Abstract
The receptor protein tyrosine phosphatase (PTPase) Dlar has an ectodomain consisting of three immunoglobulin (Ig)-like domains and nine fibronectin type III (FnIII) repeats and a cytoplasmic domain consisting of two PTPase domains, membrane-proximal PTP-D1 and C-terminal PTP-D2. A series of mutant Dlar transgenes were introduced into the Drosophila genome via P-element transformation and were then assayed for their capacity to rescue phenotypes caused by homozygous loss-of-function genotypes. The Ig-like domains, but not the FnIII domains, are essential for survival. Conversely, the FnIII domains, but not the Ig-like domains, are required during oogenesis, suggesting that different domains of the Dlar ectodomain are involved in distinct functions during Drosophila development. All detectable PTPase activity maps to PTP-D1 in vitro. The catalytically inactive mutants of Dlar were able to rescue Dlar−/− lethality nearly as efficiently as wild-type Dlar transgenes, while this ability was impaired in the PTP-D2 deletion mutants DlarΔPTP-D2 and Dlarbypass. Dlar-C1929S, in which PTP-D2 has been inactivated, increases the frequency of bypass phenotype observed in Dlar−/− genotypes, but only if PTP-D1 is catalytically active in the transgene. These results indicate multiple roles for PTP-D2, perhaps by acting as a docking domain for downstream elements and as a regulator of PTP-D1.
Protein tyrosine phosphorylation is a critically important posttranslational modification in the eukaryotic signal transduction pathways that regulate cell growth, differentiation, and development. The phosphorylation status of a given tyrosine residue is regulated by protein tyrosine kinases (PTKs) and protein tyrosine phosphatases (PTPases). The essential role of PTKs in these pathways is now well established; misregulation of PTKs can have dire consequences, including cell death, cell transformation, and tumorigenesis (see reference 8 for a review). Far less is understood about the functions of PTPases in signal transduction, especially for the receptor-like PTPases (RPTPs). While individual members of RPTP signal transduction pathways, including the kinase Abl and its substrate Enabled, have been identified genetically (50), physiologically relevant substrates and downstream signal transduction elements of most RPTPs are unknown.
The PTPase gene family is large and varied (see reference 34 for a review). LAR, Dlar, PTPδ, and PTPσ comprise a subfamily of RPTPs that have ectodomains consisting of N-terminal immunoglobulin (Ig)-like domains and membrane-proximal fibronectin type III (FnIII) repeat motifs and cytoplasmic regions consisting of two tandemly repeated PTPase-like domains (29, 37, 38, 42, 44). One of the FnIII repeat units of human LAR has been implicated in binding to the extracellular matrix (ECM) complex of laminin and nidogen (36), and Dlar cooperates with integrins to regulate actin polymerization during oogenesis by integrating signals from the ECM (3). So far, no physiologically relevant ligand has been found that binds to the Ig-like domains of mammalian LAR family members, despite the fact that Ig-like domains are typically ligand binding domains. Chicken PTPσ has recently been shown to bind to heparan sulfates in vitro (2), an interaction that requires both Ig-like domains and FnIII domains, but that finding has not yet been extended to mammalian LAR family members, and it is uncertain whether these proteoglycans are themselves ligands or function in a support role in ligand presentation (2). Known ligands of RPTPs outside of the LAR subfamily are only slightly better understood. RPTPμ and RPTPκ have been implicated in homotypic adhesion events (9, 20, 51), and RPTPζ has been shown to bind the cytokine pleiotropin, an interaction that is inhibitory to RPTPζ catalytic activity (33). It is likely that other physiologically relevant ligands for all of these RPTPs still remain to be discovered.
The PTPase domains of the LAR subfamily are extremely well conserved; the PTPase domains of Drosophila Dlar and human LAR share 74% amino acid sequence identity (44). We and others have previously reported that only the membrane-proximal PTPase-like domain (PTP-D1) of mammalian RPTPs has a physiologically significant amount of catalytic activity in vitro (28, 29, 43, 49), and that observation is extended to Drosophila PTPases in this report: all detectable in vitro PTPase catalytic activity of Dlar resides in PTP-D1.
It has been proposed that the PTPase activity of RPTPs is regulated by homodimerization, based on the finding that RPTPα-D1 crystallizes as a homodimer (5). The demonstration that RPTPα homodimerizes in vivo (24), and that RPTPα catalytic activity is inhibited in the dimeric state (25) lends support to this hypothesis. On the other hand, crystallization of the two-domain protein LAR PTP-D1D2 (35) argues against the LAR subfamily being regulated in a similar manner. The LAR-D1D2 protein is a monomer in solution (35), and the structures of LAR PTP-D1 and PTP-D2 are stabilized by a series of hydrogen-mediated interactions that make PTP-D1 and PTP-D2 quite inflexible in relation to each other, making homodimer formation very unlikely (35). The available data suggest that the LAR and RPTPα subfamilies may be regulated by distinct mechanisms.
Despite the lack of detectable PTPase catalytic activity from the C-terminal PTPase-like domain, PTP-D2, it is apparent that PTP-D2 has important functions. PTP-D2 of LAR family members interacts with and tightly binds to several cytosolic proteins, including the liprin family of coiled-coil cytoskeletal proteins, the multifunctional protein kinase/guanine nucleotide exchange factor Trio, and the Abl tyrosine kinase and its substrate Enabled (4, 13, 26, 39, 40, 50). In addition to linking RPTPs to downstream signal transduction elements, there is a growing body of evidence that PTP-D2 of LAR family members regulates the catalytic activity of PTP-D1 in vitro. For example, PTP-D2 of PTPσ binds and regulates the activity of PTP-D1 of PTPδ (47), and the PTPase domains of RPTPα and LAR can bind to each other if they are expressed as truncated fusion proteins, an interaction that appears to be regulated by oxidative stress (6, 7), although the consequence of this interaction for the PTPase catalytic activity of either RPTP is unknown.
We have previously reported that stimulation of increased catalytic activity of PTP-D1 by basic, positively charged proteins and peptides can be induced only if PTP-D2 is intact (23) and that truncation or swapping of PTP-D2 of LAR and CD45 causes changes in the substrate preference of PTP-D1 (18, 43; N. X. Krueger and H. Saito, unpublished observations). In addition, truncation of LAR PTP-D2 increases the catalytic activity of PTP-D1 (43). In this report, we demonstrate that PTP-D2 of Drosophila Dlar also regulates the catalytic activity of PTP-D1 in vitro, and we provide the first evidence that PTP-D2 may regulate PTP-D1 in vivo. Mutation of PTP-D2 has a deleterious effect on Drosophila embryonic neural development if PTP-D1 is active, but the identical mutation in PTP-D2 has no in vivo effect if PTP-D1 is mutationally inactivated.
In Drosophila, Dlar and several other RPTPs have been implicated in regulating the development of the embryonic central nervous system (CNS), axon guidance, and development of the optic lobe (12, 14, 15, 26, 30). In developing fly embryos, Dlar mRNA is supplied to the syncytial blastoderm by maternal expression that disappears around the cellularization stage. Dlar then reappears late in embryogenesis, specifically in the CNS at 12 to 16 h postfertilization (30, 45). Dlar transcripts are also expressed in adult flies during oogenesis (16). Loss-of-function mutations of the Dlar gene are lethal when homozygous (30) and cause multiple neuronal defects, including breaking and thinning of the CNS; aberrant, failed, or ectopic muscle-motoneuron synapse formation; and defective visual cognition (12, 15, 26, 30). In Dlar loss-of-function mutants, intersegmental nerve b (ISNb) and ISNd fail to properly innervate their target muscles, a phenotype that is exacerbated if other RPTP genes, including RPTP99A and RPTP10D, are simultaneously disrupted (15, 30). Nonneuronal phenotypes are also seen in Dlar loss-of-function genotypes: defects in oogenesis are observed in germ line Dlar−/− clones, and mutant embryos exhibit a loss of cell polarity, resulting in a round egg that is not oviposited (3, 17). The implication of the phenotype is that Dlar present in the follicular epithelium surrounding the oocyte collaborates with integrin receptors to organize actin polymer arrays and cell polarity in the developing egg.
In order to better understand the function of RPTPs in vivo, we have constructed a series of point mutant and deletion mutant Dlar transgenes and introduced them into the Drosophila genome. These mutant transgenes were expressed by using the bipartite GAL4-UAS transcription system, which allows tissue-specific expression of transgenes (10). We designed the transgenes to probe several unanswered questions about RPTP structure, including which portions of the complex Dlar ectodomain are required for PTPase function and which of the cytoplasmic PTPase-like domains are required for in vivo function. We report here that both the Ig-like domains and FnIII domains of the Dlar ectodomain serve important, nonredundant functions. The Ig-like domains are required for embryonic neural development, and the FnIII domains are essential during oogenesis. Surprisingly, we found that Dlar transgenes that have been mutated to inactivate the catalytic activity of PTP-D1 were able to rescue the lethality of Dlar−/− genotypes just as well as wild-type Dlar transgenes.
MATERIALS AND METHODS
Rescue of Dlar−/− lethality.
The Dlar deletion mutant constructs prepared in the plasmid pCasper were used for P-element-mediated germ line transformation of yw flies as described previously (30). These Dlar13.2/CyO; UAS-Dlar* females (where Dlar* represents either a wild-type or mutated Dlar cDNA) were mated to C155; Dlar5.5/CyO males that direct GAL4 expression to embryonic neurons. The rescue percentages were calculated as described previously (30). The male progeny do not receive the GAL4 driver in these matings.
Rescue of oocyte phenotypes.
