Abstract
Hes1 is a mammalian basic helix-loop-helix transcriptional repressor that inhibits neuronal differentiation together with corepressors of the Groucho (Gro)/Transducin-like Enhancer of split (TLE) family. The interaction of Hes1 with Gro/TLE is mediated by a WRPW tetrapeptide present in all Hairy/Enhancer of split (Hes) family members. In contrast to Hes1, the related protein Hes6 promotes neuronal differentiation. Little is known about the molecular mechanisms that underlie the neurogenic activity of Hes6. It is shown here that Hes6 antagonizes Hes1 function by two mechanisms. Hes6 inhibits the interaction of Hes1 with its transcriptional corepressor Gro/TLE. Moreover, it promotes proteolytic degradation of Hes1. This effect is maximal when both Hes1 and Hes6 contain the WRPW motif and is reduced when Hes6 is mutated to eliminate a conserved site (Ser183) that can be phosphorylated by protein kinase CK2. Consistent with these findings, Hes6 inhibits Hes1-mediated transcriptional repression in cortical neural progenitor cells and promotes the differentiation of cortical neurons, a process that is normally inhibited by Hes1. Mutation of Ser183 impairs the neurogenic ability of Hes6. Taken together, these findings clarify the molecular events underlying the neurogenic function of Hes6 and suggest that this factor can antagonize Hes1 activity by multiple mechanisms.
In the developing mammalian central nervous system (CNS), differentiated neuronal and glial cells derive from multipotent neural progenitor cells located in the proliferative zone of the neural tube. The commitment of these progenitor cells to the neuronal lineage is regulated by the antagonistic activities of a number of positively and negatively acting transcription factors containing the basic helix-loop-helix (bHLH) DNA-binding and dimerization motif (reviewed in references 2 and 18). Neurogenic bHLH factors include several evolutionarily conserved molecules related to the proneural proteins Atonal and Achaete-Scute of Drosophila (8, 13, 21). They function by forming heterodimers with the ubiquitous bHLH protein E47. These dimers bind to DNA sequences commonly referred to as E boxes (CANNTG) and transactivate the expression of genes that promote the acquisition of the neuronal fate (17, 32).
Antineurogenic bHLH factors include members of the Hairy/Enhancer of split (Hes) family (1, 26, 32). In contrast to proneural proteins, Hes factors like Hes1 and Hes5 mediate transcriptional repression and bind preferentially to DNA sequences referred to as N boxes (CACNAG) (32). They are thought to inhibit neuronal differentiation by antagonizing the neurogenic activity of the proneural proteins via multiple mechanisms, including direct involvement in the negative regulation of proneural gene expression (4, 20) and inhibition of the activity of E47-proneural protein heterodimers (1, 3, 32). Genetic perturbations that alter the normal balance of the activities of proneural and antineurogenic bHLH proteins have dramatic effects on CNS development in vivo, underscoring the importance of understanding how the functions of these factors are normally regulated (8, 16, 26, 36).
The Hes1 gene is initially expressed in proliferating neural progenitor cells and becomes down-regulated during the progenitor-to-neuron transition (32). Persistent expression of Hes1 inhibits neuronal development, whereas disruption of Hes1 function results in the premature differentiation of neuronal cells and the up-regulation of proneural genes (15, 16, 36). These observations indicate that Hes1 acts in neural progenitor cells to control the timing of neuronal differentiation. Molecular mechanisms that contribute to the positive or negative regulation of Hes1 activity in neural progenitor cells are beginning to be elucidated. In particular, studies with both invertebrate and vertebrate species show that antineurogenic Hes proteins are coexpressed, and directly interact, with general transcriptional corepressors of the Groucho/Transducin-like Enhancer of split (Gro/TLE) family (7, 12, 24, 25, 29, 34, 40). This interaction is mediated by a WRPW tetrapeptide motif present at the carboxy termini of all Hes proteins (7, 11, 24). Mutations that disrupt the Hes-Gro/TLE interactions impair the ability of Hes proteins to mediate transcriptional repression (7, 24, 29). Moreover, Hes1 activates phosphorylation mechanisms that promote the transcription repression function of Gro/TLE (25). Together, these observations identify Gro/TLE proteins as positive regulators of Hes activity and suggest that Hes1 acts by recruiting hyperphosphorylated Gro/TLE to specific DNA sites where the latter mediate transcriptional repression (25).
Another protein that has recently been implicated in the regulation of Hes1 activity is the related Hes family member Hes6 (3, 19). The Hes6 gene is expressed throughout the developing CNS, where it is found in both undifferentiated neural progenitors and differentiated neurons (3, 19, 30, 38). In contrast to Hes1, Hes6 acts as a positive regulator of neuronal differentiation in both murine retinal explants and Xenopus embryos (3, 19). Although little is known about the molecular mechanisms underlying the neurogenic ability of Hes6, a number of observations suggest that Hes6 may promote neurogenesis by antagonizing the function of Hes1. Studies with transfected nonneural cells show that Hes6 can heterodimerize with Hes1 and can inhibit the ability of Hes1 to both repress transcription from promoters containing N box sequences and suppress the activity of E47-proneural protein heterodimers (3). In addition, Hes6 does not require an intrinsic DNA-binding ability to promote neurogenesis, because mutation of the basic arm of its bHLH domain does not abolish its neurogenic ability in vivo (19). Together, these observations suggest that Hes6 may promote neuronal differentiation via DNA-binding-independent events that involve a negative regulation of Hes1 function in the CNS. Virtually nothing is known, however, about the molecular mechanisms underlying this inhibitory effect.
Here we describe experiments showing that Hes6 negatively regulates Hes1 activity by at least two mechanisms. Hes6 inhibits the interaction of Hes1 with Gro/TLE. In addition, it promotes proteolytic degradation of Hes1. This effect is maximal when both Hes1 and Hes6 contain the WRPW motif, and it is reduced by a point mutation (S183A) that removes a consensus site for phosphorylation by protein kinase CK2. In agreement with these findings, Hes6 inhibits Hes1-mediated transcriptional repression in cortical neural progenitor cells and promotes their neuronal differentiation. Moreover, the S183A mutation attenuates Hes6 phosphorylation by protein kinase CK2 and impairs the ability of Hes6 to promote neuronal differentiation. Taken together, these findings identify novel mechanisms through which Hes6 may act as a negative regulator of Hes1 activity and a positive regulator of neuronal differentiation.
MATERIALS AND METHODS
Plasmids.
PCR was used to amplify the sequences encoding full-length Hes6 (oligonucleotide primers Hes6-1 [5′-GACCATGGCTCCGTCCCA] and Hes6-2 [5′-TCACCAAGGCCTCCACACACTC]) or Hes6ΔWRPW (oligonucleotide primers Hes6-1 and Hes6-3 [5′-TCACACACTCTGAGCCCGGCGAGC]) with the full-length Hes6 cDNA Image clone W66929 as the template (5). The sequence encoding a truncated form of Hes6 lacking the first 13 amino acids [Hes6(14-224)] was also amplified by PCR (oligonucleotide primers Hes6-4 [5′-TCAGGAGGATGAGGACCGCTGGGAA] and Hes6-2); Hes6 and Hes6(14-224) behaved equally in our studies. PCR products were subcloned into the pcDNA3-GAL4bd vector digested with BamHI (followed by filling in with Klenow DNA polymerase) or into the pCMV2-HA plasmid digested with EcoRV or SmaI. The pCMV2-HA-Hes6(S183A) plasmid was obtained by first generating the sequence encoding the indicated point mutation by using a PCR-based strategy (the mutated oligonucleotide primers were Hes6-5F [5′-GACCTGTGTGCTGACCTAGAGGAGAT] and Hes6-5R [5′-TCTAGGTCAGCACACAGGTCGT]), followed by subcloning into pBluescript plasmid and DNA sequencing. The verified mutant sequence was then subcloned into pCMV2-HA-Hes6 digested with SmaI, replacing the wild-type sequence. Constructs for the bacterial expression of fusion proteins of glutathione S-transferase (GST) and Hes6 or Hes6(S183A) were obtained by digesting pCMV2-HA-Hes6 or pCMV2-HA-Hes6(S183A) with BglII and BamHI, followed by subcloning into pGEX1 digested with BamHI. The pGEX1-Hes1 DNA has been described previously (23). Constructs pEBG-Hes6 and pEBG-Hes6ΔWRPW were generated by digesting pcDNA3-GAL4bd-Hes6 or pcDNA3-GAL4bd-Hes6ΔWRPW, respectively, with EcoRI, followed by filling in with Klenow DNA polymerase and subcloning into the filled-in ClaI site of pEBG to generate plasmids for the expression of fusion proteins of GST and Hes6 or Hes6ΔWRPW in mammalian cells. Plasmid pCMV2-FLAG-Hes1ΔWRPW:Gro/TLE1 was generated by first subcloning the region encoding Hes1ΔWRPW (obtained by PCR amplification with primers Hes1-1 [5′-AATGCCAGCTGATATAATGGAG] and Hes1-2 [5′-ACATGGAGTCCGCAGTGAGCGA]) into pCMV2-FLAG digested with EcoRV. This was followed by in-frame ligation of the sequence encoding Gro/TLE1 (also obtained by PCR with primers Gro/TLE1-1 [5′-GGATGTTCCCGCAGAGCCGG] and Gro/TLE1-2 [5′-TCAGTAGATGACTTCATAGAC]) into an XbaI site located downstream of the last codon of Hes1. Ligation products were analyzed and confirmed by sequencing. Plasmids pCMV2-FLAG-Hes1, pCMV2-FLAG-Hes1ΔWRPW, pEBG-Hes1, and pEBG-Hes1ΔWRPW have been described previously (12, 23, 24). Plasmids pFOX-Luc1, pFOX-ngn3p-Luc1 (containing a portion of the neurogenin3 [ngn3] promoter extending ∼2.6 kbp upstream of the transcription start site) and pFOX-ΔN-box-ngn3p-Luc1 (containing a mutated version of the ∼2.6-kbp ngn3 promoter lacking the Hes1-binding sites located within 200 bp proximal to the transcription start site) have been described previously (20).
