Abstract
The androgen receptor (AR) binds to and activates transcription of target genes in response to androgens. In an attempt to isolate cofactors capable of influencing AR transcriptional activity, we used an immunoprecipitation method and identified a 44-kDa protein, designated p44, as a new AR-interacting protein. p44 interacts with AR in the nucleus and with an androgen-regulated homeobox protein (NKX3.1) in the cytoplasm of LNCaP cells. Transient-transfection assays revealed that p44 enhances AR-, glucocorticoid receptor-, and progesterone receptor-dependent transcription but not estrogen receptor- or thyroid hormone receptor-dependent transcription. p44 was recruited onto the promoter of the prostate-specific antigen gene in the presence of the androgen in LNCaP cells. p44 exists as a multiprotein complex in the nuclei of HeLa cells. This complex, but not p44 alone, enhances AR-driven transcription in vitro in a cell-free transcriptional system and contains the protein arginine methyltransferase 5, which acts synergistically with p44 to enhance AR-driven gene expression in a methyltransferase-independent manner. Our data suggest a novel mechanism by which the protein arginine methyltransferase is involved in the control of AR-driven transcription. p44 expression is dramatically enhanced in prostate cancer tissue compared with adjacent benign prostate tissue.
The androgen receptor (AR) mediates androgen function in the development and maintenance of normal prostate tissue (4). The growth and progression of prostate cancer are also dependent on AR. AR is a member of the nuclear receptor superfamily and, like other members of this family, contains a central DNA-binding domain (DBD), a C-terminal ligand-binding domain (LBD) with an associated activation function (AF-2) activation domain, and an N-terminal domain (NTD) containing the AF-1 activation domain (9, 20). On ligand binding, AR dissociates from heat shock proteins and chaperones, dimerizes, binds to cognate androgen response elements (AREs) in target genes, and, through its AF-1 and AF-2 domains, interacts with various coactivators that facilitate transcription by the general transcriptional machinery (9, 20). As demonstrated in studies of other activators, gene activation by AR is thought to require the general initiation factors that form preinitiation complexes on common core promoter elements (e.g., TATA) (44) and a variety of general and gene-specific coactivators that either modulate chromatin structure (26, 36) or serve as direct adaptors between activators and general initiation factors (43). A variety of cofactors have been implicated more directly in nuclear receptor function (32, 33, 54). On a growing list of cofactors that regulate nuclear receptors are the well-studied coactivators of p300/CBP, the p160 family (SRC-1, TIF-2/GRIP-1, ACTR/P-CIP) (54), p300/CREB-binding protein associated factor/GCN5 complexes (yeast SAGA and human STAGA) (5, 31), and protein arginine methyltransferases (PRMTs) (48). These cofactors have histone acetyltransferase or PRMT activities and are believed to act mainly through histone acetylation or methylation and subsequent chromatin structural perturbations but can also act through functional modification of activators (21) and coactivators (10, 55). Some exhibit ligand-dependent interactions with the AF-2 domain of receptors, whereas others interact with the AF-1 domains. The multiprotein thyroid hormone receptor-associated protein (TRAP)/Mediator complexes exhibit no intrinsic histone acetyltransferase activity (30) and show subunit-specific interactions with both nuclear receptors (TRAP220 with thyroid hormone receptor [TR], vitamin D receptor, peroxisome proliferator-activated receptor, retinoic acid receptor, retinoid X receptor, and estrogen receptor [ER] and TRAP170/vitamin D receptor-interacting protein 150 with glucocorticoid receptor [GR]) and other activators (TRAP80 with p53 and VP16). This complex, in turn, interacts with the general initiation factors and polymerase II (Pol II) and acts on DNA templates at post-chromatin-remodeling steps. Of these coactivators, p300/CBP and p160s have been shown to function with AR (1, 3, 6, 23, 29), but functions of the multicomponent STAGA (∼15 subunits) and TRAP (∼25 subunits) complexes with AR are also likely (51). Other cofactors that have been implicated in the function of AR and, in most cases, other nuclear receptors include the ARA group, ARIP3, SNURF, FHL2, cyclin D1, and AES (22, 24). Some of these factors have broader effects on basal transcription and other activators, but less is known about their mechanistic function.
Homeoboxes are conserved 61-amino-acid DBDs present in a distinct family of transcription factors, the homeodomain proteins, that play a central role in eukaryotic development, with spatial and temporal specificity (19). Consistent with their role in cell growth and differentiation, homeobox gene dysfunctions have been implicated in tumorigenesis (12). NKX3.1 is a newly discovered prostate tissue-specific and androgen-regulated gene in the homeobox gene family (41). NKX3.1 is most closely related, by virtue of 78% sequence similarity with the homeodomain region, to Drosophila NK-3. NK-3 interacts with the corepressor Groucho through the homeodomain region to repress transcription (11). Consistent with its sequence similarity to NK-3, NKX3.1 has been shown to specifically repress transcription of a luciferase reporter containing three copies of the NKX3.1-binding site upstream of a thymidine kinase core promoter (49). The chromosomal association of the NKX3.1 gene on 8p21, a region frequently deleted in prostate cancer, suggests that NKX3.1 may function as a tumor suppressor (8). Consistent with these findings, the results of recent studies of NKX3.1 knockout mice suggest that NKX3.1 exerts a growth-suppressive effect on prostate epithelial cells and controls differentiated glandular functions (2, 7, 25, 45). These findings suggest that, as a transcription factor, NKX3.1 may play an important role in prostate cell development, cell differentiation, and tumorigenesis, even though the biological and biochemical functions of NKX3.1 remain to be deciphered.
In this study, we identified a new AR-associated protein (p44) that interacts with AR directly and enhances AR-driven gene expression in vivo. We also demonstrated that in the nuclei of HeLa cells, p44 forms a multiprotein complex that functions as a coactivator of AR.
MATERIALS AND METHODS
Establishment of prostate cell lines that stably expressed a FLAG-tagged AR or NKX3.1 and immunopurification of f:AR- and f:NKX3.1-associated factors.
The mammalian expression vectors pBabe-Neo-f:AR and pBabe-Neo-f:NKX3.1 were created by subcloning FLAG-tagged human AR or NKX3.1 cDNA into the vector pBabe-Neo. The prostate cancer cell line LNCaP was purchased from the American Type Culture Collection (ATCC; Manassas, Va.) and maintained in RPMI 1640 medium plus 10% fetal bovine serum. Cells were transfected with pBabe-Neo-f:AR or pBabe-Neo-f:NKX3.1 and further incubated at 37°C for 1 to 1.5 days before being split 1:6 for G418 selection (0.5 mg/ml). The medium was changed every 3 or 4 days. Individual G418-resistant colonies, normally seen after 2 weeks, were expanded into cell lines and then characterized by Western blotting using the anti-FLAG monoclonal antibody M2. The cell lines expressing FLAG-tagged AR or FLAG-tagged NKX3.1 were further expanded and analyzed. Nuclear and cytoplasmic extracts were prepared according to our standard methods (53) and used to immunopurify the AR- or NKX3.1-containing complexes. Typically, 1 ml of nuclear or cytoplasmic extract was mixed with 20 μl of M2 resin and incubated for 3 h at 4°C with rotation. After five washes in a buffer containing 20 mM HEPES (pH 7.9), 0.2 mM EDTA, 20% glycerol, 2 mM dithiothreitol, 30 mM KCl, and 0.1% NP-40, the bound proteins were eluted from the M2 agarose by incubation at 4°C for 30 min with 20 μl of the same buffer plus 0.2 mg of the FLAG peptide (Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys) per ml. Aliquots (5 μl) of eluted proteins were mixed with equal volumes of 2× Laemmli's sample buffer and loaded onto a 10% polyacrylamide-sodium dodecyl sulfate (SDS) gel. Proteins were visualized by silver staining.
In vitro transcription and primer extension.
The basal transcription factors TFIIA, TFIIB, TFIIE, and TFIIF and PC4 were expressed in and purified from bacteria. TFIID, TFIIH, and RNA Pol II were affinity purified from stable cells expressing the corresponding FLAG-tagged subunits (56). Transcription reactions were carried out in a final volume of 25 μl and contained 90 fmol of supercoiled plasmid DNA template; the products were analyzed by the primer extension reaction as described previously (56).
cDNA cloning and Northern blot analysis.
