1 activity through the activation of ERK and the consequent stimulation of the expression and transactivating activity of c-Fos. Thus, the emerging picture is that PDGF receptors signal to c-Fos through ERK in a two-step process, initially by stimulating the activity of the c-fos promoter, thus promoting c-Fos expression, and subsequently by regulating the posttranslational processing and transactivating activity of c-Fos by promoting its direct phosphorylation (Fig. 10).
FIG. 10.
Proposed model for PDGF receptor and ERK signaling to c-Fos and AP-1-regulated transcription. PDGF (c-sis) triggers a variety of signaling pathways, including those leading to the activation of the ERK cascade (Raf, MEK, and ERK). This results in the phosphorylation of the ternary complex factor (TCF) by ERK and the stimulation of the activity of the c-fos SRE, thus promoting c-fos expression. Subsequently, ERK phosphorylates c-Fos and that stimulates the transactivating activity of c-Fos, thereby promoting the expression of AP-1-regulated genes.
A number of kinases have been reported to phosphorylate c-Fos, including protein kinases C and A, cdc2 (1), FRK (13), and a novel MAPK, ERK7 (2). c-Fos phosphorylation by ERK1 and RSK on Ser-374 and Ser-362, respectively, within the C-terminal region of c-Fos has also been described to occur soon after serum stimulation (8, 9), and while this study was in progress Murphy et al. (36) described the existence of a docking site for ERK in c-Fos which facilitates the sequential phosphorylation of c-Fos in multiple C-terminal sites upon prolonged activation of ERK. However, the functional consequences of c-Fos phosphorylation are still poorly understood. Collectively, our present findings indicate that the ERK-dependent phosphorylation of c-Fos results in a remarkable increase in its transactivating activity and that the C terminus of c-Fos plays an important role in the transcriptional activation of c-Fos by growth factors stimulating ERK.
This C-terminal region of c-Fos includes a TAD (42, 50). When fused to the DBD of the yeast transcription factor GAL4, the transcriptional activity of this c-Fos TAD was greatly increased by PDGF in an ERK-dependent manner, and direct activation of ERK resulted in an even greater increase in its transcriptional activity in this heterologous system. Similar results where obtained when the full coding region of c-fos was fused to the GAL4 DBD, supporting that the activity of ERK on the C-terminal region of c-Fos was reflected in the activation of the full-length protein. Furthermore, c-Fos expression enhanced dramatically the activation of AP-1 by PDGF, and this effect was abolished by inhibition of ERK, thus supporting the role of c-Fos and its regulation by ERK in the pathway linking PDGF stimulation to AP-1-mediated transcription.
As the TAD of c-Fos represents an effective in vivo substrate of ERK, we took advantage of the ability of PDGF and ERK activation to stimulate the c-Fos TAD to identify the most likely biologically relevant phosphorylated residues. Within the c-Fos TAD, Thr-232, -331, and -325 and Ser- 374 can be predicted as targets for ERK activity, and sequence alignment revealed that these residues are highly conserved in c-Fos sequences from distantly related species. However, when each of these sites was mutated individually, we did not observe any changes in the ability of c-Fos to be activated through ERK. In contrast, a c-Fos TAD mutant in which all sites were replaced with alanines was no longer a substrate of ERK2 in vitro, and this replacement abolished the response of the c-Fos TAD to PDGF. Of interest, this mutant c-Fos still includes Ser-362, the proposed target for RSK (8), suggesting that the sole phosphorylation by RSK may not be sufficient to stimulate the transcriptional activity of c-Fos. These observations prompted us to explore the use of an add-back approach, which consisted of investigating the nature of the relevant phosphorylation sites for ERK by returning each of the potential ERK targets one at a time back to the original residue. Interestingly, when Thr-232 was the only such potential MAPK phospho-acceptor site, the corresponding protein was not phosphorylated efficiently by ERK2. In contrast, when Thr-325, Thr-331, and Ser-374 were reintroduced in c-FosTAD-m, they each served as a substrate for ERK2. In particular, Thr-232 has been proposed to be the target of a yet-to-be-identified proline-targeted kinase, FRK, which appears to be different from ERK (13). Nonetheless, Thr-232 may serve as a substrate for ERK2 in vitro when expressed as a short polypeptide fused to GST, as reported by Bannister et al. (5). Thus, we cannot rule out that Thr-232 may be phosphorylated in vivo by ERK2 in the context of the full-length protein, or that FRK may act downstream from ERK2, as suggested by the observation that when Thr-232 was the only phospho-acceptor site in the c-Fos TAD it was sufficient to confer a partial, albeit limited, transcriptional response to PDGF and active ERK2. Similarly, each of the other sites, Thr-325 and -331 and Ser-374, was alone sufficient to promote gene transcription when stimulated by PDGF or ERK2, but less than with the wild-type c-Fos TAD. Thus, although the nature of the c-Fos Thr-232 kinase is at the present unclear, this residue is likely to act in concert with the other ERK phosphorylation sites to achieve the full activation of the TAD in vivo.
