Abstract
Histone deacetylase 1 (HDAC1) is a major regulator of chromatin structure and gene expression. Tight control of HDAC1 expression is essential for development and normal cell cycle progression. In this report, we analyzed the regulation of the mouse HDAC1 gene by deacetylases and acetyltransferases. The murine HDAC1 promoter lacks a TATA box consensus sequence but contains several putative SP1 binding sites and a CCAAT box, which is recognized by the transcription factor NF-Y. HDAC1 promoter-reporter studies revealed that the distal SP1 site and the CCAAT box are crucial for HDAC1 promoter activity and act synergistically to constitute HDAC1 promoter activity. Furthermore, these sites are essential for activation of the HDAC1 promoter by the deacetylase inhibitor trichostatin A (TSA). Chromatin immunoprecipitation assays showed that HDAC1 is recruited to the promoter by SP1 and NF-Y, thereby regulating its own expression. Coexpression of acetyltransferases elevates HDAC1 promoter activity when the SP1 site and the CCAAT box are intact. Increased histone acetylation at the HDAC1 promoter region in response to TSA treatment is dependent on binding sites for SP1 and NF-Y. Taken together, our results demonstrate for the first time the autoregulation of a histone-modifying enzyme in mammalian cells.
In eukaryotic cells, DNA is complexed with core histones and other proteins in the form of chromatin. The basic repeating unit of chromatin, the nucleosome, is built of two copies of each of the four core histones, H2A, H2B, H3, and H4, wrapped by 146 bp of DNA. This organization allows the efficient packaging of genomic DNA into the nucleus but also has a negative impact on gene expression. To overcome this nucleosomal repression, the N-terminal tails of core histones are targets for multiple modifications, such as acetylation, phosphorylation, and methylation, which can modulate chromatin compaction. The best-studied modification of core histones is the reversible acetylation of conserved lysine residues within the N termini. Acetylation results in reduced interaction between positively charged histone tails and negatively charged DNA. Histone deacetylation is believed to result in chromatin condensation, whereas acetylation correlates with increased accessibility to genes for the transcription machinery.
Two types of enzymes, the histone acetyltransferases (HATs) and the histone deacetylases (HDACs), control the acetylation of histones and other proteins. More than a dozen mammalian histone deacetylases have been identified in recent years, and they have been classified into three groups according to their homology with the yeast enzymes Rpd3, Hda1, and Sir2 (11, 19). Class I enzymes seem to be involved in more general cellular processes, whereas class II enzymes might have more tissue-specific functions. The third mammalian HDAC class is made up of enzymes with homology to the NAD-dependent deacetylase Sir2. Mammalian Sir2 was recently shown to deacetylate p53, thereby controlling stress response and cell survival (21, 24, 38).
The class I enzyme HDAC1 was the first mammalian deacetylase identified (37). Numerous transcription factors, including regulators of the cell cycle, differentiation, and development, have been shown to associate with HDAC1, thereby mediating the repression of specific target genes (1, 7, 27). Previous work from our laboratory indicated a role of mouse HDAC1 in the regulation of proliferation and development. For instance, it has been shown that the expression of HDAC1 is induced upon growth factor activation of mouse T cells and fibroblasts (3, 13). In addition, HDAC1 levels were found to be elevated in highly proliferative tissues, embryonic stem cells, and several transformed cell lines (3, 20), suggesting a link between HDAC1 function and proliferation. In accordance with this idea, disruption of the HDAC1 gene resulted in reduced proliferation of mouse embryos and embryonic stem cells (20), whereas overexpression of HDAC1 led to impaired proliferation of murine fibroblasts (3). Taken together, these results indicate that a tightly controlled cell-type-specific expression of HDAC1 is crucial for unrestricted proliferation.
Recent findings point to the existence of a control system that modulates cellular deacetylase activities via a regulatory feedback mechanism. The expression of HDAC1 and certain other mammalian histone deacetylases is increased in response to deacetylase inhibitor treatment (10, 39). Furthermore, the HDAC1 gene was recently shown to be activated by the cooperation of acetylating and phosphorylating signals, resulting in phosphoacetylation of HDAC1 promoter-associated histone H3 (13). These data demonstrated that the chromatin modifier HDAC1 is regulated by mechanisms involving changes in chromatin structure.
Here, we directly evaluate the roles of acetylases and deacetylases in the regulation of HDAC1 promoter activity. We show that binding sites important for HDAC1 promoter activity are also essential for activation of the promoter by the deacetylase inhibitor trichostatin A (TSA). Further, we demonstrate that SP1 and NF-Y transcription factors can bind to these consensus sequences and can recruit HDAC1 to its own promoter. The HDAC1-mediated repression is counteracted by histone acetyltransferases, such as p300 and P/CAF. This feedback loop provides a perfect mechanism for the precise regulation of HDAC1 levels in mammalian cells.
MATERIALS AND METHODS
Cell culture.
The cell lines Swiss 3T3, Ref52, NIH 3T3, and U2OS were cultured in Dulbecco's modified Eagle medium containing 10% (vol/vol) fetal calf serum (FCS). Embryonic stem cells were cultivated in M15 medium containing 15% (vol/vol) FCS and 103 U of leukemia inhibitory factor/ml on gelatinized culture dishes. Drosophila SL-2 cells were maintained in Schneider's insect medium. Swiss 3T3 cells were rendered quiescent by incubation in Dulbecco's modified Eagle medium containing 0.2% (vol/vol) FCS for 48 h. Growth arrest was routinely controlled by fluorescence-activated cell sorter analysis with a Partec PAS-II sorter.
To generate stable cell lines, 5 μg of linearized plasmid DNA was transfected into Swiss 3T3 fibroblasts using DAC 30 transfection reagent (Eurogentec) as recommended by the supplier. The transfected cells were selected in the presence of Geneticin (400 μg/ml; Life Technologies, Inc.). Clones were pooled after 12 to 14 days and expanded in medium containing Geneticin. The drug was removed only shortly prior to the experimental assays. For transient transfection, Ref52, NIH 3T3, and U2OS cells (8 × 104) were seeded in 3-cm-diameter petri dishes and transfected the following day with a total of 2 μg of DNA using polyethyleneimine-assisted gene transfer (2). Two microliters of polyethyleneimine were diluted in 125 μl of HEPES buffer saline and added dropwise to 2 μg of DNA diluted in 125 μl of HEPES buffer saline. After a 20-min incubation at room temperature, the transfection mixture was added to the cells, which, prior to the transfection, had their growth medium replaced by 800 μl of serum-free medium. The transfection medium was replaced by fresh medium after 6 h. After 48 h, luciferase and β-galactosidase activities were measured. Transient transfection of SL-2 cells was carried out by calcium phosphate coprecipitation as described previously (17). TSA (final concentration, 50 ng/ml = 0.16 μM) was obtained from Wako Pure Chemical Industries.
Isolation of the mouse HDAC1 promoter.
The mouse HDAC1 gene and a 365-bp upstream region were previously isolated by screening genomic libraries (18). To obtain more sequence information about the HDAC1 upstream region, we used the GenomeWalker kit from Clontech according to the manufacturer's instructions. Each PCR was performed with Clontech Advantage-GC cDNA polymerase mix. The following gene-specific primers were used: GSP1, 5′-CAC CCG CAG CTC ACC GTC GTA G-3′, and GSP2, 5′-CCC GTC AGT CTG TCC GCC CGC C-3′.
Plasmid constructs.
