Abstract
We describe a novel filamentous phage, designated VGJφ, isolated from strain SG25-1 of Vibrio cholerae O139, which infects all O1 (classical and El Tor) and O139 strains tested. The sequence of the 7,542 nucleotides of the phage genome reveals that VGJφ has a distinctive region of 775 nucleotides and a conserved region with an overall genomic organization similar to that of previously characterized filamentous phages, such as CTXφ of V. cholerae and Ff phages of Escherichia coli. The conserved region carries 10 open reading frames (ORFs) coding for products homologous to previously reported peptides of other filamentous phages, and the distinctive region carries one ORF whose product is not homologous to any known peptide. VGJφ, like other filamentous phages, uses a type IV pilus to infect V. cholerae; in this case, the pilus is the mannose-sensitive hemagglutinin. VGJφ-infected V. cholerae overexpresses the product of one ORF of the phage (ORF112), which is similar to single-stranded DNA binding proteins of other filamentous phages. Once inside a cell, VGJφ is able to integrate its genome into the same chromosomal attB site as CTXφ, entering into a lysogenic state. Additionally, we found an attP structure in VGJφ, which is also conserved in several lysogenic filamentous phages from different bacterial hosts. Finally, since different filamentous phages seem to integrate into the bacterial dif locus by a general mechanism, we propose a model in which repeated integration events with different phages might have contributed to the evolution of the CTX chromosomal region in V. cholerae El Tor.
Vibrio cholerae, the causative agent of cholera, is a gram-negative bacterium that is able to colonize the human small intestine, where it secrets cholera toxin, its main virulence attribute (30). Several filamentous phages specific for V. cholerae, including CTXφ, VSK, fs1, fs2, and 493, have been described previously (23, 26, 28, 31, 56). The most well-studied of these phages is CTXφ, which plays a crucial role in the pathogenicity of V. cholerae, since it carries the genes that encode cholera toxin (56).
Filamentous phages, unlike most bacterial viruses that are assembled in the cell's cytoplasm and subsequently released by cell lysis, are unusual in that the infected host remains viable and phages are continuously assembled and extruded from the cell in a concerted process (50, 52). Their hosts are generally gram-negative bacteria, although a report of a filamentous phage infecting the gram-positive bacterium Propionibacterium freudenreichii was recently published (7). Variable numbers of progeny are produced, ranging from less than one phage particle per cell per hour in the case of CTXφ phage of V. cholerae to hundreds of particles per cell per hour for Ff phages (f1, M13, and fd) of Escherichia coli, without a serious effect on cell growth (40, 44, 56). These phages belong to the genus Inovirus of the family Inoviridae of bacteriophages, which are long slender proteinaceous tubes that range from 0.8 to 2 μm long and are about 6 nm in diameter (39). The particles each contain a single-stranded circular DNA genome that is converted to a double-stranded replicative form (RF) in infected cells, which can be recovered as a plasmid (40, 44).
Ff phages initiate the infection process by binding one end of the phage to the F pilus of E. coli carrying an F episome (40, 44). The pilus is specifically recognized by pIII, a minor capsid protein found at one end of the phage particle (1). It is thought that the phage is then brought to the bacterial surface by retraction of the pilus (27). Subsequent translocation of phage DNA into the cytoplasm requires the product of the tolQRA genes (8, 9, 38).
Generally, the genome of each filamentous phage is organized in a modular structure, in which functionally related genes are grouped (22). Three functional modules are generally present in the best-studied members of the inovirus group that have been characterized. The replication module contains the genes coding for rolling-circle replication protein and the single-stranded DNA (ssDNA) binding protein (44). The structural module contains the major and minor coat protein-encoding genes, and the assembly and secretion module contains the genes for morphogenesis and extrusion of the virus particles (39, 51, 52). Two genes are present in the assembly and secretion module of each of the phages belonging to the E. coli Ff group, gI and gIV. Gene gI encodes an inner membrane protein, pI, which recognizes the packaging signal in the ssDNA genome of the phage and starts the assembly process (51, 52). Gene gIV encodes protein IV, which forms an aqueous channel (secretin) in the outer membrane, through which phage particles exit from the host cells (51, 52). Interaction among pIV, pI (which also regulates channel opening), and the phage coat proteins is necessary for the simultaneous assembly and extrusion of the virions (51, 52). Some phages encode their own secretins for extrusion of phage particles, but others profit from host products; for example, CTXφ uses the EpsD secretin of the type II secretion system of V. cholerae (11). Additionally, some filamentous phages encode transcriptional repressors, like RstR of CTXφ and vpf122 of Vf33, which regulate the expression of other phage genes (5, 12, 33).
