Abstract
Iron regulatory protein 1 (IRP1) binds to mRNA iron-responsive elements (IREs) and thereby controls the expression of IRE-containing mRNAs. In iron-replete cells, assembly of a cubane [4Fe-4S] cluster inhibits IRE-binding activity and converts IRP1 to a cytosolic aconitase. Earlier experiments with Saccharomyces cerevisiae suggested that phosphomimetic mutations of Ser-138 negatively affect the stability of the cluster (N. M. Brown, S. A. Anderson, D. W. Steffen, T. B. Carpenter, M. C. Kennedy, W. E. Walden, and R. S. Eisenstein, Proc. Natl. Acad. Sci. USA 95:15235-15240, 1998). Along these lines, we show here that a highly purified preparation of recombinant human IRP1 bearing a phosphomimetic S138E substitution (IRP1S138E) lacks aconitase activity, which is a hallmark of [4Fe-4S] cluster integrity. Similarly, IRP1S138E expressed in mammalian cells fails to function as aconitase. Furthermore, we demonstrate that the impairment of [4Fe-4S] cluster assembly in mammalian cells sensitizes IRP1S138E to iron-dependent degradation. This effect can be completely blocked by the iron chelator desferrioxamine or by the proteasome inhibitors MG132 and lactacystin. As expected, the stability of wild-type or phosphorylation-deficient IRP1S138A is not affected by iron manipulations. Ser-138 and flanking sequences appear to be highly conserved in the IRP1s of vertebrates, whereas insect IRP1 orthologues and nonvertebrate IRP1-like molecules contain an S138A substitution. Our data suggest that phosphorylation of Ser-138 may provide a basis for an additional mechanism for the control of vertebrate IRP1 activity at the level of protein stability.
The iron regulatory proteins IRP1 and IRP2 are involved in the coordinate posttranscriptional regulation of cellular iron metabolism by binding to mRNA iron-responsive elements (IREs). These are hairpin structures within the 5′ or 3′ untranslated regions of a growing family of mRNAs that encode proteins of iron uptake, storage, utilization, and transport, as well as energy metabolism (8, 18). Among the best-characterized IRE-containing mRNAs are those encoding the transferrin receptor, which plays a critical role in cellular iron uptake, and ferritin, a protein for intracellular iron storage. IRE-IRP interactions largely account for the reciprocal control of the transferrin receptor and ferritin expression in response to iron perturbations (25).
IRP1 and IRP2 are homologous to mitochondrial aconitase (5, 8, 23), an enzyme of the citric acid cycle, and in fact, IRP1 is its cytosolic counterpart. The active site of aconitase contains a cubane [4Fe-4S] cluster (1). In IRP1, this cluster is assembled within iron-replete cells and prevents IRE binding. However, iron starvation, nitrogen monoxide (NO), and extracellular H2O2 (5, 23) trigger the cluster's destabilization. The resulting switch to apo-IRP1 is associated with the loss of aconitase and acquisition of IRE-binding activity. The mechanism for IRP2 regulation is distinct and does not involve iron-sulfur cluster biochemistry. While IRP2 is stable in iron-starved and hypoxic cells, it undergoes proteasomal degradation in the presence of iron, oxygen (16), or the nitrosonium cation (NO+) (19).
It has been proposed that the activities of IRP1 and IRP2 may also be regulated by phosphorylation (8). The physiological significance of this finding remains elusive. Both IRP1 and IRP2 can be subjected to phosphorylation by protein kinase C (PKC) (9, 27). The rate of IRP1 phosphorylation by PKC is negatively affected by the presence of the iron-sulfur cluster (26). Experiments with synthetic oligopeptides showed that the conserved Ser-138 and Ser-711 residues are targets of phosphorylation in vitro and implied that phosphorylation may also occur in vivo. Ser-138 is located within the region involved in RNA binding and assembly of the [4Fe-4S] cluster, which spans residues 116 to 151.
More direct evidence for the involvement of Ser-138 in the control of the iron-sulfur cluster assembly was provided by experiments in an aconitase-deficient Saccharomyces cerevisiae strain. Aerobic growth of the aco1 cells depends on complementation with iron-loaded IRP1 (i.e., cytosolic aconitase). The replacement of Ser-138 in IRP1 with phosphomimetic residues (glutamic or aspartic acid) impairs aerobic growth and prevents [4Fe-4S] cluster assembly (3). The sensitivity of these mutants to oxidative destabilization of the iron-sulfur cluster in the context of yeast has been confirmed by electron paramagnetic resonance spectroscopy (4). Here, we further investigate the role of Ser-138 in the function of IRP1 by studying the properties of nonphosphorylatable IRP1S138A and phosphomimetic IRP1S138E mutants in mammalian cells.
MATERIALS AND METHODS
Materials.
Hemin, MG132, lactacystin, 3-methyladenine, and antibodies against FLAG (M2-FLAG) and β-actin were purchased from Sigma (St. Louis, Mo.). Desferrioxamine was obtained from Novartis (Dorval, Canada). Salicylaldehyde isonicotinoyl hydrazone (SIH) was a kind gift of Prem Ponka (McGill University). Fe-SIH was prepared by mixing SIH with ferric citrate in a ratio of 2:1.
Plasmid construction.
