Abstract
Recombinant adenoviruses provide a versatile system for gene expression studies and therapeutic applications. We report herein a strategy that simplifies the generation and production of such viruses. A recombinant adenoviral plasmid is generated with a minimum of enzymatic manipulations, using homologous recombination in bacteria rather than in eukaryotic cells. After transfections of such plasmids into a mammalian packaging cell line, viral production is conveniently followed with the aid of green fluorescent protein, encoded by a gene incorporated into the viral backbone. Homogeneous viruses can be obtained from this procedure without plaque purification. This system should expedite the process of generating and testing recombinant adenoviruses for a variety of purposes.
Recombinant adenoviruses currently are used for a variety of purposes, including gene transfer in vitro, vaccination in vivo, and gene therapy (1–4). Several features of adenovirus biology have made such viruses the vectors of choice for certain of these applications. For example, adenoviruses transfer genes to a broad spectrum of cell types, and gene transfer is not dependent on active cell division. Additionally, high titers of viruses and high levels of transgene expression generally can be obtained.
Decades of study of adenovirus biology have resulted in a detailed picture of the viral life cycle and the functions of the majority of viral proteins (5, 6). The genome of the most commonly used human adenovirus (serotype 5) consists of a linear, 36-kb, double-stranded DNA molecule. Both strands are transcribed and nearly all transcripts are heavily spliced. Viral transcription units are conventionally referred to as early (E1, E2, E3, and E4) and late, depending on their temporal expression relative to the onset of viral DNA replication (6). The high density and complexity of the viral transcription units poses problems for recombinant manipulation, which therefore is usually restricted to specific regions, particularly E1, E2A, E3, and E4. In most recombinant vectors, transgenes are introduced in place of E1 or E3, the former supplied exogenously. The E1 deletion renders the viruses defective for replication and incapable of producing infectious viral particles in target cells; the E3 region encodes proteins involved in evading host immunity and is dispensable for viral production per se.
Two approaches traditionally have been used to generate recombinant adenoviruses. The first involves direct ligation of DNA fragments of the adenoviral genome to restriction endonuclease fragments containing a transgene (7, 8). The low efficiency of large fragment ligations and the scarcity of unique restriction sites have made this approach technically challenging. The second and more widely used method involves homologous recombination in mammalian cells capable of complementing defective adenoviruses (“packaging lines”) (9, 10). Homologous recombination results in a defective adenovirus that can replicate in the packaging line (e.g., 293 or 911 cells) supplying the missing gene products (e.g., E1) (11). The desired recombinants are identified by screening individual plaques generated in a lawn of packaging cells (12). Though this approach has proven extremely useful, the low efficiency of homologous recombination, the need for repeated rounds of plaque purification, and the long times required for completion of the viral production process have hampered more widespread use of adenoviral vector technology.
The problems noted above have stimulated novel methods for generating adenoviral vectors. We report herein a strategy that builds on several technological and conceptual advances made in the last few years, including alternative systems for producing viral recombinants (13–16). In our system, the backbone vector, containing most of the adenoviral genome, is used in supercoiled form, obviating the need to enzymatically manipulate it. Second, the recombination is performed in Escherichia coli rather than in mammalian cells. Third, there are no ligation steps involved in generating the adenoviral recombinants, as the process takes advantage of the highly efficient homologous recombination machinery present in bacteria. Fourth, the vectors allow inclusion of up to 10 kb of transgene sequences and allow multiple transgenes to be produced from the same virus. Fifth, some of the new vectors contain a green fluorescent protein (GFP) gene incorporated into the adenoviral backbone, allowing direct observation of the efficiency of transfection and infection, processes that have been difficult to follow with adenoviruses in the past. These modifications resulted in highly efficient viral production systems that often can obviate the need for plaque purification and significantly decrease the time required to generate usable viruses.
MATERIALS AND METHODS
Cell Culture, Medium, and Reagents.
293 cells (11) were purchased from Microbix Biosystems (Toronto, Canada) or from the American Type Culture Collection, and 911 cells (17) were kindly provided by Alex J. Van der Eb of the University of Leiden. These lines were maintained in growth medium [DMEM, Life Technologies, Gaithersburg, MD, supplemented with 10% fetal bovine serum (HyClone), 100 units/ml of penicillin, and 100 μg/ml of streptomycin] at 37°C in 5% CO2.
Preparation of Competent Cells and Plasmid DNAs.
To prepare electrocompetent BJ5183 bacteria (18), the cells were grown to an OD550 of 0.8, then collected and washed twice with ice-cold 10% glycerol. Twenty-microliter aliquots of the electrocompetent BJ5183 cells were kept at −80°C. Electrocompetent DH10B cells were purchased from Life Technologies. To verify homologous recombination in bacteria, miniprep plasmid DNA was prepared by a standard alkaline lysis procedure. All other plasmids used in this study were prepared by CsCl-banding. Yields were 200–600 μg per 100 ml of Terrific Broth culture (Life Technologies) for plasmids larger than 30 kb (pAdEasy derivatives) and 400–1,000 μg for plasmids smaller than 15 kb (shuttle plasmid derivatives).
