Abstract
Transcription of luteinizing hormone receptor (LHR) gene is activated by Sp1/Sp3 at two Sp1 sites and is repressed by nuclear orphan receptors EAR2 and EAR3 through a direct-repeat (DR) motif. To elucidate the mechanism of the orphan receptor-mediated gene repression, we explored the functional connection between the orphan receptors and Sp1/Sp3 complex, and its impact on the basal transcription machinery. The Sp1(I) site was identified as critical for the repression since its mutation reduced the inhibition by EAR2 and abolished the inhibition by EAR3. Cotransfection analyses in SL2 cells showed that both Sp1 and Sp3 were required for this process since EAR3 displayed a complete Sp1/Sp3-dependent inhibitory effect. Functional cooperation between Sp1 and DR domains was further supported by mutual recruitment of EAR3 and Sp1/Sp3 bound to their cognate sites. Deletion of EAR3 N-terminal and DNA-binding domains that reduced its interaction with Sp1 impaired its inhibitory effect on human LHR (hLHR) gene transcription. Furthermore, we demonstrate interaction of TFIIB with Sp1/Sp3 at the Sp1(I) site besides its association with EAR3 and the TATA-less core promoter region. Such interaction relied on Sp1 site-bound Sp1/Sp3 complex and adaptor protein(s) present in the JAR nuclear extracts. We further demonstrated that EAR3 specifically decreased association of TFIIB to the Sp1(I) site without interfering on its interaction with the hLHR core promoter. The C-terminal region of EAR3, which did not participate in its interaction with Sp1, was required for its inhibitory function and may affect the association of TFIIB with Sp1. Moreover, perturbation of the association of TFIIB with Sp1 by EAR3 was reflected in the reduced recruitment of RNA polymerase II to the promoter. Overexpression of TFIIB counteracted the inhibitory effect of EAR3 and activated hLHR gene transcription in an Sp1 site-dependent manner. These findings therefore indicate that TFIIB is a key component in the regulatory control of EAR3 and Sp1/Sp3 on the initiation complex. Such cross talk among EAR3, TFIIB, and Sp1/Sp3 reveals repression of hLHR gene transcription by nuclear orphan receptors is achieved via perturbation of communication between Sp1/Sp3 at the Sp1-1 site and the basal transcription initiator complex.
Luteinizing hormone (LH) is a glycoprotein hormone of pituitary origin that regulates gonadal function, including steroidogenesis and gametogenesis (see references 5, 8, and 9 reviews). LH action is mediated by specific receptors that are located in the plasma membrane of specific target cells in the ovary and testis. The expression of the LH receptor (LHR) in the ovary is induced by follicle-stimulating hormone, estrogen, and growth factors in granulosa cells of the preovulatory follicles. However, the LHR gene is down-regulated after the midcycle LH surge and is subsequently increased during luteinization (8, 9). In the testis, the LHR is expressed in fetal Leydig cells and throughout adult life (8, 9). The identification of differential signaling pathways that regulate LHR gene expression, as well as the elucidation of molecular mechanism(s) of receptor regulation, is of major relevance to the understanding of normal reproductive physiology and the pathology of reproductive disorders.
Characterization of transcriptional regulatory mechanism for the LHR gene expression has been advanced by identification of the promoter region of LHR gene in different species, as well as the cis- and trans-acting elements governing the promoter activity. Our previous studies showed that the LHR gene is TATA-less and contains multiple transcriptional start sites (15, 49, 51, 52). The minimal promoter with high GC-rich sequences resides within a 5′ −180-bp region relative to the ATG start codon (+1). Two Sp1/Sp3 binding elements of central importance for the basal promoter activity have been identified in the human and the rodent genes (15, 51). Furthermore, nuclear orphan receptors EAR2 and EAR3/COUP-TFI were shown to repress the LHR gene transcription in both human and rat via binding to an imperfect direct repeat (DR) with zero spacing between the two half-sites (DR-0) (59, 60). In contrast, activation of the promoter activity by another nuclear orphan receptor TR4 through the same DR motif was observed only in the human (60). The differential regulations of the human and rat LHR (hLHR and rLHR) promoter activity by these orphan receptors were shown to result from a single-base-pair mismatch at the DR core sequence and the lack of a G residue in the 3′ sequence adjacent to the rat DR core motif (59). These findings also suggested that the relative abundance of the three nuclear orphan receptors might be significant in determining the LHR gene promoter activity in different cell types and/or at different physiological stages. This was supported by the evidence that gonadotropin-mediated down-regulation of EAR2 and EAR3/COUP-TFI expression correlated with derepression of rLHR gene transcription in rat ovary granulosa/luteal cells (59), at stages where expression of the LHR gene increase significantly in response to hormonal fluctuations during the differentiation of granulosa cells into luteal cells (35, 36, 42). On the other hand, we have recently reported that local chromatin structure modulated by histone acetylation and deacetylation plays an active role in the hLHR gene regulation (61). The Sp1(I) site of the hLHR gene promoter was identified to serve as a docking site to recruit a histone deacetylase (HDAC)-mSin3A complex to the promoter, which caused silencing of LHR gene transcription via histone hypoacetylation-induced chromatin condensation (61).
It has been recognized that protein-protein interactions between sequence-specific DNA-binding trans-acting factors is an important mechanism utilized by eukaryotes to achieve regulated expression of a target gene under specific promoter and cell context (28, 34). Moreover, transcription initiation of protein-encoding genes depends on ordered assembly of RNA polymerase II (Pol II) and other basal transcription factors (TFIIA, -B, -D, -E, -F, and -H) to form a functional preinitiation complex (PIC) at the promoter region (16, 58). Transcriptional activators and/or repressors are thought to interact with component(s) of the basal transcriptional machinery to facilitate or inhibit the assembly of a productive PIC complex (2, 7). TFIIB was shown to play an important role in the link of promoter-bound transcription factors to PIC, since direct interaction of TFIIB with nuclear hormone receptors and other regulatory proteins were observed (2, 21, 37). These include the interactions with receptors for estrogen, progesterone, thyroid, retinoid, and vitamin D, as well as interaction with EAR3/COUP-TFI- in COUP-TFI-activated chicken ovalbumin gene expression (6, 11, 13, 21, 26, 47).
We sought to elucidate here the molecular mechanism(s) of nuclear-receptor-mediated repression of LHR transcription, with particular emphasis on the impact of EAR2, EAR3, on Sp1/Sp3 function and their link to constituent(s) of the PIC. Our findings have demonstrated that transcriptional repression of the LHR gene is mediated through the interplay between orphan nuclear receptors, Sp1/Sp3, and TFIIB, in which EAR3 significantly prevents association of TFIIB at the Sp1(I) site of the LHR gene promoter.
MATERIALS AND METHODS
Reporter gene constructs and expression vectors.
All plasmids were constructed by standard recombinant DNA techniques. The wild-type hLHR and rLHR promoter reporter genes and their Sp1 site mutant constructs have been described previously (15, 51, 52). The mammalian expression plasmids of EAR2 and EAR3 were constructed as previously reported (60), and the same cDNA inserts were subcloned into insect pAc vector for their expression in Drosophila melanogaster SL2 cells. The pAc-Sp1 expression vector was a gift from Robert Tjian (Department of Molecular and Cell Biology, University of California, Berkeley). Cloning of pAc-Sp3 expression vector was previously described (20). The glutathione S-transferase (GST)/EAR3 fusion protein expression construct was prepared by inserting the full-length of EAR3 cDNA into pGEX-2T vector (Amersham Pharmacia, Piscataway, N.J.) at BamHI and EcoRI sites in frame with 5′ GST tag. The EAR3/pcDNA 3.1 expression constructs containing different deletions and C-terminal fusion of a V5 tag were generated by conventional PCR techniques. The pCMV-TFIIB mammalian expression construct was kindly provided by Danny Reinberg (Department of Biochemistry, University of Medicine and Dentistry of New Jersey, Piscataway).
Cell culture and transient transfection.
JAR cells (human placental choriocarcinoma cells; American Type Culture Collection [ATCC], Manassas, Va.) were maintained in RPMI 1640 medium supplemented with10% fetal bovine serum (FBS; Gibco, Long Island, N.Y.). CV-1 cells (African green monkey kidney cells [ATCC]) were cultured in minimal essential medium supplemented with 10% FBS and l-glutamine. Drosophila SL2 cells (ATCC) was maintained in M-3 insect medium (Quality Biological, Inc., Gaithersburg, Md.) containing 10% FBS.
Transfections of CV-1 and JAR cells were carried out by using Lipofectamine Plus reagents, and transfection of SL2 cells were performed with CellFectin reagent according to the procedures recommended by the manufacturer (Invitrogen, Carlsbad, Calif.). pCMV-SPORTS-βgal construct was used as an internal control plasmid in all transfections performed in CV-1 and JAR cells. The luciferase activities measured in CV-1 and JAR cells were normalized based on the β-galactosidase activities, whereas the luciferase activities from SL2 cells were normalized based on the relative light units (RLU) per microgram of protein. The results were expressed as means ± the standard errors (SE) from at least three independent experiments in triplicate wells.
Preparation of native nuclear extracts and Western blotting.
Nuclear extracts were prepared as described previously (19). A total of 30 μg of JAR nuclear proteins was subjected to Western blot analyses by using Sp1, Sp3, TFIIB, and TAFII 250 antibodies (Santa Cruz Biotechnology, Santa Cruz, Calif.) or purified polyclonal antibodies to EAR2 and EAR3 (60). Whole-cell lysates of JAR cells overexpressing wild-type or different deletion EAR3 constructs were prepared by using M-PER mammalian protein extraction reagent (Pierce, Rockford, Ill.). A total of 50 μg of cell lysates was analyzed in a Western blot with V5 antibody (Invitrogen).
Preparation of nuclear extracts immunodepleted of EAR3 and TFIIB.
Antibody-conjugated Sepharose was used to deplete EAR3 and TFIIB from native JAR nuclear extracts. Briefly, 15 mg of EAR3/COUP-TFI polyclonal antibody/ml was dialyzed against 0.1 M NaHCO3-0.5 NaCl, followed by centrifugation at 100,000 × g to remove aggregates. The antibody solution was then diluted with the same buffer to 5 mg/ml for use in the coupling reaction. Then, 20 ml of Sepharose slurry was washed first with 10 volumes of water and mixed with an equal volume of 0.2 M Na2CO3. Activation of Sepharose by CNBr (cyanogen bromide)-acetonitrile was carried out at room temperature for 5 min (3.2 ml of CNBr per 100 ml of Sepharose), followed by a wash with 10 volumes of ice-cold 1 M HCl and 2 volumes of ice-cold 0.1 M HCl. Coupling of the antibody to CNBr-activated Sepharose was performed by incubation of equal volumes of antibody and CNBr-activated Sepharose, and the reaction was carried out overnight at 4°C with end-over-end rotation. Then, 0.05 M ethanolamine was added to the reaction to saturate the remaining reactive group. Immunodepletion was next performed by incubation of 10 μl of Sepharose-conjugated antibody per 100 μg of JAR cell nuclear extracts at 4°C for 3 h and the supernatant was then subjected to one more round of depletion. The EAR3-depleted nuclear extracts were analyzed by Western blotting with EAR3 antibody, followed by further depletion with TFIIB antibody according to the protocol described above.
