Abstract
Transient-transfection assays have been used to identify transcription factors, and genetic analyses of these factors can allow a dissection of their mechanism of activation. Epstein-Barr nuclear antigen 1 (EBNA-1) has been shown to activate transcription from transfected templates, but its ability to activate transcription from nuclear templates has been controversial. We have established cells with integrated EBNA-1-responsive templates and have shown that EBNA-1 activates transcription from these chromatin-embedded templates dose dependently. A mutational analysis of EBNA-1 has identified a domain required for transcriptional activation of integrated templates, but not of transfected templates. The ability of EBNA-1 to activate transcription from both integrated and transfected templates can be inhibited by a derivative of EBNA-1 lacking the amino acids required for activation from integrated templates. EBNA-1's mode of activating transfected templates is therefore genetically distinct from that acting on integrated templates.
Mammalian transcriptional activators have often been identified and characterized by transfection assays that require one or more templates to be transported and assembled in the nuclei of recipient cells. Recent efforts to understand the mechanisms of transcriptional activation on defined templates in vitro have helped to define both cis-acting sequences and protein domains essential for gene regulation (23, 24, 34, 35). However, limitations of this approach include its inability to assess contributions to transcription mediated by the trafficking of the transcription factors from the cytoplasm to the nucleus, or possibly within nuclear compartments. Nor can it readily identify higher-ordered structures of the template imposed by those factors prior to or during chromatin assembly (1, 29). Here, we describe genetic analyses that identify two distinct contributions of Epstein-Barr nuclear antigen 1 (EBNA-1) to support transcription, one of which likely reflects either its trafficking or its control of template structure.
EBNA-1, encoded by Epstein-Barr virus (EBV), is a multifunctional protein essential for both EBV's extrachromosomal replication and positive and negative regulation of multiple viral promoters (9, 15, 32). EBNA-1 also positively regulates heterologous promoters when the family of repeats (FR) enhancer to which it binds is placed upstream of those promoters (5, 14, 18, 27, 38). Studies of templates with or without FR microinjected into the cytoplasm or nuclei of cells that express EBNA-1 indicate that a significant contribution of EBNA-1 to the activation of transcription occurs in the cytoplasm (14). Other studies have been interpreted to mean that EBNA-1 activates transcription on extrachromosomal but not integrated templates (11). These observations indicate that EBNA-1 likely uses multiple mechanisms to contribute to the support of transcription. It is important to know if EBNA-1 can affect the transcription of integrated templates; were it to do so, EBNA-1 might regulate cellular genes during the latent phase of the EBV life cycle and, perhaps, in EBV-associated tumors.
We have generated multiple clones of human B cells in which transcriptional templates with or without the FR enhancer are integrated into host cell DNA. We find that the introduction of vectors expressing EBNA-1 positively activates transcription of those templates in which FR is present in cis. A genetic analysis with six derivatives of EBNA-1 that all bind site specifically to this enhancer has identified 25 residues required to activate transcription of these integrated templates. A derivative lacking these residues positively activates transcription of transfected templates, indicating that its contribution may be mediated by the trafficking of EBNA-1 or by a structure imposed by EBNA-1 on its template. These results indicate that EBNA-1 encodes an element necessary for the transcription of integrated templates and one or more additional elements that promote the transcription of newly introduced templates.
MATERIALS AND METHODS
Cell culture.
The BJAB cell line, an EBV-negative Burkitt's lymphoma derivative, has been previously described (30). Cells were grown in RPMI 1640 medium (GibcoBRL) supplemented with 10% fetal bovine serum, 40 U of penicillin/ml, and 50 μg of streptomycin/ml at 37°C in a 5% CO2 humidified atmosphere.
Transfection and cloning.
Electroporation of 5 × 106 cells was performed in 500 μl of complete medium with 250 V and a capacitance of 960 μF using a Bio-Rad (Hercules, Calif.) generator. BJAB cells with integrated EBV-responsive templates were established by cotransfecting DNAs expressing the puromycin resistance gene and DNAs expressing either FR-thymidine kinase (TK)-luciferase or TK-luciferase. Forty-eight hours posttransfection, cells were plated in limiting dilutions on 96-well plates in the presence of 1 μg of puromycin/ml. The cells were grown for 2 to 3 weeks, and the proliferating cells were counted. Those microwell cultures with a >90% probability of being derived from single cells were expanded and screened for the presence of luciferase activity. All clones that demonstrated a background luciferase activity of <104 relative light units (RLU) per 105 cells were expanded and tested for responsiveness to EBNA-1. Once isolated, the clones were propagated continuously in the presence of 1 μg of puromycin/ml.
PCR.
Clones of cells resistant to puromycin were screened by PCR for the integration of FR-TK-luciferase or TK-luciferase. Colonies identified in the cloning assay were expanded, and total cellular DNA was extracted from 5 × 106 cells using the DNeasy kit from Qiagen (Valencia, Calif.). The protocol was modified to include a 4-h proteinase K digestion instead of the 15-min digestion suggested by the manufacturer. After quantification, 50 ng of DNA was subjected to PCR in the presence of 5 U of Herculase polymerase (Invitrogen, Carlsbad, Calif.) for 25 cycles in a 25-μl total volume. The primers used for the PCR were 5′-GAC GGC CAG TGC CAA GCT CG-3′ (sense sequence) and 5′-GAC GCA GGC AGT TCT ATG CGG-3′ (antisense sequence).
Plasmids.