In line with the observation that a mutated c-Fos TAD lacking all potential ERK phosphorylation sites fails to respond to PDGF, when these mutations were introduced into the full-length c-Fos the resulting mutant showed a much more limited effect on AP-1-dependent transcription in response to PDGF or ERK stimulation. On the other hand, this c-Fos mutant did not prevent the activation of AP-1 by PDGF or ERK, likely because its overexpression results in increased basal AP-1 activity, which might offset any counteracting activity resulting from preventing the activity of the phosphorylated endogenous c-Fos. Thus, although ERK phosphorylation may not be required for the basal activity of c-Fos, phosphorylation of the C-terminal TAD might be required for the transcriptional response of c-Fos to extracellular stimuli impinging on ERK activation, ultimately controlling the expression of AP-1-regulated genes.
Several lines of evidence also indicate a direct correlation between the transactivating activity of Fos proteins and their ability to transform cells (38, 50). Of interest, in NIH 3T3 cells c-Fos alone cannot induce the acquisition of a transformed phenotype, as has been reported by others (37). Instead, we observed that c-fos potentiated dramatically the focus-forming ability of c-sis, which encodes the B-chain of PDGF (18), thus supporting the notion that c-Fos represents a biologically relevant target for PDGF action. Consistent with this observation, c-fos expression has been shown to be required for cell transformation when induced by sis (35), as well as by oncogenes acting directly upstream of ERK, such as ras (31, 49) and raf (23). Similarly, the mos oncogene, which encodes a serine/threonine kinase that activates ERK, can cooperate with c-Fos to transform NIH 3T3 cells (37). Replacement of the ERK target sites in the C-terminal TAD of c-Fos, which as discussed above does not impair its basal transactivating activity, diminishes dramatically its cooperating activity with c-sis, thus suggesting that c-Fos phosphorylation by ERK may represent a biologically relevant mechanism whereby growth factors acting on receptor tyrosine kinases can control cell growth. These findings also raise the possibility that c-Fos may act downstream of ERK as an integral component of the transforming pathway elicited by a large number of oncogenes that converge in ERK activation to promote aberrant cell proliferation.
Acknowledgments
We thank Rodrigo Bravo for providing the murine c-Fos cDNA and Joan-Marc Servitja for his help in the design of primers for site-directed mutagenesis and his expert advice.