Dlar5.5/CyO; T155-GAL4 females were mated to Dlar13.2/CyO; UAS-Dlar* males to generate Dlar13.2/Dlar5.5; T155-GAL4/UAS-Dlar* progeny. Zero- to 2-day-old females of this genotype were mated to wild-type males, fed on yeast paste for 2 days, and then dissected in phosphate-buffered saline to access their ovaries. Stage 14 oocytes were examined for egg shape defects and scored either as wild type (elongated, with long dorsal appendages) or as rounded (significantly shorter and thicker, with reduced dorsal appendages) as described previously (3). All genotypes were scored blind.
Immunoblotting.
The lysates prepared from wild-type embryos (yw) or transgenic embryos (expressing Dlar transgenes under control of the C155 promoter) were immunoprecipitated with affinity-purified anti-Dlar (FnIII 1 + 2) polyclonal antibodies and subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis on a 7% gel. Following electrophoresis, the gels were blotted to nitrocellulose membranes and probed with anti-Dlar (FnIII 1 + 2) monoclonal antibody 108.3C.
PTPase assays.
Glutathione S-transferase (GST)-Dlar fusion proteins were constructed by using pGEX plasmids (Amersham Pharmacia, Piscataway, N.J.) and purified by using glutathione beads to apparent homogeneity as assessed by bromophenol blue staining of SDS-polyacrylamide gels. PTPase assays were performed with the peptide substrate Raytide, and enzyme activity was calculated, as described previously (44).
Embryonic neural histology and phenotyping.
Embryonic neural pathways were visualized by staining with anti-fasciclin II monoclonal antibody as described previously (46). Phenotypic frequencies of bypass were determined as described previously (30). Embryo staining with polyclonal anti-Dlar antibodies was performed as described for staining with anti-fasciclin II antibodies (46).
Germ line mutation detection.
Single-stranded conformational polymorphism (SSCP) electrophoresis was performed as described previously (30).
RESULTS
All detectable PTPase catalytic activity maps to PTP-D1 in vitro.
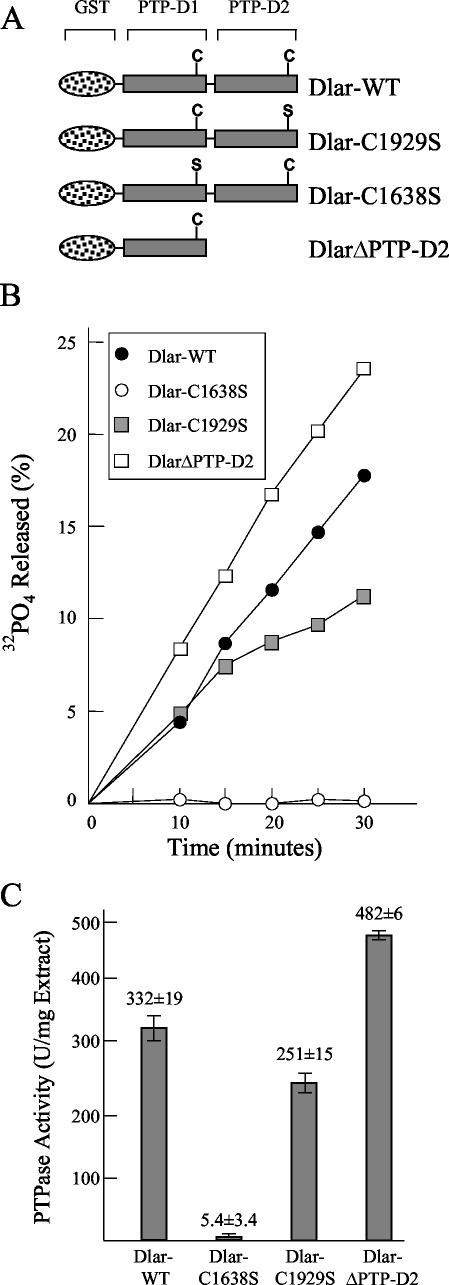
The existence of two repeated PTPase-like domains in the cytoplasmic regions of most RPTPs is one of the most puzzling aspects of RPTP structure. Assays of in vitro PTPase activity of human RPTPs LAR, CD45, PTPα, PTPɛ, PTPδ, and PTPζ indicate that all physiologically relevant PTPase activity maps to membrane-proximal PTP-D1; C-terminal PTP-D2 has little or no detectable activity (28, 29, 43, 49). To determine whether the PTPase activity of Drosophila RPTPs maps to PTP-D1 in the same manner as that of their mammalian counterparts, we constructed a series of GST-Dlar fusion proteins, in which the Dlar cytoplasmic region, with or without specific point mutations, was fused to GST (Fig. 1A). The enzyme mechanism of dephosphorylation by PTPases is known to proceed through a thiophosphate intermediate, in which a specific cysteine residue provides a sulfhydryl that covalently binds to orthophosphate before releasing it (11, 21). Mutation of this catalytically obligatory cysteine to a serine residue eliminates all PTPase activity from the mutated PTPase domain (11, 21, 28, 29, 43). We took advantage of this finding to make cysteine-to-serine point mutations in PTP-D1 and PTP-D2 in order to determine which PTPase domains were active in vitro. Figure 1B and C show the results of this analysis. Figure 1B shows a time course plot of Dlar dephosphorylation, and the data from Fig. 1B were used to calculate the specific enzyme activity shown in Fig. 1C. Mutation of Cys1638, the catalytically obligatory cysteine residue in PTP-D1, completely eliminates PTPase activity, whereas mutation of the analogous cysteine residue in PTP-D2, Cys1929, has little effect on activity. If PTP-D2 is truncated from the fusion protein, the activity is actually higher than that of the wild-type construct, which is consistent with previous data showing that PTP-D2 regulates the PTPase activity of PTP-D1 in vitro. However, the activity of the PTPase domains of Dlar, and of PTPase family members in general, varies greatly depending on the substrate assayed and stimulatory cofactors used. Determination of whether PTP-D2 regulates PTP-D1 catalytic activity against physiologically relevant substrates must await their identification.
FIG. 1.
In vitro PTPase assay of GST-Dlar fusion proteins. (A) Constructs used for PTPase assay. (B) Time course of Dlar phosphorylation by GST-Dlar fusion proteins. The fusion proteins were purified to apparent homogeneity as assessed by bromophenol blue staining of SDS-polyacrylamide gels, and the purified proteins were assayed at various time points for their ability to dephosphorylate the [32P]tyrosine-phosphorylated peptide substrate Raytide. (C) The time course data from panel B were used to calculate specific enzyme activity. The data are represented as means ± standard errors of the means.
Creation of mutated Dlar transgenes.
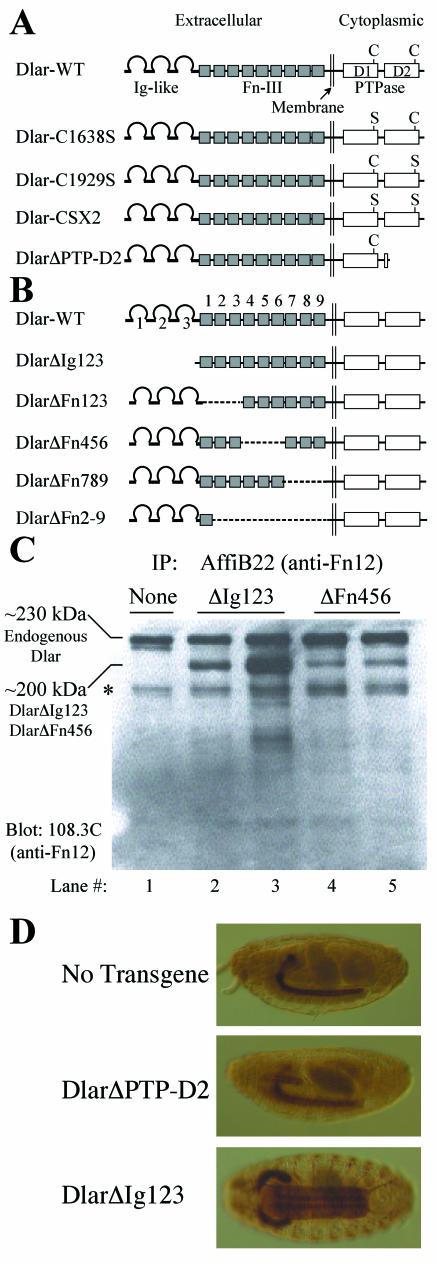
The ability of wild-type Dlar (Dlar-WT) transgenes to rescue the lethality of Dlar loss-of-function mutations gives us the opportunity to assay the roles of Dlar protein modules in vivo. A series of cytoplasmic domain (Fig. 2A) and ectodomain (Fig. 2B) constructs were made in order to characterize the in vivo function of these domains. The cytoplasmic domain constructs contain mutations similar to those used in the in vitro PTPase assays (Fig. 1) and were designed to allow us to examine the in vivo requirement for PTPase catalytic activity derived from PTP-D1 and PTP-D2. Dlar-C1638S and Dlar-C1929S mutate the catalytically obligatory cysteine residues in PTP-D1 and PTP-D2, respectively; Dlar-CSX2 is a double mutant in which both PTP-D1 and PTP-D2 are mutated; and DlarΔPTP-D2 deletes PTP-D2 from the transgene (residues 1778 to 1992 are deleted; the natural C terminus of the wild-type sequence, residues 1993 to 1997, remain intact).
FIG. 2.
