Abstract
The human ISWI-containing factor RSF (for remodeling and spacing factor) is composed of two subunits: the ATPase hSNF2H and p325 (Rsf-1), a protein encoded by a novel human gene. We previously showed that RSF mediates nucleosome deposition and generates regularly spaced nucleosome arrays. Here we report the characterization of the largest subunit of RSF, Rsf-1. We found that Rsf-1 is a highly acidic protein containing a plant homology domain. The present study includes the cloning of Rsf-1, the preparation of recombinant RSF, and the dissection of the role of each subunit in the chromatin assembly reaction. The sequence of the gene for Rsf-1 includes a recently characterized cDNA, HBXAP; postulated to be involved in the transcriptional regulation of the hepatitis B virus. HBXAP actually contains a 252-amino-acid truncation of the amino terminus of Rsf-1. Finally, comparison of HBXAP and Rsf-1 properties shows that they are functionally different.
The DNA in eukaryotes is packaged into chromatin, of which the individual chromatin subunits—the nucleosomes—are composed of 147 bp of DNA wrapped almost twice around the core histone octamers. The octamers are composed of two copies each of the four core histone proteins H2A, H2B, H3, and H4 (for a review, see reference 27). Besides its structure-related function, that is, the compaction of DNA, chromatin also regulates various aspects of DNA metabolism, including transcription. This is feasible because chromatin exists in a dynamic state.
The dynamic state of chromatin is the result of factors that perturb the state of chromatin in conjunction with factors that assemble or reposition components of chromatin. Two main families of proteins perturb chromatin: (i) enzymes that covalently modify the N-terminal tail of the histones (17, 29) and (ii) ATP-dependent chromatin remodeling complexes, which utilize the energy of ATP hydrolysis to mobilize nucleosomes or to otherwise alter chromatin structure (24, 26). These protein complexes render DNA more accessible to the binding of transcriptional regulatory factors. In addition, several factors that assemble or reposition nucleosomes have been identified and classified according to whether or not their function is coupled to DNA replication. Chromatin assembly occurs primarily during the S phase of the cell cycle, when DNA replication takes place and histone polypeptides are newly synthesized. However, outside of the S phase, chromatin assembly also occurs, although to a much lesser extent. DNA repair, histone turnover, and transcription are examples of processes during which chromatin can be temporarily altered (14, 23).
The model of nucleosome assembly proposes that histone chaperones deposit H3/H4 tetramers onto DNA in a reaction that seems to be the rate-limiting step, followed by the deposition of H2A/H2B dimers. Once histone chaperones deposit histones onto the DNA, chromatin spacing complexes mobilize the nucleosomes to produce a regularly spaced nucleosomal array. Several different chromatin spacing factors have been identified in various species (23). All of these factors share an ATPase-containing subunit, which belongs to the ISWI family. ISWI was initially identified in Drosophila melanogaster, in three different “chromatin-remodeling” complexes: ACF (10), CHRAC (25), and NURF (22). A new complex containing components of NURF, as well as TRF-2 (for TBP-related factor 2), has been identified recently (9). It has been proposed that this complex may be involved in core promoter selectivity of a subset of Drosophila genes. In yeast, there are two ISWI homologues—ISWI1 and ISWI2—each of which is a part of distinct chromatin-remodeling complexes (21). In Xenopus laevis, five ISWI complexes have been identified, one of which is the homologue of ACF (7). In humans, there are two known ISWI homologues: hSNF2H and hSNF2L. hSNF2H is part of several different chromatin-remodeling complexes, including ACF and BAZ-like complexes (2, 3, 11), CHRAC (16), NoRC (20), RSF (12), and an hSNF2H complex containing components of the cohesin and NuRD complexes (8). In contrast, hSNF2L-containing chromatin-remodeling complexes have not yet been identified. Importantly, although all of these factors share the ISWI subunit, not all of them have the ability to assemble nucleosomes.
We have previously described RSF, which is composed of two subunits, hSNF2H and p325 (Rsf-1), a protein encoded by an as-yet-uncharacterized novel gene(s). RSF was isolated from HeLa cells as an activity that allows the formation of competent RNA polymerase II transcription initiation complexes on chromatin templates as a result of its ability to mobilize nucleosomes (12). Later on, we discovered that RSF could assemble chromatin in vitro in an energy-dependent manner (13). However, in contrast to the other chromatin assembly and spacing factors, RSF-mediated chromatin assembly was found to be independent of histone chaperones (13). We describe here the isolation of a cDNA encoding Rsf-1, the largest subunit of RSF. Using complex generated with recombinant RSF, we have fully reconstituted the chromatin assembly reaction and analyzed the function of each individual subunit in the chromatin assembly reaction in vitro.
MATERIALS AND METHODS
Antibodies.
Monoclonal antibodies against the RSF subunits Rsf-1 and hSNF2H were developed by BiosChile as described previously (11). In order to generate polyclonal antibodies against the N-terminal region of Rsf-1, the first 670 bp of Rsf-1 were subcloned into the pET vector, bacterially expressed and purified by standard nickel affinity purification. Gold-conjugated antibodies were purchased from Sigma. H3-phosphorylated S28 and β-actin antibodies were purchased from Upstate Biotechnology.
Peptide sequencing of Rsf-1 by ion trap mass spectrometry.
Rsf-1 was isolated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and then subjected to reduction, carboxyamidomethylation, and digestion with trypsin. Peptides were analyzed by microcapillary reversed-phase high-pressure liquid chromatography nanoelectrospray tandem mass spectrometry on an LCQ DECA XP quadrupole ion trap mass spectrometer (ThermoFinnigan). A normalized collision energy of 30% and with an isolation width of 2.5 Da was used, with recurring ions dynamically excluded. Preliminary mapping of peptide sequences was accomplished with the SEQUEST algorithm.
Cloning of Rsf-1 and production of the corresponding protein in baculovirus.