An immunopurified f:NKX3.1-containing complex was subjected to SDS-polyacrylamide gel electrophoresis (PAGE), and peptides derived from p44 were subjected to mass spectrometric analysis (37). An expression sequence tag clone (IMAGE:785275) encoding full-length p44 was obtained from the ATCC. A 1.3-kb cDNA encoding full-length p44 was labeled with 32P using the random primed DNA labeling kit (Boehringer Mannheim GmbH) and was used to probe human multiple-tissue Northern blot membranes (BD Biosciences Clontech, Palo Alto, Calif.).
Expression and purification of recombinant proteins and antibody preparation.
Recombinant human AR was expressed in Sf9 cells via baculovirus vector pVL1393 as a FLAG-tagged fusion protein and purified on M2 agarose (56). His6-tagged p44 was expressed in bacteria via the expression vector pET15d and purified by affinity (nickel-nitrilotriacetic acid [Ni-NTA] agarose) and S-Sepharose chromatographic steps. The cDNA encoding amino acid residues 1 to 282 of human AR was subcloned into vector pET15d and expressed in bacteria. The His6-tagged AR(1-282) protein was purified through a Ni-NTA agarose column. Ten milligrams of the purified recombinant His6-tagged p44 and AR(1-282) proteins was sent to Convance Inc. (Denver, Pa.) for polyclonal antibody production in rabbits. The antisera were purified through the p44 and AR(1-282) agarose columns, respectively.
Transient transfection.
The AR, ER, GR, progesterone receptor (PR), TR, and p44 expression vectors for transfection assays were constructed by inserting their corresponding cDNA sequences into pcDNA3.1. The luciferase reporters contain the androgen, estrogen, or thyroid hormone response elements ahead of the E4 basal promoter and the luciferase gene, respectively. PC3 cells were maintained in RPMI 1640 medium plus 10% fetal bovine serum. Transfections were performed with Lipofectamine reagent (Invitrogen, Carlsbad, Calif.). Briefly, 105 cells were plated onto each well of 24-well plates approximately 24 h before transfection. After being washed with phosphate-buffered saline, cells in each well were transfected with 30 ng of an expression vector (AR, ER, GR, PR, or TR), 100 ng of the reporter plasmids, 2.5 ng of the pR-LUC internal control plasmid, and different amounts of the p44 expression vector. The total amount of DNA was adjusted to 300 ng with pcDNA3.1. Transfections were conducted in phenol-free RPMI 1640 medium; 2 h later, the medium was changed to either phenol-free RPMI 1640 plus charcoal-treated fetal bovine serum (10%) or regular medium containing 10 nM R1881, 10 nM dexamethasone, 10 nM progesterone, 1 μM β-estradiol, or 10 nM T3. Cells were cultured for another 48 h and harvested for the dual luciferase assay (Promega, Madison, Wis.).
Protein-protein interaction assay.
One microgram of recombinant glutathione S-transferase (GST) and GST fusion proteins (GST-p44, GST-NTD, GST-DBD, and GST-LBD) were expressed in bacteria and immobilized on 20 μl of glutathione-Sepharose beads. The beads were incubated with 5 μl of rabbit reticulocyte lysate containing 35S-labeled AR, NKX3.1, or PRMT5 in a final volume of 200 μl containing 20 mM HEPES (pH 7.9), 0.2 mM EDTA, 20% glycerol, 2 mM dithiothreitol, 150 or 300 mM KCl, and 0.1% NP-40. The beads were washed five times (1 ml each) with the incubation buffer, boiled in 20 μl of the SDS gel sample buffer, and analyzed by SDS-PAGE followed by autoradiography.
ChIP.
LNCaP cells were grown in phenol red-free RPMI 1640 supplemented with charcoal-dextran-stripped fetal bovine serum (10%) for 2 days and then treated with 1 nM R1881 for 16 h. Cells treated with ethanol were used as the control. Chromatin immunoprecipitation (ChIP) was performed as described previously (39) with the following modifications. Cells were cross-linked with 1% formaldehyde at room temperature for 10 min and the cross-linking reaction was stopped by addition of glycine to 0.125 M. The cross-linked chromatin was sonicated with a Branson Sonifier 450 microtip at power setting 6 for five 30-s bursts separated by cooling on ice. This treatment produced DNA fragments of average size of 700 bp. For immunoprecipitation, 2 μg of antigen-purified anti-AR or anti-p44 antibody was mixed with 300 μg of the purified cross-linked chromatin and incubated overnight at 4°C. Immunocomplexes were washed five times (10 min each) in 1 ml of the buffer containing 1% Triton X-100, 0.1% sodium deoxycholate, 0.05% SDS, 140 mM NaCl, and 1 mM phenylmethylsulfonyl fluoride; once in a solution containing 0.25 M LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA, and 10 mM Tri-HCl, pH 8.0; and twice in 1 mM EDTA-10 mM Tris-HCl, pH 8.0. After reversal and recovery of the immunoprecipitated chromatin DNA, the final DNA pellets were dissolved with 50 μl of H2O. Immunopurified DNA (5 μl) was used for a PCR (30 cycles, annealing at 50°C), with primers as follows. For prostate-specific antigen (PSA), the forward primer sequence was TCTGCCTTTGTCCGCTAGAT and the reverse primer sequence was AACCTTCATTCCCCAGGACT, which amplifies a 212-bp product from −250 to −39 upstream of the PSA transcription start site. For β-actin, the forward primer sequence was TCCTCCTCTTCCTCAATCTCG and the reverse primer sequence was AAGGCAACTTTCGGAACGG, which amplifies a 145-bp product from −118 to −974 of the β-actin gene (the A of the ATG translation start codon was arbitrarily given the number +1).
Methylation of proteins.
A cDNA (IMAGE:3833019) encoding the full-length human PRMT5 was purchased from the ATCC. The point mutant (R368A) was created by using a QuikChange site-directed mutagenesis kit (Stratagene) following the manufacturer's instructions. The mutation was confirmed by DNA sequencing analysis. The wild-type and mutant PRMT5 were expressed in bacteria via pET15d expression vector and purified through Ni2+-NTA agarose. The methylation assay was performed as follows. Two micrograms of the purified histones (27) was incubated with 0.8 μg of the purified recombinant wild-type or mutant PRMT5 and various factors in 25 μl of 50 mM Tris, pH 7.5-1 mM EDTA-1 mM EGTA-20 μCi of [3H]AdoMet (Amersham Pharmacia Biotech) at 30°C for 30 min. The reactions were stopped by the addition of 5 μl of 5× SDS sample loading buffer, and samples were resolved by SDS-15% PAGE. The gels were stained with Coomassie blue R250, destained, treated with an intensifying solution, and analyzed by autoradiography.
In situ hybridization.
We used matched normal and cancerous prostate tissues derived from radical prostatectomies of patients with prostate cancer at New York University Medical Center in an institutional review board-approved protocol. The procedure for in situ hybridization was as described previously (28). Briefly, the sections were hydrated, postfixed in 4% paraformaldehyde, and treated with proteinase K followed by deacetylation. The prehybridization and hybridization treatments were performed at 68°C using 0.3 M NaCl and 50% formamide. DNA fragments (500 bp, cDNA sequences of p44 from 1 to 500) containing both T7 and T3 promoters were generated by PCR. Corresponding 33P-labeled RNAs (sense and antisense) were generated by in vitro transcription with T7 and T3 RNA polymerases, respectively, and hybridized to the tissue sections (4 μm). After being washed, the slides were exposed to NTB-2 X-ray emulsion (Eastman-Kodak, Rochester, N.Y.) for 2 to 3 weeks and counterstained with hematoxylin-eosin. Image and statistical analyses were performed as described previously (28).
RESULTS
Immunopurification and functional analysis of FLAG-AR-associated factors.