REFERENCES
- 1.Abate, C., D. R. Marshak, and T. Curran. 1991. Fos is phosphorylated by p34cdc2, cAMP-dependent protein kinase and protein kinase C at multiple sites clustered within regulatory regions. Oncogene 6:2179-2185. [PubMed] [Google Scholar]
- 2.Abe, M. K., W. L. Kuo, M. B. Hershenson, and M. R. Rosner. 1999. Extracellular signal-regulated kinase 7 (ERK7), a novel ERK with a C-terminal domain that regulates its activity, its cellular localization, and cell growth. Mol. Cell. Biol. 19:1301-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angel, P., M. Imagawa, R. Chiu, B. Stein, R. J. Imbra, H. J. Rahmsdorf, C. Jonat, P. Herrlich, and M. Karin. 1987. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell 49:729-739. [DOI] [PubMed] [Google Scholar]
- 4.Angel, P., and M. Karin. 1991. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta 1072:129-157. [DOI] [PubMed] [Google Scholar]
- 5.Bannister, A. J., H. J. Brown, J. A. Sutherland, and T. Kouzarides. 1994. Phosphorylation of the c-Fos and c-Jun HOB1 motif stimulates its activation capacity. Nucleic Acids Res. 22:5173-5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barber, J. R., and I. M. Verma. 1987. Modification of fos proteins: phosphorylation of c-fos, but not v-fos, is stimulated by 12-tetradecanoyl-phorbol-13-acetate and serum. Mol. Cell. Biol. 7:2201-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennett, B. L., D. T. Sasaki, B. W. Murray, E. C. O'Leary, S. T. Sakata, W. Xu, J. C. Leisten, A. Motiwala, S. Pierce, Y. Satoh, S. S. Bhagwat, A. M. Manning, and D. W. Anderson. 2001. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc. Natl. Acad. Sci. USA 98:13681-13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, R. H., C. Abate, and J. Blenis. 1993. Phosphorylation of the c-Fos transrepression domain by mitogen- activated protein kinase and 90-kDa ribosomal S6 kinase. Proc. Natl. Acad. Sci. USA 90:10952-10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, R. H., P. C. Juo, T. Curran, and J. Blenis. 1996. Phosphorylation of c-Fos at the C-terminus enhances its transforming activity. Oncogene 12:1493-1502. [PubMed] [Google Scholar]
- 10.Chiariello, M., M. J. Marinissen, and J. S. Gutkind. 2000. Multiple mitogen-activated protein kinase signaling pathways connect the cot oncoprotein to the c-jun promoter and to cellular transformation. Mol. Cell. Biol. 20:1747-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cochran, B. H., J. Zullo, I. M. Verma, and C. D. Stiles. 1984. Expression of the c-fos gene and of a fos-related gene is stimulated by platelet-derived growth factor. Science 226:1080-1082. [DOI] [PubMed] [Google Scholar]
- 12.Coso, O. A., M. Chiariello, G. Kalinec, J. M. Kyriakis, J. Woodgett, and J. S. Gutkind. 1995. Transforming G protein-coupled receptors potently activate JNK (SAPK). Evidence for a divergence from the tyrosine kinase signaling pathway. J. Biol. Chem. 270:5620-5624. [DOI] [PubMed] [Google Scholar]
- 13.Deng, T., and M. Karin. 1994. c-Fos transcriptional activity stimulated by h-Ras-activated protein kinase distinct from JNK and ERK. Nature 371:171-175. [DOI] [PubMed] [Google Scholar]
- 14.English, J. M., and M. H. Cobb. 2002. Pharmacological inhibitors of MAPK pathways. Trends Pharmacol. Sci. 23:40-45. [DOI] [PubMed] [Google Scholar]
- 15.Favata, M. F., K. Y. Horiuchi, E. J. Manos, A. J. Daulerio, D. A. Stradley, W. S. Feeser, D. E. Van Dyk, W. J. Pitts, R. A. Earl, F. Hobbs, R. A. Copeland, R. L. Magolda, P. A. Scherle, and J. M. Trzaskos. 1998. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J. Biol. Chem. 273:18623-18632. [DOI] [PubMed] [Google Scholar]
