Abstract
The Caenorhabditis elegans UNC-73B protein regulates axon guidance through its ability to act as a guanine nucleotide exchange factor (GEF) for the CeRAC/MIG-2 GTPases. Like other GEFs for Rho family GTPases, UNC-73B has a Dbl homology (DH) catalytic domain, followed by a C-terminal pleckstrin homology (PH) domain. We have explored whether the PH domain cooperates with the adjacent DH domain to promote UNC-73B GEF activity and axonal pathfinding. We show that the UNC-73B PH domain binds preferentially to monophosphorylated phosphatidylinositides in vitro. Replacement of residues Lys1420 and Arg1422 with Glu residues within the PH domain impaired this phospholipid binding but did not affect the in vitro catalytic activity of the DH domain. In contrast, a mutant UNC-73B protein with a Trp1502-to-Ala substitution in the PH domain still interacted with phosphorylated phosphatidylinositides but had lost its GEF activity. UNC-73B minigenes containing these mutations were microinjected into C. elegans and transferred to unc-73(e936) mutant worms. Unlike the wild-type protein, neither PH domain mutant was able to rescue the unc-73 axon guidance defect. These results suggest that the UNC-73B PH domain plays distinct roles in targeting and promoting GEF activity towards the Rac GTPase, both of which are important for the directed movements of motorneurons in vivo.
The Rho subfamily of Ras-related GTPases mediates signaling cascades that regulate actin dynamics. In cultured Swiss 3T3 fibroblasts, Cdc42 induces formation of filopodia, while Rac and Rho proteins are required for membrane ruffling and actin stress fiber formation, respectively (23). In this context, Rho-GTPase family members play an important role during the pathfinding of axons in the developing nervous system, through their ability to couple guidance cues to modification of the actin cytoskeleton (18).
Guanine nucleotide exchange factors (GEFs) control the levels of functionally active GTP-bound, versus inactive GDP-bound, GTPases by stimulating the exchange of GDP for GTP (25, 41). Most proteins with demonstrated in vitro GEF activity for Rho, Rac, or Cdc42 contain a conserved Dbl homology (DH) domain of approximately 200 residues, which, in a number of cases, is sufficient to catalyze the exchange of GDP for GTP in vitro (23).
Regulation of GEF activity can occur through a number of different mechanisms. For example, the Vav family of Rho/Rac exchange factors can be phosphorylated on tyrosine 174 by Lck (12), resulting in activation of GEF activity through a mechanism involving displacement of the phosphorylated peptide motif from the DH domain (1). Tiam1 demonstrates specific activity toward Rac, which is enhanced by kinases, such as protein kinase C and Ca2+/calmodulin-dependent protein kinase II (19).
The DH domains of such proteins are typically followed by an adjacent pleckstrin homology (PH) domain of ∼100 residues, a signaling module that is often involved in intracellular membrane targeting. The characteristic pairing of DH and PH domains suggests that the DH-associated family of PH domains may participate in the regulation of GEF activity. PH domains could contribute binding or catalytic residues, allosterically influence catalytic activity, recruit additional factors, or position the DH domain properly with respect to the GTPase by membrane targeting (26). A crystal structure of the Tiam1-DH/PH-Rac1 indicated that the PH domain provides only simple structural stabilization of the DH domain (46), leaving open the possibility of more complex functions. However, in the case of a Dbs-DH/PH-Cdc42 crystal structure, the Dbs PH domain participates directly in binding Cdc42, through a set of interactions involving the switch 2 region of the GTPase (39).
Several reports have demonstrated that the PH domains of DH/PH cassettes can interact with phosphorylated phosphatidylinositides and thereby potentially modify the membrane association or activity of their linked DH domains (28, 43). There are a number of examples of in vitro GEF activity being regulated by interactions of PH domains with phosphatidylinositol 4,5-biphosphate [PI(4,5)P2] or PI(3,4,5)P3. These include PH domains of Vav1, Dbl, Tiam1, Sos1, P-Rex, and SWAP-70 (13, 20, 24, 37, 40, 42, 45). In the case of Vav1 and Sos1, PI(3,4,5)P3 binding to the PH domain relieves an intramolecular interaction between the DH and PH domains, allowing Rac to access the catalytic DH domain surface (16). P-Rex is also substantially activated by PI(3,4,5)P3, alone as well as synergistically with Gβγs; however, whether this mechanism involves a disruption of an interaction between the PH and DH domains is not known (45). The situation with Tiam1 is less clear, with evidence existing both for and against allosteric activation of GEF activity by PI(4,5)P2 (13, 20), and further in vitro studies indicate that the PH domain of Tiam1 may be specific for PI(3)P (43).
To date, no systematic study has been undertaken to correlate the in vitro biochemical functions (e.g., phospholipid binding and GEF activity) of DH and PH domains with in vivo biological activity in a model organism. In this regard, we have previously described the unc-73 gene from Caenorhabditis elegans (44), which encodes multiple isoforms, including an extended polypeptide (UNC-73A) with two DH/PH cassettes and a shorter N-terminal protein with only one DH/PH cassette (UNC-73B). UNC-73B has an N-terminal Sec-14 homology region followed by a spectrin repeat region, the DH and PH domains, and an SH3 domain. Mutations in unc-73, which affect the UNC-73B isoform, cause a variety of defects in axon guidance and cell motility (15). The UNC-73B DH/PH cassette specifically activates mammalian Rac1 in vitro and can stimulate actin polymerization in Rat2 fibroblasts. Indeed, the ability of unc-73 to mediate axon guidance and actin polymerization is impaired by alteration of Ser 1216 in the first DH domain, which inhibits Rac GEF activity. UNC-73B is highly related to the Trio and Kalirin family of proteins present in vertebrates (3, 17) and Drosophila (4, 5, 30, 36), and observations pertaining to UNC-73B are likely to have general implications for metazoan animals. We have, therefore, used a combination of in vitro biochemistry and functional analysis in C. elegans to examine the functional role of the PH domain of UNC-73B in Rac activation and axonal pathfinding in vivo.
MATERIALS AND METHODS
Antibodies, molecular cloning, and protein expression.
Polyclonal antibodies against UNC-73B DH/PH/SH3 were as described previously (44), while anti-green fluorescent protein (anti-GFP) (Ab290) was from Abcam. Monoclonal antibodies against Flag (M2) were from Sigma. Alexa Fluor-488 goat anti-rabbit monoclonal antibody and Texas Red phalloidin were from Molecular Probes.
The baculovirus for wild-type and mutant glutathione S-transferase (GST)/UNC-73B DH/PH/SH3 were generated by using the PharMingen Baculogold system and expressed in monolayer Sf9 cultures. The mutations created for this study are Trp1502 to Ala (the W1502A construct), Lys1424/Arg1426 to Ala/Ala (KR to AA), Lys 1420/Arg1422 to Glu/Glu (KR to EE), and ΔPH, which is the removal of residues 1398 to 1559. Mutations were created by using the Stratagene Quikchange site-directed mutagenesis kit. Mutations were carried out on wild-type cDNA (residues 1179 to 1642) present in pSL301 and subcloned into the EcoRI site of pAcGHLT-A. These UNC-73B proteins were purified as described previously (44). For mammalian expression, the wild-type and mutated sequences were amplified by PCR and subcloned into the pEGFP-C1 vector from Clontech via the BamHI site. Further details are available upon request.
The minigene construct for worm microinjections containing the wild-type sequence and C-terminal Flag3 construct was made by mutating by PCR the stop codon of the pSL301-DH/PH/SH3 construct into a BamHI site and subcloning the mutated NruI/BamHI fragment into the previously described UNC-73B minigene (44). Subsequently, an annealed pair of Flag3-expressing oligonucleotides was subcloned in frame at the BamHI/NotI sites on this minigene, and the 3′ untranslated region of UNC-73B was amplified by PCR and reintroduced into the NotI site of the minigene. Mutations were transferred into the minigene by digestion with NruI and BamHI from the pEGFP-C1 constructs. All mutations and subcloned portions of the cDNA were verified by DNA sequencing.
The pGex-CeRAC1, CeRHO, and CeCDC42 constructs were initially characterized and supplied by Louis Lim (8, 9, 10). The pGex-MIG-2 construct was a gift from Cynthia Kenyon (47). Purification of these GTPases was carried out as previously described (44).