Peptide sequences from Rsf-1 obtained by ion trap mass spectrometry identified multiple expressed sequence tag (ESTs) from the NCBI EST database. Two clones (accession numbers 545521 and 626008) were used to screen a HeLa cell (Clontech) and a human heart (Novagen) cDNA library according to the suppliers' instructions. We obtained four overlapping cDNAs that were used to construct the full-length cDNA. The Flag peptide sequence was inserted at the 5′ end, in frame, by using the PCR. In order to produce baculoviruses, the full-length Rsf-1 gene was subcloned into pVL1393 vector (Pharmingen), and the recombinant viruses were generated by using BaculoGold DNA (Pharmingen).
Northern blot.
Northern blots were carried out by using multiple-tissue Northern blots according to the supplier's instructions (Clontech). The Rsf-1 probe extended from nucleotides 1750 to 4018. The β-actin probe used was provided by the supplier.
Alkaline phosphatase treatment.
Native RSF (0.5 μg) was treated with 1 U of alkaline phosphatase (Roche) for 30 min at 37°C. The reaction was stopped with SDS sample buffer, and the proteins were resolved on an SDS-polyacrylamide gel and then silver stained.
Immunofluorescence microscopy.
HeLa cells grown on coverslips were fixed with 1% paraformaldehyde for 30 min, washed with phosphate-buffered saline (PBS), and permeabilized with 0.1% Triton X-100. Cells were stained by sequential incubation with antibodies to the proteins of interest (first antibodies) and secondary fluorescence-labeled antibodies and then visualized with a TCS (Leica) confocal imaging system.
Interphasic and mitotic nuclear extracts.
HeLa cells at 70% confluency were treated (mitotic extract) or not treated (interphasic extract) with 100 ng of nocodazole/ml for 20 h. Mitotic HeLa cells became deattached from the plate; therefore, nontreated cells were washed several times with PBS in order to eliminate mitotic cells. The medium of the treated cells, on the other hand, was collected, and the plate was washed several times to recover all mitotic cells. After a wash with PBS, the cells were resuspended in hypotonic buffer (20 mM HEPES [pH 7.8], 5 mM potassium acetate, 0.5 mM MgCl2) and Dounce homogenized 25 times. The solution was centrifuged at 4,500 rpm for 3 min, and the pellet was resuspended in the same buffer containing 1 M NaCl, followed by incubation for 1.5 h at 4°C. The suspension was centrifuged at 14,000 rpm for 20 min, and the supernatant (nuclear extract) was directly used for immunoprecipitation assays.
Protein purification.
HeLa cell core histones were purified as described previously (15). Native RSF was purified from HeLa nuclear pellet as described previously (12). Recombinant Flag-tagged HBXAP and Rsf-1 were expressed from baculovirus in Sf9 cells. Briefly, Sf9 cells were infected at a multiplicity of infection of 10 for 48 h. The cells were then pelleted, washed twice with cold PBS buffer, resuspended in lysis buffer (20 mM Tris-HCl [pH 7.9], 0.6 M NaCl, 4 mM MgCl2, 0.4 mM EDTA, 2 mM dithiothreitol, 20 mM β-glycerophosphate, 20% glycerol, 0.4 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine-HCl, 4 μg of leupeptin/ml, 2 μg of aprotinin/ml), and subjected to freeze-thaw disruption. The insoluble material was removed by centrifugation at 11,000 rpm (SLA-600TC rotor; Sorvall) for 10 min at 4°C, and the supernatant was incubated with anti-FLAG M2-agarose beads for 4 h at 4°C. The beads were washed with lysis buffer, and the bound proteins were eluted with 0.2 mg of Flag peptide/ml dissolved in lysis buffer. Recombinant RSF was expressed from baculovirus in Sf9 cells coinfected with Flag-tagged Rsf-1 and hSNF2H baculoviruses at a multiplicity of infection of 10. Recombinant RSF was purified with M2-agarose beads as described above, and the M2 column eluate was further purified by gel filtration chromatography. Recombinant hSNF2H was expressed from baculovirus in Sf9 cells and purified by chromatography on DEAE-52 resin. The protein was eluted from the DEAE-52 column with a linear ammonium sulfate gradient between 50 and 500 mM.
Immunoprecipitation.
Immunoprecipitations were performed as described previously, with several washes with a salt concentration of 500 mM KCl plus 0.05% NP-40 (13).
DNA-binding assays.
A radiolabeled 206-bp DNA fragment was incubated for 15 min at 30°C with recombinant RSF, recombinant hSNF2H, or recombinant Rsf-1 at the indicated molar ratio, in buffer consisting of 10 mM HEPES (pH 7.5), 60 mM KCl, 0.5 μg of bovine serum albumin, 0.5 mM EGTA, 10% glycerol, 1 mM dithiothreitol, and 0.2 mM phenylmethylsulfonyl fluoride. The reaction was loaded onto nitrocellulose membranes which were air dried and washed three times with TE buffer (10 mM Tris [pH 7.9], 0.1 mM EDTA). The 32P that remained on the membrane after the washing step was quantified by using a scintillation counter.
Electron microscopy.