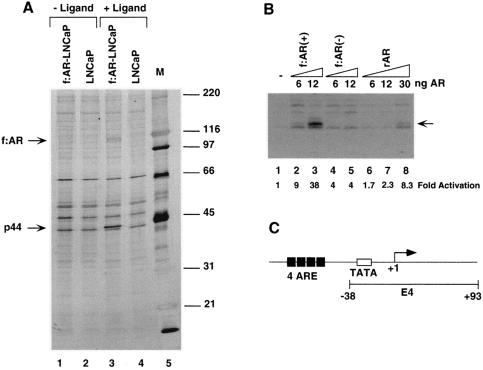
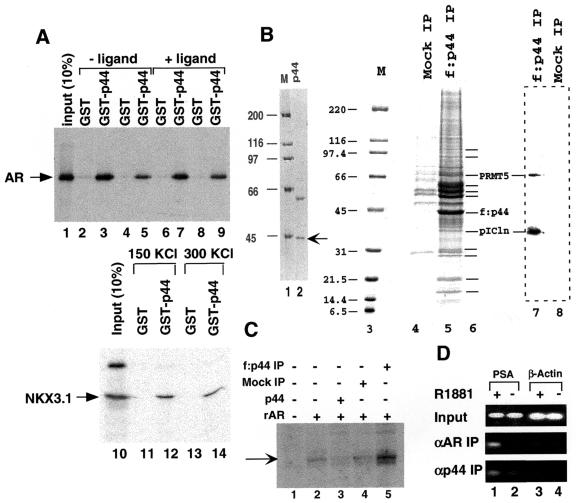
Stable cell lines expressing FLAG-epitope-tagged factors were generated previously and used to affinity purify corresponding parental complexes (52, 53, 56). To similarly identify androgen-dependent or androgen-independent AR-interacting factors, we generated a prostate cancer cell line (f:AR-LNCaP) that stably expresses a FLAG-tagged AR. Immunopurification of f:AR from nuclear extracts made from f:AR-LNCaP cells grown in the presence and absence of androgen (R1881) revealed androgen (R1881)-induced association of a 44-kDa f:AR-associated polypeptide (Fig. 1A, lane 3). The specific association of this polypeptide with f:AR is further shown by the failure of similar-sized polypeptides in extracts from control cells (not expressing f:AR) to bind to the affinity matrix (Fig. 1A, lane 4).
FIG. 1.
Effects of affinity-purified f:AR-cofactor complexes on transcription in a system reconstituted with purified factors. (A) SDS-PAGE analysis of f:AR-cofactor complexes. Lanes 1 and 3 show AR-containing complexes immunopurified from nuclear extracts made from a stably transfected, FLAG-tagged, AR-expressing LNCaP cell line grown in the presence (lane 3) or absence (lane 1) of the synthetic androgen R1881 (10 nM). The gel was stained with silver. Bands corresponding to FLAG-tagged AR (f:AR) and to the polypeptide specifically associated with AR are indicated by arrows. (B) AR- and AR-cofactor-dependent transcription. A synthetic template containing four ARE elements (C) was transcribed in the system reconstituted with purified factors (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, Pol II, and PC4) with additions of the rAR and f:AR-cofactor complexes described for panel A. The specifically initiated transcript is indicated by an arrow and was monitored by primer extension. The activation relative to levels of transcription in the absence of rAR or f:AR complexes (lane 1) is indicated at the bottom. (C) Diagram of the synthetic ARE-containing template. The template (pARE-E4) contains four tandem copies of the ARE from the PSA promoter positioned upstream of the adenovirus E4 promoter.
When assayed in the purified minimal transcription system containing TFIIA, TFIIB, TFIID, TFIIH, Pol II, and PC4 with the synthetic ARE-containing template (56) (Fig. 1C), recombinant AR (rAR) alone elicited up to 8.3-fold activation (Fig. 1B, lanes 6 to 8). The f:AR complex from nuclear extract derived from f:AR-expressing LNCaP cells grown in the presence of R1881 [f:AR(+)] elicited up to 38-fold activation (Fig. 1B, lanes 2 and 3) and at an equimolar input was 16-fold more active than rAR (Fig. 1B, lane 3 versus lane 7). In contrast, the f:AR complex from nuclear extract of f:AR-expressing LNCaP cells grown in the absence of R1881 [f:AR(−)] showed a level of activity only about 2-fold above that shown by rAR (Fig. 1B, lanes 4 and 5 versus lanes 6 and 7). The amounts (indicated in Fig. 1B) of recombinant AR and f:AR in the f:AR-containing complexes were normalized by quantitative Western blot analysis with anti-AR polyclonal antibodies. These results indicate that polypeptides associated with AR specifically in the presence of androgen can upregulate AR function; a likely candidate is the 44 kDa protein, although other minor polypeptides could also be responsible. When the cell line was further expanded, the 44-kDa polypeptide became less abundant, and an attempt to isolate a sufficient amount of the 44-kDa polypeptide for peptide sequence analysis failed.
Purification, cloning, and characterization of FLAG-NKX3.1-interacting proteins.
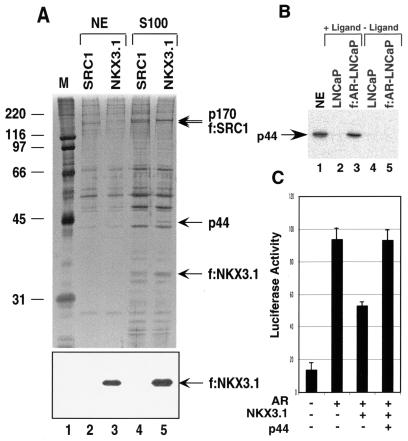
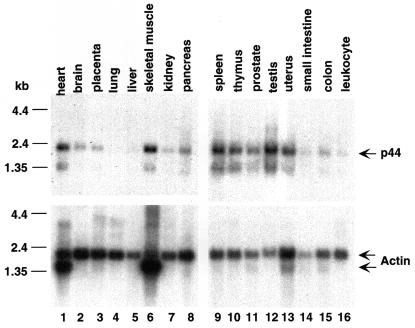
We similarly generated a prostate cancer cell line (f:NKX3.1-LNCaP) that stably expresses a FLAG-tagged NKX3.1. Immunoprecipitation of f:NKX3.1 from extracts isolated from f:NKX3.1-LNCaP cells revealed 44- and 170-kDa polypeptides that specifically associate with NKX3.1 in the cytoplasm (Fig. 2A, lane 5), but not with NKX3.1 in the nucleus (Fig. 2A, lane 3). The specific association of these polypeptides with f:NKX3.1 is further shown by the failure of similar-sized polypeptides in extracts from control cells (expressing f:SRC1) to bind to the affinity matrix (Fig. 2A, lane 4). Because f:NKX3.1 stained negatively with silver, Western blot analysis with the anti-FLAG antibody was employed to demonstrate the existence of f:NKX3.1 in immunoprecipitates derived from both nuclear (Fig. 2A, bottom panel, lane 3) and cytoplasmic (Fig. 2A, bottom panel, lane 5) extracts. After larger amounts of f:NKX3.1-associated 44-kDa protein had accumulated, we performed direct sequence analysis by mass-spectrometric methods (37). On the basis of the peptide sequence (KETPPPLVPPAAR) obtained from mass-spectrometric analysis, we obtained a cDNA encoding the 44-kDa protein. The p44 cDNA encodes a protein containing 342 amino acid residues and four putative WD-40 repeats (residues 68 to 107, 114 to 153, 157 to 196, and 280 to 319). p44 is identical in sequence to the recently identified MEP50 component of the methylosome (18) and to the WD45 subunit of the survival motor neuron (SMN) complex (35). Northern blot analysis of multiple human tissues showed that p44 mRNA is highly expressed in the heart, skeletal muscle, spleen, testis, uterus, prostate, and thymus (Fig. 3). Western blot analysis with anti-p44 antibody revealed that the f:AR preparation immunopurified from the f:AR-LNCaP cell line (Fig. 1A, lane 3) contains the same 44-kDa polypeptide (Fig. 2B, lane 3).
