Abstract
Anthranilate (2-aminobenzoate) is an important intermediate in tryptophan metabolism. In order to investigate the degradation of tryptophan through anthranilate by Burkholderia cepacia, several plasposon mutations were constructed of strain DBO1 and one mutant with the plasposon insertion in the anthranilate dioxygenase (AntDO) genes was chosen for further study. The gene sequence obtained from flanking DNA of the plasposon insertion site revealed unexpected information. B. cepacia DBO1 AntDO (designated AntDO-3C) is a three-component Rieske-type [2Fe-2S] dioxygenase composed of a reductase (AndAa), a ferredoxin (AndAb), and a two-subunit oxygenase (AndAcAd). This is in contrast to the two-component (an oxygenase and a reductase) AntDO enzyme from Acinetobacter sp. strain ADP1, P. aeruginosa PAO1, and P. putida P111. AntDO from strains ADP1, PAO1, and P111 are closely related to benzoate dioxygenase, while AntDO-3C is closely related to aromatic hydrocarbon dioxygenases from Novosphingobium aromaticivorans F199 and Sphingomonas yanoikuyae B1 and 2-chlorobenzoate dioxygenase from P. aeruginosa strains 142 and JB2. Escherichia coli cells expressing the functional AntDO-3C genes transform anthranilate and salicylate (but not 2-chlorobenzoate) to catechol. The enzyme includes a novel reductase whose absence results in less efficient transformation of anthranilate by the oxygenase and ferredoxin. AndR, a possible AraC/XylS-type transcriptional regulator, was shown to positively regulate expression of the andAcAdAbAa genes. Anthranilate was the only effector (of 12 aromatic compounds tested) that was able to induce expression of the genes.
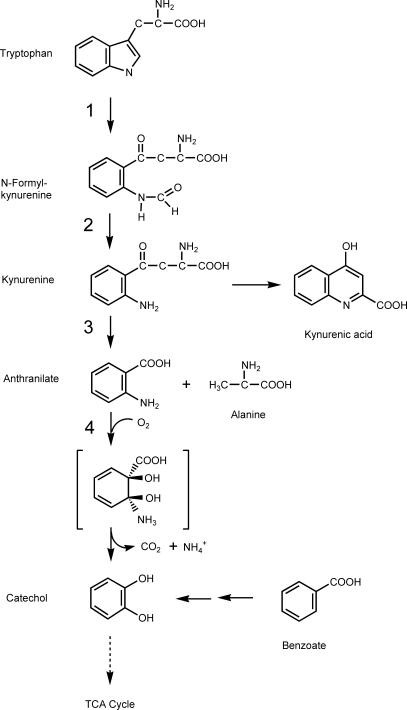
Anthranilate (2-aminobenzoate) is an important intermediate in the synthesis and degradation of tryptophan (Fig. 1) (30, 66). It is also the first aromatic intermediate in the biosynthesis of tryptophan. It has been shown or postulated to be an intermediate in the degradation of other compounds, such as o-nitrobenzoate (8, 11), quinaldine (21), carbazole (47), and those containing an indole moiety (25, 35, 41). It has been known for many years that microorganisms are able to use anthranilate as the sole source of carbon and energy aerobically or anaerobically (5, 9, 30, 58). Some eukaryotic microbes such as Trichosporon cutaneum (49) and Aspergillus niger (58) convert anthranilate to 2,3-dihydroxybenzoate by using a flavoprotein monooxygenase. Soil bacteria such as Nocardia opaca convert anthranilate to 5-hydroxyanthranilate and then to gentisate, likely by a hydroxylase (7). Pseudomonas strains convert anthranilate to catechol in a single step by using a flavoprotein dioxygenase (DO) (39, 62). The anthranilate DO (AntDO; deaminating, decarboxylating; EC 1.14.12.1) from Pseudomonas was partially purified, and two soluble protein fractions were able to convert anthranilate to catechol in the presence of Fe2+ (38). The genes for AntDO from another soil bacterium, Acinetobacter sp. strain ADP1, were cloned, and the enzymes were purified to homogeneity (6, 19). The ADP1 AntDO enzyme belongs to DO class IB, a two-component system composed of a reductase with a flavin adenine dinucleotide binding site and a plant-type [2Fe-2S] center and an oxygenase with a Rieske-type [2Fe-2S] center and a mononuclear iron as a prosthetic group (3, 42). The ADP1 AntDO enzyme is similar in sequence to the ADP1 benzoate DO enzyme (BenABC) (12), the TOL plasmid toluate DO enzyme (XylXYZ) (29), and the Burkholderia cepacia 2CBS 2-halobenzoate DO enzyme (CbdABC) (27). Other putative AntDOs found in the DNA sequence database, such as those from Pseudomonas aeruginosa PAO1 (19) and Pseudomonas putida P111 (GenBank accession number AY026914), also belong to this two-component system.
FIG. 1.
Tryptophan degradation pathways in Burkholderia and Pseudomonas (30). The enzymes for the conversion of tryptophan to catechol are labeled as follows: 1, tryptophan 2,3-DO; 2, formamidase; 3, kynureninase; 4, AntDO.
We describe here a new class of AntDO from B. cepacia DBO1, a soil bacterium originally isolated for the ability to use phthalate as the sole source of carbon and energy (10). The DBO1 AntDO enzyme belongs to DO class IIB, a three-component system very different from the two-component DO class that contains the AntDO enzyme from ADP1 and other Pseudomonas strains. The three-component system has a separate protein (ferredoxin) between the reductase and the oxygenase in the electron transfer chain. Therefore, DBO1 AntDO was designated AntDO-3C (for three components). The substrate range of AntDO-3C was also studied by using halogenated, hydroxylated, and methylated benzoates to further explore its relationship to 2-chlorobenzoate DO and other DOs. In addition, the role of an araC/xylS-type regulatory gene, found upstream of the structural genes for AntDO-3C, was studied. This is the first positively identified regulatory gene for anthranilate degradation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
B. cepacia DBO1 (ATCC 29424) is the wild-type strain (10). DBO1-Rif is a spontaneous rifampin-resistant mutant of DBO1 (R. B. Jacobi and G. J. Zylstra, unpublished data). Escherichia coli Top10F′ (Invitrogen, Carlsbad, Calif.) and E. coli DH5α (Invitrogen) were used as recipient strains in the cloning experiments. E. coli HB101 (4) was the host for helper plasmid pRK2013 (23) in mating experiments. pCR2.1-TOPO (Invitrogen) and pGEM-7Zf(−) (Promega, Madison, Wis.) were used for subcloning of DNA fragments. p34S-Km3 was used as the source of the Kmr cassette (15). The promoter probe vector pKRZ-1 has a promoterless lacZ gene cloned into broad-host-range vector pUCD615 (52). Luria-Bertani medium (LB) (40) was used as complete medium. Mineral salts basal (MSB) (57) medium was used as minimal medium. Tryptophan (10 mM), anthranilate (10 mM), or succinate (10 mM) was added to MSB medium as a carbon source. Ampicillin, kanamycin (KM), rifampin, and tetracycline were added at 100, 50, 100, and 15 μg/ml, respectively, when needed. Burkholderia strains were grown at 30°C, and E. coli strains were grown at 37°C.
Molecular techniques.
Plasposon mutagenesis was carried out by triparental mating (17). B. cepacia DBO1-Rif, E. coli DH5α containing plasposon pTnMod2-OKm3 (15, 16), and E. coli HB101 carrying pRK2013 were mixed together on LB agar plates and incubated at 30°C overnight. Cells collected were then resuspended in 10% glycerol and plated on LB-rifampin-KM plates. DNA was introduced into B. cepacia DBO1 by electrotransformation (14). Bacterial genomic DNA was prepared by the cetyltrimethylammonium bromide method (2). Plasmid DNA was purified with a NucleoSpin plasmid miniprep kit (Macherey-Nagel, Easton, Pa.). Restriction digests, ligations, transformation, and gel electrophoresis were performed by standard procedures (53). DNA extraction from agarose gels was performed with the QIAEX II gel purification kit (QIAGEN, Valencia, Calif.). PCR was performed with GeneAmp PCR system 9700 (Perkin-Elmer, Foster City, Calif.). Taq DNA polymerase (Fisher Scientific, Pittsburgh, Pa.), Pfu DNA polymerase (Stratagene, La Jolla, Calif.), and deoxynucleoside triphosphates (Perkin-Elmer) were obtained from different sources. Oligonucleotides were purchased from Invitrogen.
Cloning of the intact AntDO-3C genes directly from DBO1.
