Abstract
Nicotine adenine dinucleotide (NAD+) performs key roles in electron transport reactions, as a substrate for poly(ADP-ribose) polymerase and NAD+-dependent protein deacetylases. In the latter two processes, NAD+ is consumed and converted to ADP-ribose and nicotinamide. NAD+ levels can be maintained by regeneration of NAD+ from nicotinamide via a salvage pathway or by de novo synthesis of NAD+ from tryptophan. Both pathways are conserved from yeast to humans. We describe a critical role of the NAD+-dependent deacetylase Hst1p as a sensor of NAD+ levels and regulator of NAD+ biosynthesis. Using transcript arrays, we show that low NAD+ states specifically induce the de novo NAD+ biosynthesis genes while the genes in the salvage pathway remain unaffected. The NAD+-dependent deacetylase activity of Hst1p represses de novo NAD+ biosynthesis genes in the absence of new protein synthesis, suggesting a direct effect. The known Hst1p binding partner, Sum1p, is present at promoters of highly inducible NAD+ biosynthesis genes. The removal of HST1-mediated repression of the NAD+ de novo biosynthesis pathway leads to increased cellular NAD+ levels. Transcript array analysis shows that reduction in cellular NAD+ levels preferentially affects Hst1p-regulated genes in comparison to genes regulated with other NAD+-dependent deacetylases (Sir2p, Hst2p, Hst3p, and Hst4p). In vitro experiments demonstrate that Hst1p has relatively low affinity toward NAD+ in comparison to other NAD+-dependent enzymes. These findings suggest that Hst1p serves as a cellular NAD+ sensor that monitors and regulates cellular NAD+ levels.
NAD+ and its pyridine or dihydropyridine congeners NADP+, NADH, and NADPH play crucial roles in electron transport processes (46). NAD+ also serves as an essential cofactor in enzymatic reactions that do not involve oxidative metabolism. For example, cleavage of the glycosidic bond between nicotinamide and ADP-ribose and concomitant transfer of ADP-ribose to arginine or histidine residues of G-proteins is identified as the mechanism of action of bacterial virulence factors such as the cholera and pertussis toxins (28). A related reaction carried out by poly(ADP-ribose) polymerase couples glycosidic bond hydrolysis with formation of ADP-ribose polymers in response to DNA damage in metazoans (12). More recently, the cleavage of the glycosidic bond of NAD+ was shown to be a requisite step in the deacetylation reaction carried out by class III deacetylases of the Saccharomyces cerevisiae Sir2p family (16, 20, 39). Deacetylation is accomplished through the transfer of the acetyl group from the substrate to ADP-ribose, the NAD+ breakdown product, to generate O-acetyl ADP-ribose and the release of free nicotinamide (33, 41, 42). Yeast Sir2p, the founding member of this family of enzymes, is required for silencing at the silent mating loci, telomeres, and rRNA genes (rDNA) and is responsible for the hypoacetylated state of histones at these locations (reviewed in reference 25). In addition to silencing (38), SIR2 suppresses recombination between tandem copies of rRNA genes (10) and thus reduces the formation of extrachromosomal ribosomal DNA (rDNA) circles and their accumulation in mother cells, a function critical for maintenance of mother-cell life span (36). SIR2 homologues in higher eukaryotes have been implicated in a wide range of biological processes involving promotion of longevity in Caenorhabditis elegans (43), development and epigenetic regulation in Drosophila melanogaster (29), and modulation of stress responses and apoptosis in mammalian cells through downregulation of p53 activity (24, 44). In yeast, where it has been studied most extensively, the in vivo Sir2p enzymatic activity and its function in silencing and longevity depend on an adequate supply of NAD+ (1, 32, 39). Other cellular enzymes and processes also depend on the availability of NAD+. Under conditions of stress, such as DNA damage, NAD+ depletion in mammalian cells induced by activation of poly(ADP-ribose) polymerase is thought to perturb important essential functions and contribute to cell death (11, 14, 15). NAD+ in cells is continuously broken down and resynthesized, with a half-life in human lymphocytes of approximately 6 h (7). Because the amount of available NAD+ in cells depends on the balance of NAD+ production and destruction, homeostatic mechanisms that regulate these processes are important, particularly in times of stress.
Both yeast and mammalian cells actively import nicotinic acid (NA or niacin) from the extracellular medium and convert it into NAD+ through the salvage pathway (Fig. 1A) (reviewed in reference 13). NA produced intracellularly by hydrolytic cleavage of the glycosidic bond in NAD+ and deamidation of nicotinamide is also recycled to NAD+ through the salvage pathway. Additionally, a de novo biosynthesis or kynurenine pathway converts tryptophan to NAD+. This well-studied pathway carries out oxidative degradation of tryptophan to quinolinic acid, which is subsequently converted to NAD+. Nicotinate phosphoribosyl transferase, Npt1p, is a key enzyme in the yeast NAD+ salvage pathway and is responsible for reutilization of NA generated by NAD+ breakdown and for utilizing NA taken up from the medium (39). In the absence of NPT1, yeast cells depend on the kynurenine pathway as the sole source of NAD+. Accordingly, deletion of NPT1 causes synthetic lethality with deletion of genes in the kynurenine pathway (BNA genes [for biosynthesis of nicotinic acid]) (27). Only one of the six BNA genes, BNA3, does not show synthetic lethality with NPT1, suggesting that the enzymatic reaction carried out by BNA3 can be performed by another enzyme. Epidemiologic studies of human populations indicate that cellular NAD+ levels and the efficiency of conversion of tryptophan to niacin varies widely between individuals (17). While most of this variation is thought to be due to differences in dietary niacin and tryptophan intake, it is likely that some of the variation in NAD+ levels is genetically determined.
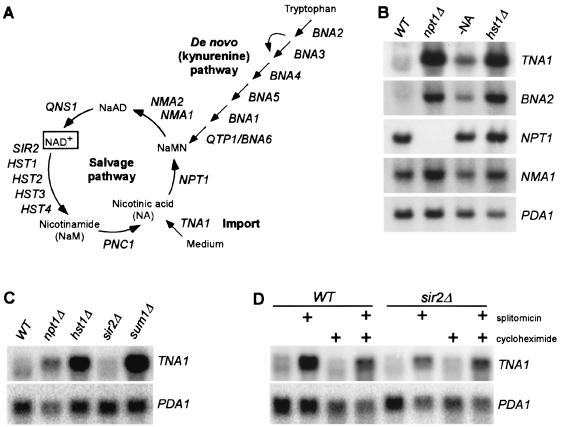
FIG. 1.
The NAD+ salvage pathway is constitutively active, whereas de novo NAD+ biosynthesis pathway is highly inducible by low NAD+ states. (A) NAD+ biosynthesis pathways in yeast. NaMN, NA mononucleotide; NaAD, desamido-NAD+. (B) Northern blot analysis of genes required for NA transport (TNA1), de novo NAD+ biosynthesis (BNA2), and the NAD+ salvage pathway (NPT1 and NMA1) in different strains and culture conditions. Wild-type BY4742 (WT) or isogenic npt1 and hst1 cells were grown in SC medium. -NA, wild-type cells grown in medium lacking NA. PDA1 transcripts were used as a loading control. (C) Northern blot analysis of the high-affinity NA transporter gene TNA1 in different mutants. Cells were grown in SC medium. PDA1 transcripts were used as a loading control. (D) Histone deacetylase activity of Hst1p is required for the repression of TNA1 transcript. Total RNA was isolated from wild-type (BY4742) cells or the isogenic sir2 mutant strains treated with the specific inhibitor of NAD+-dependent deacetylases splitomicin (75 μM) for 6 h without or with 40-min pretreatment with cycloheximide (50 μg/ml) in SC medium. PDA1 transcripts were used as a loading control.