Dlar transgenes and their expression in vivo. (A) Dlar cytoplasmic domain transgenes. Dlar-WT is shown at the top, and the Cys-to-Ser point mutants are diagrammed below. The DlarΔPTP-D2 truncation mutant is truncated at the beginning of PTP-D2. (B) Dlar ectodomain truncation mutant transgenes. The top line shows wild-type Dlar. The Ig-like domains and FnIII domains are numbered from the amino terminus (left). The missing domains in these mutants are shown by dotted lines. (C) Expression of Dlar transgenes. Extracts from wild-type embryos or embryos expressing Dlar transgenes under control of the C155 driver were immunoprecipitated (IP) with affinity-purified anti-Dlar polyclonal antibodies directed toward FnIII repeats 1 + 2 and immunoblotted with anti-Dlar monoclonal antibody (108.3C), also directed toward FnIII repeats 1 + 2. Lane 1, nontransgenic wild-type embryo (yw); lanes 2 and 3, extracts from two independent DlarΔIg123 transgenic lines; lanes 4 and 5, extracts from two independent DlarΔFn456 transgenic lines. The asterisk marks a protein isoform likely to be the proteolytically processed extracellular domain of Dlar. All transgenic embryos shown express Dlar transgenes in a wild type background. The bands corresponding to endogenous Dlar and transgenic Dlar are indicated at the left. (D) Embryo staining with anti-Dlar antibodies. (Top panel) Lateral view of a wild-type (yw) embryo stained with anti-Dlar antibodies. (Middle panel) Lateral view of a transgenic embryo expressing DlarΔPTP-D2 under control of the C155 driver. Note the characteristic salivary gland expression induced by C155. (Bottom panel) Ventral view of a transgenic embryo expressing DlarΔIg123 under C155 control. In this view, the cell bodies of ISN motoneurons can be easily visualized in the neuropils.
We also made a series of deletions in the Dlar ectodomain in order to map the portions of the ectodomain that are important in ligand-receptor interactions (Fig. 2B). DlarΔIg123 deletes the three amino-terminal Ig domains (residues 33 to 317); DlarΔFn123 deletes FnIII domains 1, 2, and 3 (residues 318 to 608); DlarΔFn456 deletes FnIII domains 4, 5, and 6 (residues 609 to 907); DlarΔFn789 deletes FnIII domains 7, 8, and 9 (residues 908 to 1193); and DlarΔFn2-9 deletes FnIII domains 2 through 9 (residues 421 to 1349).
Expression of Dlar transgenes.
These mutant transgenes were then introduced into the Drosophila genome by using P-element transformation, and their ability to rescue lethality was tested. The constructs were made by using the GAL4-UAS transcription system (10), in which a GAL4-inducible promoter is placed upstream of the Dlar cDNAs. Since a wide variety of tissue-specific GAL4 source strains are available, this allows tissue-specific expression of Dlar transgenes in essentially any tissue of interest simply by mating the appropriate fly strains.
In order to examine the expression of Dlar transgene products, we generated rabbit polyclonal and mouse monoclonal antibodies by using GST-FnIII 1 + 2 fusion proteins expressed in bacteria as antigens. Both polyclonal (affinity-purified) antibody and the monoclonal antibody 108.3C detected a polypeptide of ∼230 kDa on immunoblots of extracts prepared from wild-type fly embryos (Fig. 2C, lane 1). Both antibodies showed no staining in clones of Dlar−/− tissue, confirming their specificity (3). The 230-kDa band corresponds to the predicted molecular mass of Dlar based on its amino acid sequence. In addition, a smaller polypeptide of about 150 to 160 kDa is also detected. This smaller polypeptide could be the processed extracellular domain of Dlar, whose predicted size is about 150 kDa. The processing and shedding of the extracellular domains of mammalian LAR family RPTPs (38, 41) and a Drosophila RPTP (19) have been extensively studied. Immunoblotting of protein extracts prepared from transgenic embryos expressing the DlarΔIg123 and DlarΔFn456 transgenes revealed that the transgenes are expressed at their predicted size: proteins of 200 kDa, or ∼33 kDa smaller than the full-length Dlar (Fig. 2C, lanes 2 to 5).
To drive the expression of recombinant Dlar transgenes in all postmitotic neurons, we used the GAL4 driver C155 or one of its derivatives, elav-GAL4 (27, 31, 50); C155 is an enhancer-trap insertion into the elav locus on the X chromosome, which directs GAL4 expression to postmitotic CNS neurons and neurons of the peripheral nervous system in the developing embryo. C155/elav also induces expression in the salivary gland. elav-GAL4 has the same tissue distribution as C155; it is a version of the elav promoter that has been cloned, directly fused to GAL4, and inserted onto chromosome 3 by P-element transformation. elav-GAL4 has the same tissue distribution as C155 but expresses more stably during long-term insect culture (27, 31, 50; our unpublished observations).
The amounts of protein made by all of the Dlar transgene constructs used in this report were assessed by immunoblotting. Representative protein expression levels of selected transgenic lines are shown in Fig. 2C. Each lane in Fig. 2C shows the protein expression levels of an independently isolated Dlar transgenic line. The experiment in Fig. 2C uses embryos that express both endogenous and transgenic Dlar, which allows direct comparison of expression levels. The intensity of the 230-kDa band shows the expression level of endogenous Dlar protein, and that of the 200-kDa band shows the expression level of transgenic Dlar deletion mutant protein. In almost all cases, transgene expression levels were virtually indistinguishable from that shown for the DlarΔFn456 constructs in Fig. 2C, lanes 4 and 5. The only transgenic strain that expressed Dlar at levels that were higher than the endogenous level of expression was the DlarΔIg123 construct shown in lane 3. The expression levels of other isolated DlarΔIg123 transgenic lines are very similar to the expression level of the DlarΔIg123 strain shown in lane 2, which is virtually indistinguishable from the expression level of endogenous Dlar.
The GAL4-induced expression level of Dlar protein was also assessed in situ by staining with the anti-Dlar antibodies (see Fig. 2D). Embryos expressing the Dlar transgenes under GAL4 control were stained with anti-Dlar antibodies and examined histologically. The embryos displayed the characteristic C155/elav-induced nervous system tissue distribution. The top embryo in Fig. 2D shows the endogenous Dlar expression pattern. The embryo in the middle shows expression of the Dlar-ΔPTP-D2 transgene, and the embryo on the bottom shows the expression of the same DlarΔIg123 transgenic line blotted in lane 3 of Fig. 2C. The ventral view of the ΔIg123 embryo permits easy visualization of the CNS connectives where the ISN cell bodies reside. The staining intensities of the other transgenic lines were very similar to that of the DlarΔPTP-D2 embryo (and equivalent to the levels seen in the DlarΔFn456 embryos in the blots in Fig. 2C, lanes 4 and 5). The DlarΔIg123 embryo in Fig. 2D shows the highest level of protein expression seen in any of the transgenic lines. It should be noted that we have found no correlation between the transgene expression level of a given strain and the behavior of the transgenic line in any of the experiments described below. Thus, it is unlikely that our data can be explained simply by the level of protein produced by a given transgenic strain.
Rescue of Dlar−/− lethality by using cytoplasmic domain mutant transgenes.
The results of matings with the Dlar cytoplasmic domain constructs are shown in Table 1 and summarized in Fig. 3. At least two independently integrated P-element insertions were tested for each construct. In all of these matings, the C155 driver was used as a source of GAL4 (31). In Table 1, males bearing the C155 driver on the X chromosome were mated to females bearing the UAS-Dlar transgenes on chromosome 3, so only female offspring received both a source of GAL4 and a UAS-Dlar transgene. This provides an internal control (rescue level of male offspring) for endogenous, GAL4-independent expression of the Dlar transgene. GAL4-independent rescue was also monitored by matings 3 to 7, 15, 16, and 21 to 28 in Table 1. The reciprocal experiment, using C155 females and Dlar transgene-bearing males, generated the same amount of GAL4-dependent and GAL4-independent rescue as shown in Table 1; there is no maternal effect (data not shown). The (very low) level of GAL4-independent transgene expression is specific to the locus of insertion of each individual transgene; transgenes that are inserted into transcriptionally active loci have higher endogenous expression levels than transgenes inserted into transcriptionally quieter areas. Flies rescued by GAL4-independent Dlar transgene expression are always very small and quite sick; freshly eclosed adults usually die within 24 h. This is in clear contrast to GAL4-driven rescue, which generates fertile, healthy adults.
TABLE 1.