Expression and purification of GST and GST/EAR3 fusion proteins.
Bacterial BL21 strains transformed with pGEX-2T vector or pGEX-2T/EAR3 fusion construct were cultured at 37°C for 4 to 5 h (A600 of 0.6 to 0.7). Cells were then incubated with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside; Gibco) for another 1 h at 37°C. Cells harvested and lysed by sonication in B-PER bacterial lysis buffer (Pierce) were subjected to affinity purification by using the Amersham Pharmacia GST purification module. The purified EAR3/GST or GST protein was dialyzed against buffer D (20 mM HEPES [pH 7.9], 20% glycerol, 0.M KCl, 0.2 M EDTA, 0.5 mM dithiothreitol, 1× protease inhibitor cocktail [Roche Molecular Biochemicals, Indianapolis, Ind.]) at 4°C for 6 h before used in the DNA affinity precipitation assay (DAPA).
DAPA.
5′-Biotin end-labeled sense and antisense oligonucleotides corresponding to the wild-type (5′-AGCCAAGGGGCGGGGAGAGGG-3′) or mutant (5′-AGCCAAatctgcaGcAGAGGG-3′) Sp1-1-binding sites or to the wild-type (5′-GTCGCAGGTCAAGGCAGAGCAGACTCAG-3′) or mutant (5′-GTCGCAttTCAAaaCAGAGCAGACTCAG-3′) orphan receptor binding sites (DR) of the hLHR promoter were custom made by GeneProbe Technology, Inc. (Gaithersburg, Md.). The oligomers were annealed and gel purified by 12% polyacrylamide gel electrophoresis. Portions (50 μg) of JAR cell nuclear extracts were incubated with 0.2 μg of wild-type or mutant biotinylated probe in binding buffer (60 mM KCl, 12 mM HEPES [pH 7.9], 4 mM Tris-HCl, 5% glycerol, 0.5 mM EDTA, 1 mM dithiothreitol, 1× protease inhibitor cocktail) on ice for 45 min. For DAPA with EAR3/TFIIB-depleted JAR nuclear extracts, 20 ng of recombinant TFIIB (Promega, Madison, Wis.) and 0 to 100 ng of affinity-purified GST/EAR3 fusion protein or GST were added to the reactions. The DNA-protein complexes were then incubated with 40 μl of Tetralink avidin resin (Promega), which was preequilibrated in the binding buffer for 1 h. The incubation was continued for 1 h at 4°C with gentle rotation. DNA-protein complexes were then washed five times with the binding buffer. Next, 36 μl of 2× protein sample buffer (Invitrogen) was added to the avidin-precipitated DNA-protein complex, which was then boiled for 5 min to dissociate the complexes. The proteins were resolved by polyacrylamide gel electrophoresis, followed by Western blot detection with specific antibodies. In a modified DAPA protocol, 0 to 200 ng of purified Sp1 protein (ProteinOne, Inc., College Park, Md.) was incubated with 0.2 μg of biotinylated Sp1(I) probe for 30 min on ice, followed by incubation with 40 μl of avidin beads at 4°C for 1 h with gentle agitation. The Sp1/DNA/avidin complexes were then harvested by brief centrifugation, and the unbound form of Sp1 protein and the excess probe was washed away. The complexes were resuspended in the binding buffer and incubated with 250 ng of TFIIB (ProteinOne) for 1.5 h at 4°C. The complexes were then subjected to the regular wash procedure, followed by immunodetection of TFIIB or Sp1.
ChIP.
Chromatin immunoprecipitation (ChIP) assays were performed as previously reported (61). Briefly, JAR cells and CV-1 cells were cross-linked with 1% formaldehyde and then lysed in lysis buffer (1% sodium dodecyl sulfate [SDS], 5 mM EDTA, 50 mM Tris-HCl [pH 8.1], 1× protease inhibitor cocktail). Soluble chromatin was prepared by sonication and then diluted in dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl [pH 8.1], 1× protease inhibitor cocktail), followed by immunoclearing with salmon sperm DNA, preimmune serum, and protein A-agarose (KPL Laboratories, Gaithersburg, Md.). Immunoprecipitation was carried out for overnight with 5 μl of TFIIB or TAFII250 or RNA Pol II antibody, or 3 μl of antibody for Sp1, Sp3, or EAR3, respectively. After immunoprecipitation, protein A-agarose and salmon sperm DNA was added, and the sample was incubated for another 1 h. Precipitates were washed sequentially in TSE I (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], 150 mM NaCl), TSE II (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], 500 mM NaCl), and buffer III (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl [pH 8.1]). Precipitates were then washed three times with Tris-EDTA buffer and extracted three times with 1% SDS-0.1 M NaHCO3. Eluates were heated at 65°C for 6 h to reverse the formaldehyde cross-linking. The samples were then treated with protease K, followed by phenol extraction and ethanol precipitation. The DNA pellet was dissolved in 40 μl of Tris-EDTA buffer. Then, 1 μl of DNA was used for PCR with 20 to 25 cycles of amplification. PCR amplification of soluble chromatin prior to immunoprecipitation was used as input control for the immunoprecipitation analyses.
Co-IP.
Portions (1 mg) of whole-cell lysates prepared from JAR cells expressing wild-type or different deletion EAR3 constructs were utilized in coimmunoprecipitation (Co-IP) analyses. Briefly, the cell lysates were initially subjected to preclearing by incubation with 30 μl of protein-agarose slurry and 0.5 μg of preimmune mouse immunoglobulin G (IgG) for 30 min at 4°C with gentle agitation. The recovered supernatant was incubated with 3 μl of anti-V5 antibody for 1 h at 4°C in presence of 1× protease inhibitor cocktail. Then, 30 μl of protein A-agarose slurry was added, and the incubation was continued for overnight. Protein A-precipitated protein complex was recovered by brief centrifugation, followed by two sequential washes with 0.1% SDS and 2% SDS-radioimmunoprecipitation assay buffer (1× phosphate-buffered saline, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1 or 2% SDS). The harvested beads were resuspended in 36 μl of 2× protein sample buffer containing 2.5% of β-mercaptoethanol and were boiled for 5 min to release the bound proteins. The samples were then analyzed by Western blot with anti-Sp1 antibody.
RESULTS
The proximal Sp1 site was essential for EAR2- and EAR3/COUP-TFI-mediated repression of LHR gene promoter activity.
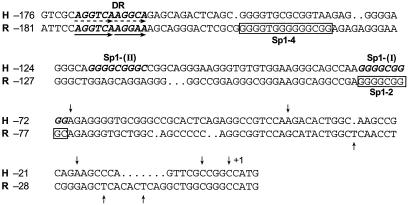
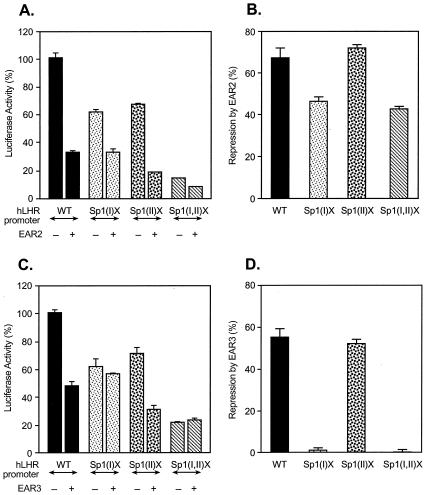
Figure 1 illustrates the sequence comparison between the human and rat LHR gene promoters, in which the functional cis elements regulating the promoter activity are indicated. The two Sp1/Sp3-binding activation domains are designated Sp1(I) and Sp1(II) for the hLHR gene and Sp1 (2) and Sp1 (4) for the corresponding elements in the rLHR gene. The DR motifs are binding sites for the nuclear orphan receptors with EAR2 and EAR3/COUP-TFI in both species and for TR4 only in humans. In order to investigate whether modulation of LHR gene transcription by these nuclear orphan receptors resulted from their influence on Sp1/Sp3-mediated function at the Sp1 sites, the nuclear orphan receptors were cotransfected in CV-1 cells with LHR promoter wild-type construct or constructs with different mutations of Sp1 sites (Fig. 2 and 3). Consistent with our previous observations in other cell lines, mutation at either Sp1 site in CV-1 cells caused significant reduction of the hLHR promoter activity [by 48% for Sp1(I)X and by 43% for Sp1(II)X], and most of the activity was abolished when both sites were mutated (Fig. 2A). Also, as shown previously, transfection of EAR2 caused marked repression of wild-type promoter activity (60). However, the EAR2-mediated inhibition was significantly inhibited in cells transfected with the Sp1(I) site-mutated promoter, and no further reduction was observed when EAR2 and the hLHR Sp1(I,II) double-mutant construct were cotransfected. In contrast, EAR2 repressed the activity of the promoter construct with a mutation at the Sp1(II) site as effectively as it did for the wild-type promoter. These findings demonstrated that the Sp1(I) site was involved in the inhibitory function of EAR2, whereas the Sp1(II) site was not relevant.
FIG. 1.
Alignment of nucleotide sequences of the promoter regions of the hLHR (H) and rLHR (R) genes. DNA sequences of the hLHR and rLHR promoters are shown, and the nucleotides are numbered relative to the translation initiation codon (ATG, +1). The transcriptional start sites are indicated by arrows. The proximal and distal functional Sp1 sites in the human, Sp1(I) and Sp1(II) (in boldface letters) and the corresponding sites in the rat, Sp1-2, and Sp1-4 (boxed letters) are indicated. The DR domains that bind nuclear orphan receptors EAR2, EAR3 (human and rat), and TR4 (human) are underlined by arrows.
FIG. 2.
Identification of the Sp1(I) site for EAR2- and EAR3/COUP-TFI-mediated inhibition of the hLHR gene transcription. The wild-type (WT) or Sp1 site mutant hLHR promoter/reporter constructs [Sp1(I)X, Sp1(II)X, and Sp1(I,II)X] was cotransfected with pcDNA3.1 vector or with expression plasmids of EAR2 (A and B) or EAR3/COUP-TFI (C and D) in CV-1 cells. The luciferase activities are expressed as the percentage of the wild-type promoter activity in absence of the orphan receptors (100%, A and C), or as percentage of repression caused by EAR2 or EAR3/COUP-TFI for the individual promoter construct (B and D). The results were normalized by β-galactosidase activity and expressed as the means ± the SE of three independent experiments in triplicate wells.
