Abstract
Hypoxia-inducible factor 1α (HIF-1α) is essential for vascular development during embryogenesis and pathogenesis. However, little is known about its role in brain development. To investigate the function of HIF-1α in the central nervous system, a conditional knockout mouse was made with the Cre/LoxP system with a nestin promoter-driven Cre. Neural cell-specific HIF-1α-deficient mice exhibit hydrocephalus accompanied by a reduction in neural cells and an impairment of spatial memory. Apoptosis of neural cells coincided with vascular regression in the telencephalon of mutant embryos, and these embryonic defects were successfully restored by in vivo gene delivery of HIF-1α to the embryos. These results showed that expression of HIF-1α in neural cells was essential for normal development of the brain and established a mouse model that would be useful for the evaluation of therapeutic strategies for ischemia, including hypoxia-mediated hydrocephalus.
Oxygen deprivation initiates a wide range of responses to increase oxygen supply, including compensation for the loss of vital energy by alternating the expression of a variety of genes. Many of these hypoxia-inducible genes appear to have a common mode of regulation that involves activation of hypoxia-inducible factor 1α (HIF-1α), an oxygen-responsive subunit member of the basic helix-loop-helix PAS (PER-ARNT-SIM) family. HIF-1α heterodimerizes with the aryl hydrocarbon receptor nuclear translocator (ARNT; HIF-1β) and plays a key role in maintaining oxygen homeostasis by signaling hypoxic exposure to genes that are involved in angiogenesis, erythropoiesis, glycolysis, and cell survival (10, 34, 37). In addition to its roles in physiological oxygen homeostasis, HIF-1α has also been implicated in the pathogenesis of various diseases, including ischemic heart disease, stroke, and cancer (11, 29, 33, 36).
HIF-1α is expressed in the developing brain (16) and modulates gene activity in response to low oxygen in a hypoxic brain in vivo. HIF-1α has also been implicated as a critical factor in the pathogenesis of brain tumor vascularization and stroke by regulating local brain hypoxia (8, 36, 38). Although these results indicate that HIF-1α is involved in angiogenesis in the brain and neuroprotection, it has not been established whether HIF-1α contributes to brain development. Systemic disruption of the Hif-1α gene leads to embryonic lethality by day of embryonic development 11 (E11) that is accompanied by cardiovascular malformation and defective cephalic vascularization, indicating that HIF-1α is essential for embryonic vascularization (14, 21, 32). However, the significance of HIF-1α in the development of the central nervous system remains unclear.
To investigate the function of HIF-1α in the central nervous system, a conditional knockout mouse was generated with the Cre/LoxP system with a nestin promoter-based neural precursor-specific Cre recombinase (12). The nestin promoter directs gene expression specifically in neural precursor cells, so that a loxP-flanked (floxed) gene can be disrupted throughout the nervous system (41). The present results show that neural cell-specific ablation of the Hif-1α gene in mice results in hydrocephalus with impaired spatial memory, identifying HIF-1 in neural cells as essential for normal development of the brain. This mouse model will be useful for elucidating the molecular mechanism of neural response to ischemia, including hydrocephalus resulting from hypoxia.
MATERIALS AND METHODS
Conditional inactivation of the Hif-1α gene in mouse brain.
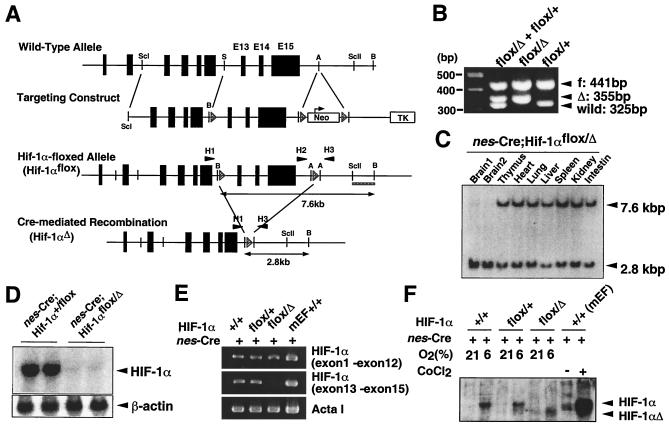
A 12-kbp BamHI-SacI genomic fragment from 129X1/SvJ mice containing exons 13 to 15 of Hif-1α was flanked with three loxP sites containing PGK-neo (Fig. 1A). The recombination frequency in embryonic stem (ES) cells (Genome Systems, St. Louis, Mo.) was 1.7%. Mice carrying this Hif-1α targeting allele were bred with mice transgenic for Cre recombinase under the control of the adenoviral EIIa promoter (24), resulting in the generation of a PGK-neo-deleted Hif-1α-floxed allele (Hif-1αflox) and Hif-1α-deleted allele (Hif-1αΔ) (Fig. 1A). The Hif-1α-floxed mice were crossed with nestin-Cre transgenic mice (12) to generate nestin-Cre;Hif-1αflox/+ and nestin-Cre;Hif-1αflox/Δ mice. The primers for genomic PCR were 5′-CTGTCTTCCCTGCTTAGGTCTTTCTAAC-3′ (H1), 5′-GAGATGGAGAAGGAGGTTAGTGTATCC-3′ (H2), and 5′-ACGTTGGCTCATGGTGTACTTTG-3′ (H3) (Fig. 1A and B).
FIG. 1.
Cre-mediated modification of Hif-1α gene. (A) Schematic representation of the mouse Hif-1α gene, the construct, floxed targeted allele, and Cre-disrupted Hif-1α gene. Hatched triangles represent loxP sites. Double-ended arrows denote BamHI restriction fragments detected with the 1.1-kbp SacII-BamHI probe (bold underline). H1, H2, and H3 (solid triangles) indicate the sites of PCR primers used for mouse screening. A, AvrII; B, BamHI; E, EcoRI; ScI, SacI; ScII, SacII; S, SalI. (B) PCR-based genotyping for Hif-1αflox/Δ (flox/Δ, mutant) and Hif-1αflox/+ (flox/+, control) mice. PCR was performed with primers H1, H2, and H3, resulting in the following products: 441 bp for athe flox allele with primers H2 and H3, 35 -bp for the deleted allele with primers H1 and H3, and 32 bp for the wild-type allele with primers H2 and H3. (C) Detection of Cre-mediated recombination in the tissues of Hif-1α-floxed/nestin-Cre mice. DNA (10 μg) from each tissue, including two independently prepared brain samples, was digested with BamHI, subjected to electrophoresis, and hybridized with the 32P-labeled SacII-BamHI fragment. (D) Expression of HIF-1α mRNA in nes-Cre;Hif-1αflox/Δ brain. Total brain RNA (10 μg) from the indicated mice was subjected to Northern blot analysis. A cDNA fragment corresponding to the targeting region was used as a 32P-labeled probe to detect Hif-1α mRNA. β-Actin signals were detected for a loading control. (E) Reverse transcription-PCR analysis of Hif-1α expression in the brain of mutant mice. cDNA samples from nes-Cre;Hif-1αflox/Δ (flox/Δ, mutant) brain, nes-Cre;Hif-1αflox/+ (flox/+, control) brain, C57BL/6N wild-type (+/+) brain, and mouse embryonic fibroblasts from wild-type mice (MEFs, +/+) were amplified for the sequences corresponding to the N-terminal (amino acids 1 to 209) and C-terminal (amino acids 693 to 822) regions of HIF-1α. The sequence encoding the C-terminal region resides within the two loxP sites, so that Cre-mediated recombination would delete this sequence in flox/Δ mutant mice. The MEFs and skeletal muscle α-actin (ActaI) were used as controls. (F) Western blot analysis of HIF-1α protein in mice maintained under normoxic (21% O2) and hypoxic (6% O2) conditions. Results shown are representative of three independent experiments. Hypoxic brain samples from nestin-Cre;Hif-1αflox/Δ (flox/Δ) mutant mice did not produce the wild-type HIF-1α signal at 120 kDa but gave a signal corresponding to the predicted nonfunctional truncated protein estimated at 107 kDa.