CTXφ, the best-studied V. cholerae-specific filamentous phage, is lysogenized in toxigenic V. cholerae strains, in which its prophage forms different arrays in the bacterial genome together with a related satellite phage, RS1 (12, 43). The CTXφ prophage is defective in classical strains but fully active in El Tor and O139 strains (13, 14), in which it can generate extrachromosomal CTXφ by a replicative process (45), giving rise to infective CTXφ particles. CTXφ uses toxin-coregulated pilus (TCP) as the primary receptor and the TolQ, -R, and -A proteins as coreceptors to inject its DNA into the host cells (20, 56). CTXφ exploits the XerC and XerD recombinases of its host to integrate into the bacterial genome (24). The integration process involves site-specific recombination between the attP site of CTXφ and the attB sequence inside the dif site of chromosome I of V. cholerae. The dif site is a 28-bp sequence at which XerCD recombinases resolve chromosome dimers (24). Other filamentous phages, such as f237 of Vibrio parahaemolyticus (25), Cf1c, Cf1t, Cf16v1, and φLf of Xanthomonas campestris, Xfφf1 of Xylella fastidiosa, and CUSφ-2 of Yersinia pestis, seem to use XerCD recombinases of their hosts to integrate into the dif locus of the bacterial genome, indicating that lysogenic filamentous phages are more common than initially thought (24).
The rest of the known V. cholerae-specific filamentous phages have been characterized less well. Phage 493 is known to use mannose-sensitive hemagglutinin (MSHA) as a receptor, and it has been suggested that this phage played some role in the emergence of the O139 epidemic serotype of V. cholerae (28, 29); however, its sequence and gene structure have not been described. Phage VSK is able to integrate into the V. cholerae chromosome (31), but the integration site has not been determined. To our knowledge, although the VSK nucleotide sequence has been deposited in international databases, the gene structure of this phage has not been discussed in any paper. Phages fs1 and fs2 have been sequenced, and their genomic structures have been described (23, 26). It has been suggested that these phages use MSHA as a receptor; however, this has not been fully demonstrated (16).
We found a novel filamentous phage, designated VGJφ, in V. cholerae O139 strain SG25-1, whose genome sequence is similar to the genome sequences of VSK and fs1. In this paper we describe a partial molecular characterization of the new phage, including its whole nucleotide sequence and genomic structure, as well as identification of the receptor that it uses to infect V. cholerae and its host range. The ability of the phage to integrate into the host chromosome and the site of integration are described. Additionally, we propose a model for the evolution of the dif locus in toxigenic El Tor strains.
MATERIALS AND METHODS
Strains, plasmids, and media.
The bacterial strains and plasmids used are described in Table 1. All strains were grown in Luria-Bertani medium (LB). Antibiotics were added, when necessary, at the following concentrations: ampicillin, 100 μg/ml; and kanamycin, 50 μg/ml.
TABLE 1.
Plasmids and primers as well as bacterial strains and their susceptibilities to VGJ-Knφ
| Strain, plasmid, or primer | Description | Source or referencea | Susceptibility to VGJ-Knφb |
|---|---|---|---|
| E. coli JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thiΔ(lac proAB) | Promega | NA |
| V. cholerae strains | |||
| 569B | O1, Classical, Inaba | Calcutta, India, 1945 | 1.6 × 108 |
| O395 | O1, Classical, Ogawa | Calcutta, India, 1964 | 8.0 × 107 |