The S138A and S138E point mutations were introduced by site-directed mutagenesis with the ExSite PCR-based protocol (Stratagene), according to the manufacturer's recommendations. The plasmid pSG5-hIRP1 (29) was utilized as the template with the following oligonucleotides: for S138A, CAA CAG AAG GGC AGA CGC CTT ACA GAA GAA TCA AG and the reverse complement CTT GAT TCT TCT GTA AGG CGT CTG CCC TTC TGT TG and for S138E, CAA CAG AAG GGC AGA CGA ATT ACA GAA GAA TCA AG and the reverse complement CTT GAT TCT TCT GTA ATT CGT CTG CCC TTC TGT TG. All mutations were confirmed by sequencing. For the generation of recombinant protein in bacteria, a 1.5-kb PstI/EcoRV fragment from pSG5-hIRP1S138A or pSG5-hIRP1S138E was subcloned into the corresponding sites of pT7-hIRP1 (13), to yield pT7-hIRP1S138A and pT7-hIRP1S138E, respectively. For the expression of His-tagged proteins in eukaryotic cells, the 2.9-kb IRP1 cassette was excised from pT7-hIRP1, pT7-hIRP1S138A, and pT7-hIRP1S138E with XbaI/HindIII and transferred, following blunt-end formation with the Klenow fragment, into the BamHI site of pUHD10-3 (29) to yield 10-3-His-hIRP1, 10-3-His-hIRP1S138A, and 10-3-His-hIRP1S138E, respectively.
Expression and purification of recombinant IRP1.
pT7-hIRP1, pT7-hIRP1S138A, and pT7-hIRP1S138E were expressed in Escherichia coli BL21(DE3), and recombinant human IRP1 was purified by affinity chromatography on Ni2+-nitrilotriacetic acid beads (13), which was followed by a second purification step with anion-exchange chromatography on a Resource Q fast-performance liquid chromatography column (Pharmacia).
Cell culture and transfections.
Human embryonic kidney HEK293 cells and murine B6 fibroblasts were grown in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum, 2 mM glutamine, 100 U of penicillin per ml, and 0.1 ng of streptomycin per ml. Transient transfections were performed with the Lipofectamine Plus reagent (Gibco BRL). HEK293 cells were cotransfected with pSG5-hIRP1S138A or pSG5-hIRP1S138E and neoCMVtBL (20) with the calcium phosphate method, and stable clones were selected in media containing 400 μg of G418/ml. B6 cells were transfected as described above with 15-1-neo (12), containing the tetracycline repressor (tTA), to generate a tTA-B6 clone by selection in the presence of 250 μg of G418/ml. The tTA-B6 cells were then cotransfected with 10-3-His-hIRP1, 10-3-His-hIRP1S138A, or 10-3-His-hIRP1S138E and puromycin-resistant pBabe (29). Stable transfectants were selected in media containing 4 μg of puromycin/ml, 250 μg of G418/ml, and 2 μg of tetracycline/ml. In preliminary transient-transfection experiments, a tight induction of IRP1 expression was observed upon the removal of tetracycline. However, all isolated clones of stable transfectants displayed leaky IRP1 expression, and the withdrawal of tetracycline had minimal effects. Therefore, the cells were maintained and cultivated for all experiments without tetracycline.
Generation of IRP1 antibodies.
Polyclonal IRP1 antibodies were raised by PRF & L, Inc. (Canadensis, Pa.), in a rabbit immunized against highly purified human recombinant IRP1 (2), kindly donated by Jean-Marc Moulis (Grenoble, France).
Western blotting.
Cytoplasmic lysates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 8% gels and transferred onto nitrocellulose filters. The blots were saturated with 10% nonfat milk in phosphate-buffered saline and probed with 1:1,000-diluted FLAG or β-actin or with 1:2,000-diluted IRP1 antibodies. Dilutions were made in phosphate-buffered saline containing 0.5% Tween 20. After being washed with this solution, the blots with (monoclonal) FLAG antibodies were incubated with peroxidase-coupled rabbit anti-mouse immunoglobulin G (1:4,000 dilution) and the blots with (polyclonal) IRP1 or β-actin antibodies were incubated with peroxidase-coupled goat anti-rabbit immunoglobulin G (1:5,000 dilution). Detection of peroxidase-coupled antibodies was performed with the enhanced chemiluminescence (ECL) method (Amersham).
Pulse-chase and immunoprecipitation.
Cells were metabolically labeled for 2 h with trans-35S label (50 μCi/ml), a 70:30 mixture of [35S]methionine-cysteine. The radioactive medium was then removed, and the cells were chased in cold media in the absence or presence of 100 μM hemin or 100 μM desferrioxamine. Endogenous IRP1 or transfected IRP1S138A and IRP1S138E were analyzed by immunoprecipitation (22) with IRP1 or FLAG antibodies, respectively. Immunoprecipitated proteins were resolved by SDS-PAGE and visualized by autoradiography. Radioactive bands were quantified by phosphorimaging.
Biochemical assays.
Cytoplasmic lysates or purified IRP1 preparations were analyzed for IRE-binding activity by electrophoretic mobility shift assay (EMSA) (21). Cytosolic extracts devoid of mitochondrial aconitase were prepared as previously described (7). The aconitase enzymatic assay was performed as described in reference 13. One unit is defined as the amount that converts 1 μmol of citrate, pH 7.4, at 25°C per min to isocitrate.
RESULTS
Recombinant IRP1S138E lacks aconitase activity.
We employed recombinant wild-type and mutant versions of IRP1 to investigate the role of Ser-138 in IRP1 function. Human wild-type IRP1, as well as the phosphorylation-deficient IRP1S138A and the phosphomimetic IRP1S138E mutants, tagged with an N-terminal His6 epitope was expressed in E. coli and purified to apparent homogeneity (Fig. 1A). All recombinant proteins were found to be active in IRE binding, but IRP1S138E displayed a weaker activity when 80 ng of purified material was analyzed by EMSA (Fig. 1B, top). However, complete IRE-binding activity could be recovered following treatment with 2-mercaptoethanol (Fig. 1B, bottom), which is commonly employed to convert cytosolic aconitase into an IRE-binding protein in vitro (5, 23). No differences in IRE-binding activity were observed when larger quantities of recombinant proteins were analyzed (data not shown).
FIG. 1.
