Abstract
Csn3 (Cops3) maps to the mouse chromosome 11 region syntenic to the common deletion interval for the Smith-Magenis syndrome, a contiguous gene deletion syndrome. It encodes the third subunit of an eight-subunit protein complex, the COP9 signalosome (CSN), which controls a wide variety of molecules of different functions. Mutants of this complex caused lethality at early development of both plants and Drosophila melanogaster. CSN function in vivo in mammals is unknown. We disrupted the murine Csn3 gene in three independent ways with insertional vectors, including constructing a ≈3-Mb inversion chromosome. The heterozygous mice appeared normal, although the protein level was reduced. Csn3−/− embryos arrested after 5.5 days postcoitum (dpc) and resorbed by 8.5 dpc. Mutant embryos form an abnormal egg cylinder which does not gastrulate. They have reduced numbers of epiblast cells, mainly due to increased cell death. In the Csn3−/− mice, subunit 8 of the COP9 complex was not detected by immunohistochemical techniques, suggesting that the absence of Csn3 may disrupt the entire COP9 complex. Therefore, Csn3 is important for maintaining the integrity of the COP9 signalosome and is crucial to maintain the survival of epiblast cells and thus the development of the postimplantation embryo in mice.
Csn3 (formerly known as Cops3) (11) encodes the third subunit of an eight-subunit complex, the COP9 signalosome (CSN), that was first identified in Arabidopsis thaliana as an essential regulator of light-mediated development (7) and is conserved from plants to mammals (43). Mutations in different subunits of the CSN usually disrupt the entire complex in plants and cause pleiotropic phenotypes such as cellular differentiation defects, stunted stature, and death at the seedling stage (25, 44). Consistent with observations in plants, disruption of the csn5 gene in Drosophila melanogaster resulted in lethality at the late larval or pupal stage (14). The COP9 signalosome was found to have protein kinase activity and regulate the deneddylation of the SCF E3 ubiquitin-ligase complex, which controls ubiquitin-dependent protein degradation. Because of these two functions, and likely others that remain to be elucidated, CSN controls a wide variety of important molecules, such as Jun, p53, and p27kip1 (3, 27, 37, 39). This potentially explains the importance of the multisubunit CSN complex in development.
Mutations in the genes encoding different subunits convey common phenotypic features. However, they can also cause distinct phenotypes, indicating that each subunit may have unique functions. D. melanogaster mutants carrying null mutations of csn4 and csn5 both displayed defects in oogenesis, embryo patterning, and larval lethality, but csn4 null flies had molting defects, while csn5 null flies developed melanotic tumors (30). Distinct functional roles for different subunits have also been observed in Schizosaccharomyces pombe (29). Reduction-of-function mutations of the Arabidopsis thaliana CSN3 gene decreased the COP9 complex level and caused defects in diverse aspects of plant development, which indicates the involvement of CSN in multifaceted developmental processes (31). The function of CSN in mammals and how alteration of CSN and subunit 3 will affect mammalian development remain unknown.
Human CSN3 maps to the Smith-Magenis syndrome common deletion interval (8, 13, 32). Smith-Magenis syndrome is a multiple congenital anomaly/mental retardation syndrome associated with developmental delay, anatomical developmental defects, and neurobehavioral abnormalities associated with del(17)(p11.2p11.2) (16). The mouse Csn3 homolog resides in the region on chromosome 11 syntenic to the human Smith-Magenis syndrome deletion interval (4, 34). We disrupted Csn3 in mice to investigate its role in mammalian development and in the Smith-Magenis syndrome phenotypes. Homozygous mice arrested after 5.5 days postcoitum (dpc) showed abnormalities in egg cylinder formation and failure of development at the gastrulation stage. Also, the CSN complex appeared to have been disrupted. This indicates the essential role of CSN in mammalian early embryonic development.
MATERIALS AND METHODS
Characterization of mouse Csn3 gene.
The gene structure of Csn3 was determined by PCR and DNA sequencing analysis (K. Walz, unpublished data) and subsequently confirmed by Blast (NCBI, http://www.ncbi.nlm.nih.gov) analysis between the complete cDNA sequence (NM-011991) (43) and the genomic sequence from bacterial artificial chromosome RP23-479J7 (AC068808) (4). Protein comparison between mouse Csn3 and those from other species was carried out with BlastP (NCBI).
To assess Csn3 gene expression, a 1.7-kb fragment containing the Csn3 full-length cDNA was isolated from the Invitrogen Pcops3 clone with BamHI and ApaI and used to probe both the adult and embryonic mouse multiple tissue Northern blot (Clontech) according to the method provided by the manufacturer.
Construction of Csn3 targeting vectors.
Mouse strain 129/SvEv genomic libraries constructed in vectors containing either the 5′ half of the human HPRT minigene (5′ HPRT vector) or 3′ half of the gene (3′ HPRT vector) were screened (48) to identify both 5′ and 3′ HPRT insertional vectors. One clone, pWY1-3′, from the 3′ HPRT library has an 8-kb genomic fragment of Csn3 and contains exons 3, 4, 5, and 6, as determined by restriction mapping and PCR analysis. The pWY1-3′ clone was digested with BglII and NdeI to delete a 3-kb fragment corresponding to exon 5 and adjacent intronic regions. After fill-in repair with Klenow fragment polymerase and subsequent religation, a BglII site was reconstructed, and this site was used to linearize the vector before introduction into embryonic stem (ES) cells. An NdeI linker was ligated into the reconstructed BglII site, and then the genomic fragment was isolated from the 3′ HPRT targeting vector with AscI and placed into the 5′ HPRT vector to get the 5′ HPRT targeting vector pWY1-5′. This NdeI site was used as a linearization site for pWY1-5′.
Targeting in embryonic stem cells and germ line transmission.
The linearized vectors pWY1-5′ and pWY1-3′ were introduced into the AB2.2 ES cells derived from Hprt-deficient 129SvEvBrd mice (28). ES cell growth, electroporation, and drug selection were performed as described previously (28, 35). The chimeric mice were mated to C57BL/6 wild-type mice, and F1 progeny were back-crossed to C57BL/6 mice to produce N2 and N3 mice. Two ES cell clones with the 5′ HPRT vector pWY1-5′ and one clone with the 3′ HPRT vector pWY1-3′ were transmitted through the germ line. Mouse strains targeted with the pWY1-5′ vector and pWY1-3′ vector are referred to as the Csn35′m strain and the Csn33′m strain, respectively, and the targeted alleles are referred to as the Csn35′m and Csn33′m alleles, respectively. Both strains were maintained by backcrossing to C57BL/6 wild-type mice. All the animals were treated in compliance with relevant animal welfare policies.
Confirmation of targeting.
After drug selection, targeted ES cells were detected by diagnostic Southern analysis with NdeI digestion. The probe was a 550-bp PCR fragment to the region that was deleted from the targeting vector by BglII and NdeI amplified with primers E5PR (5′-TCC CAT TGT AAG CCC CAC TA-3′) and E5PF (5′-GCA ATA CGG TTT ATT AGT GAT AGC C-3′). The PCR product was gel purified (Qiagen gel extraction kit) and labeled with [α-32P]dCTP by random priming (Amersham Rediprime II).