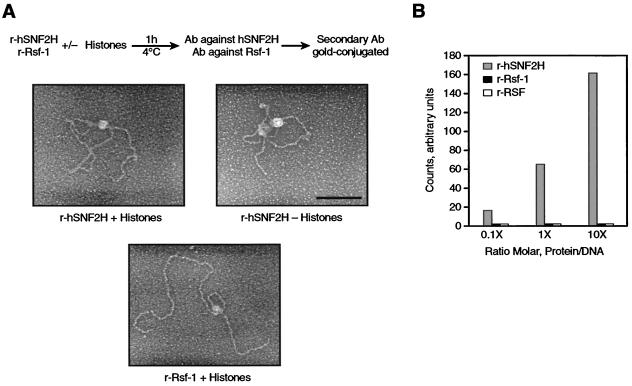
hSNF2H or Rsf-1 were incubated with 3-kb EcoRI-linearized plasmid in the presence or absence of HeLa-purified core histones in the chromatin assembly buffer. The incubation was performed for 30 min at 30°C. For the characterization of the DNA-protein complexes, the reaction was incubated with antibodies against the RSF subunit hSNF2h. After the first antibody incubation, the reaction was fixed with glutaraldehyde and purified by gel filtration (1 ml, A5 M resin; Bio-Rad). The reaction was incubated with secondary antibodies conjugated with 10-nm gold particles and then fixed with glutaraldehyde, purified, and prepared for electron miscroscopy. The protein-DNA complexes were mixed in a buffer containing 2 mM spermidine, adsorbed to glow-charged carbon-coated grids, washed with a water-graded-ethanol series, and rotary shadow cast with tungsten. Samples were examined with a Philips 420 electron microscope. Micrographs are shown in reverse contrast. A Cohu charge-coupled device camera attached to a Macintosh computer programmed with NIH IMAGE software was used to form the images. The percentage results presented were obtained by counting at least 100 DNA molecules per experiment.
ATPase.
The ATPase assay was performed as described previously (28).
Chromatin assembly and analysis.
The chromatin assembly reactions and micrococcal nuclease digestions were performed as previously described (13)
RESULTS
Isolation of the cDNA encoding the largest subunit of RSF.
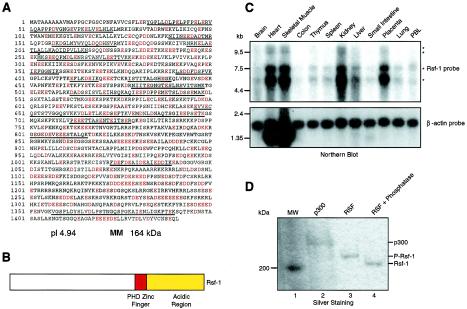
Using peptide sequences derived from the largest subunit of RSF-p325 determined by ion trap mass spectrometry, we identified two corresponding EST clones (GenBank accession numbers 545521 and 626008) in the GenBank human EST database. These EST clones were used to screen a HeLa (Clontech) and a human heart (Novagen) cDNA library. We obtained four overlapping cDNAs that together included all of the amino acids present in the isolated polypeptide (5,052 bp). The full-length cDNA encodes a highly acidic (pI 4.94), 1,441-amino-acid protein, with a predicted molecular mass of 164 kDa, although the native protein appears to be larger when subjected to electrophoresis on SDS-polyacrylamide gels (see below and Fig. 1A). The amino acid sequence analysis revealed the presence of a plant homology domain (PHD) zinc finger domain (between amino acids 893 and 942). The function of this domain is unknown, although it is found in many proteins that function through chromatin. Moreover, this domain has been proposed to be involved in mediating protein-protein interactions (1) and structural and genetic analyses of the PHD support this hypothesis (4). In addition to the acidic amino acids found throughout p325, there are several long stretches of glutamic and aspartic acids at the C-terminal region (Fig. 1A and B). In addition, p325 contains three putative nuclear localization signals (between amino acids 1084 and 1091, 1160 and 1170, and 1237 and 1244).
FIG. 1.
Rsf-1 sequence. (A) Amino acid sequence of Rsf-1. Rsf-1 is a 1,441-amino-acid protein. The peptide sequences obtained by ion trap mass spectrometry are underlined. The acidic amino acids aspartic acid (D) and glutamic acid (E) found in the Rsf-1 sequence are shown in red. The arrow indicates the first methionine of HBXAP, located at amino acid 253. (B) Schematic representation of Rsf-1. The diagram shows the position of the domains found based on the amino acid sequence of Rsf-1: the PHD and the C-terminal acidic region. MM, molecular mass. (C) Northern blots with various tissues and the Rsf-1 cDNA. Northern blots of different human tissues were analyzed with an Rsf-1 probe (upper panel). After exposure to the Rsf-1 probe, the membrane was stripped and reprobed with β-actin (lower panel). The migration of RNA markers is shown on the left side of the figure. The migrations of the different mRNA species detected with the Rsf-1 probe are denoted with asterisks. (D) Phosphatase treatment. Silver staining of an SDS-polyacrylamide gel containing native RSF treated with alkaline phosphatase. Molecular markers (lane 1), p300 (lane 2), RSF (lane 3), and alkaline phosphatase-treated RSF (lane 4) are shown.
Alignment of the nucleotide sequence against the public human genome database revealed that the gene encoding p325 is located on chromosome 11q13. Northern blot analysis of several different human tissues showed the presence of four related mRNAs, two ∼11-kb species, an ∼7-kb mRNA, and an ∼5.6-kb mRNA, which are probably alternatively spliced species since all of them hybridized with a probe to p325. p325 mRNA is expressed at different levels in the tissues analyzed; a high level of expression was observed in the heart, skeletal muscle, kidney, and placenta, whereas very low levels existed in other tissues such as the brain and colon (Fig. 1C).
During our studies on the RSF-p325 subunit a new cDNA, HBXAP (XAP-8), was deposited in the GenBank database (accession number NM016578 [gi 10835261]). HBXAP cDNA is similar to the one encoding RSF-p325 except that 252 amino acids found at the N terminus of p325 are missing (Fig. 1A). HBXAP was identified in a two-hybrid screen for proteins that interact with the transcriptional activator pX, which is encoded by the hepatitis B virus (HBV) (18). Functional assays demonstrated that HBXAP stimulated HBV transcription in a pX-dependent manner (18). Later, a larger version of HBXAP was deposited in the GenBank database (accession number AF380176 [gi 14211815]), and this species corresponded exactly to the RSF-p325 gene sequence.