Recombinant IRP1S138E fails to function as aconitase. (A) SDS-PAGE results of purified human recombinant (rec.) His-tagged wild-type IRP1 (wt), IRP1S138A, and IRP1S138E. The proteins were visualized by staining with Coomassie brilliant blue. (B) Purified recombinant proteins (80 ng) were analyzed by EMSA with a 32P-labeled IRE probe in the absence (top) or presence (bottom) of 2% 2-mercaptoethanol (2-ME). The positions of specific His-IRP1/IRE complexes and excess free probe are indicated by arrows. (C) Aconitase assay prior to (white bars) and after (black bars) treatment with ferrous sulfate-cysteine. Values correspond to results from triplicate samples; the aconitase activity is expressed as milliunits per microgram of recombinant protein. **, P < 0.01 versus values for the control (Student's t test).
The purified proteins were subsequently utilized for aconitase assays. As expected, purified wild-type IRP1 and IRP1S138A exhibit low levels of aconitase activity (Fig. 1C), reflecting the predominance of apo-IRP1 in the preparation (13). However, the aconitase activity is significantly (P < 0.01) stimulated by about three- to fivefold with the addition of ferrous sulfate and cysteine (Fig. 1C), which leads to reconstitution of the [4Fe-4S] cluster (13). By contrast, IRP1S138E is completely inactive as an aconitase, and remains so even after a treatment with ferrous sulfate and cysteine (Fig. 1C). These results are in agreement with the failure of IRP1S138E to complement aconitase activity in aco1 yeast (3) and suggest that IRP1S138E is unable to assemble a stable [4Fe-4S] cluster under aerobic conditions.
IRP1S138E expressed in mammalian cells is sensitive to iron-dependent degradation.
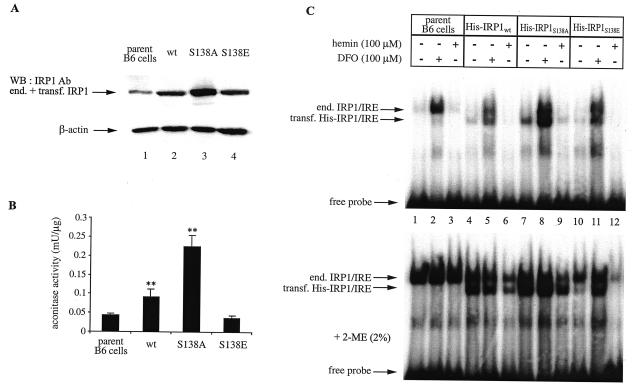
To study the function of Ser-138 in a more physiological context, His-tagged wild-type IRP1, IRP1S138A, and IRP1S138E were stably transfected into murine B6 fibroblasts. The expression of chimeric proteins was first analyzed by Western blotting with a polyclonal antibody against IRP1, which also cross-reacts with endogenous IRP1 (detection of chimeric IRP1 with commercial antibodies against the His tag was not reliable due to the poor quality of the immunoblots). The result depicted in Fig. 2A (top panel) shows that the IRP1 signal is increased by about twofold in cells transfected with wild-type IRP1 or IRP1S138E (lanes 2 and 4, respectively) and by about fourfold in cells transfected with IRP1S138A (lane 3). No differences are observed in the levels of control β-actin (bottom panel). In line with the above results, the expression of chimeric wild-type IRP1 and IRP1S138A contributes to significant (P < 0.01) increases in cytosolic aconitase activity of about twofold and fourfold, respectively (Fig. 2B). However, the aconitase activity in cytoplasmic extracts of cells expressing IRP1S138E is similar to the activity of parent cells (Fig. 2B). We conclude that IRP1S138E expressed in B6 cells fails to function as aconitase, which is in agreement with the data shown in Fig. 1C.
FIG. 2.
Iron-dependent regulation of chimeric human IRP1 constructs in B6 cells. Wild-type IRP1 (wt), IRP1S138A, and IRP1S138E, tagged with a His6 epitope at their N-termini, were stably transfected (transf.) into B6 cells. (A and B) Cytosolic extracts (devoid of mitochondrial aconitase) of nontransfected parent cells and of cells expressing chimeric wild-type IRP1, IRP1S138A, or IRP1S138E were analyzed by Western blotting (WB) (A) with antibodies (Ab) against IRP1 (top) and β-actin (bottom) and for aconitase activity (B). Values in panel B correspond to results from triplicate samples; the aconitase enzymatic activity is expressed as milliunits per microgram of total protein in the cytosolic lysate. (C) Cells were treated overnight with 100 μM desferrioxamine (DFO) or hemin. Cytoplasmic extracts of nontransfected parent cells (lanes 1 to 3) and of cells expressing chimeric wild-type IRP1 (lanes 4 to 6), IRP1S138A (lanes 7 to 9), or IRP1S138E (lanes 10 to 12) were analyzed by EMSA with a 32P-labeled IRE probe in the absence (top) or presence (bottom) of 2% 2-mercaptoethanol (2-ME). The positions of excess free probe and specific IRP1/IRE complexes, corresponding to endogenous (end.) murine IRP1 and transfected (transf.) human His-IRP1, are indicated by arrows. **, P < 0.01 versus values for the control (Student's t test).