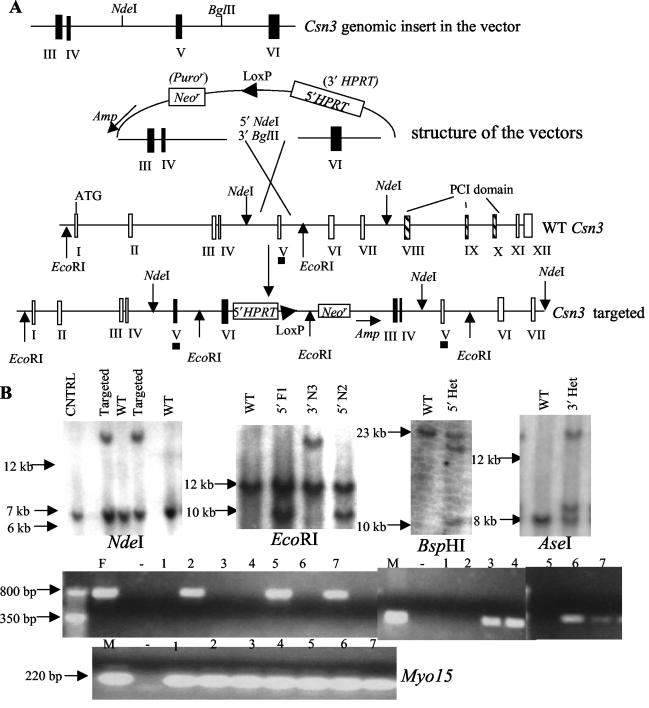
Targeted clones revealed an anticipated 18-kb fragment in addition to the 6.5-kb wild-type fragment. The targeted F1, N2, and N3 mice were confirmed by PCR and/or Southern analysis. One primer complementary to the targeting vector near the genomic fragment insertion site was synthesized, and the other primer was constructed to the genomic fragment. For the 5′ HPRT targeting vector, primers 5′ HPRT ex3-psk (pSK vector backbone) F (5′-GAC ATG TGT GCG TCT CCA TA 3′) and 5′ HPRT 2ex6-psk R (5′-CCT GAT TCT GTG GAT AAC CGT ATT A-3′) were used to yield an 800-bp product; Primers 5′ HPRT 2ex6-psk F (5′-GAC ATT CTC AGG AGC TCA GTA GGT A-3′) and 3′ HPRT ex3-psk R (5′-CGA GAA AGG AAG GGA AGA AAG-3′) are for the 3′ HPRT targeting vector, and PCR amplification with them yields a 350-bp product. Both primers are unique for their own targeting vector and do not amplify from the wild-type DNA. Primers for the Myo15 gene, Myo15 F (2) (5′-CTC TAC AAG CAC CTG CCC TC-3′) and Myo15 R (2) (5′-TAT TTC TCA AAG CTG TCA CTT TGC-3′) were used as a positive control for DNA quality. EcoRI was used in Southern analysis with the same probe used for ES cell genotyping. This probe revealed an expected 12-kb wild-type band, a 10-kb band in Csn35′m strain mice, and an 18-kb band in Csn33′m strain mice.
Construction of a 3-Mb Csn3-Zfp179 chromosomal inversion.
Zfp179 was formerly targeted with a vector from the 5′ HPRT genomic library containing a genomic fragment with all the Zfp179 exons except for the last exon. Csn3 was subsequently targeted with pWY1-3′ vector in the ES cell clone with targeted Zfp179 (42). Cre recombinase was introduced into double-targeted ES cell clones. After sodium hypoxanthine, aminopterin, the thymidine selection, inversion clones were confirmed by SacI digestion to yield the expected three bands at 28, 12.5, and 10 kb on diagnostic Southern blot analysis. The ES cell clone with the Csn3-Zfp179 chromosomal inversion was injected to produce inversion mice. Homozygous inversion mice are predicted to lose the 10-kb wild-type fragment on Southern blot analysis.
Western blotting analysis.
Proteins were extracted from the livers of wild-type and Csn35′m/+ or Csn33′m/+ littermates with Trizol following the manufacturer's instructions (Invitrogen). Total protein (10 μg) was electrophoresed on sodium dodecyl sulfate-4 to 20% polyacrylamide gels (ISC) and transferred to a nitrocellulose membrane. Anti-mouse Csn3 polyclonal antibody directed against the entire Csn3 protein was produced in a rabbit. After incubation with horseradish peroxidase-conjugated anti-rabbit immunoglobulin G antibody (Bio-Rad) and enzymatic reaction (Santa Cruz ECL system), blots were developed, and the signal was quantitated with Un-scan-it software (Silk Scientific). Antiactin antibody (Santa Cruz) was used as a loading control.
Genotyping of homozygous embryos.
BspHI was used to discriminate the heterozygous and the homozygous mice from the Csn35′m strain F1 or N2 heterozygous intercrosses with the same probe from the exon 5 described above. AseI was used for the Csn33′m strain N2 heterozygous mice intercrosses and EcoRI and PCR primers for genotyping the Csn35′m strain and Csn33′m strain were used for Csn35′m/+ and Csn33′m/+ intercrosses. DNA from 9.5 to 11.5 dpc embryos was used for genomic Southern analysis. Lysis buffer (100 μl: 50 mM KCl, 1.5 mM MgCl2, 10 mM Tris-Cl, pH 8.4, 0.5% Tween 20) was added to the yolk sac from the 9.5 dpc to 11.5 dpc embryos. After incubation at 55°C for an hour, proteinase K (1 mg/ml) (Roche) was inactivated by heating at 100°C for 15 min, and 1 or 2 μl was used for PCR analysis. For 8.5 dpc embryos, the whole embryos were lysed by adding 100 μl of lysis buffer, and 2 to 3 μl was used for PCR. For 7.5 dpc embryos, 15 μl of lysis buffer was added, and 2 to 4 μl was used for PCR. All the embryos analyzed, including those for histological analysis, were products of conception from Csn35′m/+ and Csn33′m/+ intercrosses.
Histological analysis of embryos.
Embryos at 5.5 dpc to 7.5 dpc were fixed in 4% paraformaldehyde in 1× phosphate-buffered saline (PBS) at 4°C for 4 h to overnight, embedded in paraffin, and sectioned at a thickness of 6 to 7 μm. Sections were stained with hematoxylin and eosin. Immunohistologic analysis was carried out for the selected sections. After rehydration, sections were incubated with 10% goat serum in PBS at room temperature for 1 h, followed by incubation with primary antibody (anti-Csn3 antibody or anti-Csn8 antibody) and peroxidase-conjugated secondary antibody (Vector ABC kit) in PBS with 2% goat serum at room temperature for 1 h each. Color reaction was implemented following the manufacturer's recommendation (Vector, DAB kit). These sections were counterstained with hematoxylin.
Ice-cold methanol (100%) was used to fix the 3.5 dpc blastocysts. Blastocysts were then processed through 75%, 50%, and 25% methanol in PBST (1× PBS with 0.1% Tween 20), followed by washing once in PBST. Whole-mount immunostaining was carried out as for the sections except that tetramethyl rhodamine isothiocyanate-conjugated secondary antibody was used (Sigma). A Zeiss Axioplan2 fluorescence microscope was used to detect the signal. DNA for blastocyst genotyping was prepared by adding 10μl of 1× PCR buffer (Qiagen, HotStarTaq) to the blastocyst, incubating at 95°C for 20 min, and freezing (−80°C) for 1 h, and 5 μl was used for the PCR. Primers 5′ HPRT 2Ty (tyrosinase gene) F (5′-CTG GGA GAA AAC ATA TTT TGA GAG A-3′) and 5′ HPRT TyR (5′-CCA CGA ATG CTG ACA TTC TC-3′) were used for 5′ HPRT targeting vector instead of those mentioned above.
In situ hybridization of whole-mount embryos.
A probe for the Brachyury gene was prepared as described previously (19). In situ hybridization was carried out for two litters of 7.5 dpc embryos with Wilkinson's protocol (45) with some modifications: the proteinase K treatment step was omitted, and 4% paraformaldehyde was used for all the fixation steps. After photography, embryos were processed for PCR genotyping. Primers were the same as for genotyping the blastocysts.
TUNEL analysis.
Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) staining of the selected sections was carried out according to the manufacturer's instructions (Roche, fluorescence cell death detection kit). The cells were visualized under a Zeiss Axioplan2 fluorescence microscope.
Bromodeoxyuridine incorporation analysis.
Pregnant females were injected intraperitoneally with bromodeoxyuridine (100 μg/g of body weight) and sacrificed 60 to 70 min later. Embryos were processed for histology. Bromodeoxyuridine was detected following the manufacturer's protocol (Roche, bromodeoxyuridine labeling and detection kit II) except that after dewaxing and rehydration, sections were incubated with 0.02% pepsin (Sigma) in 0.01 N HCl for 20 min at 37°C.
Blastocyst outgrowth studies.
Blastocysts at 3.5 dpc were cultured in ES cell medium (containing no leukemia inhibitory factor) in 5% CO2 at 37°C. The blastocysts were photographed after 3 days and after either 5 or 6 days in culture. The cultured embryos were processed for PCR as for 7.5 dpc embryos.
RESULTS
Characterization of the mouse Csn3 gene.
Blast analysis between the Csn3 cDNA (NM-011991) (43) and the sequence from a bacterial artificial chromosome (RP23-479J7) containing this gene (AC068808) (4) revealed that the gene contains 12 exons (Fig. 1). This is consistent with our independent analysis of Csn3 gene structure (Walz, unpublished). The translation initiation site is in exon 1 and the PCI domain (P, 26S proteasome; C, COP9 signalosome; I, initiation factor eIF3), which is found in six of eight CSN subunits (37), is encoded by exons 8 to 10. The entire Csn3 gene spans about 22 kb. Protein sequence comparison revealed high similarity of the mouse Csn3 to the CSN3 from plants and humans. It shares 99% identity with the human protein (NM-003653) (32, 38), 51% with D. melanogaster Csn3 (NM-079466) (1), and 37% with the Arabidopisis protein (AF395059) (15).