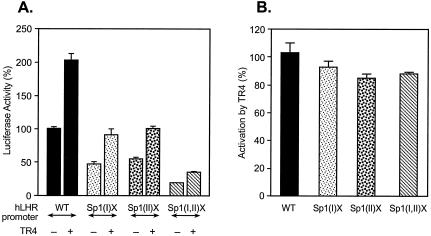
FIG. 3.
Activation of hLHR promoter activity by TR4 independent of the Sp1 site. The wild-type (WT) or Sp1 site mutant hLHR promoter/reporter constructs [Sp1(I)X, Sp1(II)X, and Sp1(I,II)X] was cotransfected with pcDNA3.1 vector only or with expression plasmid of TR4 in CV-1 cells. The luciferase activities are expressed as percentage of the wild-type promoter activity in absence of TR4 (100%), or as percentage of the activation caused by TR4 for the individual construct. The results were normalized by β-galactosidase activity and expressed as the means ± the SE of three independent experiments in triplicate wells.
When EAR3 was analyzed, it showed that repression of the hLHR promoter activity by this orphan receptor was dependent on the Sp1(I) site, since disruption of this site totally released EAR3/COUP-TFI-mediated inhibition of the promoter activity (Fig. 2C). Conversely, the inhibitory effect of EAR3/COUP-TFI was not affected by mutation of the Sp1(II) site. These results were most evident when changes were expressed as the percentage of repression for each individual promoter construct (Fig. 2B and D). Thus, the Sp1(I) site mutation elicited partial loss of the EAR2 inhibition and completely abolished EAR3/COUP-TFI repression.
In order to determine whether the TR4-activated hLHR gene transcription was also related to the Sp1(I) site, cotransfection analyses of TR4 with the hLHR promoter constructs were carried out (Fig. 3). Compared to the activation of the wild-type promoter activity by this orphan receptor, mutation at either Sp1 site or at both sites caused marginal decrease of the transcriptional stimulation induced by TR4. Thus, these findings did not reveal a site-specific Sp1 effect for TR4.
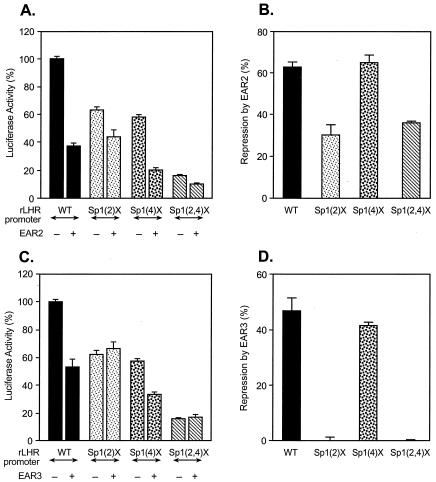
To elucidate whether the Sp1(I) site-dependent repression of the hLHR gene by EAR2 and EAR3 represented a generic mechanism in modulation of the LHR gene across species, rLHR gene promoter constructs were analyzed (Fig. 4). The rat promoter Sp1 (2) site, equivalent to the Sp1(I) site of the hLHR gene with respect to activation of the promoter activity (location and proximal functional domain of the two Sp1 elements), was found to be relevant for EAR2 and EAR3/COUP-TFI function. Mutation of this site significantly reduced the inhibition by EAR2 and abolished the repression induced by EAR3. Therefore, the results demonstrated that the Sp1 (2) site of the rat gene was a functional counterpart of the hLHR promoter Sp1(I) site in the orphan receptor-mediated repression of the rat gene transcription. Thus, in both species, the proximal Sp1 site was operative in this inhibitory regulation. In subsequent experiments, we focused our studies on the mechanism of EAR3/COUP-TFI function since this orphan receptor displayed complete dependence on the Sp1(I) site for its inhibitory effect.
FIG. 4.
Identification of the Sp1(2) site for EAR2-, EAR3/COUP-TFI-mediated repression of the rLHR gene transcription. The wild-type (WT) or Sp1 site mutant rLHR promoter/reporter constructs [Sp1(2)X, Sp1(4)X, and Sp1(2,4)X] was cotransfected with pcDNA3.1 vector only or with expression plasmids of EAR2 (A and B) or EAR3/COUP-TFI (C and D) in CV-1 cells. The luciferase activities are expressed as a percentage of the wild-type promoter activity in the absence of the orphan receptors (100% [A and C]) or as a percentage of the repression caused by EAR2 or EAR3/COUP-TFI for the individual construct (B and D). The results were normalized by β-galactosidase activity and are expressed as means ± the SE of three independent experiments in triplicate wells.
Sp1 and Sp3 were both required for inhibition of the LHR gene promoter activity by EAR3/COUP-TFI.
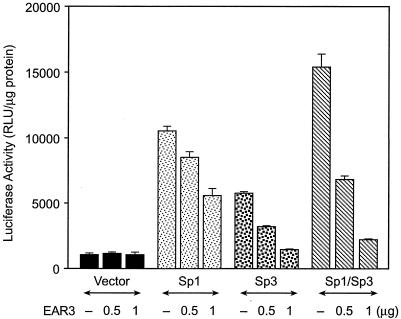
To determine whether Sp1 and Sp3, as Sp1 site binding proteins, are both involved in the repression of LHR gene transcription caused by EAR3/COUP-TFI, we conducted cotransfection studies of EAR3/COUP-TFI with the wild-type hLHR gene promoter in absence or presence of Sp1/Sp3 expression plasmids in Sp factor-deficient Drosophila SL-2 cells. As expected, the hLHR promoter displayed low basal activity in these cells in the absence of Sp1/Sp3 (Fig. 5). Moreover, cotransfection of EAR3/COUP-TFI with the hLHR promoter under this condition did not show repression of the promoter activity. On the other hand, transfection of Sp1 or Sp3 in the absence of EAR3/COUP-TFI markedly elevated the promoter activity, with the strongest activation being detected when Sp1 and Sp3 were coexpressed. This finding was consistent with our previous report that Sp1/Sp3 were potent transcriptional activators for the LHR gene (15). Furthermore, EAR3/COUP-TFI caused dose-dependent inhibition of the hLHR promoter activity when cotransfected with either Sp1 or Sp3, and potent inhibition by this orphan receptor was clearly demonstrated in the presence of both proteins. Taken together, the results indicated that both Sp1 and Sp3 participate in the silencing of transcription of hLHR gene by EAR3/COUP-TFI.
FIG. 5.
Requirement of both Sp1 and Sp3 in EAR3/COUP-TFI-mediated repression of the hLHR gene. The wild-type hLHR promoter/reporter gene construct was cotransfected with pACT2 vector only or with increasing dose of EAR3/COUP-TFI expression plasmid in the absence or presence coexpression of Sp1 or Sp3 in Sp1-deficient Drosophila SL-2 cells. The results were normalized as RLU per microgram of protein and are expressed as the means ± the SE of four independent experiments in triplicate wells.
Repression of hLHR gene transcription requires interaction of EAR3 and Sp1 in the presence of their DNA-binding sites.
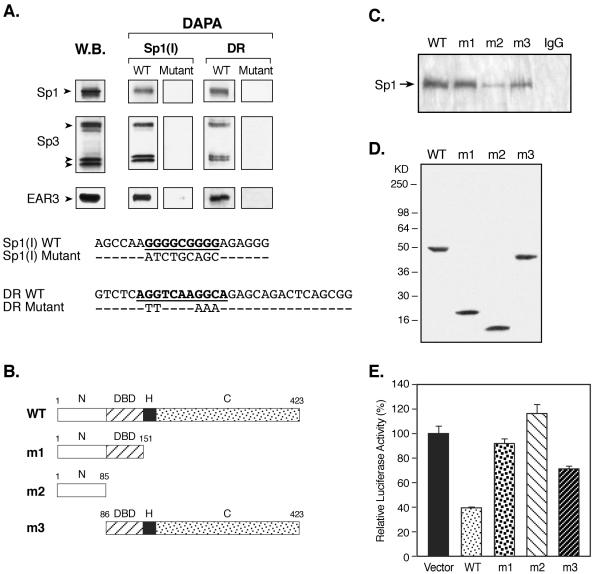
Evidence for direct protein-protein interaction between EAR3/COUP-TFI and transcription factors other than members of nuclear hormone receptor family has recently emerged, including its interaction with Sp1 (38, 44, 45). It is important to note that such interaction was not explored in COUP-TFs-mediated repression, whereas it was only detected in COUP-TFs-induced gene transactivation studies in which either a COUP-TF binding site was not present or nonfunctional within the target gene (32, 38, 41). Therefore, in order to determine whether endogenous EAR3 and Sp1/Sp3 proteins of JAR cells could interact with each other in presence of their cognate DNA-binding sites, DAPAs were performed. The probes used included double-stranded sequences corresponding to the wild-type or mutant Sp1(I) site of the hLHR promoter, and others representing the wild-type or mutant DR motifs (Fig. 6A). Expression of Sp1, Sp3, and EAR3 was also analyzed by Western blotting (Fig. 6A, lane W.B.). Sequence-specific binding of Sp1 and Sp3 to the wild-type but not to the mutant Sp1(I) probe (DAPA) was observed, as expected from our previous report (15, 61). Furthermore, association of EAR3 to the Sp1/Sp3 bound wild-type Sp1(I) site was clearly detected, and the interaction was dependent on Sp1/Sp3 binding activity since elimination of the Sp1/Sp3 binding by mutation abolished the EAR3 immunoreactive signal. On the other hand, incubation of the JAR nuclear extracts with the wild-type DR probe not only showed the binding of EAR3 but also revealed recruitment of both Sp1 and Sp3 to this DR element. The recruitment of these two non-DR binding activator proteins also required a prior binding of EAR3 at this DR site since no binding of Sp1/Sp3 was detected for the mutant DR domain. Taken together, these results have demonstrated mutual recruitment of EAR3 and Sp1/Sp3 proteins in the presence of their respective binding sites.
FIG. 6.