| CA401 | O1, Classical, Inaba | Calcutta, India, 1953 | 9.8 × 107 |
| 1395 | O1, Classical, Inaba | CIEI | 7.6 × 107 |
| CA385 | O1, Classical, Ogawa | Calcutta, India, 1953 | 1.8 × 103 |
| NIH35A3 | O1, Classical, Inaba | India, 1941 | 4.8 × 104 |
| N16961 | O1, El Tor, Inaba | Bangladesh, 1975 | 7.7 × 106 |
| C7258 | O1, El Tor, Ogawa | Peru, 1991 | 1.9 × 107 |
| 638 | C7258 A(CTXφ prophage), hap::celA | 3 | NDc |
| C6706 | O1, El Tor, Inaba | Peru, 1991 | 1.3 × 108 |
| KHT46 | C6706 Δmsh41 | 54 | ND |
| KHT52 | C6706 ΔtcpA10 | 54 | 2.1 × 108 |
| 1333 | C6706 Δ(CTXφ prophage), hap::celA | CNIC | 3.8 × 107 |
| Peru-15 | C6709 Δ(CTXφ/RSI prophages), recA::ctxB | 32 | 5.4 × 107 |
| E7946 | O1, El Tor, Ogawa | Bahrain, 1978 | 9.2 × 106 |
| Gamene | O1, El Tor, Ogawa | Institute Pasteur | 5.0 × 106 |
| 3083 | O1, El Tor, Ogawa | Viet Nam, 1964 | 2.1 × 106 |
| Malawy | O1, El Tor, Inaba | Institute Pasteur | 4.1 × 107 |
| Lima | O1, El Tor, Inaba | Lima, Peru, 1991 | 8.3 × 106 |
| SG25-1 | O139 | Calcutta, India, 1993 | 2.4 × 103 |
| 1837 | O139 | Bangladesh, 1993 | 7.6 × 106 |
| CRC262 | O139 | Bangladesh, 2000 | 2.4 × 107 |
| CRC266 | O139 | Bangladesh, 2000 | 4.7 × 107 |
| MO45 | O139 | Madras, India, 1993 | 3.2 × 104 |
| MO12C | O139 | Madurai, India, 1993 | 7.2 × 104 |
| Plasmids | |||
| pUC19 | General purpose vector | 59 | NA |
| pJM132 | Plasmid expressing wild-type mshA from C6706 | 17 | NA |
| pKOR1 | R6K origin of replication plus a Knr cassette from pUC4K | This study | NA |
| Primers | |||
| VGJphi1 | 5′-CAAGTCGAGACGAACTAAGC-3′ | This study | NA |
| VGJphi2 | 5′-CCAAGTAATACGATAAACGC-3′ | This study | NA |
| VGJphi3 | 5′-CGGCATTTTGTGGGCATCAG-3′ | This study | NA |
| VGJphi4 | 5′-CGGCCTATGGTAAAAGTTGC-3′ | This study | NA |
| VGJphi5 | 5′-TCTCTGGTATCACCGATGCG-3′ | This study | NA |
| VGJphi6 | 5′-GCGCTTCCTCCCGTGTCCTC-3′ | This study | NA |
| VGJphi7 | 5′-GCTATGGCTAGAATCAAAGGG-3′ | This study | NA |
| VGJphi8 | 5′-AAGCTAAAGAGCAAGGCG-3′ | This study | NA |
| NJ1 | 5′-GGATGTTTACGATAGC-3′ | This study | NA |
| NJ2 | 5′-TAGAACGTGTCATTGCATCG-3′ | This study | NA |
| NJ4 | 5′-GCACACAATTGACGTAAGTAC-3′ | This study | NA |
CIEI, Centro de Investigacion de Enfermedades Infecciosas, Cuernavaca, Mexico; CNIC, Centro Nacional de Investigaciones Científicas, Havana, Cuba.
Number of Kanr transductants appearing after infection of ∼108 cells of the receptor strain with 1010 Kanr transducing particles (multiplicity of infection, 100). NA, not applicable.
ND, no Kanr transductants detected.
Isolation of phage VGJφ.
SG25-1 was grown in 5 ml of LB in a 25-ml Erlenmeyer flask for 6 h with shaking (240 rpm) at 37°C. One milliliter of this culture was serially filtered through 0.45-and 0.22-μm-pore-size filters (Sartorius). An aliquot (50 μl) of the filtered supernatant was plated on solid LB to check for sterility. Another aliquot (100 μl) was mixed with 20 μl of a culture of 569B (grown under the same conditions as SG25-1) and incubated for 20 min at room temperature. The mixture was then inoculated into 100 ml of LB in a 500-ml Erlenmeyer flask and grown at 37°C with shaking (240 rpm) overnight. The culture was centrifuged (8,000 × g, 20 min), and the supernatant was filtered through a 0.45-μm-pore-size filter. Phage particles in the filtrate were precipitated by addition of NaCl and polyethylene glycol to final concentrations of 3 and 5% (wt/vol), respectively. The mixture was incubated on ice for 30 min and centrifuged at 12,000 × g for 20 min. The supernatant was discarded, and the phage-containing pellet was resuspended in 1 ml of phosphate-buffered saline.
DNA isolation and manipulation.