The full-length mouse HDAC1 promoter (pP721) was cloned into SacI/HindIII-cut pGL2Basic (Clontech) and pGL2neo vectors (9), respectively. The 5′ deletion constructs (pP365, pP303, pP210, pP125, and pP93) spanning from −1 to −700 bp of the translational start codon were generated using PCR primers containing SacI and HindIII restriction sites and cloned into the pGL2neo vector. Luciferase plasmids with mutated transcription factor binding sites were constructed by site-directed mutagenesis using an overlap extension PCR protocol (28). Two separate PCR fragments for each half of a final hybrid product were generated with mutagenesis primers (SP1mut, 5′-GAT CCC GGG GGT Gtt CGG GGC TTC GAG-3′; CCAATmut, 5′-CCG CGC ATG CCG ATT aaT TAG AGT GAG ACC-3′; and AP2mut, 5′-CCG GTC TCt ttC ACC CCT CCG CGC C-3′; lowercase letters represent mutated base pairs) and outer primers containing SacI and HindIII restriction sites. The two products were mixed, and a second PCR was performed using the two outside primers. The resulting products were cloned into SacI/HindIII-cut pGL2Basic and pGL2neo vectors, respectively. All the deletion and site-directed mutagenesis constructs were sequenced to confirm the intended deletions or mutations.
Primer extension.
Total RNA was isolated from a mouse liver, and poly(A)+ mRNA was purified using the Oligotex mRNA midi kit from Qiagen according to the manufacturer's instructions. Ten picomoles of [γ-32P]ATP-labeled oligonucleotides (SB 8, 5′-GTA ACA GAC TTT CCT CTT GG-3′) and 1 μg of poly(A)+-selected RNA were dissolved in 10 μl of water, heated to 70°C for 10 min, and chilled on ice. Reverse transcription was carried out at 42°C for 1 h using Superscript reverse transcriptase (Gibco). The same oligonucleotide was also used to sequence the mouse genomic regions cloned into the pBluescript vector pKS. The reaction products were separated on a denaturing 6% polyacrylamide gel and autoradiographed overnight.
Electrophoretic mobility shift assay.
Whole-cell extracts were prepared as described previously (9). Ten micrograms of protein extract was used for the binding reaction as described previously (33). The sequences of the top strands of the individual oligonucleotide probes were as follows: CCAAT WT, 5′-ATG CCG ATT GGT TAG AGT-3′; CCAAT mut, 5′-ATG CCG ATT aaT TAG AGT-3′; SP1 WT, 5′-GGG GGT GGG CGG GGC TTC-3′; and SP1 mut, 5′-GGG GGT Gtt CGG GGC TTC-3′. Competition experiments were performed with the corresponding unlabeled double-stranded oligonucleotides.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation assays were carried out as described previously (13) with a few modifications. Chromatin was cross-linked for 10 min using formaldehyde. The resulting chromatin solution was diluted 1:10 and precipitated with 5 μl of acetyl-specific histone antibodies, 10 μl of a monoclonal HDAC1 antibody (Upstate Biotechnology), 4 μl of polyclonal SP1 antibody (12), and 4 μl of NF-YB antibody (32). Rabbit preimmune serum (for polyclonal antibodies) and the nonspecific monoclonal antibody 10F9 (for monoclonal antibodies) were used as controls. The following day, chromatin-antibody complexes were isolated from the solution by incubation with 30 μl of protein A/G-Sepharose beads (50% slurry, 100 μg of salmon sperm DNA/ml, 500 μg of bovine serum albumin/ml) with rocking at 4°C for 2 h. The beads were harvested and washed as described previously (13). Chromatin-antibody complexes were eluted from the A/G-Sepharose beads by the addition of 2% sodium dodecyl sulfate, 0.1 M NaHCO3, and 10 mM dithiothreitol to the pellet. Cross-linking was reversed by the addition of 0.05 volume of 4 M NaCl and incubation of the eluted samples for 6 h at 65°C. The DNA was extracted with phenol-chloroform, precipitated with ethanol, and dissolved in water. The immunoprecipitated DNA was analyzed for HDAC1 promoter and histone H4 gene sequences by quantitative PCR.
PCR analysis of immunoprecipitated DNA.
All PCRs were performed on a Biometra D3 thermocycler using Promega PCR Master Mix. The linear range for each primer pair was determined empirically using different amounts of genomic DNA. PCRs with increasing amounts of genomic DNA were carried out along with the immunoprecipitated DNA. PCR products were resolved on 2% agarose gels and quantified using the ImageQuant program (Molecular Dynamics).
Primer sets used in chromatin immunoprecipitation assays.
The primer sets used in chromatin immunoprecipitation assays were as follows: histone H4 locus, upper primer (5′-GAC ACC GCA TGA AAA GAA TAG CTG-3′) and lower primer (5′-CTT TCC CAA GGC CTT TAC CAC C-3′); transfected HDAC1 promoter, upper primer (5′-GGA CTT TGG TAC AGG CCC AGG G-3′) and lower primer (5′-GCT CTC CAG CGG TTC CAT CCA C-3′); endogenous HDAC1 promoter fragment, upper primer (5′-GCC GGA CTT TGG TAC AGG CCC AGG-3′) and lower primer (5′-CTT TCC TCT TGG TGC CCT GAG TCT-3′).
RESULTS
Analysis of the mouse HDAC1 promoter.
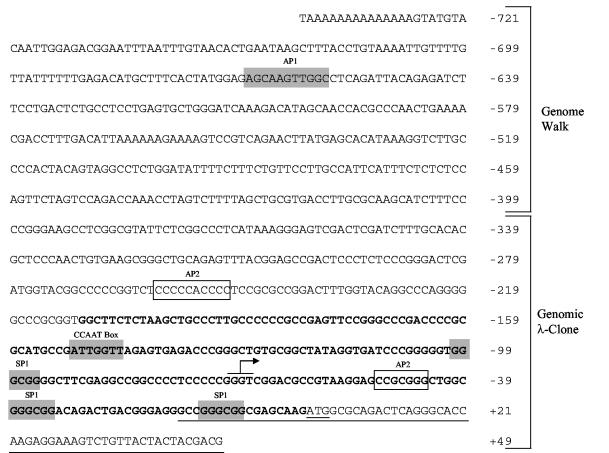
As a first step to understanding the transcriptional regulation of HDAC1, we cloned and sequenced the region upstream of the first exon of the mouse HDAC1 gene. The presumptive mouse HDAC1 promoter sequence, encompassing 721 bp upstream of the translational start codon, is shown in Fig. 1. As is typical of many housekeeping genes or genes that encode transcription factors, the DNA sequence is rich in GC nucleotides and lacks a TATA box consensus sequence. A transcription factor binding site database search, using the Genomics Computer Group software package, revealed a number of potential binding sites. Three putative SP1 binding sites are located at positions −13, −38, and −100 upstream of the translational start codon. Further, a CCAAT box at position −151 and two putative AP-2 binding sites at positions −50 and −188 could be identified. An AP1 binding site is present far upstream of the ATG start codon (−660).
FIG. 1.
Mouse HDAC1 promoter. Putative transcription factor binding sites are boxed. The core promoter sequence (P210) is shown in boldface. The sequence overlapping with the murine HDAC1 cDNA (3) and the translational start codon are underlined. The arrow indicates the major transcripitonal start site.
Determination of the HDAC1 gene transcriptional start site.