In the course of our studies on the NAD+-dependent deacetylases in yeast, we sought to identify factors that regulate cellular levels of NAD+. In this study, we report that the homologue of Sir2p, Hst1p (for homologue of SIR two) is an NAD+-dependent deacetylase with relatively low affinity for NAD+ that functions as a cellular NAD+ sensor to repress the NAD+ biosynthesis program when NAD+ levels are adequate.
MATERIALS AND METHODS
Yeast media, strains, and plasmids.
All strains were grown in synthetic complete medium (SC) or selective synthetic drop-out medium containing 2% glucose. The SC medium lacking NA was prepared according to the method of Wickersham (34) with each of the components individually added.
The list of strains used in this study is provided in Table 1. Additional deletion mutants strains npt1::kanMX, sir2::kanMX, hst1::kanMX, hst2::kanMX, hst3::kanMX, hst4::kanMX, and sum1::kanMX derived from BY4741 or BY4742 were obtained from haploid deletion sets (Saccharomyces Genome Deletion Project [SGDP], Research Genetics). YAB14145 and YAB14146 were sporulation products of diploid strains generated by matings of BY4742 sum1::kanMX (strain 13669; SGDP, Research Genetics) and BY4741 npt1::kanMX and BY4741 hst1::kanMX (strains 2645 and 1760; SGDP, Research Genetics), respectively. The double sum1::kanMX and npt1::kanMX or hst1::kanMX alleles were confirmed by PCR. All other strains were generated by one-step PCR-mediated gene replacement using the integrating pRS400 plasmid containing the kanMX gene as a template (6).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| BY4741 | MATahis3-Δ200 leu2-Δ0 ura3-Δ0 met15-Δ0 | Research Genetics |
| BY4742 | MATα his3-Δ200 leu2-Δ0 ura3-Δ0 lys2-Δ0 | Research Genetics |
| 13669 | BY4742 sum1::kanMX | Research Genetics |
| YAB14145 | MATα sum1::kanMX npt1::kanMX his3-Δ200 leu2-Δ0 ura3-Δ0 lys2-Δ0 | This study |
| YAB14146 | MATα sum1::kanMX hst1::kanMX his3-Δ200 leu2-Δ0 ura3-Δ0 lys2-Δ0 | This study |
| YMH533 | BY4742 bna2::KanMX | This study |
| YMH480 | BY4742 hst1::KanMX bna2::NAT | This study |
The glutathione S-transferase (GST)-Sir2p expression plasmid was a gift from J. Boeke (39). GST-Hst1p and GST-Hst2 expression plasmids were made by PCR amplification of the HST1 and HST2 full-length open reading frame (ORF) from genomic DNA using oligonucleotides with EcoRI and XhoI restriction sites and cloned into pGEX4T-1 (Pharmacia) as an EcoRI-XhoI fragment. The entire ORFs were sequenced to assure that no mutations were introduced during PCR amplification. The plasmids encoding 3myc epitope-tagged Sum1p, pRS415 3myc-SUM1(pJR2296), and hemagglutinin (HA) epitope-tagged Hst1p, PRS416 HST1(pJR2288), were a gift from J. Rine (30).
Antibodies, protein coimmunoprecipitation, and ChIP.
Protein coimmunoprecipitation and chromatin immunoprecipitation (ChIP) experiments were performed as described previously (9, 30). The antibodies used for protein coimunopreciptiation and immunoprecipitation of chromatin were obtained from Upstate Biotechnology (rabbit polyclonal immunoglobulin G anti-acetyl histone H3, ChIP-grade anti-acetyl histone H4, mouse monoclonal immunoglobulin G1 anti-Myc tag, clone 9E10). The sequences of the primers used for the PCR amplifications are available in the supplemental material (http://www.fhcrc.org/labs/simon/BedalovMCB.html). The anti-HA monoclonal antibody (clone 16B12) used in the Western blot analysis was obtained from Babco.
NAD+ measurements.
Total cellular NAD+ levels were measured as described previously (37), except that volumes were scaled down. Briefly, a 100-ml culture was grown to a density of 2 × 107 cells/ml, harvested by centrifugation, and washed twice in cold water. NAD+ extraction was performed in 2-ml tubes by adding 800 μl of ice-cold butanol-saturated 1 M formic acid to the cell pellet, vortexing, and incubating on ice. After 30 min, 200 μl of 100% trichloroacetic acid (TCA) was added to a tube, vortexed, and left on ice for another 15 min. Tubes were centrifuged at 14,000 × g for 10 min, and supernatant containing acid-extracted NAD+ was collected. The residual NAD+ was reextracted by adding 150 μl of 20% TCA to the pellet, vortexing, and centrifugation. The supernatant was pooled with the previously collected extract. The NAD+ content in the acid extract was determined by measuring the absorbance at 340 nm after enzymatic conversion of NAD+ to NADH as described previously.
HDA and protein purification.
A histone deacetylase assay (HDA) was performed using bacterially expressed and purified GST-Hst1p, GST-Hst2p, and GST-Sir2p as previously described (3). Briefly, histone H4 was chemically acetylated using the HDAC assay kit (Upstate Biotechnology). Hst1p, Hst2p, and Sir2p were expressed as GST fusion proteins from Pharmacia pGEX plasmids and purified according to the methods recommended by the manufacturer. For HDAs, 2 μg of GST-Hst1p, 0.2 μg of GST-HST2, and 1 μg of purified GST-Sir2p protein were incubated with [3H]acetylated histone H4 peptide (40,000 cpm) without NAD+ or with a range of NAD+ concentrations in a 20-μl reaction volume. The buffer contained 150 mM NaCl, 50 mM Tris-HCl (pH 8.0), and 1 mM dithiothreitol. Reactions were incubated at 30°C for 16 h and stopped by the addition of 25 μl of 1 N HCl-0.15 N acetic acid. Released [3H]acetate was extracted with 400 μl of ethyl acetate. The NAD+-deacetylase activity response curve was fitted, and NAD+ Km and Vmax values were calculated using Prism software (GraphPad Software).
Gene expression analysis.
cDNA microarray experiments were performed as previously described (3). Strains for the array experiments were obtained from Research Genetics (wild-type BY4741 or isogenic npt1, sir2, hst1, hst2, hst3, hst4, or sum1 deletion mutants). Several colonies from fresh cultures were inoculated into SC medium with 2% glucose, grown overnight at 30°C, diluted to 0.5 × 106 to 1 × 106 cells/ml, and grown for an additional 6 to 9 h until reaching a density of 0.5 × 107 to 1 × 107 cells/ml. For experiments with splitomicin, drug or the solvent (dimethyl sulfoxide) was added at the beginning of the final 9-h growth phase. In experiments with cycloheximide, cells were treated with 50 μg of cycloheximide/ml for 40 min prior to the addition of splitomicin. Total RNA was extracted using the hot acid phenol method.
Three competitive hybridizations for each experimental group (npt1, sir2, hst1, hst2, hst3, hst4, sum1, or hst1 gcn5 versus wild type) were performed using three separate cultures, and the log2 of the expression ratio was calculated for every ORF. To assess the intrinsic variation of expression levels for different ORFs, nine wild-type versus wild-type hybridizations were performed using nine separate cultures. Student's t test was used to assess if the difference between the log2 of the expression ratio for the ORFs in the experimental and control groups (wild type versus wild type) was significant. The spreadsheet containing the mean log2 of the expression ratios and P values is available elsewhere (data not shown) for all experiments.
RESULTS
NPT1 deletion upregulates the genes for de novo NAD+ biosynthesis and for import of NA.