Western blot analyses with monoclonal antibodies generated against the full-length RSF-p325 protein demonstrated that the cDNA clone we isolated encodes the full-length open reading frame of RSF-p325 (Fig. 3B). As mentioned above, the predicted molecular mass of p325 is 164 kDa, but it migrates above the 200-kDa marker and below the histone acetyltransferase p300 (which has a predicted molecular mass of 332 kDa) on SDS-polyacrylamide gels. This slower migration of p325 is due to its highly acidic charge and also to phosphorylation, since treatment of the protein with phosphatase resulted in the generation of a species that migrated faster than untreated p325 on SDS-polyacrylamide gels (Fig. 1D). This faster-migrating species was not a result of proteolysis since antibodies recognizing the N and C termini of p325 were reactive with this new species (data not shown). Moreover, the smallest subunit of RSF, hSNF2H, did not change its migration upon phosphatase treatment (data not shown). It is worth mentioning that the phosphorylation state of p325 does not affect the chromatin assembly reaction. RSF treated with alkaline phosphatase is able to assemble chromatin with a specific activity similar to that of the phosphorylated form (data not shown). To avoid confusion between the name of the protein and its molecular mass and to follow the nomenclature that has been used for other chromatin-remodeling factor subunits, we named p325 Rsf-1.
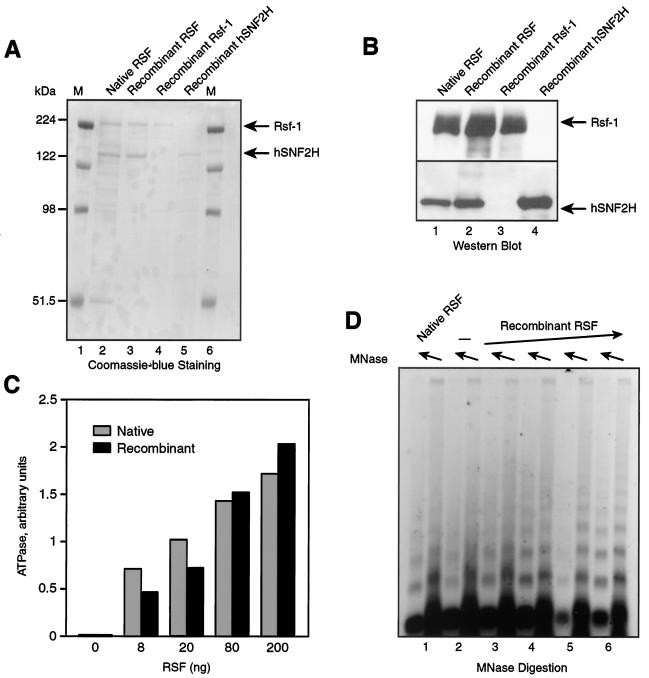
FIG. 3.
Recombinant RSF. (A) Coomassie blue staining of purified fractions of native RSF, recombinant RSF obtained by coexpression of the subunits, recombinant Rsf-1, and recombinant hSNF2H. The migration positions of the molecular markers (M) are shown on the left side of the gel. The migration of Rsf-1 and hSNF2H is indicated on the right side of the gel. (B) Western blot of recombinant RSF and its subunits. (C) ATPase activity. The graph shows the quantification of phosphate released during the ATPase reaction with increasing amounts of native and recombinant RSF obtained by coexpression of the subunits, as indicated in the figure. (D) Chromatin assembly. An agarose gel of a micrococcal nuclease digestion of a chromatin assembly reaction carried out with 0.4 μg of native RSF (lane 1), without RSF (lane 2), and with increasing amounts of recombinant RSF obtained by coexpression of the subunits (between 0.1 and 0.8 μg) (lanes 3 to 6).
Rsf-1 is a nuclear protein.
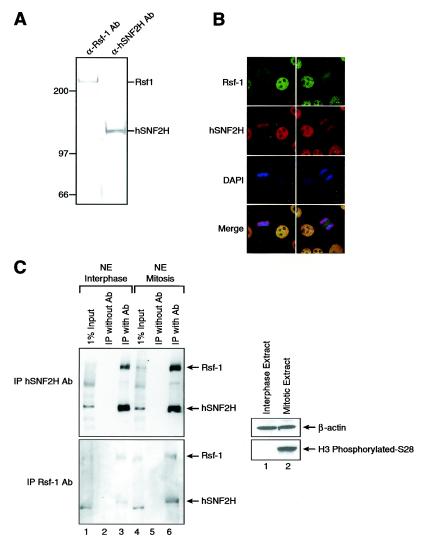
Immunofluorescence experiments in HeLa cells with highly specific antibodies to the RSF subunits Rsf-1 and hSNF2H (Fig. 2A) showed that the large subunit of RSF and hSNF2H localized to the nuclei (Fig. 2B), correlating with its purification procedure, which began with a HeLa cell-derived nuclear fraction. The staining pattern obtained with the antibodies appears to be specific; a similar pattern was obtained with antibodies directed against different regions of Rsf-1. Interestingly, during mitosis, Rsf-1 exhibited a more diffuse localization, whereas hSNF2H remained associated with mitotic chromosomes. This mitotic pattern could mean that the RSF subunits are dissociated during mitosis. In order to test this possibility, we obtained nuclear extracts from cells in interphase and in mitosis and carried out immunoprecipitation assays with antibodies against both RSF subunits (Fig. 2C). The results showed that in interphase, as well as in mitosis, the RSF subunits are interacting with each other, ruling out the possibility of dissociation of the RSF complex in mitosis. The inability to detect hSNF2H in areas other than chromosomes during mitosis is probably due to a detection problem; the very strong chromosome signal may be masking the weaker cellular signal. We suggest that another hSNF2H-containing complex remains associated with mitotic chromosomes. We speculate, based on recent studies, that this complex might be the ISWI-containing complex WICH, which is stably associated with mitotic chromosomes (3).
FIG. 2.