Rescue of Dlar−/− lethality by mutated Dlar transgenes
| Dlar Genotype | Mating | GAL4 drivera | Dlar transgene | % of expected progeny ± SD (n)b
|
|
|---|---|---|---|---|---|
| Females | Males | ||||
| Dlar5.2/Dlar5.5 | 1 | − | None | 8.3 ± 2.1 (302) | 7.9 ± 2.0 (396) |
| 2 | + | None | 5.2 ± 1.9 (276) | 10.8 ± 2.2 (367) | |
| 3 | − | P2, Dlar-WT | 22.4 ± 3.1 (507) | 22.8 ± 3.2 (488) | |
| 4 | − | P17, Dlar-C1638S | 12.9 ± 2.5 (446) | 14.7 ± 2.7 (438) | |
| 5 | − | P18, Dlar-C1638S | 20.2 ± 3.2 (447) | 12.2 ± 2.4 (470) | |
| 6 | − | P13, Dlar-C1929S | 17.2 ± 3.6 (290) | 20.2 ± 4.1 (262) | |
| 7 | − | P19, DlarΔPTP-D2 | 23.4 ± 3.8 (353) | 28.6 ± 3.9 (431) | |
| 8 | + | P2, Dlar-WT | 73.6 ± 5.9 (580) | 21.5 ± 3.3 (412) | |
| 9 | + | P17, Dlar-C1638S | 78.6 ± 6.4 (539) | 12.7 ± 2.5 (435) | |
| 10 | + | P18, Dlar-C1638S | 75.7 ± 6.7 (459) | 13.3 ± 2.7 (431) | |
| 11 | + | P13, Dlar-C1929S | 76.3 ± 8.2 (315) | 20.7 ± 4.1 (267) | |
| 12 | + | P19, DlarΔPTP-D2 | 53.6 ± 5.9 (388) | 17.9 ± 3.3 (365) | |
| Dlar13.1/Dlar5.5 | 13 | − | None | 3.5 ± 1.6 (289) | 2.0 ± 1.2 (297) |
| 14 | + | None | 0 (262) | 0.9 ± 0.9 (232) | |
| 15 | − | P4B, Dlar-WT | 8.9 ± 2.0 (448) | 8.4 ± 2.0 (445) | |
| 16 | − | P17, Dlar-C1638S | 9.9 ± 2.1 (489) | 3.6 ± 1.3 (456) | |
| 17 | + | P4B, Dlar-WT | 67.1 ± 5.6 (581) | 5.1 ± 1.5 (479) | |
| 18 | + | P17, Dlar-C1638S | 74.9 ± 6.0 (569) | 6.0 ± 1.7 (415) | |
| Dlar13.2/Dlar5.5 | 19 | − | None | 2.8 ± 1.0 (511) | 1.7 ± 0.8 (515) |
| 20 | + | None | 1.8 ± 1.0 (344) | 2.5 ± 1.3 (322) | |
| 21 | − | P2, Dlar-WT | 21.1 ± 2.5 (764) | 21.2 ± 2.4 (782) | |
| 22 | − | P17, Dlar-C1638S | 5.3 ± 1.5 (499) | 4.1 ± 1.3 (503) | |
| 23 | − | P18, Dlar-C1638S | 11.5 ± 2.7 (330) | 11.5 ± 2.6 (350) | |
| 24 | − | P13, Dlar-C1929S | 9.0 ± 3.7 (139) | 3.3 ± 2.3 (125) | |
| 25 | − | P28, Dlar-CSX2 | 36.1 ± 5.8 (255) | 8.8 ± 2.8 (236) | |
| 26 | − | P29, Dlar-CSX2 | 12.6 ± 4.8 (118) | 9.5 ± 3.9 (132) | |
| 27 | − | P20, DlarΔPTP-D2 | 11.7 ± 2.7 (344) | 18.0 ± 3.3 (363) | |
| 28 | − | P21, DlarΔPTP-D2 | 17.4 ± 3.5 (300) | 15.2 ± 3.2 (312) | |
| 29 | + | P2, Dlar-WT | 90.9 ± 5.2 (989) | 18.5 ± 2.3 (743) | |
| 30 | + | P17, Dlar-C1638S | 66.2 ± 5.4 (595) | 2.5 ± 1.0 (479) | |
| 31 | + | P18, Dlar-C1638S | 63.2 ± 6.3 (425) | 10.0 ± 2.3 (400) | |
| 32 | + | P13, Dlar-C1929S | 88.0 ± 9.8 (265) | 16.0 ± 4.3 (189) | |
| 33 | + | P28, Dlar-CSX2 | 68.9 ± 9.6 (203) | 10.5 ± 3.7 (160) | |
| 34 | + | P29, Dlar-CSX2 | 68.8 ± 9.4 (211) | 6.5 ± 2.9 (158) | |
| 35 | + | P20, DlarΔPTP-D2 | 41.2 ± 4.6 (474) | 14.8 ± 2.8 (407) | |
| 36 | + | P21, DlarΔPTP-D2 | 53.8 ± 5.7 (420) | 15.1 ± 3.0 (357) | |
The X-linked C155 driver was used in these matings. GAL4-independent expression of the Dlar transgenes is monitored by rescue percentages of males, which do not receive GAL4 in any of these matings.
One hundred percent rescue indicates that 33% of the adult progeny were homozygous for the Dlar loss-of-function alleles. n, total number of progeny counted.
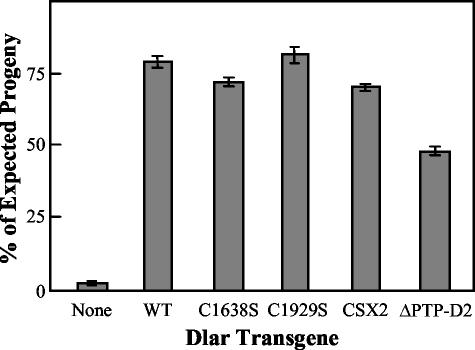
FIG. 3.
Bar graph of rescue of Dlar−/− lethality by Dlar cytoplasmic domain mutant transgenes. The bars represent the mean percentage of expected progeny observed ± standard deviation when the indicated Dlar transgene is expressed under control of the C155-GAL4 driver in a loss-of-function background. The results are pooled from rescue of several different Dlar−/− genotypes that were generated by using multiple combinations of Dlar loss-of-function alleles. At least two independently inserted Dlar transgenes were tested for each mutant shown.
Table 1, matings 8, 17, and 29, show that C155-GAL4-induced expression of Dlar-WT efficiently rescues the lethality of all Dlar−/− genotypes tested, i.e., Dlar5.2/Dlar5.5, Dlar13.1/Dlar5.5, and Dlar13.2/Dlar5.5. Since the results are equivalent for all Dlar−/− genotypes, we have conducted the bulk of our experiments with the alleles Dlar5.5 and Dlar13.2, both of which contain nonsense mutations that have been sequenced; they encode Dlar proteins truncated in the extracellular domain (30) (Fig. 4C). Surprisingly, the Dlar-C1638S transgene, which inactivates PTP-D1, the PTPase domain in which all detectable in vitro PTPase activity resides, rescues Dlar−/− lethality nearly as well as Dlar-WT (Table 1, matings 9, 10, 18, 30, and 31). Similarly, Dlar-C1929S, which would inactivate PTPase activity of PTP-D2, if any exists, also rescues lethality as well as Dlar-WT (Table 1, matings 11 and 32, and Fig. 3).
FIG. 4.
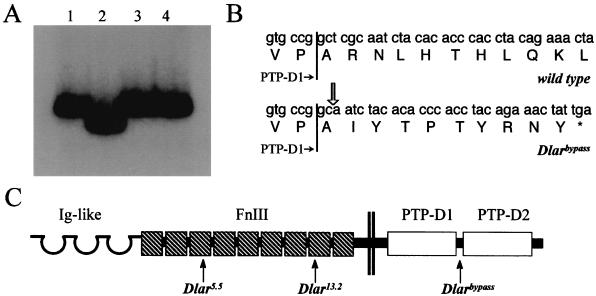
The Dlarbypass allele encodes a protein truncated after PTP-D1. (A) Autoradiograph of 32P-labled PCR products subjected to SSCP electrophoresis. Lane 1, Wild-type parental genotype (A49/A49); Lane 2, Dlarbypass/Dlarbypass; lane 3, Dlar5.5/+; lane 4, Dlar13.2/+. (B) Dlarbypass/Dlarbypass genomic DNA from the region recognized by the primers in panel A was subcloned and sequenced. Dlarbypass has a 4-bp deletion relative to the wild-type Dlar sequence. The location of the deletion is indicated by the open arrow. The asterisk indicates a stop codon. The end of the coding sequence for PTP-D1 is indicated by vertical lines. (C) Proteins encoded by Dlarbypass, Dlar5.5, and Dlar13.2. Arrows indicate the locations where the alleles are truncated by mutation.
In order to address the possibility that PTPase activity from either PTP-D1 or PTP-D2 is sufficient to restore Dlar function in vivo, we constructed Dlar-CSX2, which incorporates both the C1638S and C1929S mutations into a single transgene. Table 1, matings 33 and 34, and Fig. 3 show that Dlar-CSX2 also rescues lethality quite efficiently, indicating that a catalytically active Dlar PTPase domain is not required for survival. Interestingly, despite the fact that inactivation of catalytic activity from PTP-D2 has no effect on rescue, the PTP-D2 truncation mutant, DlarΔPTP-D2, is significantly impaired in its ability to substitute for Dlar-WT, rescuing only about half as efficiently as Dlar-C1929S (Table 1, matings 12, 35, and 36). This suggests that PTP-D2 serves an important function independent of PTPase catalysis. The results of all of our experiments rescuing Dlar−/− genotypes with the cytoplasmic domain constructs are summarized in Fig. 3, which shows that all of the cytoplasmic domain point mutant transgenes tested efficiently rescue lethality; only DlarΔPTP-D2 is impaired in its rescue capacity.
Molecular characterization of a hypomorphic Dlar allele.
In our previous genetic analysis of Dlar, we identified a partial loss-of-function allele (Dlarbypass) characterized by the same axon guidance and synaptic phenotypes that we observed in null mutants (27). However, this semilethal hypomorphic allele displays phenotypic penetrance of approximately one-third to one-half of Dlar null alleles (27), indicating that a partially functional gene product is made from the Dlarbypass allele. To determine the molecular lesion in the Dlarbypass allele, we performed SSCP analysis by electrophoresis of radiolabeled PCR products with a series of primers that scans the entire Dlar protein-coding region (30). One primer pair detected a gel shift, indicating a polymorphism (Fig. 4A). Sequence analysis of the polymorphic fragments revealed a 4-bp deletion mutation (Fig. 4B) resulting in a frameshift that truncates the cytoplasmic domain shortly before the beginning of PTP-D2 (Fig. 4C). Since the expression level of Dlar in Dlarbypass/Dlarbypass embryos is not detectably different from that in wild-type embryos (data not shown) and the only domain affected by the molecular lesion in Dlarbypass is PTP-D2, the phenotype of Dlarbypass provides more evidence that PTP-D2 performs an important function despite not being a catalytically active PTPase domain. The residual function observed in Dlarbypass and DlarΔPTP-D2 mutants indicates that while PTP-D2 is necessary for full efficiency of Dlar activity, it is not absolutely required. For several reasons, we speculate that one important function of PTP-D2 is to regulate the PTPase activity of PTP-D1 (see the next section of Results, and Discussion, for the rationale).
Effect of expression of Dlar cytoplasmic domain mutant transgenes on Dlar−/− neural phenotypes.