Some of the derivatives of EBNA-1 used in this study have been previously described (18). Mutants of EBNA-1 with deletions of amino acids 65 to 89 or 359 to 369 were constructed using whole-plasmid PCR. Amino acids 65 to 89 were replaced with a unique NgoMIV restriction site encoding a glycine and an alanine, while amino acids 359 to 369 were replaced with a unique XbaI site encoding an arginine and a serine. The PCR products were purified and digested with either NgoMIV or XbaI, ligated, and propagated in Escherichia coli. Recovered DNAs were sequenced within the EBNA-1 open reading frame to ensure accuracy of the PCR product. The amino acid composition from 396 to 455 of the shuffled joining domain (JD) derivative remained the same as the composition within wild-type EBNA-1, but its sequence had been randomized (J. Wang, personal communication).
Reporter plasmids expressing FR-TK-luciferase, TK-luciferase, or FR-TK-CAT have been previously described (21). Briefly, each plasmid with or without FR contains the herpes simplex virus TK promoter driving the expression of either the luciferase or chloramphenicol acetyltransferase (CAT) gene.
Retroviral propagation and infection.
The retroviruses used in this study are derived from a vesicular stomatitis virus G protein-pseudotyped murine leukemia virus (25). To construct a retrovirus expressing wild-type EBNA-1, the parent retroviral plasmid encoding β-galactosidase (lacZ) was digested with AgeI and AvrII and blunted with the Klenow fragment of DNA polymerase I. The desired fragment was purified and ligated with the open reading frame of EBNA-1 derived from digesting a plasmid encoding EBNA-1 with BglII and XhoI treated with the Klenow fragment. The orientation of the insert within the recovered DNA was confirmed by digestion with restriction endonucleases. Retroviruses were generated by transfecting 293-HEK cells with 3 μg of a plasmid encoding the Gag-Pol elements, 1 μg of plasmids encoding the vesicular stomatitis virus G protein, and the 5 μg of a plasmid carrying the retroviral backbone encoding either β-galactosidase or EBNA-1 using the Lipofectamine 2000 reagent (Invitrogen) as described by the manufacturer. Twenty-four hours posttransfection, the culture medium was supplemented with 50 mM HEPES. On days 2 to 4 posttransfection, the culture supernatant was collected, filtered through a 0.45-μm-pore-size filter, and stored at −80°C. The titers of the viral stocks were estimated by infecting 293-HEK cells plated at 5 × 104 per well of a 24-well plate with 1:5 serial dilutions of viral stocks. Infected cells were identified by the expression of green fluorescent protein (GFP), also encoded by each of the viruses from an internal ribosomal entry site, and titers were calculated according to a Poisson distribution.
BJAB cells growing exponentially were centrifuged and resuspended at 5 × 106 per ml prior to infection. One milliliter of cells was then infected at a multiplicity of infection of 1 by incubating the suspension in complete medium supplemented with 50 mM HEPES and 100 μg of Polybrene (Sigma, St. Louis, Mo.)/ml for 1 h at 4°C with gentle rocking. After the incubation period, the cells were collected by centrifugation, resuspended in 10 ml of complete medium, and incubated at 37°C. Forty-eight hours postinfection, the infected cells were sorted on a FACS-Vantage or FACS-DIVA (Becton Dickinson, San Jose, Calif.), enriching for the green population of cells. Luciferase assays were performed on 105 sorted green cells as described below.
Reporter assays.
Forty-eight hours posttransfection, the efficiency of transfection was estimated by counting GFP-positive cells, and the transfected cells were collected by centrifugation, washed with phosphate-buffered saline, and resuspended in cell culture lysis buffer at a concentration of 5 × 104 per μl. A total of 20 μl of lysed cells was analyzed for luciferase activity (22). RLU were normalized to the efficiency of transfection by subtracting the RLU obtained from 106 untransfected cells from the RLU obtained from 106 transfected cells and dividing by the transfection efficiency. A total of 105 cells were assayed for CAT activity using thin-layer chromatography as previously described (12). Chloramphenicol-labeled with 14C was purchased from NEN-Life Science (Boston, Mass.). The CAT activity was normalized by dividing the percent acetylated chloramphenicol by the total number of transfected cells and multiplying the resulting number by 106. The normalized numbers are referred to as acetylation units.
Prior to using the CAT assay, we established the linear range of the assay with EBV-negative BJAB cells transfected with a CAT reporter with or without vector expressing EBNA-1. A range of cellular lysates were used in the assay, and the acetylated forms of CAT were separated from the unacetylated forms by thin-layer chromatography. The thin-layer chromatography plates were exposed to a PhosphorImager screen, and the optical densities (OD) of the bands were quantified by ImageQuant version 5.0 (Molecular Dynamics, Sunnyvale, Calif.). The CAT assay was found to be linear from an OD of 104 to 106 U. The measurements of CAT activity obtained from cells transfected with various derivatives of EBNA-1 were within this linear range.
All reporter assay results are presented as increases (n-fold) in transcription. To calculate the increase, the activity detected from cells lacking the reporter gene was subtracted from the activity obtained from cells in the presence of the reporter gene. The increase in transcription was calculated by dividing the RLU or acetylation units obtained from cells transfected with the various derivatives of EBNA-1 by the RLU or acetylation units obtained from cells transfected with the empty vector.
Western blot analysis.