Immunofluorescence microscopy of RSF. (A) Western blots with RSF antibodies. Partially purified fractions of RSF derived from HeLa nuclear pellet were Western blotted with monoclonal antibodies against the RSF subunits Rsf-1 and hSNF2H. (B) Immunofluorescence microscopy. HeLa cells were fixed and permeabilized, followed by incubation with antibodies (Ab) against the RSF subunits Rsf-1 and hSNF2H. The antibody against Rsf-1 is shown in green, the antibody against hSNF2H is shown in red, and DAPI (4′,6′-diamidino-2-phenylindole) is shown in blue. The bottom panel shows the merge between all of them. Two cells are shown on the field. The cell to the left is in mitosis (metaphase), whereas the cell to the right is in interphase. (C) Interaction between Rsf-1 and hSNF2H during mitosis. Western blot analysis of immunoprecipitation analysis carried out with hSNF2H (top panel) and Rsf-1 (bottom panel) antibodies, as indicated in the left side of the figure. The input for the immunoprecipitation (IP) is indicated on top of the figure: interphasic (left side) or mitotic (right side) nuclear extract. As a negative control, the extract was incubated only with protein G-agarose and processed like the other samples. We used antibodies against histone H3 and phosphorylated S28 as a marker for mitosis and β-actin as a loading marker.
Regarding the localization of hSNF2H and Rsf-1, we have determined that almost all hSNF2H found in the nuclear pellet is associated with Rsf-1. Moreover, we have not found Rsf-1 that is not associated with hSNF2H (see Discussion). On the other hand, the hSNF2H found in nuclear extract is associated with several different complexes, including the several different forms of ACF. The levels of RSF and ACF complexes appear similar in HeLa cells (data not shown).
Recombinant RSF.
To understand the functional importance of each subunit of RSF, baculoviruses encoding each of the RSF subunits were used to infect Sf9 cells individually or together (coinfection). The procedure used to purify each of the polypeptides resulted in highly purified proteins (see Materials and Methods and Fig. 3A and B).
To test whether the recombinant RSF complex has activity, we analyzed its ability to carry out ATP hydrolysis and chromatin assembly in vitro. The nucleosome-stimulated ATPase activity of the recombinant complex, isolated by coinfection of viruses carrying individual subunits, was comparable to that of native RSF (Fig. 3C). Also, there was no apparent difference in chromatin assembly activity between recombinant and native RSF as gauged by micrococcal nuclease digestion of assembled chromatin (Fig. 3D). Moreover, the specific activity of the native and recombinant RSF complexes was similar in both assays (data not shown). From these experiments, we conclude that recombinant RSF is functionally equivalent to the native complex, at least with respect to its ATPase and chromatin assembly activities.
Function of the individual RSF components Rsf-1 and hSNF2H.
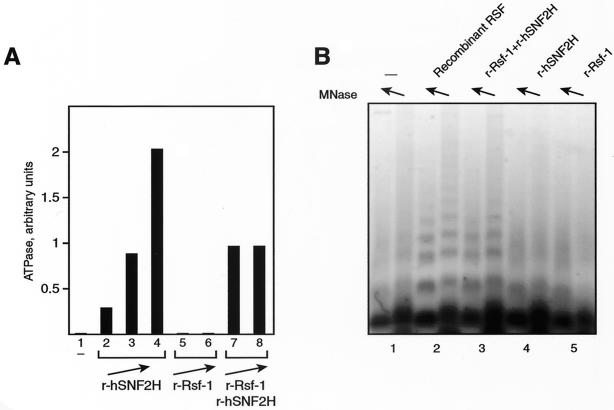
We first assessed the individual subunits for their role in generating the ATPase activity of the RSF complex. As shown previously, hSNF2H alone showed a strong ATPase activity (Fig. 4A, lanes 2 to 4), which is nucleosome dependent (data not shown). On the other hand, Rsf-1 did not display ATPase activity (Fig. 4A, lanes 5 and 6). Moreover, Rsf-1 did not affect the hSNF2H-dependent ATPase activity (Fig. 4A, lanes 7 and 8). This suggests that the ATPase activity of hSNF2H is independent of Rsf-1. This result agrees with findings for the Drosophila ISWI protein, in which Acf-1 was unable to stimulate the ATPase activity of ISWI under several different conditions (6).
FIG. 4.
Functions of the two RSF subunits. (A) ATPase activity. The graph shows the quantification of phosphate released during the ATPase reaction. Lanes show the following: no factor added (lane 1), increasing amounts of recombinant hSNF2H (between 0.2 and 0.9 μg, lanes 2 to 4), increasing amounts of recombinant Rsf-1 (0.2 and 0.4 μg, lanes 5 and 6, respectively), and the same amount of recombinant hSNF2H used in lane 3 plus increasing amounts of Rsf-1 (0.2 and 0.4 μg, lanes 7 and 8, respectively). (B) Chromatin assembly. An agarose gel shows a micrococcal nuclease digestion of chromatin assembly reactions carried out without factor (lane 1), with recombinant RSF obtained by coexpression of the subunits (lane 2), with recombinant Rsf-1 plus recombinant hSNF2H (lane 3), with recombinant hSNF2H alone (lane 4), and with recombinant Rsf-1 alone (lane 5).
We then examined the chromatin assembly activity. The reaction was carried out with recombinant RSF obtained by coinfection of the recombinant viruses encoding each subunit (Fig. 4B, lane 2). In addition, each of the subunits was tested independently (Fig. 4B, lanes 4 and 5) and together (Fig. 4B, lane 3). We found that both subunits are essential; neither hSNF2H nor Rsf-1 alone supported chromatin assembly (Fig. 4B, compare lanes 2 and 3 with lanes 4 and 5). To characterize further the role of each subunit in chromatin assembly, we analyzed various steps in the reaction. Because we showed previously that RSF binds to core histones (13), we examined whether Rsf-1 is able to interact, on its own, with core histones. To this end, we carried out immunoprecipitation experiments with recombinant Rsf-1 mixed with core histones. We found that the recombinant RSF complex coimmunoprecipitated with core histones (data not shown), and Rsf-1 alone also interacted with core histones, although to a considerably lower extent (data not shown). Since Rsf-1 does not appear to exist on its own in HeLa cells (data not shown, see above), it is likely that its interaction with hSNF2H affects the ability of Rsf-1 to interact with core histones.