The deletion of NPT1 leads to a 50 to 70% reduction in cellular NAD+ levels (39). In order to assess global transcriptional responses to NAD+ depletion and NAD+ biosynthesis genes in particular, we analyzed the transcript profile of npt1 mutant cells. Whole-genome cDNA microarray analysis demonstrated that the most highly regulated genes in npt1 cells are TNA1, a high-affinity NA transporter (23), and the BNA genes encoding a battery of enzymes in the kynurenine pathway (Table 2). All BNA genes except BNA3 were upregulated (range, 1.5- to 4.2-fold) by deletion of NPT1. The concerted upregulation of the genes for the kynurenine pathway in the npt1 mutant can be regarded as a compensatory response to NAD+ depletion that acts to restore the NAD+ levels, and it suggests the presence of a feedback mechanism that monitors NAD+ levels. Interestingly, the upregulation of NAD+ biosynthesis genes in the npt1 strain was limited to the de novo pathway, and no significant increase was detected in transcripts of genes in the salvage pathway (Table 2). Constitutive expression of salvage pathway genes was confirmed by Northern blot analysis of wild-type cells grown under conditions of adequate NAD+ (Fig. 1B). In contrast to TNA1, which was highly induced by interruption of the salvage pathway via NPT1 deletion or by growing cells in medium lacking NA, the salvage pathway genes NPT1 and NMA1 were not highly induced under the same conditions.
TABLE 2.
NAD+ biosynthesis genes analyzed by transcript arrays
| Pathway | Gene | Level of upregulationa (fold increase)
|
||||||
|---|---|---|---|---|---|---|---|---|
| npt1Δ | sir2Δ | hst1Δ | hst2Δ | hst3Δ | hst4Δ | sum1Δ | ||
| Import | TNA1 (YGR260W) | 4.9 | 1.1 | 5.9 | 1.2 | 1.3 | 1.0 | 11.7 |
| De novo | BNA2 (YJL060W) | 4.2 | 0.8 | 4.2 | 1.1 | 1.7 | 0.9 | 14.1 |
| BNA3 (YBL098W) | 1.1 | 1.1 | 1.2 | 1.2 | 1.1 | 1.0 | 1.0 | |
| BNA4 (YLR231C) | 2.6 | 1.7 | 5.2 | 1.1 | 1.2 | 1.1 | 11.7 | |
| BNA5 (YJR025C) | 1.8 | 0.9 | 2.7 | 1.0 | 1.1 | 1.0 | 4.6 | |
| BNA1 (YJR078W) | 1.6 | 0.9 | 1.6 | 1.0 | 0.9 | 0.9 | 3.9 | |
| QPT1/BNA6 (YFR047C) | 1.5 | 1.1 | 2.1 | 1.1 | 0.9 | 0.8 | 1.9 | |
| Salvage | NMA1 (YLR328W) | 0.9 | 1.0 | 0.9 | 1.0 | 0.9 | 1.1 | 1.0 |
| NMA2(YGR010W) | 0.9 | 0.9 | 1.2 | 1.0 | 1.0 | 0.9 | 1.0 | |
| QNS1 (YHR074W) | 1.1 | 0.9 | 1.0 | 1.0 | 1.0 | 1.1 | 1.3 | |
| PNC1 (YGL037C) | 0.9 | 0.9 | 1.3 | 1.1 | 1.1 | 0.8 | 0.7 | |
| NPT1 (YOR209C) | 0.0 | 0.9 | 1.2 | 1.0 | 1.0 | 1.1 | 1.3 | |
Upregulation of ≥1.5-fold is indicated in boldface.
Transcript array analysis of mutants in NAD+-dependent deacetylases revealed that HST1 represses NAD+ biosynthesis genes.
Because of the ability of NAD+-dependent histone deacetylases to respond to NAD+ levels and repress transcription, we hypothesized that members of this family of enzymes might participate in the NAD+ biosynthesis feedback loop. We therefore analyzed transcript profiles of mutants lacking different NAD+-dependent deacetylases (sir2, hst1, hst2, hst3, and hst4). This analysis revealed that a strain lacking HST1 demonstrated significant upregulation of genes in the kynurenine pathway and the gene encoding the high-affinity NA transporter TNA1. The degree of upregulation of TNA1 and BNA genes in hst1 mutants paralleled the upregulation of the same genes in the npt1 strain but was generally higher (Table 2 and Fig. 1B and C). Similarly to npt1 cells, hst1 cells did not upregulate BNA3, and there was no upregulation of genes in the salvage pathway. With two exceptions, 1.7-fold upregulation of BNA4 in sir2 mutants and of BNA2 in hst3 mutants, strains lacking other NAD+-dependent enzymes (Sir2p, Hst2p, Hst3p, or Hst4p), did not upregulate genes in the kynurenine pathway or TNA1, indicating that only Hst1p participates in this pathway.
Deacetylase activity of Hst1p represses NAD+ de novo biosynthesis genes in the absence of new protein synthesis.
In order to test whether the histone deacetylase activity of Hst1p is required for repression of NAD+ biosynthesis genes, we used a specific inhibitor of NAD+-dependent deacetylases, splitomicin (3). Conditional inactivation of Hst1p deacetylase activity with the small-molecule inhibitor combined with the protein synthesis inhibitor cycloheximide allowed us to determine if Hst1p directly represses NAD+ biosynthesis genes.
Northern blot analysis showed that treatment of wild-type cells with splitomicin resulted in upregulation of both TNA1 (Fig. 1D) and BNA2 (data not shown), confirming our earlier results with transcript arrays. Splitomicin can inhibit both Sir2p and Hst1p histone deacetylases (3), but two observations strongly suggest that its effect on TNA1 transcription is exclusively the result of the latter activity. First, unlike deletion of HST1, deletion of SIR2 did not result in upregulation of TNA1 (Table 2 and Fig. 1C). Second, deletion of SIR2 did not affect splitomicin-induced transcription of TNA1 (Fig. 1D). The induction of TNA1 transcription by splitomicin was preserved in the presence of the protein synthesis inhibitor cycloheximide (Fig. 1D), consistent with a direct effect of Hst1p on NAD+ biosynthesis gene transcription. Overall, these results support a simple feedback loop model in which NAD+ directly regulates Hst1p activity and Hst1p activity directly regulates transcription of NAD+ biosynthesis genes.
A limited supply of NAD+ selectively affects HST1-regulated transcripts.
As the enzymatic activity of all NAD+-dependent deacetylases depends on NAD+, the limited supply of NAD+ is expected to create a partial loss of function of these enzymes. Different NAD+-dependent deacetylases regulate different sets of genes, which allows the transcriptional activity of these genes to be used as a reporter of the in vivo activity of the corresponding enzyme (Fig. 2A and B). Transcript array analysis of the npt1 mutant thus allowed us to simultaneously assess the in vivo effect of reduced cellular NAD+ on the function of all NAD+-dependent deacetylases and to evaluate the sensitivity of each enzyme to a limitation of the NAD+ supply. Our transcript array analysis showed that Hst1p and its closest homologue, Sir2p, regulate different sets of genes (Fig. 2B). Although there was a significant overlap between the genes upregulated in the hst1 deletion mutant and the hst2 and the hst3 mutants (Fig. 2A), for both hst2 and hst3 mutants there was a significant number of genes whose regulation was specific to HST2 or HST3. The deletion of HST4 alters the transcription of very few genes (supplemental material).