FIG. 1.
Mouse Csn3 gene structure and targeting strategy. (A) Gene structure and the targeting vectors. The entire gene is comprised of 12 exons represented by the roman numbered boxes. Exons containing the PCI domain are depicted as vertical hatched boxes. ATG is the start codon. Exons (III, IV, V, and VI) in the targeting vector are shown as solid vertical boxes. NdeI and BglII, used to delete the 3-kb fragment corresponding to exon V and adjacent intronic regions and the linearization sites (NdeI for 5′ HPRT vector; BglII for 3′ HPRT vector) are shown. Selectable genes on the vectors are indicated (Amp, ampicillin resistance gene; Neor, neomycin resistance gene on the 5′ HPRT vector; and Puror, puromycin resistance gene on the 3′ HPRT vector). Below is shown the predicted genomic structure after targeting. Exons from the targeting vectors are in solid boxes. The restriction enzymes used for genotyping are depicted (NdeI and EcoRI). Solid horizontal bars under exon V represent the probe used in all Southern blot analyses. (B) Southern blot and PCR analyses of ES cells and mice. Genomic DNA digested with NdeI (for ES cell genotyping) or EcoRI (mice tail or embryos) or BamHI (for heterozygous [Het] and homozygous discrimination of Csn35′m strain mice) or AseI (for heterozygous and homozygous discrimination of Csn33′m strain mice) was electrophoresed and transferred to nitrocellulose membrane. Untargeted ES cell DNA was run as a control (CNTRL) for ES cell genotyping, and wild-type mouse tail DNA (WT) was run as a control for tail or embryo genotyping. PCR results for one litter of embryos are shown here (F, father; M, mother; −, negative control). Below is the amplification from the Myo15 gene as a positive control for DNA quality. The size of bands is indicated on the left side.
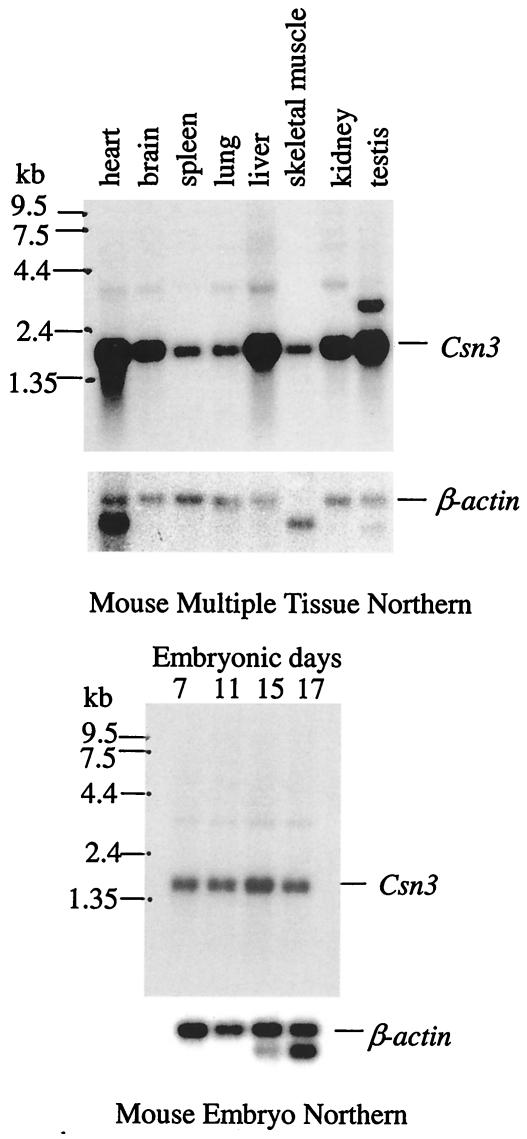
As was shown in humans (32), Csn3 is expressed ubiquitously in adult mice, with relatively high expression in the heart, brain, liver, kidney, and testis (Fig. 2). It is also expressed in 7- to 17-day embryos. With the exception of the testis, only one major transcript was detected at about 1.6 kb, which is consistent with the length of the complete cDNA sequence. In testis, a 2.7-kb transcript was also detected at a greatly reduced level compared to the 1.6-kb transcript (Fig. 2). A very faint band at ≈3.4 kb may represent an alternative splicing product or cross-reactivity.
FIG. 2.
Northern blot analysis of Csn3 gene. Both adult and embryo blots (Clontech) were probed independently with a Csn3 cDNA probe. The sizes are demarcated on the left side. The β-actin gene was detected as a loading control.
Disruption of the mouse Csn3 gene.
A targeting clone containing an 8-kb genomic fragment of the Csn3 gene was identified from the 3′ HPRT library constructed from strain 129S5 mice (48). This genomic fragment contains Csn3 exons 3, 4, 5, and 6. The same Csn3 genomic fragment was also cloned into the 5′ HPRT vector. The 3′ HPRT vector contains the 3′ half of the human HPRT minigene and puromycin (Puro) selectable gene, while the 5′ HPRT vector contains the 5′ half of the HPRT gene and the neomycin (Neo) selectable gene (Fig. 1). Both vectors are insertional vectors that can be used directly for targeting the ES cells.
Two alleles were constructed to enable PCR genotyping of early embryos. After deletion of a 3-kb fragment corresponding to exon 5 and adjacent intronic regions with BglII and NdeI, both vectors were introduced into AB2.2 ES cells (28). Homologous recombination is predicted to result in the repair of the deleted region around exon 5 (18), duplication of exons 3, 4, 5, and 6, and the insertion of the 10-kb vector (17). Such rearrangements were confirmed by PCR and Southern analysis of targeted ES cells. Because of the insertion of the 10-kb vector, the transcription of the gene should be disrupted after exon 6. Even if the Csn3 transcript can be made by skipping the entire vector, by conceptual translation, duplication of exon 3 results in a frameshift and a stop codon immediately after the first exon 6. As expected, no truncated protein was observed by Western blot analysis in the heterozygous mice with Csn3 antibody directed against the entire protein, suggesting that the truncated protein probably was not formed or was immediately degraded or that the message was degraded by nonsense-mediated decay.
Targeted ES cell clones are anticipated to have an 18-kb fragment in addition to the 6.5-kb wild-type fragment after NdeI digestion and visualized with a probe from exon 5 on diagnostic Southern blot analysis (Fig. 1). Of the 192 targeted ES cell clones screened with each targeting vector, about 20% of the clones have the expected pattern on Southern blots consistent with the predicted targeting event. Three such clones, two from 5′ HPRT vector targeting and one from 3′ HPRT vector targeting, were injected into the blastocysts from C57BL/6 mice. Chimeras were produced and germ line transmission was established. F1 mice from the chimeras were backcrossed to C57BL/6 mice to obtain N2 and N3 progeny mice. The targeted allele was confirmed in mice by Southern analysis with EcoRI, which yielded the predicted 10-kb band in the 5′ HPRT vector targeted allele (Csn35′m allele) and an 18-kb band in 3′ HPRT vector targeted allele (Csn33′m allele) in addition to the 12-kb wild-type allele and/or by PCR analysis (800 bp for Csn35′m allele, 350 bp for Csn33′m allele).
Disruption of Csn3 by chromosome inversion.
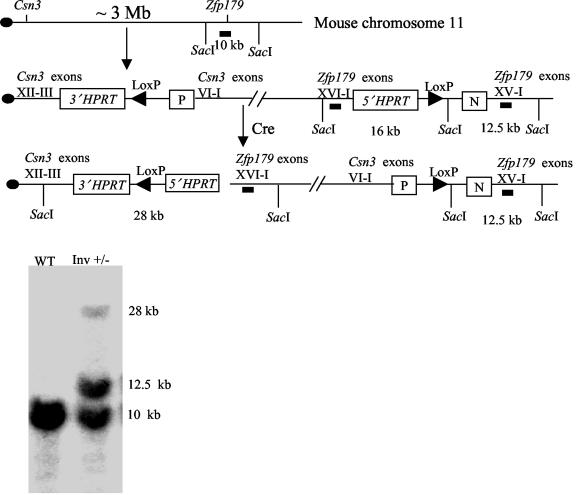
Zfp179 (47) was formerly targeted with an insertional vector from the 5′ HPRT library (48), which resulted in the duplication of the entire gene except for the last exon. Csn3 was subsequently targeted with the 3′ HPRT targeting vector in the ES cell clone containing the targeted Zfp179 gene (42). After introduction of the Cre recombinase, the entire chromosomal region between Csn3 and Zfp179 was inverted, yielding animals harboring an inv(11)17(Csn3-Zfp179) chromosome, which disrupted the Csn3 gene after exon 6 and retained the duplication of Zfp179 (Fig. 3). The inversion was confirmed by diagnostic Southern blot analysis with SacI digestion and a probe from Zfp179. Heterozygous inversion mice appeared normal. Homozygous inversion mice were absent in 32 offspring from intercrosses of the heterozygous inversion mice (Table 1), indicating embryonic lethality of the homozygous inversion. Since Zfp179 homozygous duplication mice showed no phenotype (data not shown), homozygous disruption of Csn3 most probably caused the lethality.