Transient transfections, protein-protein interaction assays, and Western blot analysis.
Human 293A cells were cultured and transfected by using the SuperFect reagent (Qiagen) as described previously (23-25). When appropriate, transfected cells were incubated for 6 h in the presence of 10 μM MG132 (Calbiochem) prior to cell lysis. Treatment of cell lysates with calf intestinal phosphatase was performed as described previously (14). To examine the effect of Hes6 on Hes1 stability, cells were transfected for 36 h with pCMV2-FLAG-Hes1/Hes1ΔWRPW (50 ng/transfection) in the absence or presence of Hes6, Hes6ΔWRPW, or Hes6(S183A) expression plasmids (200 to 800 ng/transfection). To examine the effect of Hes6 on the Hes1-Gro/TLE interaction, cells were transfected for 24 h with Hes1 or Hes1ΔWRPW expression plasmids (100 to 200 ng/transfection) in the absence or presence of Hes6 or Hes6ΔWRPW expression plasmids (100 to 200 ng/transfection). Cell lysates were prepared, and GST coprecipitation (23, 24), immunoprecipitation (14, 40), and Western blotting (6, 25, 28) studies were performed as described previously. The antibodies used were panTLE (6, 28, 34), anti-GST and anti-GAL4bd (Santa Cruz Biotechnology), antihemagglutinin (anti-HA) (Roche), or anti-FLAG (Sigma).
In vitro phosphorylation of bacterially purified Hes proteins.
Fusion proteins of GST and Hes6 or Hes6(S183A) were purified from bacteria as described previously (12, 23). Roughly 50 ng of each fusion protein was resuspended in buffer A (50 mM Tris-HCl [pH 7.6], 10 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol, 0.1% Triton X-100, 200 μm ATP) containing 200 μCi of [γ-32P]ATP per ml in the presence of 0.5 U of purified protein kinase CK2 (New England Biolabs) per μl for 15 min at 30°C. Reactions were terminated by the addition of 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and incubation at 65°C for 5 min. After gel electrophoresis, proteins were transferred to nitrocellulose and exposed to film. After autoradiography, membranes were subjected to Western blotting with anti-GST antibodies.
Telencephalic neural progenitor cell cultures.
Primary neural progenitor cell cultures were established from dorsal telencephalic cortices dissected from embryonic day 13.5 (E13.5) mouse embryos as described previously (10, 33). Cells were seeded into either four-well chamber slides (Nalge Nunc International) for immunocytochemical studies or six-well dishes (BD Labware) for transcription assays. All chambers and dishes were coated with 0.1% poly-d-lysine and 0.2% laminin (BD Biosciences). Cells were cultured in Neurobasal medium supplemented with 1% N2, 2% B27, 0.5 mM glutamine, 1% penicillin-streptomycin (Invitrogen), and 40 ng of fibroblast growth factor 2 (Collaborative Research) per ml.
Transient-transfection and transcription studies with neural progenitor cells.
Approximately 1.5 × 106 cells/ml were seeded at the start of the experiments. After 24 h in vitro (day 1), when ∼90% of the cultured cells were mitotic (10, 25, 35), transfections were performed by mixing the appropriate combinations of plasmids (the total amount of DNA was adjusted to 2.0 μg/well in each transfection) with OptiMEM medium (Invitrogen). An equal volume of OptiMEM medium was mixed separately with Lipofectamine 2000 reagent (Invitrogen) (1.25 μl/μg of DNA) and then combined with the DNA mixture and incubated for 20 min. The DNA-Lipofectamine 2000 mix was then added dropwise to each well. In each case, a pRSV-β-galactosidase DNA was cotransfected to provide a means of normalizing the assays for transfection efficiency. Twenty-four hours after transfection, cells were harvested and luciferase and β-galactosidase activities were determined as described previously (23-25). Results were expressed as mean values ± standard deviations (SD).
Immunocytochemical analysis of differentiating neural progenitor cells.
Approximately 4 × 105 cells/ml were seeded at the start of the experiments. After 48 h in vitro, cells were transfected as described above by using plasmids encoding either enhanced green fluorescent protein (GFP) alone (0.2 μg/well) or combinations of GFP and Hes6, Hes6ΔWRPW, or Hes6(S183A) (0.5 μg of Hes6 plasmid per well). The total amount of DNA was adjusted to 1.0 μg. Cells were allowed to differentiate until day 4 to 5 in vitro, when they were fixed and subjected to double-label immunocytochemical analysis of the expression of GFP, nestin (a marker of undifferentiated neural progenitor cells), or MAP2 or NeuN (markers of differentiated neurons) as described (33, 35). Antinestin (BD PharMingen), anti-MAP2 (Sigma), or anti-NeuN (Chemicon) antibodies were used. Digital image acquisition and analysis were performed with the Northern Eclipse software (Empix Inc.). Results were expressed as mean values ± SD.
RESULTS
Promotion of cortical neurogenesis by Hes6.
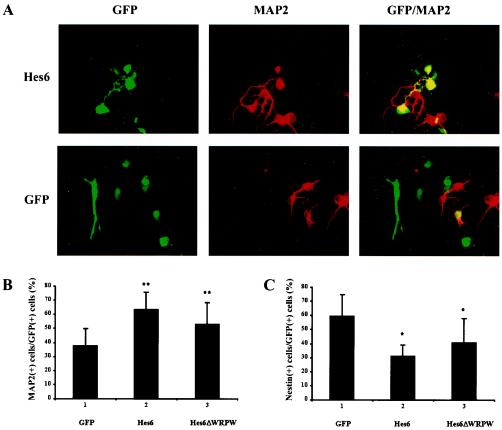
Hes6 was shown to promote neuronal differentiation in Xenopus embryos and mouse retinal explants (3, 19). To determine whether Hes6 might also promote the development of cortical neurons, we transfected exogenous Hes6 in primary cultures of neural progenitor cells isolated from the dorsal telencephalons of E13.5 mouse embryos. These cortical progenitors endogenously express Hes6 (reference 3 and data not shown), as well as Hes1 and Gro/TLE (6, 32, 40). Enhanced GFP was coexpressed to mark the transfected cells. Exogenous Hes6 expression led to a significant increase in the number of differentiated neurons compared to that with GFP alone, as revealed by immunocytochemistry with antibodies against the neuron-specific protein MAP2 (Fig. 1A and B [cf. bars 1 and 2]). This increase was correlated with a decrease in the number of undifferentiated neural progenitor cells expressing the protein nestin (Fig. 1C, cf. bars 1 and 2). These results thus show that Hes6 promotes cortical neuronal differentiation. Since previous studies have shown that the neurogenic ability of Xenopus Hes6 does not require its carboxy-terminal WRPW motif involved in Gro/TLE binding (19), we next examined whether Hes6ΔWRPW, a truncated form lacking this motif, would also promote cortical neuronal differentiation. Exogenous Hes6ΔWRPW also caused an increase in the number of differentiated neurons, although less effectively than Hes6 (Fig. 1B, cf. bars 1 to 3). Hes6 and Hes6ΔWRPW were expressed at equivalent levels (see Fig. 3A). Together, these findings strongly suggest that Hes6 promotes the differentiation of cortical progenitor cells into postmitotic neurons. They further suggest that its WRPW motif is not required for, but contributes to, a maximal neurogenic effect. This is consistent with the finding that although both Hes6 and Hes6ΔWRPW can promote neurogenesis in Xenopus embryos, the former elicited a more robust neurogenic effect than the latter (19).
FIG. 1.
Promotion of cortical neurogenesis by Hes6. (A) Primary cultures of E13.5 mouse embryonic cortical neural progenitor cells were transfected with plasmids encoding either GFP alone (bottom panels) or a combination of GFP and Hes6 (top panels). Forty-eight hours later, cells were fixed and subjected to double-labeling analysis of the expression of GFP (left panels) or MAP2 (middle panels). The combined GFP and MAP2 staining is shown in the right panels. (B and C) Quantitation of the percentage of GFP-MAP2-double-positive cells (B) or of cells in similar double-labeling experiments conducted in parallel with antibodies against nestin. Results are shown as the means ± SD (n = 5). *, P < 0.01; **, P < 0.001.
FIG. 3.
Inhibition of the coimmunoprecipitation of Gro/TLE with Hes1 by Hes6 and Hes6ΔWRPW. 293A cells were transfected with plasmids encoding Hes1 or Hes1ΔWRPW in the absence or presence of HA-Hes6 or HA-Hes6ΔWRPW, as described in Materials and Methods. One-tenth of each cell lysate was subjected to SDS-PAGE (A to C) and the remaining lysates were subjected to immunoprecipitation (IP) with anti-FLAG antibodies (D and E). Samples were analyzed by Western blotting (WB) with anti-HA (A), anti-FLAG (B and D), or anti-Gro/TLE (C and E) antibodies (Ab.). The arrow in panel D points to the position of migration of Hes1. The arrow in panel E points to the position of migration of Gro/TLE, and the arrowhead indicates a nonspecific band. IgG H., immunoglobulin G heavy chains. Positions of size standards are indicated in kilodaltons.