Behavioral study.
The spatial memory function of mice was examined with the radial maze task test as described previously (20). Mice were subjected to a maze trial (once a day) for four consecutive days, and the number of errors in the maze task was measured on each day. Spatial memory performance was estimated by the ratios of error rates on the day of experiments to the error rate on the first day. The spontaneous locomotor activity of mice was measured for 10 min in an activity box with an infrared beam (Animex; Muromachi, Tokyo, Japan) as described (20).
Morphometric analysis.
Head sections were prepared from paraformaldehyde-fixed, paraffin-embedded mice. Morphological identification of the neurons and morphometric counts of neural cell numbers in the occipital and perirhinal cortices were performed as described previously (39). Coronal and sagittal sections cut through the subcommissural organ were used to calculate the number of neural cells in brains of E16, E19, 5-day-old (P5), and 10-week-old (P70) nes-Cre;Hif-1αflox/Δ mutant mice and nes-Cre;Hif-1αflox/+ control mice (three for each section group). Glial cells and neuronal cells were identified by the characteristics of their nuclei.
For electron microscopic analysis, tissues fixed in 2.5% glutaraldehyde in 0.1 M phosphate buffer at 4°C for 2 days were immersed in 1% osmium tetroxide in 0.1 M phosphate buffer at 4°C for 2 h, dehydrated, and embedded in Epon 812 (TAAB Laboratories Equipment Limited, Berkshire, United Kingdom).
Immunohistochemistry and TUNEL analysis.
Frozen sections (16 to 20 μm) were fixed with either paraformaldehyde or acetone. The antibodies used were monoclonal anti-CD31 antibody (Becton Dickinson PharMingen, San Diego, Calif.), monoclonal anti-CD34 antibody (Cosmo Bio, Tokyo, Japan), rabbit anti-Flt-1 antibody (Santa Cruz Biotech, Santa Cruz, Calif.), rabbit anti-Flk-1 antibody (Santa Cruz Biotech, Santa Cruz, Calif.), and rabbit anti-desmin antibody (Chemicon International, Temecula, Calif.). Immunocomplexes were visualized with 3,3′-diaminobenzidine tetrahydrochloride (DAB) (Nichirei, Tokyo, Japan). Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) staining was performed with the ApopTagPlus peroxidase in situ apoptosis detection kit (Intergen Company). For double staining with TUNEL signals, DAB staining for CD31 was performed in the presence of nickel chloride.
Quantification of vessel density.
Vessel density per unit area was quantified from six randomly selected fields in the cortex (40). Images of CD31-stained sections were analyzed with Photograb-300 version 1.0 (Fuji Photo Film, Tokyo, Japan), Photoshop 5.5 (Adobe Systems Inc., San Jose, Calif.), and NIH Image 1.61 public domain software.
Laser capture microdissection.
Tissue sections (8 μm) of embryos at day 18.5 of gestation were fixed to a glass slide and briefly stained with the HistoGene LCM frozen section kit (Arcturus Co., Mountain View, Calif.). A region of the dorsal cortical plate in the brain was microdissected with a PixCell laser capture microdissector (Arcturus Co., Mountain View, Calif.). RNA from excised tissue samples representing approximately 1,000 cells of the dorsal cortical plate was reverse transcribed and amplified with the RiboAmp RNA amplification kit (Arcturus Co., Mountain View, Calif.).
Electroporation of expression vectors into telencephalic cells in utero.
A cDNA encoding the mouse Hif-1α or enhanced green fluorescent protein (EGFP) was inserted into the pEFGM mammalian expression vector, which was modified from the pEF-BOS vector (R. Kageyama, unpublished data). For the introduction of DNA, pregnant mice were deeply anesthetized, and a ventral midline incision was made to perform in utero manipulation. The expression vector (5 μg), in a solution containing 0.05% trypan blue as a tracer, was injected through the uterine wall with a glass micropipette into the telencephalic vesicle of each embryo (E13.5) in utero (31). Then 1 μg of pEFGM-EGFP was also coinjected with the expression vectors. The volume of the injected DNA solution was kept minimal (mostly about 1 μl). After injection, electroporation was carried out as described previously (7). Five days after the electroporation, at E18.5, embryos were subjected to further analysis.
Cre-adenovirus infection of primary cultured mouse embryonic fibroblasts.
A recombinant adenoviral vector including Cre recombinase gene (Ad-Cre) was constructed with the BD Adeno-X expression system (Clontech, Mountain View, Calif.) according to the manufacturer's instructions. The Cre recombinase cDNA was a kind gift from Brian Sauer at Stowers Institute for Medical Research in Kansas City. Briefly, Hif-1αflox/Δ mouse embryonic fibroblasts (MEFs) at 40 to 60% confluency were treated with Ad-Cre for 1 h at 37°C. Six hours after the infection, 100 μM CoCl2 was added to the cultured medium to mimic hypoxia, and cells were incubated for an additional 8 to 12 h.
Western blot analysis.
HIF-1α protein in the nuclear extracts from hypoxic brain tissues was detected with a chicken polyclonal antibody that was raised against peptides of amino acids 530 to 825 (4). To obtain mice under hypoxia, a hypoxic environment was produced by gradually decreasing FIO2 from 21% to 6% during an adaptation time of 1 h and kept at 6% O2 for 2 h. Embryonic fibroblasts from wild-type mice (MEFs) were treated with 200 μM CoCl2 for 24 h to mimic hypoxic conditions.
RESULTS
Generation of nes-Cre;Hif-1αflox/Δ mutant mice.
To examine the tissue-specific roles of HIF-1α, mice that carried a floxed Hif-1α allele were generated (Fig. 1A and B). LoxP sites were introduced to flank exons 13 to 15 of the Hif-1α gene (Hif-1αflox). These exons encode the COOH-terminal transactivation domain that is essential for responsiveness to hypoxia (5, 13, 23), optimal recruitment of CREB-binding protein (CBP)/p300 coactivator, and efficient nuclear localization (18, 25). This region also contains part of the bipartite nuclear localization signal of HIF-1α (28). Thus, Cre-mediated deletion would be expected to generate a gene that produces a truncated, functionally inert HIF-1α protein (Fig. 1A, see below). Hif-1αflox/flox mice were viable and appeared healthy, indicating that the floxed Hif-1α allele was functionally intact. To increase the efficiency of Cre-mediated HIF-1α disruption and prevent Cre-mediated interchromosomal recombination, Hif-1αflox/flox mice were crossed with Hif-1α+/Δ mice expressing the Cre transgene. To generate a neural cell-specific deletion, Cre-transgenic mice, in which Cre recombinase is expressed specifically in neural precursor cells under the nestin promoter/enhancer, were used (12).