We previously proposed that the binding of RSF to core histones through Rsf-1 is followed by the binding of the RSF-histone complex to the DNA (13). Therefore, we investigated whether the individual subunits of RSF could bind to DNA. Two types of experiments were performed. First, we analyzed the association of hSNF2H and Rsf-1 to DNA by electron microscopy, mixing recombinant hSNF2H or Rsf-1 along with DNA in the presence or absence of core histones. When hSNF2H was analyzed in the presence or absence of core histones, we observed ca. 30% (in the presence of histones), and the majority (in the absence of histones) of the DNA molecules contained a single large protein complex (Fig. 5A, upper panels). The large protein complex observed on the DNA was composed of hSNF2H, as demonstrated by probing the reaction with gold-conjugated antibodies to hSNF2H (Fig. 5A, upper panels). This result suggested that hSNF2H is able to interact with DNA regardless of the presence of core histones. The analysis of Rsf-1 showed the formation of similar large protein complexes in ca. 20% of the DNA molecules when the reaction was carried out in the presence of core histones (Fig. 5A, bottom panel). In contrast, when the reaction was carried out in the absence of core histones, the large protein complex bound to the DNA was not observed (data not shown).
FIG. 5.
DNA-binding activity. (A) Electron microscopic visualization of the binding of RSF subunits to DNA. The experiment was carried out as shown on the top of the figure. hSNF2H or Rsf-1 were incubated with EcoRI-linearized plasmid. The products of the reactions were first incubated with antibodies against hSNF2H or Rsf-1. This was followed by incubation with a secondary antibody conjugated with 10-nm gold, as described in Materials and Methods. In each image, the white dot found in the middle of the large protein complex corresponds to the gold particle. The top panels show the products of the reaction with hSNF2H visualized by electron microscopy in the presence (left) or absence (right) of histones. The bottom panel shows the products of the reaction with Rsf-1 in the presence of histones. Bar, 100 nm. (B) DNA filter-binding assay. Increasing amounts of recombinant hSNF2H, Rsf-1, and recombinant RSF obtained by coexpression of the subunits were incubated with radiolabeled DNA at the indicated molar ratio. The figure shows the quantification of DNA bound to the nitrocellulose filters as explained in Materials and Methods. This figure represents three independent experiments.
Nitrocellulose-based DNA-binding assays corroborated the results obtained with electron microscopy (Fig. 5B). More importantly, these studies show that the DNA-binding activity of hSNF2H was inhibited by the presence of Rsf-1, since the RSF complex failed to bind to DNA (Fig. 5B). Moreover, because the RSF complex can associate with DNA only in the presence of histones (13), our findings suggest that the association of hSNF2H with Rsf-1 affects the DNA-binding properties of hSNF2H. This change results in the specific association of RSF with DNA in a core histone-dependent manner.
Taken together, these experiments suggest that each subunit of RSF has a defined role in chromatin assembly; Rsf-1 is the histone chaperone of the complex probably allowing nucleosome formation, while hSNF2H provides the energy for the nucleosome spacing activity.
Rsf-1 versus HBXAP.
As mentioned above, while we were cloning the full-length cDNA encoding Rsf-1, a new cDNA, HBXAP was identified with a sequence identical to that of Rsf-1, except that it lacked 252 amino acids found at the N terminus of Rsf-1 (18). That report showed that HBXAP physically associates with HBV-pX and facilitates pX-dependent stimulation of HBV transcription. Therefore, we were interested in studying HBXAP in the context of the RSF complex.
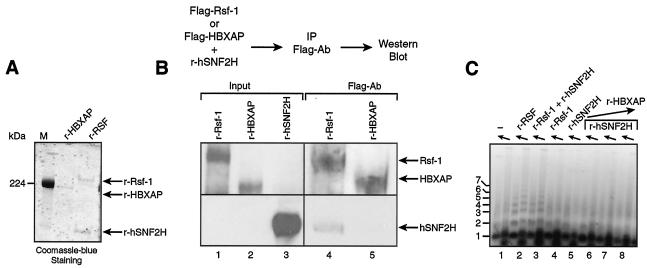
For this series of experiments, we cloned and expressed Flag-tagged HBXAP in baculovirus-infected Sf9 cells and used these cells as our source for purification of recombinant HBXAP (Fig. 6A). We first tested the ability of HBXAP to associate with hSNF2H. As a control, we used Flag-tagged Rsf-1, which, as expected, showed interaction with hSNF2H by immunoprecipitation (Fig. 6B, lane 4). In contrast, we found that HBXAP, which lacks only 252 amino acids at the N-terminal region of Rsf-1, was unable to interact with hSNF2H (Fig. 6B, lanes 5 and data not shown).
FIG. 6.
Comparison of Rsf-1 and HBXAP. (A) Coomassie blue staining of recombinant RSF obtained by coexpression of the subunits and HBXAP. The arrow indicates the migration position of the RSF subunits Rsf-1 and hSNF2H and of HBXAP. M, molecular marker. (B) Interaction between hSNF2H and HBXAP. Western blot analysis of the immunoprecipitation carried out as outlined at the top of the figure. Flag-tagged HBXAP or Flag-tagged Rsf-1 was incubated with hSNF2H and immunoprecipitated with antibody against Flag. The input shows the starting material for the immunoprecipitation before the subunits were mixed. (C) Chromatin assembly. An agarose gel of a micrococcal nuclease digestion of a chromatin assembly reaction carried out with HBXAP, as follows: without factor (lane 1), with recombinant RSF obtained by coexpression of the subunits (lane 2), with recombinant Rsf-1 and hSNF2H together (lane 3), with recombinant Rsf-1 alone (lane 4), with recombinant hSNF2H alone (lane 5), and with recombinant hSNF2H plus increasing amounts of HBXAP (between 0.16 and 1 μg) (lanes 6 to 8).