Comparison of the interaction of Hes6 or Hes1 with Gro/TLE.
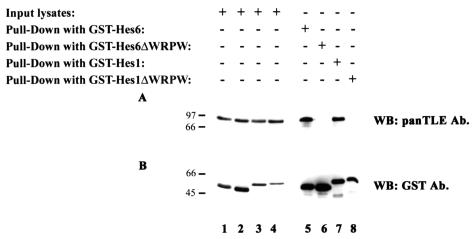
To elucidate the molecular mechanisms underlying the neurogenic activity of Hes6, we tested whether this function might involve an inhibition of the antineurogenic activity of Hes1. Both Hes1 and Hes6 bind to Gro/TLE (9, 12, 24, 25) and are coexpressed with the latter in a number of tissues (3, 6, 9, 12, 32, 34, 39). In particular, Hes1 and Hes6 are coexpressed in neural progenitor cells but not in differentiated neurons, where Hes6 continues to be expressed while Hes1 is down-regulated (3, 19, 32). This suggested that Hes6 might act as a negative regulator of Hes1 activity in neural progenitors by competing with Hes1 for binding to Gro/TLE, thus titrating away the corepressor function that Gro/TLE provides to Hes1. To examine this possibility, we first tested whether Hes6 had a higher affinity than Hes1 for Gro/TLE. 293A cells that express endogenous Gro/TLE (Fig. 2A, lanes 1 to 4) were transfected with plasmids encoding either GST-Hes6 or GST-Hes1. The precipitation of equivalent amounts of these fusion proteins (Fig. 2B, cf. lanes 5 and 7) resulted in the coprecipitation of equivalent amounts of endogenous Gro/TLE (Fig. 2A, cf. lanes 5 and 7). In contrast, expression of fusion proteins of GST and truncated forms of Hes6 or Hes1 lacking the WRPW motif (Fig. 2B, lanes 6 and 8) did not result in the coprecipitation of Gro/TLE (Fig. 2A, lanes 6 and 8), consistent with the demonstrated requirement for this motif for Gro/TLE binding (24). These findings show that Hes1 and Hes6 interact with Gro/TLE with similar affinities when they are expressed at equivalent levels.
FIG. 2.
Interaction of Hes6 and Hes1 with Gro/TLE. 293A cells were transfected with plasmids encoding the indicated GST fusion proteins. Cell lysates were collected, and the fusion proteins were isolated on glutathione-Sepharose beads. The precipitated material (Pull-Down) was subjected to SDS-PAGE (lanes 5 to 8) on a 10% gel, together with 1/10 of each input lysate collected prior to incubation with glutathione-Sepharose beads (lanes 1 to 4). This was followed by Western blotting (WB) with either antibodies (Ab.) that recognize all Gro/TLE proteins (panTLE) (A) or anti-GST antibodies (B). Positions of size standards are indicated in kilodaltons.
Effect of Hes6 on the interaction of Hes1 with Gro/TLE.
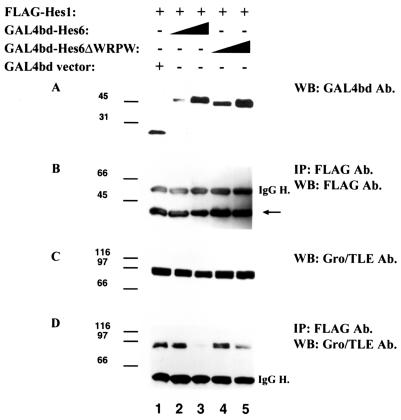
To directly test whether Hes6 might compete with Hes1 for Gro/TLE binding, we performed Hes1-Gro/TLE coimmunoprecipitation studies in the absence or presence of Hes6. 293A cells were transfected with FLAG epitope-tagged Hes1, followed by immunoprecipitation with anti-FLAG antibodies. In the absence of HA-Hes6 (Fig. 3A, lane 1), Gro/TLE coimmunoprecipitated efficiently with Hes1 (Fig. 3E, lane 1). When Hes6 was coexpressed with Hes1 (Fig. 3A, lane 2), we observed a significant decrease in the amount of Gro/TLE that coimmunoprecipitated with Hes1 (Fig. 3E, cf. lanes 1 and 2). Under these conditions (see Materials and Methods), Hes6 expression did not cause a significant decrease in the level of transfected Hes1 (Fig. 3B and D, cf. lanes 1 and 2) and had no negative effect on the expression of endogenous Gro/TLE (Fig. 3C), suggesting that the decreased Gro/TLE coimmunoprecipitation was not simply the result of decreased levels of these proteins. In this and succeeding figures, the relative intensities of the Hes1 and Hes6 immunoreactive bands do not reflect the actual relative amounts of these factors, because different antibodies were used for each protein and blots were not developed for equal lengths of time. To test whether the reduction in Hes1-Gro/TLE coimmunoprecipitation resulted from a titration effect mediated by Hes6 homodimers, the same assays were performed with Hes6ΔWRPW (Fig. 3A, lane 3). This protein also caused a decrease in Gro/TLE coimmunoprecipitation with Hes1, although this reduction was not as robust as with Hes6 (Fig. 3E, cf. lanes 1 and 3). Coexpression of Hes6ΔWRPW did not affect the levels of Hes1 or Gro/TLE (Fig. 3B and C). Similar studies were performed with fusion proteins of Hes6 and the DNA-binding domain of GAL4 (GAL4bd). Expression of increasing amounts of GAL4bd-Hes6 (Fig. 4A, lanes 2 and 3) led to a significant inhibition of the coimmunoprecipitation of Gro/TLE with Hes1 (Fig. 4D, cf. lanes 1 to 3) without significantly affecting the expression of either Hes1 (Fig. 4B, lanes 1 to 3) or Gro/TLE (Fig. 4C, lanes 1 to 3). GAL4bd-Hes6ΔWRPW had a similar effect, although it was somewhat less effective than GAL4bd-Hes6 (Fig. 4D, lanes 4 and 5).
FIG. 4.
Inhibition of the coimmunoprecipitation of Gro/TLE with Hes1 by Hes6 and Hes6ΔWRPW. 293A cells were transfected with plasmids encoding the indicated combinations of proteins, as described in Materials and Methods. Cell lysates were collected and subjected to Western blotting (WB) with anti-GAL4bd (A) or anti-Gro/TLE (C) antibodies (Ab.) or immunoprecipitation (IP) with anti-FLAG antibodies followed by Western blotting with anti-FLAG (B) or anti-Gro/TLE (D) antibodies. The arrow in panel B points to the position of migration of Hes1. IgG H., immunoglobulin G heavy chains. Positions of size standards are indicated in kilodaltons.
To extend these observations, cells were transfected with Hes1ΔWRPW, followed by immunoprecipitation with anti-FLAG antibodies. As expected, in the absence of cotransfected Hes6, Gro/TLE did not coimmunoprecipitate with Hes1ΔWRPW (Fig. 3E, lane 7). In contrast, Gro/TLE coimmunoprecipitated with Hes1ΔWRPW when the latter was cotransfected with Hes6 (Fig. 3E, lane 4) but not with Hes6ΔWRPW (Fig. 3E, lane 5). As previously reported (3), the Hes1 and Hes6 proteins heterodimerized with each other under the experimental conditions used for these assays (data not shown). Expression of Hes6 alone followed by immunoprecipitation with anti-FLAG antibodies did not result in Gro/TLE coprecipitation (Fig. 3E, lane 6). The expression of Hes1ΔWRPW was not affected by Hes6 expression (Fig. 3B and D). Taken together, these findings demonstrate that Hes6 can antagonize the interaction of Hes1 with Gro/TLE. The WRPW motif of Hes6 is not necessary for this effect, suggesting that this is not solely the result of a competition by Hes6 homodimers for Gro/TLE binding.
Effect of Hes6 on stability of Hes1.
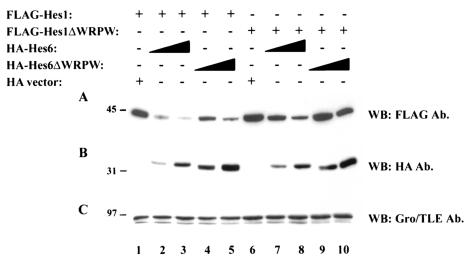
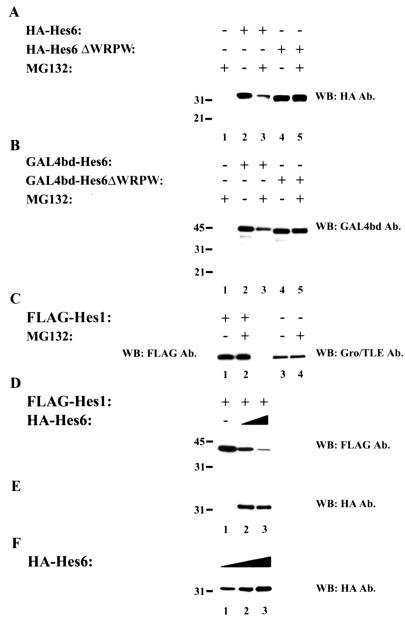
During the course of our transfection experiments, we noted that under appropriate conditions (see Materials and Methods), the coexpression of increasing levels of Hes6 caused a gradual decrease of FLAG-Hes1 immunoreactivity (Fig. 5A and B, cf. lanes 1 to 3). A similar effect was observed when Hes6ΔWRPW was expressed (Fig. 5A and B, cf. lanes 1, 4, and 5), although this truncated form appeared to cause a smaller decrease in Hes1 levels compared with Hes6. The expression of Hes1ΔWRPW was also reduced in the presence of Hes6, but not as significantly as in the case of Hes1 (Fig. 5A, cf. lanes 1 to 3 and 6 to 8). In contrast, Hes6ΔWRPW had no significant effect on Hes1ΔWRPW levels (Fig. 5A, cf. lanes 6, 9, and 10). These findings were specific, because the levels of endogenous Gro/TLE were not affected by either Hes6 or Hes6ΔWRPW (Fig. 5C). To corroborate these results and exclude any effects due to the presence of the HA epitope on Hes6, similar studies were performed with GAL4bd-Hes6. Expression of both GAL4bd-Hes6 and GAL4bd-Hes6ΔWRPW also caused a decrease in Hes1 immunoreactivity compared to the expression of GAL4bd alone (data not shown). These combined observations suggest that Hes6 promotes mechanisms that negatively regulate the stability of Hes1.