Forty-eight hours after transfection, the cells were counted and the efficiency of transfection was measured by the expression of GFP. The cells were washed with phosphate-buffered saline, resuspended at 2 × 104/μl in 1× sample buffer (0.05% sodium dodecyl sulfate, 2 mM Tris-HCl [pH 6.8], 0.1% 2-mercaptoethanol, 0.2% glycerol, and 0.02% bromophenol blue), and incubated for 30 min on ice. The lysates were sonicated and incubated at 95°C for 10 min. Protein lysates approximately equivalent to 5 × 105 cells were separated in an 8% sodium dodecyl sulfate-polyacrylamide gel, transferred electrophoretically to a nitrocellulose membrane (Bio-Rad), and blocked overnight with 5% nonfat dry milk. For the primary antibody, a rabbit antiserum affinity purified against the C terminus of EBNA-1 was used to detect wild-type EBNA-1 and the six derivatives. A murine monoclonal antibody conjugated to fluorescein isothiocyanate was used to detect β-actin (Sigma, St. Louis, Mo.). For the secondary antibody, goat anti-rabbit antibody conjugated to peroxidase was used and detected with the ECL Western blotting analysis system (Amersham Pharmacia Biotech, Piscataway, N.J.).
The intensity of each protein band was estimated using ImageQuant version 5.0 software. The loading error was corrected by normalizing to the intensity of β-actin detected in cells transfected with wild-type EBNA-1. The OD of each derivative of EBNA-1 was confirmed to be in the linear range by using different exposures of the membrane to film, and this signal was multiplied by the β-actin correction factor. The β-actin-corrected number was divided by the total number of transfected cells loaded per lane, and the amount of EBNA-1 derivative detected relative to wild-type EBNA-1 was expressed as the OD per transfected cell.
Statistical analysis.
All statistical analyses were performed using Mstat version 3.21 (N. Drinkwater, McArdle Laboratory for Cancer Research, University of Wisconsin Medical School), which is available at http://mcardle.oncology.wisc.edu/mstat. The nonparametric Wilcoxon rank sum test was used in all cases unless otherwise indicated.
RESULTS
EBNA-1 activates transcription from nuclear templates dose dependently.
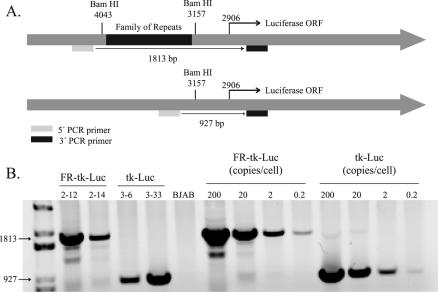
In order to determine unambiguously if EBNA-1 can activate transcription from templates within the nucleus, we established multiple clones with integrated templates. BJAB cells, an EBV-negative Burkitt's lymphoma-derived cell line, were used as host cells, and PCR was used to screen the derived clones for integration of the luciferase gene expressed from the HSV-1 TK promoter plus or minus FR (Fig. 1A). Figure 1B shows an ethidium bromide-stained 1% agarose gel that separates amplified DNA from two clones with FR-TK-luciferase and two clones with TK-luciferase integrated into the cellular DNA. A standard curve is shown, demonstrating the sensitivity of the assay to be less than one copy of DNA per cell. A total of 80 puromycin-resistant clones were initially isolated, and of these, 20 had the luciferase gene present as shown by either PCR or luciferase assay. The luciferase activity in ∼105 cells of each clone varied from 103 to 106 RLU, which is 2 to 2 × 103 times greater than that in untransfected BJAB cells (data not shown). We found that of the clones identified as having the luciferase gene integrated into the cellular genome, 25% had background luciferase activity of <104 RLU per 105 cells. We characterized multiple clones with low levels of luciferase activity to permit detection of transcription that might be activated by EBNA-1. We found that 100% of the clones isolated with background luciferase activity of <104 RLU per 105 cells were detectably responsive to EBNA-1 (data not shown).
FIG. 1.
Characterization of integrated template DNAs in clones of BJAB cells. (A) Structures of the two templates, FR-TK-luciferase and TK-luciferase, introduced into BJAB cells. The positions and sequences of the two primers used to characterize the templates and the sizes of their products generated by PCR are also shown. ORF, open reading frame. (B) Agarose gel resolving PCR products of DNAs isolated from two clones of BJAB cells into which FR-TK-luciferase (clones 2-12 and 2-14) was introduced and two clones into which TK-luciferase (clones 3-6 and 3-33) was introduced. The gel also includes the products derived by amplifying known amounts of the parental plasmids to serve as size markers and to permit estimates of the number of template molecules integrated into each cell.
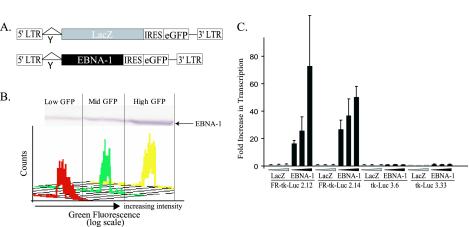
Two BJAB clones stably expressing FR-TK-luciferase and two clones stably expressing TK-luciferase were chosen for further characterization. These cells were infected with retroviral vectors expressing β-galactosidase or EBNA-1 (Fig. 2A). The infected cells were sorted to yield those with the lowest 30%, middle 30%, and highest 30% green signals, which were confirmed to express corresponding levels of EBNA-1 by Western blot analysis (Fig. 2B). EBNA-1 activated transcription dose dependently (P < 0.05; Jonckheere-Terpstra test) from those integrated templates with its binding sites in FR present in cis (Fig. 2C).
FIG. 2.
Transcription of integrated templates induced by EBNA-1 is dose dependent. (A) Structures of retroviral vectors to establish EBNA-1's dose-dependent induction of transcription. The control vector expresses β-galactosidase (LacZ), and both it and the vector expressing EBNA-1 also express enhanced GFP (eGFP), whose translation is mediated by an internal ribosomal entry site (IRES). LTR, long terminal repeat. (B) Fluorescence-activated cell sorter profiles reflecting the sorting of the infected cells as a function of the level of their expression of eGFP and the level of expression of EBNA-1 as measured by Western blotting in one set of infected, sorted cells 48 h following infection. (C) Mean RLU obtained in two independent experiments performed in duplicate were normalized by setting the RLU output from cells infected with LacZ virus expressing low GFP to 1. The differences in RLU in cells with integrated FR-TK-luciferase infected with a retrovirus expressing EBNA-1 and sorted for different levels of expression of eGFP are statistically significant (P < 0.05; Jonckhere-Terpstra test). The error bars indicate standard errors of the means.