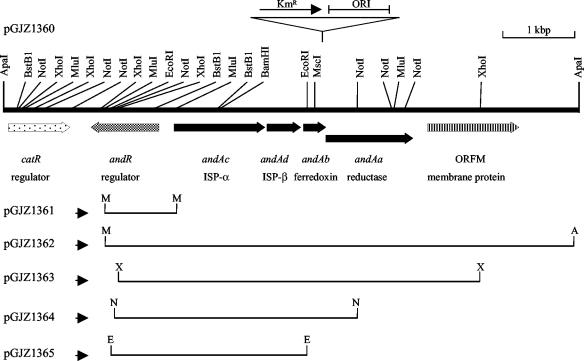
The intact AntDO-3C genes were retrieved directly from DBO1 by first inserting pCR2.1-TOPO into the upstream flanking area of the genes on the chromosome and recovering the vector and the genes later. In order to obtain the insertional DBO1 mutant, a plasmid (pGJZ1361) was constructed by cloning the 1.0-kb MluI fragment from pGJZ1360 (Fig. 2) into pCR2.1-TOPO. The MluI fragment was treated with calf intestine alkaline phosphatase (Fisher Scientific, Pittsburgh, Pa.) by the standard protocol (53) and end repaired with Taq DNA polymerase (72°C for 15 min) before the cloning. pGJZ1361 was introduced into DBO1, and transformants were selected on LB-KM. In order to confirm the insertion of pGJZ1361 into the DBO1 chromosome, total DNA of the transformants was purified as templates and PCR was performed with primers 5′-GCTCATGCCGCGAAACTG-3′ (binding site on the andR gene) and 5′-GTAAAACGACGGCCAG-3′ (M13 forward; binding site on pCR2.1-TOPO). If the desired single-crossover event occurred as predicted, a 1.1-kb product would be produced.
FIG. 2.
Restriction map of the 8.0-kb ApaI fragment containing the AntDO (AntDO-3C) genes from B. cepacia DBO1. The locations of genes, and their transcriptional directions, are shown relative to some of the restriction endonuclease recognition sites. The assigned genes are as follows: andAa, reductase gene; andAb, ferredoxin gene; andAc, ISP-α subunit gene; andAd, ISP-β subunit gene; andR, regulatory gene for the and operon; catR, putative regulatory gene for the cat operon; ORFM, putative membrane protein. The fragments that were subcloned into pCR2.1-TOPO are shown as lines. The small arrows indicate the transcription direction of the lac promoter in pCR2.1-TOPO. The insertion part of the plasposon in mutant strain PM101, including the Kmr gene, and the replication origin (ORI), are also shown. Abbreviations: A, ApaI; E, EcoRI; M, MluI; N, NotI; X, XhoI.
Construction and analysis of AntDO-3C expression clones in E. coli.
Plasmid pGJZ1362 was digested with XhoI, NotI, and EcoRI separately. The 5.1-kb XhoI, 3.4-kb NotI, and 2.7-kb EcoRI fragments (Fig. 2) were cloned into the pCR2.1-TOPO vector by the method described above. The resulting plasmids were digested with the appropriate enzymes to check the orientation of the inserts in the plasmids. The desired orientation is the one in which expression of the AntDO-3C genes is under control of the lac promoter.
The E. coli TOP10F′ strains carrying plasmids pGJZ1362, pGJZ1363, pGJZ1364, and pGJZ1365 were cultured overnight on LB-KM-tetracycline. The overnight culture was transferred to fresh medium and cultured until the optical density at 600 nm (OD600) reached 0.5. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added at a final concentration of 1.0 mM to induce expression, and the cells were harvested after 3 h by centrifugation. The collected cells were washed twice with phosphate buffer (50 mM sodium-potassium phosphate buffer, pH 6.8) and resuspended in the same buffer at an OD600 of 4.0. A reaction mixture consisting of 6 ml of cells, 20 mM glucose, and 1.0 mM anthranilate was placed in a 50-ml centrifuge tube and shaken at 200 rpm at 37°C. Culture samples (1 ml) were taken at 0, 2, and 4 h after the anthranilate was added. The supernatant was made ready for HPLC (high-performance liquid chromatography) analysis by passage through a 0.45-μm-pore-size Acrodisc 13-mm-diameter syringe filter (Pall Gelman Laboratory, East Hills, N.Y.).
Disruption of andAa and andR in strain DBO1.
In order to knock out the reductase (andAa) gene, the 5.1-kb XhoI fragment (Fig. 2) containing the andAcAdAbAa genes from plasmid pGJZ1362 was first cloned into the XhoI site in pGEM-7Zf(−). The resulting plasmid, designated pGJZ1366, was digested with NotI to remove restriction fragments inside the reductase gene (Fig. 2). The digested DNA was gel purified, and the staggered ends were repaired to blunt ends with Pfu DNA polymerase. The 0.9-kb Kmr cassette from p34S-Km3 (15) was digested with SmaI and cloned into the blunt-ended plasmid. This construct with the disrupted reductase gene was electroporated into strain DBO1. Kmr transformants were selected, and their total genomic DNA was prepared. Clones with double-crossover recombination were detected by PCR. A PCR using primers 5′-CGCGTGCGCACCTTCGG-3′ and 5′-GCCCGCCGCCGACAAAC-3′ would yield a 1.3-kb PCR product from a double-crossover mutant, while a PCR using primers 5′-GCGCCGCGCGCTATGGCTG-3′ and 5′-GATTTAGGTGACACTATAG-3′ (SP6) would yield no PCR product. The desired reductase mutant was designated HK502.
In order to knock out the andR gene, a 0.7-kb MluI-BstBI fragment (Fig. 2) containing the middle part of the andR gene was cloned into pCR2.1-TOPO as described above. The clone with the truncated andR gene's C-terminal end adjacent to the lac promoter on the plasmid was retained. The construct was electroporated into strain DBO1, and Kmr transformants were selected. The insertion of the plasmid into the 0.7-kb MluI-BstBI area in the DBO1 chromosome by single-crossover recombination was further confirmed by PCR. A 1.0-kb PCR product would be expected when primers 5′-GCTCATGCCGCGAAACTG-3′ and 5′-GTAAAACGACGGCCAG-3′ (M13 forward) and the template DNA from the desired mutant were used. The strain DBO1 andR mutant was designated HK504.
Construction and analysis of the andAc-lacZ transcriptional fusion plasmid.
The 0.48-kb SalI-XbaI fragment containing part of the andR gene, the promoter region of the andR-andAc genes, and part of the andAc gene was removed from plasmid pGJZ1361 and cloned into pKRZ-1 that has a lacZ reporter gene. The andAc-lacZ transcriptional fusion plasmid (designated pGJZ1370) was transformed into DBO1. The DBO1(pGJZ1370) strain was grown to an OD600 of 0.1 with succinate as the sole carbon source in MSB medium. The compounds tested were added to separate 3-ml cultures at a final concentration of 1.0 mM. When the cells reached an OD600 of 0.5, 0.5-ml volumes of cells were removed for the β-galactosidase assay described by Miller (44). Three separate starter cultures were used, and the assays with each compound were done in triplicate, for a total of nine assays per compound.
Sequencing and sequence analysis.
Sequencing reactions were performed as recommended by the supplier (Applied Biosystems, Inc.) and analyzed on an ABI 377 or 3100 automated DNA sequencer. The BLAST program (1) was used for similarity searches of the nonredundant National Center for Biotechnology Information (National Institutes of Health, Bethesda, Md.) sequence database. Multiple-sequence alignments were performed by MegAlign of the Lasergene software (DNAStar Inc., Madison, Wis.) by the ClustalW method (63). The tree generated by MegAlign was saved in PHYLIP format and redrawn to a radial style with the TreeView program (48).
Analysis of metabolites by HPLC.
HPLC analysis was performed with a Beckman (Fullerton, Calif.) HPLC apparatus and a Waters Spherisorb Octyl C8 column (4.6 by 150 mm) and a gradient of 0 to 50% or 0 to 100% methanol in water under acidic (0.1% acetic acid) conditions. A flow rate of 1.0 ml/min was used. A diode array detector was used to measure the absorbance of the eluting compounds at 254- and 280-nm wavelengths.
Chemicals.
Kynurenine, kynurenate, anthranilate, salicylate, 3-methylsalicylate, 4-methylsalicylate, 2-chlorobenzoate, benzoate, 2-nitrobenzoate, 2,3-dihydroxybenzoate, 2,4-dihydroxybenzoate, 2,5-dihydroxybenzote, 2,6-dihydroxybenzoate, 2,4-dimethylbenzoate, and o-toluate were obtained from Aldrich (Milwaukee, Wis.).
Nucleotide sequence accession number.
The nucleotide sequence reported here has been deposited in the GenBank database under accession no. AY223539.
RESULTS
B. cepacia DBO1 tryptophan degradation mutants.
The plasposon pTnMod2-OKm3 was introduced into B. cepacia DBO1-Rif by triparental mating. The colonies obtained were screened for growth on tryptophan or succinate as the sole source of carbon and energy on MSB medium plates. Three mutants that lacked the ability to grow on tryptophan and two mutants that could grow on tryptophan but not succinate were obtained. The tryptophan degradation mutants, designated PM100, PM101, and RL101, were selected for further study. PM101 is additionally unable to grow on anthranilate, while PM101 and RL101 retained the ability to use anthranilate as a carbon source. All of the mutant strains retain the ability to grow on benzoate, which is metabolized through catechol.