FIG. 5.
Effect of Hes6 expression on Hes1 stability. 293A cells were transfected with either FLAG-Hes1 or FLAG-Hes1ΔWRPW (50 ng/transfection), as indicated, in the absence (lanes 1 and 6) (HA vector) or presence of increasing amounts of HA-Hes6 or HA-Hes6ΔWRPW (200 ng/transfection in lanes 2, 4, 7, and 9 or 600 ng/transfection in lanes 3, 5, 8, and 10). Cell lysates were subjected to SDS-PAGE on an 11% gel, followed by sequential Western blotting (WB) with either anti-FLAG (A), anti-HA (B), or anti-Gro/TLE (C) antibodies (Ab.). Positions of size standards are indicated in kilodaltons.
To elucidate these mechanisms further, we tested whether the stability of Hes6 and/or Hes1 might be increased by inhibition of the 26S proteasome. Unexpectedly, we observed a decrease in both HA-Hes6 and GAL4bd-Hes6 immunoreactivity when cells were treated with the protease inhibitor MG132 (Fig. 6A and B, cf. lanes 2 and 3). The proteasome inhibitor lactacystin also caused a similar decrease in Hes6 immunoreactivity (data not shown). This effect was specific, because it was not observed when Hes6ΔWRPW was tested (Fig. 6A and B, cf. lanes 4 and 5). Moreover, MG132 had no effects on the expression of either Hes1 (Fig. 6C, lanes 1 and 2) or Gro/TLE (Fig. 6C, lanes 3 and 4). We also observed that the decrease in full-length HA-Hes6 or GAL4bd-Hes6 was not correlated with the appearance of smaller immunoreactive species. In particular, we did not observe bands migrating near or above the position where GAL4bd migrates (∼19 kDa) (Fig. 4A), suggesting that MG132 treatment caused extensive degradation of the Hes6 proteins. These combined findings show that Hes6 is susceptible to proteolytic mechanisms that can be mimicked or activated (rather than suppressed) by treatment with MG132. These mechanisms depend on the presence of the WRPW motif, perhaps because Hes6 is more prone to degradation when it is competent to associate with Gro/TLE or because the WRPW motif unmasks sites that are involved in degradation pathways.
FIG. 6.
Analysis of Hes6 stability. (A to C) 293A cells were transfected with the indicated combinations of proteins and then incubated in the absence or presence of MG132 as indicated, followed by cell lysis and Western blotting (WB) analysis. The levels of both HA-Hes6 (panel A, lanes 2 and 3) and GAL4bd-Hes6 (panel B, lanes 2 and 3) were reduced in the presence of MG132. In contrast, the levels of Hes6ΔWRPW (panel A, lanes 4 and 5), GAL4bd-Hes6ΔWRPW (panel B, lanes 4 and 5), Hes1 (panel C, lanes 1 and 2), and Gro/TLE (panel C, lanes 3 and 4) were not affected. Ab., antibodies. (D and E) 293A cells were transfected with increasing amounts of HA-Hes6 expression plasmid (400 ng/transfection in lane 2 or 800 ng/transfection in lane 3) in the presence of a constant amount of Hes1 (50 ng/transfection), followed by Western blotting with either anti-FLAG (D) or anti-HA (E) antibodies. (F) Cells were transfected with HA-Hes6 expression plasmid at 200 (lane 1), 400 (lane 2), or 800 (lane 3) ng/transfection in the absence of Hes1, followed by Western blotting with anti-HA antibodies. Positions of size standards are indicated in kilodaltons.
These observations raised the possibility that the susceptibility of Hes6 to proteolytic degradation might be correlated with its negative effect on the stability of Hes1. To test this, Hes1 was expressed in the absence or presence of increasingly high levels of Hes6. We found that the gradual decrease in Hes1 stability induced by transfecting increasing amounts of Hes6 DNA (Fig. 6D) was not correlated with a gradual increase in Hes6 immunoreactivity (Fig. 6E). In contrast, when Hes6 was transfected in the absence of Hes1, we observed the expected correlation between larger amounts of DNA and increasing protein levels (Fig. 6F). Taken together, these results show that Hes6 promotes degradation of Hes1 in a dose-dependent manner. They suggest further that Hes6 may become increasingly unstable when it is bound to Hes1. This in turn raises the possibility that Hes1 becomes targeted for degradation due to its association with Hes6. This process is maximally effective when both Hes1 and Hes6 contain the WRPW motif involved in Gro/TLE binding.
Inhibition of Hes1-mediated transcriptional repression by Hes6 in telencephalic neural progenitor cells.
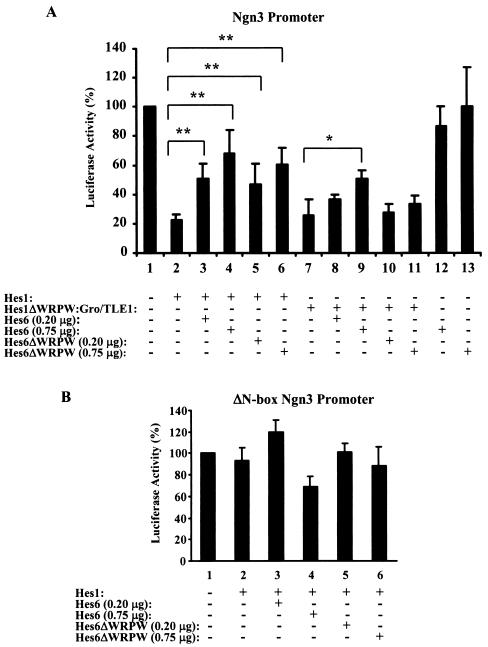
The previous results show that Hes6 can negatively regulate both the stability of Hes1 and its interaction with Gro/TLE. Since these effects are predicted to impair Hes1-mediated transcriptional repression, we next tested the possibility that Hes6 might suppress the ability of Hes1 to act as a transcriptional repressor in a cellular context where these proteins are normally coexpressed. Primary cultures of cortical neural progenitor cells were established and transfected with a reporter plasmid containing the luciferase gene under the control of the ngn3 promoter. Hes1 has been shown previously to specifically bind to this promoter and repress its activity (20). We found that the ngn3 promoter drove strong expression of the reporter gene in transfected neural progenitors and that Hes1 significantly suppressed transcription from this promoter (Fig. 7A, cf. bars 1 and 2). When increasing amounts of Hes6 were cotransfected, Hes1-mediated repression was progressively reduced (Fig. 7A, cf. bars 2 to 4). Expression of Hes6ΔWRPW also resulted in an inhibition of Hes1-mediated repression (Fig. 7A, bars 5 and 6). Control experiments showed that neither Hes6 nor Hes6ΔWRPW had an activating effect on the ngn3 promoter when transfected in the absence of Hes1 (Fig. 7A, bars 12 and 13). Moreover, no significant effects were observed when the ngn3 promoter was mutated to delete the Hes1-binding sites present within its proximal region (20) (Fig. 7B). These results show that Hes6 has the ability to inhibit transcription repression mediated by Hes1 in neural progenitor cells.
FIG. 7.
Inhibition of Hes1-mediated transcriptional repression by Hes6 and Hes6ΔWRPW. Primary cultures of neural progenitor cells isolated from the dorsal telencephalons of E13.5 mouse embryos were transfected with either the pFOX-ngn3p-Luc1 (A) or the pFOX-ΔN-box-ngn3p-Luc1 (B) reporter construct, as indicated, in the absence or presence of Hes1 or Hes1ΔWRPW:Gro/TLE1 and the indicated amounts (per transfection) of either HA-Hes6 or HA-Hes6ΔWRPW. The activity of the reporter gene in the absence of any expression plasmid was considered to be 100%. Luciferase activities were expressed as the means ± SD from at least five independent experiments performed in duplicate. *, P < 0.001; **, P < 0.0001.