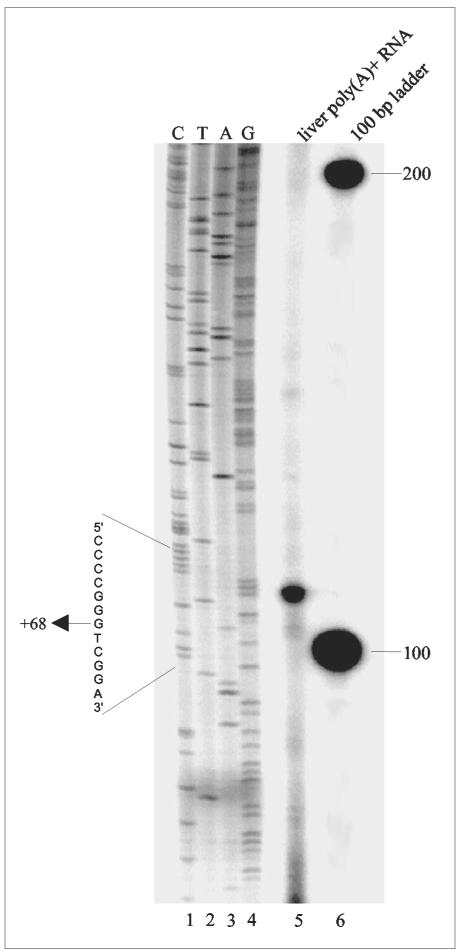
To determine the transcriptional initiation site of the HDAC1 gene, we used a reverse transcriptase primer extension assay. Primers were designed to span the putative transcription start site and then used in extension reactions with poly(A)+-selected mRNA from a mouse liver (Fig. 2). The result from one primer (SB8) revealed a strong signal, indicating a major transcription start site. In addition, several weak signals corresponding to minor initiation sites were observed. Alignment with a dideoxynucleotide sequence ladder from the same primer showed that the strong band corresponded to a guanosine within a GC-rich region. The major transcription start site is located 68 bp upstream of the ATG translational start codon (Fig. 1). The identical major start site was also found with poly(A)+ RNA isolated from the mouse T-cell line B6.1 (data not shown). In perfect agreement with these results, three recently identified expressed sequence tag clones (BY059245, BY051749, and BY058306) that are obviously derived from mouse HDAC1 mRNA extend exactly to the G triplet at +68 to +70, corresponding to the major transcription initiation site.
FIG. 2.
Determination of the 5′ ends of mouse HDAC1 transcripts. Primer extension analysis with poly(A)+-selected RNA from a mouse liver was performed using the 32P-labeled SB8 oligonucleotide (lane 5). A genomic sequencing ladder (lane 6) was created in parallel and is shown with the appropriate radiolabeled nucleotides (lanes 1 to 4). The arrow indicates the major start site of transcription.
Identification of binding sites essential for HDAC1 promoter activity.
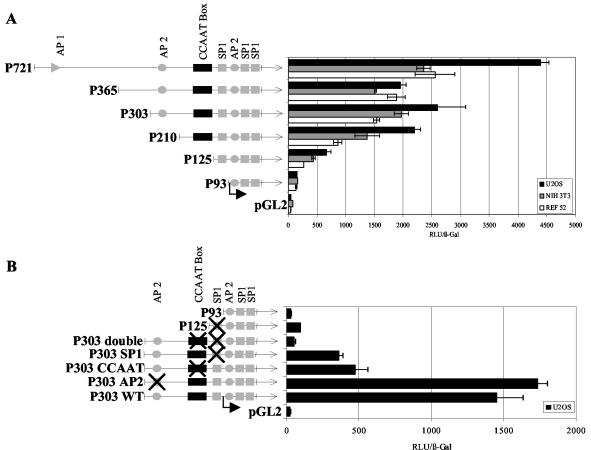
To determine transcription factor binding sites that are essential for HDAC1 promoter activity, two approaches were taken. Luciferase reporter constructs were created either with various 5′ promoter deletions or with mutated binding sites for individual transcription factors. The HDAC1 promoter-luciferase constructs were transiently transfected into various mouse and human cell lines and assayed for luciferase activity. As shown in Fig. 3A, deletion to position −210 resulted in only a moderate decrease in HDAC1 promoter activity in NIH 3T3 and Ref52 cells. In U2OS cells, the reduction of HDAC1 promoter activity was more pronounced, suggesting the presence of positive, cell-type-specific cis-acting sequences within the region. Further deletion to position −125 significantly reduced HDAC1 promoter activity, while the shortest promoter construct (P93) showed only minimal, but still significant, promoter activity. These results indicate that HDAC1 promoter activity is primarily mediated by the sequence between −93 and −210, including the distal GC box and the CCAAT box. Consequently, these motifs were individually or simultaneously mutated in the context of different promoter constructs as indicated in Fig. 3B. Furthermore, the upstream AP-2 binding site was mutated and examined for its effect on HDAC1 promoter activity. In agreement with results obtained with the 5′ deletion constructs, mutation of the CCAAT box reduced promoter activity ∼3-fold, while mutation of the distal GC box decreased promoter activity to ∼20% of that of the wild type. Mutation of the AP-2 binding site had no significant effect on luciferase activity. The same effect of point mutations on HDAC1 promoter activity was obtained in the context of the longest HDAC1 promoter construct (P721; data not shown). Constructs that contained no functional CCAAT box and distal GC box displayed very weak promoter activity (Fig. 3B, bottom). Together, these results showed that the CCAAT box and the distal GC box are crucial for the activity of the HDAC1 promoter.
FIG. 3.
Luciferase activity driven by HDAC1 promoter 5′ deletions or point mutations. Schematic representations of wild-type and mutated promoter regions of the mouse HDAC1 gene used for the production of HDAC1-luciferase constructs are shown on the left. Site-specific disruption of individual elements is indicated by crossed boxes. Reporter constructs, together with a β-galactosidase reference vector, were transfected into cells, which were harvested after 48 h and assayed for luciferase activity. Enzyme activity was normalized to β-galactosidase activity. The means ± standard deviations of three independent experiments are shown. (A) Luciferase activities of HDAC1 wild-type promoter and deletions transiently transfected into U2OS cells, NIH 3T3 fibroblasts, and Ref52 cells. (B) Luciferase activities of HDAC1 P303 promoter and corresponding constructs with mutated transcription factor binding sites in U2OS cells.
Interaction of SP1 and NF-Y transcription factors with HDAC1 promoter sequences.
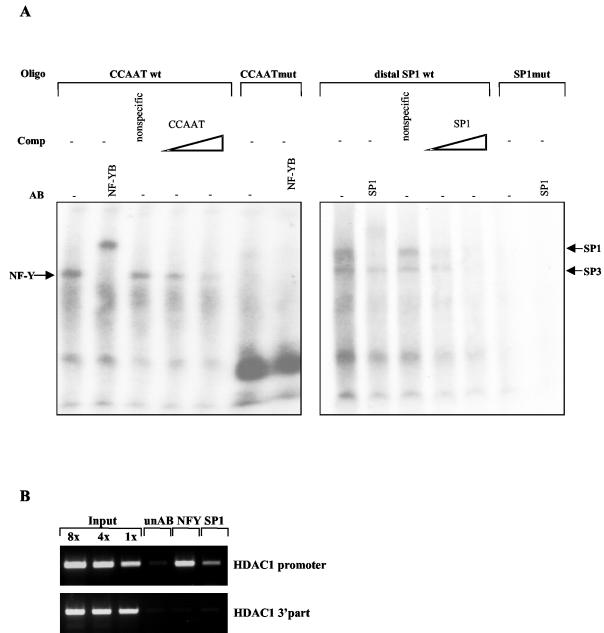
Having identified potential transcription factor binding sites on the HDAC1 promoter, we next used electrophoretic mobility shift assays to identify proteins which interact with these consensus sequences. Double-stranded oligonucleotides corresponding to the CCAAT box and to the distal GC box were incubated with protein extracts from Swiss 3T3 cells, and complexes were analyzed on a nondenaturating polyacrylamide gel electrophoresis gel (Fig. 4A). One important CCAAT box binding factor in mammalian cells is the transcriptional regulator NF-Y. NF-Y, also referred to as CP1 or CBF, consists of three subunits, A, B, and C, all of which are required for DNA binding (reviewed in reference 25). With the CCAAT box binding site, one large complex was formed which could be supershifted by the addition of NF-YB antibodies. Complex formation was inhibited by specific competitor oligonucleotides but was not affected by nonspecific competitor oligonucleotides. In contrast, no specific complexes were observed with oligonucleotides containing a mutated CCAAT box.
FIG. 4.