Both nes-Cre;Hif-1αflox/+ and nes-Cre;Hif-1αflox/Δ mice were born at the expected Mendelian frequency and were macroscopically indistinguishable at birth. The nes-Cre;Hif-1αflox/Δ mutant mice (n = 35) as well as the nes-Cre;Hif-1αflox/+ control mice (n = 53) showed no abnormality in body weight or mortality within 20 months after birth (data not shown). Southern blot analysis revealed that the 7.6-kbp band corresponding to the Hif-1α-floxed allele was not detected in the brain of nes-Cre;Hif-1αflox/Δ mice (n = 4), whereas no deletion was found in other tissues (Fig. 1C), indicating that the targeted Hif-1α exons were specifically deleted in the brain by Cre-recombinase activity. Northern blot analysis and reverse transcription-PCR indicated that nes-Cre;Hif-1αflox/Δ mutant mice indeed exhibited a brain-specific loss of wild-type Hif-1α gene expression (Fig. 1D and E). Upon exposure to 6% oxygen hypoxia, nes-Cre;Hif-1αflox/Δ mutant cells did not produce a protein corresponding to the intact HIF-1α (120 kDa), but produced a truncated protein corresponding to the recombined allele (107 kDa) (Fig. 1F).
Characterization of Hif-1αΔ allele.
As reported previously for HIF-1α-deficient mice (14, 32), the Hif-1αΔ/Δ homozygous mutations resulted in embryonic lethality (data not shown), suggesting that the truncated Hif-1α product (107 kDa) from the Hif-1αΔ allele is functionally inert. Moreover, the truncated Hif-1αΔ product does not appear to interfere with the function of endogenous HIF-1α in a dominant-negative manner, since mice heterozygous for the germ line-disrupted Hif-1α allele (Hif-1α+/Δ) generated by crossing Hif-1αflox/flox mice with EIIa-Cre-transgenic mice (24) were not developmentally disturbed.
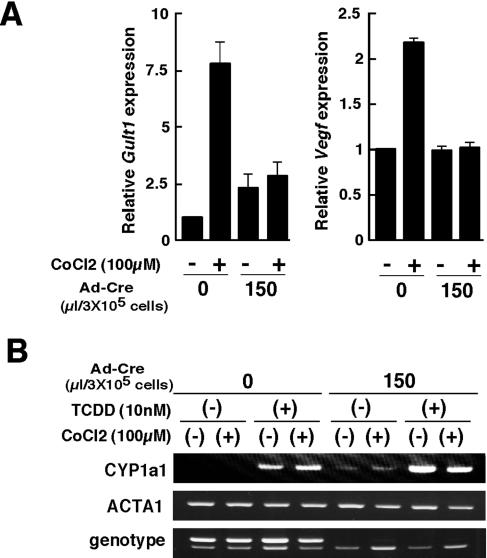
To analyze whether the truncated Hif-1α product derived from Hif-1αΔ allele may become functional under hypoxia, HIF-1α-regulated genes such as those for vascular endothelial growth factor (VEGF) and glucose transporter 1 (GLUT1) were examined in Hif-1α-floxed embryonic fibroblasts. Vegf and Glut1 expression in Hif-1αflox/Δ MEFs was induced in the presence of 100 μM CoCl2 to mimic hypoxia. However, expression of these Hif-1α downstream genes was not significantly elevated in Hif-1αflox/Δ MEFs treated with a recombinant adenovirus carrying Cre recombinase (Ad-Cre) (Fig. 2A). These results indicate that the Hif-1α Δ allele does not produce functional HIF-1α even under hypoxia.
FIG. 2.
Impaired expression of hypoxia-related genes in Hif-1α-deleted mouse embryonic fibroblasts. (A) Hif-1α-floxed MEFs (flox/Δ) were cultured with or without Ad-Cre in the absence or presence of 100 μM CoCl2 for 12 to 16 h. GLUT and VEGF expression levels were quantified by real-time reverse transcription-PCR with α-actin (ActaI) as an endogenous reference gene. Three independent experiments were performed for each genotype. Data are the means ± standard deviation of relative transcript level for the indicated genes. (B) Hif-1α-floxed MEFs with a deleted allele (flox/Δ) were cultured with or without Ad-Cre, 100 μM CoCl2, or 10 nM TCDD for 20 h. Results shown are representative of three independent experiments.
The possibility was considered that the truncated HIF-1α product derived from the Hif-1αΔ allele retains the capacity to associate with the promiscuous heterodimerization partner ARNT (also called HIF-1β) and thus may affect cellular responses by binding to ARNT and inhibiting other signal transduction pathways. This can be determined by analyzing induction of the Cyp1a1 gene, which is regulated by the AHR/ARNT signaling pathway, in the Ad-Cre-treated Hif-1αflox/Δ MEFs. Cyp1a1 expression induced by the potent AHR ligand 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) was unaffected in cells treated with Ad-Cre, even in the presence of CoCl2 (Fig. 2B). These results indicate that Cre-mediated generation of the Hif-1αΔ allele does not result in production of a protein that interferes with the functions of either HIF-1α or ARNT in a dominant negative manner, even though the Hif-1αΔ allele could produce the truncated 107-kDa product. Thus, nes-Cre;Hif-1αflox/Δ mice exhibit a functionally null mutation of the Hif-1α gene specifically in neural cells.
Hydrocephalus accompanied by reduced neuronal cell number and impaired spatial memory function in nes-Cre;Hif-1αflox/Δ mutant mice.
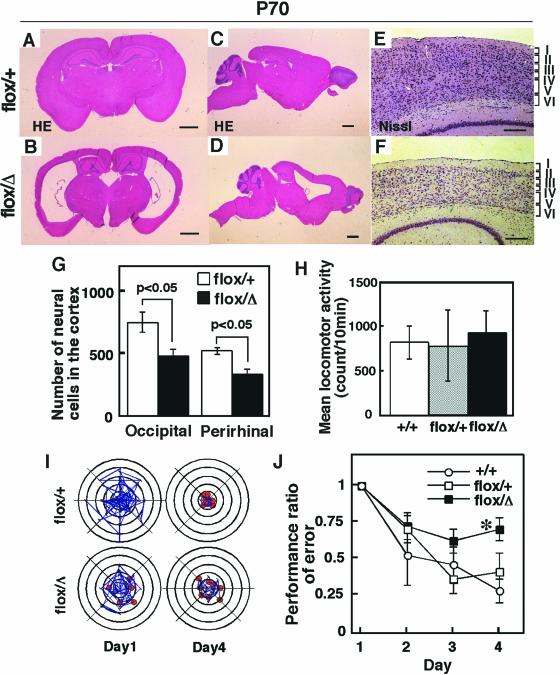
The nes-Cre;Hif-1αflox/Δ mutant mice showed no abnormality in external appearance and could not be distinguished from nes-Cre;Hif-1αflox/+ control mice. However, a striking morphological defect was found in the brain of adult nes-Cre;Hif-1αflox/Δ mutant mice. The most remarkable finding was atrophy of the cerebral cortex, with bilateral enlargement of the lateral ventricles (Fig. 3A to D). Although the six-layer structure of the cortical architecture was preserved in the mutant mice (Fig. 3E and F), neurons in the occipital and perirhinal cortices in 10-week-old (P70) mice were significantly reduced in number, by 36% ± 3.3% (P < 0.05) and 36% ± 2.6% (P < 0.05), respectively (Fig. 3G). These results indicate that the hydrocephalus observed in the adult nes-Cre;Hif-1αflox/Δ mutant mice was accompanied by reduced numbers of neural cells, although the neural structure of the cortical architecture was preserved.