FIG. 3.
Construction of an inversion chromosome inv(11) (Csn3-Zfp179). Csn3 and Zfp179 are ≈3 Mb apart on mouse chromosome 11. They were targeted with the 3′ HPRT and 5′ HPRT vectors, respectively, which resulted in the duplication of exons III to VI in Csn3, duplication of exons I to XV in Zfp179, and insertion of the entire vector (42). After the introduction of Cre recombinase, the chromosome region between two LoxP sites was inverted, which was confirmed by Southern analysis with SacI and a probe from Zfp179, depicted as solid horizontal bars. P, puromycin resistance gene; N, neomycin resistance gene; WT, wild type; Inv, inversion.
TABLE 1.
Genotypes of live-born offspring from heterozygous matings
| Mating | No. of mice
|
||
|---|---|---|---|
| Wild type | Heterozygous | Total | |
| Csn35′m strain F1 × F1 | 30 | 59 | 89 |
| Csn35′m strain N2 × N2 | 9 | 21 | 30 |
| Csn33′m strain N2 × N2 | 15 | 31 | 46 |
| Csn35′m/+ N2 × Csn33′m/+ N3 | 31 | 49 | 80 |
| Total | 85 | 160 | 245 |
| Inv × Inv | 12 | 20 | 32 |
Embryonic lethality of Csn3−/− animals.
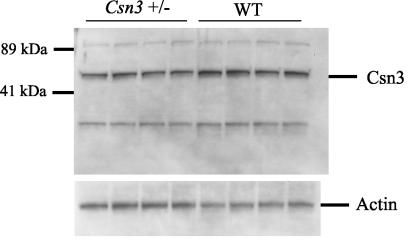
Csn35′m/+ and Csn33′m/+ mice targeted with either the 5′ or 3′ HPRT targeting vector appeared normal in comparison to their wild-type littermates. They also had normal fertility and body weight. Western blot analysis showed that the Csn3 protein level in both strains of mice was reduced by half (Fig. 4), confirming that the Csn3 gene was disrupted.
FIG. 4.
Csn3 protein level is reduced in heterozygous mice. Total protein (10 μg) from the livers of Csn35′m/+ and wild-type (WT) littermates was electrophoresed and probed with anti-Csn3 antibody. Actin was used as a loading control. Proteins are indicated on the right side and sizes on the left. The Csn3 protein is 48 kDa. Other bands represent cross-reactivity to anti-Csn3 antibody or higher-order protein complexes that include Csn3.
To discriminate the homozygous and the heterozygous mice, BspHI was used for the Csn35′m strain mice, which is predicted to yield 23-kb, 21.5-kb, and 10.5-kb fragments in heterozygous mice and to lose the 23-kb wild-type fragment in homozygous mice. AseI was used for the Csn33′m strain mice and is predicted to yield 8-kb, 8.5-kb, and 14-kb fragments in the heterozygous mice and 8.5-kb and 14-kb fragments for the homozygous mice. No PCR primers could be made to discriminate the homozygous and heterozygous embryos during early development (before 8.5 dpc) within a single allele because of the duplication of the genomic region. To analyze the consequence of disruption in homozygous Csn3 embryos, the two alleles were used to generate compound heterozygous mice. Homozygous mice from matings between the Csn35′m/+ and Csn33′m/+ mice have fragments from both alleles detected by PCR and Southern analysis with EcoRI digestion, while the heterozygous animal should have only one (Fig. 1). Of the 245 mice born from different heterozygous intercrosses (5′ by 5′; 3′ by 3′; and 5′ by 3′), no live-born homozygous mice were detected. The ratio of heterozygous mice to wild-type mice was 1.9 to 1 (Table 1). These data suggest that the Csn3−/− mutants die as embryos.
Phenotype of Csn3−/− mice in early embryogenesis.
To determine the time of developmental arrest during embryogenesis, timed matings were performed with crosses between Csn35′m/+ and Csn33′m/+ mice (Table 2; Fig. 5). From 9.5 to 11.5 dpc, 41 embryos were examined, 29 of which (71%) were morphologically normal. Of these 29 embryos, 21 were heterozygous and were indistinguishable from the wild-type littermates. No homozygous embryos were observed. The remaining 12 embryos were being resorbed at these stages, and DNA could not be recovered for genotyping. At 8.5 dpc, 8 out of 31 embryos examined were abnormal, 7 of which were starting to be resorbed. One Csn3−/− homozygous embryo was detected which had developed a head anlage, but this was much smaller than normal. It appeared transparent and did not have other structures that should have developed at that time (e.g., somites) (data not shown). Since we only observed one such embryo, it probably represents an exception. Twenty-three 7.5 dpc embryos were examined. Seven were homozygous, and each was one quarter to one fifth of the size of the normal embryos. They had an elongated shape similar to the normal embryos but appeared to be arresting prior to gastrulation (Fig. 5A and B). Therefore, Csn3−/− embryos arrested development before 8.5 dpc and were mostly resorbed by 8.5 dpc.
TABLE 2.
Genotypes of embryos from heterozygous matings
| Time (dpc) | No. of embryos
|
||||
|---|---|---|---|---|---|
| Wild type | Heterozygous | Homozygous | Othera | Total | |
| 7.5 | 5 | 10 | 7 | 1 | 23 |
| 8.5 | 7 | 16 | 1 | 7 | 31 |
| 9.5-11.5 | 8 | 21 | 0 | 12 | 41 |
Embryos resorbed or genotype not determined.
FIG. 5.