Protein complexes interacting with binding sites in the HDAC1 promoter. (A) Electrophoretic mobility shift assays were performed by incubating protein extracts from exponentially growing Swiss 3T3 cells with the indicated oligonucleotide probes (Oligo). Antibodies (AB) and double-stranded competitor oligonucleotides (Comp; 10- and 100-fold molar excess) were added as indicated (−, not added). Specific complexes are marked by arrows. (B) NF-Y and SP1 are associated with the proximal HDAC1 promoter. Formaldehyde-cross-linked chromatin was prepared from proliferating Swiss 3T3 cells and precipitated with NF-YB antibodies (NFY), SP1 antibodies (SP1), or nonspecific antibodies (unAB). DNA from the antibody-bound fraction and total input DNA isolated from chromatin used for the immunoprecipitation were analyzed by quantitative PCR using primers specific for the HDAC1 promoter or the 3′ part of the mouse HDAC1 gene (exon 12).
The GC-containing oligonucleotide probe produced two shifted complexes, which could be competed with specific oligonucleotides. One of the complexes could be supershifted with antibodies against SP1, whereas a mutated GC box probe gave no complexes at all. These results suggested that NF-Y and a member of the SP1 transcription factor family recognize the corresponding binding sites within the HDAC1 promoter. Therefore, we next asked whether SP1 and NF-Y are also associated in vivo with the mouse HDAC1 promoter. As shown in Fig. 4B, chromatin immunoprecipitation assays revealed that the transcription factors NF-Y and SP1 bind specifically to the HDAC1 promoter region. In contrast, SP1 and NF-Y were absent from the 3′ part of the HDAC1 gene, indicating a site-specific recruitment of these transcription factors to the HDAC1 upstream region.
SP1 and NF-Y transcription factors are involved in activation of the HDAC1 promoter.
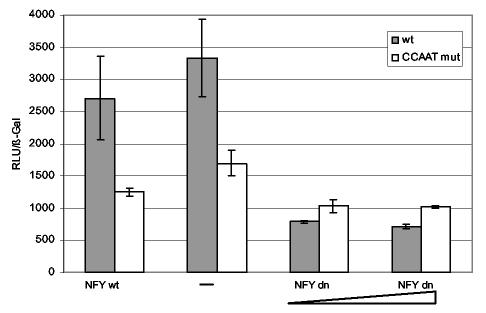
To investigate the role of NF-Y in HDAC1 promoter activity in vivo, a dominant-negative NF-YA subunit was cotransfected with HDAC1 promoter reporter plasmids into U2OS cells. As shown in Fig. 5, HDAC1 promoter activity was significantly reduced by cotransfection of increasing amounts of the dominant-negative NF-YA subunit, but not by cotransfection of wild-type NF-YA, compared to an empty vector. On the other hand, the dominant-negative NF-YA protein had only a minor effect on an HDAC1 promoter with a mutated CCAAT box.
FIG. 5.
HDAC1 promoter activity is repressed by a dominant-negative mutant of NF-YA. U2OS cells were transiently transfected with 0.5 μg of HDAC1 pP721 wild-type reporter plasmid (wt) or the corresponding CCAAT mutant reporter plasmid (CCAAT mut) without (−) or together with 0.5 or 1 μg of expression vector encoding dominant-negative NF-YA (NFY dn) and a β-galactosidase reference vector. As a control, the reporter plasmids were cotransfected with 1 μg of the expression plasmid pNF-YA13 encoding wild-type NF-YA (NFY wt). pCIneo empty vector was used to bring the total amount of the DNA mixture to 2 μg. The activity (in relative light units [RLU]) of luciferase relative to that of β-galactosidase is presented. The means ± standard deviations of three independent experiments are shown.
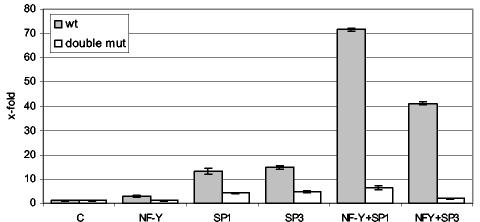
Next, we examined the effects of SP family members and NF-Y on HDAC1 promoter activity using Drosophila SL-2 cells (Fig. 6). This cell line is devoid of endogenous SP family members and NF-Y (41). Transfection of the wild-type HDAC1 promoter construct alone resulted in no significant promoter activity, whereas cotransfection with all three NF-Y subunits induced promoter activity ∼3-fold. Cotransfection of the wild-type promoter together with SP1 or SP3 increased promoter activity >10-fold. However, inclusion of NF-Y and SP1 or SP3 dramatically enhanced HDAC1 promoter activity by 70- and 40-fold, respectively. In contrast, these effects were not observed with the double-mutated HDAC1 promoter construct lacking the NF-Y site and the distal GC box. These results suggest that both SP family members and NF-Y can act synergistically to activate transcription from the mouse HDAC1 promoter.
FIG. 6.
Synergistic activation of the HDAC1 promoter by Sp1/Sp3 and NF-Y in Drosophila SL-2 cells. Drosophila SL-2 cells were cotransfected with 0.5 μg of wild-type (wt) or double-mutated (double mut) HDAC1 reporter plasmids (pP721 or pP721double) with or without expression vectors encoding Sp1/Sp3 or NF-Y (100 ng each). The total amount of DNA (10 μg) was adjusted by the addition of salmon sperm DNA. The data are presented as stimulation (x-fold), where the value of luciferase activity normalized to total cell protein for the reporter alone is set at 1. The means ± standard deviations of three independent experiments are shown.
Repression of the HDAC1 promoter by HDAC1 itself can be relieved by TSA.
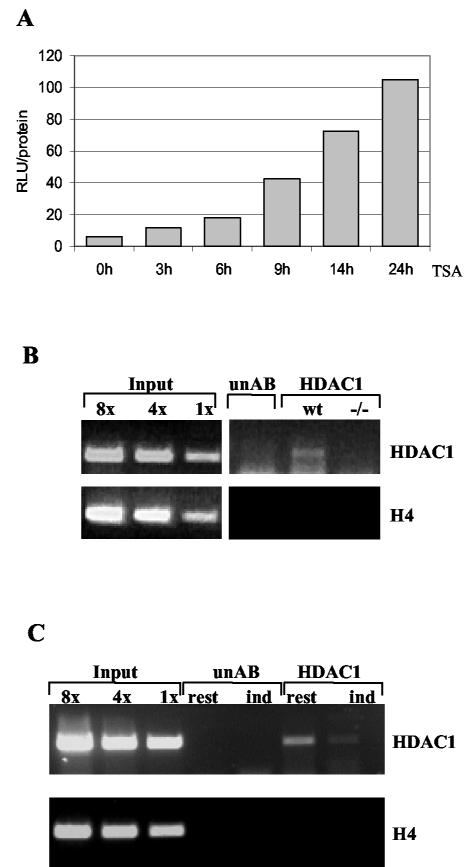
Previously, it was shown by run-on assays that the HDAC1 gene is transcriptionally activated by the histone deacetylase inhibitor TSA (13). To show that the upstream sequence of the mouse HDAC1 gene is sensitive to TSA, the wild-type HDAC1 promoter-reporter construct was stably transfected into Swiss 3T3 cells arrested for 48 h and treated with TSA for various periods. As shown in Fig. 7A, luciferase expression driven by the HDAC1 promoter increased up to 10-fold following TSA treatment. This result suggests that histone deacetylases might be involved in the repression of HDAC1 transcription in resting fibroblasts.
FIG. 7.
Autoregulation of the HDAC1 promoter. (A) Activation of stably integrated HDAC1 promoter P721 by TSA. Stably transfected Swiss 3T3 cells were serum arrested for 48 h and treated with TSA for the indicated periods. The cells were harvested, and luciferase activity (in relative light units [RLU]) was determined and normalized to the corresponding amount of protein. (B) HDAC1 is associated with its own promoter region. Formaldehyde-cross-linked chromatin was prepared from undifferentiated proliferating wild-type (wt) and HDAC1−/− (−/−) ES cells and immunoprecipitated with an HDAC1 antibody (HDAC1) or a nonspecific antibody (unAB). DNA from the antibody-bound fraction and total input DNA isolated from chromatin used for the immunoprecipitation were analyzed by quantitative PCR for the presence of the HDAC1 promoter region and the histone H4 control gene. (C) Specific recruitment of HDAC1 to the HDAC1 promoter region in resting Swiss 3T3 fibroblasts. Cross-linked chromatin was prepared from resting fibroblasts before (rest) and after (ind) restimulation with 20% FCS for 18 h and precipitated with the monoclonal HDAC1 antibody or an unrelated control antibody (unAB). The precipitated DNA was analyzed as described for panel B.