Genetic analysis of EBNA-1 transcriptional activation.
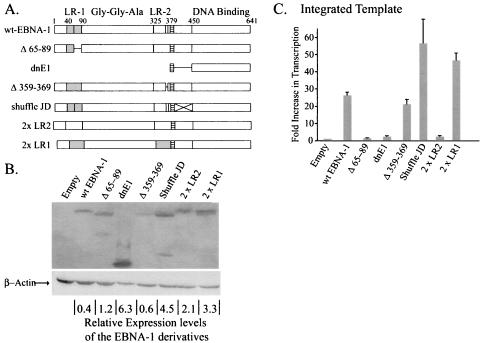
We analyzed six derivatives of EBNA-1 to identify elements within it that could contribute to its transcription of either integrated or newly introduced templates (Fig. 3A). All derivatives were constructed to contain both the EBNA-1 nuclear localization signal and its DNA-binding and dimerization domain to insure that they would have the ability to home to the nucleus and to bind DNAs site specifically. A rabbit antibody to the EBNA-1 DNA-binding and dimerization domain was used in order to detect each of the derivatives with equal efficiency (Fig. 3B). Expression vectors for wild-type EBNA-1 and these six derivatives were introduced into a clone of BJAB cells in which FR-TK-luciferase is integrated, and the levels of luciferase activity were measured (Fig. 3C). Three derivatives (Δ359-369, shuffle JD, and 2×LR1) stimulated luciferase activity to the same level as did wild-type EBNA-1 (P < 0.05). These derivatives lack the charged, unique sequences in linking region 2 (LR2), have had the amino acids of the putative JD randomized, and lack all of LR2, respectively. The last derivative, 2×LR1, has LR1 substituted for LR2. LR1 and LR2 are the linking regions of EBNA-1 and have been shown to contribute to the activation of transcription and replication mediated by EBNA-1 (18, 38). In vitro the linking regions are essential for linking DNAs to which EBNA-1 binds site specifically (5, 17-19, 21). In vivo they are essential for tethering EBNA-1 to chromosomes (20). That the derivative shuffled JD activated transcription like wild-type EBNA-1 demonstrated that the EBNA-1 sequence between 379 and 450 is not critical for this function. All of the derivatives that activated transcription as efficiently as did wild-type EBNA-1 had LR1 intact (Fig. 3A). A derivative of EBNA-1 with the unique sequences within LR1 deleted, Δ65-89, had lost the ability to promote transcription (Fig. 3C). In fact, both of the derivatives lacking intact LR1 (Δ65-89 and 2×LR2) behaved as did the formerly characterized dominant-negative derivative of EBNA-1, dnE1 (13). This derivative of EBNA-1, which also lacks LR1, failed to activate transcription from integrated templates (Fig. 3C) significantly over the level of the empty-vector control (increase over empty vector, 2.4-fold; P = 0.33). This failure demonstrates that DNA binding by EBNA-1 to a template is not sufficient for transcriptional activation and that the amino terminus is therefore required for activity from an integrated template. The signals measured with dnE1 are statistically the same as with the empty vector, and both are robust enough to ensure that the signals measured with Δ65-89 and 2×LR2 are real but indistinguishable from these negative controls. Importantly, we found that the derivatives dnE1, 2×LR1, and 2×LR2 affected transcription in a second clone of BJAB cells carrying integrated FR-TK-luciferase, as did the clones documented in Fig. 3C with the same rank order of activities (data not shown). This finding confirms that transcription of the integrated FR-TK-luciferase template mediated by the various derivatives of EBNA-1 is independent of integration events. Taken together, these measurements identify LR1 and pinpoint its unique residues, 65 to 89, as being critical for EBNA-1’s activation of the transcription of integrated templates.
FIG. 3.
Mutational analysis of EBNA-1 identifies a transcriptional activation domain within LR1. (A) Derivatives of EBNA-1 depicted schematically. EBNA-1 has two highly charged regions within its amino terminus, LR1 (shaded boxes) and LR2. The nuclear localization signal of EBNA-1 is found adjacent to a putative flexible linker domain or JD and is represented in all derivatives by the hatched boxes. The Gly-Gly-Ala repeats span ∼225 residues in the B95-8 strain of EBV. The derivative used in these studies contains only three Gly-Gly-Ala repeats. wt, wild type. (B) Western blot demonstrating expression levels of the various derivatives relative to that of wild-type EBNA-1. The membrane was simultaneously probed with antibodies to EBNA-1 and β-actin, which served as a loading control. The relative levels of expression of the transfected derivatives of EBNA-1 were corrected for loading error, and OD units obtained from ImageQuant analysis are represented as OD units per transfected cell at the bottom of each lane. (C) Mean luciferase results corrected for transfection efficiency obtained from at least three independent transfections performed in duplicate are depicted graphically. The increases in transcription over empty vector (mean increase [n-fold] over BJAB cells lacking integrated FR-TK-luciferase, 10; average number of RLU, 5,425 ± 231) mediated by wt-EBNA-1 (mean increase [n-fold] over empty vector, 26 [P = 5.3 × 10−6]; average number of RLU, 85,000 ± 3,780), Δ359-369 (mean increase [n-fold] over empty vector, 21 [P = 0.009]; average number of RLU, 95,900 ± 17,800), shuffled JD (mean increase [n-fold] over empty vector, 56 [P = 0.05]; average number of RLU, 116,000 ± 10,700), and 2×LR1 (mean increase [n-fold] over empty vector, 46.7 [P = 0.007]; average number of RLU, 21,2000 ± 31,000) are significant. The apparent increases in transcription over wild-type EBNA-1 mediated by the shuffled JD and 2×LR1 derivatives are not statistically significant (P > 0.05). The derivatives Δ65-89 (mean increase [n-fold] over empty vector, 1.5 [P = 0.85]; average number of RLU, 6,510 ± 491), 2×LR2 (mean increase [n-fold] over empty vector, 2.5 [P = 0.21]; average number of RLU, 9,169 ± 1,070), and dnE1 (mean increase [n-fold] over empty vector, 2.4 [P = 0.33]; average number of RLU, 8,957 ± 1,800) were found to have no effect on transcription from the integrated template.