FIG. 2.
p44 associates with NKX3.1 in the cytoplasm. (A) SDS-PAGE analysis of purified f:NKX 3.1-containing complex. Immunoprecipitation was performed with nuclear extracts (NE) (lane 3) and cytoplasmic extracts (lane 5) made from a stably transfected, FLAG-tagged, NKX3.1-expressing cell line. Bands corresponding to FLAG-tagged NKX3.1 and polypeptides specifically associated with NKX3.1 (p44 and p170) are indicated by arrows. The specific association of these polypeptides with f:NKX3.1 is further shown by the failure of similarly sized polypeptides in extracts from control cells (expressing f:SRC1) to bind to the affinity matrix (lanes 2 and 4). Lane 1, standard molecular weight markers (Bio-Rad). The bottom panel is a Western blot of the same samples with anti-FLAG monoclonal antibody. (B) Western blot analysis of the f:AR-complexes with anti-p44 antibody. Lane 1 contains 5 μl of nuclear extract made from LNCaP cells. (C) NKX3.1 partially represses AR-dependent gene expression, and the overexpression of p44 relieves this repression. PC3 cells were transfected with 100 ng of 4× ARE-E4-luc reporter plasmid, 30 ng of pcDNA-AR, 60 ng of pcDNA NKX3.1, and 100 ng of p44, as indicated. Cells were grown in the presence of 10 nM R1881 for 48 h after transfection and then harvested for luciferase activity assays.
FIG. 3.
Northern blot analysis of p44 expression. The membranes were probed with 32P-labeled p44 cDNA (top) and 32P-labeled β-actin cDNA (bottom).
p44 specifically enhances AR-dependent transcription in vivo.
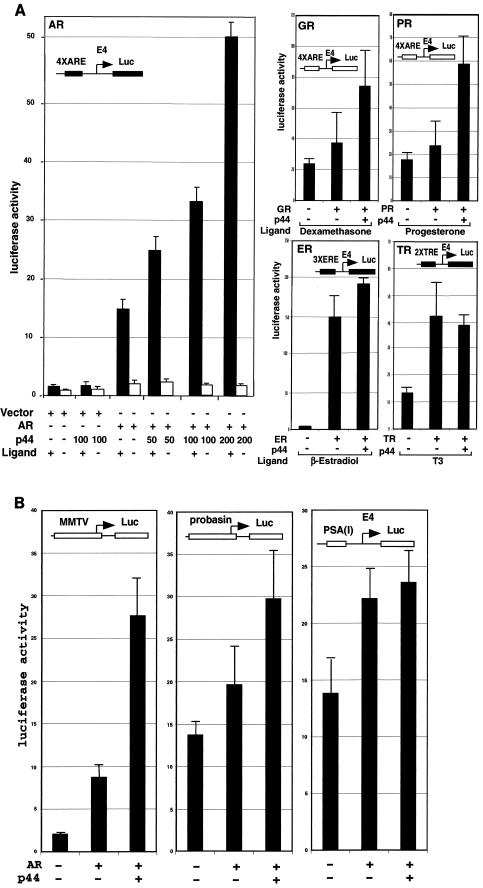
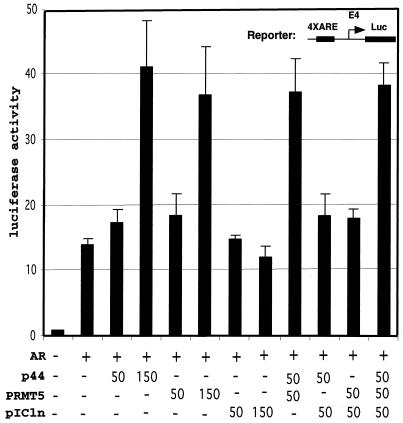
To investigate the effect of p44 on AR-dependent transcription in vivo, a luciferase reporter (4× ARE-E4-Luc) containing four tandem PSA promoter AREs (−152 to −174) (14) and the minimal adenovirus E4 promoter (−38 to + 93) upstream of the luciferase gene was cotransfected with expression vectors for AR and p44 into PC3 cells in the absence or presence of ligand (R1881). As shown in Fig. 4A, AR activated the reporter gene about 12-fold in the presence of ligand, and coexpressed p44 showed a strong (up to 3.3-fold) enhancement of this activity that is likely restricted in magnitude by contributions from endogenous p44 (data not shown). p44 did not influence reporter gene activity in the absence of cotransfected AR or ligand (R1881), indicating that the enhancing effect of p44 on AR-dependent gene expression was caused by an effect on the E4 promoter. To investigate the receptor specificity of p44, we examined the effects of p44 on transcription of reporters containing the same E4 core promoter under the control of GR, PRb, ERα, and TR. As shown in Fig. 4A, p44 also enhanced GR- and PR-driven gene expression and, in contrast, showed no effect on TR- or ER-mediated transcription. Hence, p44 shows some nuclear receptor-specific effects in vivo. p44 also enhanced AR-driven transcription from natural ARE-containing mouse mammary tumor virus (15) and probasin (−244 to + 12) (13) promoters but had no obvious effects on the promoter derived from the PSA enhancer (−4354 to −3858) (46) (Fig. 4B). These results suggest that p44 has promoter specificity.
FIG.4.
p44 specifically enhances AR-mediated transcription in vivo. (A) p44 enhanced AR-, GR-, and PR-mediated transcription. PC3 cells were transfected with 100 ng of 4× ARE-, 3× ERE-, or 2× TRE-E4-luc reporter plasmid, 30 ng of pcDNA-AR, -GR, -PR, -ER, or -TR, and the indicated amounts of pcDNA-p44 expression plasmid. Cells were grown in the absence or presence of 10 nM R1881, 10 nM dexamethasone, 10 nM progesterone, 1 μM estradiol, or 10 nM T3 for 48 h after transfection and then harvested for luciferase activity assays. (B) p44 selectively affected AR-mediated luciferase gene expression from different promoters. PC3 cells were transfected with 100 ng of MMTV-, probasin-, or PSA(I)-luc reporter plasmid, 30 ng of pcDNA-AR, and 150 ng of pcDNA-p44 expression plasmid. Cells were grown in the presence of 10 nM R1881 for 48 h after transfection and then harvested for luciferase activity assays.
p44 interacts directly with AR and NKX3.1.
Consistent with the observed intracellular association of p44 with f:NKX3.1 (Fig. 2A, lane 5), p44 interacted directly and strongly with NKX3.1 in a salt-insensitive manner in a GST pull-down assay (Fig. 5A, lanes 10 to 14). Recombinant AR was also found to interact strongly (but in a salt-sensitive manner) with a GST-p44 fusion protein but not with GST alone (Fig. 5A, lanes 1 to 9), which was consistent with the intracellular association of f:AR with p44 (Fig. 1A, lane 3). The ligand-independent in vitro interaction of purified p44 with purified AR contrasts with the ligand-dependent intracellular association of p44 with AR. However, other AR-associated proteins (such as heat shock proteins) whose interactions are reversed by androgen, thus facilitating p44 interaction, may explain this dependency. Our identification of a common interacting protein (p44) prompted studies of the effect of NKX3.1 on AR function, which showed that overexpression of NKX3.1 represses AR-driven gene expression in vivo (Fig. 2C). One possible explanation is that NKX3.1 may sequester p44 in the cytoplasm, thus inhibiting AR-driven gene expression. Overexpression of ectopic p44 was shown to completely overcome the NKX3.1-mediated repression, which supports this theory (Fig. 2C). Since p44 enhances AR activity in either the presence (in LNCaP cells) or absence (in PC3 cells) of NKX3.1, the mechanism of enhancement by p44 may be completely independent of NKX3.1 action.