The degradation pathway of tryptophan in Burkholderia strains (Fig. 1), as well as in Pseudomonas strains, is thought to proceed through N-formyl-kynurenine, kynurenine, and anthranilate to catechol (30). However, kynurenine is not expected to accumulate in the supernatant because it converts to kynurenate spontaneously. The culture supernatants of PM100, PM101, and RL101 grown on succinate plus tryptophan were analyzed by HPLC. Accumulating compound retention times were compared with the retention times of the peaks of the standard compounds kynurenine (17.1 min), kynurenate (17.8 min), and anthranilate (22.3 min). The data showed that the mutant PM101 accumulates kynurenate and anthranilate from tryptophan, PM100 accumulates only kynurenate, and RL101 shows a major peak other than kynurenate or anthranilate (data not shown). RL101 most likely accumulates N-formyl-kynurenine, which is not commercially available as a standard. The results suggest that PM101 has a block in the step for the conversion of anthranilate to catechol because of the insertion of the plasposon, PM100 lacks a functional kynureninase that converts kynurenine to anthranilate, and RL101 possibly lacks a functional formamidase.
Cloning and sequencing of the AntDO genes from strain DBO1.
PM101 must have a plasposon insertion in the genes for AntDO (or their regulator) because the mutant unable to grow on tryptophan or anthranilate is able to grow on succinate and benzoate and accumulates anthranilate when growth on succinate plus tryptophan. This being the case, PM101's genomic DNA was isolated and separately digested with the restriction enzymes NotI, PstI, SalI, and ApaI. The digested DNA fragments were self-ligated, and the circular plasmids were transformed into E. coli DH5α with selection on LB-KM. These plasmids contain the self-cloning plasposon and the genomic DNA next to the plasposon insertion site (Fig. 2). The sizes of the plasmids obtained from PM101 are 2.5 kb from the NotI digestion, 3.5 kb from the PstI digestion, 3.4 kb from the SalI digestion, and 9.8 kb from the ApaI digestion, inclusive of the plasposon vector. The 9.8-kb ApaI plasmid clone was designated pGJZ1360. These plasmids and subclones derived from them were used to determine the complete nucleotide sequence of the genomic ApaI fragment.
Identification of genes in the sequenced region.
Sequence analysis of the ApaI fragment (Fig. 2) revealed seven open reading frames (ORFs). ORF3 to ORF6 were designated andAc, andAd, andAb, and andAa on the basis of their homology to a number of DO-encoding genes. The plasposon insertion site is in the 3′ end of the andAd gene. The amino acid sequences of AndAc (423 amino acids [aa], 48 kDa) and AndAd (161 aa, 18.9 kDa) show similarity to large (α) and small (β) subunits of terminal oxygenases from multicomponent primary DOs. AndAb (108 aa, 11.7 kDa) features a ferredoxin protein. AndAa (406 aa, 42.9 kDa) shows similarity to reductase components of DO systems. It shows that the strain DBO1 AntDO system is a three-component system that is very different from the Acinetobacter two-component AntDO system. The DBO1 AntDO enzyme was designated AntDO-3C to distinguish it from the two-component AntDO enzyme. The consensus sequence Cys-X-His-X15-17-Cys-X2-His for Rieske-type [2Fe-2S] clusters is found in the iron sulfur protein α subunit (ISP-α; Cys95-X-His97-X17-Cys115-X2-His118 in AndAc) and the ferredoxin component (Cys49-X-His51-X16-Cys68-X2-His71 in AndAb). The residues His223, His228, and Asp370 in AndAc, located after the domain for binding of the [2Fe-2S] cluster, are conserved in other ISP-α subunits. Their important roles in binding of the mononuclear iron center was demonstrated in naphthalene DO (36). Three residues, Asp26, Arg71, and Gly126, in AndAd are conserved in other ISP-β subunits. The reductase AndAa contains conserved domains for NADH binding and flavin adenine dinucleotide binding (residues 7 to 288).
Tables 1 through 3 compare the similarity of AndAc, AndAd, and AndAa to other DO components. BphA1cA2c and BphA1dA2d from Novosphingobium aromaticivorans F199 possibly have the reductase (BphA4) and the ferredoxin (BphA3) in common with other sets of oxygenases found on plasmid pNL1 (51). NagG and NagH (the α and β subunits of salicylate-5-hydroxylase) from Ralstonia sp. strain U2 (24) could have NagAa (reductase) and NagAb (ferredoxin) in common with NagAcAd (naphthalene DO). OhbB and OhbA (the α and β subunits of 2-chlorobenzoate DO) from P. aeruginosa strains 142 and JB2 (31, 64) have no associated reductase or ferredoxin that has been found so far. AndAa shows the greatest similarity to the putative ferredoxin reductase MocF (accession no. AAD53005) from Rhizobium leguminosarum, ThcD (rhodocoxin reductase) from Rhodococcus erythropolis (45), and CarAd (carbazole catabolism-related protein) from Sphingomonas sp. strain CB3 (55). AndAa and NagAa from Ralstonia sp. strain U2 are very different, because they are from different classes of DO. The ferredoxin AndAb shows similarity to other ferredoxins, such as HybD (65%) from P. aeruginosa JB2 (32), NagAb (60%) from Ralstonia sp. strain U2 (68), and NahAb (63%) from P. putida NCIB9816 (56).
TABLE 1.
Similarity between large subunits of terminal DOs
| Protein | % Similarity (identity) with:
|
||||
|---|---|---|---|---|---|
| AndAc | BphA1c | BphA1d | OhbB | NagG | |
| AndAc | 100 (100) | 67 (52) | 64 (42) | 61 (46) | 56 (39) |
| BphA1c | 100 (100) | 67 (49) | 70 (49) | 58 (42) | |
| BphA1d | 100 (100) | 70 (41) | 55 (38) | ||
| OhbB | 100 (100) | 59 (42) | |||
| NagG | 100 (100) | ||||
TABLE 3.
Similarity between reductase components
| Protein | % Similarity (identity) with:
|
||||
|---|---|---|---|---|---|
| AndAa | MocF | ThcD | BphA4 | CarAd | |
| AndAa | 100 (100) | 57 (43) | 55 (42) | 51 (38) | 50 (38) |
| MocF | 100 (100) | 53 (40) | 48 (35) | 55 (40) | |
| ThcD | 100 (100) | 50 (35) | 51 (33) | ||
| BphA4 | 100 (100) | 57 (41) | |||
| CarAd | 100 (100) | ||||
From the recently obtained genomic sequence of the related strain Burkholderia fungorum LB400 (referred to as LB400), two gene clusters each with five ORFs (Bcep_p_7244 to Bcep_p_7248, accession number NZ_AAAC01000305; Bcep_p_7642 to Bcep_p_7646, accession number NZ_AAAC01000308) in LB400 show a high level of similarity to the AntDO-3C gene cluster in DBO1. The two gene clusters from LB400 are 90% identical to each other on the basis of a DNA sequence comparison. The oxygenase and ferredoxin components of AntDO-3C from LB400 and DBO1 have an average of 90% similarity, while the reductase components have a lower level of similarity (71%).
A putative regulatory gene (designated andR) located upstream of the andAc gene is transcribed in the opposite direction from the AntDO-3C structural genes. AndR (318 aa, 35.1 kDa) has a conserved helix-turn-helix domain for DNA binding at its C-terminal end. It shows similarity to the transcriptional regulators in the AraC/Xyls family (Table 4). Most of the characterized proteins of this family are positive transcriptional regulators (26). The putative regulators located upstream of the two putative AntDO-3C gene clusters in LB400 show 64% similarity to AndR. AndR shows relatively less similarity to XylS from P. putida MT-2 (33), IpbR from P. putida RE204 (18), CbdS from Burkholderia sp. strain TH2 (61), and BenR from P. putida PRS2000 (13). Another putative regulatory gene is located downstream of the andR gene. This gene was designated catR because its product (296 aa, 32.5 kDa) shows similarity to transcriptional activators CatR2 (85%) and CatR1 (71%) from Burkholderia sp. strain TH2 (60) and its downstream sequence shows a high level of similarity to the cis,cis-muconate lactonizing enzyme gene (catB; data not shown). These regulatory proteins have a DNA binding domain in their N-terminal part and a LysR substrate binding domain following the previous domain. They belong to the LysR regulatory protein family. CatR from DBO1 is 86% similar to the putative protein Bcep_p_1743 (334 aa) from LB400. However, the gene for Bcep_p_1743 is not physically associated with the putative AntDO-3C genes in LB400. The DBO1 CatR protein also shows similarity to other transcriptional regulatory proteins, such as CatR (66%) from Pseudomonas sp. strain CA10 (47) and BenM (63%) from Acinetobacter sp. strain ADP1 (12). An ORF (designated ORFM) encoding a 48.3-kDa protein (428 aa) is located downstream of the andAa gene; however, no obvious ribosomal binding site or alternative translation initiation site was observed in front of ORFM. ORFM encodes a protein with 72% similarity to the putative inner membrane protein YjiN (423 aa) from Salmonella enterica serovar Typhimurium LT2 (43).