We attempted to use the Dlar cytoplasmic domain mutant transgenes to rescue the observed neural abnormalities seen in Dlar−/− embryos. We previously reported that Dlar-WT rescues ISNb and ISNd defects in the developing Dlar−/− mutant embryos, if embryos that express transgenic Dlar mRNA are examined, as assessed by in situ hybridization with Dlar cDNA probes (30). In these experiments, when transgene-expressing embryos are selected on the basis of Dlar mRNA expression in the peripheral nervous system (where the C155/elav promoters drive expression in addition to the CNS), we see very efficient rescue. However, we have been unable to show statistically significant rescue of the neural defects at the population level in every genetically transgenic embryo, even though we attempted to do so with several different GAL4 drivers. We found that if embryos are examined without preselection by in situ hybridization, embryos expressing Dlar-WT under control of elav-GAL4 in the Dlar13.2/Dlar5.5 background have an ISNb bypass at a frequency of 34.6% ± 3.1% (n = 350 A2-A7 embryonic segments counted), compared to an ISNb bypass frequency of 32.3% ± 3.0% (n = 369) seen in Dlar13.2/Dlar5.5 embryos expressing no transgene at all (Fig. 5).
FIG. 5.
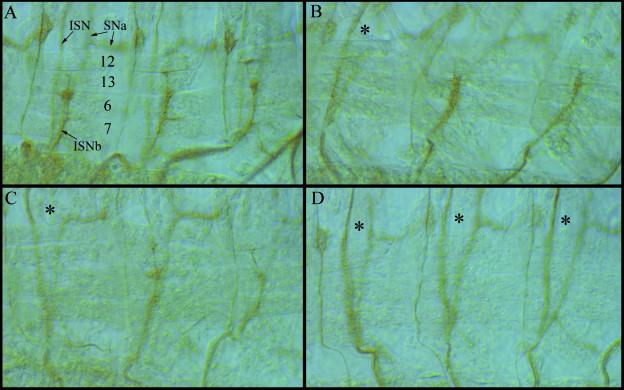
The frequency of bypass phenotypes is increased in Dlar−/Dlar− embryos by overexpression of Dlar-C1929S. Embryonic motoneuron pathways were visualized in stage 16 embryos by staining with anti-fasciclin II monoclonal antibody. The numbers in panel A indicate specific muscles within each hemisegment; ISN, ISNa, and ISNb are indicated by arrows. In all panels, asterisks indicate hemisegments with a bypass phenotype. The frequencies of bypass phenotype observed in this experiment are 32.3% ± 3.0% (n = 369 hemisegments scored) for Dlar5.5/13.2 embryos expressing no transgene, 34.6% ± 3.1% (n = 350) for Dlar5.5/13.2 embryos expressing the Dlar-WT transgene, 38.9% ± 4.9% (n = 162) for Dlar5.5/13.2 embryos expressing Dlar-C1638S, 54.8% ± 3.6% (n = 429) for Dlar5.5/13.2 embryos expressing Dlar-C1929S, and 25.3% ± 3.2% (n = 249) for Dlar5.5/13.2 embryos expressing Dlar-CSX2. Expression of Dlar-WT does not generate statistically significant rescue of the bypass phenotype, but expression of Dlar-C1929S significantly increases bypass frequency. (A) A wild-type embryo. Note the wild-type pattern of innervation of muscles 6, 7, 12, and 13 by ISNb. (B) A Dlar5.5/13.2 embryo expressing no transgenes. One hemisegment is bypass and two are wild type in this photograph. (C) A Dlar5.5/13.2 embryo expressing Dlar-WT. (D) A Dlar5.5/13.2 embryo expressing Dlar-C1929S. All three hemisegments shown exhibit bypass phenotypes.
There are several possible explanations for the lack of significant rescue at the population level. (i) The GAL4 system is well known to be subject to mosaicism, often showing inconsistent levels of expression from cell to cell or embryo to embryo. The level of Dlar transgene mRNA induced by the GAL4 system can be very heterogeneous, with some embryos expressing much more mRNA than others. Note that despite these high mRNA levels, we never see levels of Dlar protein more than twofold greater than the endogenous level of wild-type Dlar (Fig. 2C and D), indicating that expression of Dlar protein is tightly regulated posttranscriptionally. (ii) Efficient expression of transgenic proteins is often dependent on RNA splicing or untranslated region sequences, both of which are absent in the cDNA construct that we used here. Thus, normal levels of protein may be difficult to achieve unless mRNA levels are well above normal. This problem is notorious in some systems (such as mouse), and this may be an issue here. (iii) Differential splicing of LAR has been demonstrated in vertebrates, and our unpublished data suggest that alternative splice forms may also exist in Drosophila. Thus, the cDNAs that we used for the rescue may not represent isoforms of Dlar that act most efficiently for motor axon guidance. (iv) There is a temporal discrepancy between the endogenous Dlar expression pattern and the expression induced by elav. Endogenous Dlar is expressed early in developing CNS neurons, whereas elav-induced expression is limited to postmitotic neurons. Therefore, there may not be sufficient time for Dlar protein to accumulate in bioactive concentrations.
In the course of the neural analysis, we found that elav-GAL4-driven expression of one Dlar cytoplasmic domain mutant transgene, Dlar-C1929S, causes a significant increase of the bypass phenotype in Dlar−/− backgrounds. When Dlar-C1929S is expressed under elav-GAL4 control in Dlar13.2/Dlar5.5 embryos, an ISNb bypass frequency of 54.8% ± 3.6% (n = 429) is observed, which is a much higher frequency of bypass than is ever observed in Dlar13.2/Dlar5.5 embryos not expressing transgenes or in Dlar13.2/Dlar5.5 embryos expressing any other Dlar transgene tested (Fig. 5). Interestingly, no similar increase in the frequency of bypass is seen in Dlar13.2/Dlar5.5 embryos expressing Dlar-CSX2 (ISNb bypass frequency, 25.3% ± 3.2%; n = 259), despite the presence of the same C1929S mutation in this transgene. The finding that a mutation within PTP-D2 has no detectable phenotype unless PTP-D1 is catalytically active suggests that PTP-D2 affects neural development indirectly via regulation of the catalytic activity of PTP-D1. If this interpretation is correct, these data represent the first evidence that PTP-D2 regulates the PTPase activity of PTP-D1 in vivo.
It should be noted that there is no phenotype observed when Dlar-C1929S is expressed in insects that have a wild-type copy of Dlar. We have not ever observed any dominant effects from expressing Dlar transgenes in a wild-type background in any of the assays described in this paper. The only time a gain-of-function phenotype is observed is in Dlar−/− embryos, meaning that the Dlar-C1929S gain-of-function phenotype is most accurately described as recessive.
Rescue of the Dlar−/− lethal phenotype by Dlar ectodomain mutant transgenes.
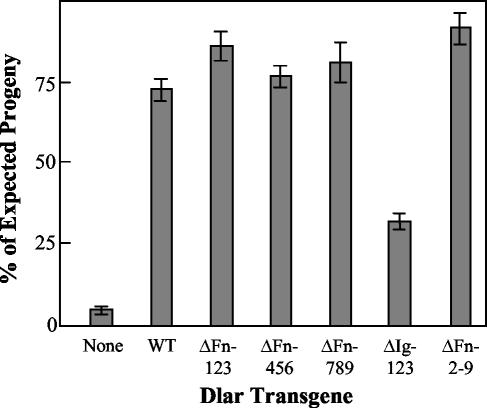
The Dlar ectodomain constructs were also assayed for their capacity to rescue Dlar−/− lethality. The results of these experiments are summarized in Fig. 6; multiple transgenic lines were tested for each construct. The same types of control matings that are shown in Table 1 were performed for the ectodomain rescue experiments, but those results are omitted here for clarity. We found that in terms of survival, the FnIII domains of the Dlar ectodomain are dispensable. DlarΔFn123, DlarΔFn456, DlarΔFn789, and DlarΔFn2-9 all rescue lethality as efficiently as Dlar-WT. On the other hand, DlarΔIg123 is severely impaired in its capacity to rescue lethality despite the fact that DlarΔIg123 is expressed at higher levels than Dlar-WT and all of the ΔFn transgenes (Fig. 2C and D). These results demonstrate that while the Ig-like domains of Dlar are necessary for efficient function in the Drosophila nervous system, the FnIII domains are dispensable in this context.
FIG. 6.
Rescue of Dlar−/− lethality by Dlar ectodomain deletion mutant transgenes. The ability of Dlar transgenes to rescue the lethality of the genotype Dlar13.2/Dlar5.5 was assessed. The represent the mean percentage of expected progeny observed ± standard deviation when the indicated Dlar transgene is expressed under control of the C155-GAL4 driver in a Dlar13.2/Dlar5.5 background. The results shown are pooled from multiple independent transgenic lines for each transgene tested.
Rescue of the Dlar−/− oocyte phenotype by Dlar ectodomain mutant transgenes.
We also assayed the capacity of the Dlar ectodomain constructs to rescue the observed oocyte phenotype seen in Dlar−/− embryos. Although mutations in Dlar are largely lethal, careful nurturing of crosses between the alleles Dlar5.5 and Dlar13.2 can yield a small number of Dlar5.5/Dlar13.2 escapers (see Table 1, matings 19 and 20, for escaper frequencies). Analysis of these adults, or of homozygous mutant clones in a wild-type background, reveals a requirement for Dlar in the follicular epithelium surrounding the oocyte (3, 17). In this context, Dlar collaborates with integrin receptors to orchestrate accurate cell polarity and the organization of actin polymer arrays; in the absence of Dlar, epithelial polarity fails, resulting in a round egg that is not oviposited (3). To determine which components of the Dlar extracellular domain are required for Dlar function in oogenesis, we tested the ability of the ectodomain deletion constructs to rescue the round-egg defect of Dlar homozygous escapers. In the absence of transgenes, Dlar5.5/Dlar13.2 mutants show a moderate penetrance of abnormally round stage 14 oocytes in dissected ovaries (14.7%) (Table 2). Expression of a wild-type Dlar transgene in this background with the epithelial driver T155-GAL4 (3) rescues this defect to near-wild-type levels; the data in Table 2 represent a 91% rescue of the oocyte phenotype. Although our analysis of Drosophila viability demonstrated a requirement for Dlar Ig-like domains, in the follicular epithelium they are dispensable; the transgene lacking all 3 Ig-like domains, DlarΔIg123, rescued as efficiently as the wild-type transgene. However, expression of DlarΔFn2-9 in Dlar13.2/Dlar5.5 embryos not only failed to rescue egg-shape defects, it caused nearly a fourfold increase in the penetrance of round stage 14 oocytes observed. Analyses of the smaller FnIII deletion constructs show that sequences contained within FnIII domains 1 to 6 are required for this function of Dlar, while FnIII domains 7 to 9 are dispensable (Table 2). Thus, the Ig-like domains are required for survival, but not during oogenesis, and the FnIII domains are required during oogenesis.