Analyses of interaction of EAR3 and the Sp1/Sp3 complex in regulation of hLHR gene transcription. (A) In the upper panel are shown DAPAs of mutual recruitment of EAR3 and Sp1/Sp3 complex in the presence of their cognate binding sites (DAPA). JAR nuclear extracts were incubated with 5′ biotin-labeled wild-type Sp1(I) or orphan receptor-binding DR probe or with the corresponding mutant probes (Mutant) devoid of Sp1/Sp3 or EAR3 binding activities. The avidin-precipitated complexes were analyzed by immunodetection with antibodies to Sp1, Sp3, and EAR3. Endogenous expression of the relevant transcription factors was also analyzed by Western blot (W.B.). In the lower panel are shown DNA sequences of the probes utilized in the DAPAs. (B) Schematic diagram of the wild-type (WT) and various deletion constructs of EAR3 (m1, m2, and m3) used in the following Co-IP and functional studies. The numbers represent the amino acid position. Abbreviations: N, N-terminal region; C, C-terminal region; H, hinge region. (C) Co-IP studies of interaction between EAR3 and Sp1. Lysates isolated from JAR cells overexpressing the wild-type (lane WT) or mutant EAR3 constructs (lanes m1, m2, and m3) were used in the Co-IP studies. Monoclonal anti-V5 antibody was utilized to coprecipitate Sp1 protein, followed by immunodetection of Sp1. Normal mouse IgG (lane IgG) was also included as a negative control. (D) Western blot analyses of expression of the wild-type (WT) and mutant EAR3 constructs (m1, m2, and m3) in JAR cells with anti-V5 antibody. (E) Cotransfection studies of the wild-type hLHR gene promoter/reporter gene construct with the wild-type and EAR3 mutant constructs in CV-1 cells. The luciferase activities were expressed as a percentage of the hLHR promoter activity in the absence of EAR3 (100%). The results were normalized by β-galactosidase activity and expressed as the means ± the SE of three independent experiments in triplicate wells.
To further explore the role of direct interaction of EAR3 and Sp1 in the Sp1(I) site-dependent repression of hLHR gene transcription by EAR3, three deletion constructs of EAR3 with C-terminal fusion of a V5 tag were generated and analyzed in both protein-protein interaction and functional studies (Fig. 6B). One construct contains N-terminal 151 amino acids of EAR3, which harbors its extreme N terminus and DNA-binding domain (DBD) but without the hinge region and putative C-terminal ligand-binding domain (Fig. 6B, m1, N and DBD). The second EAR3 mutant construct was derived through further deletion of the DBD thus it contains only N-terminal 85 amino acids (Fig. 6B, m2). Another deletion construct employed covers most of the sequences of EAR3 including the DBD and the C-terminal region, but it bears a truncation of its most N-terminal region (Fig. 6B, m3). Co-IP was then performed with anti-V5 antibody to coprecipitate Sp1 protein in JAR cells overexpressing wild-type or truncated forms of EAR3 (Fig. 6C, WT, m1, m2, and m3). Strong immunoreactive signals of Sp1 with similar intensities were demonstrated for both the wild-type and m1 deletion constructs. These results are consistent with the previous notion that the N-terminal DBD-containing region of EAR3 was sufficient to support interaction between EAR3 and Sp1, whereas the hinge and the C-terminal domains were dispensable in this regard (32, 38). In contrast, analysis of the m2 construct revealed only residual binding of Sp1, indicating that further removal of the DBD domain of EAR3 (compared to m1) largely abolished its association with Sp1. Analysis of the m3 construct confirmed that the EAR3 DBD domain was critical for its interaction with Sp1, since it was effectively coprecipitated when the DBD domain was present in this construct. Also, there was significantly reduced association between Sp1 and the m3 construct in which the N-terminal region was absent. This indicates that this region of EAR3 was required for its optimal interaction with Sp1. Moreover, the observed differences of EAR3/Sp1 interaction were not attributed to variation at protein expression levels of the constructs used, since similar expression of wild-type and truncated forms of EAR3 were observed in Western blot analyses (Fig. 6D). Functional studies of these deletion constructs and the wild-type EAR3 in regulation of hLHR gene transcription was next pursued in cotransfection studies with the wild-type hLHR promoter/reporter gene construct in CV-1 cells, which as indicated previously lack EAR3 expression. Coexpression of the wild-type EAR3 with the hLHR gene promoter caused marked repression of the hLHR gene transcription compared to that in presence of the vector only (Fig. 6E, WT, by 61.7%). When the m2 construct was tested, it was found that deletion of the most of the C-terminal sequences of EAR3 including the DBD abolished such inhibitory effect (m2). These results were in agreement with our previous reports demonstrating that occupancy by EAR3 of the DR motif was a prerequisite for its repression of the hLHR gene transcription, since mutation of the DR motif to disrupt the EAR3 binding abolished the inhibition of hLHR gene promoter activity (60). Moreover, no repression was observed for the m1 construct although this deletion form of EAR3 associated with Sp1 protein similarly as the wild-type EAR3 when analyzed in the Co-IP assays (Fig. 6D and E). Therefore, truncation of the C-terminal region of EAR3 in the m1 construct removed its intrinsic inhibitory activity possibly through disruption of the interaction of EAR3 with putative cofactor protein(s), which may impact on TFIIB-mediated Sp1 activation of the hLHR gene (see below). Also, the repression of the hLHR gene transcription by EAR3 was alleviated upon deletion of its most N-terminal region in the m3 construct (Fig. 6E, by 28.4% for m3 versus by 61.7% for WT), which was shown to be necessary for optimal interaction of EAR3 with Sp1 (Fig. 6C, m3). These results therefore demonstrate that the decreased interaction between EAR3 and Sp1 (m2 and m3) caused no or minimal repression of the hLHR gene promoter activity. Furthermore, our findings indicate that various regions of EAR3 support its inhibitory activity, and their functional impact was observed even in presence of unimpaired physical interaction between Sp1 and EAR3 (m1).
Recruitment of TFIIB to the TATA-less hLHR promoter revealed multiple mechanisms.
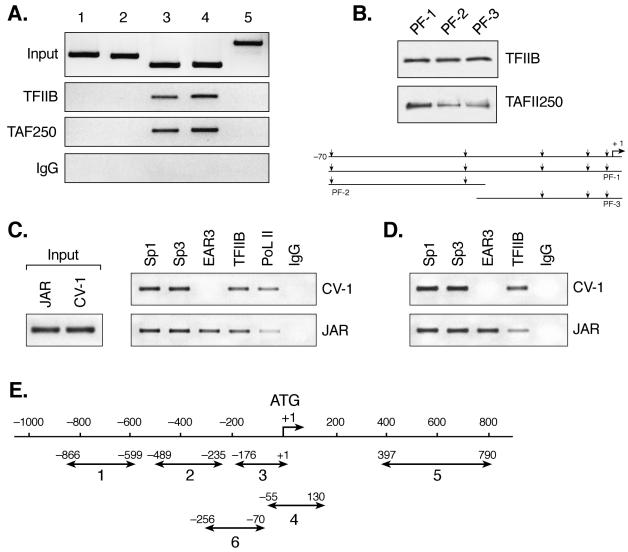
Several lines of evidence have indicated that members of nuclear hormone receptor family, including orphan receptors, modulate target gene expression by facilitating or interfering with the formation of the PIC (16, 46). This notion has been derived in part from the evidence of direct interaction between nuclear receptors and components of the basal transcription machinery, e.g., TFIIB or TFIID (23, 54). Therefore, we performed studies to determine whether the Sp1 site-dependent repression of hLHR gene transcription by EAR3 relies on the participation of basal transcription factor(s) that may associate with the Sp1 site to relay their modulation signal to the PIC. This is extremely relevant when addressed within the hLHR gene promoter context, since our understanding of regulation of TATA-less genes is limited. ChIP was initially carried out to investigate the association of TFIIB to the hLHR gene promoter, in which six regions of the gene were analyzed as shown in Fig. 7E. TAFII 250, the largest subunit of the TFIID complex, was also examined. Occupancy of TFIIB to the full promoter region (positions −176 to + 1) was clearly demonstrated. In contrast, no binding was detected for the two segments 5′ to the promoter (Fig. 7A, lanes 1, 2, and 3). TFIIB showed also significant binding to region 4, which encompasses the transcription start site region and a short stretch of coding sequence but without upstream regulatory Sp1 sites and the DR element. Moreover, no binding of TFIIB to an internal coding region of the hLHR gene was observed (region 5). Similar results were also obtained for TAFII 250. These findings demonstrated that TFIIB and TAFII 250 were capable of binding to the hLHR TATA-less core promoter region without requisite binding of Sp1/Sp3 or the orphan receptors at their response elements.
FIG. 7.
Recruitment of basal and trans-transcription factors to the hLHR gene promoter in JAR and CV-1 cells. (A) Recruitment of basal transcription factors, TFIIB and TAFII 250, to the hLHR gene promoter in ChIP assays. Soluble chromatin from JAR cells was precipitated with antibodies against TFIIB, TAFII 250, or preimmune rabbit or mouse immunoglobulin (row IgG). The DNA regions analyzed in PCR are schematically represented in panel E as follows: lanes 1 and 2 refer to 5′ flanking sequences to the promoter; lane 3 covers the full promoter region (positions −176 to +1); lane 4 encompasses the core promoter region only without upstream Sp1 and DR regulatory elements, and a short stretch of coding sequence downstream of ATG (+1); and lane 5 covers solely a part of coding region of the hLHR gene. The results of amplification of soluble chromatin before precipitation were shown as control (Input), and only the negative control with mouse IgG was shown as representative. (B) DAPAs of association of TFIIB or TAFII 250 to the hLHR gene core promoter region. The 5′ biotin-labeled probes utilized encompass DNA sequences of positions −70 to +1 (promoter fragment 1 [PF-1]), positions −70 to −30 (PF-2), and positions −34 to +1 (PF-3), respectively. The probes were incubated with JAR nuclear extracts, followed by immunodetection for bound TFIIB and TAFII 250. The region of the hLHR gene core promoter with arrows indicating the multiple transcription start sites is also shown. (C) ChIP analyses of the recruitment of Sp1, Sp3, EAR3, TFIIB, and RNA Pol II to the hLHR promoter region (region 3 in panel E) in CV-1 and JAR cells. Rabbit IgG was also included as negative control. (D) ChIP analyses of recruitment of Sp1, Sp3, EAR3, and TFIIB to region 6 (in panel E) in CV-1 and JAR cells. Region 6 covers the Sp1 sites and the DR motif of the hLHR gene promoter and some 5′ flanking sequences to the promoter but does not contain the core promoter region. (E) Schematic representation of DNA regions of hLHR gene analyzed in ChIP assays.
To further determine binding of these two basal transcription factors to the hLHR promoter, DAPA was performed with probes that covered only the TATA-less core promoter regions (designated promoter fragments PF-1, -2, and -3, Fig. 7B). TFIIB showed comparable binding to all three DNA segments, whereas TAFII 250 binding to the shorter probes was lower compared to the PF-1 region. This implied differences in recruitment of these two basal transcription factors to the hLHR gene core promoter, where multiple transcription start sites and initiator elements reside (15, 50). Taken together, the DAPA results combined with those from ChIP assays demonstrated that TFIIB and TAFII 250 were recruited to the TATA-less hLHR gene promoter in the presence or absence of upstream Sp1 and DR regulatory domains.