Establishment of an Adenoviral E4-Expressing Cell Line.
A plasmid that constitutively expresses tet repressor in the same transcription unit as a geneticin-resistance marker was transfected into 911 cells. After growth in geneticin (0.4 mg/ml, Life Technologies), a clone stably expressing the tet repressor, 911tet, was chosen for further manipulation. A second vector that expressed adenoviral E4 under the control of tet responsive promoter was constructed by cloning a fragment containing adenoviral nucleotides 35,468–32,828 into the pBI vector (CLONTECH), resulting in pBI-E4. The pBI-E4 plasmid was cotransfected with linearized pCEP4 (Invitrogen) into 911tet cells. Stable clones were generated through selection in 0.4 mg/ml of geneticin, 0.1 mg/ml of hygromycin B (Calbiochem), and 100 ng/ml of doxycyclin (Sigma). A single clone, called 911-E4, was chosen for viral production based on its tight regulation of E4 protein expression. Expression of adenoviral E4 after removal of doxycyclin was confirmed by immunohistochemical analysis using a mAb against E4ORF6, kindly provided by P. Hearing (State University of New York, Stony Brook) (19).
Construction of Vectors for Homologous Recombination in Bacteria.
The adenoviral plasmids (pAdEasy-1 and pAdEasy-2) and the shuttle vectors (pShuttle, pShuttle-CMV, pAdTrack, and pAdTrack-CMV) were constructed through multiple rounds of subcloning of PCR products or of restriction endonuclease fragments. All PCR-derived fragments were sequenced to confirm their predicted composition. Detailed information about the constructions is available from the authors upon request.
Adenoviral backbone vectors.
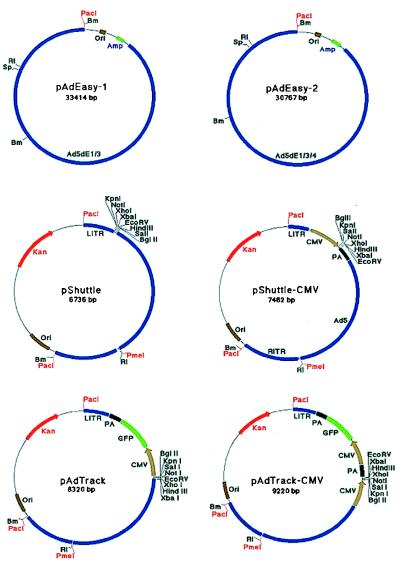
The pAdEasy-1 adenoviral plasmid contains all Ad5 sequences except nucleotides 1–3,533 (encompassing the E1 genes) and nucleotides 28,130–30,820 (encompassing E3).
The pAdEasy-2 vector is identical to pAdEasy-1 except that it contains an additional deletion of Ad5 nucleotides 32,816–35,462 (encompassing E4).
Shuttle vectors.
Vector pShuttle is used for expression of transgenes when no GFP tracer is desired. It contains a polylinker for insertion of exogenous transgenes. This site is surrounded by adenoviral sequences (“arms”) that allow homologous recombination with pAdEasy-1. The left arm contains Ad5 nucleotides 34,931–35,935, which mediate homologous recombination with pAdEasy vectors in E. coli, plus inverted terminal repeat (ITR) and packaging signal sequences (nucleotides 1–480 of Ad5) required for viral production in mammalian cells. The right arm contains Ad5 nucleotides 3,534–5,790, which mediate homologous recombination with pAdEasy vectors. Artificially created PacI sites surround both arms. The pShuttle plasmid also contains a kanamycin resistance gene from pZero 2.1 (Invitrogen) and the origin of replication from pBR322 (Life Technologies). We have found, as have others (14, 15), that the relatively low copy number of plasmids generated with this origin is essential for the stability of large constructs in E. coli.
The pShuttle-CMV vector is identical to pShuttle except for the addition of a cytomegalovirus (CMV) promoter and polyadenylation site (both from pEGFP-C1, CLONTECH). A polylinker is present between the CMV promoter and polyadenylation site.
The pAdTrack vector is used for production of GFP-trackable viruses containing transgenes under the control of a chosen promoter. It was constructed by subcloning the gene encoding enhanced GFP from pEGFP-C1 into pShuttle.
The pAdTrack-CMV vector is identical to pAdTrack except for the addition of a CMV promoter and polyadenylation site (as in pShuttle-CMV).
Vectors encoding both β-galactosidase (β-gal) and GFP.