TABLE 2.
Rescue of Dlar−/− oocyte phenotype
| Dlar transgene | % Round oocytes ± SD (n) |
|---|---|
| None | 14.7 ± 2.2 (299) |
| DLAR-WT | 1.0 ± 0.7 (200) |
| DLARΔIg123 | 0 (217) |
| DLARΔFn2-9 | 57.6 ± 5.4 (198) |
| DLARΔFn123 | 23.7 ± 2.8 (299) |
| DLARΔFn456 | 42.1 ± 3.9 (271) |
| DLARΔFn789 | 1.7 ± 1.0 (176) |
DISCUSSION
The functional mapping of multidomain proteins is important to determine the structural basis of their interactions, and in the present context this type of analysis has allowed us to study one of the most striking structural features of RPTPs: the existence of two repeated PTPase-like domains in the cytoplasmic region. The in vitro PTPase assay data strongly support the idea that PTP-D1 contains all physiologically relevant PTPase catalytic activity; no physiologically relevant PTPase catalysis can be detected from wild-type PTP-D2 of any RPTP tested. We have extended these findings to Dlar in this study: all PTPase activity maps to PTP-D1 in vitro. We attempted to demonstrate that PTPase activity from PTP-D1 is essential for survival, but to our surprise, mutant Dlar transgenes that inactivate catalytic activity derived from either PTP-D1, PTP-D2, or both simultaneously are able to rescue Dlar−/− lethality nearly as well as the wild-type Dlar. Of the cytoplasmic domain constructs tested, only the PTP-D2 truncation mutant, DlarΔPTP-D2, is significantly impaired in its rescue capacity. The simplest explanation of these data is that the functions of Dlar that are essential for insect survival do not require PTPase catalytic activity. This finding is contrary to what has been observed for another Drosophila PTPase, DPTP69D, which has been shown to require a catalytically active PTP-D1 in order to rescue DPTP69D−/− lethality (19). Given the exquisite conservation of LAR family PTPase domains through evolution, the lack of an essential role for PTPase catalysis in Dlar function is a profoundly surprising finding.
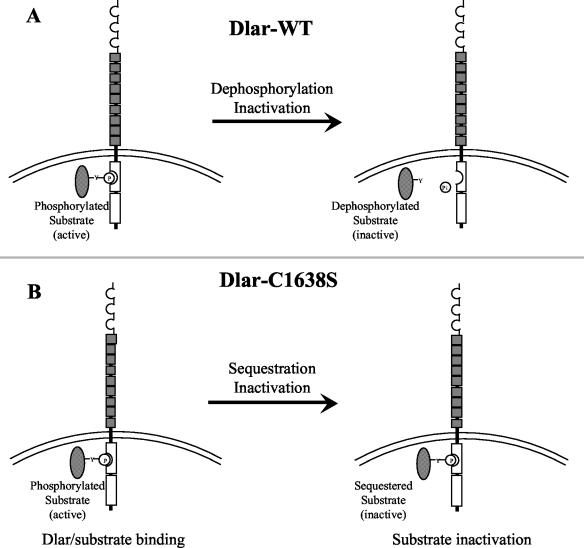
There remain other possible interpretations of these data. It is possible that the Cys-to-Ser mutants, while unable to dephosphorylate, generate a PTPase domain that is still able to recognize and bind to its substrate. Indeed, we have previously reported the ability of the Cys-to-Ser-mutated PTP-D1 of CD45 to tightly and stably bind the substrate CD3ζ (18). If this type of binding occurs for Dlar-C1638S, then it is possible that by binding to substrate, Dlar-C1638S sequesters substrate and prevents it from functioning in downstream signaling events. Figure 7 shows a schematic of this speculative model. There is precedence for this type of interaction to have biological effects. In the case of the Drosophila cytosolic PTPase corkscrew (csw), it has been demonstrated genetically that csw-derived PTPase activity is essential for proper photoreceptor development and that a Cys-to-Ser mutant of csw has the capacity to partially restore csw function in csw loss-of-function backgrounds (1). If this model is correct, the implication for Dlar signal transduction is that the in vivo effect of dephosphorylation by Dlar is the functional inactivation of its substrate. It is also possible that a variant of this substrate sequestration model is occurring. Genetics have already shown that both Dlar and PTP69D are functionally redundant and that they have interchangeable cytoplasmic domains (15, 32). It is therefore possible that PTP69D catalysis is sufficient in a Dlar−/− background, as long as substrates are recruited and bound to the Dlar-C1638S PTPase domains.
FIG. 7.
Speculative model of the mechanism of rescue of lethality by Dlar-C1638S. The ability of Dlar-C1638S to rescue lethality nearly as well as Dlar-WT does not necessarily mean that PTPase catalytic activity is not required for Dlar function. While they are unable to catalyze dephosphorylation, it is possible for Cys-to-Ser PTPase domain mutants to stably bind to substrates (1, 18). If the in vivo function of Dlar is to functionally inactivate its substrate via dephosphorylation, as diagrammed in panel A, then panel B shows that by binding and sequestering substrate, Dlar-1638S could mimic the function of Dlar-WT without being catalytically active.
The interpretation of these data would have been greatly simplified by analysis of a Dlar transgene that completely lacked a cytoplasmic domain, DlarΔPTP-D1D2. However, repeated and intensive efforts to express this transgene were unsuccessful. It is possible that expression of this transgene is deleterious for insect survival. Indeed, it has been observed that expression of proteins similar to the DlarΔPTP-D1D2 proteins that we tried to express generate dramatic dominant gain-of-function phenotypes at neuromuscular junctions (26).
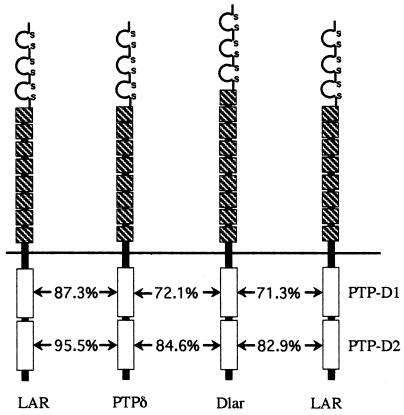
Despite the fact that PTP-D2 does not appear to be a catalytically active PTPase domain, there is now a growing body of evidence that PTP-D2 performs several vitally important functions in vivo. Indeed, within the LAR PTPase subfamily, the sequence of PTP-D2 is better conserved than that of PTP-D1 (Fig. 8), indicating a strong evolutionary pressure for conservation of PTP-D2 structure and function. Our data clearly indicate that PTP-D2 is vitally important for survival: truncation of PTP-D2 in the Dlarbypass allele results in a semilethal phenotype, and the DlarΔPTP-D2 transgene is impaired in its capacity to rescue Dlar−/− lethality. Taken in the context of the capacity of Dlar-C1929S to efficiently rescue Dlar−/− lethality, the data strongly argue for the idea that PTP-D2 performs essential functions independent of PTPase catalysis. One of these functions is to serve as a docking domain for downstream signal transduction elements. PTP-D2 of LAR subfamily members interacts physically and functionally with a variety of cytoplasmic proteins, including several members of the liprin family, Trio, Ena, and Abl (4, 13, 26, 39, 40, 50). Another function of PTP-D2 appears to be regulation of the PTPase activity of PTP-D1. Expression of the Dlar-C1929S transgene in a Dlar−/− background significantly increases the observed frequency of the bypass phenotype in developing embryos (Fig. 5), but the identical C1929S mutation has no effect on neural development if it is expressed in tandem with the PTP-D1-inactivating C1638S mutation in the Dlar-CSX2 transgene. Thus, we hypothesize that PTP-D2 regulates the catalytic activity of PTP-D1. This hypothesis is based on the reasoning that if PTP-D2 exerts its effect on neural development indirectly via regulation of PTP-D1 PTPase activity, inactivation of PTP-D1 catalysis should abolish the phenotypic consequences of mutations in PTP-D2. There are other possible mechanisms for the effect of the C1929S mutation. It is also possible that the C1929S mutation generates a PTP-D2 capable of sequestering a substrate in the manner that we suggest in Fig. 7, generating a gain-of-function phenotype, or that the C1929S mutation causes a conformational change that results in lack of interaction with substrates or docking proteins. In any of these cases, it seems quite likely that the C1929S mutation has an additional effect on PTP-D2 beyond simply inactivating potential PTP-D2 catalytic activity (if, in fact, any relevant PTP-D2 catalytic activity exists at all).
FIG. 8.
Amino acid sequence comparison in the LAR subfamily of RPTPs. The percentages of amino acid identity between the indicated PTPase domains are shown. Within the LAR subfamily, catalytically inactive PTP-D2 has a greater degree of sequence conservation than catalytically active PTP-D1.