- 16.Franchimont, N., D. Durant, S. Rydziel, and E. Canalis. 1999. Platelet-derived growth factor induces interleukin-6 transcription in osteoblasts through the activator protein-1 complex and activating transcription factor-2. J. Biol. Chem. 274:6783-6789. [DOI] [PubMed] [Google Scholar]
- 17.Galang, C. K., C. J. Der, and C. A. Hauser. 1994. Oncogenic Ras can induce transcriptional activation through a variety of promoter elements, including tandem c-Ets-2 binding sites. Oncogene 9:2913-2921. [PubMed] [Google Scholar]
- 18.Gazit, A., H. Igarashi, I. M. Chiu, A. Srinivasan, A. Yaniv, S. R. Tronick, K. C. Robbins, and S. A. Aaronson. 1984. Expression of the normal human sis/PDGF-2 coding sequence induces cellular transformation. Cell 39:89-97. [DOI] [PubMed] [Google Scholar]
- 19.Grover-Bardwick, A., E. Adamson, and D. Mercola. 1994. Transformation-specific pattern of phosphorylation of c-Jun, Jun-B, Jun-D and Egr-1 in v-sis transformed cells. Carcinogenesis 15:1667-1674. [DOI] [PubMed] [Google Scholar]
- 20.Gupta, P., and R. Prywes. 2002. ATF1 phosphorylation by the ERK MAPK pathway is required for epidermal growth factor-induced c-jun expression. J. Biol. Chem. 277:50550-50556. [DOI] [PubMed] [Google Scholar]
- 21.Hill, C. S., and R. Treisman. 1995. Transcriptional regulation by extracellular signals: mechanisms and specificity. Cell 80:199-211. [DOI] [PubMed] [Google Scholar]
- 22.Hunter, T., and M. Karin. 1992. The regulation of transcription by phosphorylation. Cell 70:375-387. [DOI] [PubMed] [Google Scholar]
- 23.Jamal, S., and E. Ziff. 1990. Transactivation of c-fos and beta-actin genes by raf as a step in early response to transmembrane signals. Nature 344:463-466. [DOI] [PubMed] [Google Scholar]
- 24.Karin, M. 1995. The regulation of AP-1 activity by mitogen-activated protein kinases. J. Biol. Chem. 270:16483-16486. [DOI] [PubMed] [Google Scholar]
- 25.Karin, M., Z. Liu, and E. Zandi. 1997. AP-1 function and regulation. Curr. Opin. Cell Biol. 9:240-246. [DOI] [PubMed] [Google Scholar]
- 26.Kerr, L. D., J. T. Holt, and L. M. Matrisian. 1988. Growth factors regulate transin gene expression by c-fos-dependent and c-fos-independent pathways. Science 242:1424-1427. [DOI] [PubMed] [Google Scholar]
- 27.Kovary, K., and R. Bravo. 1991. The Jun and Fos protein families are both required for cell cycle progression in fibroblasts. Mol. Cell. Biol. 11:4466-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kruijer, W., J. A. Cooper, T. Hunter, and I. M. Verma. 1984. Platelet-derived growth factor induces rapid but transient expression of the c-fos gene and protein. Nature 312:711-716. [DOI] [PubMed] [Google Scholar]
- 29.Kyriakis, J. M., P. Banerjee, E. Nikolakaki, T. Dai, E. A. Rubie, M. F. Ahmad, J. Avruch, and J. R. Woodgett. 1994. The stress-activated protein kinase subfamily of c-Jun kinases. Nature 369:156-160. [DOI] [PubMed] [Google Scholar]
- 30.Lamph, W. W., P. Wamsley, P. Sassone-Corsi, and I. M. Verma. 1988. Induction of proto-oncogene JUN/AP-1 by serum and TPA. Nature 334:629-631. [DOI] [PubMed] [Google Scholar]
- 31.Ledwith, B. J., S. Manam, A. R. Kraynak, W. W. Nichols, and M. O. Bradley. 1990. Antisense-fos RNA causes partial reversion of the transformed phenotypes induced by the c-Ha-ras oncogene. Mol. Cell. Biol. 10:1545-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lucibello, F. C., and R. Muller. 1991. Proto-oncogenes encoding transcriptional regulators: unravelling the mechanisms of oncogenic conversion. Crit. Rev. Oncog. 2:259-276. [PubMed] [Google Scholar]