There was no correlation between the levels of expression of the EBNA-1 derivatives and whether they stimulated luciferase activity. Abrogation of transcriptional activity mediated by EBNA-1 could not be attributed to low levels of expression, as five out of the six derivatives studied (Δ65-89, dnE1, shuffled JD, 2×LR2, and 2×LR1) were found to be expressed at higher levels than wild-type EBNA-1. Two mutants (shuffled JD and 2×LR1) were found to support transcription as efficiently as the wild type (Fig. 3C). The other three derivatives of EBNA-1, Δ65-89, dnE1, and 2×LR2, failed to activate transcription appreciably over the control transfected cells (Fig. 3B and C). The last derivative studied, Δ359-369, was expressed at levels similar to those of wild-type EBNA-1 and activated transcription from integrated templates as efficiently as did wild-type EBNA-1 (Fig. 3B and C).
A derivative of EBNA-1 can selectively activate transcription of transfected templates.
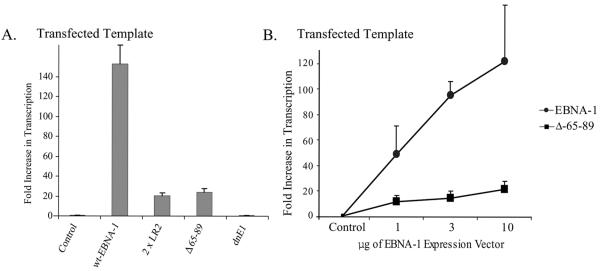
We wished to determine if the domains within EBNA-1 required to activate transcription of integrated templates were also required to activate the transcription of transfected templates. LR1 and, in particular, its unique residues, 65 to 89, were necessary to activate transcription from an integrated template (Fig. 3C). Plasmids encoding EBNA-1 or one of its derivatives were transfected into EBV-negative cells along with a plasmid expressing FR-TK-CAT. We normalized the signals from these experiments to the CAT activity expressed from FR-TK-CAT in the presence of an empty vector rather than to TK-CAT, because the latter vector has a higher spontaneous level of activity than does FR-TK-CAT (27, 39). EBNA-1 lacking all of LR1 (2×LR2) and EBNA-1 lacking only amino acids 65 to 89 failed to activate the transcription of integrated templates (Fig. 3C) but activated the transcription of the transfected template ∼10-fold over the control value (Fig. 4A; P = 0.009). The dominant-negative derivative, dnE1, failed to activate either template and thus served as a negative control in the experiments. To confirm these observations, BJAB cells were transfected with a plasmid DNA encoding FR-TK-luciferase with increasing amounts of plasmid DNAs encoding either wild-type EBNA-1 or the derivative of EBNA-1 lacking amino acids 65 to 89 (Fig. 4B). We found that the Δ65-89 derivative activated transcription from a transfected template dose dependently (P < 0.005; Jonckheere-Terpstra test). EBNA-1 therefore encodes a function that activates the transcription of transfected templates but not of integrated templates.
FIG. 4.
The ability of EBNA-1 to activate transcription from transfected templates is independent of its ability to activate transcription from integrated templates. (A) The mean CAT activities obtained in three independent experiments performed in duplicate normalized to the CAT activity obtained in cells transfected with FR-TK-CAT and an empty vector control are depicted graphically. The CAT activity obtained from cells transfected with a vector encoding wild-type (wt) EBNA-1 was 150-fold higher than that obtained from cells transfected with a control plasmid (P = 0.002). The CAT activity obtained from cells transfected with a vector encoding Δ65-89 was 24-fold higher than that obtained from cells transfected with a control plasmid (P = 0.001). The derivative 2×LR2 was also found to increase transcription from the transfected template 20-fold over cells transfected with a control plasmid (P = 0.002), while the dnE1 derivative had no effect on transcription from this template (P = 0.3). (B) The Δ65-89 derivative of EBNA-1 activates transcription from transfected templates in a dose-dependent fashion (P = 0.003; Jonckhere-Terpstra test). BJAB cells were transfected with 50 ng of a plasmid encoding FR-TK-luciferase with increasing amounts of plasmids encoding either wild-type EBNA-1 or Δ65-89. Luciferase activity was assayed 48 h posttransfection and is represented on the y axis as induction (n-fold) over cells transfected with a vector encoding FR-TK-luciferase in the absence of EBNA-1. The error bars indicate standard errors of the means.
The ability EBNA-1 to support transcription is inhibited by coexpression of Δ65-89.