To examine whether HDAC1 itself binds and thereby represses its own promoter, we performed chromatin immunoprecipitation assays of wild-type ES cells and HDAC1−/− ES cells using a monoclonal HDAC1 antibody (Fig. 7B). Chromatin immunoprecipitation assays revealed that in wild-type ES cells, HDAC1 promoter DNA was immunoprecipitated with the HDAC1 antibody, while no DNA was recovered from ES cells lacking HDAC1. Further, no DNA was immunoprecipitated from the H4 gene locus. This result indicates that the HDAC1 protein is involved in the regulation of its own gene.
Expression of the HDAC1 gene was previously shown to be induced by growth factors (3). In growth factor-deprived fibroblasts, inhibition of deacetylases by TSA relieves the transcriptional repression of the HDAC1 promoter. Therefore, it is likely that deacetylases become specifically recruited to the HDAC1 upstream region when cells are deprived of growth factors. To test this hypothesis, we analyzed the association of HDAC1 with the HDAC1 promoter in resting and serum-activated Swiss 3T3 cells. Indeed, HDAC1 was associated with its own promoter in resting fibroblasts (Fig. 7C). Stimulation of Swiss 3T3 cells with high serum concentrations resulted in significant dissociation of the deacetylase from the HDAC1 promoter. Thus, reduced activity of the HDAC1 gene in resting fibroblasts correlated with recruitment of HDAC1 to its promoter.
The distal SP1 binding site and the CCAAT box are necessary for promoter activation by TSA and recruitment of HDAC1.
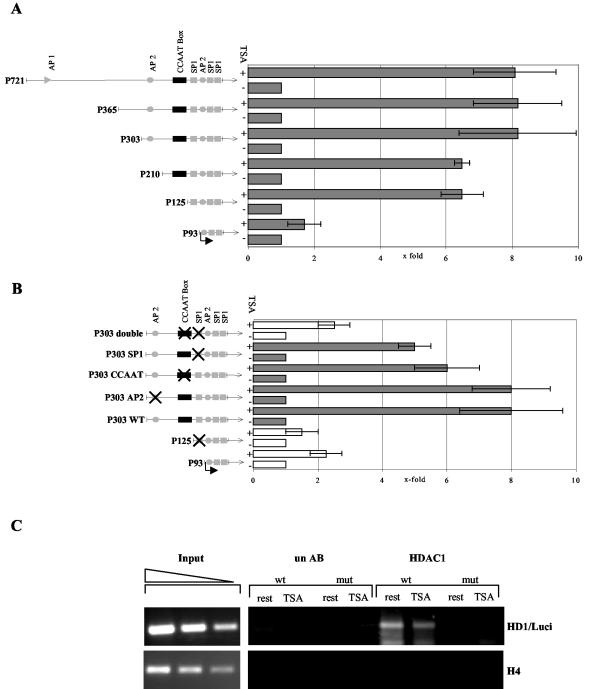
To determine transcription factor binding sites that are essential for promoter activation by TSA, we established stable Swiss 3T3 cell lines carrying the various 5′ promoter deletion constructs or the HDAC1 promoter point mutation constructs stably integrated into their genomes. Deletion or point mutations within the different promoter constructs in stably transfected Swiss 3T3 fibroblast lines had basically the same effect as in transiently transfected cell lines (Fig. 3 and data not shown). Cells were synchronized in G0 by serum deprivation for 48 h and treated with TSA for 20 h, and luciferase activity was determined. As shown in Fig. 8A, deletion to position −125 did not significantly effect TSA-mediated activation of the HDAC1 promoter, but deletion to position −93 nearly abolished activation by TSA, suggesting that the distal GC box is necessary for the TSA effect on the HDAC1 promoter.
FIG. 8.
The distal SP1 binding site and the CCAAT box are crucial for TSA-mediated activation and recruitment of HDAC1 to its promoter region. (A) Schematic representations of wild-type and mutated promoter regions of the mouse HDAC1 gene are on the left. Site-specific disruption of individual elements is indicated by crossed boxes. Stably transfected Swiss 3T3 cells containing the HDAC1 5′ deletion mutants were serum arrested for 48 h and treated with TSA. After a further 20 h, the cells were harvested and assayed for luciferase activity. Enzyme activity was normalized to the corresponding protein concentration and plotted relative to the untreated value set as 1. The means ± standard deviations of three independent experiments are shown. (B) Stably transfected Swiss 3T3 cells containing the HDAC1 point mutants were treated as described for panel A and examined for luciferase activity. (C) Chromatin immunoprecipitation analysis of stably transfected HDAC1 promoter P721 for the presence of HDAC1. Cross-linked chromatin of resting (rest) or TSA-treated (TSA) stable cell lines carrying the wild-type HDAC1 promoter (wt) or the double-mutated HDAC1 promoter (mut) was immunoprecipitated with an HDAC1 antibody (HDAC1) or a nonspecific antibody (un AB). DNA from the antibody-bound fraction and total input DNA isolated from chromatin used for the immunoprecipitation were analyzed by quantitative PCR using a primer specific for the transfected promoter (HD1/Luci).
Surprisingly, mutation of this site had only a marginal effect on TSA-mediated activation, similar to single point mutations within the CCAAT box, whereas mutation of the AP2 binding site had no effect at all (Fig. 8B). In contrast, constructs containing no functional CCAAT box and distal GC box (P93, P125 SP1, and P303 double) all showed significantly reduced TSA-mediated induction (Fig. 8B). From this, we concluded that activation of the HDAC1 promoter by TSA requires either the distal SP1 binding site or the CCAAT box.
These data suggest that histone deacetylases such as HDAC1 are recruited to the promoter by SP1 and NF-Y. Mutation of both binding sites should abolish binding of HDAC1 to its promoter region. This assumption was verified by chromatin immunoprecipitation assays using stably transfected cell lines containing either the wild-type promoter or the double-mutated promoter. Cells were arrested for 48 h and treated with TSA for another 20 h, and chromatin was immunoprecipitated with monoclonal HDAC1 antibodies or nonspecific control antibodies. The results, shown in Fig. 8C, revealed that HDAC1 is associated only with the wild-type HDAC1 promoter region but not with the promoter containing no functional CCAAT box and distal SP1 binding site. No HDAC1 was detected at the histone H4 gene locus. Further, TSA treatment reduced but did not abolish binding of HDAC1 to the promoter (see Discussion).
HDAC1 promoter is regulated by the balanced action of HATs and HDACs involving histone acetylation at the HDAC1 promoter.