TABLE 4.
Similarity between regulators in the AraC/XylS family
| Protein | % Similarity (identity) with:
|
||||
|---|---|---|---|---|---|
| AndR | XylS | IpbR | CbdS | BenR | |
| AndR | 100 (100) | 41 (27) | 40 (24) | 41 (22) | 38 (22) |
| XylS | 100 (100) | 57 (41) | 57 (44) | 73 (59) | |
| IpbR | 100 (100) | 54 (37) | 65 (51) | ||
| CbdS | 100 (100) | 59 (43) | |||
| BenR | 100 (100) | ||||
Cloning of the intact AntDO-3C genes directly from strain DBO1.
The genes for AntDO-3C were obtained by direct cloning of a genomic region surrounding the plasposon insertion point. In order to perform functional studies on the encoded proteins, the intact (nondisrupted) genes are needed. A novel strategy was used to clone the genes directly from the DBO1 chromosome in such a way as to immediately allow their controlled expression in E. coli. The 1.0-kb MluI fragment (Fig. 2), containing part of andR, the region between andR and andAc, and part of andAc, was cloned into pCR2.1-TOPO (see Materials and Methods). The resulting plasmid, with the lac promoter reading into the andAc gene fragment, was designated pGJZ1361 and transformed into DBO1. Since pGJZ1361 cannot replicate in strain DBO1, the Kmr transformants would be the result of a single crossover in the 1.0-kb MluI area. Several colonies were picked, and the insertion of plasmid pGJZ1361 into their chromosomes was confirmed by PCR (see Materials and Methods). The mutant strain with the insertion (designated HK501) still maintains the ability to grow on anthranilate. Total genomic DNA purified from HK501 was digested with the restriction enzyme ApaI, self-ligated, and transformed into E. coli TOP10F′ cells. The resulting plasmid (designated pGJZ1362) has the pCR2.1-TOPO vector and the desired 8.0-kb insert containing the truncated putative regulator gene and intact andAc, andAd, andAb, andAa, and ORFM genes (Fig. 2).
Expression of AntDO-3C in E. coli.
Expression of the andAcAdAbAa genes on plasmid pGJZ1362 is under control of the lac promoter. The lacIq gene on the F′ factor in E. coli TOP10F′ represses the expression of these genes. The IPTG-induced E. coli cells carrying pGJZ1362 readily converted anthranilate to catechol on either LB or MSB medium (data not shown). Different subclones were constructed from plasmid pGJZ1362 (Fig. 2). The 5.1-kb XhoI fragment containing the andAcAdAbAa genes, the 3.4-kb NotI fragment containing the andAcAdAb genes, and the 2.7-kb EcoRI fragment containing only the andAcAd genes were cloned into pCR2.1-TOPO, and the resulting plasmids were designated pGJZ1363, pGJZ1364, and pGJZ1365 (Fig. 2). The lac promoter also controls the expression of the genes on these plasmids.
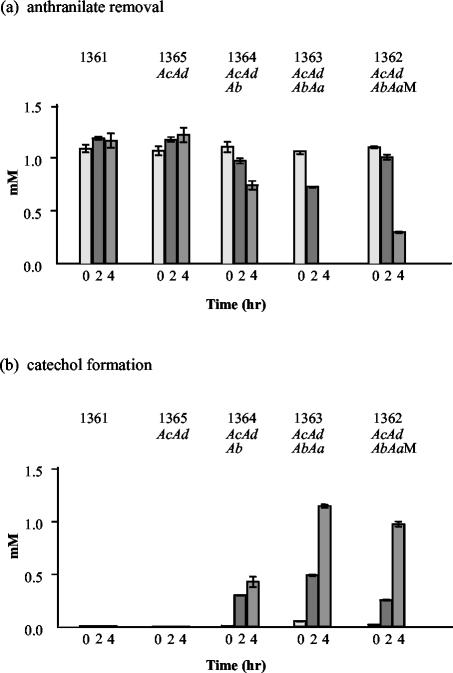
The ability of E. coli Top10F′ carrying pGJZ1362, pGJZ1363, pGJZ1364, and pGJZ1365 after IPTG induction to transform anthranilate to catechol was analyzed as described in Materials and Methods. E. coli Top10F′(pGJZ1363) completely transformed 1 mM anthranilate to catechol in 4 h (Fig. 3). E. coli Top10F′(pGJZ1362) oxidized 85% of the anthranilate in the same time interval. Both clones have the genes for the oxygenase, ferredoxin, and reductase components, with pGJZ1362 additionally containing ORFM. Transformation of anthranilate is thus slightly inhibited by the presence of ORFM. E. coli Top10F′(pGJZ1364) encoding only the oxygenase and ferredoxin components transformed 37% of the anthranilate to catechol. These data suggest that the ferredoxin has the ability to accept electrons from unidentified reductase sources in E. coli, with a much lower efficiency than from the natural reductase (AndAa). E. coli Top10F′(pGJZ1365) encoding only the oxygenase component and the negative control TOP10F′(pGJZ1361) do not transform anthranilate at all. These data demonstrate that the andAcAdAbAa genes encode a specific ferredoxin and reductase that act together with an oxygenase component to oxidize anthranilate to catechol.
FIG. 3.
Anthranilate consumption (a) and catechol formation (b) by IPTG-induced E. coli strains carrying the andAcAd genes (pGJZ1365), the andAcAdAb genes (pGJZ1364), and the andAcAdAbAa genes (pGJZ1362 and pGJZ1363). pGJZ1362 has a putative membrane protein gene (ORFM) right after the andAcAdAbAa genes. The E. coli strain carrying pGJZ1361 was used as the negative control. Transformation of anthranilate to catechol was monitored by HPLC analysis. The initial concentration of anthranilate was 1.0 mM.
Substrate range of AntDO-3C.
E. coli cells with functional AntDO-3C were tested for the ability to convert substrates other than anthranilate. E. coli Top10F′(pGJZ1363) with the intact andAcAdAbAa genes was selected for this study. Salicylate, 3-methylsalicylate, 4-methylsalicylate, 2-chlorobenzoate, benzoate, 2-nitrobenzoate, 2,3-dihydroxybenzoate, 2,4-dihydroxybenzoate, 2,5-dihydroxybenzote, 2,6-dihydroxybenzoate, 2,4-dimethylbenzoate, and o-toluate were among the compounds tested. Of the 12 aromatic compounds tested, noticeable transformation by AntDO-3C was only observed for salicylate and benzoate. Over the 4-h incubation period, 20% of the salicylate was transformed to catechol. Only 10% of the benzoate was transformed, and a peak of increasing intensity was observed on the HPLC chromatogram. Benzoate is presumably converted into benzoate dihydrodiol by AntDO-3C because the functionally similar AntDO enzyme from ADP1 transforms benzoate to benzoate dihydrodiol (19). The 1-ml culture supernatant containing the putative benzoate dihydrodiol was treated with 100 μl of 1 N HCl at 100°C for 10 min. Following this treatment, the HPLC peak for the putative dihydrodiol disappeared and compounds with HPLC retention times identical to those of phenol and salicylate were detected. Since phenol and salicylate are the expected breakdown products of the benzoate dihydrodiol, this confirms the ability of AntDO-3C to oxidize benzoate to a dihydrodiol. These results indicate that AntDO-3C has a narrow substrate range.
Disruption of andAa and andR in DBO1.
As described above, expression of AntDO-3C in E. coli requires the presence of the reductase and ferredoxin for full enzymatic activity. The importance of the reductase was further demonstrated by inserting a kanamycin cassette into the andAa gene in the DBO1 chromosome (see Materials and Methods). The mutant DBO1 strain with the disrupted reductase gene is unable to grow on anthranilate, demonstrating the importance of this specific reductase in AntDO-3C enzymatic activity.
The andR gene in the DBO1 chromosome was disrupted by the following strategy. A 0.7-kb MluI-BstBI fragment (Fig. 2) containing the middle part of the andR gene was first cloned into pCR2.1-TOPO with the correct direction for insertion (see Materials and Methods). The DBO1 transformant with insertion of the constructed plasmid into the 0.7-kb MluI-BstBI area in the chromosome by a single-crossover event was designated HK504 (see Materials and Methods). HK504 has two truncated copies of the andR gene. One copy lost 70 aa from the C-terminal end that is the hypothetical DNA binding site. The other copy is missing the N-terminal 18 aa and the promoter area of the andR gene. HK504 with the two incomplete copies of andR lost the ability to grow on anthranilate, confirming that the andR gene is involved in the regulation of anthranilate degradation.
Expression of the AntDO-3C genes is tightly regulated.