To test various aspects of these systems, two vectors (pGFP+GAL-1 and -2) containing β-gal and GFP genes were constructed. Each contained the β-gal gene from pUT651 (Cayla, Toulouse, France). The only difference between the two vectors was the presence in pGFP+GAL-2 of a “stuffer” fragment from human genomic DNA. pGFP+GAL-2 thereby contained the maximum amount of foreign sequences (≈10 kb) possible to be included in the adenovirus systems described here. Both pGFP+GAL-1 and pGFP+GAL-2 contained two independent CMV-driven transcription units (one for GFP and one for β-gal).
Generation of Recombinant Adenoviral Plasmids by Homologous Recombination in E. coli.
High competence of bacteria cells is desired to achieve efficient recombination. Typically, 0.5–1.0 μg of a shuttle vector plasmid (≈one-fifth of a miniprep) was linearized with PmeI, purified by phenol/chloroform extraction and ethanol precipitation, and mixed with 0.1 μg of supercoiled pAdEasy-1 or pAdEasy-2 in a total volume of 6.0 μl. Twenty microliters of electrocompetent E. coli BJ5183 cells were added, and electroporation was performed in 2.0 mm cuvettes at 2,500 V, 200 ohms, and 25 μF in a Bio-Rad Gene Pulser electroporator. The cells were immediately placed in 500 μl of l-Broth (Life Technologies) and grown at 37°C for 20 min. One hundred twenty-five microliters of the cell suspension then was inoculated onto each of four 10-cm Petri dishes containing l-agar plus 50 μg/ml of kanamycin. After 16–20 hr growth at 37°C, 10–25 colonies per dish generally were obtained. The smaller colonies (which usually represented the recombinants) were picked and grown in 2 ml of l-Broth containing 50 μg/ml of kanamycin. Clones were first screened by analyzing their supercoiled sizes on agarose gels, comparing them to pAdEasy-1 or pAdEasy-2 controls. Those clones that had inserts were further tested by restriction endonuclease digestions, generally PacI, SpeI, and BamHI. (Recombinations sometimes occurred between the plasmid Ori sequences shared between the shuttle and pAdEasy vectors; such recombinants were as useful as those generated by homologous recombination of the “left arm” sequences, but resulted in slightly different restriction patterns; see map in Fig. 1.) Once confirmed, supercoiled plasmid DNA was transformed into DH10B cells for large-scale amplification by electroporation. In such cases, 1.0 μl of plasmid DNA (≈100 ng) in 15 μl of water was mixed with 5.0 μl of electrocompetent DH10B cells in a total volume of 20.0 μl, and electroporation was performed as described above.
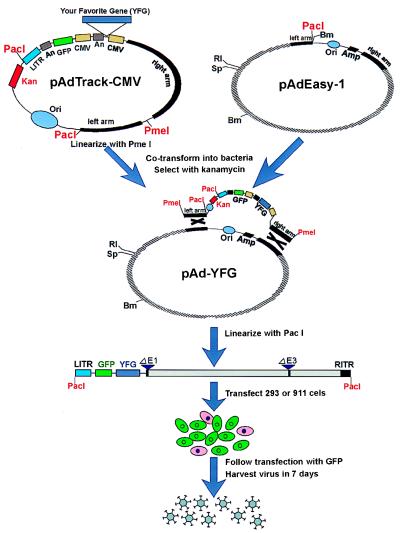
Figure 1.
Schematic outline of the AdEasy system. The gene of interest is first cloned into a shuttle vector, e.g., pAdTrack-CMV. The resultant plasmid is linearized by digesting with restriction endonuclease PmeI, and subsequently cotransformed into E. coli BJ5183 cells with an adenoviral backbone plasmid, e.g., pAdEasy-1. Recombinants are selected for kanamycin resistance, and recombination was confirmed by multiple restriction endonuclease analyses. Finally, the linearized recombinant plasmid is transfected into adenovirus packaging cell lines, e.g., 911 or 293 cells. Recombinant adenoviruses typically are generated within 7–10 days. The “left arm” and “right arm” represent the regions mediating homologous recombination between the shuttle vector and the adenoviral backbone vector (see Materials and Methods for sequence composition). An, polyadenylation site; Bm, BamHI; RI, EcoRI; LITR, left-hand ITR and packaging signal; RITR, right-hand ITR; Sp, SpeI.
Production of Adenoviruses in Mammalian Cells.