FIG. 3.
Hydrocephalus and impaired spatial memory in nes-Cre;Hif-1αflox/Δ adult mice. (A to D) Adult brain coronal (A and B) and sagittal (C and D) sections from 10-week-old mice (P70) stained with hematoxylin and eosin. The nes-Cre;Hif-1αflox/Δ (flox/Δ) mutant mice exhibit enlarged lateral ventricles (B and D). Results shown are representative of three independent mice for each genotype. (E and F) Nissl staining of the nucleus indicates generation of the six-layer structure of the cortex (denoted by roman numerals) in the nes-Cre;Hif-1αflox/Δ mutant mice. (G) Neuronal cell numbers were measured on the coronal and sagittal sections through the subcommissural organ in brains from nes-Cre;Hif-1αflox/Δ (flox/Δ, black bar, n = 3) mutant and nes-Cre;Hif-1αflox/+ (flox/+, white bar, n = 3) control mice. Nissl staining was used for calculation of the neuronal cell numbers in occipital and perirhinal cortices in P70 mice (G). P values, calculated by Mann-Whitney U test, indicate significant reduction of neuronal cell numbers. (H) The spontaneous locomotor activity in an open field is indicated (mean ± standard error, n = 15 for each group). (I) Representative data for eight-arm radial maze task test. Blue line indicates amount of mouse movement for visiting the arms, and a red circle indicates the success of mouse's visiting an arm and eating food in the arm, as described in a previous report (20). (J) Performance error ratios in the radial maze task test were measured on the indicated days. Values (means ± standard error, n = 10 for each group) normalized to the comparable data on day 1 were measured for nes-Cre;Hif-1αflox/Δ (flox/Δ, solid squares), nes-Cre;Hif-1αflox/+ (flox/+, open squares), and C57BL/6N wild-type (+/+, open circles) mice. *, P < 0.05. Bars: 1 mm (A to D), 200 μm (E and F).
To evaluate the consequence of the morphological defects, adult nes-Cre;Hif-1αflox/Δ mutant mice were examined for learning and memory functions by use of the radial maze task test (20). Examination of spontaneous locomotion before the radial maze task test exhibited no significant difference between the activity of nes-Cre;Hif-1αflox/Δ mutant mice (933.8 ± 60.5 counts/10 min, n = 15) and nes-Cre;Hif-1αflox/+ control mice (774.8 ± 102.45 counts/10 min, n = 15) (Fig. 3H). Spatial memory task was examined by measuring the day-by-day decrease in the error rate of visiting wrong arms of the radial maze when a food pellet was placed behind each arm (20). It was previously shown that the initial storage of spatial memory is developed within 4 days in normal mice (20). We found that nes-Cre;Hif-1αflox/+ control mice (n = 10) normally learned the maze well enough to find a food pellet within 4 days of the test (Fig. 3I and J). On the other hand, nes-Cre;Hif-1αflox/Δ mutant mice (n = 10) exhibited significantly impaired memory consolidation on day 4 (error ratio = 0.699 ± 0.082, P < 0.05 versus 0.450 ± 0.126 in the nes-Cre;Hif-1αflox/+ heterozygous control mice) (Fig. 3J). Thus, nes-Cre;Hif-1αflox/Δ mutant mice exhibit deficient brain function.
Neural cell apoptosis and vascular regression in nes-Cre;Hif-1αflox/Δ mice during embryogenesis.
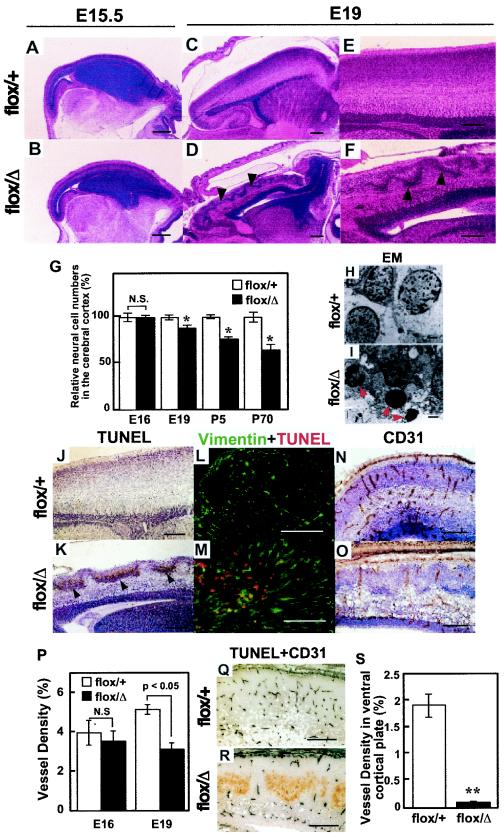
During embryogenesis, no gross morphological difference was detected until E15.5 between nes-Cre;Hif-1αflox/Δ mutant mice and controls (Fig. 4A and B). However, it was apparent at E19 that the cortex of the brain in the mutant mice was thinner than that of the control mice and that dense nuclei accumulated in a wavy architecture in the cortical telencephalon of the mutant mice (Fig. 4C to F). The neural cell numbers at a cerebral cortical area on the dorsal side of the hippocampus in nes-Cre;Hif-1αflox/Δ mutant mice were significantly lower by 12% ± 3.3% (n = 3, P < 0.05), 15% ± 2.6% (n = 3, P < 0.05), and 36% ± 2.6% (n = 3, P < 0.05) at E19, P5, and P70, respectively, than those in nes-Cre;Hif-1αflox/+ control mice (Fig. 4G). These results suggest that the embryonic reduction of neuronal cells in the brain results in the hydrocephalus seen in adult nes-Cre;Hif-1αflox/Δ mutant mice.
FIG. 4.