We then investigated whether this inhibitory effect was the result of either a promotion of Hes1 degradation or the prevention of Hes1-Gro/TLE complex formation (or a combination of both). We hypothesized that transcriptional repression mediated by a chimeric protein in which Hes1 was constitutively associated with Gro/TLE might be suppressed by Hes6 if that involved a proteolysis of Hes1 but not if it required an inhibition of Hes1-Gro/TLE interaction. A fusion protein (Hes1ΔWRPW:Gro/TLE1) in which the WRPW motif of Hes1 was removed and the entire sequence of Gro/TLE1 was subcloned in its place was engineered. This chimeric protein repressed transcription driven by the ngn3 promoter in neural progenitor cells, and its repressive activity was comparable to that of Hes1 (Fig. 7A, cf. bars 2 and 7). We found that cotransfection of increasing amounts of Hes6 had a derepression effect on Hes1ΔWRPW:Gro/TLE1, although this was somewhat weaker than its inhibitory effect on Hes1 (Fig. 7A, cf. bars 2 to 4 and 7 to 9). These findings indicate that Hes6 can antagonize Hes1 transcriptional repression activity even when Hes1 is constitutively bound to Gro/TLE, strongly suggesting that an inhibition of the Hes1-Gro/TLE interaction is not the only mechanism utilized by Hes6 to suppress Hes1. In turn, this implicates mechanisms involving the promotion of Hes1 degradation in this event. Importantly, although Hes6ΔWRPW had an inhibitory effect on Hes1 (Fig. 7A, cf. bars 2, 5, and 6), it had no significant effect on Hes1ΔWRPW:Gro/TLE1 (Fig. 7A, cf. bars 7, 10, and 11).
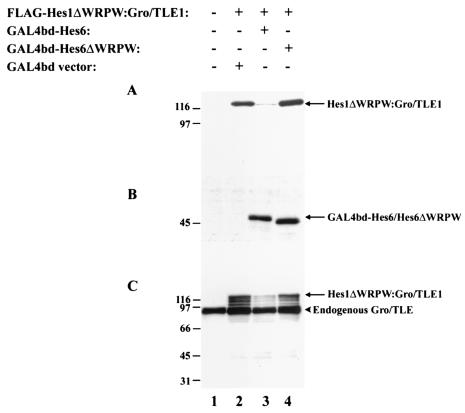
In agreement with these findings, examination of the expression of Hes1ΔWRPW:Gro/TLE1 by using antibodies against the amino-terminal FLAG epitope showed that Hes6 caused a significant reduction in immunoreactivity compared to controls (Fig. 8A, cf. lanes 2 and 3), indicating that Hes6 promotes degradation of Hes1ΔWRPW:Gro/TLE1. Both GAL4bd-Hes6 and HA-Hes6 had the same effect on the expression of Hes1ΔWRPW:Gro/TLE1 (data not shown). In contrast, Hes6ΔWRPW did not affect the expression of this fusion protein (Fig. 8A and B, cf. lanes 2 and 4), consistent with the lack of a negative effect of Hes6ΔWRPW on the transcription repression ability of Hes1ΔWRPW:Gro/TLE1 described above. Reprobing with anti-Gro/TLE antibodies directed against the carboxy-terminal domain of this fusion protein confirmed that Hes6ΔWRPW did not decrease the expression of Hes1ΔWRPW:Gro/TLE1 like Hes6 did (Fig. 8C, cf. lanes 2 to 4). Moreover, using these antibodies, we noticed that coexpression of Hes6 was not correlated with detectable immunoreactive species migrating between endogenous Gro/TLEs (Fig. 8C) and full-length Hes1ΔWRPW:Gro/TLE1 (Fig. 8C) or lower forms of smaller size. These observations suggest that Hes6 expression caused a general proteolysis of the Hes1ΔWRPW:Gro/TLE1 fusion protein and not solely a confined degradation of its amino-terminal portion. Taken together, these findings show that Hes6 inhibits Hes1-mediated transcriptional repression in neural progenitor cells and strongly suggest that the promotion of Hes1 proteolysis by Hes6 is important for this inhibitory effect.
FIG. 8.
Effect of Hes6 on expression of Hes1ΔWRPW:Gro/TLE1. 293A cells were transfected with plasmids encoding the indicated combinations of proteins, followed by preparation of cell lysates and Western blotting with either anti-FLAG (A), anti-GAL4bd (B), or anti-Gro/TLE (C) antibodies. Positions of size standards are indicated in kilodaltons.
Involvement of Ser183 in the ability of Hes6 to promote Hes1 degradation and neuronal differentiation.
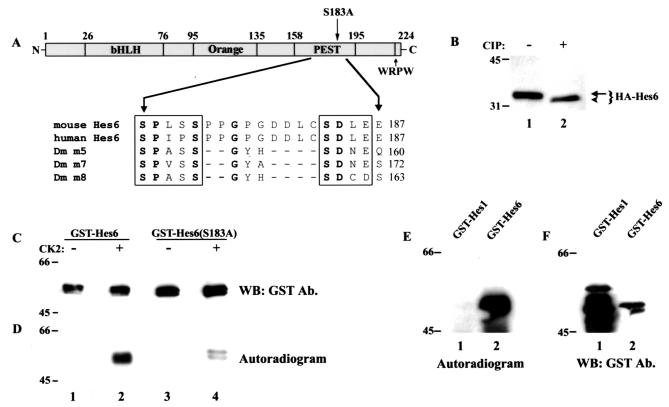
Previous studies (37) have shown that the Drosophila Hes family members Enhancer of split m5, m7, and m8 contain an evolutionarily conserved sequence motif characterized by a carboxy-terminal consensus site for phosphorylation by protein kinase CK2, defined as (S/T)(D/E)X(D/E), preceded at a short distance by the sequence SP(A/V)SS. This sequence, hereafter referred to as the SPXSS-SDXE motif is located within a region with a high PEST score (37). PEST-rich sequences behave as cis-acting signals that regulate protein turnover and have been suggested to be activated via phosphorylation (27, 31). The Drosophila m5, m7, and m8 proteins were shown to associate with and be phosphorylated by protein kinase CK2 at their conserved SPXSS-SDXE sequences. This phosphorylation is believed to activate their PEST domains and result in decreased stability (37).
Using the program PESTfind (http://at.embnet.org/embnet/tools/bio/PESTfind), we identified a conserved potential PEST sequence at the carboxy termini of mouse and human Hes6 proteins (Fig. 9A) (PEST score, +13.02; PEST scores of greater than +5 are considered significant). This region contains a conserved sequence similar to the SPXSS-SDXE motif found in the PEST domain of Drosophila m5, m7, and m8 (Fig. 9A). This raised the possibility that Hes6 might be phosphorylated by protein kinase CK2 and that this event may regulate its stability through modulation of PEST sequence activity. To test this, we first determined whether Hes6 is a phosphorylated protein. Lysates from cells transfected with Hes6 were incubated in the absence or presence of calf intestinal phosphatase, followed by gel electrophoresis. After this treatment, Hes6 exhibited a faster electrophoretic mobility, indicating that it is a phosphorylated protein (Fig. 9B, cf. lanes 1 and 2). In addition, purified protein kinase CK2 directly phosphorylated a fusion protein of GST and Hes6 isolated from bacteria (Fig. 9C and D, lanes 2). Importantly, an S183A mutation within the SPXSS-SDXE motif significantly attenuated phosphorylation of Hes6 by protein kinase CK2 even when Hes6(S183A) was present at higher levels than wild-type Hes6 (Fig. 9C and D, cf. lanes 2 and 4). Hes1, which does not contain an SPXSS-SDXE motif, was not phosphorylated by protein kinase CK2 (Fig. 9E, lane 1) even when expressed at significantly higher levels than Hes6 (Fig. 9F, cf. lanes 1 and 2). Taken together, these findings identify Hes6 as a specific target of protein kinase CK2 and strongly suggest that this kinase can phosphorylate Hes6 at Ser183.
FIG. 9.
Analysis of Hes6 phosphorylation. (A) Schematic representation of the domain structure of Hes6. Indicated are the bHLH domain, the Orange domain predicted to form helices 3 and 4, the PEST region containing the SPXSS-SDXE motif and its resident Ser183, and the WRPW tetrapeptide. Shown in detail are the sequences of the SPXSS-SDXE elements from mouse and human Hes6 (3) and Drosophila Enhancer of split m5, m7, and m8 (37). Invariant residues are indicate in boldface. (B) 293A cells were transfected with HA-Hes6, and cell lysates were incubated in the absence or presence of calf intestinal phosphatase (CIP), followed by Western blotting with anti-HA antibodies. (C and D) The indicated GST fusion proteins were purified and subjected to in vitro phosphorylation in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of purified protein kinase CK2, followed by autoradiography (D) and Western blotting (WB) with anti-GST antibodies (Ab.) (C). (E and F) The indicated GST fusion proteins were purified and subjected to in vitro phosphorylation in the presence of purified protein kinase CK2, followed by autoradiography (E) and Western blotting with anti-GST antibodies (F). Positions of size standards are indicated in kilodaltons in panels B to F.
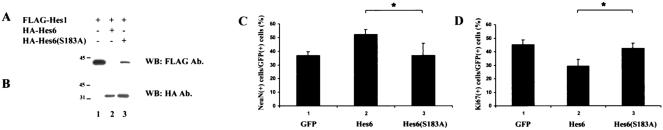
Based on these observations, we tested whether Ser183 might be important for the ability of Hes6 to cause a reduced stability of Hes1. 293A cells were transfected with Hes1 alone or in the presence of Hes6 or Hes6(S183A). Hes6 caused a dramatic decrease in Hes1 expression, whereas Hes6(S183A) had a weaker, although still detectable, effect (Fig. 10A and B, cf. lanes 1 to 3). These findings suggest that phosphorylation of Ser183 plays a positive role in the ability of Hes6 to promote Hes1 degradation. In turn, this raised the possibility that Hes6(S183A) might have a weaker neurogenic activity than wild-type Hes6 due to its reduced ability to decrease Hes1 stability. To examine this possibility, cortical progenitor cells were transfected with Hes6 or Hes6(S183A), and the transfected cells were examined for the expression of markers of either proliferating cells (the Ki67 protein) or differentiated neurons (the NeuN protein), as described previously (33). We found that exogenous Hes6 led to the differentiation of supernumerary neurons (Fig. 10C, cf. bars 1 and 2) and a decrease in undifferentiated progenitors (Fig. 10D, cf. bars 1 and 2). In contrast, Hes6(S183A) did not promote similar effects (Fig. 10C and D, bars 3). Taken together, these findings identify an important role for Ser183 in the neurogenic activity of Hes6 and show a correlation between phosphorylation of this residue by protein kinase CK2 and the ability of Hes6 to negatively regulate Hes1 functions and promote neuronal development.