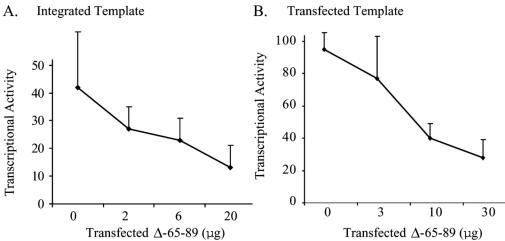
One derivative of EBNA-1, dnE1, has been well characterized as a dominant-negative mutant of EBNA-1 that inhibits both EBNA-1’s activation of transcription and its support of replication, at least in part by competing for wild-type site-specific DNA binding (13). We speculated that the mutant of EBNA-1 with the transcriptional activation domain found within amino acids 65 to 89 deleted would inhibit transcription mediated by EBNA-1. We found that expression of increasing amounts of the Δ65-89 derivative resulted in significant inhibition of the ability of EBNA-1 to activate transcription from both integrated (Fig. 5A) and transfected (Fig. 5B) templates (P ≤ 0.004; Jonckheere-Terpstra test).
FIG. 5.
The derivative of EBNA-1 with amino acids 65 to 89 deleted inhibits wild-type EBNA-1's transcription function in a dominant-negative manner. (A) Graphic representation of the results of three independent experiments performed in duplicate using BJAB cells stably transfected with FR-TK-luciferase plus 2 μg of a vector encoding EBNA-1 and either no vector or increasing amounts of a vector encoding Δ65-89. (B) Graphic representation of the results of three independent experiments performed in duplicate in which BJAB cells were transiently transfected with a plasmid encoding FR-TK-luciferase along with 3 μg of one encoding EBNA-1 and no vector or increasing amounts of the vector encoding Δ65-89. The decrease in luciferase activity mediated by Δ65-89 is statistically significant in both panels (P ≤ 0.004; Jonckhere-Terpstra test). The error bars indicate standard errors of the means.
DISCUSSION
We have established multiple clones of EBV-negative cells with EBNA-1-responsive templates integrated into the cellular DNA. The transcription of these integrated templates can be increased by EBNA-1 in all clones isolated with <104 RLU per 105 cells at baseline (data not shown); the increase in transcription mediated by EBNA-1 in these clones was proportional to the level of expression of EBNA-1 (Fig. 2). Such clones constituted ∼25% of those isolated. A recent study also analyzed EBNA-1 activation of transcription from integrated templates but failed to detect it (11). That study used uncloned populations of cells in which the high basal levels of reporter expression likely masked the stimulation that EBNA-1 would yield in the responsive cells, thereby limiting the ability to detect transcriptional activity from integrated templates (11). Our study of six derivatives of EBNA-1 showed that amino acids 65 to 89 are essential to activate transcription from integrated templates. This domain of EBNA-1 contributes to transcription from transfected templates but is not required for the activation of gene expression from these templates (Fig. 3 and 4). A derivative of EBNA-1 lacking its first 330 residues, which include LR1 and its Gly-Gly-Ala repeats, also failed to activate transcription from an integrated template (data not shown) but has been found to activate transcription of transfected templates (13). We have further shown that a derivative of EBNA-1 lacking amino acids 65 to 89 inhibits the ability of wild-type EBNA-1 to activate transcription from both integrated and transfected templates (Fig. 5).
The different contributions to transcription of EBNA-1 may play distinct roles in the EBV life cycle. DNAs of herpesviruses are packaged free of nucleosomes in the viral capsid (16). Following infection of cells, the viral DNA must enter the nucleus and be packaged into chromatin (7, 16). EBNA-1, therefore, likely affects gene expression both from naked viral DNA, represented by the transfected templates, and from chromatin-embedded viral DNA, represented by the integrated template in our experiments.
These findings indicate that EBNA-1 contributes to transcription by two genetically separable mechanisms which can be distinguished by using integrated templates or those introduced by transfection. These two templates differ in that the transfected templates enter the cell as naked DNA and need to be shuttled into the nucleus while the integrated templates are present in the nucleus bound as chromatin. Previous studies have found that EBNA-1-responsive templates microinjected into the cytoplasm of cells had a 100-fold increase in transcriptional activity, while those injected into the nucleus had a 5- to 20-fold increase in the presence of EBNA-1 (14). These results indicate that EBNA-1 apparently contributes to transcription from nuclear templates, but much of its ability to activate transcription might come from its ability to localize plasmids to the nucleus (14). In our studies, EBNA-1 activated transcription from integrated templates by up to 70-fold and activated transcription from transfected templates by up to 120-fold over background (Fig. 2 and 4). One derivative we studied, dnE1, has both the EBNA-1 nuclear localization sequence and its DNA-binding domain intact but failed to affect the transcription of transfected templates (Fig. 3C). These two domains of EBNA-1 should be sufficient to shuttle FR-positive DNAs into the nucleus. That dnE1 failed to activate transcription leads us to favor the hypothesis that EBNA-1 contributes to the activation of transcription by modulating the formation of chromatin on naked templates to which it can bind. This hypothesis is supported by recent work by Avolio-Hunter et al. in which EBNA-1 was shown to destabilize nucleosomes in vitro (2). The derivative of EBNA-1 lacking amino acids 65 to 89 failed to activate transcription from integrated templates (Fig. 3C) but was able to activate transcription from a transfected template by at least 24-fold (17% of wild-type function) dose dependently (Fig. 4A and B). A similar mutant of EBNA-1 with a slightly different deletion (residues 61 to 83) increases the transcription of transfected templates by only 1% (38). These findings indicate that EBNA-1 contributes to transcription from both transfected and integrated templates but has at least one function that acts only on transfected templates.