Neuronal cell death and vascular regression in nes-Cre;Hif-1αflox/Δ embryonic mice. (A to F) Hematoxylin and eosin staining for sagittal sections of embryonic brains from the indicated mice at E15.5 (A and B) and E19 (C to F). Arrowheads indicate the dense nuclei accumulated in a wavy architecture in nes-Cre;Hif-1αflox/Δ mutant telencephalon (D and F). (G) Relative neural cell numbers in the cerebral cortex werecalculated in brains from nes-Cre;Hif-1αflox/Δ (flox/Δ, black bar, n = 3) mutant and nes-Cre;Hif-1αflox/+ (flox/+, white bar, n = 3) control mice at E16, E19, postnatal day 5 (P5), and P70. *, P < 0.05. P values, calculated by Mann-Whitney U test, indicate significant reduction of neuronal cell numbers. (H and I) Transmission electron photomicrographs in the cortex of E19 embryos. Arrowheads indicate apoptotic cells observed in TUNEL-positive area in nes-Cre;Hif-1αflox/Δ telencephalon (I). (J to M) TUNEL (J and K), double staining for vimentin and TUNEL (L and M) and CD31, a marker of endothelial cells, (N and O) staining of sagittal sections from the nestin-Cre;Hif-1αflox/+ (flox/+, control) and nestin-Cre;Hif-1αflox/Δ (flox/Δ, mutant) E19 brains. Visible TUNEL-positive area (K, arrowheads), loss of vimentin-positive (green) cells in TUNEL-positive (red) area (M), and loss of CD31-positive area (O, brown) in the flox/Δ mutant cortex. (P) CD31-positive vessel density in the cortex was measured on E15.5 and E19 in brains from nes-Cre;Hif-1αflox/Δ (flox/Δ, black bar, n = 6) and nes-Cre;Hif-1αflox/+ (flox/+, white bar, n = 6) mice. Vessel density was calculated as a percentage of the CD31-positive area within the cortex. P < 0.05 at E19, not significant (N.S.) at E15.5. (Q and R) Discrete expressions of CD31 (blue) and TUNEL (brown) in sagittal sections of E19 brain cortices from nes-Cre;Hif-1αflox/Δ mice (R). (S) Vessel density of the ventral cortical plate in E19 mouse brains (n = 3 for each group) were measured. **, P < 0.005. Bars: 200 μm (A to F, J to O, Q, and R), 1 μm (H and I).
Electron microscopic analysis and TUNEL analysis indicated that the abnormal cells in the mutant brain were apoptotic neural cells (Fig. 4H to K). Their localization predominantly in the cortical plate suggested that the apoptotic cells were immature neural cells (Fig. 4K). To examine whether these apoptotic cells in the mutant mice included radial glia cells, which are essential for normal corticogenesis, brain tissues were double stained for TUNEL and vimentin, a cell marker for mesenchyme, including radial glia cells (Fig. 4L). These studies revealed that most TUNEL-positive cells at the ventral cortical plate in nes-Cre;Hif-1αflox/Δ mutant mice were negative for vimentin (Fig. 4M). In addition, some TUNEL-positive cells were positive for vimentin in the mutant brain, indicating that apoptotic cells in the mutant brain include glial cells as well as neural cells. These results indicate that HIF-1α in neural cells is essential for normal development of the mouse brain.
Interestingly, the density of CD31-positive endothelial vessels was significantly reduced in the telencephalon of mutant brains at E19 (Fig. 4N to P). Double staining for TUNEL and CD31 indicated that apoptotic neural cells and endothelial vessels in the telencephalon were localized discretely with each other (Fig. 4Q and R). The density of the vessels within the cortical plate was significantly lower in nes-Cre;Hif-1αflox/Δ mutant brains (n = 6) than in nes-Cre;Hif-1αflox/+ control brain (P < 0.005, 0.08% ± 0.02% versus 1.915% ± 0.22%) (Fig. 4S). The colocalization of apoptotic neural cells and deficient angiogenesis suggests that neural apoptosis in the HIF-1α-deficient embryonic brain is at least partly due to a lack of blood supply and thus resembles ischemia.
Expression of angiogenic factors in nes-Cre;Hif-1αflox/Δ embryonic brains.
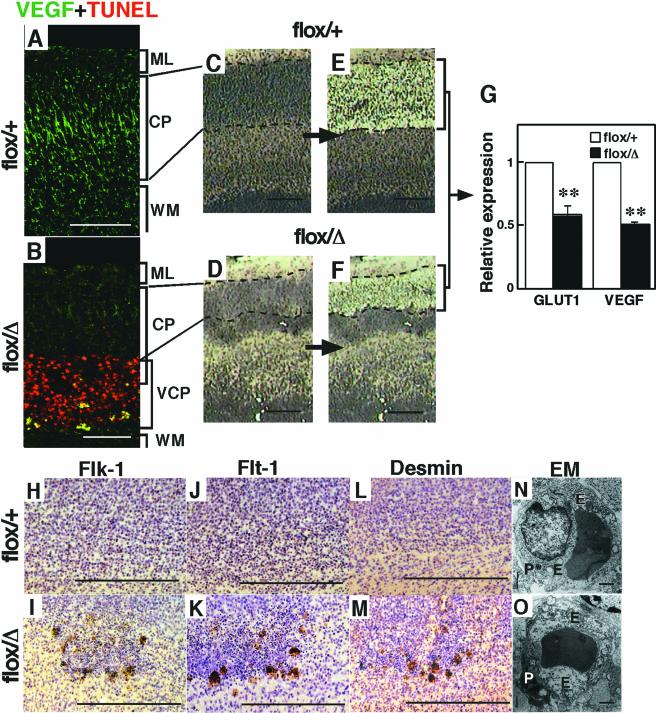
To determine the functional loss of HIF-1α activity in the Hif-1α mutant brains, the expression of hypoxia-mediated angiogenic factors such as VEGF and GLUT-1 was examined. In the neural cells at the dorsal cortical plate at E19, the expression of VEGF protein was markedly reduced in tthe nes-Cre;Hif-1αflox/Δ mutant brain compared to the nes-Cre;Hif-1αflox/+ control brain (Fig. 5A and B). Quantitative PCR analysis of mRNA expression levels in the microdissected dorsal cortical plate (Fig. 5C to F) indicated that the transcripts derived from the Hif-1α-regulated genes, such as Vegf and Glut1, were significantly reduced in nes-Cre;Hif-1αflox/Δ mutant brain (Fig. 5G). These results indicate that the dorsal cortical plate in the nes-Cre;Hif-1αflox/Δ mutant brain is deficient in the expression of HIF-1α-responsive genes during embryogenesis.
FIG. 5.
Reduction of HIF-1α-dependent Vegf expression at dorsal cortical plate, and HIF-1α-independent expression of angiogenic factors at the ventral cortical plate in nes-Cre;Hif-1αflox/Δ mutant mice. (A and B) Reduction of VEGF (green) expression at the dorsal cortical plate and increase at the ventral area of the surrounding TUNEL-positive cells in the nes-Cre;Hif-1αflox/Δ (flox/Δ, n = 3) mutant brains (B) compared with the nes-Cre;Hif-1αflox/+ (flox/+, n = 3) control brains (A). (C and D) Sagittal brain sections before and after microdissection of the region of the dorsal cortical plate in flox/+ (C) and flox/Δ (D) brains. (E) Glut1 and Vegf expression determined by a real-time reverse transcription-PCR method with α-actin as an endogenous reference gene. Reduction of expression levels of Glut1 and Vegf in HIF-1α-deficient dorsal cortical plate. **, P < 0.005. (F to K) Flk-1+ (G) and Flt-1+ (I) cells, expressed on a developing endothelial and vascular muscle cells, and desmin-positive cells (K), a marker of vascular smooth muscle cells, are localized around the apoptotic area in flox/Δ mutant cortex (n = 3 for each group). (L and M) Electron microscopic observation of endothelial cells in the E19 cortex, indicating that the cytoplasm of endothelial cells around the apoptotic areas of the flox/Δ mutant cortex were swollen with a hydropic appearance where polyribosomes were disaggregated (n = 3 for each group) (M). E, endothelial cell; P, pericyte. Bars: 200 μm (A to D, F to K), 1 μm (L and M).