- 33.Marinissen, M. J., M. Chiariello, and J. S. Gutkind. 2001. Regulation of gene expression by the small GTPase Rho through the ERK6 (p38 gamma) MAP kinase pathway. Genes Dev. 15:535-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marinissen, M. J., M. Chiariello, M. Pallante, and J. S. Gutkind. 1999. A network of mitogen-activated protein kinases links G protein-coupled receptors to the c-jun promoter: a role for c-Jun NH2-terminal kinase, p38s, and extracellular signal-regulated kinase 5. Mol. Cell. Biol. 19:4289-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mercola, D., A. Rundell, J. Westwick, and S. A. Edwards. 1987. Antisense RNA to the c-fos gene: restoration of density-dependent growth arrest in a transformed cell line. Biochem. Biophys. Res. Commun. 147:288-294. [DOI] [PubMed] [Google Scholar]
- 36.Murphy, L. O., S. Smith, R. H. Chen, D. C. Fingar, and J. Blenis. 2002. Molecular interpretation of ERK signal duration by immediate early gene products. Nat. Cell Biol. 4:556-564. [DOI] [PubMed] [Google Scholar]
- 37.Okazaki, K., and N. Sagata. 1995. The Mos/MAP kinase pathway stabilizes c-Fos by phosphorylation and augments its transforming activity in NIH 3T3 cells. EMBO J. 14:5048-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piechaczyk, M., and J. M. Blanchard. 1994. c-fos proto-oncogene regulation and function. Crit. Rev. Oncol. Hematol. 17:93-131. [DOI] [PubMed] [Google Scholar]
- 39.Rydziel, S., D. Durant, and E. Canalis. 2000. Platelet-derived growth factor induces collagenase 3 transcription in osteoblasts through the activator protein 1 complex. J. Cell. Physiol. 184:326-333. [DOI] [PubMed] [Google Scholar]
- 40.Shaulian, E., and M. Karin. 2002. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4:E131-E136. [DOI] [PubMed] [Google Scholar]
- 41.Shaulian, E., and M. Karin. 2001. AP-1 in cell proliferation and survival. Oncogene 20:2390-2400. [DOI] [PubMed] [Google Scholar]
- 42.Sutherland, J. A., A. Cook, A. J. Bannister, and T. Kouzarides. 1992. Conserved motifs in Fos and Jun define a new class of activation domain. Genes Dev. 6:1810-1819. [DOI] [PubMed] [Google Scholar]
- 43.Teramoto, H., P. Salem, K. C. Robbins, X. R. Bustelo, and J. S. Gutkind. 1997. Tyrosine phosphorylation of the vav proto-oncogene product links FcɛRI to the Rac1-JNK pathway. J. Biol. Chem. 272:10751-10755. [DOI] [PubMed] [Google Scholar]
- 44.Tong, L., S. Pav, D. M. White, S. Rogers, K. M. Crane, C. L. Cywin, M. L. Brown, and C. A. Pargellis. 1997. A highly specific inhibitor of human p38 MAP kinase binds in the ATP pocket. Nat. Struct. Biol. 4:311-316. [DOI] [PubMed] [Google Scholar]
- 45.Treisman, R. 1996. Regulation of transcription by MAP kinase cascades. Curr. Opin. Cell Biol. 8:205-215. [DOI] [PubMed] [Google Scholar]
- 46.Treisman, R. 1994. Ternary complex factors: growth factor regulated transcriptional activators. Curr. Opin. Genet. Dev. 4:96-101. [DOI] [PubMed] [Google Scholar]
- 47.Whitmarsh, A. J., and R. J. Davis. 1996. Transcription factor AP-1 regulation by mitogen-activated protein kinase signal transduction pathways. J. Mol. Med. 74:589-607. [DOI] [PubMed] [Google Scholar]
- 48.Whitmarsh, A. J., P. Shore, A. D. Sharrocks, and R. J. Davis. 1995. Integration of MAP kinase signal transduction pathways at the serum response element. Science 269:403-407. [DOI] [PubMed] [Google Scholar]
- 49.Wick, M., F. C. Lucibello, and R. Muller. 1992. Inhibition of Fos- and Ras-induced transformation by mutant Fos proteins with structural alterations in functionally different domains. Oncogene 7:859-867. [PubMed] [Google Scholar]
- 50.Wisdon, R., and I. M. Verma. 1993. Transformation by Fos proteins requires a C-terminal transactivation domain. Mol. Cell. Biol. 13:7429-7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhan, Y., S. Kim, H. Yasumoto, M. Namba, H. Miyazaki, and H. Iwao. 2002. Effects of dominant-negative c-Jun on platelet-derived growth factor-induced vascular smooth muscle cell proliferation. Arterioscler. Thromb. Vasc. Biol. 22:82-88. [DOI] [PubMed] [Google Scholar]