An andAc-lacZ transcriptional fusion plasmid was constructed as described in Materials and Methods. Anthranilate, salicylate, benzoate, 2-chlorobenzoate, 2-nitrobenzoate, 2,3-dihydroxybenzoate, 2,4-dihydroxybenzoate, 2,5-dihydroxybenzoate, 2,6-dihydroxybenzoate, 2,4-dimethylbenzoate, o-toluate, and catechol were tested separately for the ability to induce the expression of the lacZ gene on the plasmid in DBO1. The LacZ activity of permeabilized cells was compared with that of the negative control, to which no effector was added. Although 12 related compounds were tested, only anthranilate is an effective inducer. Anthranilate-induced LacZ activity (27.5 ± 2.0 Miller units) was approximately 31 times that of uninduced cells (0.9 Miller unit). The LacZ activity of cells exposed to the structurally related compounds listed above is close to the negative control value. These results demonstrate that expression of the AntDO-3C genes in DBO1 is tightly regulated. Even though benzoate and salicylate can serve as fortuitous substrates for the DO, they are not able to induce expression of the AntDO-3C genes.
DISCUSSION
AntDO-3C is a class IIB DO.
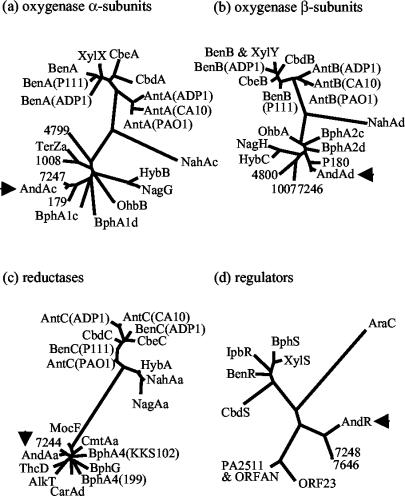
The sequence analysis shows that AntDO-3C belongs to the class IIB family of DOs that consist of three components (reductase, ferredoxin, and a two-subunit oxygenase). AntDO-3C is very different from previously studied two-component AntDOs from Acinetobacter and Pseudomonas strains (6, 19). The AntDO-3C protein sequences were aligned with those of several similar proteins. Phylogenetic trees constructed by such alignments show that the oxygenase components from the same class are clustered together (Fig. 4a and b). AntDO-3C forms a cluster with several other terminal oxygenases that are distant from the rest of the members of the family of primary oxygenases. In this cluster, AntDO-3C forms a tighter internal cluster with two putative proteins from LB400 (only one pair of the putative AntDO-3C oxygenase components, Bcep_p_7246 and Bcep_p_7247, was selected for comparison). The distance between the reductase component cluster containing AndAa and the other two reductase clusters (Fig. 4c) is far greater than the distances between different clusters of oxygenase components (Fig. 4a and b).
FIG. 4.
Phylogenetic trees of the oxygenase α subunits (a), β subunits (b), reductase (c), and regulators (d). Abbreviations: AlkT, alkane monooxygenase reductase of P. putida TF4-1L (accession no. CAB54063); AndAaAbAcAd, anthranilate DO of B. cepacia DBO1 (accession no. AY223539); AndR, regulator of andAaAbAcAd of DBO1 (accession no. AY223539); AntABC(ADP1), AntDO of Acinetobacter sp. strain ADP1 (accession no. AF071556); AntABC(PAO1), putative AntDO of P. aeruginosa strain PAO1 (accession no. NC_002516); AntABC(CA10), putative AntDO of Pseudomonas sp. strain CA10 (accession no. AB047548); AraC, regulator of the l-arabinose operon of E. coli K-12 (accession no. CAA23508); BenABC, benzoate DO of P. putida PRS2000 (accession no. AF218267); BenR, positive regulator of benABC of P. putida PRS2000 (accession no. AAF63447); BenABC(ADP1), benzoate DO of Acinetobacter sp. strain ADP1 (accession no. AF009224); BenABC(P111), benzoate DO of P. aeruginosa P111 (accession no. AY026914); BphA1cA1dA2cA2d, aromatic oxygenase of N. aromaticivorans F199 (accession no. NC_002033); BphA4(F199), aromatic oxygenase ferredoxin reductase of N. aromaticivorans F199 (accession no. NP_049182); BphA4(KKS102), ferredoxin reductase of Pseudomonas sp. strain KKS102 (accession no. BAA04112); BphG, biphenyl DO of B. fungorum LB400 (accession no. AAB63429); BphS, putative regulator of the meta-pathway operon of Pseudomonas sp. strain IC (accession no. AAC34589); CarAd, carbazole initial DO reductase subunit of Sphingomonas sp. strain CB3 (accession no. AAC38619); CbdABC, 2-halobenzoate 1,2-DO of Burkholderia sp. strain TH2 (accession no. AB035324); CbdS, positive regulator of cbdABC of Burkholderia sp. strain TH2 (accession no. BAB21583); CbeABC, 2-halobenzoate 1,2-DO of Burkholderia sp. strain NK8 (accession no. AB024746); CmtAa, p-cumate DO ferredoxin reductase subunit of P. putida F1 (accession no. AAB62284); HybABC, salicylate 5-hydroxylase of P. aeruginosa JB2 (accession no. AAC69483); IpbR, positive regulator of ipb operon of P. putida RE204 (accession no. AAC034350); MocF, putative ferredoxin reductase of Sinorhizobium meliloti LB-30 (accession no. AAD53005); NagGH, salicylate 5-hydroxylase of Ralstonia sp. strain U2 (accession no. AF036940); NagAa, ferredoxin reductase of Ralstonia sp. strain U2 (accession no. AAD12606); NahAaAcAd, naphthalene DO of P. putida 9816-4 (accession no. U49496); OhbAB, 2-halobenzoate DO of P. aeruginosa 142 (accession no. AF121970); OrfAN, unknown protein of P. putida P111 (accession no. AAK52294); PA2511, probable transcriptional regulator of P. aeruginosa PAO1 (accession no. 251201); 179 and 180, hypothetical proteins Bcep_p_179 and Bcep_p_180 of B. fungorum LB400 (accession no. NZ_AAAC01000057); 1007 and 1008, hypothetical proteins Bcep_p_1007 and Bcep_p_1008 of B. fungorum LB400 (accession no. NZ_AAAC01000150); 4799 and 4800, hypothetical proteins Bcep_p_4799 and Bcep_p_4800 of B. fungorum LB400 (accession no. NZ_AAAC01000272); 7244, 7246, 7247, and 7248, hypothetical proteins Bcep_p_7244, Bcep_p_7246, Bcep_p_7247, and Bcep_p_7248 of B. fungorum LB400 (accession no. NZ_AAAC01000305); 7646, hypothetical protein of B. fungorum LB400 (accession no. ZP_00034749); TerZa, terephthalate 1,2-DO oxygenase large subunit of Delftia sp. strain T7 (accession no. BAC15591); ThcD, ferredoxin reductase of R. erythropolis NI86/21 (accession no. AAC45752); XylXY, toluate 1,2-DO of P. putida PaW630 (accession no. AF134348); XylS, positive regulator of xylDLEGF of P. putida MT-2 (accession no. P07859).
The reductase component is important for the activity of AntDO-3C.
It appears that no specific electron transfer components are required for the activity of the oxygenases closely related to AntDO-3C (Fig. 4a to c). For example, the genes for OhbAB from P. aeruginosa 142 are not physically associated with electron transfer component genes, and the enzyme activity in recombinant E. coli and Pseudomonas strains is manifested by the ohbAB-encoded terminal oxygenase alone, implying the use of electron transfer components provided by the recipient cells (64). The bphA2cA1c genes from N. aromaticivorans F199 and Sphingomonas yanoikuyae B1 are preceded by a Rieske-type [2Fe-2S] ferredoxin gene; however, the bphA1dA2d genes are not physically associated with any ferredoxin gene. There are six ISP genes (bphA1a to -f and bphA2a to -f) on plasmid pNL1 from F199 and in the chromosome of B1 but only one reductase gene (bphA4) and one ferredoxin gene (bphA3). The ferredoxin and reductase components in B1 have been shown to be involved with more than one oxygenase (37). Sharing of the same set of electron transfer genes by two ISPs (reductase-ISPx-ferredoxin-ISPy) has been observed in Ralstonia sp. strain U2 (24), Comamonas testosteroni GZ42 (24), and Burkholderia sp. strain DNT (59). For example, in Ralstonia sp. strain U2, NagAa (reductase) and NagAb (ferredoxin) could be shared by oxygenases NagGH (salicylate 5-hydroxylase) and NagAcAd (naphthalene DO). NagAa and NagAb are not necessarily required because E. coli expressing NagG and NagH alone was able to convert salicylate to gentisate, although with a low level of activity. It has been proposed that loose association with electron transfer components might be the characteristic for this cluster of DOs (64). The sequence analysis suggested coevolution of the ISP-α and ISP-β genes, whereas electron transfer components may not be obligatory for these oxygenases. Other examples can also be found in the proteins distantly related to AntDO-3C. E. coli expressing NagAc and NagAd along with the ferredoxin NagAb from Ralstonia sp. strain U2 was able to oxidize indole to indigo, while the reductase NagAa was apparently not required (24). Deletion of dntAa (reductase) from the Burkholderia sp. strain DNT 2,4-dinitrotoluene DO gene cluster did not eliminate the ability of E. coli containing such deletion derivatives to transform 2,4-dinitrotoluene (59). The expression of toluene-4-monooxygenase activity in E. coli and other Pseudomonas strains did not require the reductase component (TmoF), although the activity was enhanced at least twofold by its presence (67).