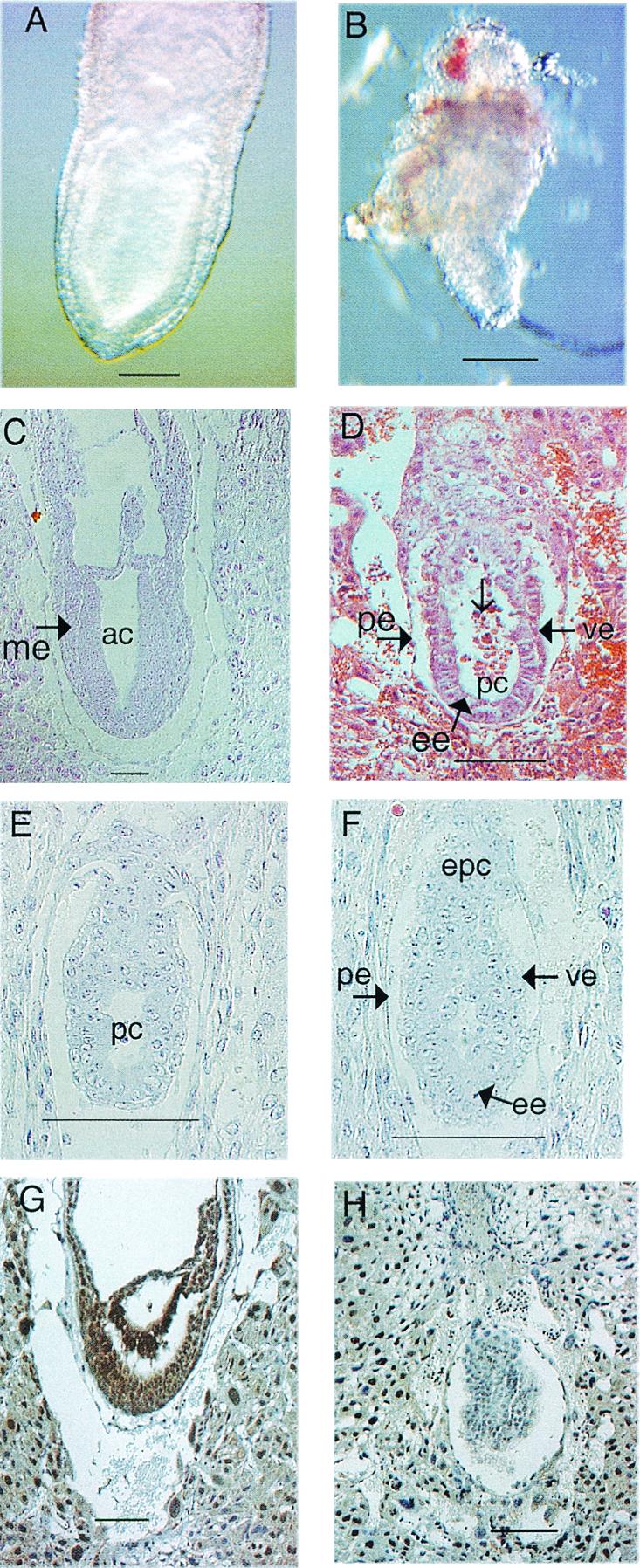
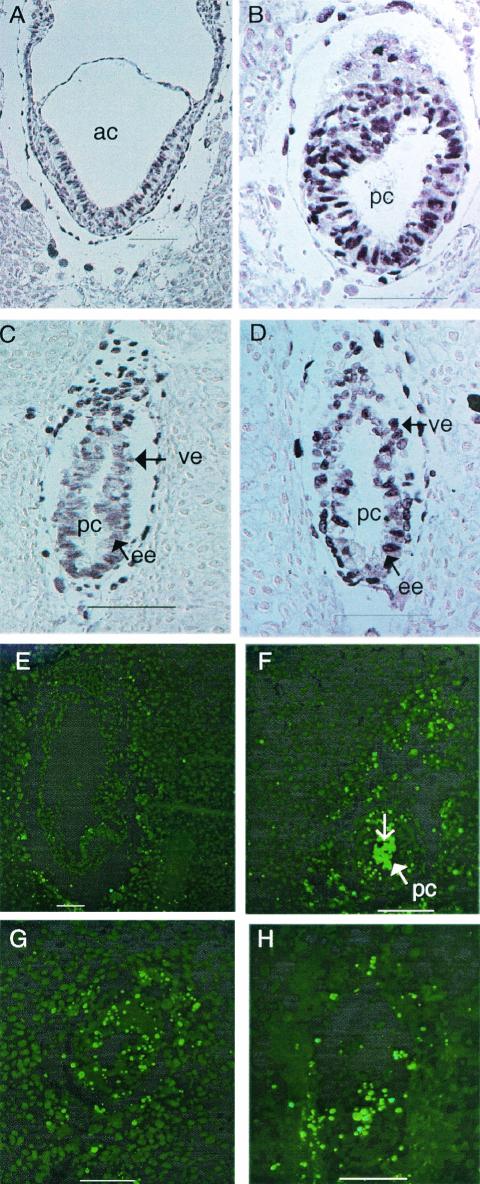
Morphology and histological analysis of normal and mutant embryos from 5.5 dpc to 7.5 dpc. (A and B) Morphology of normal (A) and homozygous Csn3−/− (B) 7.5 dpc embryos. (C and D) Hematoxylin- and eosin-stained normal (C) and abnormal (D) 7.5 dpc embryos. The normal embryo has undergone gastrulation and has mesoderm (me). The abnormal embryo still has two layers of cells. No mesoderm was observed. Note the disorganized epiblast cells (ee) and seemingly dead cells inside the proamniotic cavity (pc) (open arrow). (E and F) Hematoxylin- and eosin-stained 5.5 dpc normal (E) and abnormal (F) embryos. (G and H) Immunohistological analysis of 7.5 dpc embryos with Csn3 antibody. (G) Normal embryo; (H) mutant in which there is some background staining but no real nuclear staining as seen in panel G. ac, amniotic cavity; ee, embryonic ectoderm (or epiblast cells); epc, ectoplacental cone; pc, proamniotic cavity; pe, parietal endoderm; ve, visceral endoderm. Bars, 100 μm.
To further investigate the phenotype of homozygous mice, histological analyses were performed on 5.5, 6.5, and 7.5 dpc embryos (Fig. 5, 7, and 9; Table 3). At 5.5 dpc, eight mutants defined by visual abnormalities were similar to their normal littermates in size and shape. They had morphologically normal visceral and parietal endoderm, and the ectoplacental cone was forming. However, the epiblast cells were slightly disorganized, and the proamniotic cavity was not obvious in comparison to the normal embryos (Fig. 5E and F). At 6.5 dpc, 10 abnormal embryos were detected out of 61 embryos examined. These exhibited the expected egg cylinder shape but were slightly growth retarded. While the abnormal embryos exhibited two germ layers, the number of epiblast cells appeared to be greatly reduced and the cells were disorganized. The proamniotic cavity in the 6.5 dpc abnormal embryos was more obvious compared to the abnormal embryos at 5.5 dpc, and dead cells could be detected (see Fig. 7C and D and 9A and B). At 7.5 dpc, normal embryos should have gastrulated and formed mesoderm, but 13 out of 77 embryos were much smaller and still consisted of two layers of cells as seen in the 6.5 dpc embryos, and there was no indication of mesoderm formation. Compared to the presumptive mutants at 6.5 dpc, those at 7.5 dpc were bigger, with more epiblast cells; more dead cells were found in the proamniotic cavity, and cells were more disorganized (Fig. 5C and D). Resorbed embryos were also found at various stages.
FIG. 7.
Bromodeoxyuridine and TUNEL analysis. (A, B, C, and D) Bromodeoxyuridine labeling of normal (A and C) and mutant (B and D) 7.5 dpc (A and B) and 6.5 dpc (C and D) embryos. (E, F, G, and H) TUNEL staining of 7.5 (E, F, and G) and 6.5 dpc (H) embryos. There is almost no signal in the normal control embryo (E). (F, G, and H) Mutants. Note strong staining inside the proamniotic cavity (open arrow). Embryos in panels A and B are from the same litter, as are those in panels E and F. ac, amniotic cavity; ee, embryonic ectoderm (or epiblast cells); pc, proamniotic cavity; ve, visceral endoderm. Bars, 100 μm.
FIG. 9.
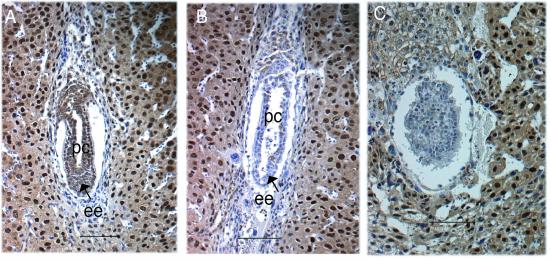
Immunohistological analysis with Csn8 antibody. (A) A 6.5 dpc normal embryo. (B and C) Mutants at 6.5 dpc (B) and 7.5 dpc (C). Panels A and B show embryos from the same litter. Panel C and panel H in Fig. 5 show different sections of the same embryo. ee, embryonic ectoderm (or epiblast cells); pc, proamniotic cavity. Bars, 100 μm.
TABLE 3.
Histological analysis of 5.5 dpc to 7.5 dpc embryosa
| Time (dpc) | No. of embryos
|
||||||
|---|---|---|---|---|---|---|---|
| Morphology
|
Genotype
|
Total | |||||
| Abnormal | Normal | Other | −/− | +/− or +/+ | Other | ||
| 5.5 | 4 | 11 | 1 | 4 | 13 | 1 | 34 |
| 6.5 | 4 | 19 | 1 | 6 | 31 | 0 | 61 |
| 7.5 | 8 | 49 | 3 | 5 | 12 | 0 | 77 |
| Total | 16 | 79 | 5 | 15 | 56 | 1 | 172 |
Morphological analysis was carried out for some embryos but the genotype was not determined. Genotype was determined by immunohistological analysis. Other, embryo resorbed or genotype not determined.
Immunohistology analysis was carried out to genotype the embryos with an anti- Csn3 antibody, which mainly stains the nucleus. The Csn3 protein was expressed ubiquitously in the 5.5, 6.5, and 7.5 dpc wild-type embryos, indicating the importance of this gene in early embryonic development (Fig. 5G and data not shown). In the mutants, Csn3 antibody appeared to have some background staining, likely resulting from cross-reactivity (Fig. 4), but the nonspecific staining appeared quantitatively different from the specific nuclear staining (Fig. 5G and H). In total, eight litters were genotyped. Abnormal embryos were confirmed to be the homozygous mutants.
In order to assess the effects of homozygous disruption of Csn3 on preimplantation embryos, immunohistochemistry with anti-Csn3 antibody was performed on 3.5 dpc blastocysts. Fourteen blastocysts were stained, two of which were confirmed to be mutants by PCR (data not shown). The mutants were normal in morphology, which was expected because there was only a slight difference between the mutants and normal embryos at 5.5 dpc. The Csn3 staining was indistinguishable between the mutant and the wild-type blastocysts, consistent with maternal protein deposition in the blastocysts.