To investigate whether differential recruitment of transcription factors to the hLHR gene is operative as a mechanism utilized in repression of hLHR gene transcription by the nuclear orphan receptor, recruitment of Sp1/Sp3 and EAR3 to the hLHR gene in JAR and CV-1 cells was assessed by ChIP assays. Our previous studies demonstrated that hLHR promoter is bound at the DR element by endogenous EAR3 in JAR cells when analyzed by electrophoretic mobility shift assays (EMSAs) (15, 60). Moreover, the DR motif bound by EAR3 conferred potent repression of the hLHR gene promoter activity in these cells (15, 60). In contrast, EMSA did not show specific binding of the DR motif with nuclear extracts isolated from CV-1 cells, which lack endogenous expression of EAR3. This was consistent with the evidence that the DR motif did not function as an inhibitory site for hLHR gene transcription in CV-1 cells in the absence of overexpressed exogenous EAR3 protein (60). Analyses of recruitment of Sp1/Sp3 and EAR3 to the hLHR gene promoter in these two cells therefore could provide insights into the occupancy of the hLHR gene promoter by these transcription factors at DR-mediated repressed versus nonrepressed states. In both cells, recruitment of Sp1 and Sp3 to the hLHR gene promoter was present (Fig. 7C and E, region 3). This was consistent with our previous findings from EMSA analyses showing that Sp1/Sp3 bound the hLHR promoter similarly in both cell types (15, 60). Binding of EAR3 to the hLHR promoter region in JAR cells was also shown (Fig. 7C, region 3), indicating Sp1/Sp3 and EAR3 co-occupy the hLHR gene promoter in vivo. In contrast, binding of EAR3 in CV-1 cells was not detectable under the same experimental conditions (21 PCR cycles). Furthermore, no apparent differences in TFIIB recruitment to the hLHR gene promoter were observed in these two cells. However, a decreased association of RNA Pol II to the promoter was observed in JAR cells compared to its binding in CV-1 cells. To focus on the role of Sp1 sites in the hLHR gene repression, ChIP assays were further extended to examine the recruitment of these transcription factors to a region that covers Sp1 and DR elements and some of its 5′-flanking sequences with exclusion of the hLHR core promoter region (Fig. 7D and E, region 6). The studies revealed that the association of TFIIB to this region was reduced in JAR cells compared to CV-1 cells. These results therefore indicate that interaction of TFIIB with a DNA region that harbors DR and Sp1 elements was subject to changes when endogenous binding of EAR3 at the DR motif was present. Further analyses of TFIIB association with DR-bound EAR3 and Sp1 site-bound Sp1/Sp3 were therefore explored in DAPAs.
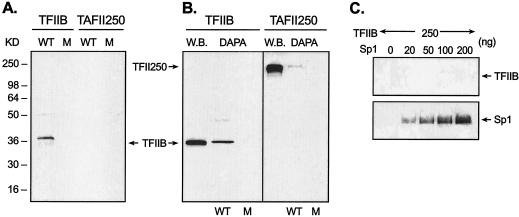
An association of TFIIB with the wild-type DR motif-bound form of EAR3 was observed (Fig. 8A, WT). In contrast, mutation of the DR element devoid of EAR3 binding abolished the TFIIB association (M, mutant). The results are also in agreement with previous findings that EAR3 and TFIIB interacted with each other in a GST pull-down assay (21). Moreover, the lack of detection of TAFII 250 indicated the selective anchoring of TFIIB to the DR motif. Further analyses of TFIIB association with Sp1/Sp3-bound Sp1(I) site in DAPA revealed robust binding of TFIIB to the Sp1(I) site (repeatedly observed within a few seconds of exposure time). The association was sequence specific since no binding was detected when the Sp1(I) site was mutated. As control, recruitment of TAFII250 to the same site was barely observed, and only a faint signal of TAFII 250 was developed after a long time of exposure (30 min). To our knowledge, these results provide the first evidence of TFIIB association with an Sp1 binding site. To further delineate the interaction between TFIIB and Sp1, purified Sp1 and TFIIB proteins were incubated with the wild-type Sp1(I) probe in a modified DAPA, in which excess unbound Sp1 protein was washed away prior to TFIIB addition to ensure interaction of TFIIB with DNA-bound form of Sp1 protein (see details in Materials and Methods). However, no direct interaction was observed between TFIIB and Sp1, whereas binding of Sp1 protein to this site at the indicated doses was shown as a control (Fig. 8C). Similar findings were obtained by using conventional DAPAs, and interaction analyses by Co-IP approaches with JAR nuclear extracts or purified Sp1/TFIIB proteins also gave negative results (data not shown). In addition, a previous report showed no detectable interaction between Sp1 and TFIIB in a GST pull-down assay (2). Thus, our findings demonstrating effective interaction of TFIIB with the Sp1/Sp3-DNA complex in the presence of JAR nuclear extracts, which contrast to the lack of direct interaction with purified materials, indicate that the DNA-bound Sp1 structure and a component from the nuclear extracts are required for the interaction with TFIIB. Taken together, our findings indicate that the association of TFIIB protein with the Sp1(I) site was dependent on currently unidentified adaptor protein(s) rather than by direct interaction of TFIIB/Sp1. It is clear that EAR3 does not act as a TFIIB-anchoring protein in this process, since further evidence demonstrated that TFIIB remained attached to the Sp1(I) site in the absence of EAR3 (see below and Fig. 9A). In summary, our results have revealed that more than one mechanism exists for recruitment of TFIIB to the TATA-less hLHR promoter: besides binding of TFIIB to the core promoter region and the orphan receptor-bound DR motif, the Sp1/Sp3-bound Sp1(I) site was identified as a docking site to tether this PIC component to the hLHR gene promoter. Furthermore, in presence of EAR3-mediated repression, reduced recruitment of RNA Pol II to the hLHR gene promoter and decreased binding of TFIIB to the Sp1/DR regulatory region was observed. These results imply that an active interplay among EAR3, TFIIB, and Sp1/Sp3 has functional consequences in the regulation of hLHR gene transcription.
FIG. 8.
Association of TFIIB with the DR motif and Sp1(I) site of hLHR gene promoter. (A and B) DAPAs were performed to analyze the association of endogenous TFIIB or TAFII 250 to the hLHR gene DR motif (A) or to the Sp1(I) site (B). JAR nuclear extracts were incubated with 5′ biotin-labeled probes, which include the wild-type (WT) and mutant DR elements, and the wild-type and mutant Sp1(I) site. Immunodetection was then carried out with antibodies against TFIIB or TAFII 250. Endogenous expression of TFIIB and TAFII 250 in JAR cells is also shown in Western blot analyses (B [W.B.]). (C) Analyses of protein-protein interaction between TFIIB and Sp1 in DAPAs. Purified Sp1 protein at the indicated doses was incubated with the biotinylated Sp1(I) probe. The excess unbound Sp1 was washed away, and only the DNA-bound form of Sp1 was incubated with 250 ng of purified TFIIB protein. The avidin-precicipated complexes were subject to immunodetection with against TFIIB or Sp1.
FIG. 9.
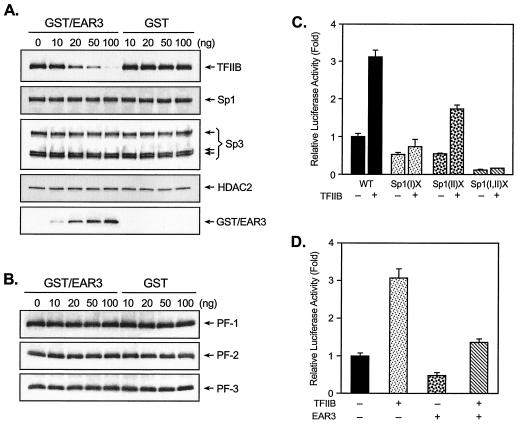
Functional analyses of cross talk among EAR3, Sp1/Sp3, and TFIIB in the regulation of transcription of the hLHR gene. (A) DAPAs were carried out with incubation of JAR nuclear extracts, depleted of TFIIB and EAR3, with 5′ biotin-labeled wild-type Sp1(I) probe, in which a constant amount of 20 ng of recombinant TFIIB protein was added in the presence of 0 to 100 ng of affinity-purified GST/EAR3 fusion protein or GST tag protein only. The avidin-precipitated complexes were subjected to Western blot analyses for immunodetection for TFIIB, Sp1, Sp3, HDAC2, and GST/EAR3. (B) DAPAs of association of TFIIB to the biotinylated PF-1, PF-2, and PF-3 probes (see Fig. 7) in the presence of increasing doses of GST/EAR3 or GST protein. (C) The wild-type (WT) or Sp1 site mutant hLHR promoter constructs [Sp1(I)X, Sp1(II)X, and Sp1(I,II)X] was transfected into JAR cells in absence or presence of pCMV-TFIIB. (D) The wild-type hLHR gene promoter was cotransfected in JAR cells with pCMV-TFIIB, pcDNA3.1-EAR3 expression plasmid, or both. Luciferase activities are expressed as the increase in wild-type promoter activity in the absence of TFIIB and EAR3 (onefold). The results were normalized by β-galactosidase activity and are expressed as the means ± the SE of four independent experiments in triplicate wells.
Functional interplay among EAR3, TFIIB, and Sp1/Sp3 in repression of hLHR gene transcription.
It is reasonable therefore to investigate whether the observed interactions among EAR3, TFIIB, and Sp1/Sp3 constitute a dynamic interplay where a change at one interface could elicit a change at another interface. In this regard, we initially focused on the impact of EAR3 in the recruitment of TFIIB to the Sp1(I) site in order to decipher the critical requirement of Sp1(I) site in EAR3-repressed hLHR gene transcription. JAR cell nuclear extracts that had been immunodepleted of endogenous EAR3 and TFIIB were incubated with the Sp1(I) probe, to which a constant amount of recombinant TFIIB was added in absence or presence of increasing dose of GST/EAR3 fusion protein or GST only (Fig. 9A). Association of TFIIB to this site in the absence of EAR3 was clearly demonstrated, with an intensity similar to what we had observed previously with JAR nuclear proteins containing both native TFIIB and EAR3 (Fig. 8). The results indicated that EAR3 was not necessarily the bridging molecule tethering TFIIB to the Sp1/Sp3 complex. However, GST/EAR3 fusion protein exhibited marked inhibition of the TFIIB association in a dose-dependent manner in which, upon addition of 100 ng of GST/EAR3, the binding was hardly detectable (TFIIB). In contrast, GST alone did not have affect for the TFIIB association since the binding signals remained unchanged regardless of the dose of GST protein. Furthermore, the observed decrease of TFIIB association at the Sp1(I) site might result from either EAR3 causing a decreased association of this factor with Sp1/Sp3-DNA complex via a change at the protein-protein level or exerting a direct negative impact on the Sp1/Sp3 binding activities and therefore causing a overall reduction in recruitment of their associating proteins. The similar binding activities of Sp1/Sp3 to the Sp1(I) site in the absence or presence of GST/EAR3 excluded EAR3-induced reduction of Sp1/Sp3 binding activities (Sp1 and Sp3). Also, the unchanged association of HDAC2 with the Sp1(I) site in the absence or presence of EAR3/GST confirmed the specific negative effect of EAR3 on TFIIB interaction. The binding of GST/EAR3 or GST proteins for the Sp1(I) site was also analyzed and shown (GST/EAR3).