In DBO1, the genes for the AntDO-3C oxygenase component are physically associated with the genes for the electron transport components. This type of linkage is also observed in the putative AntDO-3C homologs in LB400. The AntDO-3C oxygenase component expressed alone cannot endow E. coli with enzyme activity for the transformation of anthranilate to catechol. The oxygenase component along with the ferredoxin component in E. coli was able to transform anthranilate to catechol, but with only one-third of the activity compared with the E. coli strain expressing all three components of AntDO-3C. The importance of the reductase was further demonstrated by disruption of the andAa gene in DBO1. The strain with the disrupted andAa gene lost the ability to grow at the expense of anthranilate. By comparison, in Acinetobacter species strain ADP1, a disrupted reductase gene (antC) does not result in a mutant strain unable to grow on anthranilate because BenC (benzoate 1,2-DO reductase) can substitute for AntC in anthranilate oxidation (6).
AntDO-3C has a narrow substrate range.
Among the compounds tested, the enzyme can only transform anthranilate, benzoate, and salicylate but not structurally related compounds such as 2-chlorobenzoate and o-toluate. The functionally similar ADP1 AntDO enzyme also has a narrow substrate range, being unable to transform 2-chlorobenzoate and only transforming 13% of the o-toluate tested when purified enzymes were used in vitro (19). The activity of ADP1 AntDO toward benzoate was 69% of the activity toward anthranilate (19), while the activity of AntDO-3C toward benzoate was only 10% of the activity toward anthranilate. This suggests that ADP1 AntDO has a more relaxed substrate range than AntDO-3C. Although ADP1 AntDO could convert benzoate to benzoate-dihydrodiol, the ADP1 mutant strain lacking the functional structural gene encoding BenDO did not grow on benzoate (19). The phenotype may reflect the inability of benzoate to induce AntDO, insufficient production of the benD-encoded diol dehydrogenase in vivo, and/or problems with the rate of benzoate transformation by AntDO (19). The same may be true of strain DBO1 because benzoate is not an inducer of the AntDO genes and the enzyme transforms benzoate with low efficiency. The OhbAB protein from P. aeruginosa 142 could also transform benzoate with 37% of the activity toward its native substrate (50). Some benzoate DO enzymes exhibit the ability to transform anthranilate to catechol, such as the benzoate DO from Pseudomonas arvilla C-1 (65), and BopXY from Rhodococcus sp. strain 19070 (28). However, BenABC from ADP1, with a narrow substrate range, cannot transform anthranilate (46).
AntDO-3C can also transform salicylate to catechol. Some other proteins closely related to AntDO-3C, such as OhbAB and BphA1cA2c, may have the same ability. Although salicylate was not reported as a substrate for OhbAB from P. aeruginosa 142, it is reasonable to assume that OhbAB could transform salicylate because of its broad substrate range. Benzoate, 4-chloroanthranilate, 5-chloroanthranilate, o-trifluoromethylbenzoate, 2,4-dichlorobenzoate, 2,5-dichlorobenzoate, and other compounds are the substrates for OhbAB (50, 54). In fact, the functionally similar but phylogenetically different 2-cholorobenzoate DO enzyme from P. cepacia 2CBS was shown to transform salicylate to catechol (22). BphA1cA2c from S. yanoikuyae B1 has been shown to transform salicylate to catechol by preliminary data (O. Cho, S. Hwang, S. W. Kim, and E. Kim, Abstr. 102nd Gen. Meet. Am. Soc. Microbiol. 2002, abstr. K-12, p. 270, 2002). This function of BphA1cA2c is important because the proposed naphthalene degradation pathway of S. yanoikuyae B1 goes through salicylate to catechol (69) and the typical salicylate hydroxylase (NahG) homolog for the conversion of salicylate to catechol was not found in the nucleotide sequence.
Evolutionary relationship between AntDO-3C and 2-chlorobenzoate DO.
Anthranilate and 2-halobenzoate have been linked together because of their structural similarity. Anthranilate has even been suggested to be a useful substrate for pre-enrichment of bacteria that can liberate halide from 2-halobenzoate by dioxygenation (20). It has been proposed that AntDO and 2-halobenzoate 1,2-DO may be closely related enzymes because of the observation that purified or partially purified 2-halobenzoate 1,2-DO was able to effectively convert anthranilate to catechol (54). The similarity between AntDO-3C and OhbAB from P. aeruginosa 142 provides more clues about this hypothesis. The ohbAB genes are embedded in a transposon-like context, implying the likely involvement of horizontal gene transfer in the evolution of 2-chlorobenzoate DO activity. IS elements may play an important role in the transfer of the genes. It has been shown that similar ohbAB genes from P. aeruginosa strain JB2 can be transferred to other bacteria (31). It is interesting that B. fungorum LB400 not only has two copies of AntDO-3C homologs but also has almost the exact copy of OhbAB (100% identity for OhbA, 99% identity for OhbB; the sequence for the ohbB gene is not complete; only the sequence for the first 318 aa is in the database). Although the question of the origin of the ohbAB genes remains to be answered, one could speculate that ohbAB evolved from AntDO-3C-like genes, since tryptophan is a more common substrate than 2-chlorobenzoate. During evolution, OhbAB not only acquired a broader substrate range but also became more independent from the electron donor components.
Gene organization and evolution.
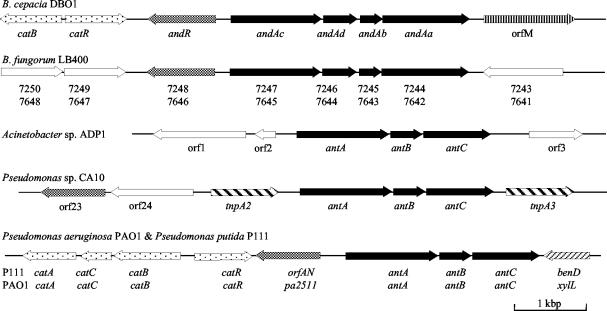
Figure 5 shows the organization of the known AntDO genes from two different evolutionary origins. The ADP1 antABC genes are separated from the two supraoperonic gene clusters containing the ben and cat genes by approximately one-third of the ADP1 chromosome (6). In ADP1, the genes are organized as ORF1-ORF2-antABC-ORF3 (Fig. 5). In Pseudomonas sp. strain CA10, the antABC genes are accompanied on both sides by two IS elements containing transposases (tnpA2-antABC-tnpA3). In P. aeruginosa PAO1 and P. putida P111, ant loci map to a region between the ben and cat genes (Fig. 5). A gene encoding a putative transcriptional activator belonging to the AraC/XylS family was located upstream of the antABC genes. It may be that the antABC genes, together with the regulatory gene, were originally with the other genes involved in aromatic compound degradation and were moved to other loci in ADP1 and CA10, in the latter case participating in the carbazole degradation pathway. The DBO1 AntDO-3C genes are near the genes for catechol degradation, just as the antABC genes are close to the cat genes in PAO1 and P111. However, the two copies of AntDO-3C homologs genes in LB400 are not physically linked to the genes for catechol degradation.
FIG. 5.
Organizations of the AntDO genes from B. cepacia DBO1, B. fungorum LB400 (accession no. NZ_AAAC01000305 and NZ_AAAC01000308), Acinetobacter sp. strain ADP1 (accession no. AF071556), Pseudomonas sp. strain CA10 (accession no. AB047548), P. aeruginosa PAO1 (accession no. NC_002516), and P. putida P111 (accession no. AY026914). Abbreviations: andAaAbAcAd, AntDO-3C genes; andR, positive regulator of and operon; antABC, AntDO genes; catA, catechol 1,2-DO gene; catB, muconate cycloisomerase I gene; catC, muconolactone δ-isomerase gene; catR, transcriptional regulatory gene for cat operon; benD, benzoate diol dehydrogenase gene; orfAN, putative protein gene; orfM, putative membrane protein gene; pa2511, putative protein gene; tnpA, transposase gene; xylL, benzoate diol dehydrogenase gene. The structural genes for the DOs are illustrated as solid black lines, andR and the putative AntDO regulatory genes are illustrated as shaded lines, and the genes for the cat operon are illustrated as dots.
AndR positively regulates expression of the AntDO-3C genes.