Approximately 1.5 × 106 cells (911, 293, or 911E4) were plated in 25 cm2 flasks 24 hr before transfection, by which time they reached 50–70% confluency. Cells were washed once with 3 ml of OptiMEM (Life Technologies), then 2.5 ml of OptiMEM was added to each flask and the flasks returned to the CO2 incubator for 15–30 min before transfection. Four micrograms of recombinant adenoviral vector DNA, digested with PacI and ethanol-precipitated, were used for transfection of each 25 cm2 flask. A transfection mix was prepared by adding 4 μg of linearized plasmid DNA and 20 μl of Lipofectamine (Life Technologies) to 500 μl of OptiMEM (Life Technologies) according to the manufacturer’s instructions. After incubation at room temperature for 15–30 min, the transfection mix was added to the cells. After 4–6 hr at 37°C, the media containing the transfection mix was removed, and 6 ml of growth medium was added. For transfections of 911E4 cells, doxycyclin was removed from the growth media ≈24 hr after transfection. Transfected cells were monitored for GFP expression and collected 7–10 days after transfection by scraping cells off flasks and pelleting them along with any floating cells in the culture. All but 3 ml of the supernatant was removed. After three cycles of freezing in a methanol/dry ice bath and rapid thawing at 37°, 1 ml of viral lysate was used to infect 3–5 × 106 cells in a 25 cm2 flask. The efficiency of such infections could be conveniently followed with GFP. Three to four days later, viruses were harvested as described above. At this point, viral titers were often high enough to use for gene transfer experiments in cultured cells. To generate higher titer viral stocks, packaging cells were infected at a multiplicity of infection (MOI) of 0.1 to 1 and grown for 3–4 days, at which time viruses were harvested as described above. This process was repeated 1–3 times, with a final round using a total of 5 × 108 packaging cells in fifteen 75 cm2 flasks and an MOI of 1–5. After 3–5 days, 50% lysis was observed, and the resultant viruses were purified by CsCl banding; final yields were generally 1011 to 1012 plaque-forming units. Procedures for CsCl banding and viral plaquing are described in ref. 20.
RESULTS AND DISCUSSION
Generation of Adenoviral Recombinants: General Considerations.
The overall strategy developed here is diagrammed in Fig. 1 and involves three steps. First, the gene of interest is cloned into a shuttle vector (e.g., pAdTrack-CMV, Fig. 2). Second, the resultant construct is cleaved with a restriction endonuclease to linearize it and transformed together with a supercoiled adenoviral vector (e.g., pAdEasy-1) into E. coli strain BJ5183. Recombinants are selected with kanamycin and screened by restriction endonuclease digestion. Third, the recombinant adenoviral construct is cleaved with PacI to expose its inverted terminal repeats and transfected into a packaging cell line (e.g., 293 or 911 cells) (11, 17). In the past, validation of successful virus production at early stages of the process has been one of the most technically demanding aspects of adenoviral vector production. In some of the systems described here, the process of viral production can be directly and conveniently followed in the packaging cells by visualization of the GFP reporter that is incorporated into the viral backbone. After 7–10 days, viruses are harvested and either used directly for experimentation or amplified by infecting packaging cells.
Figure 2.
Shuttle vectors and adenoviral plasmids. Abbreviations are defined in the legend to Fig. 1. See text for details.
Important points about this approach include the following: (i) Several different shuttle vectors were constructed. Some, like pAdTrack and pAdTrack-CMV, allow convenient tracing of all steps in viral production through an incorporated GFP reporter. Others, like pShuttle, are used when particularly large transgenes must be expressed. Table 1 lists optimum combinations of shuttle and backbone vectors for various purposes. (ii) The homologous recombination step is mediated by a restriction endonuclease-cleaved shuttle vector (like pAdTrack-CMV) and an intact supercoiled adenoviral vector (like pAdEasy-1). The ability to use intact adenoviral plasmids, uncleaved by restriction endonucleases, proved critical for efficiently generating desired recombinants. An additional advantage of using supercoiled adenoviral vectors is that preparation of a single, laboratory-scale batch of the adenoviral vector DNA will yield sufficient material for hundreds of different recombinants. (iii) The selection of recombinants is afforded by kanamycin resistance provided by the shuttle vector. Because the restriction-cleaved shuttle vector yields only a low background of kanamycin-resistant colonies, the homologous recombination system had a high signal-to-noise ratio. (iv) The E. coli strain BJ5183 is not recA but is deficient in other enzymes that mediate recombination in bacteria. It was chosen, from among several strains mutated in recA, recBCD, recJ, or recF (21, 22), because of its higher efficiency of transformation and stable propagation of plasmid DNA in pilot experiments. Once recombination is achieved and verified, the adenoviral recombinant DNA can be simply transferred to a recA, endA strain (such as DH10B) for greater yields of DNA if desired. (Because of its recA status, DH10B cannot be used to generate adenoviral recombinants by homologous recombination.) (v) For viruses containing two independent transcription units driven by the same promoter, we found it important to place them in head-to-tail orientation, rather than head-to-head, to avoid undesired recombination events in bacteria. (vi) The packaging cell lines (293, 911, or 911E4) are each highly transfectable by lipid-DNA complexes. The 293 and 911 cells constitutively express the E1 gene products required for propagation of all recombinant adenoviruses, whereas the 911E4 cells express the E1 and E4 gene products required for pAdEasy-2-derived constructs.