Interestingly, high expression of VEGF was detected in a fraction of TUNEL-positive cells at the ventral cortical plate in the mutant brain (Fig. 5B). In addition, the VEGF receptors Flk-1 and Flt-1, mostly expressed by endothelial and vascular smooth muscle cells (1), were found to accumulate around the apoptotic areas in the mutant brain (Fig. 5H to K). Furthermore, desmin, a cell marker of vascular smooth muscle cells (35), was also localized around the apoptotic cells in the mutant brain (Fig. 5L and M). The vessels around the apoptotic cells appeared to be swollen and hydropic (Fig. 5N and O). These results suggest that angiogenesis is arrested at the ventral cortical plate around apoptotic neural cells during the embryogenesis of nes-Cre;Hif-1αflox/Δ mutant mice.
Restoration of neural cell apoptosis and vascular regression by the introduction of Hif-1α into embryonic nes-Cre;Hif-1αflox/Δ mutant brains.
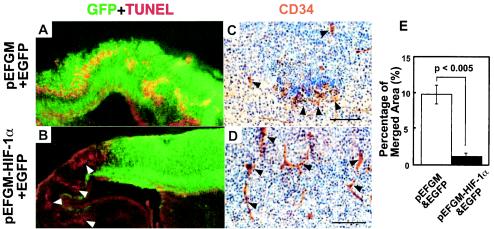
Deprivation of oxygen leads to a severe dysfunction of the central nervous system from fetus to adulthood. The defective brain in nes-Cre;Hif-1αflox/Δ mutant mice represents a useful experimental model to evaluate therapeutic strategies for ischemic brain insults, including hypoxia-mediated hydrocephalus. To explore this possibility, an HIF-1α expression vector was introduced into telencephalic cells of mutant embryos by electroporation in utero. Introduction of a control EGFP vector into the brain revealed colocalization of the transfected cells and TUNEL-positive apoptotic cells in a wavy pattern within the cerebral cortex of the mutant brain (Fig. 6A). However, upon introduction of Hif-1α along with EGFP, virtually all EGFP-positive cells became TUNEL negative (Fig. 6B and E). In addition to preventing cortical neurons from undergoing apoptosis, Hif-1α gene transfer to the mutant telencephalon also restored the thickness of the telencephalon (Fig. 6B) and vessel formation in situ (Fig. 6C and D). These results indicate that Hif-1α gene transfer in vivo could restore the development of the brain in neuronal cell-specific HIF-1α-deficient mice.
FIG. 6.
Restoration of neuronal cell death and vascular defects in the telencephalon of nes-Cre;Hif-1αflox/Δ mutant mice by Hif-1α transfection. (A and B) The telencephalons of nes-Cre;Hif-1αflox/Δ mice were transfected in utero at E13.5 with the EGFP expression vector (1 μg) along with either the control expression vector pEFGM (5 μg, A) or HIF-1α expression vector pEFGM-HIF-1α (5 μg, B). Brain sections at E18.5 were stained for TUNEL (red) and EGFP (green). TUNEL-positive red signals merged in EGFP-positive green cells (yellow signals as in A) were significantly reduced by the codelivery of Hif-1α (B and E, n = 3). The TUNEL-positive signals were found in the area without gene delivery (B, white arrowheads), indicating that neuronal apoptosis was specifically rescued by Hif-1α gene delivery. (C and D) The telencephalon sections from nes-Cre;Hif-1αflox/Δ E18.5 embryos that had been transfected with HIF-1α- and EGFP-expressing vectors were stained for CD34 (brown) and hematoxylin (blue). The sparse distribution of CD34+ endothelial cells (arrowheads) around the apoptotic area with vehicle and EGFP gene delivery (C) was specifically restored and formed vessel structures in the area of Hif-1α- and EGFP-transfected cells (D). (E) Ratios of merged TUNEL-positive (yellow) to EGFP-positive (green) area in the telencephalon transfected with DNA of Hif-1α (n = 3, black bar) or vehicle (n = 3, white bar) together with EGFP were measured at E18.5 in nes-Cre;Hif-1αflox/Δ mice (C and D). Bars, 50 μm.
DISCUSSION
HIF-1α was shown to be essential for early mammalian development by the analysis of systemic HIF-1α knockout mice, which die by E11 of gestation (14, 21). However, it remains unclear how HIF-1α contributes to the development and functions of the mouse brain at subsequent stages and whether HIF-1α in neural cells might be involved in the development of the central nervous system. In the present study, a neural cell-specific HIF-1α-deficient mouse was established, and the developing brain was characterized during embryogenesis and after birth. These results demonstrate that HIF-1α in neural cells is essential for the normal development of a functional brain. In the absence of HIF-1α in neural cells, mice were viable to adulthood and appeared indistinguishable from wild-type mice. However, the neural cell-specific HIF-1α-deficient mice exhibited severe hydrocephalus accompanied by impaired memory. These results show that HIF-1α in neural cells is indispensable for optimal development of a functional brain and that other hypoxia-responsive proteins are incapable of fully compensating for HIF-1α deficiency in neural cells.
To delete the floxed Hif-1α gene in neural precursor cells of the developing nervous system, transgenic mice that expressed Cre recombinase under the control of the nestin promoter and an additional neural precursor-specific enhancer were used (12). A previous report established that this transgenic line expresses Cre in the ventricular zone of the developing central nervous system, such as the telencephalon and spinal cord, at E11.5 of gestation (12). The Cre transgene is also expressed in the peripheral nervous system, such as the dorsal root ganglia, but not in endothelial cells of blood vessels (R. Kageyama, unpublished data). Thus, the nes-Cre;Hif-1αflox/Δ mutant genes generated in this study could be specifically targeted Hif-1α genes in neural cells and not in endothelial cells. In fact, Tie2-Cre-mediated HIF-1α-deficient mice, in which the Hif-1α gene is deleted in endothelial cells of blood vessels, did not show phenotypes similar to those of nes-Cre;Hif-1αflox/Δ mutant mice, including hydrocephalus accompanied by apoptotic neural cells and vascular regression (F. J. Gonzalez and S. H. Yim, unpublished data). The Tie2-Cre gene used in this study is expressed specifically in endothelial cells (19). Thus, the defective vessel formation in the mutant brain is likely due to the lack of angiogenic signals from HIF-1α-deficient neural cells as recently suggested (30).