Nucleotide exchange assays.
A fluorescence assay in which the C. elegans GTPases were preloaded with 12.5 μM mant-GDP (Molecular Probes) by incubation with the fluorescently labeled nucleotide in the presence of 6.25 mM EDTA for 30 min at room temperature was used (1). The loading was then quenched by adding MgCl2 to a final concentration of 50 mM; unbound nucleotide was removed, and protein was transferred to a buffer containing 20 mM HEPES (pH 7), 50 mM NaCl, and 2 mM MgCl2 by using a NAP-10 gel filtration column. For each reaction, 0.5 μM GTPase and 0.2 μM GEF were added together with 2.5 μM GTP in a volume of 800 μl. The time course of fluorescence was monitored with an LS-50B spectrophotometer (Perkin-Elmer) with λexcitation at 350 nm, λemission at 440 nm (slits 5/15 nm), and an emission filter in place at 390 nm.
To test for the effect of phospholipids on GEF activity, various water-soluble diethyl-phosphorylated phosphatidylinositides (Echelon) were added prior to the beginning of each reaction at a final concentration of 25 μM with respect to phosphatidylinositide concentration.
Phospholipid dot blot.
PIP strips were obtained from Echelon, and dot blots were carried out according to the manufacturer's suggested protocol with a few modifications. Strips were blocked overnight at 4°C in Tris-buffered saline containing 10% glycerol (TBSG) and 3% bovine serum albumin. Baculovirus-produced GST/UNC-73 DH/PH/SH3 wild-type and mutant fusion proteins were purified with glutathione-Sepharose (Pharmacia) as before and then washed in kinase buffer and labeled with [32P]ATP by using the catalytic subunit of protein kinase A (Sigma) as suggested by the manufacturer's protocol. The 32P-labeled fusion proteins were then washed with phosphate-buffered saline, eluted once with 10 mM glutathione in phosphate-buffered saline, and then applied to the PIP strips and incubated at a final concentration of 1 μM in 5 ml 3% BSA-TBSG for 50 min at room temperature. Each blot was then washed five times for 10 min each in TBSG and developed with a phosphorimager.
C. elegans strains and manipulations.
The unc-73 mutant allele e936 has been previously described (44). Transformation was performed as described previously (35) by coinjecting the UNC-73B minigenes with the plasmid pAC12 (20 ng/μl), which contains the motorneuron-specific promoter region of UNC-129 fused to GFP (11). This allowed us to assay the circumferential navigation of the DA and DB neurons, which migrate dorsally from ventral cell bodies to form the dorsal nerve cord, into e936/dyp5 balanced heterozygotes and maintained as extrachromosomal arrays. Homozygous e936 worms carrying the various arrays were then collected. Four different concentrations of the UNC-73B minigene (40, 10, 5, and 1 ng/μl) were tested for rescue. The ability of the UNC-73B wild-type and KR-to-AA minigenes and the inability of the KR-to-EE, W1502A, and ΔPH minigenes to rescue the unc-73(e936) worms occurred consistently over the range of concentrations tested. Animals injected at 5 ng/μl were counted for axon guidance defects. At all concentrations of the W1502A minigene, homozygous e936 animals were sterile, preventing maintenance of that line in the mutant worms. Thus, in this case, homozygous e936 animals from heterozygous e936/dpy5 worms carrying the W1502A array were counted. Although there is the possibility of maternal contribution of UNC-73 to these animals, this contribution in itself is not enough to rescue the axon guidance defects in these worms. Living animals were mounted on a slide in a small drop of 10-mg/ml levamisole on a 2% agarose pad for microscopy. Axons that exhibited more than a single morphology were scored as containing the more severe defect (i.e., in descending severity, outgrowth defect > longitudinal defect > branching defect > oblique migration > normal migration) (22, 38).
RESULTS
The UNC-73B DH domain activates CeRAC1 and MIG-2.
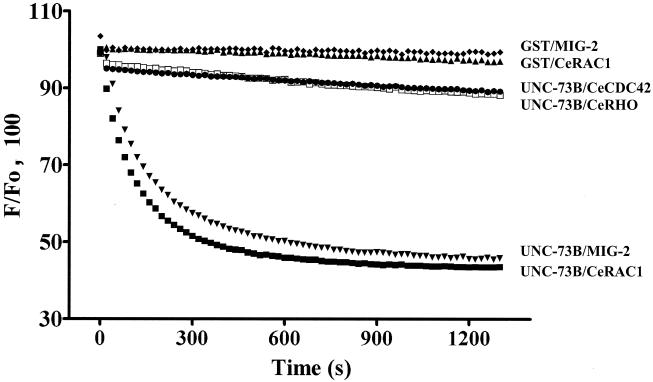
Previously, we demonstrated that the DH domain of UNC-73B is a specific GEF for the mammalian Rac1 GTPase in vitro. To determine whether this specificity is maintained for the C. elegans GTPases, we purified bacterially expressed GST fusion proteins of CeRAC1, CeRHO, and CeCDC42 and carried out fluorescent GEF assays with a polypeptide containing the DH/PH and SH3 domains of UNC-73B, expressed by using a baculovirus vector in insect cells. By monitoring for release of mant-GDP (Fig. 1), we found that the UNC-73B protein containing the DH domain demonstrated specific activity towards CeRAC1, and in agreement with previous findings with the mammalian GTPases, did not show GEF activity towards CeRHO and CeCDC42.
FIG. 1.
UNC-73B DH is an exchange factor for CeRAC and MIG-2. The normalized change in fluorescence (F) upon addition either of GST/UNC-73B DH/PH/SH3 to preloaded (mant-GDP) CeRAC (▪), MIG-2 (▾), CeRHO (□), and CeCDC42 (•) or of GST to preloaded CeRAC (▴) and MIG-2 (⧫) is shown. Proper loading of mant-GDP to CeRHO and CeCDC42 was demonstrated by monitoring release of the fluorescent nucleotide by EDTA (data not shown).
The mig-2 gene also encodes a C. elegans Rho family GTPase; interestingly, both activating and loss-of-function mutations in mig-2 result in defects in cell migrations and axon guidance, similar to those displayed by unc-73 mutants (47). Furthermore, unc-73 and ced-10 (rac-1)/mig-2 are in the same genetic pathway for a number of cell migrations (33; G. Dalpé and J. Culotti, unpublished data), and the Drosophila Trio is an exchange factor for the Drosophila homolog of MIG-2, Mtl (36). UNC-73B was able to release preloaded mant-GDP from MIG-2 in vitro, suggesting that UNC-73B can directly activate the MIG-2 GTPase as well as CeRAC1 (Fig. 1).
The UNC-73B PH domain interacts with phosphorylated phosphatidylinositides.
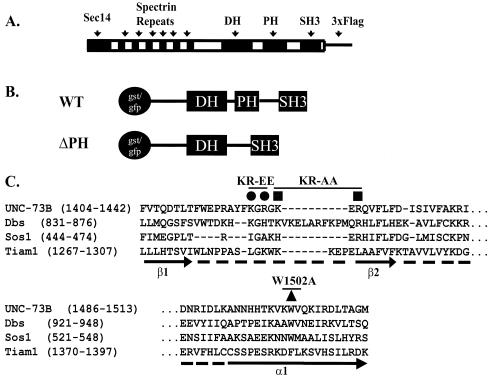
To investigate the role of the PH domain in UNC-73B function, we generated four mutants with changes in the PH domain-coding sequence. To evaluate the possible effects of phosphatidylinositide binding (see below), we altered positively charged amino acids located in the β1/β2 loop of the UNC-73B PH domain, which correspond to residues involved in phosphatidylinositide binding in other PH domains. Based on the sequence alignment of the UNC-73B PH domain with the Dbs, Sos1, and Tiam1 PH domains, Lys1424 and Arg1426 are the most conserved basic residues in these proteins and were, therefore, initially targeted for simultaneous replacement by Ala (Fig. 2). However, since a double mutation of both residues to alanine (hereafter referred to as KR to AA) did not abolish phospholipid binding (see below), a second double mutant was made, changing both Lys1420 and Arg1422 to negatively charged Glu residues (referred to as KR to EE) (Fig. 2). Also, another pair of mutations was created to investigate other potential roles for the PH domain, such as direct or indirect involvement in Rac binding and activation, as well as a potential role in other protein-protein interactions. One mutation changed the conserved Trp1502 to Ala (W1502A) in the C-terminal α-helix portion of the PH domain, which was expected to have a global effect on PH domain folding and function. Finally, a truncated UNC-73B protein in which the entire PH domain was deleted was created.