Table 1.
Selection of vector systems
| Shuttle plasmid | Adenoviral backbone | Packaging cells | Maximum insert size | GFP tracer | Use |
|---|---|---|---|---|---|
| pAdTrack-CMV | pAdEasy-1 | 293 or 911 | 5.0 kb | Yes | Standard for expression of transgene under CMV promoter |
| pAdTrack-CMV | pAdEasy-2 | 911E4 | 7.7 kb | Yes | Used only for expression of large genes under CMV promoter |
| pAdTrack | pAdEasy-1 | 293 or 911 | 5.9 kb | Yes | Expression of transgene(s) under a chosen (non-CMV) promoter |
| pAdTrack | pAdEasy-2 | 911E4 | 8.6 kb | Yes | Used only for expression of a large gene(s) under a chosen (non-CMV) promoter |
| pShuttle-CMV | pAdEasy-1 | 293 or 911 | 6.6 kb | No | Standard for expression of transgene under CMV promoter |
| pShuttle-CMV | pAdEasy-2 | 911E4 | 9.3 kb | No | Used only for expression of large genes under CMV promoter |
| pShuttle | pAdEasy-1 | 293 or 911 | 7.5 kb | No | Expression of transgene(s) under a chosen (non-CMV) promoter |
| pShuttle | pAdEasy-2 | 911E4 | 10.2 kb | No | Used only for expression of a large gene(s) under a chosen (non-CMV) promoter |
Generation of Adenoviral Recombinants: Practical Considerations.
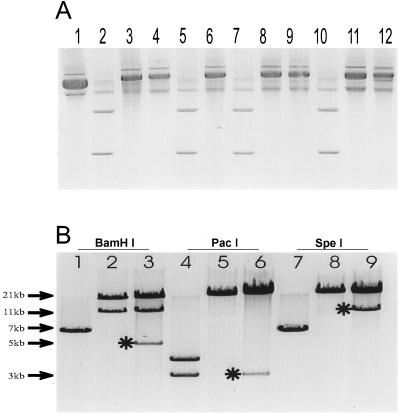
The results obtained while generating a virus encoding β-gal provides a representative example of the yields and practical considerations involved with the approach described herein. As described in Materials and Methods, a β-gal cDNA was placed in the polylinker of pAdTrack-CMV to generate the shuttle vector pGFP+GAL. To make pAdEasy-GFP+GAL, 1 μg of linearized pGFP+GAL was cotransformed with 0.1 μg of supercoiled circular pAdEasy-1 into E. coli BJ5183 cells (see vector diagrams in Fig. 2). The transformation yielded ≈100 kanamycin-resistant clones, of which ≈two-thirds contained recombinants based on the sizes of undigested miniprep plasmid DNA (Fig. 3A). Candidate clones were digested with several restriction endonucleases to verify proper recombination. As shown in Fig. 3B, the expected restriction fragments were generated in each case. For example, with BamHI, a 5.1-kb fragment containing the GFP gene was produced from pAdEasy-GFP+GAL (lane 3) in addition to the 11.7- and 21.7-kb fragments generated from pAdEasy-1 sequences (lane 2). When digested with PacI, a 3.0-kb fragment was produced (Fig. 3B, lane 6).
Figure 3.
Generation of stable recombinants in bacterial cells. (A) DNA from recombinant pAdEasy-GFP+GAL constructs derived from homologous recombination of pAdTrack-CMV-βgal and pAdEasy-1in BJ5183 cells was purified from minipreps. The DNA was analyzed in supercoiled form by electrophoresis through an 0.8% agarose gel and ethidium bromide staining. Lane 1, pAdEasy-1 control; lane 2, pAdTrack-GFP+GAL control; lanes 3–12, different pAdEasy-GFP+GAL clones. Based on the migration rates, the clones in lanes 3, 4, 6, 8, 9, 11, and 12 were potential valid recombinants. (B) Representative digestions with BamHI (lanes 1–3), PacI (lanes 4–6), and SpeI (lanes 7–9). Plasmids pAdTrack-CMV (lanes 1, 4, and 7), pAdEasy-1 (lanes 2, 5, and 8) and a pAdEasy-GFP+GAL recombinant (lanes 3, 6, and 9) are shown. ∗ indicate the diagnostic fragments obtained with each enzyme.
Plasmids could be produced directly from E. coli BJ5183 cells, but the yields were relatively low (<0.5 μg from 2 ml of culture). Therefore, miniprep DNA from E. coli BJ5183 cells was used to transform DH10B cells, a recA strain in which high-quality and high yields of plasmid DNA can be obtained more easily. Yields of supercoiled pAdEasy-derived vectors averaged 2–5 μg per ml from DH10B cells.