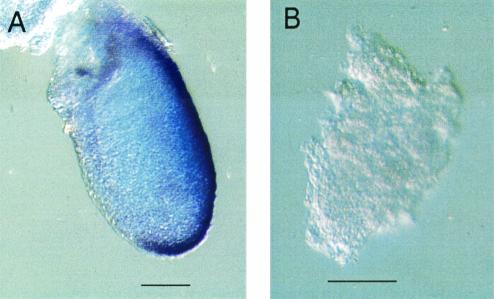
A distinct and obvious mesoderm layer was apparently not formed in Csn3−/− embryos, but the disorganized epiblast cells may have obscured the discrimination of mesodermal cells. In order to examine mesoderm development in the mutants, the expression pattern of the Brachyury gene was evaluated. Brachyury is one of the earliest markers for mesodermal cells. At 7.5 dpc it is expressed in the primitive streak, in which the mesoderm formation begins, in the node, and in the notochordal plate extending anteriorly from the node (9, 19). Normal embryos showed the expected expression pattern, whereas the Csn3−/− mutant embryos showed no Brachyury expression (Fig. 6). This indicates that the primitive streak and the mesoderm were not formed in the Csn3−/− embryos, consistent with our histological observations. In summary, Csn3−/− mice arrested at early embryonic development after 5.5 dpc and fail to undergo gastrulation.
FIG. 6.
No Brachyury expression in 7.5 dpc Csn3−/− mutant embryos. (A) Normal embryo. (B) Csn3−/− embryo. Bars, 100 μm.
Apoptotic cell death in Csn3−/− embryos.
The failure of the homozygous embryos to develop could result from increased cell death, decreased proliferation, or both. To investigate these possibilities, TUNEL staining and bromodeoxyuridine labeling were performed (Fig. 7). TUNEL detects nuclear fragmentation, which is one of the characteristic features of programmed cell death. In normal embryos, cell death was rarely detected, while in four mutants at 6.5 or 7.5 dpc, death of epiblast cells was greatly increased. Consistent with the histological observations, those cells inside the proamniotic cavity were confirmed to have undergone apoptotic cell death by TUNEL assay (Fig. 7E, F, G, and H). Six embryos at 6.5 or 7.5 dpc were subjected to bromodeoxyuridine analysis. The percentage of cells that incorporated bromodeoxyuridine was similar between wild-type embryos and the mutants (Fig. 7A, B, C, and D), and therefore, it is likely that the arrest of embryonic development was mainly because of increased cell death in the epiblast.
Blastocysts outgrowth studies.
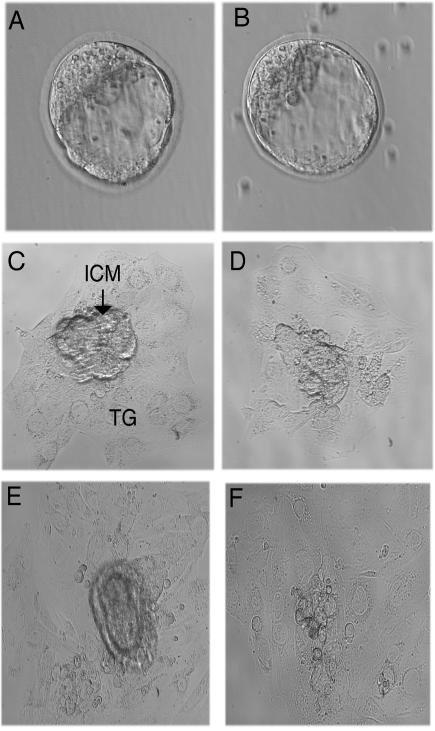
As an alternative approach to investigate postimplantation development, we examined blastocyst outgrowth in vitro. In total, 11 blastocysts were examined, of which two were documented to be Csn3−/− mutants by genotyping. The blastocysts of the mutants were morphologically normal (Fig. 8). After 3 days of culture, the inner cell mass (ICM) growth of the mutants was comparable to that of the normal littermates. The number of ICM cells was greatly reduced after 5 to 6 days of culture in the mutants. The trophoblastic cells grew similarly in both mutant and normal embryos. It appears that the ICM initially developed and then there was reduced cell growth. These findings are consistent with our in vivo observations that epiblast cells proliferate normally until 6.5 dpc in Csn3−/− embryos, but the cell number is reduced secondary to cell death.
FIG. 8.
Outgrowth of blastocysts in vitro. Morphologically abnormal blastocysts were not observed in 33 blastocysts. Wild-type blastocyst (A) and Csn3−/− mutant blastocyst (B) are morphologically similar. After 3 days of culture, the inner cell mass (ICM) developed in both mutant and wild-type embryos (C and D). After 5 days of culture, the ICM of wild-type embryos developed more in comparison to the 3-day culture (E), while the ICM of mutant embryos in the 6-day culture appeared greatly reduced in cell number (F). TG, trophoblast giant cells. Magnification: 400× for blastocysts; 200× for cultures.
Decreased CSN stability in Csn3−/− mice.
Since the disruption of individual CSN subunits was reported to disrupt the entire complex in plants (44), we examined embryos for the expression of the Csn8 subunit of CSN. In normal embryos, the expression pattern of Csn8 was similar to that of Csn3, as determined by anti-Csn8 antibody staining (Fig. 9A and data not shown). Both were expressed ubiquitously at 5.5, 6.5, and 7.5 dpc and mainly associated with the nucleus. In the Csn3−/− mice, Csn8 was not detected (Fig. 9B and C), an observation consistent with disruption of the entire CSN complex.
DISCUSSION
The COP9 signalosome (CSN) is an eight-subunit complex. Mutations in different subunits, including the third subunit in A. thaliana, all result in severely retarded seedling development and lethality after the seedling stage (25). In D. melanogaster, disruption of csn4 and csn5 caused lethality at the late larval stage, with a maternal contribution (30). Obviously, CSN and its subunits are important for early development in both plants and D. melanogaster. Here we demonstrate that Csn3 is essential for early mouse embryonic development.
In D. melanogaster, maternal Csn4 or Csn5 protein deposited in the csn4 or csn5 null embryos supported embryonic development until the late larval stage. Without maternally contributed proteins, oogenesis was arrested (30). In mice, mutant embryos can grow with maternal deposition (40), but maternal RNAs are usually degraded by 6 dpc (36), and embryos after that stage require at least some de novo gene expression (20). In Csn3−/− blastocysts, the Csn3 staining was similar to that in the normal embryos, which indicates that there is maternal protein deposition. At 5.5 dpc, the Csn3 protein level was greatly reduced, Csn3−/− showed slight defects at 5.5 dpc and grew through 6.5 and 7.5 dpc. It is difficult to determine whether the survival to this stage can be entirely attributed to a maternal effect or whether the Csn3 protein requirements for development are less before 6.5 dpc because of the slower rate of cell proliferation compared to that after 6.5 dpc, or perhaps a combination of both possibilities.
Two biochemical functions have been found for the CSN. One is the protein kinase function, and the other is controlling the E3 ubiquitin ligases, through which CSN controls the degradation of a wide variety of proteins, including those involved in the cell signaling pathway, such as c-Jun, and those in cell cycle control, such as p53 and p27kip1 (3, 37, 41). Microinjection of the CSN complex into HeLa cells inhibited p27kip1 degradation and impeded G1-S phase progression (46). Obviously, CSN controls cell growth and proliferation, but how disruption of the complex will affect cells is largely unknown.
In Csn3−/− mice, bromodeoxyuridine incorporation was similar to that in wild-type mice. However, cell death appeared to play a major role in the developmental arrest of the Csn3−/− mice. The cell death may be explained by the accumulation of molecules involved in the apoptotic pathway such as p53 or the reduction of certain factors important for the survival of the cell. Recently, it was demonstrated that Csn5 is important for the repair of DNA double-strand breaks, and its absence during meiosis activates a DNA damage checkpoint in D. melanogaster (12). In mammalian cells, p53 responds to DNA damage by delaying the cell cycle or inducing programmed cell death (22, 24). To elucidate whether the cell death in Csn3−/− mice is induced by double-strand breaks requires a further understanding of CSN function. Epiblast cells undergo rapid cell division just prior to gastrulation, which may explain why they were primarily affected.