FIG. 10.
Effects of S183A mutation on Hes6 functions. (A and B) 293A cells were transfected with FLAG-Hes1 (50 ng/transfection) in the absence (lane 1) or presence of either HA-Hes6 (lane 2) or HA-Hes6(S183A) (lane 3) (600 ng/transfection). Cell lysates were subjected to SDS-PAGE, followed by Western blotting (WB) with either anti-FLAG (A) or anti-HA (B) antibodies (Ab.). Shown is a representative example of results from four separate experiments that gave the same results. Positions of size standards are indicated in kilodaltons. (C and D) E13.5 mouse embryonic cortical progenitor cells were transfected with plasmids encoding either GFP alone or a combination of GFP and Hes6 or GFP and Hes6(S183A). Forty-eight hours later, cells were fixed and subjected to double-labeling analysis of the expression of GFP, NeuN, or Ki67. Shown is the quantitation of the percentage of GFP-NeuN (C)- or GFP-Ki67 (D)-double-positive cells. Results are shown as the means ± SD (n = 4). *, P < 0.01.
DISCUSSION
Involvement of Hes6 in neuronal differentiation.
Previous studies with mouse and Xenopus have revealed that Hes6 expression is correlated with the transition of neural progenitor cells to the neuronal fate (3, 19, 30, 38). In Xenopus, Hes6 activation follows the expression of neuronal determination genes such as ngn family members and overlaps with neuronal differentiation genes such as NeuroD (19). In mice, Hes6 expression was detected in both the proliferative zone containing neural progenitor cells and areas containing postmitotic neurons (3). Taken together with the demonstration that Xenopus Hes6 expression is not activated by the Notch signaling pathway, which plays an antineurogenic role, but rather appears to be driven by neurogenic bHLH proteins (19), these observations first suggested an involvement of Hes6 in mechanisms that positively regulate neurogenesis. This possibility was confirmed by ectopic expression studies with Xenopus embryos and murine retinal explants that revealed that Hes6 promotes neuronal differentiation (19). Importantly, those studies also suggested that Hes6 may act primarily by promoting the differentiation of progenitors that already express proneural proteins, perhaps by antagonizing the functions of inhibitors of the latter. By removing this inhibition, Hes6 may allow proneural proteins to perform their neurogenic functions more effectively, leading to enhanced neuronal differentiation. In an effort to clarify how Hes6 may antagonize inhibitory activities that negatively regulate proneural protein functions, we have focused on the Hes1 protein, a well-characterized member of a family of bHLH proteins that act as inhibitors of proneural proteins in both invertebrates and vertebrates (18). In particular, Hes1 inhibits transcription from proneural gene promoters (4, 20) and the expression of proneural genes is prematurely activated in Hes1 nullizygous mice (16), suggesting that Hes1 acts as a negative regulator of proneural proteins in vivo. Hes1 and Hes6 are coexpressed in differentiating neural progenitor cells (3, 19, 32), and they can heterodimerize in transfected cells and in vitro (3). Moreover, Hes6 was shown to reduce the ability of Hes1 to repress transcription from an artificial promoter in NIH 3T3 cells (3). These observations raised the possibility that Hes6 acts as a negative regulator of the antineurogenic activity of Hes1. However, they did not clarify the molecular mechanisms that underlie this function. To address this important question, we have performed a combination of molecular and cellular investigations that have characterized two complementary mechanisms that Hes6 may utilize to negatively regulate Hes1 activity and positively regulate neuronal differentiation.
Inhibition of Hes1-Gro/TLE interaction by Hes6.
Our studies have shown that the interaction of Hes1 with its transcriptional corepressor Gro/TLE is reduced when Hes6 is coexpressed at levels that do not have a significant effect on the stability of either Hes1 or Gro/TLE. This effect is unlikely to result solely from a competition for Gro/TLE between Hes6 and Hes1 homodimers, because a truncated form of Hes6 that is unable to bind to Gro/TLE also inhibits the interaction of Hes1 with the latter. Our finding that Hes1ΔWRPW-Hes6 heterodimers, which have only one WRPW motif, appear to interact with Gro/TLE like Hes1-Hes6 heterodimers, which have both WRPW motifs, suggests instead that Hes1-Hes6 heterodimers interact more poorly with Gro/TLE than homodimers of either protein. Reasons for this reduced affinity may include the fact that the folding of these heterodimers may not allow a proper alignment of the WRPW motifs of Hes1 and Hes6. Gro/TLE proteins exist as tetramers, so the correct alignment of WRPW motifs may be critical for the establishment of a strong interaction between Hes factors and Gro/TLE. A weaker association may be caused by differential posttranslational modifications, including phosphorylation of Ser183 of Hes6 (see below). Alternatively, other cofactors that may interact selectively with either Hes1 or Hes6 may not allow a strong interaction between Gro/TLE and Hes1-Hes6 heterodimers. In either case, the formation of Hes1-Hes6 heterodimers that interact poorly with Gro/TLE is likely to prevent or reduce the interaction of Hes1 homodimers with Gro/TLE, thereby depriving Hes1 of its critical transcriptional corepressor and negatively regulating its functions. As discussed below, this situation may lead, under conditions of increasing Hes6 expression, to an additional mechanism of Hes1 suppression, namely, the targeting of Hes1-Hes6 dimers for proteolytic degradation.
Regulation of Hes1 stability by Hes6.
Our investigations have shown for the first time that expression of increasing amounts of Hes6 causes a gradual decrease of Hes1 stability resulting in a loss of full-length protein. This finding raises the interesting possibility that Hes6 may act as a negative regulator of Hes1 activity by regulating the stability of the latter. Such a situation may occur, for instance, in determined neural progenitor cells, in which increased proneural protein activity may promote an up-regulation of Hes6 expression. In turn, Hes6 may cause inactivation of Hes1 by affecting its turnover, thereby contributing to the mechanisms that will drive those progenitors into the neuronal lineage. Such a situation might explain not only the ability of Hes6 to suppress Hes1-mediated repression but also the previous observation that Hes6 can also suppress the ability of Hes1 to inhibit the activity of E2A-proneural protein heterodimers (3). Inhibition of proneural protein activity by Hes1 is thought to involve the formation of heterodimers between Hes1 and ubiquitous bHLH proteins such as E47, thus titrating away the latter from the proneural proteins. Proteolytic degradation of Hes1 would therefore be expected to prevent these interactions and inhibit this effect.
We have shown that Hes6 is intrinsically susceptible to proteolytic degradation events that can be uncovered by exposure to the protease inhibitor MG132. The mechanisms underlying the effect of MG132 on Hes6 are still unclear and likely involve indirect effects resulting from either the MG132-mediated activation of genes that encode factors that may destabilize Hes6, the inhibition of proteolytic pathways that may normally degrade factors that reduce Hes6 stability, or the inhibition of pathways leading to the expression of factors that render Hes6 more stable. Regardless of the exact nature of the events induced by MG132, the observation that Hes6 is prone to proteolytic degradation is in agreement with the presence of an evolutionarily conserved PEST domain containing an SPXSS-SDXE subdomain that includes a resident consensus protein kinase CK2 phosphorylation site at Ser183. The presence of PEST domains is characteristic of proteins that undergo increased turnover, and phosphorylation of PEST sequences by protein kinase CK2 was shown to negatively affect intrinsic protein stability (22, 27, 31). The Drosophila Hes family members Enhancer of split m5, m7, and m8 share with Hes6 a similar SPXSS-SDXE motif within a carboxy-terminal region characterized by a high PEST score. They were shown to bind directly to protein kinase CK2 and to be phosphorylated by this kinase at their conserved SDXE site. This phosphorylation is believed to decrease their stability (37). In agreement with those results, we have shown that Hes6, but not Hes1, is phosphorylated by protein kinase CK2 at Ser183 within the SDXE motif, suggesting a previously unrecognized relatedness of Hes6 to the m5-m7-m8 subgroup of Drosophila Enhancer of split proteins.
Our studies have also shown that maximal Hes6-mediated degradation of Hes1 is correlated with a decreased stability of Hes6 itself. This observation suggests that the formation of Hes1-Hes6 heterodimers may increase the intrinsic susceptibility of Hes6 to degradation, causing the recruitment of Hes1 into the same proteolytic mechanisms. Although the molecular events underlying such a process remain to be fully elucidated, our investigations have revealed important roles for both the protein kinase CK2 phosphorylation site at Ser183 of Hes6 and the WRPW motif. We have shown that mutation of Ser183 into Ala attenuates, albeit does not eliminate, the destabilizing effect of Hes6 on Hes1. This finding suggests that the SPXSS-SDXE motif of Hes6 and its resident Ser183 may contribute to the mechanisms that activate the PEST domain of Hes6. Heterodimerization with Hes1 may render this region more accessible to such mechanisms, thereby promoting the degradation of Hes6 and Hes1. Alternatively, the phosphorylation of Ser183 may cause a misalignment of the WRPW motifs of Hes1 and Hes6 when these factors heterodimerize, leading to a conformation that results in suboptimal Gro/TLE binding compared to homodimers of either protein. This may lead to the formation of misfolded Hes1-Hes6-Gro/TLE ternary complexes that may be recognized as defective and targeted for removal via proteolytic degradation. Mutation of Ser183 may allow Hes1-Hes6 dimers to interact better with Gro/TLE, resulting in the formation of properly folded complexes with increased stability.