FIG. 2.
UNC-73B and various UNC-73B mutants and constructs. (A) The modular structure of the UNC-73B protein (1,638 amino acids) with the C-terminal Flag3 used in C. elegans transgenic studies is shown along with the DH/PH/SH3 (amino acids 1179 to 1642) and ΔPH (Δ1398-1559 with an insertion of an Asn) fragments used for GEF assays and phospholipid binding studies. (B and C) Sequence alignment of various PH domains, β1 and β2 strands, and the β1/β2 loop and α1 helix of the UNC-73B PH (accession number AAC71109), human Sos1 (Q07889), mouse Dbs (AAB33461), and mouse Tiam1 (NP_033410) PH domains. •, basic residues mutated to Glu (KR-to-EE construct); ▪, basic residues mutated to Ala (KR to AA); ▴ conserved tryptophan mutated to alanine (W1502A). WT, wild type.
To qualitatively determine if the UNC-73B PH domain, like a number of other PH domains, interact with phospholipids, we used an established dot blot assay (27). A GST fusion protein containing the wild-type UNC-73B DH/PH/SH3 domains was expressed in insect cells, purified, radiolabeled with protein kinase A on a site within the GST fusion linker region, and then applied to nitrocellulose filters that were previously spotted with various phospholipids. The use of 32P-labeled UNC-73B proved to be a sensitive method to determine the interactions of the wild-type and mutant proteins with phospholipids, indicating that such labeling does not have a deleterious affect on phospholipid binding. Since the purified wild-type or mutant proteins were labeled by phosphorylation to the same extent, this method can therefore be used to accurately assess the relative phospholipid binding properties of the different mutants.
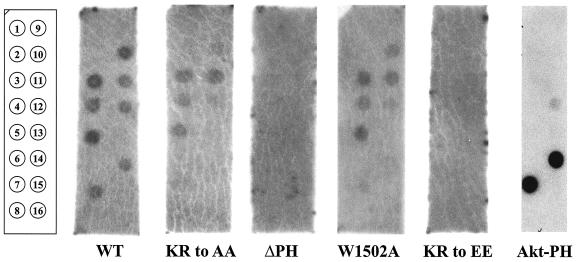
As shown in Fig. 3, the DH/PH/SH3 fragment of UNC-73B recognized several phospholipids, binding preferentially to the monophosphorylated phosphatidylinositides [PI(3)P, PI(4)P, and PI(5)P], more weakly to the bisphosphorylated lipids tested, and poorly to PI(3,4,5)P3. The PKB/Akt PH domain, as a positive control, demonstrated strong and specific interaction with PI(3,4)P2 and PI(3,4,5)P3. GST protein alone did not display binding to any phospholipid (data not shown). This profile of interaction for UNC-73B is very similar to that seen for the PH domains of intersectin and Dbs (43).
FIG. 3.
The wild-type (WT) UNC-73B PH domain demonstrates promiscuous binding to phosphorylated phosphatidylinositides. Radiolabeled wild-type GST/UNC-73B DH/PH/SH3, mutant fusion proteins, and the isolated AKT-PH domain were applied individually to PIP strips (Echelon) containing various phospholipids, as indicated. The numbered positions of the strip on the far left represent dots containing the following: 1, phosphatidylinositide; 2, Ins(1,3,4,5)P4; 3, PI(3)P; 4, PI(4)P; 5, PI(5)P; 6, phosphatidylethanolamine; 7, PI(3,4)P2; 8, blank; 9, phosphatidylcholine; 10, PI(3,5)P2; 11, phosphatidic acid; 12, PI(4,5)P2; 13, phosphatidylserine; 14, PI(3,4,5)P3; 15, solvent; 16, blank.
The various mutants of the UNC-73B PH domain were also expressed in insect cells as GST fusion proteins and characterized for their ability to interact with phospholipids. The ΔPH construct (GST/UNC-73B DH/SH3) showed no binding to any phospholipid, confirming that the PH domain of UNC-73B is responsible for its interaction with phosphorylated phosphatidylinositides. The KR-to-EE mutation also abolished the PH domain interaction with phospholipids, while the KR-to-AA mutation, in contrast, was still able to bind with a profile similar to that of the wild type (Fig. 3). The KR-to-EE mutation therefore had a more deleterious effect on binding the negatively charged phospholipids than did the KR-to-AA mutation, and this allowed us to investigate the effects of altering PH domain-phospholipid binding on the in vivo function of UNC-73B.
Surprisingly, the W1502A mutant was also able to interact with phospholipids. This substitution, based on structural studies of other PH domains, might be expected to destabilize the PH domain. PH domain mutants tested previously for in vitro binding to phospholipids have focused on the β1/β2 and β3/β4 loop regions of the PH domain, which were identified as the binding face for phospholipid interaction. Based on the previous work describing the Dbs-DH/PH-Cdc42 structure (39), the C-terminal α-helix of the PH domain containing the invariant Trp and the phospholipid binding sites are on the opposing sides of the domain, and it is conceivable that replacement of the invariant Trp with Ala spares phospholipid binding. Our results show that the W1502A mutant retains an ability to interact with acidic phospholipids, which suggests that (at least for the UNC-73B PH domain) the resulting destabilization of the PH domain structure is not sufficient to prevent lipid binding.
Alteration of the UNC-73B PH domain affects in vitro GEF activity of the DH domain.
Structural studies with Dbs and Tiam1 in complexes with Cdc42 and Rac1, respectively, show a role for their PH domains in DH domain stability and, in the case of Dbs, direct contact of the PH domain with the GTPase (39, 46). We therefore investigated the consequences of the UNC-73B PH domain mutations on the catalytic efficiency of the neighboring DH domain and on actin polymerization in a cell-based assay.
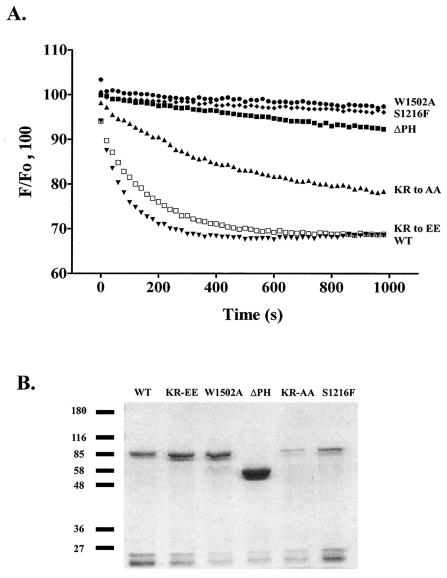
To quantify the effect of the PH domain mutations on the catalytic efficiency of the DH GEF activity, we used the fluorescent GEF assay. As a negative control, we used a protein with the S1216F substitution in the UNC-73 DH domain, induced by the unc-73(rh40) mutation, which causes severe axon guidance and cell migration defects due to its disruption of the integrity of the DH domain. This mutant failed to release mant-GDP from bacterially expressed, preloaded CeRAC1 (Fig. 4A) (44). We also found that the W1502A PH mutation adversely affected the activity of the DH domain (Fig. 4A), resulting in a loss of GEF activity very similar to that observed with the S1216F mutation. Likewise, the ΔPH mutant also demonstrated a large reduction in GEF activity. These results, taken together with microinjection data (see below), show that the PH domain plays a positive role in activation of CeRAC by the UNC-73B DH domain.
FIG. 4.