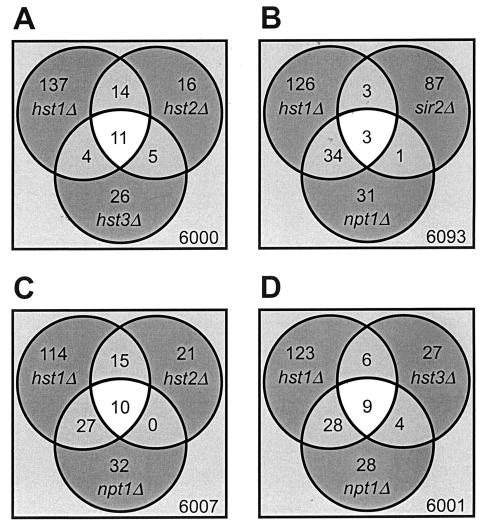
FIG. 2.
NPT1 deletion preferentially affects HST1-regulated genes. The Venn diagrams compare genes upregulated 1.6-fold or more relative to wild type in sir2, hst1, hst2, and hst3 cells.
Using sensitive reporter assays, deletion of NPT1 has been previously shown to result in upregulation of genes at the rDNA and telomeres and, to a lesser degree, the silent mating loci, the three sites regulated by SIR2 (39). Our transcript arrays, however, demonstrated that at sites outside of the rDNA, deletion of NPT1 had significantly less effect on genes regulated by SIR2 than on genes regulated by HST1. The deletion of NPT1 resulted in upregulation of only 4% of SIR2-repressed genes (4 of 94 transcripts) (Fig. 2B), with only one of the four overlapping transcripts specific to SIR2. In contrast, a significantly higher proportion of HST1-repressed genes (37 of 166, or 22%) were upregulated in response to NPT1 deletion, with NAD+ biosynthesis genes among the most highly regulated transcripts (supplemental material). Although 22% and 28% of HST2-responsive (Fig. 2 C) and HST3-responsive (Fig. 2D) genes were upregulated by deletion of NPT1, the overlap between NPT1- and HST2-regulated or between NPT1- and HST3-regulated transcripts was entirely (for HST2) or largely (for HST3) limited to genes upregulated in hst1 mutants. Thus, NPT1 deletion selectively affected HST1-regulated genes. Conversely, greater than 50% of all transcriptional upregulation in the npt1 strain could be attributed to a loss of Hst1p function. The finding that HST1-regulated genes readily respond to relatively small changes in the cellular NAD+ level (the deletion of NPT1 induced an approximately 50% reduction in total cellular NAD+ under our assay conditions) and to a greater degree than genes regulated by other NAD+-dependent deacetylases supports the idea that Hst1p serves as a primary cellular NAD+ sensor.
The NAD+ Km of Hst1p is higher than that of Sir2p or Hst2p, which is consistent with its role as a cellular NAD+ sensor.
The difference in sensitivity to NPT1 deletion between SIR2-, HST2-, and HST1-responsive genes could be a result of different binding affinities for NAD+ between Hst1p, Hst2p, and Sir2p. We directly tested this possibility by measuring the enzymatic rate constants of recombinant Hst1p, Hst2p, and Sir2p. Sir2p is, like Hst1p, a nuclear enzyme, and Hst2p accounts for a mostly NAD+-dependent deacetylase in yeast whole-cell extracts. The NAD+ Km for Hst1p was 94.2 ± 5.4 μM (mean ± standard deviation), more than threefold higher than the NAD+ Km for Sir2p (29.7 ± 2.7 μM) or for Hst2p (15.0 ± 1.9 μM) (Fig. 3). Because Hst1p has lower NAD+ binding affinity than Sir2p, the drop in deacetylase activity in response to NAD+ depletion, and hence derepression of target genes, is expected to be more pronounced for Hst1p-regulated genes than for Sir2p- or Hst2p-regulated genes. The relatively low binding affinity of Hst1p toward NAD+ is consistent with its proposed role as a physiological NAD+ sensor and regulator of NAD+ biosynthesis.
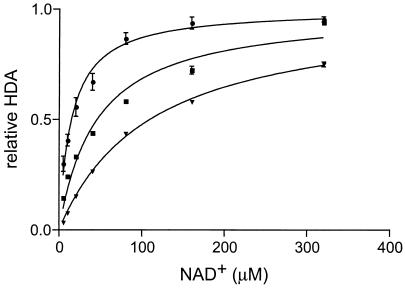
FIG. 3.
Hst1p has lower affinity toward NAD+ than Sir2p or Hst1p. Shown are HDA activities of recombinant Hst1p (triangles), Hst2p (circles), and Sir2p (squares). Bacterially expressed and purified GST-Hst1p, GST-Hst2p, and GST-Sir2p proteins were incubated with chemically [3H]acetylated histone H4 peptide (40,000 cpm/reaction mixture) and different concentrations of NAD+. NAD+-dependent deacetylase activity, normalized for the Vmax of each protein, is plotted. The Vmax for Sir2p was 375 ± 12 cpm/h, Vmax for Hst2p was 331 ± 12 cpm/h, and Vmax for Hst1p was 170 ± 4 cpm/h. The NAD+ Km for Sir2p was 29.7 ± 2.7 μM, the NAD+ Km for Hst2p was 15.0 ± 1.9 μM, and the NAD+ Km for Hst1p was 94.2 ± 5.4 μM.
The previously reported NAD+ Km for Hst2p was 70 μM (41). We cannot explain the reason for the difference between the previously reported NAD+ Km for Hst2p and our result, except to note that the two measurement were obtained using different Hst2p protein tagging methods and protein preparations, different deacetylation substrates, and different reaction conditions.
The Hst1p recruitment factor Sum1p is present at the promoters of NAD+ biosynthesis genes irrespective of the cellular NAD+ levels.
The Sum1p transcription factor binds Hst1p and is known to recruit Hst1p to the promoters of midsporulation genes (49). In order to assess if the same interacting partner recruits Hst1p to the promoters of NAD+ biosynthesis genes, we analyzed the transcription profiles of a sum1 mutant and compared them with the profile of hst1 cells. In addition to midsporulation genes, which were upregulated in both mutants (supplemental material), NAD+ biosynthesis genes were also highly upregulated in both strains (Table 2 and Fig. 1C). This result suggests that, in addition to recruiting Hst1p to the promoters of midsporulation genes, Sum1p also recruits Hst1p to the promoters of NAD+ biosynthesis genes. We confirmed the presence of myc-tagged Sum1p at the promoters of TNA1 and BNA2 in wild-type cells in ChIP experiments (Fig. 4A). Furthermore, Sum1p could be detected at the promoters of the TNA1 and BNA2 genes, even under conditions of low cellular NAD+ in npt1 cells. Sum1p binding to these promoters was also detected in hst1 cells. In addition, protein coimmunoprecipitation experiments showed that Sum1p and Hst1p interaction was unaffected by low cellular NAD+ in the npt1 mutant (Fig. 4B). The finding that Sum1p occupancy of the promoters of NAD+ biosynthesis genes does not depend on cellular NAD+ level supports a model whereby the degree of repression of NAD+ biosynthesis genes is achieved through modulation of the deacetylase activity of Hst1p and not by regulation of Sum1p-mediated recruitment of Hst1p to the promoters. Although Sum1p and Hst1p interacted in protein coimmunoprecipitation studies (Fig. 4B), we were unable to demonstrate the presence of Hst1p in the promoters of TNA1 or NAD+ biosynthesis genes in the ChIP experiments (data not shown). Since Hst1p does not bind to DNA directly, but rather via Sum1p, it is possible that cross-linking of Hst1p to DNA is not sufficient for detection of this interaction in immunoprecipitation experiments, as has been seen with the midsporulation genes also known to be regulated by Hst1p (30).