In agreement with the results presented above showing undetectable interaction between HBXAP and hSNF2H and in contrast to Rsf-1, hSNF2H was unable to mediate chromatin assembly in the presence of HBXAP (Fig. 6C).
Taken together, these results suggest that the failure of HBXAP to assemble chromatin is due to the inability of HBXAP to interact with hSNF2H. This conclusion suggests that the activities of each of the RSF subunits (that is, the ability of Rsf-1 to interact with core histones and the ATPase activity of hSNF2H) are tightly coupled during chromatin assembly.
DISCUSSION
The reconstitution of RSF allowed us to characterize the function of each of its subunits in the chromatin assembly reaction. We found that the ATPase activity of RSF depends entirely on hSNF2H. Rsf-1, on the other hand, is the histone chaperone of the complex. We also showed that hSNF2H binds to DNA independently of histones. However, in the context of RSF, there is no binding to DNA unless histones are present. This result suggests that Rsf-1 modulates the DNA-binding activity of hSNF2H, perhaps by blocking the DNA-binding domain of hSNF2H. As expected, our results showed that both subunits are required for chromatin assembly.
The largest subunit of RSF, Rsf-1, turned out to be a larger version of the recently identified protein HBXAP. Northern blot analyses revealed the existence of several species of Rsf-1 and HBXAP mRNAs (19) (Fig. 1C). Consequently, it is likely that a family of Rsf-1/HBXAP proteins exists.
HBXAP is involved in pX-dependent activation of HBV transcription (18) and also displayed a transcriptional repressive activity in vitro when targeted to a reporter gene by fusion to the Gal4 DNA-binding domain (19). The RSF complex, on the other hand, was initially identified as a chromatin-remodeling complex and, due to its ability to mobilize nucleosomes in vitro, allows the binding of transcriptional activators to their target sites. As a result, competent RNA polymerase II preinitiation complexes can be formed on chromatin templates (12). The apparent discrepancy between the roles of HBXAP and Rsf-1 may be due to the inability of HBXAP to interact with hSNF2H and, therefore, with chromatin remodeling complexes. Consistent with this is our observation that HBXAP is unable to support chromatin assembly in vitro. Taken together, these results suggest that Rsf-1 and HBXAP have distinct functions in the cell. In order to analyze further Rsf-1 and HBXAP, we developed antibodies against the N-terminal region of Rsf-1 that is missing in HBXAP. We used these antibodies for localization studies in HeLa cells with immunofluorescence analyses, but we could not observe any differences when the resultant immunofluorescence patterns were compared to those generated with antibodies that recognize both Rsf-1 and HBXAP (data not shown).
It is possible that the histone chaperone activity of Rsf-1 is, in part, a consequence of its acidic properties and the presence of glutamic and aspartic acid stretches at the C-terminal region. We searched for sequence homology with other histone chaperone proteins, such as CAF-1 and NAP-1, but we were unable to find any clear similarities. Homology searches between Rsf-1 and other subunits of the ISWI chromatin remodeling complexes, such as Acf-1 (as well as the BAZ family of proteins) and NURF-301, yielded no matches. A search for homologues in other species showed a candidate in Drosophila, the CG8677 gene product.
Regarding the hSNF2H and Rsf-1 association, we have found that almost all hSNF2H found in the HeLa nuclear pellet fraction is associated with Rsf-1 (data not shown). In contrast, the hSNF2H found in the nuclear extract fraction is associated with several forms of ACF and BAZ-containing complexes. With respect to Rsf-1, we have not found Rsf-1 that is not associated with hSNF2H (data not shown), suggesting that Rsf-1 exists in the cell as RSF.
RSF and other members of the ISWI family of chromatin remodeling factors are abundant in HeLa cells. We found that there are similar amounts of RSF and ACF in HeLa cells (11). An obvious question, then, is why does the cell need so much of and so many kinds of these factors? One likely explanation is that each complex has a specific function and that its activity is modulated by the other components of the complexes. Unfortunately, most of the biochemical analyses carried out thus far showed that the complexes have similar properties (for example, ATPase, chromatin assembly, and chromatin remodeling). Therefore, the development of specific assays will be necessary for the analysis of each of these complexes. As an example, RNA interference has been used recently for the analysis of the function of ACF in vivo. The experiments suggest that ACF has a role in DNA replication of heterochromatin at pericentromeric regions, allowing DNA replication to proceed through highly condensed chromatin regions (5).
Consequently, one of the challenges of future investigations is to decipher the mechanism of action of these factors within the cell, as well as to discover the functional differences between the various chromatin-remodeling factors.
Acknowledgments
We thank Lynne Vales for helpful comments on the manuscript and Kathy Jones and Anton Tutter for the gift of baculovirus-expressing hSNF2H. We also thank members of the Reinberg laboratory for helpful suggestions.
This work was supported by a grant from the National Institutes of Health (GM37120) and by the Howard Hughes Medical Institute.