The PH domain of UNC-73B promotes GEF activity of the accompanying DH domain in vitro. (A) GEF assay of UNC-73B PH mutants. The change in fluorescence (F) of preloaded mant-GDP/CeRAC on addition of wild-type (WT) GST/UNC-73B DH/PH/SH3 (▾); the KR-to-EE (□), KR-to-AA (▴), ΔPH (▪), and W1502A (•) mutants; and the previously described S1216F mutant (⧫) is shown. (B) Sodium dodecyl sulfate-8% polyacrylamide gel electrophoresis and Coomassie blue staining of baculovirus-produced and affinity-purified GST-UNC-73B DH/PH/SH3 wild-type and PH domain mutant fusion proteins (as indicated) used for the in vitro GEF assays for panel A and Table 1. Protein samples were purified by glutathione-Sepharose affinity purification, eluted with 10 mM glutathione, and concentrated by using the Millipore Ultrafree Centrifugal Filter and Tube, and subsequently an equal volume of protein solution was applied per lane for an initial approximation of concentration and purity. After dialysis, final protein concentrations were determined by UV spectrophotometry (absorbance at 280 nm) and 0.2 μM was used per GEF reaction (see panel A and Table 1). Molecular weight markers (in thousands) are indicated on the left.
The effects of mutating the basic residues within the PH domain on the catalytic activity of UNC-73B were also assessed quantitatively. The KR-to-EE mutant, which is unable to interact with phospholipids, was able to catalyze the release of mant-GDP at a rate similar to that of the wild type. Thus, a mutation within the β1/β2 loop that did affect phospholipid binding did not affect the in vitro catalytic efficiency of the UNC-73B DH domain to any significant degree (Table 1). However, the KR-to-AA PH domain alteration did cause a reduced ability to catalyze the release of nucleotide. Thus, although this fusion protein was able to interact with phosphorylated phosphatidylinositides, the replacement of K1424 and R1426 by Ala moderately impaired DH activity.
TABLE 1.
Rate constants of guanine nucleotide exchange reactions catalyzed by wild-type and mutant UNC-73B proteins on CeRAC
| UNC-73B | kobs (s−1, 103)a | Fold stimulationb |
|---|---|---|
| None | 0.096 ± 0.01 | |
| Wild type | 11.2 ± 0.21 | 117 |
| KR-to-AA mutant | 3.16 ± 0.026 | 32 |
| KR-to-EE mutant | 5.54 ± 0.10 | 58 |
| ΔPH mutant | 0.27 ± 0.013 | 2.8 |
| W1502A mutant | 0.12 ± 0.01 | 1.3 |
| S1216F mutant | 0.12 ± 0.01 | 1.3 |
The rates of release of mant-GDP from unmodified CeRAC (kobs) stimulated by approximately 0.2 μM wild-type and mutant UNC-73B proteins were determined by fitting the data from Fig. 4A as single exponential decays. Results are expressed as means ± standard deviations.
The fold stimulation reflects the ratio of the kobs measured for the GEF-containing reaction to that for the unstimulated reaction containing no GEF (none).
The quantitative analysis of GEF activity presented in Fig. 4A depends on the relevant proteins being stable. Figure 4B demonstrates that the level of purified intact proteins generated from the baculovirus expression system was sufficient for their biochemical analysis in Fig. 4A and Table 1. Interestingly, the ΔPH domain construct consistently expressed at much higher levels than the wild-type protein.
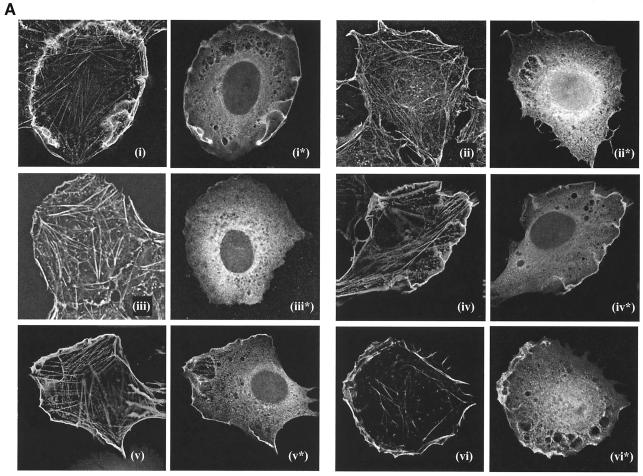
As reported previously, nuclear microinjection of Rat2 cells with an expression construct encoding the wild-type UNC-73B DH/PH/SH3 fused to GFP induced high levels of polymerized actin at the plasma membrane (44). This is consistent with the ability of the UNC-73B DH domain to activate Rac, which in turn induces actin polymerization (Fig. 5A, panel i). The cellular activity was dependent on the PH domain, as the ΔPH construct failed to produce lamellipodia above basal levels (Fig. 5A, panel ii). Likewise, the W1502A PH mutant was also inactive in this assay. In contrast, mutants with substitutions in the β1/β2 loop of the PH domain retained an ability to induce actin polymerization (Fig. 5A, panels iv and v). Thus, although the PH domain is required for GEF activity, abolishing the phospholipid binding properties of the PH domain (KR-to-EE mutant) did not adversely alter actin-polymerizing activity when the mutant protein was overexpressed in Rat2 cells. This is potentially due to the high level of expression from microinjection of the plasmid into Rat2 cells.
FIG.5.
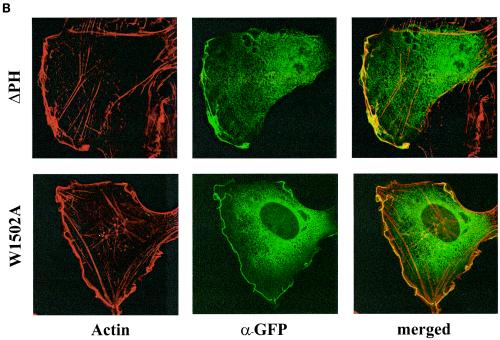
Function and localization of wild-type UNC-73B and PH domain mutants in a cell culture-based assay. (A) Effects of UNC-73B mutants on actin polymerization in Rat2 fibroblasts. Rat2 fibroblasts were injected into the nucleus with plasmid DNA encoding wild-type GFP/UNC-73B DH/PH/SH3 (panels i and i*); the ΔPH (panels ii and ii*), W1502A (panels iii and iii*), KR-to-AA (panels iv and iv*), and KR-to-EE (panels v and v*) mutants; and Flag-V12 Rac (panels vi and vii*). Three hours after injection, cells were fixed and subsequently stained with anti-GFP (Abcam Ab290) or M2 (anti-Flag) antibody for identification of injected cells and filamentous actin (Texas Red phalloidin). Actin staining is present in the initial panel for each injected construct, while the anti-GFP or anti-Flag staining is present in the asterisk-labeled panels, as appropriate. (B) Localization of ΔPH and W1502A in Rac-activated cells. GFP constructs of the two mutants of UNC-73B PH domain (ΔPH and W1502A) were coinjected in the nuclei of Rat2 cells with Flag-V12 Rac in the presence of serum to induce membrane ruffling. Cells were fixed and stained for actin and anti-GFP (α-GFP) as for panel A. Actin and anti-GFP staining, as well as the merged images of the two, are shown.
Immunofluorescence analysis demonstrated a colocalization of the expressed GFP/UNC-73B DH/PH/SH3 wild-type fusion protein with polymerized actin, along with a significant amount of cytoplasmic staining (Fig. 5A, panels i and i*). This colocalization also occurred with the KR-to-AA and KR-to-EE mutants (Fig. 5A, panels iv* and v*), demonstrating that the KR-to-EE mutant protein can interact with membrane ruffles despite its inability to interact with phospholipids in vivo.
To determine whether the PH domain was necessary for this colocalization in Rat2 fibroblasts, we determined the localization of the ΔPH and W1502A proteins in fibroblasts containing an activated form of Rac. To do this, we coinjected these two constructs with the constitutively active Flag V12-Rac construct and determined their localization by staining for anti-GFP. Figure 5B shows an overlapping of staining of ΔPH and W1502A with the polymerized actin, indicating that colocalization of UNC-73B with lamellipodia occurs independent of the PH domain, potentially through its SH3 domain and/or DH domain.
GEF activity of UNC-73B DH/PH/SH3 is not affected by phospholipids.