Analyses of additional derivatives of EBNA-1 indicate that LR1 is critical to activate transcription of integrated templates. The derivative 2×LR2 lacks LR1 and activates transcription on transfected templates better than does dnE1 (Fig. 4A) (18), but is no better than dnE1 in stimulating transcription on integrated templates (Fig. 3C). The LR1 domain of EBNA-1 also clearly contributes to the transcription of transfected templates; a derivative in which LR2 is replaced with LR1 supported transcription of transfected templates at wild-type levels (Fig. 4A) (18). The idea that LR1 contributes to transcription on both integrated and transfected templates is also supported by the deletion of its unique residues, 65 to 89, dominantly inhibiting wild-type EBNA-1 activation of each kind of template (Fig. 5). However, LR2 can also contribute to transcription on transfected templates. A derivative of EBNA-1 in which LR1 is replaced with LR2 stimulates transcription of transfected templates by 20-fold, which is ∼13% of wild-type activity (Fig. 4A).
The fact that EBNA-1's contributions to the transcription of transfected or integrated templates can be separated genetically may have an unexpected benefit. Vectors derived from EBNA-1 are being considered for gene therapy in people (3, 4, 6, 8, 10, 26, 28, 31, 37). However, EBNA-1 has been found to be associated with an increased risk of tumor development in some strains of transgenic mice (36). This risk has been attributed in part to EBNA-1's transcriptional activation of host genes (33). The Δ65-89 derivative of EBNA-1 supports long-term extrachromosomal replication of oriP plasmids, as does wild-type EBNA-1 (unpublished observations), but should not be able to activate the expression of host cell genes. It may prove to be an ideal partner in human gene therapy using oriP vectors.
Acknowledgments
We thank Paul Ahlquist, Norman Drinkwater, Richard Krauss, JinDong Wang, and Ngan Lam for critically reviewing the manuscript. We thank Julia D'Alo and Jackie Perrigoue for their technical assistance.
Our work was supported by Public Health Service grants CA-22443, CA-07175, and T32-CA-009614-13.
REFERENCES
- 1.Archer, T. K., P. Lefebvre, R. G. Wolford, and G. L. Hager. 1992. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science 255:1573-1576. (Erratum, 256:161.) [DOI] [PubMed]
- 2.Avolio-Hunter, T. M., P. N. Lewis, and L. Frappier. 2001. Epstein-Barr nuclear antigen 1 binds and destabilizes nucleosomes at the viral origin of latent DNA replication. Nucleic Acids Res. 29:3520-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee, S., E. Livanos, and J. M. Vos. 1995. Therapeutic gene delivery in human B-lymphoblastoid cells by engineered non-transforming infectious Epstein-Barr virus. Nat. Med. 1:1303-1308. [DOI] [PubMed] [Google Scholar]
- 4.Calos, M. P. 1996. The potential of extrachromosomal replicating vectors for gene therapy. Trends Genet. 12:463-466. [DOI] [PubMed] [Google Scholar]
- 5.Ceccarelli, D., and L. Frappier. 1998. Separation of the DNA replication and transactivation activities of EBNA-1, the origin binding protein of Epstein-Barr virus. Gene Ther. Mol. Biol. 3:1-10. [Google Scholar]
- 6.Cui, F. D., T. Kishida, S. Ohashi, H. Asada, K. Yasutomi, E. Satoh, T. Kubo, S. Fushiki, J. Imanishi, and O. Mazda. 2001. Highly efficient gene transfer into murine liver achieved by intravenous administration of naked Epstein-Barr virus (EBV)-based plasmid vectors. Gene Ther. 8:1508-1513. [DOI] [PubMed] [Google Scholar]
- 7.Dyson, P. J., and P. J. Farrell. 1985. Chromatin structure of Epstein-Barr virus. J. Gen. Virol. 66:1931-1940. [DOI] [PubMed] [Google Scholar]
- 8.Franken, M., A. Estabrooks, L. Cavacini, B. Sherburne, F. Wang, and D. T. Scadden. 1996. Epstein-Barr virus-driven gene therapy for EBV-related lymphomas. Nat. Med. 2:1379-1382. [DOI] [PubMed] [Google Scholar]
- 9.Gahn, T. A., and B. Sugden. 1995. An EBNA-1-dependent enhancer acts from a distance of 10 kilobase pairs to increase expression of the Epstein-Barr virus LMP gene. J. Virol. 69:2633-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harada, Y., M. Iwai, S. Tanaka, T. Okanoue, K. Kashima, H. Maruyama-Tabata, H. Hirai, E. Satoh, J. Imanishi, and O. Mazda. 2000. Highly efficient suicide gene expression in hepatocellular carcinoma cells by Epstein-Barr virus-based plasmid vectors combined with polyamidoamine dendrimer. Cancer Gene Ther. 7:27-36. [DOI] [PubMed] [Google Scholar]
- 11.Kang, M. S., S. C. Hung, and E. Kieff. 2001. Epstein-Barr virus nuclear antigen 1 activates transcription from episomal but not integrated DNA and does not alter lymphocyte growth. Proc. Natl. Acad. Sci. USA 98:15233-15238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaykas, A., K. Worringer, and B. Sugden. 2001. CD40 and LMP-1 both signal from lipid rafts but LMP-1 assembles a distinct, more efficient signaling complex. EMBO J. 20:2641-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirchmaier, A. L., and B. Sugden. 1997. Dominant-negative inhibitors of EBNA-1 of Epstein-Barr virus. J. Virol. 71:1766-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langle-Rouault, F., V. Patzel, A. Benavente, M. Taillez, N. Silvestre, A. Bompard, G. Sczakiel, E. Jacobs, and K. Rittner. 1998. Up to 100-fold increase of apparent gene expression in the presence of Epstein-Barr virus oriP sequences and EBNA1: implications of the nuclear import of plasmids. J. Virol. 72:6181-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, M. A., M. E. Diamond, and J. L. Yates. 1999. Genetic evidence that EBNA-1 is needed for efficient, stable latent infection by Epstein-Barr virus. J. Virol. 73:2974-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leinbach, S. S., and W. C. Summers. 1980. The structure of herpes simplex virus type 1 DNA as probed by micrococcal nuclease digestion. J. Gen. Virol. 51:45-59. [DOI] [PubMed] [Google Scholar]