To demonstrate lack of function of the modified Hif-1α allele, MEFs derived from the floxed mice were analyzed. Hif-1αΔ/Δ homozygous mutant MEFs were incapable of responding to CoCl2, as revealed by lack of induction of HIF-1α-responsive genes such as Vegf and Glut-1. In addition, Hif-1αΔ/Δ homozygous mutations were embryonic lethal, in reported for other HIF-1α-deficient mouse lines (14, 32), suggesting that the Hif-1αΔ allele was functionally inert. However, it was previously shown that a deletion mutant of HIF-1α that nearly corresponds to our Hif-1αΔ-derived 107-kDa product slightly mediates the hypoxic induction of the N-terminal transactivation domain-dependent transcription of the reporter constructs (5, 17). A splicing variant of human HIF-1α lacking part of the 13th and 14th exons also had a markedly attenuated transactivation activity (9). More recently, it was demonstrated that the COOH-terminal transactivation domain contains a conserved and critical Asn that requires hydroxylation by a 2-oxoglutarate-dependent dioxygenase (2, 6, 13, 15) for recruitment of the coactivator p300 (25; reviewed by Bruick and McKnight [3]). Thus, while it is likely that the truncated 107-kDa protein produced by the recombined floxed Hif-1α allele is not functional, we cannot totally exclude the possibility that it may retain an unknown marginal function, even though we did not detect any in vivo activity.
The possibility exists that the truncated protein exhibits dominant negative activity. In this connection, neither the nes-Cre;Hif-1αflox/+ mice nor the Hif-1α+/Δ mice were developmentally disturbed, and neither the presence nor the expression of Hif-1αΔ/Δ alleles interfered with the TCDD-induced ARNT-mediated expression of Cyp1a1. These results indicate that Cre-mediated generation of the Hif-1αΔ allele does not produce a protein that interferes with the functions of either HIF-1α or ARNT in a dominant negative manner, even though the Hif-1αΔ allele could produce the truncated 107-kDa product.
Mouse mutations that genetically cause hydrocephalus have been reported, created by manipulating genes involved in the growth, survival, adhesion, cell structure, and migration of neural cells as well as cerebrospinal fluid regulation (22, 26, 27). However, these phenotypes were not relevant to abnormal hypoxic responses. By producing a situation in which hypoxic responses in the brain are disturbed, the HIF-1α-deficient model described in the present study provides a new tool to study hypoxia-mediated hydrocephalus, which occurs in humans upon hypoxic encephalopathy, and could be of great value in assessing the therapeutic effects of angiogenic and/or neuroprotective factors on ischemic damage in the brain.
Acknowledgments
We thank H. B. Jiang and J. Ding for maintenance of the mice and S. Kimura and S. H. Yim for critical review of the manuscript.
REFERENCES
- 1.Breier, G., M. Clauss, and W. Risau. 1995. Coordinate expression of vascular endothelial growth factor receptor-1 (flt-1) and its ligand suggests a paracrine regulation of murine vascular development. Dev. Dyn. 204:228-239. [DOI] [PubMed] [Google Scholar]
- 2.Bruick, R. K., and S. L. McKnight. 2001. A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294:1337-1340. [DOI] [PubMed] [Google Scholar]
- 3.Bruick, R. K., and S. L. McKnight. 2002. Transcription. Oxygen sensing gets a second wind. Science 295:807-808. [DOI] [PubMed] [Google Scholar]
- 4.Camenisch, G., M. Tini, D. Chilov, I. Kvietikova, V. Srinivas, J. Caro, P. Spielmann, R. H. Wenger, and M. Gassmann. 1999. General applicability of chicken egg yolk antibodies: the performance of IgY immunoglobulins raised against the hypoxia-inducible factor 1α. FASEB J. 13:81-88. [DOI] [PubMed] [Google Scholar]
- 5.Ema, M., K. Hirota, J. Mimura, H. Abe, J. Yodoi, K. Sogawa, L. Poellinger, and Y. Fujii-Kuriyama. 1999. Molecular mechanisms of transcription activation by HLF and HIF1α in response to hypoxia: their stabilization and redox signal-induced interaction with CBP/p300. EMBO J. 18:1905-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Epstein, A. C., J. M. Gleadle, L. A. McNeill, K. S. Hewitson, J. O'Rourke, D. R. Mole, M. Mukherji, E. Metzen, M. I. Wilson, A. Dhanda, Y. M. Tian, N. Masson, D. L. Hamilton, P. Jaakkola, R. Barstead, J. Hodgkin, P. H. Maxwell, C. W. Pugh, C. J. Schofield, and P. J. Ratcliffe. 2001. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107:43-54. [DOI] [PubMed] [Google Scholar]
- 7.Funahashi, J., T. Okafuji, H. Ohuchi, S. Noji, H. Tanaka, and H. Nakamura. 1999. Role of Pax-5 in the regulation of a mid-hindbrain organizer's activity. Dev. Growth Differ. 41:59-72. [DOI] [PubMed] [Google Scholar]
- 8.Giordano, F. J., and R. S. Johnson. 2001. Angiogenesis: the role of the microenvironment in flipping the switch. Curr. Opin. Genet. Dev. 11:35-40. [DOI] [PubMed] [Google Scholar]
- 9.Gothie, E., D. E. Richard, E. Berra, G. Pages, and J. Pouyssegur. 2000. Identification of alternative spliced variants of human hypoxia-inducible factor-1α. J. Biol. Chem. 275:6922-6927. [DOI] [PubMed] [Google Scholar]
- 10.Hofer, T., H. Wenger, and M. Gassmann. 2002. Oxygen sensing, HIF-1α stabilization and potential therapeutic strategies. Pflugers Arch. 443:503-507. [DOI] [PubMed] [Google Scholar]
- 11.Hopfl, G., R. H. Wenger, U. Ziegler, T. Stallmach, O. Gardelle, R. Achermann, M. Wergin, B. Kaser-Hotz, H. M. Saunders, K. J. Williams, I. J. Stratford, M. Gassmann, and I. Desbaillets. 2002. Rescue of hypoxia-inducible factor-1α-deficient tumor growth by wild-type cells is independent of vascular endothelial growth factor. Cancer Res. 62:2962-2970. [PubMed] [Google Scholar]
- 12.Isaka, F., M. Ishibashi, W. Taki, N. Hashimoto, S. Nakanishi, and R. Kageyama. 1999. Ectopic expression of the bHLH gene Math1 disturbs neural development. Eur. J. Neurosci. 11:2582-2588. [DOI] [PubMed] [Google Scholar]
- 13.Ivan, M., K. Kondo, H. Yang, W. Kim, J. Valiando, M. Ohh, A. Salic, J. M. Asara, W. S. Lane, and W. G. Kaelin, Jr. 2001. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292:464-468. [DOI] [PubMed] [Google Scholar]