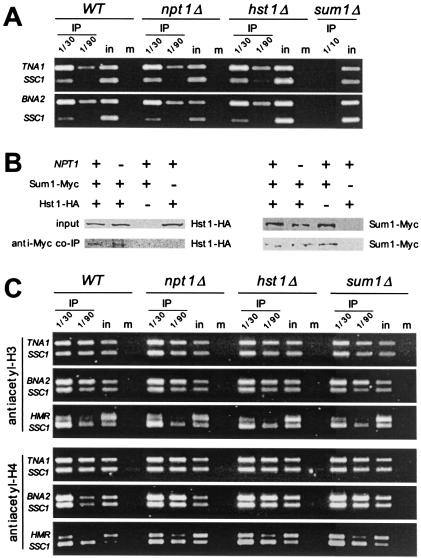
FIG. 4.
(A) Sum1p associated with TNA1 and BNA2 promoters regardless of the NAD+ status of the cell. Cell lysates were prepared from formaldehyde cross-linked wild-type (WT) or npt1- or hst1-containing myc epitope-tagged SUM1 or sum1 cells which served as a negative control. DNA immunoprecipitated with anti-myc antibody was analyzed by simultaneous PCR amplification of TNA1 or BNA2 promoter and the SSC1 control gene. One-thirtieth and 1/90 of the immunoprecipitated DNA (IP), 1/50,000 of the input DNA (in), or 1/10 of mock-immunoprecipitated (no antibody used) DNA (m) was used for PCR amplification. (B) Sum1p and Hst1p interact regardless of the NAD+ status of the cell. Cell lysates prepared from wild-type, npt1, and sum1 hst1 cells containing plasmids with myc epitope-tagged SUM1 and/or HA epitope-tagged Hst1p were immunoprecipitated using anti-myc antibody. Whole-cell lysates and anti-myc immunoprecipitates were analyzed for the presence of Hst1-HA and Sum1-Myc by immunoblotting. (C) HST1- and SUM1-mediated repression of the TNA1 and BNA2 genes is not associated with global histone H3 and H4 deacetylation. Cell lysates were prepared from formaldehyde cross-linked WT, npt1, hst1, and sum1 cells. DNA immunoprecipitated with anti-acetyl-H3 or anti-acetyl-H4 antibody was analyzed by simultaneous amplification of TNA1, BNA2, or the silent HMR loci and the control SSC1 promoter. One-thirtieth and 1/90 of the immunoprecipitated DNA (IP), 1/50,000 of the input DNA (in), or 1/10 of mock immunoprecipitated (no antibody used) DNA (m) was used for PCR amplification.
Chromatin at silent mating loci and at telomeres is hypoacetylated by the action of Sir2p. Experiments in which Hst1p was inappropriately recruited to silent mating type loci (using a SUM1-1 mutation) demonstrate that if recruited Hst1p, similarly to Sir2p, is capable of deacetylating large regions of chromatin at these loci (30, 40). Because we hypothesized that Hst1p acts at the promoters of NAD+ biosynthesis genes, we wanted to determine whether Hst1p likewise deacetylates large regions of chromatin at these loci and whether the acetylation status of these promoters can be altered by depleting cellular NAD+ levels. Therefore, we examined the promoters of TNA1 and BNA2 by ChIP with anti-acetyl-H3 and anti-acetyl-H4 antibodies. We did not detect differences in the global acetylation status of chromatin at these promoters when comparing wild-type cells with hst1, npt1, or sum1 cells (Fig. 4C). Control experiments confirmed the hypoacetylated state of chromatin at the silent HMR locus, a known effect of Sir2p (Fig. 4C). Rusche and Rine (30) were likewise unable to detect differences in acetylation between the wild type and a sum1 or hst1 mutant in the midsporulation genes. These results may suggest that histone deacetylation by Hst1p at NAD+ biosynthesis genes may be restricted to small regions of chromatin or that it targets only specific histones within larger regions, specific lysines, or proteins other than histones.
HST1 deletion increases steady-state NAD+ levels through the upregulation of a de novo biosynthesis pathway.
The supply of NAD+ through the kynurenine pathway is sufficient to support yeast growth in mutants with a defective NAD+ salvage pathway. Because hst1 cells, like an npt1 mutant, upregulate the enzymes in the kynurenine pathway, we reasoned that HST1 deletion might augment NAD+ biosynthesis and increase the steady-state NAD+ levels. To test this possibility we measured and compared the NAD+ levels in wild-type and hst1 cells. Cells lacking NPT1 are known to have low NAD+ levels (39) and were included as controls. For NAD+ measurements, cells were grown in standard SC medium (containing 100 μM tryptophan and 3.5 μM NA), the same conditions known to induce derepression of NAD+ biosynthesis genes in hst1 cells in transcript array experiments. We also measured NAD+ levels in cells grown without the NAD+ precursors NA and/or tryptophan. Tryptophan is the starting material for the de novo NAD+ biosynthesis, and the amount of available tryptophan is known to affect the flux through the kynurenine pathway (reviewed in reference 13). When no tryptophan was added to the medium, endogenously synthesized tryptophan was the only source of this amino acid. As previously shown (39), the deletion of NPT1 in cells grown in complete medium resulted in a 50% reduction in NAD+ at steady state (Fig. 5A). The deletion of HST1 in cells grown in complete medium led to a small but reproducible increase in the steady-state NAD+ levels (23% increase; P = 0.04) (Fig. 5A and B). When grown in media without tryptophan, NA, or both, cells lacking HST1 had a higher increase in NAD+ levels (52 to 71%) than wild-type cells (Fig. 5B). The augmented effect of HST1 deletion on NAD+ levels in media lacking tryptophan and/or niacin is due to a 20 to 30% decrease in NAD+ levels in wild-type cells grown without these nutrients. In contrast to wild-type cells, hst1 cells showed very little or no decrease in NAD+ levels when grown in media lacking NAD+ precursors compared to growth in medium with tryptophan and niacin.
FIG. 5.
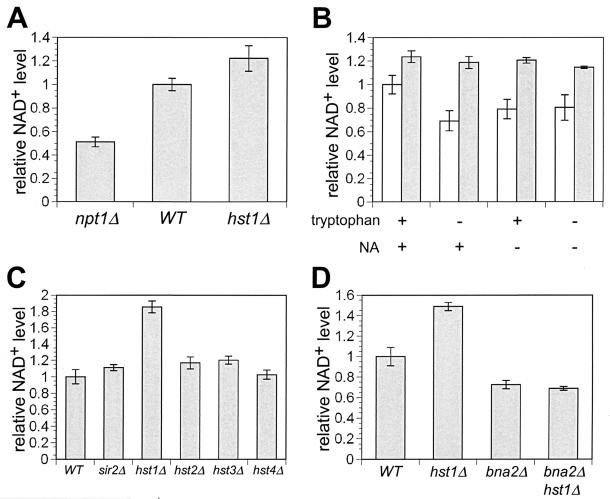
HST1 deletion increases total cellular NAD+ levels through the upregulation of NAD+ biosynthesis genes. (A) Relative steady-state NAD+ levels in wild-type (WT) and npt1 hst1 mutant cells grown in SC medium. (B) The lack of NAD+ precursors in the medium augments the difference in NAD+ levels between wild-type cells (white bars) and hst1 mutant cells (gray bars). Relative steady-state NAD+ levels in wild-type (wt) and hst1 cells grown in medium with or without added NAD+ precursors NA and/or tryptophan are shown. (C) Among mutants in NAD+-dependent deacetyalse genes, only hst1 has significantly elevated NAD+ levels. Relative steady-state NAD+ levels are shown for wild-type and NAD+-dependent deacetylase mutants in medium lacking tryptophan. (D) Increased cellular NAD+ levels in hst1 mutants depend on an intact NAD+ de novo pathway. Relative steady-state NAD+ levels in wild-type (WT) and hst1, bna2, and bna2 hst1 mutants in medium lacking tryptophan are shown.