V. cholerae total DNA was prepared as described by Ausubel et al. (2). Plasmid DNA was prepared by using the Wizard plus SV system (Promega), and genomic ssDNA from phage particles was prepared as described by Sambrook et al. (53). DNA restriction and modification enzymes were used according to the manufacturer's instructions (Promega). The DNA was electrophoresed on 0.8% (wt/vol) agarose gels and was visualized with ethidium bromide (1 μg/ml) or with acridine orange (1 μg/ml), which fluoresces green with double-stranded DNA and orange with ssDNA (42). Southern blot analysis was performed with the following digoxigenin-labeled probes: the whole ssDNA genome of VGJφ was used as the VGJφ-specific probe, and a 2.9-kb EcoRI-PstI fragment containing the RS1 element from plasmid pURS1 (4) was used as the RS1-specific probe. Sequencing reactions were performed with a Thermo Sequenase CyS dye terminator kit (Amersham Pharmacia Biotech), and the sequences were determined with an ALFexpress DNA sequencer obtained from Amersham Pharmacia Biotech. The whole sequence of the VGJφ genome was assembled by combining data obtained from sequencing of cloned fragments of the RF in the pUC19 vector with modified universal and reverse primers and from direct sequencing of the purified genomic ssDNA of the phage (plus strand) with the primers shown in Table 1.
Nucleotide and protein sequence analysis and alignment.
Sequences were assembled and analyzed by using ALFwin Sequence Analyzer (version 2.10; Amersham Pharmacia Biotech) and Vector NTI 6000 (InforMax, Inc.). The BLAST program was used to search for homologous DNA or protein sequences in databases. The ClustalW program was used for alignment analysis. Prediction of transmembrane regions and orientation were performed with the TMpred software (http://www.ch.embnet.org/software/TMPRED_form.html).
Electron microscopy.
Purified phage particles were negatively stained with 4% (wt/vol) uranyl acetate and mounted on freshly prepared Formvar grids, which were examined with a JEOL JEM-2000EX transmission electron microscope (JEOL, Tokyo, Japan). Several individual phage particles were measured, and the average size was calculated.
Infection assay.
VGJφ or VGJ-Knφ donor strains were grown in LB until the required optical density depending on the use. One milliliter of a culture was filtered through a 0.22-μm-pore-size filter (Sartorius). Aliquots (100 μl) of pure or serially diluted cell-free supernatant of the donor were mixed with 20 μl (∼108 cells) of a culture of the receptor strain grown in LB for 6 h at 37°C and 240 rpm. The mixture was incubated for 20 min to allow infection and then plated onto a soft LB agar (0.4%) overlay in the case of VGJφ or onto solid LB plates supplemented with kanamycin (LBK) in the case of VGJ-Knφ. The number of opaque plaques or Knr CFU was determined at 20 h. Alternatively, the infection mixture was seeded directly into LB broth if necessary.
Tagging VGJφ with a kanamycin resistance cassette.
The RF of VGJφ (pVGJφ) was linearized by using its unique XbaI site (Fig. 1A). Plasmid pKORI (1.6 kb), which contained the R6K replication origin and a Knr cassette from pUC4K, was also linearized with XbaI and inserted into the same site of pVGJφ to obtain pVGJ-Knφ. This insertion did not interrupt any of the identified open reading frames (ORFs) of VGJφ. pVGJ-Knφ was electroporated into V. cholerae 569B. The phage produced from pVGJ-Knφ RF was designated VGJ-Knφ.
FIG. 1.
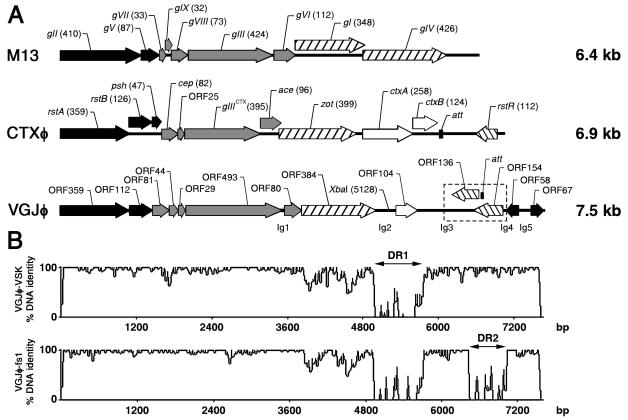
Genomic organization of VGJφ phage. (A) Linear ORF maps of M13, CTXφ, and VGJφ phages, aligned by using the first base of the replication initiator gene. ORFs or genes are represented by arrows oriented in the direction of transcription. The replication module is represented by solid arrows, the structural module is represented by gray arrows, and the assembly and secretion modules are represented by arrows with left cross-hatching. ORFs encoding putative repressors of VGJφ and rstR of CTXφ are represented by arrows with right cross-hatching. ORFs of VGJφ were designated according to the number of amino acids specified. M13 and CTXφ genes were designated by usual designations, and the lengths of the products encoded (in amino acids) are indicated in parentheses. Intergenic regions of VGJφ (Ig) and the region containing the att sequences are also indicated. The XbaI site where a Knr cassette is inserted in VGJ-Knφ is indicated. The region enclosed in a box (dashed lines) is a region that is magnified in Fig. 2B. Most of the gene function assignments for VGJφ are still putative. (B) Similarity of VGJφ DNA with VSK and fs1 phage DNAs. The distinctive regions in VGJφ which are absent in VSK and/or fs1 are indicated (DR1 and DR2). Levels of identity were determined by moving a 15-bp window along the alignment in 5-bp increments.