FIG. 5.
p44-containing complex enhances AR-driven transcription. (A) p44 interacts directly with AR and NKX3.1. GST-p44 fusion protein expressed in bacteria was immobilized on glutathione agarose beads. Beads were incubated with 35S-labeled AR (lanes 1 to 9) or NKX3.1 (lanes 10 to 14) in BC150-0.1% NP-40 (lanes 2, 3, 6, 7, 11, and 12) or BC300-0.1% NP-40 (lanes 4, 5, 8, 9, 13, and 14) in the absence (lanes 2 to 5 and 10 to 14) or presence (lanes 6 to 9) of 50 nM R1881 for 2 h at 4°C. After being washed with the incubation buffer, the beads were boiled with SDS sample buffer and subjected to SDS-PAGE followed by autoradiography. (B) SDS-PAGE analysis of purified p44 and p44-containing complexes. Lane 2 shows recombinant p44 expressed in bacteria and purified on an Ni-NTA agarose affinity column; lanes 5 and 4 show p44-containing complexes immunopurified from nuclear extracts made from a stably transfected, FLAG-tagged, p44-expressing HeLa cell line and immunoprecipitate from extracts made from control cells (not expressing f:p44), respectively. The gels were stained with Coomassie blue R250. The band corresponding to p44 is indicated by the arrow at the right. Polypeptides specifically associated with p44 are indicated by short lines at the right (lane 6). Lanes 1 and 3, standard molecular weight markers (Bio-Rad); lanes 7 and 8, Western blot analysis of the immunoprecipitate isolated from p44-expressing cells (lane 7) and control cells (lane 8) using anti-PRMT5 and anti-pICln antibodies. (C) The p44-containing complex enhances AR-dependent transcription. A synthetic template, pARE-E4, was transcribed in the system reconstituted with purified factors (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, Pol II, and PC4) with additions of rAR, p44, and the f:p44-containing complex described for panel B. The specifically initiated transcript is indicated by an arrow and was monitored by primer extension. (D) p44 was recruited on the PSA promoter in the presence of the androgen. LNCaP cells were grown in the absence (lanes 2 and 4) or presence (lanes 1 and 3) of 1 nM R1881. A ChIP assay was performed with antigen-purified anti-AR (middle panel) or anti-p44 (top panel) antibodies. The purified-protein-DNA cross-links were reversed, and the resulting DNA was amplified by a PCR with two specific primers derived from promoter regions of PSA (lanes 1 and 2) or β-actin (lanes 3 and 4). The same set of PCRs (top panel) was performed with chromatin DNA (Input) used for the ChIP assay.
p44-containing complex enhances AR-driven transcription in vitro.
To further explore the function and regulation of p44, we established a HeLa cell line that stably expresses FLAG-tagged p44 and used it to immunopurify p44-containing complexes. SDS-PAGE analysis revealed a large number of polypeptides (more abundant in the 50- to 60-kDa range and less abundant in the 70- to 100-kDa and 10- to 30-kDa ranges) (Fig. 5B, lane 5). The recombinant p44 expressed in bacteria (Fig. 5B, lane 2) inhibited AR-dependent transcription from the synthetic ARE-E4 promoter (Fig. 5C, lane 3 versus lane 2). This repression contrasted with the activation observed in vivo (transient transfection) (Fig. 4). One possible reason for this is that p44 activation of AR-driven gene expression requires additional factors. In the absence of these factors in the in vitro-reconstituted transcription system, p44 might sequester some transcription factors (such as AR) through direct interactions and thus repress transcription. This hypothesis is supported by our finding that the p44-containing complex purified from the FLAG-tagged stable cell line enhanced AR-driven transcription from the same promoter (Fig. 5C, lane 5). The p44-containing complex did not affect GAL4-VP16-driven transcription in the same in vitro transcription system (data not shown).
The occupancy of specific DNA sites by specific DBDs (e.g., AR) and associated proteins can be established by the ChIP assay (38). This assay is a direct and powerful method of assessing in vivo protein-DNA interactions. AR binds to the PSA promoter region in the presence of androgen (R1881) (Fig. 5D, middle panel, lane 1). As negative controls, the products amplified by PCR at the same time from the β-actin promoter were not changed in response to the addition of androgen (Fig. 5D, middle panel, lanes 3 and 4). This observation is consistent with the fact that the PSA promoter is directly targeted by AR (57). We performed this assay more than 10 times and consistently observed the androgen-dependent recruitment of AR to the PSA proximal promoter in LNCaP cells. Cofactors can also be cross-linked by formaldehyde treatment to chromatin through their interactions with DNA-binding factors in living cells. Therefore, the ChIP assay is also a direct way to determine cofactor occupancy on AR target genes. Figure 5D shows the androgen-dependent recruitment of p44 onto the PSA promoter (bottom panel, lane 1 versus lane 2). The ChIP assay with anti-p44 antibody was independently performed twice, and the results were consistent. We found that when larger amounts (twofold) of DNA were used in our standard PCRs (30 cycles), we still observed the androgen-dependent recruitment of p44 to the PSA promoter, although the background was slightly higher. However, fewer amplification cycles (27 cycles) in the PCR gave better results when the larger amounts of DNA were used. These results suggest that p44 functions on the AR target gene in vivo.
Others have reported that MEP50 and WD45 form complexes with PRMT5 and pICln in the methylosome and SMN complexes, respectively (18, 35). To further establish whether the latter two proteins are present in our p44-containing complex, Western blot analysis with anti-PRMT5 and anti-pICln antibodies was performed. Both PRMT5 and pICln proteins were present in the p44-containing complex (Fig. 5B, lane 7). To determine whether PRMT5 and pICln are involved in AR-driven gene expression, we subcloned cDNAs encoding PRMT5 and pICln (IMAGE:3836445) (ATCC) into the expression vector pcDNA3.1. To investigate the effect of PRMT5 and pICln on AR-dependent transcription in vivo, an ARE-containing luciferase reporter was cotransfected with expression vectors for AR, PRMT5, pICln, or different combinations into prostate cancer PC3 cells in the presence of ligand (R1881). As shown in Fig. 6, AR activated the reporter gene about 12-fold, and coexpressed PRMT5 showed a strong (up to 2.5-fold) enhancement of this activity. PRMT5 did not influence reporter gene activity in the absence of cotransfected AR or ligand (R1881) (data not shown), indicating that the enhancing effect of PRMT5 on AR-dependent gene expression was caused by an effect on the E4 promoter. To investigate the effect of PRMT5 plus p44 in the same assay, we cotransfected PC3 cells with limited amounts (50 ng) of PRMT5 and p44 alone or in combination. Figure 6 shows that 50 ng of PRMT5 or p44 had little effect on AR-dependent transcription. However, the same amounts of combined PRMT5 plus p44 resulted in strong (threefold) activation, indicating that PRMT5 and p44 function synergistically. In contrast, pICln alone or in combination with p44, PRMT5, or both had no significant effect on AR-dependent transcription (Fig. 6). Western blot analysis indicated that f:AR complex (Fig. 1A, lane 3) also contains PRMT5 and pICln (data not shown).
FIG. 6.
PRMT5 synergizes with p44 to enhance AR-driven gene expression. PC3 cells were transfected with 100 ng of ARE-E4-luc reporter plasmid, 30 ng of pcDNA-AR, and indicated amounts (in nanograms) of pcDNA-p44, -PRMT5, or -pICln expression plasmid. Cells were grown in the absence or presence of 10 nM R1881 for 48 h after transfection and then harvested for dual luciferase activity assays.
The methyltransferase activity of PRMT5 is not required for the enhanced transactivation of AR.