Since Hst1p, like other NAD+-dependent enzymes, hydrolyzes NAD+, we wanted to investigate if decreased consumption of NAD+ plays any role in the observed increase in cellular NAD+ levels in the hst1 mutant. We first measured NAD+ levels in mutants lacking different NAD+-dependent histone deacetylases (sir2, hst1, hst2, hst3, and hst4). This analysis showed that only the hst1 mutant had steady-state NAD+ levels significantly higher than those in wild-type cells (Fig. 5C). To determine if the increased NAD+ level in the hst1 mutant is due to upregulation of the NAD+ biosynthesis pathway and to assess the potential contribution of decreased NAD+ consumption, we examined the effect of HST1 deletion in a bna2 strain, which is deficient in the de novo NAD+ biosynthesis pathway. We found that NAD+ levels in the bna2 strain were lower than in wild-type cells (Fig. 5D). However, the NAD+ levels in the bna2 hst1 double mutant were the same as those in the bna2 mutant. This result demonstrates that increased NAD+ levels in the hst1 strain are solely due to the upregulation of NAD+ biosynthesis and not to decreased NAD+ consumption. The result that removal of HST1-mediated repression of NAD+ biosynthesis genes in the hst1 mutant leads to an increased steady-state NAD+ level firmly establishes Hst1p as a regulator of NAD+ biosynthesis.
DISCUSSION
Given the variety of enzymatic processes that require NAD+, it is important for cells to maintain adequate levels of NAD+. Living cells have elaborate systems that monitor and respond to internal and external environments in order to maintain homeostasis. Genetic analysis in yeast has been successfully used to dissect the pathways that participate in glucose, nitrogen, and phosphate regulatory circuits (reviewed in reference 48). However, despite the detailed knowledge of the genetic components of the regulatory circuitry, the actual sensing molecules are often unknown.
In this report, we describe a feedback mechanism that monitors and regulates cellular NAD+ levels. We found that the NAD+-dependent histone deacetylase Hst1p plays a key role in this process, acting as a sensor and regulator of cellular NAD+ levels. NAD+ can be generated by a salvage pathway that reutilizes nicotinamide formed by the turnover of NAD+ and NADP+ or from NA and nicotinamide provided in the diet (Fig. 1). Alternatively, NAD+ can be synthesized de novo from tryptophan via the kynurenine pathway (reviewed in reference 13). The analysis of transcript arrays of the npt1 strain, which has low NAD+ levels due to a defect in the NAD+ salvage pathway, suggests that salvage pathway genes are constitutively active while genes encoding the kynurenine pathway and NA transport are repressed when NAD+ levels are adequate. These results, as well as prior reports that NA represses the high-affinity NA transporter gene TNA1 (23), suggested the existence of a feedback mechanism that senses and represses NAD+ biosynthesis genes when NAD+ levels are adequate. The transcript array analysis of mutants of all yeast NAD+-dependent deacetylases (sir2, hst1, hst2, hst3, and hst4) demonstrated that HST1 actively represses de novo NAD+ biosynthesis genes as well as the NA transporter TNA1. The Sum1p transcription factor, a known interacting partner of Hst1p (30, 49), also represses the NAD+ biosynthesis genes. Our results suggest a simple feedback loop whereby Hst1p, recruited to the NAD+ biosynthesis gene via Sum1p, senses NAD+ levels and, through modulation of its deacetylase activity, directly regulates transcription of NAD+ biosynthesis genes. Using splitomicin, an inhibitor of NAD+-dependent deacetylases (3), we first demonstrated that deacetylase activity of Hst1p is required for repression of NAD+ biosynthesis genes. Furthermore, the observation that pharmacologic inhibition of Hst1p deacetylase activity derepressed NAD+ biosynthesis genes in the absence of new protein synthesis supports the idea that deacetylase activity directly represses the NAD+ biosynthesis gene promoters. Sum1p was present at the promoters of NAD+ biosynthesis genes in ChIP experiments in wild-type cells, where NAD+ levels are adequate, and under low-NAD+ conditions with the npt1 mutant. We propose that NAD+ levels modulate the deacetylase activity of Hst1p, which is constitutively present at the promoters of NAD+ biosynthesis genes, thus adjusting the degree of repression of NAD+ biosynthesis genes.
The comparison of transcript array profiles of the npt1 mutant, deficient in the NAD+ salvage pathway, and mutants in NAD+-dependent histone deacetylases (sir2, hst1, hst2, hst3, and hst4) demonstrates that reduced cellular NAD+ levels preferentially affect HST1-regulated transcripts. The Hst1p NAD+ Km of 94.2 μM is more than threefold higher than the Km of the other major nuclear NAD+-dependent deacetylase, Sir2p, or of the major cytoplasmic deacetylase, Hst2p. The relatively high NAD+ Km for Hst1p is consistent with its proposed role as an NAD+ sensor and a regulator of NAD+ biosynthesis and explains the observation that NAD+ depletion, induced by deletion of NPT1, affects a higher proportion of HST1-regulated genes than of SIR2-regulated genes. The low NAD+ binding affinity of Hst1p assures that the NAD+ biosynthesis program is activated and adequate NAD+ levels are restored before other NAD+-dependent processes are compromised.
In order for Hst1p to be an effective NAD+ sensor, its NAD Km should be within the range of physiological concentrations of free nuclear NAD+. Using our own measurements of total cellular NAD+ as well as the results of others (32) and assuming a volume of distribution equal to total cellular volume (70 μm3), the cellular NAD+ concentration can be estimated to be about 1.5 to 2 mM. However, because most the cellular NAD+ and NADH exist in the bound form (35), the concentrations of free NAD+ are likely to be significantly lower (reviewed in reference 22). The study by Zhang et al. (50) in mammalian cells revealed that only approximately 10% of total cellular NAD(P)H is in the free form. These authors used the combination of two-photon excitation microscopy for measurements of free NADH and a conventional technique for measuring the free pool of NAD+ to NADH (through the pyruvate/lactate ratio) and estimated the concentration of free nuclear NAD+ to be about 85 μM. Assuming similar concentrations of free NAD+ in the yeast nucleus, the measured NAD+ Km of Hst1p would be very suitable for an NAD+ sensor.
Our model suggests that Hst1p needs to continuously counteract the activity of histone acetyltransferase (HAT) at the promoters of NAD+ biosynthesis genes to maintain a transcriptionally inactive state. The transcription factor Gcn4p and the Gcn4p-associated histone acetyltransferase Gcn5p were implicated in upregulation of NAD+ biosynthesis genes in response to amino acid depletion (26). However, we found that Gcn5p HAT was not required for upregulation of NAD+ biosynthesis genes in an hst1 mutant, since the gcn5 hst1 double mutant upregulated NAD+ biosynthesis genes as much as the hst1 single mutant (supplemental material). A different HAT may be targeted to the promoters of NAD+ biosynthesis genes by other transcription factors or may be part of a global acetylation maintenance system (47). For midsporulation genes, the other large group of genes that are repressed with Hst1p, the transcriptional activator Ndt80p is induced during sporulation and mediates their activation during the sporulation program (reviewed in reference 45). However, our observation that the pharmacologic inhibition of Hsp1p deacetylase activity leads to induction of NAD+ biosynthesis in the presence of cycloheximide suggests that no new transcription factor or associated HAT needs to be synthesized for activation of NAD+ biosynthesis genes.
In contrast to SIR2-mediated silencing, which spans large chromosome domains, our transcript arrays suggest that HST1-mediated repression is restricted to specific genes and does not extend to flanking DNA regions (supplemental material). The limited size of deacetylated chromatin may be the reason for the lack of the observed difference in global acetylation states of histones H3 and H4 at the promoter of NAD+ biosynthesis genes between wild-type cells and an hst1 mutant in ChIP experiments. The alternative explanation is that the relevant targets of Hst1p at the promoters of NAD+ biosynthesis genes may be proteins other than histones. However, our biochemical data as well as prior in vivo data (Hst1p can replace Sir2p in deacetylating histones at the silent mating loci in a SUM1-1 mutant [30]) demonstrate that Hst1p is a histone deacetylase.