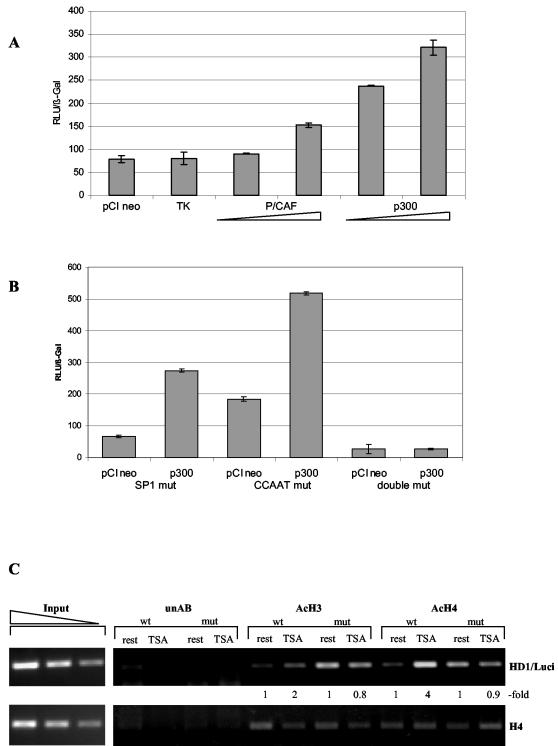
Our results strongly suggest that activation of the HDAC1 promoter by TSA involves a change in the balance between acetylating and deacetylating activities at the HDAC1 promoter. To strengthen this observation, cellular HAT activity was increased by cotransfection of constructs expressing p300, a known histone acetyltransferase, together with either the wild-type HDAC1 promoter construct or mutated HDAC1 promoter constructs. Expression of increasing amounts of p300 significantly induced HDAC1 promoter activity, while cotransfection with a vector encoding the thymidine kinase (TK) protein showed no effect on HDAC1 promoter activity compared to an empty vector (Fig. 9A). A single mutation within the distal SP1 binding site or the CCAAT box did not affect transactivation by p300, but simultaneous mutation of both sites abolished activation by p300 (Fig. 9B). In agreement with these data, ectopic expression of the adenoviral protein E1A, a known inhibitor of p300, decreased HDAC1 promoter activity and abolished the coactivator function of p300 (data not shown). Coexpression of P/CAF, another histone-acetylating enzyme, also led to significant, albeit less pronounced, induction of HDAC1 promoter activity. These results strongly suggest that histone deacetylases and acetyltransferases are recruited by the same factors to the HDAC1 promoter.
FIG.9.
Involvement of histone acetyltransferases in the activation of the HDAC1 gene. (A) U2OS cells were transiently cotransfected with 0.5 μg of HDAC1 wild-type reporter plasmid (pP721) or with 0.5 or 1 μg of expression vectors encoding p300 or P/CAF, together with a β-galactosidase reference vector. As a control, the reporter plasmid was cotransfected with 1 μg of expression plasmids encoding the TK protein. pCIneo empty vector was used to bring the total amount of the DNA mixture to 2 μg. The activity of luciferase (in relative light units [RLU]) to relative to that of β-galactosidase is presented. The means ± standard deviations of three independent experiments are shown. (B) Cotransfection was carried out as described for panel A using HDAC1 point mutants (mut) as reporter constructs. (C) Chromatin immunoprecipitation performed with antibodies against acetylated histone H3 and H4 using chromatin isolated from stably transfected Swiss 3T3 cells carrying either the wild-type HDAC1 promoter (wt) or the double-mutated HDAC1 promoter (mut) before (rest) and after (TSA) TSA treatment. DNA from the antibody-bound fraction and total input DNA isolated from chromatin used for the immunoprecipitation were analyzed by quantitative PCR using a primer specific for the transfected promoter (HD1/Luci; see Materials and Methods). PCR products were quantified using the ImageQuant program, and relative signal intensities (-fold) are indicated.
To examine whether activation of the HDAC1 promoter by changes in the HAT-HDAC balance involves hyperacetylation of histones at the HDAC1 promoter, we again carried out chromatin immunoprecipitation experiments with antibodies directed against acetylated histone isoforms. Stable cell lines containing either the wild-type promoter or the double-mutated promoter were arrested by serum deprivation and treated with TSA as previously described. Hyperacetylated chromatin was immunoprecipitated with acetyl-specific histone antibodies. As shown in Fig. 9C, wild-type HDAC1 promoter DNA was enriched twofold in anti-acetyl H3 immunoprecipitates and fourfold in anti-acetyl H4 immunoprecipitates after TSA treatment. Mutation of the distal SP1 site and the CCAAT box abolished TSA-induced hyperacetylation at the HDAC1 promoter region (Fig. 9C, top). Interestingly, in the absence of the deacetylase inhibitor, the mutated promoter showed elevated basal histone acetylation levels that might be due to loss of the binding sites for HDAC1 recruiting factors. No significant changes in histone acetylation after TSA treatment were observed for a control locus, the histone H4 gene (Fig. 9C, bottom). These data are in perfect agreement with the observation that both histone-modifying enzymes (HATs and HDACs) are recruited to the HDAC1 promoter by the distal SP1 site and the CCAAT box.
DISCUSSION
Organization of the mouse HDAC1 promoter.
In this report, we analyzed the murine HDAC1 promoter and its regulation by acetylating and deacetylating enzymes. The organization of the murine HDAC1 gene is very similar to that of the HDAC2 gene, suggesting a duplication of the ancestral gene (18, 43). In contrast, the upstream region of the HDAC1 gene does not show significant sequence homology to the mouse HDAC2 promoter. The HDAC1 promoter region is very GC rich and lacks a TATA box. The most important regulatory elements are located within a small region upstream of the major transcription start site of the HDAC1 gene.
Activation of the mouse HDAC1 promoter.
The 210-bp HDAC1 promoter fragment that confers essentially full promoter activity contains a CCAAT box and a GC box upstream of the transcription start site and two GC boxes and an AP2 binding site downstream of the transcription start site. Mutation analysis showed that the HDAC1 promoter requires an intact CCAAT box and the distal GC box for full promoter activity. A number of nuclear transcription factors, including C/EBP, NF-Y, MSY1, and CTF/NF-1, have been shown to bind to the CCAAT box (23). GC-rich motifs are very common in many promoters, and several SP1 family members can bind to these sequences (36).
Electrophoretic mobility shift assays identified NF-Y as the transcription factor that binds to the CCAAT box and SP1 as the transcription factor recognizing the distal GC box. Further evidence that the transcription factor NF-Y is functional at the mouse HDAC1 promoter was obtained with the help of a dominant-negative mutant that inhibited expression of an HDAC1 promoter-reporter construct. We further showed that NF-Y does not work in isolation but rather operates in conjunction with SP1 or SP3 to regulate HDAC1 transcription. Both transcription factors were found to be associated with the endogenous HDAC1 promoter by chromatin immunoprecipitation experiments. Mutation of the CCAAT box and SP1 site in combination reduced HDAC1 promoter activity to minimal levels, whereas expression of both SP1/SP3 and NF-Y in Drosophila SL-2 cells led to amplification of HDAC1 promoter activity beyond that seen with either transcription factor alone. A functional synergism between SP1 and NF-Y has also been shown for other genes, including the genes for rat fatty acid synthase, human p27(Kip1), hamster TK, rat pyruvate kinase M, and the transforming growth factor β type II receptor (14, 15, 30, 33, 41, 45). In addition, a physical interaction of these transcription factors has been demonstrated (23, 31, 41). Taken together, our results strongly suggest that efficient HDAC1 expression depends upon functional interaction between the two transcription factors NF-Y and SP1.
Feedback regulation of the HDAC1 promoter.
Activation of the HDAC1 promoter by the histone deacetylase inhibitor TSA suggests a role for histone-modifying enzymes in the regulation of the HDAC1 gene. Very recently, it was shown that HDAC1 expression is induced by TSA in resting mouse fibroblasts (13), and similarly, HDAC inhibitors have been shown to affect the expression of class I HDACs in Hep3G cells and human lymphocytes (8, 10). These findings suggest a potential negative-feedback loop controlling HDAC1 expression. We show here by chromatin immunoprecipitation assays that HDAC1 can indeed bind to its promoter region, thereby repressing its own expression. Further, we demonstrate that the distal GC box or the CCAAT box, which are also involved in TSA-mediated activation of the HDAC1 promoter, can bind cofactors that recruit HDAC1 to the promoter region. This would result in targeted histone deacetylation and consequent silencing of the HDAC1 gene. This mechanism allows the cell to fine tune HDAC1 levels in response to changes in cellular deacetylase activity.