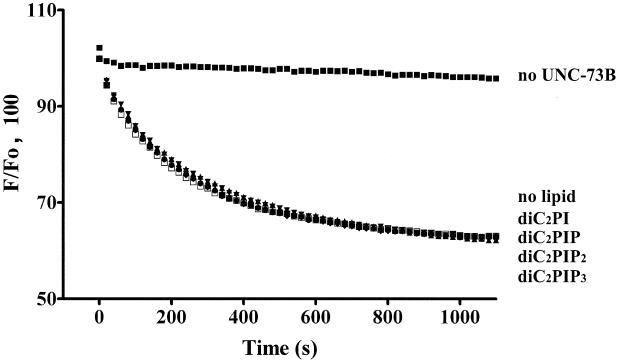
The ability of the UNC-73B PH domain to associate with phosphatidylinositides, coupled with previous reports of phosphatidylinositide-induced modulation of GEF catalysis in solution (13, 16, 24, 45), led us to evaluate the effects of phosphatidylinositides on UNC-73B-catalyzed GEF activity. To avoid contaminating the reaction with organic solvents required to solubilize long-chain phospholipids, which has been shown to inhibit Dbs GEF activity (43), we used water-soluble diethyl analogs of phosphatidylinositides as used for the investigation of Vav and Sos dependence of phospholipids on activity (16). The addition of a 25 μM concentration of either diethyl-PI, diethyl-PI(4)P, diethyl-PI(4,5)P2 or diethyl-PI(3,4,5)P3 did not significantly affect catalysis by UNC-73B DH/PH/SH3 on the preloaded mant-GDP/CeRAC complex (Fig. 6). This result corresponds to previous observations that phosphatidylinositides failed to alter nucleotide exchange by intersectin, Dbs, and Tiam-1 on mammalian Rac1 or Cdc42 (43). The effect of the water-soluble lipids on the PH domain mutants of UNC-73B was also tested, and no significant change in activity for any of the mutants in the presence or absence of these lipids was detected (data not shown). Therefore, unlike Vav and Sos1, the UNC-73B GEF activity is not obviously altered by soluble phosphorylated phosphatidylinositides, and the DH domain catalytic function is not apparently influenced by an allosteric mechanism involving PH domain inhibition by phosphatidylinositides.
FIG. 6.
Water-soluble phosphorylated phosphatidylinositides do not affect UNC-73B GEF activity on bacterially expressed nonmodified CeRAC. The change in fluorescence (F) of wild-type GST/UNC-73B DH/PH/SH3 in the presence of 25 μM diethyl-phosphorylated phosphatidylinositides is shown. Samples without UNC-73B in the reaction mixture [in the presence of diethyl-PI(4)P] or without any lipid also were used.
The UNC-73B PH domain is required for axon guidance.
To explore the involvement of the UNC-73B PH domain in axon guidance, we used in vivo analysis with C. elegans to determine the biological activities of the PH domain mutants described above. To this end, transgenic animals containing either the wild-type UNC-73B minigene or variants with mutations affecting the PH domain as extrachromosomal arrays were created. Previously identified unc-73 mutants exhibit a variety of defects in axon guidance and cell motility (44). To quantitate the abilities of the different transgenes to rescue the unc-73 mutant phenotype, the UNC-73B minigenes were coinjected with an unc-129::gfp construct that expresses GFP specifically in the DA and DB motorneurons. In wild-type N2 animals these neurons exhibit a simple morphology, in that they migrate as a single axon circumferentially from a ventral cell body to the dorsal nerve cord, where they fasciculate with other dorsal nerve cord axons and migrate either anteriorly, posteriorly, or both (Fig. 7A).
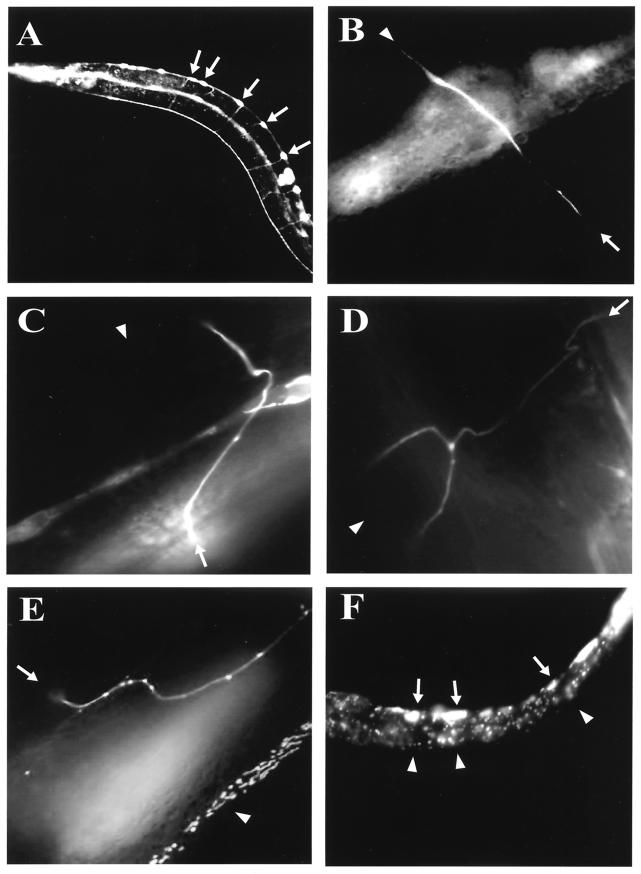
FIG.7.
Categorization of axon guidance and growth defects in unc-73(e936) mutants. (A) Image of a wild-type young adult hermaphrodite expressing GFP in the DA and DB motorneurons driven by the neuronal promoter region of unc-129. The arrows indicate the cell bodies of these neurons on the ventral side of the worm. (B) A motorneuron rescued by the UNC-73B wild-type minigene coexpressed with unc-129::gfp in the unc-73(e936) worm. The arrow indicates the position of the ventral cell body, while the dorsal side of the worm, at approximately a right angle to the ventral cell body, is indicated by the arrowhead. (C to F) Examples of misguided axons expressing the UNC-73B minigene-containing mutations within the PH domain coinjected with the unc-129::gfp plasmid in unc-73(e936) animals. Arrows represent the position of the cell body, and arrowheads indicate the position at the dorsal cord of the axon's proper destination. (C) An axon migrating towards the dorsal cord and then setting off at an oblique angle before straightening and completing its journey to the dorsal cord. (D) Branched axon morphology of the motorneuron before it reaches the dorsal cord. (E) Demonstration of a longitudinal defect of the axon, as it extends towards the dorsal side of the worm, migrates anteriorly, and then migrates dorsally before finally migrating anteriorly prior to reaching the dorsal cord. (F) An outgrowth defect, with no axon projecting out of the cell bodies.
Axon morphologies were categorized as (i) normal (these exit the cell body at right angles to the ventral cord, reach, and then extend to and along the dorsal cord), (ii) oblique phenotypes (axons that travel at much less than right angles from the ventral cell bodies towards the dorsal cord), (iii) branched defects (axons contain multiple branches as they extend towards the dorsal cord), (iv) longitudinal defects (these axons travel anteriorly or posteriorly, parallel to the ventral and dorsal cords at any point between the ventral and dorsal cords), and (v) outgrowth defects (axons that fail to project out of the cell body) (Fig. 7) (31).
The unc-129::gfp marker was injected alone to score for the degree of misguidance that occurs in unc-73(e936) animals. These animals have an inactive Unc phenotype, are small and dumpyish, and demonstrate axon guidance defects. As shown in Fig. 8, a majority of motorneuron axons are severely misguided, with 64% of them exhibiting either a branching morphology, a longitudinal trajectory, or outgrowth defects. Establishment of a stable extrachromosomal array in unc-73 (e936) animals carrying the wild-type UNC-73B minigene rescued the Unc phenotype of these animals. The wild-type extrachromosomal transgene array also caused a reduction in the fraction of severely misguided axons to 22.5%.
FIG. 8.
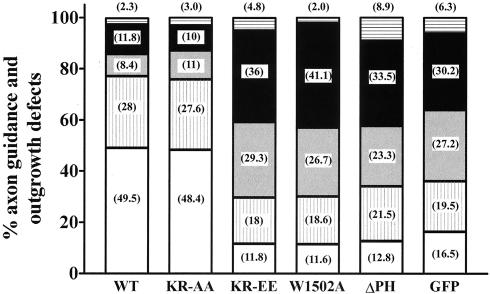
Quantitation of axon guidance and growth defects in unc-73(e936) animals expressing various UNC-73B minigenes. Individual neurons were visualized, categorized, and scored for the specific guidance errors and growth defects as defined for Fig. 7. In parentheses within the bars are the actual percentages recorded for each phenotype. A minimum of 96 worms were counted, including of at least 380 neurons for each minigene. White bars, axons with normal morphology; vertically striped bars, axons migrating at an oblique trajectory; grey bars, axons with branching defects; black bars, axons displaying a longitudinal trajectory; horizontally striped bars, axons with outgrowth defects.