To evaluate the IRE-binding activities of the chimeric proteins and assess how they respond to iron manipulations, cytoplasmic extracts were analyzed by EMSA (Fig. 2C). In this assay, the activity of chimeric IRP1 (lanes 4 to 12) can be easily distinguished from that of endogenous IRP1, which is apparent when extracts of nontransfected parent cells are analyzed (lanes 1 to 3), due to the faster electrophoretic mobilities of human IRP1/IRE complexes. Overnight exposure of B6 cells to the iron chelator desferrioxamine results in activation of endogenous IRP1 and of the transfected wild-type IRP1, IRP1S138A, and IRP1S138E. Likewise, overnight administration of hemin leads to a complete loss of IRE-binding activity. A treatment of cytoplasmic lysates with 2-mercaptoethanol prior to EMSA is routinely performed to monitor total IRP1 levels. The experimental results depicted in Fig. 2C (bottom panel) reveal that while the IRE-binding activity of endogenous IRP1 and transfected wild-type IRP1 and IRP1S138A in extracts of untreated or iron-loaded cells can be recovered in vitro by 2-mercaptoethanol, this is not the case for IRP1S138E (lanes 10 to 12). Thus, an apparent iron-replete state of untreated cells (lane 10) or further iron loading by treatment with exogenous hemin (lane 12) seems to result in an irreversible inactivation of IRP1S138E. It is also evident that the recovery of endogenous IRP1 activity from iron-loaded B6 transfectants by 2-mercaptoethanol is relatively poor (see Discussion).
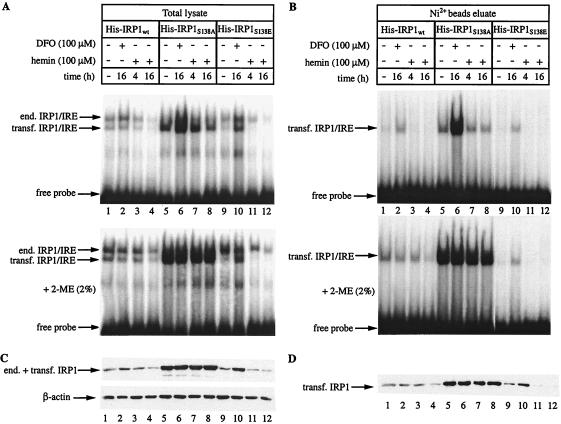
To better characterize the effects of hemin on IRP1S138E, we utilized the His tag for purification of the transfected proteins from the crude cytoplasmic extract by means of Ni2+ affinity chromatography (Fig. 3). Analysis of the total lysate (Fig. 3A) and of the purified chimeric proteins (Fig. 3B) by EMSA confirms that the IRE-binding activity of IRP1S138E obtained from iron-loaded cells cannot be recovered in vitro with 2-mercaptoethanol (compare lanes 1 to 8 with lanes 9 to 12 in panels A and B). All affinity-purified proteins were inactive as aconitases, possibly due to the instability of this activity upon dilution. Finally, the purified material was subjected to Western blotting with the IRP1 antibody. Analysis of the total lysate (Fig. 3C) reveals a hemin-induced reduction in the intensity of the signal from IRP1S138E-expressing cells (lanes 11 and 12), while desferrioxamine produced the opposite effect (lane 10). Notably, the signal from untreated cells (lane 9) is weaker than that from iron-starved cells (lane 10). The iron manipulations leave the expression of wild-type IRP1 or IRP1S138A largely unaffected (lanes 1 to 8). Probing the filter with an antibody against β-actin (bottom) provides a loading control. The negative effects of hemin in IRP1S138E expression are clearly demonstrated by the immunoblot of the affinity-purified proteins (Fig. 3D), where the background of endogenous IRP1 is eliminated (compare lanes 9 to 12 in panels C and D).
FIG. 3.
Hemin-dependent decrease of IRP1S138E expression in B6 cells. Cells expressing human wild-type IRP1, IRP1S138A, or IRP1S138E were treated with 100 μM desferrioxamine (DFO) or hemin for the indicated time intervals. His-tagged chimeric IRP1 was purified from cytoplasmic extracts by affinity chromatography with Ni2+-nitrilotriacetic acid beads. (A and B) Total lysate (A) or affinity-purified chimeric IRP1 (B) was analyzed by EMSA with a 32P-labeled IRE probe in the absence (top) or presence (bottom) of 2% 2-mercaptoethanol (2-ME). The positions of excess free probe and specific IRP1/IRE complexes, corresponding to endogenous (end.) murine IRP1 and transfected (transf.) human His-IRP1, are indicated by arrows. (C and D) Western blotting of total lysates (C) or affinity-purified chimeric IRP1 (D) with antibodies against IRP1 and β-actin (C, bottom).
The above results were obtained with N-terminally tagged wild-type and mutant IRP1s, transfected into mouse B6 fibroblasts. To exclude the possibility that the decrease in IRP1S138E expression in untreated and hemin-treated cells might be related to the manipulation at the protein's N terminus by the His tag, we generated plasmids encoding IRP1S138A or IRP1S138E tagged at its C terminus with a FLAG epitope. These constructs were stably transfected into HEK293 cells, and the chimeric proteins were analyzed in the absence or presence of hemin. In addition, a construct encoding FLAG-tagged wild-type IRP1 was utilized for similar analysis following transient transfection into HEK293 cells. The cells were exposed to hemin for 4 or 16 h, and protein levels were determined by Western blotting. Probing with antibodies against IRP1 or FLAG shows no significant effects from hemin on the steady-state levels of chimeric wild-type IRP1 (Fig. 4A) and IRP1S138A (Fig. 4B). However, this assay reveals a significant hemin-dependent reduction in the signal obtained from IRP1S138E-expressing cells with the IRP1 antibody, which corresponds to both endogenous IRP1 and chimeric IRP1S138E (Fig. 4C, top). Following 16-h exposure of the cells to the drug, the decrease in signal intensity is ∼50% (lane 3). Probing with a FLAG antibody (middle panel) demonstrates that this decrease is due to an almost complete loss of transfected IRP1S138E, since under these conditions the background of endogenous IRP1 is eliminated. No differences in the levels of control β-actin are observed (bottom panel). Notably, in untreated cells IRP1S138E is expressed at relatively lower levels than those of IRP1S138A and wild-type chimeric IRP1 (compare lanes 1 in Fig. 4).
FIG. 4.