It was previously known that the anthranilate degradation pathway in ADP1 is inducible because cells previously grown with succinate degraded anthranilate with a delay of 2 h (6). Studies of regulatory mutants suggested that neither of the LysR-type activators known to control benzoate and catechol degradation, BenM and CatM, controls ant gene expression (6). It was also suggested that anthranilate-to-catechol conversion in ADP1 appears to be tightly regulated because neither catechol nor cis,cis-muconate can induce expression of the antA-lacZ transcriptional fusion construct (6). Disruption of the xylS-like ORF3 gene next to the antA gene has no effect on anthranilate degradation. In P. aeruginosa 142, the ohbR gene, encoding a protein similar to the IclR family of transcriptional repressors, was located upstream of the ohbA gene (64). However, the function of OhbR was not defined. We showed in the present work by transcriptional fusion assays that AndR, encoded by a gene found upstream of andAc that is transcribed in the opposite direction, regulates expression of the AntDO-3C genes in strain DBO1. This is the first regulatory protein experimentally shown to be involved in regulation of the expression of anthranilate degradation. The and operon is positively regulated by the andR gene, and induction is very specific. Among the effectors tested, only anthranilate effectively activated the and gene promoter.
A comparison of the sequences of AndR and other members of the AraC/XylS family shows that AndR is located outside the cluster that contains the well-studied XylS and BenR proteins and the cluster containing the putative regulators (PA2511 and OrfAN) for the AntDO genes from P. aeruginosa PAO1 and P. putida P111 (Fig. 4d). ORF23, located upstream of the antABC genes in Pseudomonas sp. strain CA10, codes for a protein that forms a close cluster with PA2511 and OrfAN. The upstream promoter sequences for the AntDO-3C gene operons from DBO1 and LB400 show 31% identity. The promoter sequences of the two putative LB400 AntDO-3C genes show 92% identity. In the promoters for the AraC/XylS family, two direct tandem repeats located upstream of the −35 area with the sequence TGCAN7GGNTA are presumed to be the binding site for regulator dimers (34). Two very similar repeats with the sequence TC(C/G)(C/G)-N7-8-GGATA are located in the strain DBO1 and operon promoter area. The two repeats are usually separated by a spacer of 7 nucleotides, but in DBO1, they are separated by a spacer of 14 nucleotides. Further experiments are needed to determine whether these are the binding sites for AndR.
TABLE 2.
Similarity between small subunits of terminal DOs
| Protein | % Similarity (identity) with:
|
||||
|---|---|---|---|---|---|
| AndAd | OhbA | BphA2d | BphA2c | NagH | |
| AndAd | 100 (100) | 66 (48) | 66 (45) | 62 (37) | 58 (33) |
| OhbA | 100 (100) | 64 (45) | 63 (50) | 52 (27) | |
| BphA2d | 100 (100) | 65 (48) | 57 (34) | ||
| BphA2c | 100 (100) | 51 (28) | |||
| NagH | 100 (100) | ||||
Acknowledgments
This work was supported by National Science Foundation grants MCB-0078465 and CHE-9810248.
We thank C. Alan Kachel for DNA sequencing assistance.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1991. Current protocols in molecular biology, vol. I. John Wiley & Sons, Inc., New York, N.Y.
- 3.Batie, C. J., D. P. Ballou, and C. J. Correll. 1991. Phthalate dioxygenase reductase and related flavin-iron-sulfur containing electron transferases, p. 544-554. In F. Müller (ed.), Chemistry and biochemistry of flavoenzymes. CRC Press, Inc., Boca Raton, Fla.
- 4.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 5.Braun, K., and D. T. Gibson. 1984. Anaerobic degradation of 2-aminobenzoate (anthranilic acid) by denitrifying bacteria. Appl. Environ. Microbiol. 48:102-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bundy, B. M., A. L. Campbell, and E. L. Neidle. 1998. Similarities between the antABC-encoded anthranilate dioxygenase and the benABC-encoded benzoate dioxygenase of Acinetobacter sp. strain ADP1. J. Bacteriol. 180:4466-4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cain, R. B. 1968. Anthranilic acid metabolism by microorganisms: formation of 5-hydroxyanthranilate as an intermediate in anthranilate metabolism by Nocardia opaca. Antonie Van Leeuwenhoek 34:17-32. [PubMed] [Google Scholar]
- 8.Cain, R. B. 1966. Induction of an anthranilate oxidation system during the metabolism of ortho-nitrobenzoate by certain bacteria. J. Gen. Microbiol. 42:197-217. [DOI] [PubMed] [Google Scholar]
- 9.Cain, R. B. 1966. Utilization of anthranilic and nitrobenzoic acids by Nocardia opaca and a flavobacterium. J. Gen. Microbiol. 42:219-235. [DOI] [PubMed] [Google Scholar]
- 10.Chang, H. K., and G. J. Zylstra. 1998. Novel organization of the genes for phthalate degradation from Burkholderia cepacia DBO1. J. Bacteriol. 180:6529-6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chauhan, A., and R. K. Jain. 2000. Degradation of o-nitrobenzoate via anthranilic acid (o-aminobenzoate) by Arthrobacter protophormiae: a plasmid-encoded new pathway. Biochem. Biophys. Res. Commun. 267:236-244. [DOI] [PubMed] [Google Scholar]
- 12.Collier, L. S., G. L. Gaines III, and E. L. Neidle. 1998. Regulation of benzoate degradation in Acinetobacter sp. strain ADP1 by BenM, a LysR-type transcriptional activator. J. Bacteriol. 180:2493-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cowles, C. E., N. N. Nichols, and C. S. Harwood. 2000. BenR, a XylS homologue, regulates three different pathways of aromatic acid degradation in Pseudomonas putida. J. Bacteriol. 182:6339-6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dennis, J. J., and P. A. Sokol. 1995. Electrotransformation of Pseudomonas. Mol. Biol. Methods 47:125-133. [DOI] [PubMed]
- 15.Dennis, J. J., and G. J. Zylstra. 1998. Improved antibiotic-resistance cassettes through restriction site elimination using Pfu DNA polymerase PCR. BioTechniques 25:772-774, 776. [DOI] [PubMed] [Google Scholar]
- 16.Dennis, J. J., and G. J. Zylstra. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 64:2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eaton, R. W., and K. N. Timmis. 1986. Characterization of a plasmid-specified pathway for catabolism of isopropylbenzene in Pseudomonas putida RE204. J. Bacteriol. 168:123-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eby, D. M., Z. M. Beharry, E. D. Coulter, D. M. Kurtz, Jr., and E. L. Neidle. 2001. Characterization and evolution of anthranilate 1,2-dioxygenase from Acinetobacter sp. strain ADP1. J. Bacteriol. 183:109-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engesser, K. H., and P. Schulte. 1989. Degradation of 2-bromo-, 2-chloro- and 2-fluorobenzoate by Pseudomonas putida CLB 250. FEMS Microbiol. Lett. 51:143-147. [DOI] [PubMed] [Google Scholar]
- 21.Fetzner, S. 2000. Enzymes involved in the aerobic bacterial degradation of N-heteroaromatic compounds: molybdenum hydroxylases and ring-opening 2,4-dioxygenases. Naturwissenschaften 87:59-69. [DOI] [PubMed] [Google Scholar]
- 22.Fetzner, S., R. Muller, and F. Lingens. 1992. Purification and some properties of 2-halobenzoate 1,2-dioxygenase, a two component enzyme system from Pseudomonas cepacia 2CBS. J. Bacteriol. 174:279-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuenmayor, S. L., M. Wild, A. L. Boyes, and P. A. Williams. 1998. A gene cluster encoding steps in conversion of naphthalene to gentisate in Pseudomonas sp. strain U2. J. Bacteriol. 180:2522-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujioka, M., and H. Wada. 1968. The bacterial oxidation of indole. Biochim. Biophys. Acta 158:70-78. [DOI] [PubMed] [Google Scholar]