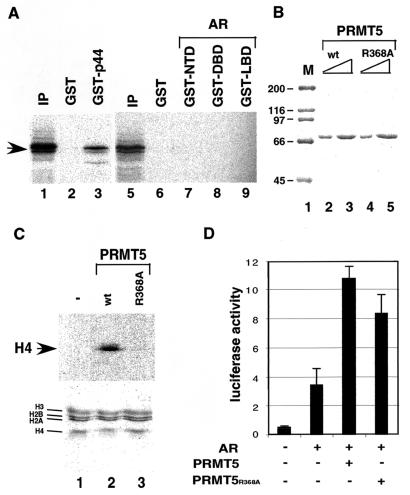
We further studied the interactions among AR, p44, and PRMT5. In vitro-produced, 35S-labeled PRMT5 was incubated with immobilized GST, GST-p44, GST-NTD, GST-DBD, and GST-LBD. After being washed, bound proteins were resolved by SDS-PAGE and visualized by autoradiography. p44 directly interacted with PRMT5 (Fig. 7A, lane 3). In contrast, PRMT5 did not bind any domain (NTD, DBD, or LBD) of AR (Fig. 7A, lanes 7 to 9). These results indicate that PRMT5 is recruited to AR target genes through its direct interaction with p44. The conserved arginine residue (R368) is essential for the methyltransferase activity of PRMT5 (40). The wild-type (Fig. 7B, lanes 2 and 3) and R368A mutant (lanes 4 and 5) PRMT5 were expressed in and purified from bacteria. Incubations of histones purified from HeLa cells with [3H]AdoMet plus recombinant PRMT5 resulted in transfer of the radioactive methyl groups to the histone H4 (Fig. 7C, top panel, lane 2). In contrast to previous observations (40), we did not detect the methylation of the histone H2A by PRMT5. This discrepancy might be due to the different preparations of histones used for the methylation assay. The preparation of individual histones was used in the previous study, but purified natural histones (containing the octamer of 2H2A, 2H2B, 2H3, and 2H4) were used in our study. The R368A mutation on PRMT5 dramatically reduced the methyltransferase activity in vitro (Fig. 7C, top panel, lane 3 versus lane 2). However, this mutation did not significantly decrease the PRMT5-mediated transactivation of AR in vivo (Fig. 7D), indicating that the methyltransferase activity of PRMT5 is not required. Western blot analysis with anti-PRMT5 antibody revealed that the levels of the wild-type and mutant PRMT5 in the transfected PC3 cells are same (data not shown). However, the same mutation on PRMT5 impaired its activity as transcriptional corepressor on the cyclin E1 promoter (16).
FIG. 7.
(A) p44 physically interacted with PRMT5. The 35S-labeled PRMT5 were incubated with the indicated GST fusion proteins. After being washed, bound proteins were resolved by SDS-10% PAGE and visualized by autoradiography. Lanes 1 and 5 contain 10% of the labeled PRMT5 used in binding reactions. (B) SDS-PAGE analysis of recombinant wild-type (lanes 2 and 3) and R368A mutant (lanes 4 and 5) PRMT5 expressed in bacteria and purified on an Ni-NTA agarose affinity column. The gel was stained with Coomassie blue R250. Lane 1, standard molecular weight markers (M) (Bio-Rad). (C) In vitro methyltransferase assay. The methyltransferase assay was performed as described in Materials and Methods. (Top) Autoradiography of the gel; (bottom) Coomassie blue staining of the same gel. Individual histones are indicated on the left. (D) Methyltransferase activity is not required for PRMT5 function on AR-driven gene expression. PC3 cells were transfected with 100 ng of 4× ARE-E4-luc reporter plasmid, 30 ng of pcDNA-AR, and 150 ng of pcDNA-PRMT5 or pcDNA-PRMT5(R368A) expression plasmid, as indicated. Cells were grown in the presence of 10 nM R1881 for 48 h after transfection and then harvested for luciferase activity assays.
Overexpression of p44 correlates with prostate tumorigenesis.
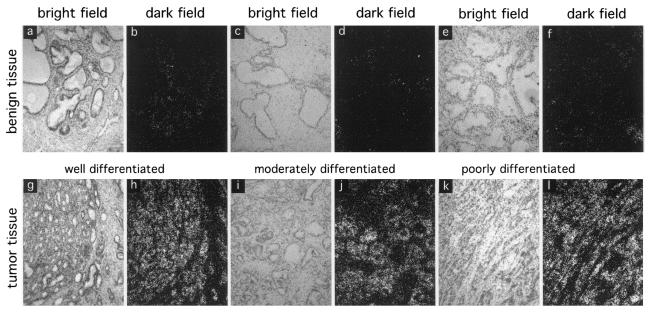
In order to investigate the possibility that p44 might be important for modulating AR function in prostate cancer, we investigated the expression of p44 at the mRNA level using quantitative in situ hybridization methods in 43 primary prostate cancers with different degrees of differentiation. Expression of p44 was detected only in prostate epithelial cells and was significantly up-regulated in 36% of well-differentiated prostate tumors (Table 1; Fig. 8a, b, g, and h), in more than 80% of moderately differentiated prostate tumors (Table 1; Fig. 8c, d, i, and j), and in more than 60% of poorly differentiated prostate tumors (Table 1; Fig. 8e, f, k, and l). The average increase was 5.1-fold. These results indicate that changes in p44 expression (and possibly in expression of associated proteins) might play an important role in affecting the deregulation of normal AR and NKX3.1 functions in prostate tumorigenesis.
TABLE 1.
Quantitative data for the in situ hybridization analysis
| Tumor grade | No. of cases | No. with increase in p44a | % |
|---|---|---|---|
| Low | 11 | 4 | 36 |
| Moderate | 23 | 19 | 82 |
| High | 10 | 6 | 60 |
Number of tumors with a 2- to 10-fold increase in p44 expression.
FIG. 8.
Enhanced expression of p44 in prostate tumor tissues. Frozen sections (4 μm thick) of prostate tissues were prepared and kept frozen until used. The frozen tissue sections were fixed in 4% paraformaldehyde for 30 min, dehydrated with ethanol, and hybridized with antisense p44 RNA probes labeled in vitro with [α-33P]UTP. The slides were washed first with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at room temperature and then twice with 0.2× SSC at 45°C for 20 min. The slides were exposed and evaluated with a Nikon microscope with a digital camera interfaced to a computer.
DISCUSSION
In this study, we describe the isolation of p44 as a new AR- and NKX3.1-interacting protein both in vitro and in vivo. Transient-transfection assays demonstrated that p44 increased AR transcriptional activity in an androgen-dependent manner. p44 forms a multiprotein complex that enhanced AR-dependent transcription in a cell-free transcriptional system.
A novel cofactor complex functions as an AR coactivator.
Increasing numbers of cofactors have been indicated in the function of AR (22, 24). They are identified through their physical interactions with AR and enhanced or repressed AR-mediated transcription in vivo. Our attempt to isolate the AR-associated proteins from stable cell lines resulted in the identification of p44. p44 and p44-containing complex enhanced AR-dependent transcription in vivo and in vitro, respectively. The protein sequence of p44 is identical to that of a component (MEP50) of the methylosome (18) and a subunit (WD45) of the SMN complex (34). The methylosome complex contains PRMT5/JBP1, pICln, and Sm proteins and mediates the assembly of spliceosomal snRNP (17). MEP50 is important for methylosome activity and binds to PRMT5/JBP1 and to a subset of Sm proteins (18). SMN is part of a complex that contains the Sm proteins and PRMT5 and is necessary and sufficient for assembly of spliceosomal U-richsnRNP (35, 47). The methylosome and SMN complexes were isolated from the cytoplasm of HeLa cells, and the p44-containing complex was purified from HeLa cell nuclear extract (17, 35). Thus, p44 may form distinct complexes with different proteins in the cytoplasm and in the nucleus for different roles (transcription versus splicing and/or translocation). The apparent size of MEP50 (above that of the 45-kDa bovine serum albumin) revealed by SDS-PAGE (17, 18) is larger than that of p44 and WD45 (below that of the 45-kDa bovine serum albumin), indicating that posttranslational modifications may exist in MEP50.
PRMT5 is present within the p44-containing complex.
Two types of PRMT activities have been identified in mammalian cells (58). PRMT1, PRMT2, and PRMT4/CARM1 have been found to participate in nuclear receptor transcriptional activation (42, 48, 50). The methylation of histones H3 and H4 by PRMT1 and PRMT4/CARM1 correlates with transcriptional activation, suggesting that they act by modifying chromatin structure. More recently, PRMT5 was identified as a corepressor of cyclin E1 transcription (16). Forced expression of PRMT5 negatively affected cyclin E1 promoter activity, which required the methyltransferase activity of PRMT5 (16). In contrast, our results demonstrate that PRMT5 is a positive AR cofactor that functions in a methyltransferase activity-independent manner in transient-transfection assays. Since the reporter gene in the transient transfection is likely not well packaged into chromatin, we cannot rule out the involvement of the methyltransferase activity of PRMT5 in AR function on the genes integrated into chromatin. Similarly, PRMT2 was identified as a methyltransferase based on the protein sequence and functioned as a positive cofactor for ERα, but so far its methyltransferase activity has not been identified with substrates including histones and ERα (42). The enhancement of AR-dependent transcription by PRMT5 might result from activation domains existing within PRMT5 protein or from the structural role of PRMT5 required for assembling the p44-containig cofactor complex. The former possibility is not supported by that fact that no activation was observed when PRMT5 was tethered to DNA through the DBD of GAL4 (data not shown).
p44 is overexpressed in prostate cancer.