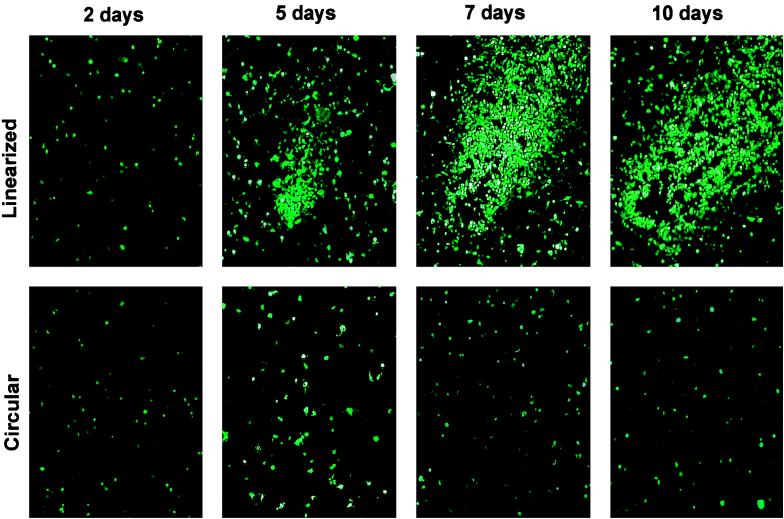
To produce viruses, 4 μg of pAdEasy-GFP+GAL was digested with PacI to liberate linear adenoviral genomes, then transfected into 293 cells. It was critical to linearize the vectors at the PacI sites, as transfection of circular plasmids yielded no viruses, consistent with previous results (14, 23, 24). To assess how soon the packaged viral particles could be observed, transfected cells were monitored by GFP expression. As shown in Fig. 4, GFP expression was visible 24 hr after transfection in 20–30% of the cells, representing the fraction of the population that was transfected. In cells transfected with nonlinearized pAdEasy-GFP+GAL, this expression slowly faded over 1 week. In cells transfected with linearized pAdEasy-GFP+GAL, however, this expression never faded and comet-like foci, visualized with GFP fluorescence but invisible by phase contrast microscopy, began to appear at 4–5 days after transfection (Fig. 4). Cells in the center of foci often were lysed a week after transfection, though the foci were still very difficult to see without the aid of GFP fluorescence. Interestingly, only 10–50 comet-like plaques were observed per 25 cm2 flask, whereas >105 cells expressed GFP after transfection. This finding was surprising in view of the fact that every 293 cell theoretically should have the capacity to produce virus from the transfected vector. Evidently, degradation of the exogenous DNA or other factors that limit the efficiency of viral production drastically decrease the number of cells that produce virus. These results may explain the difficulties of achieving efficient viral production after homologous recombination (rather than direct transfection) in mammalian cells.
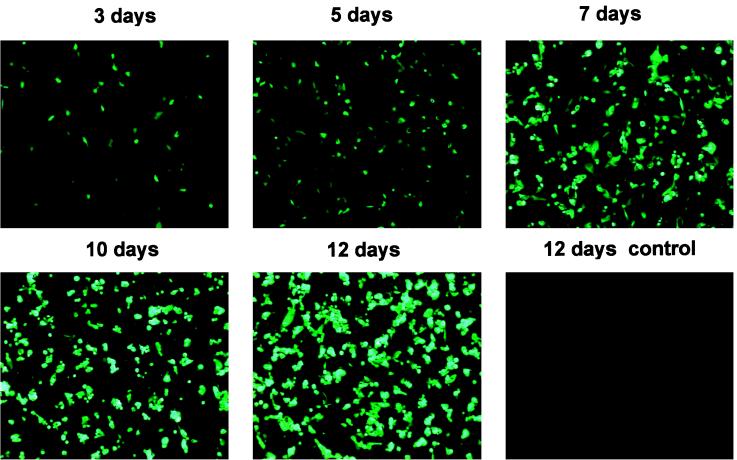
Figure 4.
Adenovirus-producing foci after transfection of 293 cells. PacI-digested pAdEasy-GFP-GAL was transfected into 293 cells and GFP expression was visualized by fluorescence microscopy at the indicated times thereafter. Comet-like adenovirus-producing foci became apparent at 4–5 days. No such foci were observed in the cells transfected with circular (i.e., not PacI-digested) pAdEasy-GFP-Gal.
As another way to assess viral production after transfection of 293 cells, cells were collected and lysed at various times after transfection and the lysates assessed for viral production through transfer of GFP or β-gal expression. In each case, 2% of the viruses harvested from a single transfection were used to infect ≈105 recipient 293 cells. As shown in Fig. 5, significant amounts of virus were present as early as 3 days after transfection, concordant with the appearance of first observable viral foci (Fig. 4). Viral titers increased substantially over the next week (Fig. 5). Importantly, β-gal expression perfectly paralleled GFP expression, as assessed in three ways. First, the titers of virus, assessed by X-Gal (5-bromo-4-chloro-3-indolyl β-d-galactoside) staining of infected cells, was identical to that determined from GFP expression of the same cultures before X-Gal staining. Second, when GFP expressing cells were marked before staining with X-Gal, every cell that expressed GFP also was found to express β-gal and vice versa. And third, standard plaque assays demonstrated that virtually all (>95%) plaques expressed both GFP and β-gal (data not shown). This homogeneity among the plaques was important for another reason: it obviated the need, in general, to plaque-purify viruses. Such plaque purification represents one of the most time-consuming steps in classical adenovirus vector production.