- 14.Iyer, N. V., L. E. Kotch, F. Agani, S. W. Leung, E. Laughner, R. H. Wenger, M. Gassmann, J. D. Gearhart, A. M. Lawler, A. Y. Yu, and G. L. Semenza. 1998. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 α. Genes Dev. 12:149-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaakkola, P., D. R. Mole, Y. M. Tian, M. I. Wilson, J. Gielbert, S. J. Gaskell, A. Kriegsheim, H. F. Hebestreit, M. Mukherji, C. J. Schofield, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292:468-472. [DOI] [PubMed] [Google Scholar]
- 16.Jain, S., E. Maltepe, M. M. Lu, C. Simon, and C. A. Bradfield. 1998. Expression of ARNT, ARNT2, HIF1 α, HIF2 α and Ah receptor mRNAs in the developing mouse. Mech. Dev. 73:117-123. [DOI] [PubMed] [Google Scholar]
- 17.Jiang, B. H., J. Z. Zheng, S. W. Leung, R. Roe, and G. L. Semenza. 1997. Transactivation and inhibitory domains of hypoxia-inducible factor 1α. Modulation of transcriptional activity by oxygen tension. J. Biol. Chem. 272:19253-19260. [DOI] [PubMed] [Google Scholar]
- 18.Kallio, P. J., K. Okamoto, S. O'Brien, P. Carrero, Y. Makino, H. Tanaka, and L. Poellinger. 1998. Signal transduction in hypoxic cells: inducible nuclear translocation and recruitment of the CBP/p300 coactivator by the hypoxia-inducible factor-1α. EMBO J. 17:6573-6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kisanuki, Y. Y., R. E. Hammer, J. Miyazaki, S. C. Williams, J. A. Richardson, and M. Yanagisawa. 2001. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev. Biol. 230:230-242. [DOI] [PubMed] [Google Scholar]
- 20.Komatsu, H., J. Nogaya, N. Kuratani, M. Ueki, S. Yokono, and K. Ogli. 1998. Repetitive post-training exposure to enflurane modifies spatial memory in mice. Anesthesiology 89:1184-1190. [DOI] [PubMed] [Google Scholar]
- 21.Kotch, L. E., N. V. Iyer, E. Laughner, and G. L. Semenza. 1999. Defective vascularization of HIF-1α-null embryos is not associated with VEGF deficiency but with mesenchymal cell death. Dev. Biol. 209:254-267. [DOI] [PubMed] [Google Scholar]
- 22.Kume, T., K. Y. Deng, V. Winfrey, D. B. Gould, M. A. Walter, and B. L. Hogan. 1998. The forkhead/winged helix gene Mf1 is disrupted in the pleiotropic mouse mutation congenital hydrocephalus. Cell 93:985-996. [DOI] [PubMed] [Google Scholar]
- 23.Kung, A. L., S. Wang, J. M. Klco, W. G. Kaelin, and D. M. Livingston. 2000. Suppression of tumor growth through disruption of hypoxia-inducible transcription. Nat. Med. 6:1335-1340. [DOI] [PubMed] [Google Scholar]
- 24.Lakso, M., J. G. Pichel, J. R. Gorman, B. Sauer, Y. Okamoto, E. Lee, F. W. Alt, and H. Westphal. 1996. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. USA 93:5860-5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lando, D., D. J. Peet, D. A. Whelan, J. J. Gorman, and M. L. Whitelaw. 2002. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science 295:858-861. [DOI] [PubMed] [Google Scholar]
- 26.Liedtke, W., W. Edelmann, P. L. Bieri, F. C. Chiu, N. J. Cowan, R. Kucherlapati, and C. S. Raine. 1996. GFAP is necessary for the integrity of CNS white matter architecture and long-term maintenance of myelination. Neuron 17:607-615. [DOI] [PubMed] [Google Scholar]
- 27.Lindeman, G. J., L. Dagnino, S. Gaubatz, Y. Xu, R. T. Bronson, H. B. Warren, and D. M. Livingston. 1998. A specific, nonproliferative role for E2F-5 in choroid plexus function revealed by gene targeting. Genes Dev. 12:1092-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo, J. C., and M. Shibuya. 2001. A variant of nuclear localization signal of bipartite-type is required for the nuclear translocation of hypoxia inducible factors (1α, 2α and 3α). Oncogene 20:1435-1444. [DOI] [PubMed] [Google Scholar]
- 29.Maxwell, P. H., C. W. Pugh, and P. J. Ratcliffe. 2001. Activation of the HIF pathway in cancer. Curr. Opin. Genet. Dev. 11:293-299. [DOI] [PubMed] [Google Scholar]
- 30.Ogunshola, O. O., A. Antic, M. J. Donoghue, S. Y. Fan, H. Kim, W. B. Stewart, J. A. Madri, and L. R. Ment. 2002. Paracrine and autocrine functions of neuronal vascular endothelial growth factor (VEGF) in the central nervous system. J. Biol. Chem. 277:11410-11415. [DOI] [PubMed] [Google Scholar]
- 31.Ohtsuka, T., M. Sakamoto, F. Guillemot, and R. Kageyama. 2001. Roles of the basic helix-loop-helix genes Hes1 and Hes5 in expansion of neural stem cells of the developing brain. J. Biol. Chem. 276:30467-30474. [DOI] [PubMed] [Google Scholar]
- 32.Ryan, H. E., J. Lo, and R. S. Johnson. 1998. HIF-1 α is required for solid tumor formation and embryonic vascularization. EMBO J. 17:3005-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryan, H. E., M. Poloni, W. McNulty, D. Elson, M. Gassmann, J. M. Arbeit, and R. S. Johnson. 2000. Hypoxia-inducible factor-1α is a positive factor in solid tumor growth. Cancer Res. 60:4010-4015. [PubMed] [Google Scholar]
- 34.Schipani, E., H. E. Ryan, S. Didrickson, T. Kobayashi, M. Knight, and R. S. Johnson. 2001. Hypoxia in cartilage: HIF-1α is essential for chondrocyte growth arrest and survival. Genes Dev. 15:2865-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmid, E., M. Osborn, E. Rungger-Brandle, G. Gabbiani, K. Weber, and W. W. Franke. 1982. Distribution of vimentin and desmin filaments in smooth muscle tissue of mammalian and avian aorta. Exp. Cell Res. 137:329-340. [DOI] [PubMed] [Google Scholar]
- 36.Semenza, G. L. 2000. HIF-1 and human disease: one highly involved factor. Genes Dev. 14:1983-1991. [PubMed] [Google Scholar]
- 37.Semenza, G. L. 1999. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu. Rev. Cell. Dev. Biol. 15:551-578. [DOI] [PubMed] [Google Scholar]
- 38.Semenza, G. L. 2000. Surviving ischemia: adaptive responses mediated by hypoxia-inducible factor 1. J. Clin. Investig. 106:809-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimada, A., A. Ohta, I. Akiguchi, and T. Takeda. 1992. Inbred SAM-P/10 as a mouse model of spontaneous, inherited brain atrophy. J. Neuropathol. Exp. Neurol. 51:440-450. [DOI] [PubMed] [Google Scholar]
- 40.Swift, M. E., H. K. Kleinman, and L. A. DiPietro. 1999. Impaired wound repair and delayed angiogenesis in aged mice. Lab. Investig. 79:1479-1487. [PubMed] [Google Scholar]
- 41.Zimmerman, L., B. Parr, U. Lendahl, M. Cunningham, R. McKay, B. Gavin, J. Mann, G. Vassileva, and A. McMahon. 1994. Independent regulatory elements in the nestin gene direct transgene expression to neural stem cells or muscle precursors. Neuron 12:11-24. [DOI] [PubMed] [Google Scholar]