In the LAR crystal structure, the overall structures of PTP-D1 and PTP-D2 are remarkably similar (35). The main difference in the structure between the two PTPase domains is that the phosphotyrosine binding pocket is covered in PTP-D2; the cysteine residue analogous to Dlar Cys1929 in LAR PTP-D2 is buried in the interior of the protein and is not accessible to the external environment (35), which provides a structural explanation for why PTP-D2 is not catalytically active. However, we have demonstrated that LAR PTP-D2 can become a highly active PTPase in vitro if two specific point mutations (Leu1644Tyr and Glu1779Asp) are made within PTP-D2 (35). Thus, it is possible that if the correct substrate was assayed or if LAR protein with the appropriate posttranslational modifications was isolated, physiologically relevant amounts of PTPase activity derived from PTP-D2 would be detected. However, the capacity of the Dlar-C1929S transgene to fully rescue lethality indicates that, unlike the essential docking and regulatory functions of PTP-D2, any PTPase catalytic activity that may be derived from PTP-D2 is nonessential for survival.
RPTPs are characterized by the structurally diverse extracellular domains, including Ig-like and FnIII domains, that are also present in several cell adhesion molecules. The combination of these cell adhesion molecule-like extracellular domains with intracellular PTPase domains enables RPTPs to directly couple extracellular adhesion-mediated events to intracellular signaling to modulate phosphotyrosine levels. Identification of physiologically relevant ligands for RPTPs has proven to be difficult, and delineation of functional domains may help identification of RPTP ligands. The large Dlar ectodomain clearly participates in multiple distinct interactions. The three Ig-like domains of Dlar are required for survival, and the FnIII domains are critical for regulating oogenesis, suggesting that different sets of receptor-ligand interactions are involved in different contexts. The data in this report are consistent with the idea that the Dlar ectodomain binds to more than one individual ligand or, at minimum, that Dlar binds to a single ligand at multiple different sites in its ectodomain. The lethality rescue data show that in the absence of FnIII domains, interaction of ligand with the Ig-like domains alone is sufficient to completely substitute for Dlar-WT. Alternatively, it is also possible that the Ig-like domains and/or the Fn domains participate in the formation and stability of Dlar homodimers or in heterodimer formation with other cell surface protein(s) and that deletion of these domains prevents the assembly of a functional multiprotein receptor complex.
The removal of Ig-like domains dramatically reduces survival frequency, but ∼30% of adults do survive despite the lack of Ig-like domains. There are several possible mechanisms for the residual activity of the DlarΔIg123 mutant. First, the relevant ligand may bind to both Ig-like and FnIII domains, but with a higher affinity to Ig-like domains. In the absence of Ig-like domains, however, low-affinity interaction of the ligand with the FnIII domains may be sufficient to elicit low-level Dlar function. Such differential ligand binding affinity has been observed in binding of the fibroblast growth factor receptor with its ligands (48). Second, expression of the intact cytoplasmic PTPase domains present in these mutants may elicit low-level Dlar function even in the absence of any extracellular ligand binding. Third, Dlar may form a heterodimer with a coreceptor, and its ligand may still bind the coreceptor with a reduced efficiency.
In contrast to the rescue-of-lethality data, rescue of the Dlar−/− oogenesis phenotype indicates that the Ig-like domains are not required for Dlar functional regulation of oocyte elongation, whereas the FnIII domains play a major role. A subset of the FnIII domains, Fn1 to Fn6, are required for proper Dlar function in oogenesis, suggesting potential heterogeneity in ligand presentation in different developmental contexts. The basal surface of follicle cells is closely associated with ECM that is rich in laminin (22). Given the findings that an FnIII domain of human LAR binds to an ECM complex of laminin and nidogen (36) and that Dlar and integrins integrate ECM signals to regulate actin polymerization during oogenesis (3), it is possible that a similar interaction occurs between the FnIII domains of Dlar and the laminin-rich ECM during oogenesis. Indeed, it is also plausible that the Fn domains may function in neural growth cones as a sensor-receptor for specific elements of the ECM during axon pathfinding. Alternatively, other extracellular ligands or cis-interacting proteins on follicle cell membranes may require FnIII domains for proper function in this context.
Expression of the DlarΔFn2-9 transgene not only failed to rescue egg-shape defects, it also caused a dramatic increase in the penetrance of Dlar−/− round stage 14 oocytes observed. We believe that DlarΔFn2-9 causes this phenotype as a result of the Dlar cytoplasmic domain titrating away downstream factors common to multiple signaling pathways, since a truncation mutant lacking all of the Dlar extracellular domain has the same effect (3). This interpretation is consistent with observations of the embryonic nervous system, where Dlar cooperates with a family of RPTPs to regulate axon guidance and outgrowth (15). Multiple RPTPs are also expressed during oogenesis (16) and may function in parallel with Dlar to regulate follicle cell polarity. It is likely that Dlar lacking a functional extracellular domain could titrate important downstream factors from other phosphatases, interfering with their function.
It is clear from the data in this report that Dlar participates in signal transduction pathways in several distinct contexts. The Ig-like domains appear to function independently of the FnIII domains in ligand recognition, and the PTPase domains also perform distinct functional roles. Given the genetic and biochemical evidence that the multifunctional protein Trio, which contains Ig-like domains, coiled-coil domains, Rac guanine nucleotide exchange factor domains, Rho guanine nucleotide exchange factor domains, and a serine/threonine kinase-like domain, is part of LAR/Dlar signal transduction pathways, it is becoming apparent that LAR subfamily RPTPs participate in multiple and complex signal transduction pathways, making identification of physiologically relevant substrates and ligands all the more difficult and increasingly important.
Acknowledgments
Neil X. Krueger and R. Sreekantha Reddy contributed equally to this work.
We thank David Luyimbazi for excellent technical assistance.
This work was supported by grants from the NIH (GM53415), the International Human Frontier Science Program (RG0122), and the Ministry of Education, Culture, Sports, Science and Technology of Japan to H.S. and by NIH grants (NS40043 and NS35909) to D.V.V. D.V.V. is a Leukemia and Lymphoma Society Scholar. J.B. was supported by an NSERC fellowship.
REFERENCES
- 1.Allard, J. D., R. Herbst, P. M. Carroll, and M. A. Simon. 1998. Mutational analysis of the SRC homology 2 domain protein-tyrosine phosphatase Corkscrew. J. Biol. Chem. 273:13129-13135. [DOI] [PubMed] [Google Scholar]
- 2.Aricescu, A. R., I. W. McKinnell, W. Halfter, and A. W. Stoker. 2002. Heparan sulfate proteoglycans are ligands for receptor protein tyrosine phosphatase σ. Mol. Cell. Biol. 22:1881-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bateman, J., R. S. Reddy, H. Saito, and D. Van Vactor. 2001. The receptor tyrosine phosphatase Dlar and integrins organize actin filaments in the Drosophila follicular epithelium. Curr. Biol. 11:1317-1327. [DOI] [PubMed] [Google Scholar]
- 4.Bateman, J., H. Shu, and D. Van Vactor. 2000. The guanine nucleotide exchange factor trio mediates axonal development in the Drosophila embryo. Neuron 26:93-106. [DOI] [PubMed] [Google Scholar]
- 5.Bilwes, A. M., J. den Hertog, T. Hunter, and J. P. Noel. 1996. Structural basis for inhibition of receptor protein-tyrosine phosphatase-α by dimerization. Nature 382:555-559. [DOI] [PubMed] [Google Scholar]
- 6.Blanchetot, C., L. G. Tertoolen, and J. den Hertog. 2002. Regulation of receptor protein-tyrosine phosphatase α by oxidative stress. EMBO J. 21:493-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanchetot, C., L. G. Tertoolen, J. Overvoorde, and J. den Hertog. 2002. Intra- and intermolecular interactions between intracellular domains of receptor protein-tyrosine phosphatases. J. Biol. Chem. 277:47263-47269. [DOI] [PubMed] [Google Scholar]
- 8.Blume-Jensen, P., and T. Hunter. 2001. Oncogenic kinase signalling. Nature 411:355-365. [DOI] [PubMed] [Google Scholar]
- 9.Brady-Kalnay, S. M., A. J. Flint, and N. K. Tonks. 1993. Homophilic binding of PTPμ, a receptor-type protein tyrosine phosphatase, can mediate cell-cell aggregation. J. Cell Biol. 122:961-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brand, A. H., and N. Perrimon. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401-415. [DOI] [PubMed] [Google Scholar]
- 11.Cho, H., R. Krishnaraj, E. Kitas, W. Bannwarth, C. T. Walsh, and K. S. Anderson. 1992. Isolation and structural elucidation of a novel phosphocysteine intermediate in the LAR protein tyrosine phosphatase enzymatic pathway. J. Am. Chem. Soc. 114:7296-7298. [Google Scholar]
- 12.Clandinin, T. R., C. H. Lee, T. Herman, R. C. Lee, A. Y. Yang, S. Ovasapyan, and S. L. Zipursky. 2001. Drosophila LAR regulates R1-R6 and R7 target specificity in the visual system. Neuron 32:237-248. [DOI] [PubMed] [Google Scholar]