- 26.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haak, B., S. Fetzner, and F. Lingens. 1995. Cloning, nucleotide sequence, and expression of the plasmid-encoded genes for the two-component 2-halobenzoate 1,2-dioxygenase from Pseudomonas cepacia 2CBS. J. Bacteriol. 177:667-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haddad, S., D. M. Eby, and E. L. Neidle. 2001. Cloning and expression of the benzoate dioxygenase genes from Rhodococcus sp. strain 19070. Appl. Environ. Microbiol. 67:2507-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harayama, S., M. Rekik, A. Bairoch, E. L. Neidle, and L. N. Ornston. 1991. Potential DNA slippage structures acquired during evolutionary divergence of Acinetobacter calcoaceticus chromosomal benABC and Pseudomonas putida TOL pWW0 plasmid xylXYZ, genes encoding benzoate dioxygenases. J. Bacteriol. 173:7540-7548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayaishi, O., and R. Y. Stanier. 1951. The bacterial oxidation of tryptophan. III. Enzymatic activity of cell-free extracts from bacteria employing the aromatic pathway. J. Bacteriol. 62:691-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hickey, W. J., and G. Sabat. 2001. Integration of matrix-assisted laser desorption ionization-time of flight mass spectrometry and molecular cloning for the identification and functional characterization of mobile ortho-halobenzoate oxygenase genes in Pseudomonas aeruginosa strain JB2. Appl. Environ. Microbiol. 67:5648-5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hickey, W. J., G. Sabat, A. S. Yuroff, A. R. Arment, and J. Perez-Lesher. 2001. Cloning, nucleotide sequencing, and functional analysis of a novel, mobile cluster of biodegradation genes from Pseudomonas aeruginosa strain JB2. Appl. Environ. Microbiol. 67:4603-4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inouye, S., A. Nakazawa, and T. Nakazawa. 1986. Nucleotide sequence of the regulatory gene xylS on the Pseudomonas putida TOL plasmid and identification of the protein product. Gene 44:235-242. [DOI] [PubMed] [Google Scholar]
- 34.Kaldalu, N., U. Toots, V. de Lorenzo, and M. Ustav. 2000. Functional domains of the TOL plasmid transcription factor XylS. J. Bacteriol. 182:1118-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamath, A. V., and C. S. Vaidyanathan. 1990. New pathway for the biodegradation of indole in Aspergillus niger. Appl. Environ. Microbiol. 56:275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kauppi, B., K. Lee, E. Carredano, R. E. Parales, D. T. Gibson, H. Eklund, and S. Ramaswamy. 1998. Structure of an aromatic-ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase. Structure 6:571-586. [DOI] [PubMed] [Google Scholar]
- 37.Kim, E., and G. J. Zylstra. 1999. Functional analysis of genes involved in biphenyl, naphthalene, phenanthrene, and m-xylene degradation by Sphingomonas yanoikuyae B1. J. Ind. Microbiol. Biotechnol. 23:294-302. [DOI] [PubMed] [Google Scholar]
- 38.Kobayashi, S., and O. Hayaishi. 1970. Anthranilic acid conversion to catechol. Methods Enzymol. 17A:505-510. [Google Scholar]
- 39.Kobayashi, S., S. Kuno, N. Itada, and O. Hayaishi. 1964. O18 studies on anthranilate hydroxylase-a novel mechanism of double hydroxylation. Biochem. Biophys. Res. Commun. 16:556-561. [DOI] [PubMed] [Google Scholar]
- 40.Lennox, E. S. 1955. Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1:190-206. [DOI] [PubMed] [Google Scholar]
- 41.Madsen, E. L., and J. M. Bollag. 1989. Pathway of indole metabolism by a denitrifying microbial community. Arch. Microbiol. 151:71-76. [Google Scholar]
- 42.Mason, J. R., and R. Cammack. 1992. The electron-transport proteins of hydroxylating bacterial dioxygenases. Annu. Rev. Microbiol. 46:277-305. [DOI] [PubMed] [Google Scholar]
- 43.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 44.Miller, J. H. 1972. Experiment 48: assay of β-galactosidase, p. 352-355. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 45.Nagy, I., G. Schoofs, F. Compernolle, P. Proost, J. Vanderleyden, and R. de Mot. 1995. Degradation of the thiocarbamate herbicide EPTC (S-ethyl dipropylcarbamothioate) and biosafening by Rhodococcus sp. strain NI86/21 involve an inducible cytochrome P-450 system and aldehyde dehydrogenase. J. Bacteriol. 177:676-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neidle, E. L., M. K. Shapiro, and L. N. Ornston. 1987. Cloning and expression in Escherichia coli of Acinetobacter calcoaceticus genes for benzoate degradation. J. Bacteriol. 169:5496-5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nojiri, H., H. Sekiguchi, K. Maeda, M. Urata, S. Nakai, T. Yoshida, H. Habe, and T. Omori. 2001. Genetic characterization and evolutionary implications of a car gene cluster in the carbazole degrader Pseudomonas sp. strain CA10. J. Bacteriol. 183:3663-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 49.Powlowski, J. B., S. Dagley, V. Massey, and D. P. Ballou. 1987. Properties of anthranilate hydroxylase (deaminating), a flavoprotein from Trichosporon cutaneum. J. Biol. Chem. 262:69-74. [PubMed] [Google Scholar]
- 50.Ramanov, V., and R. P. Hausinger. 1994. Pseudomonas aeruginosa 142 uses a three-component ortho-halobenzoate 1,2-dioxygenase for metabolism of 2,4-dichloro- and 2-chlorobenzoate. J. Bacteriol. 176:3368-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Romine, M. F., L. C. Stillwell, K. K. Wong, S. J. Thurston, E. C. Sisk, C. Sensen, T. Gaasterland, J. K. Fredrickson, and J. D. Saffer. 1999. Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J. Bacteriol. 181:1585-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rothmel, R. K., D. L. Shinabarger, M. R. Parsek, T. L. Aldrich, and A. M. Chakrabarty. 1991. Functional analysis of the Pseudomonas putida regulatory protein CatR: transcriptional studies and determination of the catR DNA-binding site by hydroxyl-radical footprinting. J. Bacteriol. 173:4717-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 54.Selifonov, S. A., J. E. Gurst, and L. P. Wackett. 1995. Regioselective dioxygenation of ortho-trifluoromethylbenzoate by Pseudomonas aeruginosa 142: evidence for 1,2-dioxygenation as a mechanism in ortho-halobenzoate dehalogenation. Biochem. Biophys. Res. Commun. 213:759-767. [DOI] [PubMed] [Google Scholar]
- 55.Shepherd, J. M., and G. Lloyd-Jones. 1998. Novel carbazole degradation genes of Sphingomonas CB3: sequence analysis, transcription, and molecular ecology. Biochem. Biophys. Res. Commun. 247:129-135. [DOI] [PubMed] [Google Scholar]
- 56.Simon, M. J., T. D. Osslund, R. Saunders, B. D. Ensley, S. Suggs, A. Harcourt, W. C. Suen, D. L. Cruden, D. T. Gibson, and G. J. Zylstra. 1993. Sequences encoding naphthalene dioxygenase in Pseudomonas putida strain 67, NCIB 9816-4. Gene 127:31-37. [DOI] [PubMed] [Google Scholar]
- 57.Stanier, R. Y., N. J. Palleroni, and M. Duodoroff. 1966. The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol. 43:159-271. [DOI] [PubMed] [Google Scholar]
- 58.Subba Rao, P. V., K. Moore, and G. H. N. Towers. 1967. The conversion of tryptophan to 2,3-dihydroxybenzoic acid and catechol by Aspergillus niger. Biochem. Biophys. Res. Commun. 28:1008-1012. [PubMed] [Google Scholar]
- 59.Suen, W. C., B. E. Haigler, and J. C. Spain. 1996. 2,4-Dinitrotoluene dioxygenase from Burkholderia sp. strain DNT: similarity to naphthalene dioxygenase. J. Bacteriol. 178:4926-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suzuki, K., A. Ichimura, N. Ogawa, A. Hasebe, and K. Miyashita. 2002. Differential expression of two catechol 1,2-dioxygenases in Burkholderia sp. strain TH2. J. Bacteriol. 184:5714-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suzuki, K., N. Ogawa, and K. Miyashita. 2001. Expression of 2-halobenzoate dioxygenase genes (cbdSABC) involved in the degradation of benzoate and 2-halobenzoate in Burkholderia sp. TH2. Gene 262:137-145. [DOI] [PubMed] [Google Scholar]
- 62.Taniuchi, H., M. Hatanaka, S. Kuno, O. Hayaishi, M. Nakajima, and N. Kurihara. 1964. Enzymatic formation of catechol from anthranilic acid. J. Biol. Chem. 229:2204-2211. [PubMed] [Google Scholar]
- 63.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsoi, T. V., E. G. Plotnikova, J. R. Cole, W. F. Guerin, M. Bagdasarian, and J. M. Tiedje. 1999. Cloning, expression, and nucleotide sequence of the Pseudomonas aeruginosa 142 ohb genes coding for oxygenolytic ortho dehalogenation of halobenzoates. Appl. Environ. Microbiol. 65:2151-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamaguchi, M., and H. Fujisawa. 1980. Purification and characterization of an oxygenase component in benzoate 1,2-dioxygenase system from Pseudomonas arvilla C-1. J. Biol. Chem. 255:5058-5063. [PubMed] [Google Scholar]
- 66.Yanofsky, C. 1956. The enzymatic conversion of anthranilic acid to indole. J. Biol. Chem. 223:171-184. [PubMed] [Google Scholar]
- 67.Yen, K. M., and M. R. Karl. 1992. Identification of a new gene, tmoF, in the Pseudomonas mendocina KR1 gene cluster encoding toluene-4-monooxygenase. J. Bacteriol. 174:7253-7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou, N. Y., S. L. Fuenmayor, and P. A. Williams. 2001. nag Genes of Ralstonia (formerly Pseudomonas) sp. strain U2 encoding enzymes for gentisate catabolism. J. Bacteriol. 183:700-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zylstra, G. J., and E. Kim. 1997. Aromatic hydrocarbon degradation by Sphingomonas yanoikuyae B1. J. Ind. Microbiol. Biotechnol. 19:408-414. [DOI] [PubMed] [Google Scholar]