The comparison of steady-state NAD+ levels in wild-type cells and an hst1 mutant indicates that the derepression of NAD+ biosynthesis and TNA1 transporter genes increases cellular NAD+ levels. However, the degree of augmentation varies (20 to 70%) depending on the availability of the NAD+ precursors NA and tryptophan. The NAD+ levels in wild-type cells grown in media lacking NAD+ precursors are about 20 to 30% lower than those in wild-type cells grown in media supplemented with standard concentrations of NA (3.5 μM) and tryptophan (100 μM). These results suggest that the kynurenine pathway is not entirely repressed in wild-type cells grown in standard concentrations of tryptophan and NA and contributes to the NAD+ supply. Consistent with this idea, disruption of the kynurenine pathway trough BNA2 deletion leads to a significant decrease in cellular NAD+ levels in media lacking tryptophan as well as in complete medium (data not shown). The deletion of HST1 induces an approximately 20% increase in NAD+ levels in cells grown in complete medium. However, when the supply of tryptophan and NA is scarce (and levels of NAD+ are lower), the upregulation of the kynurenine pathway caused by the deletion of HST1 increases cellular NAD+ about 50 to 70%. These levels are similar to hst1 mutants grown in SC medium. The observed plateau of NAD+ levels in hst1 cells grown in SC medium may be due to product inhibition of specific enzymes or other nontranscriptional regulatory mechanisms.
Bernstein et al. (4) proposed a regulatory role for Sir2p in NAD+ biosynthesis on the basis of increased levels of the BNA1 transcript in the sir2 mutant. In their experiments, however, the remaining BNA genes and TNA1 were unaffected by SIR2 deletion. Additionally, deletion of SIR2 has no effect on steady-state NAD+ levels (reference 32 and our results). Our experiments prove that, unlike the SIR2 deletion, HST1 deletion results in upregulation of the majority of de novo NAD+ biosynthesis genes and, more importantly, increases steady-state NAD+ levels.
Our results demonstrating that Hst1p is a cellular NAD+ sensor and a transcriptional regulator of the NAD+ biosynthesis pathway raise the possibility that it may serve similar functions in other cellular processes. The largest group of genes repressed by Hst1p are midsporulation genes. Intriguingly, midsporulation genes are upregulated similarly to NAD+ biosynthesis genes in npt1 mutants (data not shown). The sporulation program is initiated when diploid yeast cells are exposed to unfavorable growth conditions: lack of glucose and nitrogen and a nonfermentable carbon source (reviewed in reference 45). Our preliminary results suggest that transfer of cells to sporulation medium results in a significant drop in NAD+ levels (data not shown). This observation is consistent with a model where a change in the availability of key nutrients results in decreased NAD+ levels which, through modulation of Hst1p activity, serve as an input for the regulatory network that controls progression through meiosis and the sporulation program.
Caloric restriction (CR) extends life span in many species, including rodents, nematodes, fruit flies, and yeast (reviewed in reference 18). In yeast, where CR can be modeled by reducing glucose content of the medium from 2 to 0.5%, SIR2 and an intact NAD+ salvage pathway were shown to be required for CR-induced life span extension (21). NAD+, a major cofactor in various aspects of cellular metabolism, was proposed to serve as a regulatory effector that enabled Sir2p to sense the metabolic rate of cells and link it to transcriptional silencing and longevity. However, the environmental manipulations, such as exposure to low glucose levels (21), or genetic interventions, such as overexpression of NPT1 or other genes in the salvage pathway (1), that led to increased life span were not accompanied by increased total cellular NAD+ levels. Compartmental increase of NAD+ in the nucleus (without a change in total cellular NAD+), increased flux through the salvage pathway, or enhanced removal of the NAD+ metabolites (e.g., nicotinamide) which may inhibit Sir2p deacetylase activity (2, 5) were proposed as explanations for this discrepancy. Our results describing a regulatory circuit that monitors and regulates cellular NAD+ levels suggest that any genetic or environmental manipulation that alters cellular NAD+ might be first encountered and modified by this homeostatic mechanism.
Sir2p and other NAD+-dependent deacetylases comprise a subset of NAD+-dependent transcriptional regulators. In fact CtBP, a transcription cofactor implicated in metazoan development and oncogenesis also binds NAD+ (reviewed in reference 8) and was recently shown to possess NAD+-dependent dehydrogenase activity (19). Furthermore, DNA binding of Clock and NPAS2, heterodimeric transcription factors involved in circadian rhythm control, was shown to be regulated by cellular redox states through the NAD+/NADH ratio (31). Thus, it appears that NAD+ may have inputs into several nuclear pathways. The description of the regulatory circuitry that monitors cellular NAD+ levels presents a step forward in understanding the relationship between cell metabolic state and nuclear functions.
Acknowledgments
We are grateful to D. Gottschling, L. Guarente, and J. Rine for plasmids and yeast strains, members of the Gottschling and Simon laboratories for valuable discussions, and E. Foss, D. Gottschling, F. van Leeuwen, and S. Parkhurst for comments on the manuscript.
This work was supported by a National Heart, Lung and Blood Institute grant (HL04211) and Ellison Medical Foundation Award (to A.B.) and a National Cancer Institute grant (CA78746, to J.S.).