REFERENCES
- 1.Aasland, R., T. J. Gibson, and A. F. Stewart. 1995. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20:56-59. [DOI] [PubMed] [Google Scholar]
- 2.Bochar, D. A., J. Savard, W. Wang, D. W. Lafleur, P. Moore, J. Cote, and R. Shiekhattar. 2000. A family of chromatin remodeling factors related to Williams syndrome transcription factor. Proc. Natl. Acad. Sci. USA 97:1038-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bozhenok, L., P. A. Wade, and P. Varga-Weisz. 2002. WSTF-ISWI chromatin remodeling complex targets heterochromatic replication foci. EMBO J. 21:2231-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Capili, A. D., D. C. Schultz, I. F. Rauscher, and K. L. Borden. 2001. Solution structure of the PHD domain from the KAP-1 corepressor: structural determinants for PHD, RING and LIM zinc-binding domains. EMBO J. 20:165-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins, N., R. A. Poot, I. Kukimoto, C. Garcia-Jimenez, G. Dellaire, and P. D. Varga-Weisz. 2002. An ACF1-ISWI chromatin-remodeling complex is required for DNA replication through heterochromatin. Nat. Genet. 32:627-632. [DOI] [PubMed] [Google Scholar]
- 6.Eberharter, A., S. Ferrari, G. Langst, T. Straub, A. Imhof, P. Varga-Weisz, M. Wilm, and P. B. Becker. 2001. Acf1, the largest subunit of CHRAC, regulates ISWI-induced nucleosome remodeling. EMBO J. 20:3781-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guschin, D., T. M. Geiman, N. Kikyo, D. J. Tremethick, A. P. Wolffe, and P. A. Wade. 2000. Multiple ISWI ATPase complexes from xenopus laevis: functional conservation of an ACF/CHRAC homolog. J. Biol. Chem. 275:35248-35255. [DOI] [PubMed] [Google Scholar]
- 8.Hakimi, M. A., D. A. Bochar, J. A. Schmiesing, Y. Dong, O. G. Barak, D. W. Speicher, K. Yokomori, and R. Shiekhattar. 2002. A chromatin remodeling complex that loads cohesin onto human chromosomes. Nature 418:994-998. [DOI] [PubMed] [Google Scholar]
- 9.Hochheimer, A., S. Zhou, S. Zheng, M. C. Holmes, and R. Tjian. 2002. TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature 420:439-445. [DOI] [PubMed] [Google Scholar]
- 10.Ito, T., M. Bulger, M. J. Pazin, R. Kobayashi, and J. T. Kadonaga. 1997. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 90:145-155. [DOI] [PubMed] [Google Scholar]
- 11.LeRoy, G., A. Loyola, W. S. Lane, and D. Reinberg. 2000. Purification and characterization of a human factor that assembles and remodels chromatin. J. Biol. Chem. 275:14787-14790. [DOI] [PubMed] [Google Scholar]
- 12.LeRoy, G., G. Orphanides, W. S. Lane, and D. Reinberg. 1998. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science 282:1900-1904. [DOI] [PubMed] [Google Scholar]
- 13.Loyola, A., G. LeRoy, Y. H. Wang, and D. Reinberg. 2001. Reconstitution of recombinant chromatin establishes a requirement for histone-tail modifications during chromatin assembly and transcription. Genes Dev. 15:2837-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mello, J. A., and G. Almouzni. 2001. The ins and outs of nucleosome assembly. Curr. Opin. Genet. Dev. 11:136-141. [DOI] [PubMed] [Google Scholar]
- 15.Orphanides, G., G. LeRoy, C. H. Chang, D. S. Luse, and D. Reinberg. 1998. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92:105-116. [DOI] [PubMed] [Google Scholar]
- 16.Poot, R. A., G. Dellaire, B. B. Hulsmann, M. A. Grimaldi, D. F. Corona, P. B. Becker, W. A. Bickmore, and P. D. Varga-Weisz. 2000. HuCHRAC, a human ISWI chromatin remodeling complex contains hACF1 and two novel histone-fold proteins. EMBO J. 19:3377-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70:81-120. [DOI] [PubMed] [Google Scholar]
- 18.Shamay, M., O. Barak, G. Doitsh, I. Ben-Dor, and Y. Shaul. 2002. Hepatitis B virus pX interacts with HBXAP, a PHD finger protein to coactivate transcription. J. Biol. Chem. 277:9982-9988. [DOI] [PubMed] [Google Scholar]
- 19.Shamay, M., O. Barak, and Y. Shaul. 2002. HBXAP, a novel PHD-finger protein, possesses transcription repression activity. Genomics 79:523-529. [DOI] [PubMed] [Google Scholar]
- 20.Strohner, R., A. Nemeth, P. Jansa, U. Hofmann-Rohrer, R. Santoro, G. Langst, and I. Grummt. 2001. NoRC-a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J. 20:4892-4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsukiyama, T., J. Palmer, C. C. Landel, J. Shiloach, and C. Wu. 1999. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 13:686-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsukiyama, T., and C. Wu. 1995. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell 83:1011-1020. [DOI] [PubMed] [Google Scholar]
- 23.Tyler, J. K. 2002. Chromatin assembly. Cooperation between histone chaperones and ATP-dependent nucleosome remodeling machines. Eur. J. Biochem. 269:2268-2274. [DOI] [PubMed] [Google Scholar]
- 24.Varga-Weisz, P. 2001. ATP-dependent chromatin remodeling factors: nucleosome shufflers with many missions. Oncogene 20:3076-3085. [DOI] [PubMed] [Google Scholar]
- 25.Varga-Weisz, P. D., M. Wilm, E. Bonte, K. Dumas, M. Mann, and P. B. Becker. 1997. Chromatin-remodeling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature 388:598-602. [DOI] [PubMed] [Google Scholar]
- 26.Vignali, M., A. H. Hassan, K. E. Neely, and J. L. Workman. 2000. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 20:1899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolffe, A. 1998. Chromatin: structure and function. Academic Press, Inc., San Diego, Calif.
- 28.Zhang, Y., G. LeRoy, H. P. Seelig, W. S. Lane, and D. Reinberg. 1998. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95:279-289. [DOI] [PubMed] [Google Scholar]
- 29.Zhang, Y., and D. Reinberg. 2001. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 15:2343-2360. [DOI] [PubMed] [Google Scholar]