So far, many genes have been found to be important for early embryonic development. Fliih and Cgbp were essential for development at implantation (4.5 dpc) (5, 6). Hnf-4 homozygous mice showed defects at 7.5 dpc (9). The phenotypes of our mutants are more similar to those of Rad51 homozygous knockout mice, which also start to show defects at 5.5 dpc and are resorbed by 8.5 dpc (26). Both mutants formed the proamniotic cavity, failed to go through gastrulation, and had increased cell death in the epiblast. However, decreased cell proliferation was more obvious in Rad51−/− embryos than in our Csn3−/− animals. Interestingly, mouse Rad51 is homologous to Rad51 in Saccharomyces cerevisiae, which repairs double-strand breaks by recombination. Mouse Rad51 may also be associated with double-strand break repair because trophectoderm-derived cells from homozygous mutants were sensitive to gamma radiation, and the mutant embryos lived longer and developed further in a p53 mutant background. It will be interesting to see if there is any relation between CSN and Rad51.
In Arabidopsis thaliana, mutations in most of the CSN subunits, including CSN3, can lead to the failure of CSN complex formation (31, 44). CSN3 has previously been demonstrated to interact directly with CSN8 (15). In our case, Csn8 was undetectable in the Csn3−/− embryos, suggesting that the entire complex was disrupted. As a result, at least certain functions of the complex, such as the SCF deneddylation activity that requires the entire complex (10), were most likely to be deficient in the Csn3−/− cells. Csn4 and Csn5 were both found not only in the CSN complex but also in other forms in D. melanogaster, and in csn5 null mutants the complex was formed (23, 30). It appears that some of the subunits have their own functions independent of the CSN complex. Overexpression of Csn5 promoted the degradation of p27kip1 in mammalian cells (41), while high levels of CSN complex inhibited p27kip1 degradation (46). The human CSN3 was also found in two to three larger complexes than the CSN complex (38). It is unknown if those larger complexes include CSN. It is likely that the phenotype observed in the Csn3−/− mice resulted from the disruption of both the CSN complex and the Csn3-specific functions.
In addition to an important role in early embryonic development, Csn3 may be required in later development and in different tissue functions. Reduction-of-function lines of CSN3 in A. thaliana showed diverse developmental defects (31), and Northern analysis showed ubiquitous expression in adult tissues of mice. To elucidate the role played by Csn3 in later development and in different organ systems, conditional knockout mice will be required.
Smith-Magenis syndrome is caused by a microdeletion on 17p11.2, and clinical manifestations include mental retardation, skeletal abnormalities, sleep disorder, and neurobehavioral anomalies (16). The phenotype is thought to result from the haploinsufficiency of one or more genes. Since heterozygous Csn3 knockout mice showed no obvious phenotypes, it may not be the gene responsible for the Smith-Magenis syndrome phenotypes. Previously, Csn3 was speculated to be possibly responsible for the sleep disorder because it controls light-mediated development in plants (33). Sleep studies in heterozygous Df(11) (Csn3-Zfp179) mice (42) are ongoing.
Another important reagent developed in our study was an inv(11)17(Csn3-Zfp179) inversion chromosome. Balancer chromosomes are used widely in D. melanogaster for stock maintenance and mutagenesis screens (2). Recently, a balancer chromosome was introduced into mouse genetics and used for a large-scale mouse mutagenesis screen (21, 49). The ≈3-Mb inversion covering the Smith-Magenis syndrome mouse syntenic region may be used to maintain individual mutations in genes that lie within this interval.
In conclusion, we demonstrated the importance of Csn3 in CSN complex assembly and its essential role in early mouse embryonic development. It will be interesting to investigate further the potential relationships between CSN and cell death and CSN and double-strand break repair and other molecules involved in double-strand break response and repair, such as Rad51. This may enable new insight into CSN function.
Acknowledgments
We thank Kathryn Hentges for assistance with embryo analysis and Dena Mansouri for technical support.
This work was supported in part by a grant from the National Cancer Institute, NIH (PO1CA75719).
REFERENCES
- 1.Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne, P. G. Amanatides, S. E. Scherer, P. W. Li, R. A. Hoskins, R. F. Galle, et al. 2000. The genome sequence of Drosophila melanogaster. Science 287:2185-2195. [DOI] [PubMed] [Google Scholar]
- 2.Ashburner, M. 1989. Drosophila: a Laboratory handbook. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 3.Bech-Otschir, D., R. Kraft, X. Huang, P. Henklein, B. Kapelari, C. Pollmann, and W. Dubiel. 2001. COP9 signalosome-specific phosphorylation targets p53 to degradation by the ubiquitin system. EMBO J. 20:1630-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bi, W., J. Yan, P. Stankiewicz, S.-S. Park, K. Walz, C. F. Boerkoel, L. Potocki, L. G. Shaffer, K. Devriendt, M. J. M. Nowaczyk, K. Inoue, and J. R. Lupski. 2002. Genes in a refined Smith-Magenis syndrome critical deletion interval on chromosome 17p11.2 and the syntenic region of the mouse. Genome Res. 12:713-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, H. D., S. Fountain, I. S. McLennan, L. A. Berven, M. F. Crouch, D. A. Davy, J. A. Hooper, K. Waterford, K.-S. Chen, J. R. Lupski, B. Ledermann, I. G. Young, and K. I. Matthaei. 2002. Fliih, a gelsolin-related cytoskeletal regulator essential for early mammalian embryonic development. Mol. Cell. Biol. 22:3518-3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlone, D. L., and D. G. Skalnik. 2001. CpG binding protein is crucial for early embryonic development. Mol. Cell. Biol. 21:7601-7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chamovitz, D. A., N. Wei, M. T. Osterlund, A. G. von Arnim, J. M. Staub, M. Matsui, and X.-W. Deng. 1996. The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 86:115-121. [DOI] [PubMed] [Google Scholar]
- 8.Chen, K.-S., P. Manian, T. Koeuth, L. Potocki, Q. Zhao, A. C. Chinault, C. C. Lee, and J. R. Lupski. 1997. Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat. Genet. 17:154-163. [DOI] [PubMed] [Google Scholar]
- 9.Chen, W. S., K. Manova, D. C. Weinstein, S. A. Duncan, A. S. Plump, V. R. Prezioso, R. F. Bachvarova, and J. E. Darnell, Jr. 1994. Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev. 8:2466-2477. [DOI] [PubMed] [Google Scholar]
- 10.Cope, G. A., G. S. B. Suh, L. Aravind, S. E. Schwarz, S. L. Zipursky, E. V. Koonin, and R. J. Deshaies. 2002. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 298:608-611. [DOI] [PubMed] [Google Scholar]
- 11.Deng, X.-W., W. Dubiel, N. Wei, K. Hofmann, K. Mundt, J. Colicelli, J. Kato, M. Naumann, D. Segal, M. Seeger, M. Glickman, D. A. Chamovitz, and A. Carr. 2000. Unified nomenclature for the COP9 signalosome and its subunits: an essential regulator of development. Trends Genet. 16:202-203. [DOI] [PubMed] [Google Scholar]
- 12.Doronkin, S., I. Djagaeva, and S. K. Beckendorf. 2002. CSN5/Jab1 mutations affect axis formation in the Drosophila oocyte by activating a meiotic checkpoint. Development 129:5053-5064. [DOI] [PubMed] [Google Scholar]
- 13.Elsea, S. H., K. Mykytyn, K. Ferrell, K. L. Coulter, P. Das, W. Dubiel, P. I. Patel, and J. E. Metherall. 1999. Hemizygosity for the COP9 signalosome subunit gene, SGN3, in the Smith-Magenis syndrome. Am. J. Med. Genet. 87:342-348. [PubMed] [Google Scholar]
- 14.Freilich, S., E. Oron, Y. Kapp, Y. Nevo-Caspi, S. Orgad, D. Segal, and D. A. Chamovitz. 1999. The COP9 signalosome is essential for development of Drosophila melanogaster. Curr. Biol. 9:1187-1190. [DOI] [PubMed] [Google Scholar]