- 13.Debant, A., C. Serra-Pages, K. Seipel, S. O'Brien, M. Tang, S. H. Park, and M. Streuli. 1996. The multidomain protein Trio binds the LAR transmembrane tyrosine phosphatase, contains a protein kinase domain, and has separate rac-specific and rho-specific guanine nucleotide exchange factor domains. Proc. Natl. Acad. Sci. USA 93:5466-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desai, C. J., J. G. Gindhart, Jr., L. S. B. Goldstein, and K. Zinn. 1996. Receptor tyrosine phosphatases are required for motor axon guidance in the Drosophila embryo. Cell 84:599-609. [DOI] [PubMed] [Google Scholar]
- 15.Desai, C. J., N. X. Krueger, H. Saito, and K. Zinn. 1997. Competition and cooperation among receptor tyrosine phosphatases control motoneuron growth cone guidance in Drosophila. Development 124:1941-1952. [DOI] [PubMed] [Google Scholar]
- 16.Fitzpatrick, K. A., S. M. Gorski, Z. Ursuliak, and J. V. Price. 1995. Expression of protein tyrosine phosphatase genes during oogenesis in Drosophila melanogaster. Mech. Dev. 53:171-183. [DOI] [PubMed] [Google Scholar]
- 17.Frydman, H. M., and A. C. Spradling. 2001. The receptor-like tyrosine phosphatase LAR is required for epithelial planar polarity and for axis determination within Drosophila ovarian follicles. Development 16:3209-3220. [DOI] [PubMed] [Google Scholar]
- 18.Furukawa, F., M. Itoh, N. X. Krueger, M. Streuli, and H. Saito. 1994. Specific interaction of the CD45 protein-tyrosine phosphatase with tyrosine-phosphorylated CD3 ζ chain. Proc. Natl. Acad. Sci. USA 91:10928-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garrity, P. A., C. H. Lee, I. Salecker, H. C. Robertson, C. J. Desai, K. Zinn, and S. L. Zipursky. 1999. Retinal axon target selection in Drosophila is regulated by a receptor protein tyrosine phosphatase. Neuron 22:707-717. [DOI] [PubMed] [Google Scholar]
- 20.Gebbink, M. F., G. C. Zondag, R. W. Wubbolts, R. L. Beijersbergen, I. van Etten, and W. H. Moolenaar. 1993. Cell-cell adhesion mediated by a receptor-like protein tyrosine phosphatase. J. Biol. Chem. 268:16101-16104. [PubMed] [Google Scholar]
- 21.Guan, K., and J. E. Dixon. 1991. Evidence for protein-tyrosine-phosphatase catalysis proceeding via a cysteine-phosphate intermediate. J. Biol. Chem. 266:17026-17030. [PubMed] [Google Scholar]
- 22.Gutzeit, H. O., W. Eberhardt, and E. Gratwohl. 1991. Laminin and basement membrane-associated microfilaments in wild-type and mutant Drosophila ovarian follicles. J. Cell Sci. 100:781-788. [DOI] [PubMed] [Google Scholar]
- 23.Itoh, M., M. Streuli, N. X. Krueger, and H. Saito. 1992. Purification and characterization of the catalytic domains of the human receptor-linked protein tyrosine phosphatases HPTPβ, leukocyte common antigen (LCA), and leukocyte common antigen-related molecule (LAR). J. Biol. Chem. 267:12356-12363. [PubMed] [Google Scholar]
- 24.Jiang, G., J. den Hertog, and T. Hunter. 2000. Receptor-like protein tyrosine phosphatase α homodimerizes on the cell surface. Mol. Cell. Biol. 20:5917-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang, G., J. den Hertog, J. Su, J. Noel, J. Sap, and T. Hunter. 1999. Dimerization inhibits the activity of receptor-like protein-tyrosine phosphatase-α. Nature 401:606-610. [DOI] [PubMed] [Google Scholar]
- 26.Kaufmann, N., J. DeProto, R. Ranjan, H. Wan, and D. Van Vactor. 2002. Drosophila liprin-α and the receptor phosphatase Dlar cooperate during synapse morphogenesis. Neuron 34:27-38. [DOI] [PubMed] [Google Scholar]
- 27.Kaufmann, N., Z. P. Wills, and D. Van Vactor. 1998. Drosophila Rac1 controls motor axon guidance. Development 125:453-461. [DOI] [PubMed] [Google Scholar]
- 28.Krueger, N. X., and H. Saito. 1992. A human transmembrane protein-tyrosine-phosphatase, PTPζ, is expressed in brain and has an N-terminal receptor domain homologous to carbonic anhydrases. Proc. Natl. Acad. Sci. USA 89:7417-7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krueger, N. X., M. Streuli, and H. Saito. 1990. Structural diversity and evolution of human receptor-like protein tyrosine phosphatases. EMBO J. 9:3241-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krueger, N. X., D. Van Vactor, H. I. Wan, W. M. Gelbart, C. S. Goodman, and H. Saito. 1996. The transmembrane phosphatase DLAR controls motor axon guidance in Drosophila. Cell 84:611-622. [DOI] [PubMed] [Google Scholar]
- 31.Lin, D. M., and C. S. Goodman. 1994. Ectopic and increased expression of fasciclin II alters motoneuron growth cone guidance. Neuron 13:507-523. [DOI] [PubMed] [Google Scholar]
- 32.Maurel-Zaffran, C., T. Suzuki, G. Gahmon, J. E. Treisman, and B. J. Dickson. 2001. Cell-autonomous and -nonautonomous functions of LAR in R7 photoreceptor axon targeting. Neuron 32:225-235. [DOI] [PubMed] [Google Scholar]
- 33.Meng, K., A. Rodriguez-Pena, T. Dimitrov, W. Chen, M. Yamin, M. Noda, and T. F. Deuel. 2000. Pleiotrophin signals increased tyrosine phosphorylation of β-catenin through inactivation of the tyrosine phosphatase β/ζ. Proc. Natl. Acad. Sci. USA 97:2603-2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mustelin, T., G. S. Feng, N. Bottini, A. Alonso, N. Kholod, D. Birle, J. Merlo, and H. Huynh. 2002. Protein tyrosine phosphatases. Front. Biosci. 7:85-142. [DOI] [PubMed] [Google Scholar]
- 35.Nam, H.-J., F. Poy, N. X. Krueger, H. Saito, and C. A. Frederick. 1999. Crystal structure of the tandem phosphatase domains of RPTP LAR. Cell 97:449-457. [DOI] [PubMed] [Google Scholar]
- 36.O'Grady, P., T. C. Thai, and H. Saito. 1998. The laminin-nidogen complex is a ligand for a specific splice isoform of the transmembrane protein tyrosine phosphatase LAR. J. Cell Biol. 141:1675-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan, M.-G., K. P. Rim, T. Lu, T. Florio, and P. J. Stork. 1993. Cloning and expression of two structurally distinct receptor-linked protein-tyrosine phosphatases generated by RNA processing from a single gene. J. Biol. Chem. 268:19284-19291. [PubMed] [Google Scholar]
- 38.Pulido, R., N. X. Krueger, C. Serra-Pages, H. Saito, and M. Streuli. 1995. Molecular characterization of the human transmembrane protein-tyrosine phosphatase δ. J. Biol. Chem. 270:6722-6728. [DOI] [PubMed] [Google Scholar]
- 39.Serra-Pages, C., N. L. Kedersha, L. Fazikas, Q. Medley, A. Debant, and M. Streuli. 1995. The LAR transmembrane protein tyrosine phosphatase and a coiled-coil LAR-interacting protein co-localize at focal adhesions. EMBO J. 14:2827-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Serra-Pages, C., Q. G. Medley, M. Tang, A. Hart, and M. Streuli. 1998. Liprins, a family of LAR transmembrane protein-tyrosine phosphatase-interacting proteins. J. Biol. Chem. 273:15611-15620. [DOI] [PubMed] [Google Scholar]
- 41.Streuli, M., N. X. Krueger, P. D. Ariniello, M. Tang, J. M. Munro, W. A. Blattler, D. A. Adler, C. M. Disteche, and H. Saito. 1992. Expression of the receptor-linked protein tyrosine phosphatase LAR: proteolytic cleavage and shedding of the CAM-like extracellular region. EMBO J. 11:897-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Streuli, M., N. X. Krueger, L. R. Hall, S. F. Schlossman, and H. Saito. 1988. A new member of the immunoglobulin superfamily that has a cytoplasmic region homologous to the leukocyte common antigen. J. Exp. Med. 168:1523-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Streuli, M., N. X. Krueger, T. Thai, M. Tang, and H. Saito. 1990. Distinct functional roles of the two intracellular phosphatase like domains of the receptor-linked protein tyrosine phosphatases LCA and LAR. EMBO J. 9:2399-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Streuli, M., N. X. Krueger, A. Y. M. Tsai, and H. Saito. 1989. A family of receptor-linked protein tyrosine phosphatases in humans and Drosophila. Proc. Natl. Acad. Sci. USA 86:8698-8702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tian, S. S., P. Tsoulfas, and K. Zinn. 1991. Three receptor-linked protein-tyrosine phosphatases are selectively expressed on central nervous system axons in the Drosophila embryo. Cell 67:675-685. [DOI] [PubMed] [Google Scholar]
- 46.Van Vactor, D., H. Sink, D. Fambrough, R. Tsoo, and C. S. Goodman. 1993. Genes that control neuromuscular specificity in Drosophila. Cell 73:1137-1153. [DOI] [PubMed] [Google Scholar]
- 47.Wallace, M. J., C. Fladd, J. Batt, and D. Rotin. 1998. The second catalytic domain of protein tyrosine phosphatase δ binds to and inhibits the first catalytic domain of PTPσ. Mol. Cell. Biol. 18:2608-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, F., M. Kan, G. Yan, J. Xu, and W. L. McKeehan. 1995. Alternately spliced NH2-terminal immunoglobulin-like Loop I in the ectodomain of the fibroblast growth factor (FGF) receptor 1 lowers affinity for both heparin and FGF-1. J. Biol. Chem. 270:10231-10235. [DOI] [PubMed] [Google Scholar]
- 49.Wang, Y., and J. C. Pallen. 1991. The receptor-like protein tyrosine phosphatase HPTPα has two active catalytic domains with distinct substrate specificities. EMBO J. 10:3231-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wills, Z., J. Bateman, C. A. Korey, A. Comer, and D. Van Vactor. 1999. The tyrosine kinase Abl and its substrate Enabled collaborate with the receptor phosphatase Dlar to control motor axon guidance. Neuron 22:301-312. [DOI] [PubMed] [Google Scholar]
- 51.Zondag, G. C., G. M. Koningstein, Y. P. Jiang, J. Sap, W. H. Moolenaar, and M. F. Gebbink. 1995. Homophilic interactions mediated by receptor tyrosine phosphatases μ and κ. A critical role for the novel extracellular MAM domain. J. Biol. Chem. 270:14247-14250. [DOI] [PubMed] [Google Scholar]