In resting fibroblasts, where HDAC1 gene expression is repressed, deacetylase activity would be dominant at the HDAC1 promoter. Inhibition of deacetylases would change the local equilibrium between acetyltransferases and deacetylases and favor transcription of the HDAC1 gene. In agreement with this scenario, TSA treatment of resting Swiss 3T3 cells led to histone hyperacetylation at the HDAC1 promoter and concomitant transcriptional activation, indicating the presence of acetyltransferases at the HDAC1 promoter. Further evidence for the role of HATs in the regulation of HDAC1 expression was provided by the observation that overexpression of p300 increases HDAC1 promoter activity in a dose-dependent manner. Mutation analysis suggests that histone acetyltransferases are recruited to the HDAC1 promoter by the same transcription factors that target HDAC1 to its promoter region. The fact that mutation of either the CCAAT box or the distal GC box alone did not abolish TSA-mediated activation of the HDAC1 promoter suggests that each transcription factor can independently recruit HATs to the HDAC1 promoter. However, both transcription factors together might be more efficient in the recruitment of coactivators, since the luciferase reporter assays clearly demonstrate cooperativity between NF-Y and SP1 in the activation of the HDAC1 promoter.
The binding sites for NF-Y and SP1 are required not only for activation of the HDAC1 promoter by acetyltransferases but also for recruitment of HDAC1, indicating a dual regulatory role of these transcription factors. Both transcription factors have been implicated in activation and repression of transcription. For instance, SP1 can serve as an anchor protein for positive and negative regulators of the mouse TK promoter (9) and the human p21/CIP1/WAF1 gene (20). SP1 can directly bind to HDAC1 (9) and was shown to associate with the acetyltransferase p300 (4). Similarly, NF-Y can recruit the transcriptional coactivators P/CAF and p300 (16, 22) but was also found to be involved in negative regulation of gene expression (26, 40, 42). A series of studies has described acetylation-mediated gene regulation in context with the transcription factor NF-Y or SP1 (16, 29, 34, 44, 45).
Interestingly, the presence of HDAC1 at the HDAC1 promoter was reduced in the presence of the histone deacetylase inhibitor TSA. Strikingly, a TSA-mediated dissociation of the SP1-HDAC1 complex was recently observed in the context of the regulation of the TβRII gene in human pancreatic cancer cells (45). Together, these data suggest that posttranslational modifications could regulate the recruitment of deacetylases. Indeed, several recent reports show that the regulatory functions of members of the SP1 transcription factor family are modulated by acetylation and phosphorylation (5, 6, 35).
In summary, our results demonstrate that transcription of the mouse HDAC1 gene is regulated by acetyltransferases and deacetylases, which are recruited by the transcription factors NF-Y and SP1 to the HDAC1 promoter. To our knowledge, this is the first report that describes the feedback regulation of a mammalian histone-modifying enzyme. Several findings indicate that the acetylation-dependent regulation of HDACs is a more general phenomenon. For instance, histones associated with the HDAC2 promoter were found to be hyperacetylated in HDAC1-null cells, suggesting that HDAC1 regulates not only its own expression but also that of other class I deacetylases (20). In agreement with this idea, levels of HDAC2 and HDAC3 are increased in HDAC1-deficient embryonic stem cells (20). Accordingly, the expression of endogenous HDAC1, HDAC2, and HDAC3 is decreased in HDAC1-overexpressing Swiss 3T3 fibroblasts (B. Schuettengruber and K. Kroboth, unpublished data). Similar to the autoregulatory circuits controlling the phosphorylation of proteins in signal transduction pathways, the activities of deacetylases and acetylases might be adjusted by feedback regulation.
Acknowledgments
We thank G. Zupkovitz for cultivating the ES cells, G. Weitzer for the GenomeWalker kit, G. Suske and R. Mantovani for antibodies, T. F. Osborne for the Drosophila expression constructs, E. Wintersberger for the NF-Y constructs and antibodies, H. Rotheneder for antibodies, T. Sauer for fluorescence-activated cell sorter analyses, and E. Wintersberger, H. Rotheneder, and R. Moriggl for helpful comments on the manuscript.
This work was supported by the Austrian FWF (grant P14909-GEN) and the Herzfelder Family Foundation.
REFERENCES
- 1.Ahringer, J. 2000. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 16:351-356. [DOI] [PubMed] [Google Scholar]
- 2.Baker, A., M. Saltik, H. Lehrmann, I. Killisch, V. Mautner, G. Lamm, G. Christofori, and M. Cotten. 1997. Polyethylenimine (PEI) is a simple, inexpensive and effective reagent for condensing and linking plasmid DNA to adenovirus for gene delivery. Gene Ther. 4:773-782. [DOI] [PubMed] [Google Scholar]
- 3.Bartl, S., J. Taplick, G. Lagger, H. Khier, K. Kuchler, and C. Seiser. 1997. Identification of mouse histone deacetylase 1 as a growth factor-inducible gene. Mol. Cell. Biol. 17:5033-5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Billon, N., D. Carlisi, M. B. Datto, L. A. van Grunsven, A. Watt, X. F. Wang, and B. B. Rudkin. 1999. Cooperation of Sp1 and p300 in the induction of the CDK inhibitor p21WAF1/CIP1 during NGF-mediated neuronal differentiation. Oncogene 18:2872-2882. [DOI] [PubMed] [Google Scholar]
- 5.Braun, H., R. Koop, A. Ertmer, S. Nacht, and G. Suske. 2001. Transcription factor Sp3 is regulated by acetylation. Nucleic Acids Res. 29:4994-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi, H., J. Lee, J. Park, and Y. Lee. 2002. Trichostatin A, a histone deacetylase inhibitor, activates the IGFBP-3 promoter by upregulating Sp1 activity in hepatoma cells: alteration of the Sp1/Sp3/HDAC1 multiprotein complex. Biochem. Biophys. Res. Commun. 296:1005-1012. [DOI] [PubMed] [Google Scholar]
- 7.Cress, W. D., and E. Seto. 2000. Histone deacetylases, transcriptional control, and cancer. J. Cell. Physiol. 184:1-16. [DOI] [PubMed] [Google Scholar]
- 8.Dangond, F., and S. R. Gullans. 1998. Differential expression of human histone deacetylase mRNAs in response to immune cell apoptosis induction by trichostatin A and butyrate. Biochem. Biophys. Res. Commun. 247:833-837. [DOI] [PubMed] [Google Scholar]
- 9.Doetzlhofer, A., H. Rotheneder, G. Lagger, M. Koranda, V. Kurtev, G. Brosch, E. Wintersberger, and C. Seiser. 1999. Histone deacetylase 1 can repress transcription by binding to Sp1. Mol. Cell. Biol. 19:5504-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray, S. G., and T. J. Ekstrom. 1998. Effects of cell density and trichostatin A on the expression of HDAC1 and p57Kip2 in Hep 3B cells. Biochem. Biophys. Res. Commun. 245:423-427. [DOI] [PubMed] [Google Scholar]
- 11.Gray, S. G., and T. J. Ekström. 2001. The human histone deacetylase family. Exp. Cell Res. 262:75-83. [DOI] [PubMed] [Google Scholar]
- 12.Hagen, G., S. Muller, M. Beato, and G. Suske. 1994. Sp1-mediated transcriptional activation is repressed by Sp3. EMBO J. 13:3843-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hauser, C., B. Schuettengruber, S. Bartl, G. Lagger, and C. Seiser. 2002. Activation of the HDAC1 gene by cooperative histone phosphorylation and acetylation. Mol. Cell. Biol. 22:7820-7830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu, Z., S. Jin, and K. W. Scotto. 2000. Transcriptional activation of the MDR1 gene by UV irradiation. Role of NF-Y and Sp1. J. Biol. Chem. 275:2979-2985. [DOI] [PubMed] [Google Scholar]