The transgene encoding the KR-to-AA mutation within the PH domain was also able to rescue the Unc phenotype of the e936 homozygous animals. As noted above, the KR-to-AA mutant protein is still able to bind phospholipids, as well as cooperate with the DH domain in GEF catalysis in vitro. Quantitation showed that this mutant minigene reduced the number of DA and DB motorneurons exhibiting defects as well as the wild type (24%, versus 22.5% for wild type). This is despite the wild-type protein having significantly more robust Rac GEF activity in vitro than the KR-to-AA mutant protein (Fig. 4A and Table 1).
In contrast, the transgene array encoding the KR-to-EE mutation was not able to rescue the Unc phenotype of the e936 homozygous animals, even though this mutant can stimulate GEF catalysis in the context of the DH/PH cassette, both in GEF assays on modified and unmodified CeRAC and by microinjection into Rat2 fibroblasts (Fig. 8). This result suggests the importance of the positive electrostatic surface of UNC-73B's PH domain, a characteristic of a large number of PH domains, for UNC-73B biological activity. Since the KR-to-EE mutant can still enhance GEF activity in vitro, these data also indicate that the UNC-73B PH domain has an additional function in vivo that is important for activation of CeRAC and axon guidance, which correlates with its phospholipid-binding ability.
The minigenes containing the PH deletion and the W1502A point mutation were also unable to rescue axon guidance in the unc-73(e936) mutant, despite the ability of the W1502A construct to interact with phosphorylated phosphatidylinositides (Fig. 8). This is consistent with the finding that proteins carrying these PH domain mutations were inactive in UNC-73B DH domain-directed GEF catalysis in vitro.
Previous work has shown that both a dominant active form of mig-2 and loss of mig-2 function result in similar axon guidance defects in C. elegans (47); in addition, mutations in the PH domain of Vav have been reported to give dominant active mutants (34). It was, therefore, important to determine whether the UNC-73B mutants that failed to rescue axon guidance in vivo exert a dominant active effect. To distinguish between dominant negative and dominant positive mutations, we injected the unc-73 minigenes into e936/dpy5 heterozygous worms, which contain the wild type unc-73 allele tightly linked to the mutant dpy5 allele and which are phenotypically wild type. Thus, expression from a potential dominant active transgene plus the contribution from the wild-type allele would result in worms that exhibit a more severe Unc phenotype than they would have in the e936 homozygous state.
By this assay, none of the transgenes that failed to rescue the unc-73 defect were dominant active mutations. Rather, they appeared to have a dominant negative effect, as indicated in Fig. 8, since the KR-to-EE, W1502A, and ΔPH transgenes gave a more severe phenotype in vivo with respect to axon guidance than GFP alone in the e936 homozygous background. This is potentially explained by the fact that the e936 allele is a mutation of the splice donor site of intron 16, which results in severe reduction, but not complete removal, of wild-type UNC-73B protein (44). Thus, the mutant transgenes may interfere with the function of the low levels of endogenous wild-type UNC-73B produced in the unc-73(e936) homozygous mutant background.
DISCUSSION
Here, we provide evidence that the UNC-73B PH domain has two distinct functions in regulating the activation of CeRAC1 and MIG-2 and thus in controlling the guidance of axon migrations in C. elegans. One apparent role for the UNC-73B PH domain is to directly cooperate with the adjacent DH domain to promote GEF activity, while a second is to bind phosphoinositides and thus potentially position the active DH/PH cassette in an appropriate location relative to CeRAC1/MIG-2 GTPases.
With respect to GEF catalytic activity, deleting the UNC-73B PH domain results in a marked decrease in DH domain function in vitro, as noted previously with Trio (32). However, we have also found that mutating the highly conserved Trp1502 to alanine in the PH domain yields a mutant UNC-73B protein that is still capable of binding phosphorylated phosphatidylinositides but lacks GEF activity. Furthermore, a W1502A mutant UNC-73B minigene could not rescue the axon guidance defects of worms with a loss-of-function mutation in the endogenous unc-73 gene. These data indicate that the PH domain may have a direct effect on GEF activity that is important for in vivo function.
The PH domain could aid in GEF activity by stabilizing the DH domain structure, by direct binding to CeRAC, or both. Of interest is that recent crystallographic data for Cdc42 in complex with Dbs demonstrates that the β1 strand and the β3/β4 loop of the Dbs PH domain directly interact with switch 2 of the GTPase (39). Those authors noted that the PH domain residues involved in the GTPase interaction are conserved in a subset of Dbl family proteins, of which Trio and UNC-73B are members. The in vitro GEF activities of Dbs, UNC-73B, and the N-terminal Trio DH domains are all enhanced very significantly by the presence of their PH domains. Also, the nuclear magnetic resonance structure of the Trio N-terminal DH domain demonstrates that it is properly folded in the absence of its PH domain, adopting a structure very similar to that of other DH domains (32, 2). Since the DH domain structure does not seem to require the PH domain, a plausible explanation for the ability of the Trio, UNC-73B, and Dbs PH domains to accelerate in vitro GEF activity is that the PH domain makes direct contact with the GTPase and assists in catalysis. It is apparent that some GEFs (Vav, for example) can function efficiently in the absence of their PH domains, but this ability is not universally shared by all DH/PH-containing proteins. Our results provide functional in vitro and in vivo data in support of the view, suggested by the Dbs-DH/PH-Cdc42 crystal structure, that the PH domain of a subset of DH/PH proteins contributes to exchange activity in a fashion that is physiologically relevant.
A well-recognized role of PH domains in general is to target proteins to specific sites in cellular membranes by interacting with phosphorylated phosphatidylinositides (29). The UNC-73B PH domain recognizes a number of phosphorylated phosphatidylinositides in vitro, although these data do not exclude the possibility that the UNC-73B PH domain has other binding partners. As in other PH domains, the β1/β2 loop is at least partly responsible for this interaction, since mutating two basic residues within this loop to glutamate abolished phospholipid binding. Phospholipid binding by the UNC-73B PH domain, however, is dispensable for Rac activation when ectopically overexpressed in cultured cells, since the KR-to-EE mutation (which disrupts phosphatidylinositide binding) caused actin accumulation at the plasma membrane in Rat2 fibroblasts and could promote the release of mant-GDP from CeRAC in vitro. However, the UNC-73B minigene containing the KR-to-EE mutation was incapable of rescuing unc-73 mutants in vivo, indicating that the binding of ligands such as phosphatidylinositides is important to the function of the UNC-73B PH domain motor axons in the intact worm. This may reflect the significant demands in correctly localizing a protein within the growth cone of an extending axon. A scheme consistent with our data is that UNC-73B can localize to the vicinity of the membrane through regions other than the PH domain, and this is followed by an interaction of the PH domain with phosphorylated phosphatidylinositides, allowing for the proper spatial organization of the DH/PH domain with the membrane-bound Rac (39) and resulting in the formation of a complex which is necessary for efficient CeRAC1/MIG-2 activation and correct axon guidance in vivo.
Although we do not know which phosphorylated phospholipid (or combination of phosphoinositides) is critical for in vivo activity of UNC-73B, there is a preference for the UNC-73B PH domain to bind monophosphorylated phosphoinositides in vitro. The lack of specificity that UNC-73B PH domain shows towards any single phosphatidylinositide is not unique and has been demonstrated with both the Dbs and intersectin PH domains. Most PH domains, including the UNC-73B PH domain, characteristically have a surface with a strong positive electrostatic potential that can mediate the interaction with the negatively charged phospholipids (7). The importance of this charged surface is supported by our data for UNC-73B. We also found no evidence for an allosteric enhancement of UNC-73B GEF activity by phosphorylated phosphatidylinositides in vitro. It is clear that PH domains in a number of GEFs exhibit different phospholipid specificities, with the advantage that this would allow for the unique temporal and spatial regulation of different GEFs and thus for different developmental processes to be regulated by the Rac GTPases (21). Indeed, activated Rac has been shown to interact with and activate type 1α PI(4)P 5 kinase (14), leading us to speculate that UNC-73B could be involved in formation of both PI(4,5)P2 and PI(3,4,5)P3, causing the subsequent activation of other GEFs during the signaling cascade involved in axon guidance, growth, and cell motility.