The observation that certain cofactors are abnormally expressed in some prostate cancers indicates the importance of nuclear receptor cofactors in transcriptional control of AR function and also points to their possible role in neoplastic conversion (28). Overexpression of p44 in prostate cancer tissues indicates that it may play an important role in prostate tumorigenesis, and there is well documented evidence that abnormal NKX3.1 expression is involved in prostate tumorigenesis. Our finding that p44 interacts with both NKX3.1 and AR suggests that it might play a role in coregulating these two pathways.
In summary, our results point to a novel cofactor complex in the regulation of AR-dependent transcription. AR is an important regulatory factor in the development, differentiation, and maintenance of male reproductive functions, as well as in the regulation of other sexually dimorphic processes ranging from the development of neural tissues to the modulation of immune function. Thus, the p44-containing complex may play a pivotal role in these biological processes by modulating the transcriptional activity of AR.
Acknowledgments
We thank Ann Sutton for her critical editorial review and Lola Lopez for expert assistance in the preparation of the manuscript. We thank Donald J. Tindall for generously providing the human NKX 3.1 cDNA clone.
This work was supported in part by U.S. Department of the Army grant DAMS17-01-1-0097, CaP CURE, Cancer Center Support Core grant CA16672, and SPORE in Prostate Cancer grant CA90270 from the National Cancer Institute, National Institutes of Health.
K. Hosohata and P. Li contributed equally to this work.
REFERENCES
- 1.Aarnisalo, P., J. J. Palvimo, and O. A. Janne. 1998. CREB-binding protein in androgen receptor-mediated signaling. Proc. Natl. Acad. Sci. USA 95:2122-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdulkadir, S. A., J. A. Magee, T. J. Peters, Z. Kaleem, C. K. Naughton, P. A. Humphrey, and J. Milbrandt. 2002. Conditional loss of Nkx3.1 in adult mice induces prostatic intraepithelial neoplasia. Mol. Cell. Biol. 22:1495-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alen, P., F. Claessens, G. Verhoeven, W. Rombauts, and B. Peeters. 1999. The androgen receptor amino-terminal domain plays a key role in p160 coactivator-stimulated gene transcription. Mol. Cell. Biol. 19:6085-6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bentel, J. M., and W. D. Tilley. 1996. Androgen receptors in prostate cancer. J. Endocrinol. 151:1-11. [DOI] [PubMed] [Google Scholar]
- 5.Berger, S. L. 1999. Gene activation by histone and factor acetyltransferases. Curr. Opin. Cell Biol. 11:336-341. [DOI] [PubMed] [Google Scholar]
- 6.Berrevoets, C. A., P. Doesburg, K. Steketee, J. Trapman, and A. O. Brinkmann. 1998. Functional interactions of the AF-2 activation domain core region of the human androgen receptor with the amino-terminal domain and with the transcriptional coactivator TIF2 (transcriptional intermediary factor 2). Mol. Endocrinol. 12:1172-1183. [DOI] [PubMed] [Google Scholar]
- 7.Bhatia-Gaur, R., A. A. Donjacour, P. J. Sciavolino, M. Kim, N. Desai, P. Young, C. R. Norton, T. Gridley, R. D. Cardiff, G. R. Cunha, C. Abate-Shen, and M. M. Shen. 1999. Roles for Nkx3.1 in prostate development and cancer. Genes Dev. 13:966-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowen, C., L. Bubendorf, H. J. Voeller, R. Slack, N. Willi, G. Sauter, T. C. Gasser, P. Koivisto, E. E. Lack, J. Kononen, O. P. Kallioniemi, and E. P. Gelmann. 2000. Loss of NKX3.1 expression in human prostate cancers correlates with tumor progression. Cancer Res. 60:6111-6115. [PubMed] [Google Scholar]
- 9.Brinkmann, A. O., L. J. Blok, P. E. de Ruiter, P. Doesburg, K. Steketee, C. A. Berrevoets, and J. Trapman. 1999. Mechanisms of androgen receptor activation and function. J. Steroid Biochem. Mol. Biol. 69:307-313. [DOI] [PubMed] [Google Scholar]
- 10.Chen, H., R. J. Lin, W. Xie, D. Wilpitz, and R. M. Evans. 1999. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell 98:675-686. [DOI] [PubMed] [Google Scholar]
- 11.Choi, C. Y., Y. H. Kim, H. J. Kwon, and Y. Kim. 1999. The homeodomain protein NK-3 recruits Groucho and a histone deacetylase complex to repress transcription. J. Biol. Chem. 274:33194-33197. [DOI] [PubMed] [Google Scholar]
- 12.Cillo, C., A. Faiella, M. Cantile, and E. Boncinelli. 1999. Homeobox genes and cancer. Exp. Cell Res. 248:1-9. [DOI] [PubMed] [Google Scholar]
- 13.Claessens, F., P. Alen, A. Devos, B. Peeters, G. Verhoeven, and W. Rombauts. 1996. The androgen-specific probasin response element 2 interacts differentially with androgen and glucocorticoid receptors. J. Biol. Chem. 271:19013-19016. [DOI] [PubMed] [Google Scholar]
- 14.Cleutjens, K. B., C. C. van Eekelen, H. A. van der Korput, A. O. Brinkmann, and J. Trapman. 1996. Two androgen response regions cooperate in steroid hormone regulated activity of the prostate-specific antigen promoter. J. Biol. Chem. 271:6379-6388. [DOI] [PubMed] [Google Scholar]
- 15.De Vos, P., F. Claessens, B. Peeters, W. Rombauts, W. Heyns, and G. Verhoeven. 1993. Interaction of androgen and glucocorticoid receptor DNA-binding domains with their response elements. Mol. Cell. Endocrinol. 90:R11-R16. [DOI] [PubMed] [Google Scholar]
- 16.Fabbrizio, E., S. El Messaoudi, J. Polanowska, C. Paul, J. R. Cook, J. H. Lee, V. Negre, M. Rousset, S. Pestka, A. Le Cam, and C. Sardet. 2002. Negative regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO Rep. 3:641-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friesen, W. J., S. Paushkin, A. Wyce, S. Massenet, G. S. Pesiridis, G. Van Duyne, J. Rappsilber, M. Mann, and G. Dreyfuss. 2001. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol. Cell. Biol. 21:8289-8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friesen, W. J., A. Wyce, S. Paushkin, L. Abel, J. Rappsilber, M. Mann, and G. Dreyfuss. 2002. A novel WD repeat protein component of the methylosome binds Sm proteins. J. Biol. Chem. 277:8243-8247. [DOI] [PubMed] [Google Scholar]
- 19.Gehring, W. J., M. Affolter, and T. Burglin. 1994. Homeodomain proteins. Annu. Rev. Biochem. 63:487-526. [DOI] [PubMed] [Google Scholar]
- 20.Gelmann, E. P. 2002. Molecular biology of the androgen receptor. J. Clin. Oncol. 20:3001-3015. [DOI] [PubMed] [Google Scholar]
- 21.Gu, W., and R. G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90:595-606. [DOI] [PubMed] [Google Scholar]
- 22.Heinlein, C. A., and C. Chang. 2002. Androgen receptor (AR) coregulators: an overview. Endocr. Rev. 23:175-200. [DOI] [PubMed] [Google Scholar]
- 23.Ikonen, T., J. J. Palvimo, and O. A. Janne. 1997. Interaction between the amino- and carboxyl-terminal regions of the rat androgen receptor modulates transcriptional activity and is influenced by nuclear receptor coactivators. J. Biol. Chem. 272:29821-29828. [DOI] [PubMed] [Google Scholar]
- 24.Janne, O. A., A. M. Moilanen, H. Poukka, N. Rouleau, U. Karvonen, N. Kotaja, M. Hakli, and J. J. Palvimo. 2000. Androgen-receptor-interacting nuclear proteins. Biochem. Soc. Trans. 28:401-405. [PubMed] [Google Scholar]