The impact of EAR3 on the association of TFIIB to the core promoter region was further investigated (Fig. 9B). In contrast to the EAR3-mediated reduction of association of TFIIB to the Sp1(I) site, we show that the binding of TFIIB to the three core promoter regions, PF-1, PF-2, and PF-3, which lack Sp1 and DR regulatory sequences (see Fig. 7), was not affected by GST/EAR3 addition. These results indicate that EAR3 perturbs the interaction of TFIIB with Sp1/Sp3 complexes at the Sp1(I) site without changing its association with the basal transcriptional machinery at the core promoter sequence. Furthermore, these observations are in agreement with data obtained from the ChIP assays, in which the recruitment of TFIIB to the hLHR promoter region remained unaffected, whereas its recruitment to the Sp1 site region was significantly decreased in JAR cells when binding of EAR3 was present (Fig. 7, region 6).
Our observation of the interplay among EAR3, TFIIB, and Sp1/Sp3 complex at the protein level prompted us to investigate their functional contributions to the regulation of hLHR gene promoter activity. Cotransfection studies of wild-type (Fig. 9, WT) or Sp1 site mutant hLHR promoter constructs [Sp1(I)X, Sp1(II)X, or Sp1(I,II)X] with TFIIB expression plasmid in JAR cells demonstrated that overexpression of TFIIB significantly activated the wild-type promoter activity (by twofold, Fig. 9C). Furthermore, the TFIIB-induced promoter activity was abolished upon mutation of the Sp1(I) but was not affected by mutation at the Sp1(II) site. This indicates that the Sp1(I) site is critical for TFIIB induction of the hLHR gene promoter activity. Moreover, these results are consistent with our previous observation of the obligatory role of the Sp1(I) site in EAR3-mediated repression of hLHR gene transcription (Fig. 2). Overexpression of TFIIB in JAR cells caused increased promoter activity, presumably by release of the basal inhibition by endogenous EAR3 in these cells (Fig. 9C). Cotransfection of EAR3 and the wild-type hLHR gene promoter in JAR cells caused significant inhibition of promoter activity (Fig. 9D). However, such inhibition was overcome by coexpression of TFIIB with EAR3, where TFIIB increased the promoter activity by twofold over the repressed activity induced by EAR3. These results indicate that TFIIB counteracted the repression of hLHR gene transcription induced by endogenous and overexpressed EAR3. Taken together, our findings have demonstrated that EAR3 specifically targets TFIIB so that it decreases the association of TFIIB with Sp1/Sp3 complex at the Sp1(I) site without affecting the binding of TFIIB to the hLHR gene core promoter region. This provides a molecular basis for the functional connection of these transcriptional factors in the regulation of hLHR gene promoter activity, in which mutation of the Sp1(I) site abolished both the EAR3-elicited repression and TFIIB-induced activation of the hLHR gene transcription.
DISCUSSION
Our current studies have investigated the molecular mechanism of the nuclear orphan receptors-mediated repression of LHR gene transcription. We have demonstrated that, in both humans and rats, the proximal Sp1 site of the LHR gene promoter was critical for EAR2-, EAR3/COUP-TFI-mediated inhibition of promoter activity. Transactivator proteins Sp1 and Sp3, which bind to this site and are essential for the basal promoter activity, were both required for this repression. Moreover, the interaction between EAR3 and Sp1/Sp3 in the presence of their binding elements provides evidence to support the Sp1 site-dependent repression of LHR gene by EAR3. It was further evident that deletion of the EAR3 N-terminal region and DBD decreased and abolished, respectively, the EAR3-Sp1 interaction, and repression of the hLHR gene by this orphan receptor was minimal or absent. The C-terminal region of EAR3 that did not affect its interaction with Sp1 was found to be required for expression of the EAR3 inhibitory function. Furthermore, multiple domains were utilized to recruit TFIIB to the hLHR gene, including the TATA-less core promoter region, the DR domain, and the Sp1(I) site. More significantly, EAR3 was shown to cause marked reduction of the association of this basic transcription factor to the Sp1(I) site without changing its interaction with the hLHR gene core promoter. Functionally, TFIIB was shown to reverse the inhibitory effect of EAR3 and to activate hLHR gene transcription in an Sp1(I)-dependent manner. Taken together, our results have demonstrated that TFIIB is an important target in the repression of the LHR gene by the nuclear orphan receptor, in which EAR3 inhibits interaction between the Sp1(I) site and the PIC.
Transcriptional modulation of target gene expression by nuclear orphan receptors, particularly by EAR3/COUP-TFI, has been widely studied and significant knowledge has been generated (see references 31 and 48 for reviews). Initially identified as an activator for chicken ovalbumin gene transcription (30, 55), EAR3/COUP-TFI was later found to suppress expression of an array of genes that are involved in diverse aspects of biological functions (31, 48). Several models have been proposed for COUP-TFs-mediated gene silencing, which includes passive mechanisms via direct competition for the binding sites with most nonsteroid hormone receptors (also known as type II nuclear hormone receptors, RARs, RXRs, TRs, VDR, and PPAR), or through heterodimerization with RXR to quench this common partner for the type II hormone receptors (22, 48). Moreover, active repression by COUP-TFs was exhibited by its interaction with corepressors NcoR (nuclear receptor corepressor) and SMRT (silencing mediator for retinoid and thyroid hormone receptor), through which HDACs are recruited to induce an inhibitory chromatin environment (1, 29, 43).
However, our previous results have indicated that these defined models do not comply with the strong repression of LHR gene by EAR2 and EAR3/COUP-TFI. The first line of evidence is derived from the observation that the imperfect DR-0 motif within the LHR gene promoter was not a functional recognition site for the type II nuclear receptors mentioned above (60). This finding ruled out the passive repression mechanisms employed in COUP-TFs-repressed hormone responsive genes. We have recently shown that hLHR gene transcription is subject to regulation by histone acetylation and deacetylation, in which the Sp1(I) site was identified to tether the HDAC-mSin3A complex to the hLHR gene (61). Furthermore, it was found that EAR3 bound to the DR-0 motif did not participate in the histone hypoacetylation-induced silencing event (61). This indicates that the mechanism of silencing hLHR gene transcription through alteration of HDAC activities is independent of the pathway involving unliganded hormone receptors or orphan receptors, corepressors NcoR/SMRT, and HDAC complexes (1, 29, 43).
The present study revealed a novel mechanism for COUP-TF-mediated target gene silencing, in which cross talk of EAR3/COUP-TFI with both the Sp1/Sp3-bound Sp1(I) site and TFIIB was demonstrated. Protein-protein interactions between COUP-TFs and DNA-binding transcription factors other than members of the nuclear hormone receptor family have been observed in the regulation of several target genes. These include interaction of COUP-TF with Oct1 and Oct2 proteins to positively regulate the vHNF1 gene (hepatocyte nuclear factor 1 [33]) and with Sp1 protein at Sp1 site to potentiate Sp1-activated transcription of human immunodeficiency virus long terminal repeat and NGFI-A genes (32, 38). However, it is relevant that in these cases only COUP-TF-mediated gene transactivation was observed, and direct binding of COUP-TF to its response cis element was either absent or dispensable for the transactivation effect of COUP-TF. To our knowledge, the present study provides the first evidence demonstrating that repression of a target gene by EAR2, EAR3/COUP-TFI is Sp1 site dependent. In agreement with previous findings, the DBD was identified in the present study as a major interaction surface for EAR3 association with Sp1 (38). Moreover, the most N-terminal region of EAR3 also played an active role in such interaction since deletion of this region resulted in a decreased association of EAR3/Sp1 and a comparable reduced inhibitory effect of EAR3 on hLHR gene transcription. The N-terminal domain, in addition to its intrinsic weak Sp1 binding activity, could contribute to maintaining the appropriate conformation of the EAR3 DBD domain for its optimal interaction with Sp1. Furthermore, the finding that the C-terminal domain was required for the EAR3 repression indicated that various regions, which were not limited to the Sp1-interacting DBD, participated in the inhibition of the hLHR gene transcription by EAR3. These studies provide evidence supporting the view that the C-terminal domain of EAR3 possesses important functions to facilitate an EAR3 interplay with other regulatory proteins. Therefore, regulation of hLHR gene transcription by EAR3 could be achieved by a mechanism in which EAR3, through its DBD anchored to Sp1 at the Sp1(I) site, exerts its negative function by its C-terminal region to affect cofactor protein(s) or basal transcriptional machinery component(s), e.g., TFIIB. In addition, our previous reports have shown that repression of the LHR gene depends on the binding of EAR2 and EAR3 to the DR-0 domain, since mutations that compromised their binding activities abolished repression of the LHR gene (59, 60).
Our evidence that Sp1/Sp3 participates in the inhibitory effect of EAR2 and EAR3/COUP-TFI reveals novel functions for these transcription factors, in addition to their role in the basic activation of the LHR gene transcription. Although Sp1/Sp3 were originally regarded as constitutive regulatory proteins essential for the expression of many housekeeping genes under basal conditions, they have been found to have an active role in the conveyance of specific signals to target genes, some of which mediate a broad spectrum of physiological responses (see reference 3 for a review). Studies on the interactions of Sp1 with transcription factors p53, estrogen receptor, and Smad proteins are among a number of reports showing that the Sp1 site is important in conveying distinct modulations to a target gene (10, 17, 53). Such Sp1-conferred specificity thus provides insight into our understanding of gene regulation. This particularly brings attention to a target gene such as LHR, for which the promoter activity is controlled by ubiquitous Sp1/Sp3 factors, but meanwhile its expression displays strict temporal and spatial specificity during gonadal maturation and steroid biosynthesis (see references 35 and 36 for reviews). The critical requirement of the proximal Sp1 site in silencing of the LHR gene transcription elicited by either orphan receptors or chromatin condensation indicates that Sp1/Sp3 are key effectors in integrating different silencing mechanisms, yielding repression of the LHR gene transcription.