Hemin-dependent decrease of IRP1S138E expression in HEK293 cells. The cells were either transiently transfected with pSG5-hIRP1 or stably transfected with pSG5-hIRP1S138A or pSG5-hIRP1S138E to express chimeric wild-type IRP1 (IRP1wt) (A), IRP1S138A (B), or IRP1S138E (C), respectively. All chimeric proteins are tagged with a FLAG epitope at their C termini. The cells were treated with 100 μM hemin for the indicated time intervals, and cell lysates were analyzed by Western blotting (WB) with antibodies (Ab) against IRP1 (top), FLAG (middle), or β-actin (bottom). end., endogenous; transf., transfected.
The above results raised the possibility that IRP1S138E may be destabilized in iron-replete cells. This issue was directly addressed with a pulse-chase experiment. HEK293 cells expressing IRP1S138A or IRP1S138E were metabolically labeled with [35S]methionine-cysteine for 2 h. Subsequently, after the label was washed, the cells expressing IRP1S138A were chased for 4 to 24 h in the absence (Fig. 5A, top) or presence (bottom) of 100 μM hemin. Similarly, the cells expressing IRP1S138E were chased for 4 to 24 h without any iron perturbations (Fig. 5B, top) or in the presence of 100 μM hemin (middle) or 100 μM desferrioxamine (bottom). Analysis of the transfected proteins by immunoprecipitation with the FLAG antibody and quantification of the data with phosphorimaging show that IRP1S138A is fairly stable (half-life, >12 h) and that its turnover remains largely unaffected by hemin (Fig. 5A). In contrast, the stability of IRP1S138E is subjected to iron-dependent regulation (Fig. 5B). Thus, in untreated control cells, IRP1S138E has a half-life of ∼6 h (top). In the presence of hemin, the half-life of IRP1S138E further decreases to ∼4 h (middle). The decay of IRP1S138E can be completely inhibited by desferrioxamine (bottom). We conclude that IRP1S138E undergoes iron-dependent degradation in apparently iron-replete control cells or in iron-overloaded, hemin-treated cells but that it is stabilized in iron-deficient cells treated with desferrioxamine. As an additional control, we performed a pulse-chase experiment with untransfected parent HEK293 (Fig. 5C) and B6 (Fig. 5D) cells to analyze endogenous IRP1. This experiment confirms previous findings (22, 28) by showing that the half-life of endogenous IRP1 is >12 h. It also demonstrates that IRP1 stability remains unaffected by a treatment with hemin in both cell types, in agreement with the data obtained with IRP1S138A (Fig. 5A).
FIG. 5.
(A and B) IRP1S138E, but not IRP1S138A, is sensitive to iron-mediated degradation. HEK293 cells were metabolically labeled for 2 h with [35S]methionine-cysteine and chased with cold media for the indicated time intervals in the absence of iron perturbations (control [ctrl]) or in the presence of 100 μM hemin (+hemin) or 100 μM desferrioxamine (+DFO). The half-life (t1/2) of IRP1S138A (A) or IRP1S138E (B) was assessed by quantitative immunoprecipitation of 500 μg of cytoplasmic extracts with 8.8 μg of FLAG antibody. (C and D) Hemin does not accelerate the turnover of endogenous IRP1 in HEK293 or B6 cells. HEK293 (C) or B6 cells (D) were treated as described for panels A and B, and the decay of endogenous IRP1 was assessed by quantitative immunoprecipitation of 500 μg of cytoplasmic extracts with 20 μl of IRP1 antiserum. Immunoprecipitated material was analyzed by SDS-PAGE on 8% gels, and proteins were visualized by autoradiography. Radioactive bands were quantified by phosphorimaging, and the indicated half-lives were calculated by plotting the data (data not shown).
Proteasomal inhibitors block iron-dependent degradation of IRP1S138E.
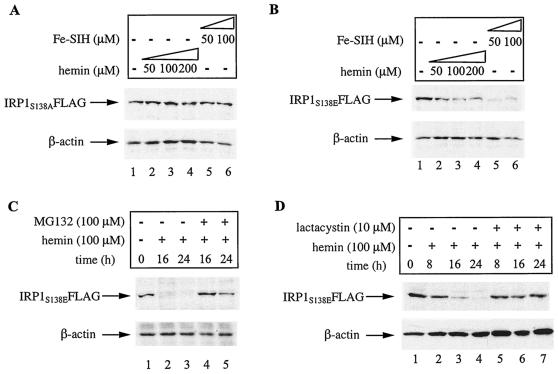
To investigate whether IRP1S138E is specifically subjected to degradation by hemin, or iron in general, we compared the effects of hemin and Fe-SIH, an alternative iron donor, on IRP1 expression (24). The HEK293 stable clones were exposed to different doses of hemin or Fe-SIH for 8 h, and the expression of chimeric IRP1 was analyzed by Western blotting with the FLAG antibody. As expected, these drugs have no effects on the steady-state levels of IRP1S138A (Fig. 6A, top) and control β-actin (bottom). However, both hemin and Fe-SIH inhibit the expression of IRP1S138E (Fig. 6B, top) without affecting control β-actin (bottom). Thus, IRP1S138E is sensitive to degradation by both heme and nonheme iron.
FIG. 6.
Iron-dependent degradation of IRP1S138E is blocked by proteasomal inhibitors. (A and B) HEK293 cells expressing IRP1S138A or IRP1S138E were treated with the indicated concentrations of hemin or Fe-SIH for 16 h. The expression levels of IRP1S138A (A) and IRP1S138E (B) were analyzed by Western blotting with the FLAG antibody (top). (C and D) HEK293 cells expressing IRP1S138E were treated for the indicated time intervals with 100 μM hemin in the presence or absence of 100 μM MG132 (C) or 10 μM lactacystin (D). The expression of IRP1S138E was analyzed by Western blotting with the FLAG antibody (top). All blots shown were also hybridized with an antibody against β-actin (bottom). The cells were viable under all experimental conditions.