- 25.Kim, M. J., R. D. Cardiff, N. Desai, W. A. Banach-Petrosky, R. Parsons, M. M. Shen, and C. Abate-Shen. 2002. Cooperativity of Nkx3.1 and Pten loss of function in a mouse model of prostate carcinogenesis. Proc. Natl. Acad. Sci. USA 99:2884-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kingston, R. E., and G. J. Narlikar. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339-2352. [DOI] [PubMed] [Google Scholar]
- 27.Kundu, T. K., Z. Wang, and R. G. Roeder. 1999. Human TFIIIC relieves chromatin-mediated repression of RNA polymerase III transcription and contains an intrinsic histone acetyltransferase activity. Mol. Cell. Biol. 19:1605-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, P., X. Yu, K. Ge, J. Melamed, R. G. Roeder, and Z. Wang. 2002. Heterogeneous expression and functions of androgen receptor co-factors in primary prostate cancer. Am. J. Pathol. 161:1467-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma, H., H. Hong, S. M. Huang, R. A. Irvine, P. Webb, P. J. Kushner, G. A. Coetzee, and M. R. Stallcup. 1999. Multiple signal input and output domains of the 160-kilodalton nuclear receptor coactivator proteins. Mol. Cell. Biol. 19:6164-6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malik, S., and R. G. Roeder. 2000. Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem. Sci. 25:277-283. [DOI] [PubMed] [Google Scholar]
- 31.Martinez, E., V. B. Palhan, A. Tjernberg, E. S. Lymar, A. M. Gamper, T. K. Kundu, B. T. Chait, and R. G. Roeder. 2001. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol. 21:6782-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKenna, N. J., and B. W. O'Malley. 2002. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108:465-474. [DOI] [PubMed] [Google Scholar]
- 33.McKenna, N. J., J. Xu, Z. Nawaz, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1999. Nuclear receptor coactivators: multiple enzymes, multiple complexes, multiple functions. J. Steroid Biochem. Mol. Biol. 69:3-12. [DOI] [PubMed] [Google Scholar]
- 34.Meister, G., C. Eggert, D. Buhler, H. Brahms, C. Kambach, and U. Fischer. 2001. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr. Biol. 11:1990-1994. [DOI] [PubMed] [Google Scholar]
- 35.Meister, G., and U. Fischer. 2002. Assisted RNP assembly: SMN and PRMT5 complexes cooperate in the formation of spliceosomal UsnRNPs. EMBO J. 21:5853-5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neely, K. E., and J. L. Workman. 2002. The complexity of chromatin remodeling and its links to cancer. Biochim. Biophys. Acta 1603:19-29. [DOI] [PubMed] [Google Scholar]
- 37.Ogryzko, V. V., T. Kotani, X. Zhang, R. L. Schiltz, T. Howard, X. J. Yang, B. H. Howard, J. Qin, and Y. Nakatani. 1998. Histone-like TAFs within the PCAF histone acetylase complex. Cell 94:35-44. [DOI] [PubMed] [Google Scholar]
- 38.Orlando, V. 2000. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem. Sci. 25:99-104. [DOI] [PubMed] [Google Scholar]
- 39.Orlando, V., and R. Paro. 1993. Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell 75:1187-1198. [DOI] [PubMed] [Google Scholar]
- 40.Pollack, B. P., S. V. Kotenko, W. He, L. S. Izotova, B. L. Barnoski, and S. Pestka. 1999. The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyltransferase activity. J. Biol. Chem. 274:31531-31542. [DOI] [PubMed] [Google Scholar]
- 41.Prescott, J. L., L. Blok, and D. J. Tindall. 1998. Isolation and androgen regulation of the human homeobox cDNA, NKX3.1. Prostate 35:71-80. [DOI] [PubMed] [Google Scholar]
- 42.Qi, C., J. Chang, Y. Zhu, A. V. Yeldandi, S. M. Rao, and Y. J. Zhu. 2002. Identification of protein arginine methyltransferase 2 as a coactivator for estrogen receptor alpha. J. Biol. Chem. 277:28624-28630. [DOI] [PubMed] [Google Scholar]
- 43.Roeder, R. G. 1998. Role of general and gene-specific cofactors in the regulation of eukaryotic transcription. Cold Spring Harbor Symp. Quant. Biol. 63:201-218. [DOI] [PubMed] [Google Scholar]
- 44.Roeder, R. G. 1996. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21:327-335. [PubMed] [Google Scholar]
- 45.Schneider, A., T. Brand, R. Zweigerdt, and H. Arnold. 2000. Targeted disruption of the Nkx3.1 gene in mice results in morphogenetic defects of minor salivary glands: parallels to glandular duct morphogenesis in prostate. Mech. Dev. 95:163-174. [DOI] [PubMed] [Google Scholar]
- 46.Schuur, E. R., G. A. Henderson, L. A. Kmetec, J. D. Miller, H. G. Lamparski, and D. R. Henderson. 1996. Prostate-specific antigen expression is regulated by an upstream enhancer. J. Biol. Chem. 271:7043-7051. [DOI] [PubMed] [Google Scholar]
- 47.Shen, E. C., M. F. Henry, V. H. Weiss, S. R. Valentini, P. A. Silver, and M. S. Lee. 1998. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev. 12:679-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stallcup, M. R., D. Chen, S. S. Koh, H. Ma, Y. H. Lee, H. Li, B. T. Schurter, and D. W. Aswad. 2000. Co-operation between protein-acetylating and protein-methylating co-activators in transcriptional activation. Biochem. Soc. Trans. 28:415-418. [PubMed] [Google Scholar]
- 49.Steadman, D. J., D. Giuffrida, and E. P. Gelmann. 2000. DNA-binding sequence of the human prostate-specific homeodomain protein NKX3.1. Nucleic Acids Res. 28:2389-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, H., Z. Q. Huang, L. Xia, Q. Feng, H. Erdjument-Bromage, B. D. Strahl, S. D. Briggs, C. D. Allis, J. Wong, P. Tempst, and Y. Zhang. 2001. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 293:853-857. [DOI] [PubMed] [Google Scholar]
- 51.Wang, Q., D. Sharma, Y. Ren, and J. D. Fondell. 2002. A coregulatory role for the TRAP/Mediator complex in androgen receptor mediated gene expression. J. Biol. Chem. 277:42852-42858. [DOI] [PubMed] [Google Scholar]
- 52.Wang, Z., and R. G. Roeder. 1998. DNA topoisomerase I and PC4 can interact with human TFIIIC to promote both accurate termination and transcription reinitiation by RNA polymerase III. Mol. Cell 1:749-757. [DOI] [PubMed] [Google Scholar]
- 53.Wang, Z., and R. G. Roeder. 1997. Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes Dev. 11:1315-1326. [DOI] [PubMed] [Google Scholar]
- 54.Xu, L., C. K. Glass, and M. G. Rosenfeld. 1999. Coactivator and corepressor complexes in nuclear receptor function. Curr. Opin. Genet. Dev. 9:140-147. [DOI] [PubMed] [Google Scholar]
- 55.Xu, W., H. Chen, K. Du, H. Asahara, M. Tini, B. M. Emerson, M. Montminy, and R. M. Evans. 2001. A transcriptional switch mediated by cofactor methylation. Science 294:2507-2511. [DOI] [PubMed] [Google Scholar]
- 56.Yu, X., P. Li, R. G. Roeder, and Z. Wang. 2001. Inhibition of androgen receptor-mediated transcription by amino-terminal enhancer of split. Mol. Cell. Biol. 21:4614-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, J., S. Zhang, P. E. Murtha, W. Zhu, S. S. Hou, and C. Y. Young. 1997. Identification of two novel cis-elements in the promoter of the prostate-specific antigen gene that are required to enhance androgen receptor-mediated transactivation. Nucleic Acids Res. 25:3143-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang, Y., and D. Reinberg. 2001. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 15:2343-2360. [DOI] [PubMed] [Google Scholar]