The exclusive involvement of the Sp1(I) site in this repression of the LHR gene indicates that individual Sp1 sites could be differentially utilized to exert different functions in the control of the expression of a target gene. This phenomenon was also noted in the regulation of several genes with multiple Sp1 sites. A well-known example is the cyclin-dependent kinase (Cdk) inhibitor p21WAF1/Cip1 gene, for which responses to transforming growth factor β, phorbol esters, and various HDAC inhibitors were linked differentially to six Sp1 sites within its promoter (see reference 14 for a review). Such site-specific effect may be due to a prerequisite formation of a specific DNA configuration that, in turn, serves as a platform for harboring interactions among proteins or protein complexes, since DNA bending and the relative position of an enhancer domain from the transcription start site are thought to be important for proper expression of a target gene (4, 24, 39).
The precise mechanism whereby the regulatory signal exerted from DNA-specific trans-acting factors is transduced to RNA Pol II is currently the focus of intensive studies. For classical nuclear hormone receptors, this process is triggered by ligand binding to the receptors (12, 46). Interactions of TFIIB with various transcription factors have been observed, including nuclear hormone receptors (6, 11, 13, 21, 26, 47), the acidic activator VP16 (25), HNF4 (27), proteins of the Rel/NF-κB family (40, 56), and several others (54, 57). These results suggest that TFIIB serves as a bridge between the basal machinery and specific activators. It is also plausible that TFIIB may play a key function in gene transactivation, since recruitment of TFIIB to PIC was shown to be a rate-limiting step in the initiation of the gene transcription (13, 18, 58). However, on the other hand, the mechanism by which transcriptional repression by orphan receptors is operated in this regard was largely unexplored.
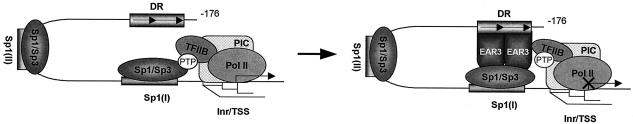
Recruitment of TFIIB to the hLHR gene promoter reveals that the TATA-less core promoter region, the EAR3-bound DR-0 motif, and the Sp1/Sp3-bound Sp1(I) site all participate in anchoring this basic transcription factor to the gene promoter. Furthermore, robust binding of TFIIB to the Sp1(I) site has indicated that the proximal Sp1/Sp3 binding domain could serve as a platform to accommodate an interplay among EAR3, TFIIB, and Sp1/Sp3. The lack of direct interaction between TFIIB and Sp1 implies that the association of TFIIB with the Sp1(I) site could involve an adaptor protein(s) (see model in Fig. 10). In this regard, EAR3 is not required as a bridging molecule since the binding of TFIIB was clearly present in absence of EAR3 by using EAR3-depleted nuclear extracts. More significantly, the demonstration that EAR3 dose dependently inhibited the association of TFIIB at the Sp1(I) site without affecting its binding to the hLHR core promoter region indicates that EAR3 specifically targets the communication between the TFIIB and the Sp1/Sp3 complex rather than causing a change in recruitment of the TFIIB to the basal transcriptional machinery. The TFIIB activation of hLHR gene transcription in a Sp(I) site-dependent manner, which correlated with the release of the EAR3 inhibition, has provided insights into the mechanism(s) of the antagonist action of EAR3 and TFIIB in the control of hLHR gene transcription. The negative impact of EAR3 through disruption of the stimulatory signal of Sp1 resulted in a nonproductive or less-productive form of the PIC. In this regard, decreased recruitment of RNA Pol II to the hLHR gene promoter was observed in JAR cells when EAR3-mediated repression through the DR-0 motif was present. Taken together, these findings have provided a molecular basis for the Sp1(I) site-dependent repression of the LHR gene by EAR3, in which EAR3 inhibits the LHR gene transcription through compromising interaction between TFIIB and the Sp1/Sp3 at their cognate binding site.
FIG. 10.
Model for the mechanism of Sp1(I) site-dependent silencing of the hLHR gene transcription by EAR3. EAR3 bound to the DR motif interacts with Sp1/Sp3 bound to the Sp1(I) site. Such interaction significantly prevents the robust association of TFIIB to the Sp1(I) site without affecting the recruitment of TFIIB to the hLHR gene core promoter region. Anchoring of the TFIIB at the Sp1(I) site does not require prior binding of EAR3 to its cognate site, since the association is clearly present in the absence of EAR3. We propose that interaction of TFIIB with Sp1/Sp3 is indirectly bridged by a currently unidentified protein(s), as indicated by an open circle (putative tethering protein [PTP]). The EAR3-reduced association of TFIIB to the Sp1/Sp3-DNA complex may induce a nonproductive or less-productive form of PIC, in which the recruitment of RNA Pol II to the hLHR promoter was decreased when the hLHR gene was subjected to a repressed state by EAR3 in JAR cells. Inr, initiator element; TSS, transcriptional start site.
Acknowledgments
We thank Robert Tjian (Department of Molecular and Cell Biology, University of California, Berkeley) and Danny Reinberg (Department of Biochemistry, University of Medicine and Dentistry of New Jersey, Piscataway) for kindly providing the pAc-Sp1 expression plasmid and the pCMV-TFIIB construct, respectively.
REFERENCES
- 1.Bailey, P. J., D. H. Dowhan, K. Franke, L. J. Burke, M. Downes, and G. E. Muscat. 1997. Transcriptional repression by COUP-TF II is dependent on the C-terminal domain and involves the N-CoR variant, RIP13delta1. J. Steroid Biochem. Mol. Biol. 63:165-174. [DOI] [PubMed] [Google Scholar]
- 2.Baniahmad, A., I. Ha, D. Reinberg, S. Tsai, M. J. Tsai, and B. W. O'Malley. 1993. Interaction of human thyroid hormone receptor beta with transcription factor TFIIB may mediate target gene derepression and activation by thyroid hormone. Proc. Natl. Acad. Sci. USA 90:8832-8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black, A. R., J. D. Black, and J. Azizkhan-Clifford. 2001. Sp1 and Kruppel-like factor family of transcription factors in cell growth regulation and cancer. J. Cell Physiol. 188:143-160. [DOI] [PubMed] [Google Scholar]
- 4.Campbell, A. 2002. DNA binding and bending to initiate packaging of phage lambda DNA. Mol. Cell 9:928-929. [DOI] [PubMed] [Google Scholar]
- 5.Catt, K. J., and M. L. Dufau. 1991. Gonadotropic hormones: biosynthesis, secretion, receptors and action, p. 105-155. In S. S. C. Yen and R. B. Jaffe (ed.), Reproductive endocrinology. The W. B. Saunders Co., Philadelphia, Pa.
- 6.Chen, H. W., and M. L. Privalsky. 1997. Retinoid X and retinoic acid receptors interact with transcription factor II-B by distinct mechanisms. Mol. Cell Endocrinol. 129:55-61. [DOI] [PubMed] [Google Scholar]
- 7.Choy, B., and M. R. Green. 1993. Eukaryotic activators function during multiple steps of preinitiation complex assembly. Nature 366:531-536. [DOI] [PubMed] [Google Scholar]
- 8.Dufau, M. L. 1998. The luteinizing hormone receptor. Annu. Rev. Physiol. 60:461-496. [DOI] [PubMed] [Google Scholar]
- 9.Dufau, M. L., C. H. Tsai-Morris, Z. Z. Hu, and E. Buczko. 1995. Structure and regulation of the luteinizing hormone receptor gene. J. Steroid Biochem. Mol. Biol. 53:283-291. [DOI] [PubMed] [Google Scholar]
- 10.Feng, X. H., X. Lin, and R. Derynck. 2000. Smad2, Smad3, and Smad4 cooperate with Sp1 to induce p15Ink4B transcription in response to TGF-β. EMBO J. 19:5178-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fondell, J. D., F. Brunel, K. Hisatake, and R. G. Roeder. 1996. Unliganded thyroid hormone receptor alpha can target TATA-binding protein for transcriptional repression. Mol. Cell. Biol. 16:281-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fondell, J. D., H. Ge, and R. G. Roeder. 1996. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl. Acad. Sci. USA 93:8329-8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fondell, J. D., A. L. Roy, and R. G. Roeder. 1993. Unliganded thyroid hormone receptor inhibits formation of a functional preinitiation complex: implications for active repression. Genes Dev. 7:1400-1410. [DOI] [PubMed] [Google Scholar]
- 14.Gartel, A. L., and A. L. Tyner. 1999. Transcriptional regulation of the p21WAF1/CIP1 gene. Exp. Cell Res. 246:280-289. [DOI] [PubMed] [Google Scholar]
- 15.Geng, Y., C. H. Tsai-Morris, Y. Zhang, and M. L. Dufau. 1999. The human luteinizing hormone receptor gene promoter: activation by Sp1 and Sp3 and inhibitory regulation. Biochem. Biophys. Res. Commun. 263:366-371. [DOI] [PubMed] [Google Scholar]
- 16.Gill, G., and R. Tjian. 1992. Eukaryotic coactivators associated with the TATA box binding protein. Curr. Opin. Genet. Dev. 2:236-242. [DOI] [PubMed] [Google Scholar]
- 17.Gualberto, A., and A. S. Baldwin, Jr. 1995. p53 and Sp1 interact and cooperate in the tumor necrosis factor-induced transcriptional activation of the HIV-1 long terminal repeat. J. Biol. Chem. 270:19680-19683. [DOI] [PubMed] [Google Scholar]
- 18.Hisatake, K., R. G. Roeder, and M. Horikoshi. 1993. Functional dissection of TFIIB domains required for TFIIB-TFIID-promoter complex formation and basal transcription activity. Nature 363:744-747. [DOI] [PubMed] [Google Scholar]
- 19.Hu, Z., L. Zhuang, X. Guan, J. Meng, and M. L. Dufau. 1997. Steroidogenic factor-1 is an essential transcriptional activator for gonad-specific expression of promoter I of the rat prolactin receptor gene. J. Biol. Chem. 272:14263-14271. [DOI] [PubMed] [Google Scholar]
- 20.Hu, Z. Z., L. Zhuang, J. Meng, and M. L. Dufau. 1998. Transcriptional regulation of the generic promoter III of the rat prolactin receptor gene by C/EBPβ and Sp1. J. Biol. Chem. 273:26225-26235. [DOI] [PubMed] [Google Scholar]