Finally, we examined the potential of various inhibitors to antagonize the degradation of IRP1S138E. The proteasomal inhibitors MG132 (Fig. 6C) and lactacystin (Fig. 6D) completely protect IRP1S138E from degradation following treatment of the cells with hemin. By contrast, lysosomal inhibitors (ammonium chloride and 3-methyladenine) fail to do so (data not shown). These results suggest that iron promotes the degradation of IRP1S138E by the proteasome.
Ser-138 is highly conserved in vertebrate IRP1 molecules.
Ser-138 and its flanking sequences constitute a PKC phosphorylation site, which was earlier found to be conserved in mammalian (human, rabbit, rat, and mouse) IRP1 molecules (9). In the last few years, IRP1 sequence data from additional vertebrate and invertebrate species have been obtained. Multiple sequence alignment of all IRP1 molecules currently deposited in Swiss-Prot (Fig. 7) reveals that as in mammals (Homo sapiens, Oryctolagus cuniculus, Rattus norvegicus, and Mus musculus), Ser-138 is also conserved in chicken (Gallus gallus) IRP1. However, Ser-138 is replaced with an alanine in IRP1 proteins from crayfish (Pacifastacus leniusculus) and insects (Drosophila melanogaster and Manduca sexta) and in IRP1-like proteins from plants (Arabidopsis thaliana, Citrus limon, and Curcubita maxima) and the malaria parasite (Plasmodium falciparum). Interestingly, the stretches of amino acids upstream and downstream of mammalian and chicken Ser-138 sequences shown in Fig. 7 are almost identical (with the exception of the D131H and A136S substitutions in rat and chicken Ser-138 sequences, respectively). There is also some degree of conservation in the sequences surrounding Ala-138 of invertebrate IRP1 molecules, but this is clearly less prominent than in the vertebrate counterparts and restricted to fewer positions (for example, Q129, V130, N142, and E146). Taken together, Ser-138 and flanking sequences are highly conserved only in the IRP1 molecules of vertebrates, not invertebrates.
FIG. 7.
Multiple sequence alignment of amino acids 129 to 147 in IRP1s of various species currently deposited in Swiss-Prot. Ser-138 (asterisk) is conserved only in the IRP1 proteins from vertebrates. The genInfo identifier (gi) numbers of all molecules are indicated on the right. The alignment was made by the Clustal W algorithm (MacVector software, version 6.5.3).
DISCUSSION
Genetic (3) and biochemical (4) experiments in yeast aco1 cells have suggested that phosphomimetic replacements of Ser-138 impair the process of [4Fe-4S] cluster assembly in chimeric rabbit IRP1 proteins under aerobic conditions. Consequently, this mutation abolishes the aconitase function of IRP1. Here we confirm these findings after studying the aconitase activity of highly purified human recombinant IRP1S138E. The aconitase assay (Fig. 1C) clearly suggests that while wild-type IRP1 and IRP1S138A are capable of reconstituting (at least partially) their [4Fe-4S] clusters following treatment with iron and cysteine, IRP1S138E fails to do so. The activating effect of 2-mercaptoethanol in the IRE-binding activity of IRP1S138E (Fig. 1B) is consistent with the presence of an [4Fe-4S] cluster in the protein. However, purified recombinant IRP1S138E is fully active as an IRE-binding protein at higher concentrations. On the basis of this observation and of the aconitase data (Fig. 1C), we speculate that IRP1S138E may be sensitive to oxidation in the absence of the [4Fe-4S] cluster. All results shown in Fig. 1 were from experiments performed under aerobic conditions. Considering the genetic data from yeast (3), we cannot exclude the possibility that IRP1S138E may recover its ability to assemble a cluster in an anaerobic environment.
Since the IRE/IRP system is not conserved in yeast, we investigated the function of IRP1S138E in the more physiologically relevant context of mammalian cells. In B6 transfectants, the IRE-binding activity of IRP1S138E can be induced following iron chelation with desferrioxamine (Fig. 2C and 3A and B). This finding suggests that in apparently iron-replete B6 cells, IRP1S138E may assemble a [4Fe-4S] cluster, which is expected to render the protein to cytosolic aconitase and inactivate IRE binding. To directly address this possibility, we performed aconitase assays with affinity-purified wild-type and mutant IRP1s, but we were unable to detect any enzymatic activity. Conceivably, aconitase activity is more sensitive to dilution than IRE-binding activity. Nevertheless, the results with the crude lysates (Fig. 2B) are in agreement with the notion that IRP1S138E is unable to function as an aconitase in B6 transfectants. The positive effect of desferrioxamine in the IRE-binding activity of IRP1S138E may suggest that in iron-replete but not overloaded B6 cells, this mutant behaves as a null form of IRP1, possibly due to partial assembly of the iron-sulfur cluster (15). In addition, the increase in IRP1S138E steady-state levels (lanes 10 in Fig. 3C and D) indicates that desferrioxamine may stabilize IRP1S138E (see also below).
We were intrigued by the failure of 2-mercaptoethanol to recover the IRE-binding activity in extracts of iron-loaded IRP1S138E-expressing cells (Fig. 2C and 3A and B). A similar effect was previously observed with (endogenous) IRP1 in B6 cells in response to menadione-induced oxidative stress (10). Although under these conditions the loss of activity was not associated with any significant alterations in IRP1 levels (10), we show here that iron loading leads to the accelerated turnover of IRP1S138E. The iron-dependent degradation of IRP1S138E is documented for two different cell lines of mouse (Fig. 2 and 3) and human (Fig. 4 to 6) origins and for two types of modified constructs carrying an N- or C-terminal epitope tag. It should be noted that wild-type IRP1 has a relatively long half-life (>12 h), which is not dramatically affected by iron (22, 28) (Fig. 5C and D). These findings also hold true for IRP1S138A (Fig. 5A). In contrast, our data suggest that iron promotes the degradation of IRP1S138E by the proteasome (Fig. 5B and 6C and D), in a fashion akin to the well-established iron-dependent degradation of IRP2 (14, 17). The degradation kinetics appear to correlate with the level of iron loading. In apparently iron-replete control cells, the half-life of IRP1S138E is ∼6 h (Fig. 5B). Further iron loading with hemin accelerates the turnover of IRP1S138E and reduces its half-life to ∼4 h. Importantly, the degradation of IRP1S138E can be efficiently blocked by the iron chelator desferrioxamine.