The UNC-73B PH domain may potentially be involved in intramolecular or intermolecular protein interactions necessary to target UNC-73B to the plasma membrane to allow it to carry out catalysis. Trio has been shown to interact with the actin binding protein filamin through its PH domain (6). We were able to reproduce this interaction of Trio with filamin in vitro (data not shown); however, attempts to demonstrate an interaction with UNC-73B and human filamin as well as a C. elegans form of filamin proved to be unsuccessful.
In conclusion, with the combined use of biochemical techniques and C. elegans as a model organism, we identified two functions of the UNC-73B PH domain that appear to be important in axon guidance in vivo. First, the PH domain has a direct effect on the ability of the DH domain to activate Rac GTPases. We infer that the UNC-73B PH domain is required to provide residues that facilitate binding to CeRAC/MIG-2 and/or maintain the structural integrity of the accompanying DH domain and thus to generate GTP-bound CeRAC/MIG-2 in the growth cones of specific neurons. Second, the positively charged face of the PH domain is important for binding to phosphoinositides and for axonal pathfinding, although phospholipid binding does not appear to allosterically affect DH domain GEF activity. The latter interaction may help to locate UNC-73B to specific sites in the extending axonal membrane where Rac activation is regulated by guidance cues. It will be of interest to determine how signals from guidance receptors regulate UNC-73B catalytic function in neurons.
Acknowledgments
We thank Li Shu for help in maintaining transgenic worm lines and Rob Steven for the UNC-73B minigene.
This work was supported by grants from the Canadian Institutes of Health Research (CIHR) and the National Cancer Institute of Canada. J.C. was supported by a grant (NS41397) from the U.S. National Institutes of Health. T.P. is a distinguished investigator of the CIHR.
REFERENCES
- 1.Aghazadeh, B., W. E. Lowry, X.-Y. Huang, and M. K. Rosen. 2000. Structural basis for relief of autoinhibition of the Dbl homology domain of proto-oncogene Vav by tyrosine phosphorylation. Cell 102:625-633. [DOI] [PubMed] [Google Scholar]
- 2.Aghazadeh, B., K. Zhu, T. J. Kubiseski, G. A. Liu, T. Pawson, Y. Zheng, and M. K. Rosen. 1998. Structure and mutagenesis of the Dbl homology domain. Nat. Struct. Biol. 12:1098-1107. [DOI] [PubMed] [Google Scholar]
- 3.Alam, M. R., R. C. Johnson, D. N. Darlington, T. A. Hand, R. E. Mains, and B. A. Eipper. 1997. Kalirin, a cytosolic protein with spectrin-like and GDP/GTP exchange factor-like domains that interacts with peptidylglycine α-amidating monooxygenase, an integral membrane peptide-processing enzyme. J. Biol. Chem. 272:12667-12675. [DOI] [PubMed] [Google Scholar]
- 4.Awasaki, T., M. Saitoh, M. Sone, E. Suzuki, R. Sakai, K. Ito, and C. Hama. 2000. The Drosophila Trio plays and essential role in patterning of axons by regulating their directional extension. Neuron 26:119-131. [DOI] [PubMed] [Google Scholar]
- 5.Bateman, J., H. Shu, and D. Van Vactor. 2000. The guanine nucleotide exchange factor Trio mediates axonal development in the Drosophila embryo. Neuron 26:93-106. [DOI] [PubMed] [Google Scholar]
- 6.Bellanger, J.-M., C. Astier, C. Sardet, Y. Ohta, T. P. Stossel, and A. Debant. 2000. The Rac1- and RhoG-specific GEF domain of Trio targets filamin to remodel cytoskeletal actin. Nat. Cell Biol. 2:888-892. [DOI] [PubMed] [Google Scholar]
- 7.Blomberg, N., E. Baraldi, M. Sattler, M. Saraste, and M. Nilges. 2000. Structure of a PH domain from the C. elegans muscle protein UNC-89 suggests a novel function. Structure 8:1079-1087. [DOI] [PubMed] [Google Scholar]
- 8.Chen, W., H. H. Lim, and L. Lim. 1993. The CDC42 homologue from Caenorhabditis elegans. Complementation of yeast mutation. J. Biol. Chem. 268:13280-13285. [PubMed] [Google Scholar]
- 9.Chen, W., H. H. Lim, and L. Lim. 1993. A new member of the ras superfamily, the rac1 homologue from Caenorhabditis elegans. Cloning and sequence analysis of cDNA, pattern of developmental expression, and biochemical characterization of the protein. J. Biol. Chem. 268:320-324. [PubMed] [Google Scholar]
- 10.Chen, W., and L. Lim. 1994. The Caenorhabditis elegans small GTP-binding protein RhoA is enriched in the nerve ring and sensory neurons during larval development. J. Biol. Chem. 269:32394-32404. [PubMed] [Google Scholar]
- 11.Colavita, A., S. Krishna, H. Zheng, R. W. Padgett, and J. G. Culotti. 1998. Pioneer axon guidance by UNC-129, a C. elegans TGF-β. Science 281:706-709. [DOI] [PubMed] [Google Scholar]
- 12.Crespo, P., K. E. Schuebel, A. A. Ostrom, J. S. Gutkind, and X. R. Bustelo. 1997. Phosphotyrosine-dependent activation of Rac1 GDP/GTP exchange by the Vav proto-oncogene product. Nature 385:169-172. [DOI] [PubMed] [Google Scholar]
- 13.Crompton, A. M., L. H. Foley, A. Wood, W. Roscoe, D. Stokoe, F. McCormick, M. Symons, and G. Bollag. 2000. Regulation of Tiam1 nucleotide exchange activity by pleckstrin domain binding ligands. J. Biol. Chem. 275:25751-25759. [DOI] [PubMed] [Google Scholar]
- 14.Cullen, P. J., G. E. Crozier, G Banting, and H. Mellor. 2001. Modular phosphoinositide-binding domains—their role in signalling and membrane trafficking. Curr. Biol. 11:R882-R893. [DOI] [PubMed] [Google Scholar]
- 15.Culotti, J. G. 1994. Axon guidance mechanisms in Caenorhabditis elegans. Curr. Opin. Genet. Dev. 4:587-595. [DOI] [PubMed] [Google Scholar]
- 16.Das, B., X. Shu, G.-J. Day, J. Han, U. M. Krishna, J. R. Falck, and D. Broek. 2000. Control of intramolecular interactions between the pleckstrin homology and Dbl homology domains of Vav and Sos1 regulates Rac binding. J. Biol. Chem. 275:15074-15081. [DOI] [PubMed] [Google Scholar]
- 17.Debant, A., C. Serra-Pages, K. Seipel, S. O'Brien, M. Tang, S. H. Park, and M. Streuli. 1996. The multidomain protein Trio binds the LAR transmembrane tyrosine phosphatase, contains a protein kinase domain, and has separate rac-specific and rho-specific guanine nucleotide exchange factor domains. Proc. Natl. Acad. Sci. USA 93:5466-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dickson, B. J. 2001. Rho GTPases in growth cone guidance. Curr. Opin. Neurobiol. 11:103-110. [DOI] [PubMed] [Google Scholar]
- 19.Fleming, I. N., C. M. Elliot, and J. H. Exton. 1998. Phospholipase C-γ, protein kinase C and Ca2+/calmodulin-dependent protein kinase II are involved in platelet derived growth factor-induced phosphorylation of Tiam1. FEBS Lett. 429:229-233. [DOI] [PubMed] [Google Scholar]