Figure 5.
Adenoviral titer monitored by GFP expression. Linearized pAdEasy-GFP+GAL was transfected into 293 cells as described in Fig. 4, and cells were harvested at the indicated times after transfection. Two percent of a freeze/thaw lysate of these cells was used to infect 293 cells, and fluorescence microscopy of the infected cells was performed 24 hr later. No viruses were generated in 12 days after the transfection of circular (i.e., not cleaved with PacI) pAdEasy-GFP+GAL (labeled “control”).
The titers of the homogeneous viruses produced 7–12 days after transfection of 911 or 293 cells ranged from 106 to 108 expression-forming units (efu)/ml on 293 cells. In the experiment shown in Fig. 5, the efu was 107/ml. The titer generally was proportional to the efficiency of transfection of the packaging line. Titers determined by plaque assays (expressed in standard plaque-forming units) were equivalent. These viruses could be used to achieve gene expression in a variety of cell lines of human, mouse, and hamster origin. The levels of efu on these additional lines were similar to that achieved with adenoviruses made by classical methods, and differences in expression likely reflected differences in adenovirus receptors and processing among the various lines. In the human colorectal cancer cell line HCT116, efu was ≈20- to 200-fold lower than that achieved in 293 or 911 cells.
We carried out similar experiments with a virus containing GFP and β-gal expression units plus a “stuffer.” The total foreign sequences contained in this virus were 10.1 kb, necessitating use of the pAd-Easy-2 adenoviral vector and a packaging line expressing adenoviral E4 plus E1 genes (see Materials and Methods and Table 1). In general, viral production using the pAdEasy-2-based system was somewhat slower (10–14 days to produce viral titers equivalent to those produced in 7–10 days in 911 or 293 cells) and the final viral titers about 10-fold lower than with pAdEasy-1-based systems. Therefore, pAdEasy-2 and 911-E4 cells were used only to produce viruses containing transgenes too large to produce with pAdEasy-1 (Table 1). In general, we have found that 911 cells are the preferred producers for pAdEasy-1-derived viruses, though 293-derived cells also produced acceptable results (17).
We have generated more than 20 different adenoviruses, with inserts ranging up to 10 kb, with the systems described here (25). Though several systems for generating recombinant viruses through Cre-mediated or homologous recombination in yeast or bacteria have been described in the literature (13–15, 26), the system described herein has several advantages in terms of ease and speed. The fact that the adenoviral components of the system can be used in supercoiled form poses advantages in terms of the reproducibility and stability of the derived recombinants. The ability to recover reasonable quantities of homogeneous viruses, without plaque purification, represents a major practical advantage. And the GFP tracer makes it possible to follow all stages of the viral production process in a convenient fashion. In the case of cells that are inefficiently infected by adenoviruses, the GFP tracer additionally makes it possible to isolate expressing cells through fluorescence-activated cell sorting and thereby facilitates several kinds of experiment. Finally, the system described herein is efficient enough so that small libraries of transgenes produced in adenoviruses can be envisioned. Viruses with a particular modification of a transgene (produced by degenerate PCR, for example) could be selected in vivo from a pool of viruses on the basis of functional assays, and the sequence of the selected virus determined by sequencing appropriate PCR products.
Investigators who wish to obtain any of the vectors described in this work, along with more detailed protocols for their production and analysis, should contact the authors at the following e-mail address: tche@welchlink.welch.jhu.edu.