- 15.Fu, H., N. Reis, Y. Lee, M. H. Glickman, and R. D. Vierstra. 2001. Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. EMBO J. 20:7096-7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenberg, F., V. Guzzetta, R. Montes de Oca-Luna, R. E. Magenis, A. C. M. Smith, S. F. Richter, I. Kondo, W. B. Dobyns, P. I. Patel, and J. R. Lupski. 1991. Molecular analysis of the Smith-Magenis syndrome: a possible contiguous-gene syndrome associated with del(17) (p11.2). Am. J. Hum. Genet. 49:1207-1218. [PMC free article] [PubMed] [Google Scholar]
- 17.Hasty, P., J. Rivera-Pérez, C. Chang, and A. Bradley. 1991. Target frequency and integration pattern for insertion and replacement vectors in embryonic stem cells. Mol. Cell. Biol. 11:4509-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasty, P., J. Rivera-Pérez, and A. Bradley. 1995. Gene conversion during vector insertion in embryonic stem cells. Nucleic Acids Res. 23:2058-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herrmann, B. G. 1991. Expression pattern of the Brachyury gene in whole-mount TWis/TWis mutant embryos. Development. 113:913-917. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, M. H. 1981. The molecular and cellular basis of preimplantation mouse development. Biol. Rev. Camb. Phil. Soc. 56:463-498. [DOI] [PubMed] [Google Scholar]
- 21.Justice, M. J., J. K. Noveroske, J. S. Weber, B. Zheng, and A. Bradley. 1999. Mouse ENU mutagenesis. Hum. Mol. Genet. 8:1955-1963. [DOI] [PubMed] [Google Scholar]
- 22.Kastan, M. B., O. Onyekwere, D. Sidransky, B. Vogelstein, and R. W. Craig. 1991. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 51:6304-6311. [PubMed] [Google Scholar]
- 23.Kim, T.-H., K. Hofmann, A. G. von Arnim, and D. A. Chamovitz. 2001. PCI complexes: pretty complex interactions in diverse signaling pathways. Trends Plant Sci. 6:379-386. [DOI] [PubMed] [Google Scholar]
- 24.Kuerbitz, S. J., B. S. Plunkett, W. V. Walsh, and M. B. Kastan. 1992. Wild-type p53 is a cell cycle checkpoint determinant following irradiation. Proc. Natl. Acad. Sci. USA 89:7491-7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwok, S. F., R. Solano, T. Tsuge, D. A. Chamovitz, J. R. Ecker, M. Matsui, and X.-W. Deng. 1998. Arabidopsis homologs of a c-Jun coactivator are present both in monomeric form and in the COP9 complex, and their abundance is differentially affected by the pleiotropic cop/det/fus mutations. Plant Cell 10:1779-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim, D.-S., and P. Hasty. 1996. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol. 16:7133-7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyapina, S., G. Cope, A. Shevchenko, G. Serino, T. Tsuge, C. Zhou, D. A. Wolf, N. Wei, A. Shevchenko, and R. J. Deshaies. 2001. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 292:1382-1385. [DOI] [PubMed] [Google Scholar]
- 28.Matzuk, M. M., M. J. Finegold, J.-G. J. Su, A. J. W. Hsueh, and A. Bradley. 1992. α-inhibin is a tumour-suppressor gene with gonadal specificity in mice. Nature 360:313-319. [DOI] [PubMed] [Google Scholar]
- 29.Mundt, K. E., C. Liu, and A. M. Carr. 2002. Deletion mutants in COP9/signalosome subunits in fission yeast Schizosaccharomyces pombe display distinct phenotypes. Mol. Biol. Cell 13:493-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oron, E., M. Mannervik, S. Rencus, O. Harari-Steinberg, S. Neuman-Silberberg, D. Segal, and D. A. Chamovitz. 2002. COP9 signalosome subunits 4 and 5 regulate multiple pleiotropic pathways in Drosophila melanogaster. Development 129:4399-4409. [DOI] [PubMed] [Google Scholar]
- 31.Peng, Z., G. Serino, and X.-W. Deng. 2001. A role of Arabidopsis COP9 signalosome in multifaceted developmental processes revealed by the characterization of its subunit 3. Development 128:4277-4288. [DOI] [PubMed] [Google Scholar]
- 32.Potocki, L., K.-S. Chen, and J. R. Lupski. 1999. Subunit 3 of the COP9 signal transduction complex is conserved from plants to humans and maps within the Smith- Magenis syndrome critical region in 17p11.2. Genomics 57:180-182. [DOI] [PubMed] [Google Scholar]
- 33.Potocki, L., D. Glaze, D.-X. Tan, S.-S. Park, C. D. Kashork, L. G. Shaffer, R. J. Reiter, and J. R. Lupski. 2000. Circadian rhythm abnormalities of melatonin in Smith-Magenis syndrome. J. Med. Genet. 37:428-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Probst, F. J., K.-S. Chen, Q. Zhao, A. Wang, T. B. Friedman, J. R. Lupski, and S. A. Camper. 1999. A physical map of the mouse shaker-2 region contains many of the genes commonly deleted in Smith-Magenis syndrome(del17p11.2p11.2). Genomics 55:348-352. [DOI] [PubMed] [Google Scholar]
- 35.Ramírez-Solis, R., A. C. Davis, and A. Bradley. 1993. Gene targeting in embryonic stem cells. Methods Enzymol. 225:855-878. [DOI] [PubMed] [Google Scholar]
- 36.Sawicki, J. A., T. Magnuson, and C. J. Epstein. 1981. Evidence for expression of the paternal genome in the two-cell mouse embryo. Nature 294:450-451. [DOI] [PubMed] [Google Scholar]
- 37.Schwechheimer, C., and X.-W. Deng. 2001. COP9 signalosome revisited: a novel mediator of protein degradation. Trends Cell Biol. 11:420-426. [DOI] [PubMed] [Google Scholar]
- 38.Seeger, M., R. Kraft, K. Ferrell, D. Bech-Otschir, R. Dumdey, R. Schade, C. Gordon, M. Naumann, and W. Dubiel. 1998. A novel protein complex involved in signal transduction possessing similarities to 26S proteasome subunits. FASEB J. 12:469-478. [PubMed] [Google Scholar]
- 39.Seeger, M., C. Gordon, and W. Dubiel. 2001. Protein stability: the COP9 signalosome gets in on the act. Curr. Biol. 11:R643-646. [DOI] [PubMed] [Google Scholar]
- 40.Shen-Li, H., R. C. O'Hagan, H. Jr. Hou, J. W. II. Horner, H.-W. Lee, and R. A. DePinho. 2000. Essential role for Max in early embryonic growth and development. Genes Dev. 14:17-22. [PMC free article] [PubMed] [Google Scholar]
- 41.Tomoda, K., Y. Kubota, and J. Kato. 1999. Degradation of the cyclin-dependent- kinase inhibitor p27Kip1 is instigated by Jab1. Nature 398:160-165. [DOI] [PubMed] [Google Scholar]
- 42.Walz, K., S. Caratini-Rivera, W. Bi, P. Fonseca, D. L. Mansouri, J. Lynch, H. Vogel, J. L. Noebels, A. Bradley, and J. R. Lupski. 2003. Modeling del(17) (p11.2p11.2) and dup(17) (p11.2p11.2) contiguous gene syndromes by chromosome engineering in mice: phenotypic consequences of gene dosage imbalance. Mol. Cell. Biol. 23:3646-3655. [DOI] [PMC free article] [PubMed]
- 43.Wei, N., T. Tsuge, G. Serino, N. Dohmae, K. Takio, M. Matsui, and X.-W. Deng. 1998. The COP9 complex is conserved between plants and mammals and is related to the 26S proteasome regulatory complex. Curr. Biol. 8:919-922. [DOI] [PubMed] [Google Scholar]
- 44.Wei, N., and X.-W. Deng. 1999. Making sense of the COP9 signalosome. A regulatory protein complex conserved from Arabidopsis to human. Trends Genet. 15:98-103. [DOI] [PubMed] [Google Scholar]
- 45.Wilkinson, D. G. 1992. Whole mount in situ hybridization of vertebrate embryos, p. 75-83. In D. G. Wilkinson (ed.), In situ hybridization: a practical approach. Oxford University Press, Oxford, UK.
- 46.Yang, X., S. Menon, K. Lykke-Andersen, T. Tsuge, D. Xiao, X. Wang, R. J. Rodriguez-Suarez, H. Zhang, and N. Wei. 2002. The COP9 signalosome inhibits p27kip1 degradation and impedes G1-S phase progression via deneddylation of SCF Cul1. Curr. Biol. 12:667-672. [DOI] [PubMed] [Google Scholar]
- 47.Zhao, Q., K.-S. Chen, B. A. Bejjani, and J. R. Lupski. 1998. Cloning, genomic structure, and expression of mouse ring finger protein gene, Znf179. Genomics 49:394-400. [DOI] [PubMed] [Google Scholar]
- 48.Zheng, B., A. A. Mills, and A. Bradley. 1999. A system for rapid generation of coat color-tagged knockouts and defined chromosomal rearrangements in mice. Nucleic Acids Res. 27:2354-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng, B., M. Sage, W.-W. Cai, D. M. Thompson, B. C. Tavsanli, Y.-C. Cheah, and A. Bradley. 1999. Engineering a mouse balancer chromosome. Nat. Genet. 22:375-378. [DOI] [PubMed] [Google Scholar]