We also observed that the recovery of endogenous IRE-binding activity in extracts of hemin-treated B6 transfectants by 2-mercaptoethanol was relatively poor (for example, Fig. 2C, lanes 6, 9, and 12). However, IRP1 could be completely recovered by 2-mercaptoethanol from hemin-treated parent B6 cells (Fig. 2C, lane 3). We speculate that the weak effect of 2-mercaptoethanol on endogenous IRP1 in the transfectants may be related to nonreversible inactivation of the protein, possibly via posttranslational modifications (perhaps oxidation), but the reason for this is unclear. Notably, similar effects have previously been observed in a different setting (10). In support of this interpretation, the pulse-chase experiments with B6 and HEK293 cells in the absence or presence of hemin (Fig. 5C and D) rule out the possibility that hemin may generally promote the degradation of IRP1. Thus, our results do not support a general model for heme-mediated degradation of IRP1, which had been proposed in the past (11) well before it became clear that the mechanism for IRP1 regulation involves an iron-sulfur cluster switch and that IRP2 is regulated at the level of protein stability. Nevertheless, in light of the data presented here, it might be worth considering Ser-138 phosphorylation as a possibility underlying the heme-mediated degradation of IRP1 described previously (11).
In conclusion, IRP1S138E expressed in mammalian cells under normoxic conditions not only is unable to assemble a [4Fe-4S] cluster but also undergoes iron-mediated degradation via the proteasome. Considering that Ser-138 may be a target of phosphorylation by PKC (9), our data raise the possibility for a new mode of IRP1 regulation at the level of protein stability. Preliminary experiments with HL-60 cells aiming to assess the effect of iron on the stability of endogenous IRP1 after treatment with phorbol 12-myristate 13-acetate (PMA) to activate PKC pathways were negative. The effectiveness of this treatment was monitored by immunoblot detection of PMA-induced p21WAF (6). However, it should be noted that the evidence for Ser-138 phosphorylation within IRP1 in PMA-treated HL-60 cells remains thus far indirect and has primarily been based on in vitro phosphorylation assays with oligopeptides as substrates (9). It is conceivable that in the context of the entire protein, Ser-138 may be a target for modification by other kinases. Alternatively, our culture conditions may have favored the predominance of iron-loaded [4Fe-4S] IRP1, which is not a preferable substrate for phosphorylation by PKC (26). Generation and employment of phospho-specific antibodies are expected to help towards identifying conditions for Ser-138 phosphorylation within IRP1 in vivo and studying the responses of the phosphorylated protein to iron.
The conservation of Ser-138 and the flanking sequences in the IRP1 proteins of vertebrates is indicative of the functional significance of this residue. If this assumption proves correct, the S138A substitution in crayfish and insect IRP1s and in the IRP1-like molecules of plants and the malaria parasite (Fig. 7) suggests that Ser-138 in vertebrate IRP1s has evolved to integrate a more complex repertoire of regulatory signals.
Acknowledgments
C.F. is a recipient of a postdoctoral fellowship from the Fonds de la Recherche en Santé du Quebéc. This work was supported by a grant from the Canadian Institutes for Health Research.
REFERENCES
- 1.Beinert, H., R. H. Holm, and E. Münck. 1997. Iron-sulfur clusters: nature's modular, multipurpose structures. Science 277:653-659. [DOI] [PubMed] [Google Scholar]
- 2.Brazzolotto, X., J. Gaillard, K. Pantopoulos, M. W. Hentze, and J.-M. Moulis. 1999. Human cytoplasmic aconitase (iron regulatory protein 1) is converted into its [3Fe-4S] form by hydrogen peroxide in vitro but is not activated for iron-responsive element binding. J. Biol. Chem. 274:21625-21630. [DOI] [PubMed] [Google Scholar]
- 3.Brown, N. M., S. A. Anderson, D. W. Steffen, T. B. Carpenter, M. C. Kennedy, W. E. Walden, and R. S. Eisenstein. 1998. Novel role of phosphorylation in Fe-S cluster stability revealed by phosphomimetic mutations at Ser-138 of iron regulatory protein 1. Proc. Natl. Acad. Sci. USA 95:15235-15240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, N. M., M. C. Kennedy, W. E. Antholine, R. S. Eisenstein, and W. E. Walden. 2002. Detection of a [3Fe-4S] cluster intermediate of cytosolic aconitase in yeast expressing iron regulatory protein 1: insights into the mechanism of Fe-S cluster cycling. J. Biol. Chem. 277:7246-7254. [DOI] [PubMed] [Google Scholar]
- 5.Cairo, G., and A. Pietrangelo. 2000. Iron regulatory proteins in pathobiology. Biochem. J. 352:241-250. [PMC free article] [PubMed] [Google Scholar]
- 6.Das, D., G. Pintucci, and A. Stern. 2000. MAPK-dependent expression of p21(WAF) and p27(kip1) in PMA-induced differentiation of HL60 cells. FEBS Lett. 472:50-52. [DOI] [PubMed] [Google Scholar]