- 21.Ing, N. H., J. M. Beekman, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1992. Members of the steroid hormone receptor superfamily interact with TFIIB (S300-II). J. Biol. Chem. 267:17617-17623. [PubMed] [Google Scholar]
- 22.Leng, X., A. J. Cooney, S. Y. Tsai, and M. J. Tsai. 1996. Molecular mechanisms of COUP-TF-mediated transcriptional repression: evidence for transrepression and active repression. Mol. Cell. Biol. 16:2332-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leong, G. M., K. S. Wang, M. J. Marton, J. C. Blanco, I. M. Wang, R. J. Rolfes, K. Ozato, and J. H. Segars. 1998. Interaction between the retinoid X receptor and transcription factor IIB is ligand-dependent in vivo. J. Biol. Chem. 273:2296-2305. [DOI] [PubMed] [Google Scholar]
- 24.Lim, F. L., A. Hayes, A. G. West, A. Pic-Taylor, Z. Darieva, B. A. Morgan, S. G. Oliver, and A. D. Sharrocks. 2003. Mcm1p-induced DNA bending regulates the formation of ternary transcription factor complexes. Mol. Cell. Biol. 23:450-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin, Y. S., I. Ha, E. Maldonado, D. Reinberg, and M. R. Green. 1991. Binding of general transcription factor TFIIB to an acidic activating region. Nature 353:569-571. [DOI] [PubMed] [Google Scholar]
- 26.MacDonald, P. N., D. R. Sherman, D. R. Dowd, S. C. Jefcoat, Jr., and R. K. DeLisle. 1995. The vitamin D receptor interacts with general transcription factor IIB. J. Biol. Chem. 270:4748-4752. [DOI] [PubMed] [Google Scholar]
- 27.Malik, S., and S. K. Karathanasis. 1996. TFIIB-directed transcriptional activation by the orphan nuclear receptor hepatocyte nuclear factor 4. Mol. Cell. Biol. 16:1824-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKenna, N. J., R. B. Lanz, and B. W. O'Malley. 1999. Nuclear receptor coregulators: cellular and molecular biology. Endocrinol. Rev. 20:321-344. [DOI] [PubMed] [Google Scholar]
- 29.Nagy, L., H. Y. Kao, D. Chakravarti, R. J. Lin, C. A. Hassig, D. E. Ayer, S. L. Schreiber, and R. M. Evans. 1997. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89:373-380. [DOI] [PubMed] [Google Scholar]
- 30.Pastorcic, M., H. Wang, A. Elbrecht, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1986. Control of transcription initiation in vitro requires binding of a transcription factor to the distal promoter of the ovalbumin gene. Mol. Cell. Biol. 6:2784-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pereira, F. A., M. J. Tsai, and S. Y. Tsai. 2000. COUP-TF orphan nuclear receptors in development and differentiation. Cell Mol. Life Sci. 57:1388-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pipaon, C., S. Y. Tsai, and M. J. Tsai. 1999. COUP-TF upregulates NGFI-A gene expression through an Sp1 binding site. Mol. Cell. Biol. 19:2734-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Power, S. C., and S. Cereghini. 1996. Positive regulation of the vHNF1 promoter by the orphan receptors COUP-TF1/Ear3 and COUP-TFII/Arp1. Mol. Cell. Biol. 16:778-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Privalsky, M. L. 2001. Regulation of SMRT and N-CoR corepressor function. Curr. Top. Microbiol. Immunol. 254:117-136. [DOI] [PubMed] [Google Scholar]
- 35.Richards, J. S. 1980. Maturation of ovarian follicles: actions and interactions of pituitary and ovarian hormones on follicular cell differentiation. Physiol. Rev. 60:51-89. [DOI] [PubMed] [Google Scholar]
- 36.Richards, J. S., and K. Kersey. 1989. Changes in theca and granulosa cell function in antral follicles developing during pregnancy in the rat: gonadotropin receptors, cyclic AMP, and estradiol-17b. Biol. Reprod. 21:1185-1201. [DOI] [PubMed] [Google Scholar]
- 37.Roberts, S. G., I. Ha, E. Maldonado, D. Reinberg, and M. R. Green. 1993. Interaction between an acidic activator and transcription factor TFIIB is required for transcriptional activation. Nature 363:741-744. [DOI] [PubMed] [Google Scholar]
- 38.Rohr, O., D. Aunis, and E. Schaeffer. 1997. COUP-TF and Sp1 interact and cooperate in the transcriptional activation of the human immunodeficiency virus type 1 long terminal repeat in human microglial cells. J. Biol. Chem. 272:31149-31155. [DOI] [PubMed] [Google Scholar]
- 39.Scaffidi, P., and M. E. Bianchi. 2001. Spatially precise DNA bending is an essential activity of the sox2 transcription factor. J. Biol. Chem. 276:47296-47302. [DOI] [PubMed] [Google Scholar]
- 40.Schmitz, M. L., G. Stelzer, H. Altmann, M. Meisterernst, and P. A. Baeuerle. 1995. Interaction of the COOH-terminal transactivation domain of p65 NF-κB with TATA-binding protein, transcription factor IIB, and coactivators. J. Biol. Chem. 270:7219-7226. [DOI] [PubMed] [Google Scholar]
- 41.Schwartz, C., P. Catez, O. Rohr, D. Lecestre, D. Aunis, and E. Schaeffer. 2000. Functional interactions between C/EBP, Sp1, and COUP-TF regulate human immunodeficiency virus type 1 gene transcription in human brain cells. J. Virol. 74:65-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Segaloff, D. L., H. Y. Wang, and J. S. Richards. 1990. Hormonal regulation of luteinizing hormone/chorionic gonadotropin receptor mRNA in rat ovarian cells during follicular development and luteinization. Mol. Endocrinol. 4:1856-1865. [DOI] [PubMed] [Google Scholar]
- 43.Shibata, H., Z. Nawaz, S. Y. Tsai, B. W. O'Malley, and M. J. Tsai. 1997. Gene silencing by chicken ovalbumin upstream promoter-transcription factor I (COUP-TFI) is mediated by transcriptional corepressors, nuclear receptor-corepressor (N-CoR) and silencing mediator for retinoic acid receptor and thyroid hormone receptor (SMRT). Mol. Endocrinol. 11:714-724. [DOI] [PubMed] [Google Scholar]
- 44.Smirnov, D. A., S. Hou, and R. P. Ricciardi. 2000. Association of histone deacetylase with COUP-TF in tumorigenic Ad12-transformed cells and its potential role in shut-off of MHC class I transcription. Virology 268:319-328. [DOI] [PubMed] [Google Scholar]
- 45.Stroup, D., and J. Y. Chiang. 2000. HNF4 and COUP-TFII interact to modulate transcription of the cholesterol 7α-hydroxylase gene (CYP7A1). J. Lipid Res. 41:1-11. [PubMed] [Google Scholar]
- 46.Tong, G. X., M. R. Tanen, and M. K. Bagchi. 1995. Ligand modulates the interaction of thyroid hormone receptor beta with the basal transcription machinery. J. Biol. Chem. 270:10601-10611. [DOI] [PubMed] [Google Scholar]
- 47.Tsai, S. Y., I. Sagami, H. Wang, M. J. Tsai, and B. W. O'Malley. 1987. Interactions between a DNA-binding transcription factor (COUP) and a non-DNA binding factor (S300-II). Cell 50:701-709. [DOI] [PubMed] [Google Scholar]
- 48.Tsai, S. Y., and M. J. Tsai. 1997. Chick ovalbumin upstream promoter-transcription factors (COUP-TFs): coming of age. Endocrinol. Rev. 18:229-240. [DOI] [PubMed] [Google Scholar]
- 49.Tsai-Morris, C. H., Y. Geng, E. Buczko, and M. L. Dufau. 1995. Characterization of diverse functional elements in the upstream Sp1 domain of the rat luteinizing hormone receptor gene promoter. J. Biol. Chem. 270:7487-7494. [DOI] [PubMed] [Google Scholar]
- 50.Tsai-Morris, C. H., Y. Geng, E. Buczko, and M. L. Dufau. 1998. A novel human luteinizing hormone receptor gene. J. Clin. Endocrinol. Metab. 83:288-291. [DOI] [PubMed] [Google Scholar]
- 51.Tsai-Morris, C. H., Y. Geng, X. Z. Xie, E. Buczko, and M. L. Dufau. 1994. Transcriptional protein binding domains governing basal expression of the rat luteinizing hormone receptor gene. J. Biol. Chem. 269:15868-15875. [PubMed] [Google Scholar]
- 52.Tsai-Morris, C. H., X. Xie, W. Wang, E. Buczko, and M. L. Dufau. 1993. Promoter and regulatory regions of the rat luteinizing hormone receptor gene. J. Biol. Chem. 268:4447-4452. [PubMed] [Google Scholar]
- 53.Wang, F., I. Samudio, and S. Safe. 2001. Transcriptional activation of rat creatine kinase B by 17β-estradiol in MCF-7 cells involves an estrogen responsive element and GC-rich sites. J. Cell Biochem. 84:156-172. [DOI] [PubMed] [Google Scholar]
- 54.Wang, I. M., J. C. Blanco, S. Y. Tsai, M. J. Tsai, and K. Ozato. 1996. Interferon regulatory factors and TFIIB cooperatively regulate interferon-responsive promoter activity in vivo and in vitro. Mol. Cell. Biol. 16:6313-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, L. H., S. Y. Tsai, R. G. Cook, W. G. Beattie, M. J. Tsai, and B. W. O'Malley. 1989. COUP transcription factor is a member of the steroid receptor superfamily. Nature 340:163-166. [DOI] [PubMed] [Google Scholar]
- 56.Xu, X., C. Prorock, H. Ishikawa, E. Maldonado, Y. Ito, and C. Gelinas. 1993. Functional interaction of the v-Rel and c-Rel oncoproteins with the TATA-binding protein and association with transcription factor IIB. Mol. Cell. Biol. 13:6733-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu, L., P. M. Loewenstein, Z. Zhang, and M. Green. 1995. In vitro interaction of the human immunodeficiency virus type 1 Tat transactivator and the general transcription factor TFIIB with the cellular protein TAP. J. Virol. 69:3017-3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zawel, L., and D. Reinberg. 1993. Initiation of transcription by RNA polymerase II: a multi-step process. Prog. Nucleic Acids Res. Mol. Biol. 44:67-108. [DOI] [PubMed] [Google Scholar]
- 59.Zhang, Y., and M. L. Dufau. 2001. EAR2 and EAR3/COUP-TFI regulate transcription of the rat LH receptor. Mol. Endocrinol. 15:1891-1905. [DOI] [PubMed] [Google Scholar]
- 60.Zhang, Y., and M. L. Dufau. 2000. Nuclear orphan receptors regulate transcription of the gene for the human luteinizing hormone receptor. J. Biol. Chem. 275:2763-2770. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, Y., and M. L. Dufau. 2002. Silencing of transcription of the human luteinizing hormone receptor gene by histone deacetylase-mSin3A complex. J. Biol. Chem. 277:33431-33438. [DOI] [PubMed] [Google Scholar]