- 20.Fleming, I. N., A. Gray, and C. P. Downes. 2000. Regulation of the Rac1-specific exchange factor Tiam1 involves both phosphoinositide 3-kinase-dependent and -independent components. Biochem. J. 351:173-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gulli, M.-P., and M. Peter. 2001. Temporal and spatial regulation of Rho-type guanine-nucleotide exchange factors: the yeast perspective. Genes Dev. 15:365-379. [DOI] [PubMed] [Google Scholar]
- 22.Hakeda-Suzuki, S., J. Ng, J. Tzu, G. Dietzl, Y. Sun, M. Harms, T. Nardine, L. Luo, and B. J. Dickson. 2002. Rac function and regulation during Drosophila development. Nature 416:438-442. [DOI] [PubMed] [Google Scholar]
- 23.Hall, A. 1998. Rho GTPases and the actin cytoskeleton. Science 279:509-514. [DOI] [PubMed] [Google Scholar]
- 24.Han, J., K. Luby-Phelps, B. Das, X. Shu, Y. Xia, R. D. Mosteller, M. Krishna, J. R. Falck, M. A. White, and D. Broek. 1998. Role of substrates and products of PI-3 kinase in regulating activation of Rac-related guanosine triphosphatases by Vav. Science 279:558-560. [DOI] [PubMed] [Google Scholar]
- 25.Hart, M. J., A. Eva, D. Zangrilli, S. Aaronson, T. Evans, R. A. Cerione, and Y. Zheng. 1994. Cellular transformation and guanine nucleotide exchange activity are catalyzed by a common domain on the dbl oncogene product. J. Biol. Chem. 269:62-65. [PubMed] [Google Scholar]
- 26.Hoffman, G. R., and R. A. Cerione. 2002. Signaling to the Rho GTPases: networking with the DH domain. FEBS Lett. 513:85-91. [DOI] [PubMed] [Google Scholar]
- 27.Kavran, J. M., D. E. Klein, A. Lee, M. Falasca, S. J. Isakoff, E. Y. Skolnik, and M. A. Lemmon. 1998. Specificity and promiscuity in phosphoinositide binding by pleckstrin homology domains. J. Biol. Chem. 273:30497-30508. [DOI] [PubMed] [Google Scholar]
- 28.Kubiseski, T. J., Y. M. Chook, W. E. Parris, M. Rozakis-Adcock, and T. Pawson, T. 1997. High affinity binding of the pleckstrin homology domain of mSos1 to phosphatidylinositol (4,5)-bisphosphate. J. Biol. Chem. 272:1799-1804. [DOI] [PubMed] [Google Scholar]
- 29.Lemmon, M. A., K. M. Ferguson, and C. S. Abrams. 2002. Pleckstrin homology domains and the cytoskeleton. FEBS Lett. 513:71-76. [DOI] [PubMed] [Google Scholar]
- 30.Liebl, E. C., D. J. Forstohoefel, L. S. Franco, S. H. Sample, J. E. Hess, J. A. Cowger, M. P. Chandler, A. M. Shupert, and M. A. Seeger. 2000. Dosage-sensitive, reciprocal genetic interactions between the Abl tyrosine kinase and the putative GEF trio reveal trio's role in axon pathfinding. Neuron 26:107-118. [DOI] [PubMed] [Google Scholar]
- 31.Lim, Y.-S., S. Mallapur, G. Kao, X.-C. Ren, and W. G. Wadsworth. 1999. Netrin UNC-6 and the regulation of branching and extension of motorneuron axons from the ventral nerve cord of Caenorhabditis elegans. J. Neurosci. 19:7048-7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, X., H. Wang, M. Eberstadt, A. Schnuchel, E. T. Olejniczak, R. P. Meadows, J. M. Schkeryantz, D. A. Janowick, J. E. Harlan, E. A. S. Harris, D. E. Staunton, and S. W. Fesik. 1998. NMR structure and mutagenesis of the N-terminal Dbl homology domain of the nucleotide exchange factor Trio. Cell 95:269-277. [DOI] [PubMed] [Google Scholar]
- 33.Lundquist, E. A., P. W. Reddien, E. Hartwieg, H. R. Horvitz, and C. I. Bargmann. 2001. Three C. elegans Rac proteins and several alternative Rac regulators control axon guidance, cell migration and apoptotic cell phagocytosis. Development 128:4475-4488. [DOI] [PubMed] [Google Scholar]
- 34.Ma, A. D., A. Metjian, S. Bagrodia, S. Taylor, and C. S. Abrams. 1998. Cytoskeletal reorganization by G protein-coupled receptors is dependent on phosphoinositide 3-kinase, a Rac guanosine exchange factor, and Rac. Mol. Cell. Biol. 18:4744-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mello, C. C., J. M. Kramer, D. Stinchcomb, and V. Ambros. 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10:3959-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newsome, T. P., S. Schmidt, G. Dietzl, K. Keleman, B. Asling, A. Debant, and B. J. Dickson. 2000. Trio combines with Dock to regulate Pak activity during photoreceptor axon pathfinding in Drosophila. Cell 101:283-294. [DOI] [PubMed] [Google Scholar]
- 37.Nimunal, A. S., B. A. Yatsula, and D. Bar-Sagi. 1998. Coupling of Ras and Rac guanosine triphosphatase through the Ras exchanger Sos. Science 279:560-563. [DOI] [PubMed] [Google Scholar]
- 38.Ng, J., T. Nardine, M. Harms, J. Tzu, A. Goldstein, Y. Sun, G. Dietzl, B. J. Dickson, and L. Luo. 2002. Rac GTPases control axon growth, guidance and branching. Nature 416:442-447. [DOI] [PubMed] [Google Scholar]
- 39.Rossman, K. L., D. K. Worthylake, J. T. Snyder, D. P. Siderovski, S. L. Campbell, and J. Sondek. 2002. A crystallographic view of interactions between Dbs and Cdc42: PH domain-assisted guanine nucleotide exchange. EMBO J. 21:1315-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russo, C., Y. Gao, P. Mancin, C. Vanni, M. Porotto, M. Falasca, M. R. Torrisi, Y. Zheng, and A. Eva. 2001. Modulation of oncogenic DBL activity by phosphoinositol phosphate binding to pleckstrin homology domain. J. Biol. Chem. 276:19524-19531. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt, A., and A. Hall. 2002. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 16:1587-1609. [DOI] [PubMed] [Google Scholar]
- 42.Shinohara, M., Y. Terada, A. Iwamatsu, A. Shinohara, N. Mochizuki, M. Higuchi, Y. Gotho, S. Ihara, S. Nagata, H. Itoh, Y. Fukui, and R. Jessberger. 2002. SWAP-70 is a guanine-nucleotide-exchange factor that mediates signalling of membrane ruffling. Nature 416:759-763. [DOI] [PubMed] [Google Scholar]
- 43.Snyder, J. T., K. L. Rossman, M. A. Baumeister, W. M Pruitt, D. P. Siderovski, C. J. Der, M. A. Lemmon, and J. Sondek. 2001. Quantitative analysis of the effect of phosphoinositide interactions on the function of Dbl family proteins. J. Biol. Chem. 276:45868-45875. [DOI] [PubMed] [Google Scholar]
- 44.Steven, R., T. J. Kubiseski, H. Zheng, S. Kulkarni, J. Mancillas, A. R. Morales, C. W. V. Hogue, T. Pawson, and J. Culotti. 1998. UNC-73 activates the Rac GTPase and is required for cell and growth cone migrations in C. elegans. Cell 92:785-795. [DOI] [PubMed] [Google Scholar]
- 45.Welch, H. C. E., W. J. Coadwell, C. D. Ellson, G. J. Ferguson, S. R. Andrews, H. Erdjument-Bromage, P. Tempst, P. T. Hawkins, and L. R. Stephens. 2002. P-Rex1, a PtdIns(3, 4, 5)P3- and Gβγ-regulated guanine-nucleotide exchange factor for Rac. Cell 108:809-821. [DOI] [PubMed] [Google Scholar]
- 46.Worthylake, D. K., K. L. Rossman, and J. Sondek. 2000. Crystal structure of Rac1 in complex with the guanine nucleotide exchange region of Tiam1. Nature 408:682-688. [DOI] [PubMed] [Google Scholar]
- 47.Zipkin, I. D., R. M. Kindt, and C. J. Kenyon. 1997. Role of a new Rho family member in cell migration and axon guidance in C. elegans. Cell 90:883-894. [DOI] [PubMed] [Google Scholar]