Acknowledgments
We thank Y.-N. Chang and B. Nelkin for critical reading of the manuscript, A. J. Van der Eb of the University of Leiden for generously providing 911 cells, D. Hanahan of the University of California, San Francisco for providing E. coli BJ5183 cells, and P. Hearing of the State University of New York, Stony Brook for providing adenovirus E4ORF6 antibody. We are also grateful to C. Lengauer and B. Tombal of the Johns Hopkins Oncology Center for their assistance with GFP fluorescence detection. The work was supported by Grant CA35494 from the National Institutes of Health. B.V. is an Investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- β-gal
β-galactosidase
- CMV
cytomegalovirus
- efu
expression-forming units
- GFP
green fluorescent protein
- ITR
inverted terminal repeat
References
- 1.Miller A D. Nature (London) 1992;357:455–460. doi: 10.1038/357455a0. [DOI] [PubMed] [Google Scholar]
- 2.Morgan R A, Anderson F A. Annu Rev Biochem. 1993;62:191–217. doi: 10.1146/annurev.bi.62.070193.001203. [DOI] [PubMed] [Google Scholar]
- 3.Graham F L, Prevec L. Methods Mol Biol. 1991;7:109–128. doi: 10.1385/0-89603-178-0:109. [DOI] [PubMed] [Google Scholar]
- 4.Berkner K L. BioTechniques. 1988;6:616–629. [PubMed] [Google Scholar]
- 5.Shenk T. In: Fields Virology. Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Philadelphia: Lippincott; 1996. pp. 2111–2148. [Google Scholar]
- 6.Horwitz M S. In: Fields Virology. Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Philadelphia: Lippincott; 1996. pp. 2149–2171. [Google Scholar]
- 7.Ballay A, Leverero M, Buendia M A, Tiollais P, Perricaudet M. EMBO J. 1985;4:3861–3865. doi: 10.1002/j.1460-2075.1985.tb04158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenfeld M A, Siegfried W, Yoshimura K, Yoneyama K, Fukayama M, Stier L F, Paakko P K, Gilardi P, Stratford-Perricaudet L D, Perricaudet M, et al. Science. 1991;252:431–434. doi: 10.1126/science.2017680. [DOI] [PubMed] [Google Scholar]
- 9.Mittal S K, McDermott M R, Johnson D C, Prevec L, Graham F L. Virus Res. 1993;28:67–90. doi: 10.1016/0168-1702(93)90090-a. [DOI] [PubMed] [Google Scholar]
- 10.Stratford-Perricaudet L D, Makeh I, Perricaudet M, Briand P. J Clin Invest. 1992;90:626–630. doi: 10.1172/JCI115902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham F L, Smiley J, Russel W C, Nairn R. J Gen Virol. 1977;36:59–72. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 12.Becker T C, Noel R J, Coats W S, Gomez-Foix A M, Alam T, Gerard R D, Newgard C B. Methods Cell Biol. 1994;43:161–189. doi: 10.1016/s0091-679x(08)60603-2. [DOI] [PubMed] [Google Scholar]
- 13.Ketner G, Spencer F, Tugendreich S, Connelly C, Hieter P. Proc Natl Acad Sci USA. 1994;91:6186–6190. doi: 10.1073/pnas.91.13.6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chartier C, Degryse M, Gantzer M, Dieterie A, Pavirani A, Mehtali M. J Virol. 1996;70:4805–4810. doi: 10.1128/jvi.70.7.4805-4810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crouzet J, Naudin L, Orsini C, Vigne E, Ferrero L, Roux A L, Benolt P, Latta M, Torrent C, Denefle P, et al. Proc Natl Acad Sci USA. 1997;94:1414–1419. doi: 10.1073/pnas.94.4.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prasher D, Eckenrode V, Ward W, Prendergast F, Cormier M. Gene. 1992;111:229–233. doi: 10.1016/0378-1119(92)90691-h. [DOI] [PubMed] [Google Scholar]
- 17.Fallaux F J, Kranenberg O, Creamer S J, Houweling A, van Ormondt H, Hoeben R C, van der Eb A J. Hum Gene Ther. 1996;7:215–222. doi: 10.1089/hum.1996.7.2-215. [DOI] [PubMed] [Google Scholar]
- 18.Hanahan D. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 19.Obert S, O’Connor R J, Schmid S, Hearing P. Mol Cell Biol. 1994;14:1333–1346. doi: 10.1128/mcb.14.2.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Becker T C, Noel R J, Coats W S, Gomez-Foix A M, Alam T, Gerard R D, Newgard C B. Methods Cell Biol. 1994;43:161–189. doi: 10.1016/s0091-679x(08)60603-2. [DOI] [PubMed] [Google Scholar]
- 21.West S. Cell. 1994;76:9–15. doi: 10.1016/0092-8674(94)90168-6. [DOI] [PubMed] [Google Scholar]
- 22.Camerini-Otero R D, Hsieh P. Annu Rev Genet. 1995;29:509–552. doi: 10.1146/annurev.ge.29.120195.002453. [DOI] [PubMed] [Google Scholar]
- 23.Berkner K L, Sharp P A. Nucleic Acids Res. 1983;11:6003–6020. doi: 10.1093/nar/11.17.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanahan D, Gluzman Y. Mol Cell Biol. 1984;4:302–309. doi: 10.1128/mcb.4.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hermeking H, Lengauer C, Polyak K, He T-C, Zhang L, Thiagalingam S, Kinzler K W, Vogelstein B. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 26.Hardy S, Kitamura M, Harris-Stansil T, Dai Y, Phipps M L. J Virol. 1997;71:1842–1849. doi: 10.1128/jvi.71.3.1842-1849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]