- 17.Mackey, D., T. Middleton, and B. Sugden. 1995. Multiple regions within EBNA1 can link DNAs. J. Virol. 69:6199-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mackey, D., and B. Sugden. 1999. The linking regions of EBNA1 are essential for its support of replication and transcription. Mol. Cell. Biol. 19:3349-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mackey, D., and B. Sugden. 1997. Studies on the mechanism of DNA linking by Epstein-Barr virus nuclear antigen 1. J. Biol. Chem. 272:29873-29879. [DOI] [PubMed] [Google Scholar]
- 20.Marechal, V., A. Dehee, B. R. Chikhi, T. Piolot, M. M. Coppey, and J. C. Nicolas. 1999. Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J. Virol. 73:4385-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Middleton, T., and B. Sugden. 1992. EBNA1 can link the enhancer element to the initiator element of the Epstein-Barr virus plasmid origin of DNA replication. J. Virol. 66:489-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitchell, T., and B. Sugden. 1995. Stimulation of NF-κB-mediated transcription by mutant derivatives of the latent membrane protein of Epstein-Barr virus. J. Virol. 69:2968-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Natarajan, K., B. M. Jackson, H. Zhou, F. Winston, and A. G. Hinnebusch. 1999. Transcriptional activation by Gcn4p involves independent interactions with the SWI/SNF complex and the SRB/mediator. Mol. Cell 4:657-664. [DOI] [PubMed] [Google Scholar]
- 24.Neely, K. E., A. H. Hassan, C. E. Brown, L. Howe, and J. L. Workman. 2002. Transcription activator interactions with multiple SWI/SNF subunits. Mol. Cell. Biol. 22:1615-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ory, D. S., B. A. Neugeboren, and R. C. Mulligan. 1996. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc. Natl. Acad. Sci. USA 93:11400-11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phillips, J. E., B. Thyagarajan, and M. P. Calos. 1999. Epstein-Barr virus plasmid model system for analyzing recombination in human cells. Plasmid 41:198-206. [DOI] [PubMed] [Google Scholar]
- 27.Reisman, D., and B. Sugden. 1986. trans activation of an Epstein-Barr viral transcriptional enhancer by the Epstein-Barr viral nuclear antigen 1 Mol. Cell. Biol. 6:3838-3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sclimenti, C. R., and M. P. Calos. 1998. Epstein-Barr virus vectors for gene expression and transfer. Curr. Opin. Biotechnol. 9:476-479. [DOI] [PubMed] [Google Scholar]
- 29.Smith, C. L., and G. L. Hager. 1997. Transcriptional regulation of mammalian genes in vivo. A tale of two templates. J. Biol. Chem. 272:27493-27496. [DOI] [PubMed] [Google Scholar]
- 30.Steinitz, M., and G. Klein. 1975. Comparison between growth characteristics of an Epstein-Barr virus (EBV)-genome-negative lymphoma line and its EBV-converted subline in vitro. Proc. Natl. Acad. Sci. USA 72:3518-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stoll, S. M., C. R. Sclimenti, E. J. Baba, L. Meuse, M. A. Kay, and M. P. Calos. 2001. Epstein-Barr virus/human vector provides high-level, long-term expression of alpha1-antitrypsin in mice. Mol. Ther. 4:122-129. [DOI] [PubMed] [Google Scholar]
- 32.Sugden, B., and N. Warren. 1989. A promoter of Epstein-Barr virus that can function during latent infection can be transactivated by EBNA-1, a viral protein required for viral DNA replication during latent infection. J. Virol. 63:2644-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsimbouri, P., M. E. Drotar, J. L. Coy, and J. B. Wilson. 2002. bcl-xL and RAG genes are induced and the response to IL-2 enhanced in EmuEBNA-1 transgenic mouse lymphocytes. Oncogene 21:5182-5187. [DOI] [PubMed] [Google Scholar]
- 34.Utley, R. T., C. C. Adams, M. Vettese-Dadey, J. L. Workman, and J. Cote. 1994. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. EMBO J. 13:6031-6040.7813441 [Google Scholar]
- 35.Wallberg, A. E., K. E. Neely, A. H. Hassan, J. A. Gustafsson, J. L. Workman, and A. P. Wright. 2000. Recruitment of the SWI-SNF chromatin remodeling complex as a mechanism of gene activation by the glucocorticoid receptor tau1 activation domain. Mol. Cell. Biol. 20:2004-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson, J. B., J. L. Bell, and A. J. Levine. 1996. Expression of Epstein-Barr virus nuclear antigen-1 induces B cell neoplasia in transgenic mice. EMBO J. 15:3117-3126. [PMC free article] [PubMed] [Google Scholar]
- 37.Wohlgemuth, J. G., S. H. Kang, G. H. Bulboaca, K. A. Nawotka, and M. P. Calos. 1996. Long-term gene expression from autonomously replicating vectors in mammalian cells. Gene Ther. 3:503-512. [PubMed] [Google Scholar]
- 38.Wu, H., P. Kapoor, and L. Frappier. 2002. Separation of the DNA replication, segregation, and transcriptional activation functions of Epstein-Barr nuclear antigen 1. J. Virol. 76:2480-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wysokenski, D. A., and J. L. Yates. 1989. Multiple EBNA1-binding sites are required to form an EBNA1-dependent enhancer and to activate a minimal replicative origin within oriP of Epstein-Barr virus. J. Virol. 63:2657-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]