The possibility that enhanced proteolysis of Hes1 and Hes6 may be caused by their association into incorrectly folded complexes is also suggested by our observation that the formation of Hes1-Hes6 heterodimers does not appear to be sufficient to activate proteolytic degradation of these proteins by itself, because removal of the WRPW motif from either Hes1, Hes6, or both progressively attenuates Hes1 degradation promoted by Hes6. Moreover, Hes6ΔWRPW had no detectable effect on the stability of the chimeric protein Hes1ΔWRPW:Gro/TLE1, in contrast to the significant degradation induced by full-length Hes6. In addition, heterodimers of Hes1 and Hes6 do not efficiently coimmunoprecipitate with Gro/TLE, regardless of whether they contain one or two WRPW motifs, suggesting that they may not be able to form stable complexes. Since removal of the WRPW motif does not impair the ability of Hes1 and Hes6 to heterodimerize (data not shown), these observations suggest that heterodimers of Hes1 and Hes6 may be more susceptible to degradation if they are associated with Gro/TLE through their WRPW motifs. Heterodimers lacking this motif, and thus unable to interact with Gro/TLE, may be able to fold more properly and avoid extensive degradation. Based on these combined observations, we propose that Hes1-Hes6 heterodimers are prone to increased degradation when they form complexes with Gro/TLE. This situation may be due to specific structural features of these proteins that may not allow the formation of properly folded complexes with Gro/TLE, in turn resulting in the activation of proteolytic mechanisms involving Ser183 of Hes6. Conversely, it may be phosphorylation of Ser183 that causes a misfolding of the carboxy termini of these heterodimers and an impaired ability to bind to Gro/TLE, resulting in degradation as a secondary effect to remove the misfolded complexes. Future studies will be aimed at distinguishing between these possibilities. In either case, it appears that Ser183 plays an important role in Hes6 functions, as further indicated by the inability of Hes6(S183A) to promote neuronal differentiation (see below for further details).
We recognize that other mechanisms are also possible. For instance, the WRPW motif of Hes6 may promote the instability of Hes1-Hes6 heterodimers in a Gro/TLE-independent manner, possibly by acting as a binding site for proteins other than Gro/TLE, resulting in the direct or indirect recruitment of proteolytic enzymes. However, it remains to be determined whether the WRPW motif mediates interactions with proteins other than Gro/TLE. In addition, we cannot rule out the possibility that the destabilizing effect of Hes6 on Hes1 is the result of the activity of Hes6 as a transcriptional repressor. Hes6 may directly suppress the expression of factors that promote the stability of Hes1. This seems unlikely, however, because Hes6ΔWRPW, which cannot recruit the Gro/TLE corepressor and was shown to be unable to mediate transcriptional repression when fused to GAL4bd (9), also promotes Hes1 degradation. In addition, the in vivo neurogenic activity of Hes6 does not appear to be DNA-binding dependent, arguing against mechanisms that are based solely on direct transcriptional functions (19). It remains possible, though, that Hes6 mediates as-yet-uncharacterized transcriptional mechanisms that may affect Hes1 expression in a dose-dependent manner.
Characterization of the molecular mechanisms underlying the suppression of Hes1-mediated transcriptional repression by Hes6.
To begin to elucidate whether different mechanisms of Hes1 inhibition are used by Hes6 in combination (to achieve maximal effects) or separately (perhaps depending on particular cellular and/or developmental conditions), we have examined the effect of Hes6 on the ability of Hes1 to mediate transcriptional repression in cortical progenitor cells, where these proteins are coexpressed. We have found that Hes6 suppresses Hes1-mediated repression. Both Hes6 and Hes6ΔWRPW have a similar inhibitory effect. This observation does not suggest that the suppression of Hes1 activity derives from the Hes6-mediated repression of a gene(s) encoding a positive regulator(s) of Hes1, because previous studies have shown that Hes6 requires its WRPW motif to repress transcription when targeted to DNA as a fusion protein with GAL4bd (9). Moreover, this finding also argues against a mechanism involving solely a competition for Gro/TLE between Hes1 and Hes6 homodimers. To determine whether Hes1 suppression was the result of the inhibition of the interaction of Hes1 with Gro/TLE or the promotion of Hes1 proteolysis (or a combination of both), we have examined the effect of Hes6 on a chimeric protein in which Hes1 is constitutively bound to Gro/TLE. This fusion protein represses transcription in cortical progenitor cells like full-length Hes1, and its repressive ability should not be affected by conditions that would otherwise inhibit Gro/TLE recruitment. Our investigations have revealed that Hes6 still has an inhibitory effect on Hes1ΔWRPW:Gro/TLE1, although this is weaker than its effect on Hes1. These findings thus suggest that the promotion of Hes1 degradation plays an important role in the inhibitory effect of Hes6 on Hes1-mediated repression. In agreement with this possibility, we have found that Hes6ΔWRPW, which does not promote a significant proteolysis of Hes1ΔWRPW:Gro/TLE1, does not have a negative effect on repression mediated by the latter. Together, these findings clarify mechanisms that underlie the ability of Hes6 to act as a negative regulator of Hes1 in cortical neural progenitor cells.
Promotion of cortical neurogenesis by Hes6.
To determine if Hes6 is involved in the regulation of neuronal differentiation in the mammalian forebrain, we have examined the consequence of exogenous Hes6 expression in primary cultures of cortical neural progenitor cells. In our studies, Hes6 induced a decrease in the number of undifferentiated progenitor cells and an increase in the number of differentiated neurons arising from these progenitors, showing that Hes6 promotes cortical neuronal differentiation. This effect is likely the result of the recruitment of supernumerary progenitors into the neuronal lineage. Because neural progenitor cells of the dorsal telencephalon express proneural proteins such as Ngn1 and -2, our results are consistent with previous studies on Xenopus suggesting that Hes6 promotes the neuronal differentiation of Ngn-expressing neural progenitor cells (19). Based on these results and our demonstration that Hes6 efficiently suppresses Hes1-mediated transcriptional repression in cortical progenitors, we propose that the inhibition of Hes1 activity is at least one of the mechanisms utilized by Hes6 to promote neuronal differentiation. In possible agreement with this, we have found that the mutated protein Hes6(S183A) had an attenuated negative effect on the stability of Hes1 compared to wild-type Hes6 and did not promote neuronal differentiation. These observations suggest a correlation between a reduced ability to promote Hes1 degradation and reduced Hes6 neurogenic activity. We found that Hes6(S183A) was able to cause a detectable decrease of Hes6 stability in 293A cells but failed to promote the neuronal differentiation of cortical progenitors. This situation may reflect that the observed residual levels of Hes1 may be sufficient to inhibit neuronal differentiation or that Hes6(S183A) may have a weaker effect on Hes1 in neural progenitors compared to 293A cells. It is entirely possible, however, that additional mechanisms involving Ser183 may be important for the neurogenic activity of Hes6. Further elucidation of the mechanisms underlying Hes6 activity will clarify important events regulating vertebrate neurogenesis.
Acknowledgments
We thank R. St.-Arnaud for reagents, R. Lo and K. Joachim for excellent technical assistance, S. Morris for help and suggestions, R. Bremner for constructive comments, and H. Nuthall, W. Song, and J. Nadeau for help during this work.
These studies were supported by grants from the Canadian Institutes of Health Research (CIHR) (MOP-13957 and GR-14971) to S.S. E.T. was supported by an MNI JTC Fellowship and is a CIHR Fellow. F.M.T. is supported by a studentship from the Fonds de la Recherche en Sante du Quebec (FRSQ). S.S. is an FRSQ Senior Scholar.