- 7.Drapier, J. C., H. Hirling, J. Wietzerbin, P. Kaldy, and L. C. Kühn. 1993. Biosynthesis of nitric oxide activates iron regulatory factor in macrophages. EMBO J. 12:3643-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eisenstein, R. S. 2000. Iron regulatory proteins and the molecular control of mammalian iron metabolism. Annu. Rev. Nutr. 20: 627-662. [DOI] [PubMed] [Google Scholar]
- 9.Eisenstein, R. S., P. T. Tuazon, K. L. Schalinske, S. A. Anderson, and J. A. Traugh. 1993. Iron-responsive element-binding protein: phosphorylation by protein kinase C. J. Biol. Chem. 268:27363-27370. [PubMed] [Google Scholar]
- 10.Gehring, N., M. W. Hentze, and K. Pantopoulos. 1999. Inactivation of both RNA binding and aconitase activities of iron regulatory protein-1 by quinone-induced oxidative stress. J. Biol. Chem. 274:6219-6225. [DOI] [PubMed] [Google Scholar]
- 11.Goessling, L. S., S. Daniels-McQueen, M. Bhattacharyya-Pakrasi, J.-J. Lin, and R. E. Thach. 1992. Enhanced degradation of the ferritin repressor protein during induction of ferritin messenger RNA translation. Science 256:670-673. [DOI] [PubMed] [Google Scholar]
- 12.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gray, N. K., S. Quick, B. Goossen, A. Constable, H. Hirling, L. C. Kühn, and M. W. Hentze. 1993. Recombinant iron regulatory factor functions as an iron-responsive element-binding protein, a translational repressor and an aconitase: a functional assay for translational repression and direct demonstration of the iron switch. Eur. J. Biochem. 218:657-667. [DOI] [PubMed] [Google Scholar]
- 14.Guo, B., J. D. Phillips, Y. Yu, and E. A. Leibold. 1995. Iron regulates the intracellular degradation of iron regulatory protein 2 by the proteasome. J. Biol. Chem. 270:21645-21651. [DOI] [PubMed] [Google Scholar]
- 15.Haile, D. J., T. A. Rouault, J. B. Harford, M. C. Kennedy, G. A. Blondin, H. Beinert, and R. D. Klausner. 1992. Cellular regulation of the iron-responsive element binding protein: disassembly of the cubane iron-sulfur cluster results in high-affinity RNA binding. Proc. Natl. Acad. Sci. USA 89:11735-11739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanson, E. S., and E. A. Leibold. 1999. Regulation of the iron regulatory proteins by reactive nitrogen and oxygen species. Gene Expr. 7:367-376. [PMC free article] [PubMed] [Google Scholar]
- 17.Iwai, K., R. D. Klausner, and T. A. Rouault. 1995. Requirements for iron-regulated degradation of the RNA binding protein, iron regulatory protein 2. EMBO J. 14:5350-5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johansson, H. E., and E. C. Theil. 2002. Iron-response element (IRE) structure and combinatorial RNA regulation, p. 237-253. In D. M. Templeton (ed.), Molecular and cellular iron transport. Marcel Dekker, Inc., New York, N.Y.
- 19.Kim, S., and P. Ponka. 2002. Nitrogen monoxide-mediated control of ferritin synthesis: implications for macrophage iron homeostasis. Proc. Natl. Acad. Sci. USA 99:12214-12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwon, H., N. Pelletier, C. DeLuca, P. Genin, S. Cisternas, R. Lin, M. A. Wainberg, and J. Hiscott. 1998. Inducible expression of IκBα repressor mutants interferes with NF-κB activity and HIV-1 replication in Jurkat T cells. J. Biol. Chem. 273:7431-7440. [DOI] [PubMed] [Google Scholar]
- 21.Mueller, S., and K. Pantopoulos. 2002. Activation of iron regulatory protein-1 (IRP1) by oxidative stress. Methods Enzymol. 348:324-337. [DOI] [PubMed] [Google Scholar]
- 22.Pantopoulos, K., N. Gray, and M. W. Hentze. 1995. Differential regulation of two related RNA-binding proteins, iron regulatory protein (IRP) and IRPB. RNA 1:155-163. [PMC free article] [PubMed] [Google Scholar]
- 23.Pantopoulos, K., and M. W. Hentze. 2000. Nitric oxide, oxygen radicals and iron metabolism, p. 293-313. In L. Ignarro (ed.), Nitric oxide. Academic Press, San Diego, Calif.
- 24.Ponka, P., and H. M. Schulman. 1985. Acquisition of iron from transferrin regulates reticulocyte heme synthesis. J. Biol. Chem. 260:14717-14721. [PubMed] [Google Scholar]
- 25.Rouault, T., and J. B. Harford. 2000. Translational control of ferritin synthesis, p. 655-670. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Schalinske, K. L., S. A. Anderson, P. T. Tuazon, O. S. Chen, M. C. Kennedy, and R. S. Eisenstein. 1997. The iron-sulfur cluster of iron regulatory protein 1 modulates the accessibility of RNA binding and phosphorylation sites. Biochemistry 36:3950-3958. [DOI] [PubMed] [Google Scholar]
- 27.Schalinske, K. L., and R. S. Eisenstein. 1996. Phosphorylation and activation of both iron regulatory proteins 1 and 2 in HL-60 cells. J. Biol. Chem. 271:7168-7176. [DOI] [PubMed] [Google Scholar]
- 28.Tang, C. K., J. Chin, J. B. Harford, R. D. Klausner, and T. A. Rouault. 1992. Iron regulates the activity of the iron-responsive element binding protein without changing its rate of synthesis or degradation. J. Biol. Chem. 267:24466-24470. [PubMed] [Google Scholar]
- 29.Wang, J., and K. Pantopoulos. 2002. Conditional derepression of ferritin synthesis in cells expressing a constitutive IRP1 mutant. Mol. Cell. Biol. 22:4638-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]