REFERENCES
- 1.Anderson, R. M., K. J. Bitterman, J. G. Wood, O. Medvedik, H. Cohen, S. S. Lin, J. K. Manchester, J. I. Gordon, and D. A. Sinclair. 2002. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J. Biol. Chem. 277:18881-18890. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, R. M., K. J. Bitterman, J. G. Wood, O. Medvedik, and D. A. Sinclair. 2003. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature 423:181-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bedalov, A., T. Gatbonton, W. P. Irvine, D. E. Gottschling, and J. A. Simon. 2001. Identification of a small molecule inhibitor of Sir2p. Proc. Natl. Acad. Sci. USA 98:15113-15118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein, B. E., J. K. Tong, and S. L. Schreiber. 2000. Genomewide studies of histone deacetylase function in yeast. Proc. Natl. Acad. Sci. USA 97:13708-13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bitterman, K. J., R. M. Anderson, H. Y. Cohen, M. Latorre-Esteves, and D. A. Sinclair. 2002. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J. Biol. Chem. 277:45099-45107. [DOI] [PubMed] [Google Scholar]
- 6.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 7.Carson, D. A., S. Seto, and D. B. Wasson. 1987. Pyridine nucleotide cycling and poly(ADP-ribose) synthesis in resting human lymphocytes. J. Immunol. 138:1904-1907. [PubMed] [Google Scholar]
- 8.Chinnadurai, G. 2002. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol. Cell 9:213-224. [DOI] [PubMed] [Google Scholar]
- 9.Dudley, A. M., C. Rougeulle, F. Winston, and A. T. Dudley. 1999. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 13:2940-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gottlieb, S., and R. E. Esposito. 1989. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell 56:771-776. [DOI] [PubMed] [Google Scholar]
- 11.Ha, H. C., and S. H. Snyder. 1999. Poly(ADP-ribose) polymerase is a mediator of necrotic cell death by ATP depletion. Proc. Natl. Acad. Sci. USA 96:13978-13982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hageman, G. J., and R. H. Stierum. 2001. Niacin, poly(ADP-ribose) polymerase-1 and genomic stability. Mutat. Res. 475:45-56. [DOI] [PubMed] [Google Scholar]
- 13.Henderson, L. M.1983. Niacin. Annu. Rev. Nutrit. 3:289-307. [DOI] [PubMed] [Google Scholar]
- 14.Herceg, Z., and Z. Q. Wang. 1999. Failure of poly(ADP-ribose) polymerase cleavage by caspases leads to induction of necrosis and enhanced apoptosis. Mol. Cell. Biol. 19:5124-5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herceg, Z., and Z. Q. Wang. 2001. Functions of poly(ADP-ribose) polymerase (PARP) in DNA repair, genomic integrity and cell death. Mutat. Res. 477:97-110. [DOI] [PubMed] [Google Scholar]
- 16.Imai, S., C. M. Armstrong, M. Kaeberlein, and L. Guarente. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795-800. [DOI] [PubMed] [Google Scholar]
- 17.Jacobson, E. L., and M. K. Jacobson. 1997. Tissue NAD as a biochemical measure of niacin status in humans. Methods Enzymol. 280:221-230. [DOI] [PubMed] [Google Scholar]
- 18.Koubova, J., and L. Guarente. 2003. How does calorie restriction work? Genes Dev. 17:313-321. [DOI] [PubMed] [Google Scholar]
- 19.Kumar, V., J. E. Carlson, K. A. Ohgi, T. A. Edwards, D. W. Rose, C. R. Escalante, M. G. Rosenfeld, and A. K. Aggarwal. 2002. Transcription corepressor CtBP is an NAD+-regulated dehydrogenase. Mol. Cell 10:857-869. [DOI] [PubMed] [Google Scholar]
- 20.Landry, J., A. Sutton, S. T. Tafrov, R. C. Heller, J. Stebbins, L. Pillus, and R. Sternglanz. 2000. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. USA 97:5807-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin, S. J., P. A. Defossez, and L. Guarente. 2000. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289:2126-2128. [DOI] [PubMed] [Google Scholar]
- 22.Lin, S. J., and L. Guarente. 2003. Nicotinamide adenine dinucleotide, a metabolic regulator of transcription, longevity and disease. Curr. Opin. Cell Biol. 15:241-246. [DOI] [PubMed] [Google Scholar]
- 23.Llorente, B. 2000. Transcriptional regulation of the Saccharomyces cerevisiae DAL5 gene family and identification of the high affinity nicotinic acid permease TNA1. FEBS Lett. 475:237-241. [DOI] [PubMed] [Google Scholar]
- 24.Luo, J., A. Y. Nikolaev, S. Imai, D. Chen, F. Su, A. Shiloh, L. Guarente, and W. Gu. 2001. Negative control of p53 by Sir2α promotes cell survival under stress. Cell 107:137-148. [DOI] [PubMed] [Google Scholar]
- 25.Moazed, D. 2001. Enzymatic activities of Sir2 and chromatin silencing. Curr. Opin. Cell Biol. 13:232-238. [DOI] [PubMed] [Google Scholar]
- 26.Natarajan, K., M. R. Meyer, B. M. Jackson, D. Slade, C. Roberts, A. G. Hinnebusch, and M. J. Marton. 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21:4347-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panozzo, C., M. Nawara, C. Suski, R. Kucharczyka, M. Skoneczny, A. M. Becam, J. Rytka, and C. J. Herbert. 2002. Aerobic and anaerobic NAD+ metabolism in Saccharomyces cerevisiae. FEBS Lett. 517:97-102. [DOI] [PubMed] [Google Scholar]
- 28.Passador, L., and W. Iglewski. 1994. ADP-ribosylating toxins. Methods Enzymol. 235:617-631. [DOI] [PubMed] [Google Scholar]
- 29.Rosenberg, M. I., and S. M. Parkhurst. 2002. Drosophila Sir2 is required for heterochromatic silencing and by euchromatic Hairy/E(Spl) bHLH repressors in segmentation and sex determination. Cell 109:447-458. [DOI] [PubMed] [Google Scholar]
- 30.Rusche, L. N., and J. Rine. 2001. Conversion of a gene-specific repressor to a regional silencer. Genes Dev. 15:955-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rutter, J., M. Reick, L. C. Wu, and S. L. McKnight. 2001. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science 293:510-514. [DOI] [PubMed] [Google Scholar]
- 32.Sandmeier, J. J., I. Celic, J. D. Boeke, and J. S. Smith. 2002. Telomeric and rDNA silencing in Saccharomyces cerevisiae are dependent on a nuclear NAD+ salvage pathway. Genetics 160:877-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sauve, A. A., I. Celic, J. Avalos, H. Deng, J. D. Boeke, and V. L. Schramm. 2001. Chemistry of gene silencing: the mechanism of NAD+-dependent deacetylation reactions. Biochemistry 40:15456-15463. [DOI] [PubMed] [Google Scholar]
- 34.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 35.Sies, H. 1982. Metabolic compartmentation. Academic Press, London, England.
- 36.Sinclair, D. A., and L. Guarente. 1997. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell 91:1033-1042. [DOI] [PubMed] [Google Scholar]
- 37.Smith, J. S., J. Avalos, I. Celic, S. Muhammad, C. Wolberger, and J. D. Boeke. 2002. SIR2 family of NAD+-dependent protein deacetylases. Methods Enzymol. 353:282-300. [DOI] [PubMed] [Google Scholar]
- 38.Smith, J. S., and J. D. Boeke. 1997. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 11:241-254. [DOI] [PubMed] [Google Scholar]
- 39.Smith, J. S., C. B. Brachmann, I. Celic, M. A. Kenna, S. Muhammad, V. J. Starai, J. L. Avalos, J. C. Escalante-Semerena, C. Grubmeyer, C. Wolberger, and J. D. Boeke. 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. USA 97:6658-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sutton, A., R. C. Heller, J. Landry, J. S. Choy, A. Sirko, and R. Sternglanz. 2001. A novel form of transcriptional silencing by Sum1-1 requires Hst1 and the origin recognition complex. Mol. Cell. Biol. 21:3514-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanner, K. G., J. Landry, R. Sternglanz, and J. M. Denu. 2000. Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc. Natl. Acad. Sci. USA 97:14178-14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanny, J. C., and D. Moazed. 2001. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: evidence for acetyl transfer from substrate to an NAD breakdown product. Proc. Natl. Acad. Sci. USA 98:415-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tissenbaum, H. A., and L. Guarente. 2001. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410:227-230. [DOI] [PubMed] [Google Scholar]
- 44.Vaziri, H., S. K. Dessain, E. Ng Eaton, S. I. Imai, R. A. Frye, T. K. Pandita, L. Guarente, and R. A. Weinberg. 2001. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107:149-159. [DOI] [PubMed] [Google Scholar]
- 45.Vershon, A. K., and M. Pierce. 2000. Transcriptional regulation of meiosis in yeast. Curr. Opin. Cell Biol. 12:334-339. [DOI] [PubMed] [Google Scholar]
- 46.Voet, D., and J. G. Voet. 1995. Biochemistry, p. 334-337. John Wiley & Sons, New York, N.Y.
- 47.Vogelauer, M., J. Wu, N. Suka, and M. Grunstein. 2000. Global histone acetylation and deacetylation in yeast. Nature 408:495-498. [DOI] [PubMed] [Google Scholar]
- 48.Wilson, W. A., and P. J. Roach. 2002. Nutrient-regulated protein kinases in budding yeast. Cell 111:155-158. [DOI] [PubMed] [Google Scholar]
- 49.Xie, J., M. Pierce, V. Gailus-Durner, M. Wagner, E. Winter, and A. K. Vershon. 1999. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 18:6448-6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, Q., D. W. Piston, and R. H. Goodman. 2002. Regulation of corepressor function by nuclear NADH. Science 295:1895-1897. [DOI] [PubMed] [Google Scholar]