REFERENCES
- 1.Akazawa, C., Y. Sasai, S. Nakanishi, and R. Kageyama. 1992. Molecular characterization of a rat negative regulator with a basic helix-loop-helix structure predominantly expressed in the developing nervous system. J. Biol. Chem. 267:21879-21885. [PubMed] [Google Scholar]
- 2.Anderson, D. J. 2001. Stem cells and pattern formation in the nervous system: the possible versus the actual. Neuron 30:19-35. [DOI] [PubMed] [Google Scholar]
- 3.Bae, S. K., Y. Bessho, M. Hojo, and R. Kageyama. 2000. The bHLH gene Hes6, an inhibitor of Hes1, promotes neuronal differentiation. Development 127:2933-2943. [DOI] [PubMed] [Google Scholar]
- 4.Chen, H. A., A. Thiagalingam, H. Chopra, M. W. Borges, J. N. Feder, B. D. Nelkin, S. B. Baylin, and D. W. Ball. 1997. Conservation of the Drosophila lateral inhibition pathway in human lung cancer: a hairy-related protein (HES-1) directly represses achaete-scute homolog-1 expression. Proc. Natl. Acad. Sci. USA 94:5355-5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cossins, J., A. E. Vernon, Y. Zhang, A. Philpott, and P. H. Jones. 2002. Hes6 regulates myogenic differentiation. Development 129:2195-2207. [DOI] [PubMed] [Google Scholar]
- 6.Dehni, G., Y. Liu, J. Husain, and S. Stifani. 1995. TLE expression correlates with mouse embryonic segmentation, neurogenesis, and epithelial determination. Mech. Dev. 53:369-381. [DOI] [PubMed] [Google Scholar]
- 7.Fisher, A. L., S. Ohsako, and M. Caudy. 1996. The WRPW motif of the Hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Mol. Cell. Biol. 16:2670-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fode, C., Gradwohl, X. Morin, A. Dierich, M. LeMeur, C. Goridis, and F. Guillemot. 1998. The bHLH protein NEUROGENIN2 is a determination factor for epibranchial placode-derived sensory neurons. Neuron 20:483-494. [DOI] [PubMed] [Google Scholar]
- 9.Gao, X., T. Chandra, M. O. Gratton, I. Quelo, J. Prud'homme, S. Stifani, and R. St.-Arnaud. 2001. HES6 acts as a transcriptional repressor in myoblasts and can induce the myogenic differentiation program. J. Cell Biol. 154:1161-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh, A., and M. E. Greenberg. 1995. Distinct roles for bFGF and NT-3 in the regulation of cortical neurogenesis. Neuron 15:89-103. [DOI] [PubMed] [Google Scholar]
- 11.Grbavec, D., and S. Stifani. 1996. Molecular interaction between TLE1 and the carboxyl-terminal domain of HES-1 containing the WRPW motif. Biochem. Biophys. Res. Commun. 223:701-705. [DOI] [PubMed] [Google Scholar]
- 12.Grbavec, D., R. Lo, Y. Liu, and S. Stifani. 1998. Transducin-like Enhancer of split 2, a mammalian homologue of Drosophila Groucho, acts as a transcriptional repressor, interacts with Hairy/Enhancer of split proteins, and is expressed during neuronal development. Eur. J. Biochem. 258:339-349. [DOI] [PubMed] [Google Scholar]
- 13.Guillemot, F., L. C. Lo, J. E. Johnson, A. Auerbach, D. J. Anderson, and A. L. Joyner. 1993. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell 75:463-476. [DOI] [PubMed] [Google Scholar]
- 14.Husain, J., R. Lo, D. Grbavec, and S. Stifani. 1996. Affinity for the nuclear compartment and expression during cell differentiation implicate phosphorylated Groucho/TLE1 forms of higher molecular mass in nuclear functions. Biochem. J. 317:523-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishibashi, M., K. Moriyoshi, Y. Sasai, K. Shiota, S. Nakanishi, and R. Kageyama. 1994. Persistent expression of helix-loop-helix factor HES-1 prevents mammalian neural differentiation in the central nervous system. EMBO J. 13:1799-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishibashi, M., S. L. Ang, K. Shiota, S. Nakanishi, R. Kageyama, and F. Guillemot. 1995. Targeted disruption of mammalian hairy and Enhancer of split homolog 1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 9:3136-3148. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, J. E., S. J. Birren, T. Saito, and D. J. Anderson. 1992. DNA binding and transcriptional regulatory activity of mammalian achaete-scute homologous (MASH) proteins revealed by interaction with a muscle specific enhancer. Proc. Natl. Acad. Sci. USA 89:3596-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kageyama, R., and S. Nakanishi. 1997. Helix-loop-helix factors in growth and differentiation of the vertebrate nervous system. Curr. Opin. Genet. Dev. 7:659-665. [DOI] [PubMed] [Google Scholar]
- 19.Koyano-Nakagawa, N., J. Kim, D. Anderson, and C. Kintner. 2000. Hes6 acts in a positive feedback loop with the neurogenins to promote neuronal differentiation. Development 127:4203-4216. [DOI] [PubMed] [Google Scholar]
- 20.Lee, J. C., S. B. Smith, H. Watada, J. Lin, D. Scheel, J. Wang, R. G. Mirmira, and M. S. German. 2001. Regulation of the pancreatic pro-endocrine gene Neurogenin3. Diabetes 50:928-936. [DOI] [PubMed] [Google Scholar]
- 21.Lee, J. E., S. M. Hollenberg, L. Snider, D. L. Turner, N. Lipnick, and H. Weintraub. 1995. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science 268:836-844. [DOI] [PubMed] [Google Scholar]
- 22.Liu, Z. P., R. L. Galindo, and S. A. Wasserman. 1997. A role for CKII phosphorylation of the cactus PEST domain in dorsoventral patterning of the Drosophila embryo. Genes Dev. 11:3413-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLarren, K. W., R. Lo, D. Grbavec, K. Thirunavukkarasu, G. Karsenty, and S. Stifani. 2000. The mammalian basic helix loop helix protein HES-1 binds to and modulates the transactivating function of the Runt-related factor Cbfa1. J. Biol. Chem. 275:530-538. [DOI] [PubMed] [Google Scholar]
- 24.McLarren, K. W., F. Theriault, and S. Stifani. 2001. Association with the nuclear matrix and interaction with Groucho and RUNX proteins regulate the transcription repression ability of the basic helix loop helix factor Hes1. J. Biol. Chem. 276:1578-1584. [DOI] [PubMed] [Google Scholar]
- 25.Nuthall, H., J. Husain, K. W. McLarren, and S. Stifani. 2002. Role for Hes1-induced phosphorylation in Groucho-mediated transcriptional repression. Mol. Cell. Biol. 22:389-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohtsuka, T., M. Ishibashi, G. Gradwhohl, S. Nakanishi, F. Guillemot., and R. Kageyama. 1999. Hes1 and Hes5 as Notch effectors in mammalian neuronal differentiation. EMBO J. 18:2196-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Packman, L. C., K. Kubota, J. Parker, and N. J. Gay. 1997. Casein kinase II phosphorylates Ser468 in the PEST domain of the Drosophila IkappaB homologue cactus. FEBS Lett. 400:45-50. [DOI] [PubMed] [Google Scholar]
- 28.Palaparti, A., A. Baratz, and S. Stifani. 1997. The Groucho/Transducin-like Enhancer of split transcriptional repressors interact with the genetically defined amino-terminal silencing domain of histone H3. J. Biol. Chem. 272:26604-26610. [DOI] [PubMed] [Google Scholar]
- 29.Paroush, Z., R. L. Finley, T. Kidd, S. M. Wainwright, P. W. Ingham, R. Brent, and D. Ish-Horowicz. 1994. Groucho is required for Drosophila neurogeneis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell 79:805-815. [DOI] [PubMed] [Google Scholar]
- 30.Pissarra, L., D. Henrique, and A. Duarte. 2000. Expression of hes6, a new member of the Hairy/Enhancer of split family, in mouse development. Mech. Dev. 95:275-278. [DOI] [PubMed] [Google Scholar]
- 31.Rechsteiner, M., and S. W. Rogers. 1996. PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 21:267-271. [PubMed] [Google Scholar]
- 32.Sasai, Y., R. Kageyama, Y. Tagawa, R. Shigemoto, and S. Nakanishi. 1992. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 5:2620-2634. [DOI] [PubMed] [Google Scholar]
- 33.Seidah, N., S. Benjannet, L. Wickham, J. Marcinkiewicz, S. Belanger Jasmin, S. Stifani, A. Basak, A. Prat, and M. Chretien. 2003. The secretory proprotein convertase neural apoptosis-regulated convertase 1 (NARC-1): liver regeneration and neuronal differentiation. Proc. Natl. Acad. Sci. USA 100:928-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stifani, S., C. M. Blaumueller, N. J. Redhead, R. E. Hill, and S. Artavanis-Tsakonas. 1992. Human homologs of a Drosophila Enhancer of split gene product define a novel family of nuclear proteins. Nat. Genet. 2:119-127. [DOI] [PubMed] [Google Scholar]
- 35.Toma, J. G., H. El-Bizri, F. Barnabe-Heider, R. Aloyz, and F. D. Miller. 2000. Evidence that helix-loop-helix proteins collaborate with retinoblastoma tumor suppressor protein to regulate cortical neurogenesis. J. Neurosci. 20:7648-7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomita, K., M. Ishibashi, K. Nakahara, S. L. Ang, S. Nakanishi, M. Asashima, and R. Kageyama. 1996. Mammalian hairy and Enhancer of split homolog 1 regulates differentiation of retinal neurons and is essential for eye morphogenesis. Neuron 16:723-734. [DOI] [PubMed] [Google Scholar]
- 37.Trott, R. L., K. Madhavi, Z. Paroush, and A. P. Bidwai. 2001. Drosophila melanogaster casein kinase II interacts with and phosphorylates the basic helix-loop-helix proteins m5, m7, and m8 derived from the Enhancer of split complex. J. Biol. Chem. 276:2159-2167. [DOI] [PubMed] [Google Scholar]
- 38.Vasiliauskas, D., and C. Stern. 2000. Expression of mouse HES-6, a new member of the Hairy/Enhancer of split family of bHLH transcription factors. Mech. Dev. 98:133-137. [DOI] [PubMed] [Google Scholar]
- 39.Yao, J., Y. Liu, R. Lo, I. Tretjakoff, A. Peterson, and S. Stifani. 2000. Disrupted development of the cerebral hemispheres in transgenic mice expressing the mammalian Groucho homologue Transducin-like Enhancer of split 1 in postmitotic neurons. Mech. Dev. 93:105-115. [DOI] [PubMed] [Google Scholar]
- 40.Yao, J., E. Lai, and S. Stifani. 2001. The winged-helix protein brain factor 1 interacts with Groucho and Hes proteins to repress transcription. Mol. Cell. Biol. 21:1962-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]