- 15.Inoue, T., J. Kamiyama, and T. Sakai. 1999. Sp1 and NF-Y synergistically mediate the effect of vitamin D(3) in the p27(Kip1) gene promoter that lacks vitamin D response elements. J. Biol. Chem. 274:32309-32317. [DOI] [PubMed] [Google Scholar]
- 16.Jin, S., and K. W. Scotto. 1998. Transcriptional regulation of the MDR1 gene by histone acetyltransferase and deacetylase is mediated by NF-Y. Mol. Cell. Biol. 18:4377-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karlseder, J., H. Rotheneder, and E. Wintersberger. 1996. Interaction of Sp1 with the growth- and cell cycle-regulated transcription factor E2F. Mol. Cell. Biol. 16:1659-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khier, H., S. Bartl, B. Schuettengruber, and C. Seiser. 1999. Cloning and characterization of the mouse histone deacetylase 1 gene: integration of a retrovirus in 129SV mice. Biochim. Biophys. Acta 1489:365-373. [DOI] [PubMed] [Google Scholar]
- 19.Khochbin, S., and H. Y. Kao. 2001. Histone deacetylase complexes: functional entities or molecular reservoirs. FEBS Lett. 494:141-144. [DOI] [PubMed] [Google Scholar]
- 20.Lagger, G., D. O'Carroll, M. Rembold, H. Khier, J. Tischler, G. Weitzer, B. Schuettengruber, C. Hauser, R. Brunmeir, T. Jenuwein, and C. Seiser. 2002. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 21:2672-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langley, E., M. Pearson, M. Faretta, U. M. Bauer, R. A. Frye, S. Minucci, P. G. Pelicci, and T. Kouzarides. 2002. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 21:2383-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, Q., M. Herrler, N. Landsberger, N. Kaludov, V. V. Ogryzko, Y. Nakatani, and A. P. Wolffe. 1998. Xenopus NF-Y pre-sets chromatin to potentiate p300 and acetylation-responsive transcription from the Xenopus hsp70 promoter in vivo. EMBO J. 17:6300-6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang, F., F. Schaufele, and D. G. Gardner. 2001. Functional interaction of NF-Y and Sp1 is required for type a natriuretic peptide receptor gene transcription. J. Biol. Chem. 276:1516-1522. [DOI] [PubMed] [Google Scholar]
- 24.Luo, J., A. Y. Nikolaev, S.-I. Imai, A. Shiloh, L. Guarente, and W. Gu. 2001. Negative control of p53 by Sir2α promotes cell survival under stress. Cell 107:137-148. [DOI] [PubMed] [Google Scholar]
- 25.Maity, S. N., and B. de Crombrugghe. 1998. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem. Sci. 23:174-178. [DOI] [PubMed] [Google Scholar]
- 26.Manni, I., G. Mazzaro, A. Gurtner, R. Mantovani, U. Haugwitz, K. Krause, K. Engeland, A. Sacchi, S. Soddu, and G. Piaggio. 2001. NF-Y mediates the transcriptional inhibition of the cyclin B1, cyclin B2, and cdc25C promoters upon induced G2 arrest. J. Biol. Chem. 276:5570-5576. [DOI] [PubMed] [Google Scholar]
- 27.Ng, H. H., and A. Bird. 2000. Histone deacetylases: silencers for hire. Trends Biochem. Sci. 25:121-126. [DOI] [PubMed] [Google Scholar]
- 28.Pan, J., and R. P. McEver. 1993. Characterization of the promoter for the human P-selectin gene. J. Biol. Chem. 268:22600-22608. [PubMed] [Google Scholar]
- 29.Peng, Y., and N. Jahroudi. 2003. The NFY transcription factor inhibits von Willebrand factor promoter activation in non-endothelial cells through recruitment of histone deacetylases. J. Biol. Chem. 278:8385-8394. [DOI] [PubMed] [Google Scholar]
- 30.Roder, K., S. S. Wolf, K. F. Beck, and M. Schweizer. 1997. Cooperative binding of NF-Y and Sp1 at the DNase I-hypersensitive site, fatty acid synthase insulin-responsive element 1, located at −500 in the rat fatty acid synthase promoter. J. Biol. Chem. 272:21616-21624. [DOI] [PubMed] [Google Scholar]
- 31.Roder, K., S. S. Wolf, K. J. Larkin, and M. Schweizer. 1999. Interaction between the two ubiquitously expressed transcription factors NF-Y and Sp1. Gene 234:61-69. [DOI] [PubMed] [Google Scholar]
- 32.Salsi, V., G. Caretti, M. Wasner, W. Reinhard, U. Haugwitz, K. Engeland, and R. Mantovani. 2003. Interactions between p300 and multiple NF-Y trimers govern cyclin B2 promoter function. J. Biol. Chem. 278:6642-6650. [DOI] [PubMed] [Google Scholar]
- 33.Sorensen, P., and E. Wintersberger. 1999. Sp1 and NF-Y are necessary and sufficient for growth-dependent regulation of the hamster thymidine kinase promoter. J. Biol. Chem. 274:30943-30949. [DOI] [PubMed] [Google Scholar]
- 34.Sowa, Y., T. Orita, S. Minamikawa, K. Nakano, T. Mizuno, H. Nomura, and T. Sakai. 1997. Histone deacetylase inhibitor activates the WAF1/Cip1 gene promoter through the Sp1 sites. Biochem. Biophys. Res. Commun. 241:142-150. [DOI] [PubMed] [Google Scholar]
- 35.Sun, J. M., H. Y. Chen, M. Moniwa, D. W. Litchfield, E. Seto, and J. R. Davie. 2002. The transcriptional repressor Sp3 is associated with CK2 phosphorylated histone deacetylase 2. J. Biol. Chem. 277:35783-35786. [DOI] [PubMed] [Google Scholar]
- 36.Suske, G. 1999. The Sp-family of transcription factors. Gene 238:291-300. [DOI] [PubMed] [Google Scholar]
- 37.Taunton, J., C. A. Hassig, and S. L. Schreiber. 1996. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272:408-411. [DOI] [PubMed] [Google Scholar]
- 38.Vaziri, H., S. K. Dessain, E. Ng Eaton, S. I. Imai, R. A. Frye, T. K. Pandita, L. Guarente, and R. A. Weinberg. 2001. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107:149-159. [DOI] [PubMed] [Google Scholar]
- 39.Verdel, A., and S. Khochbin. 1999. Identification of a new family of higher eukaryotic histone deacetylases—coordinate expression of differentiation-dependent chromatin modifiers. J. Biol. Chem. 274:2440-2445. [DOI] [PubMed] [Google Scholar]
- 40.Xu, Y., D. Banville, H. F. Zhao, X. Zhao, and S. H. Shen. 2001. Transcriptional activity of the SHP-1 gene in MCF7 cells is differentially regulated by binding of NF-Y factor to two distinct CCAAT-elements. Gene 269:141-153. [DOI] [PubMed] [Google Scholar]
- 41.Yamada, K., T. Tanaka, K. Miyamoto, and T. Noguchi. 2000. Sp family members and nuclear factor-Y cooperatively stimulate transcription from the rat pyruvate kinase M gene distal promoter region via their direct interactions. J. Biol. Chem. 275:18129-18137. [DOI] [PubMed] [Google Scholar]
- 42.Yun, J., H. D. Chae, H. E. Choy, J. Chung, H. S. Yoo, M. H. Han, and D. Y. Shin. 1999. p53 negatively regulates cdc2 transcription via the CCAAT-binding NF-Y transcription factor. J. Biol. Chem. 274:29677-29682. [DOI] [PubMed] [Google Scholar]
- 43.Zeng, Y. Y., C. M. Tang, Y. L. Yao, W. M. Yang, and E. Seto. 1998. Cloning and characterization of the mouse histone deacetylase-2 gene. J. Biol. Chem. 273:28921-28930. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, Y., and M. L. Dufau. 2002. Silencing of transcription of the human luteinizing hormone receptor gene by histone deacetylase-mSin3A complex. J. Biol. Chem. 277:33431-33438. [DOI] [PubMed] [Google Scholar]
- 45.Zhao, S., K. Venkatasubbarao, S. Li, and J. W. Freeman. 2003. Requirement of a specific Sp1 site for histone deacetylase-mediated repression of transforming growth factor beta type II receptor expression in human pancreatic cancer cells. Cancer Res. 63:2624-2630. [PubMed] [Google Scholar]