Integration of VGJ-Knφ.
Attenuated strain 1333 and toxigenic strain N16961 were infected with VGJ-Knφ. Two groups of 15 BALB/c suckling mice were orogastrically inoculated with 105 cells of each infected strain diluted in 50 μl of phosphate-buffered saline. The mice were not fed for 4 h before and after inoculation and then were put back with their mothers. Two mice from each group were sacrificed on day 7 postinoculation, and the small intestines were removed, homogenized, and plated on solid LBK to recover colonizing vibrios that retained the Knr phenotype. Twelve Knr CFU were used to assess integration of VGJ-Knφ into each strain. Analysis of the integration into a predicted site was carried out by Southern blotting, and the results were confirmed by PCR performed with primers that hybridized on both sides of the novel junctions between the phage and the bacterial DNA. Primers NJ1 and NJ2 (Table 1) were used to amplify the DNA region containing the left novel junction, while primers VGJphi3 and NJ4 (Table 1) were used to amplify the DNA region containing the right novel junction. The amplified products were sequenced with the same primers.
Protein methods.
Preparations of total cell or phage proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) or Tris-Tricine SDS-PAGE as necessary (2). Identification of proteins by mass spectrometry was carried out with a QTOF-2 spectrometer (Micromass, Manchester, United Kingdom). The mass spectra were processed and analyzed with MassLinx v 3.5 and MaxEntropy v 3.0 software (Micromass). The proteins were identified by using ProFound v 3.2 (http://prowl.rockefeller.edu/cgi-bin/ProFound), Pepsea (MDS Proteomics), and MASCOT (Matrix Science Ltd.) software.
Nucleotide sequence accession numbers.
The nucleotide sequences of VGJφ and the novel junctions of the VGJφ prophage have been deposited in the GenBank database under accession numbers AY242528, AY242529, and AY242530.
RESULTS
Purification and characterization of VGJφ.
We previously observed the presence of an extrachromosomal genetic element in total DNA preparations from V. cholerae SG25-1, an O139 strain from India. This element could be transmitted to the indicator strain 569B by contact with cell-free culture supernatants of SG25-1 (data not shown). Transduction of the extrachromosomal element without direct cell-cell contact indicated that this episome corresponded to the RF of a bacteriophage. A restriction analysis of this RF revealed a restriction map (data not shown) and genome size different from those of all previously described phages of V. cholerae. Therefore, we designated the new phage VGJφ.
We purified VGJφ phage particles from culture supernatants of infected 569B; this strain was used because it contains a defective CTXφ prophage, which eliminated the possibility of contamination with CTXφ particles. The nucleic acid extracted from purified particles showed resistance to treatment with RNases and with all restriction enzymes tested but sensitivity to treatment with mung bean and S1 nucleases (data not shown), and it fluoresced orange when it was electrophoresed in the presence of acridine orange (data not shown). These results indicated that the nucleic acid extracted from phage particles was ssDNA. A VGJφ-specific probe (see Materials and Methods) hybridized with the RF purified from infected cells, as well as with the ssDNA, indicating identity between the viral genome and the RF (data not shown). The single-stranded nature of the phage genomic DNA suggested that the phage was a filamentous bacteriophage. Electron microscopy preparations of the isolated phage confirmed the presence of filamentous structures that were approximately 7 nm wide and 1,000 nm long that were not present in uninfected control cells (data not shown).
Since both SG25-1 and 569B are toxigenic strains containing CTXφ prophages with active genes, we tested Peru-15 (32), which lacks all CTXφ genes, to rule out the possibility of any contribution of CTXφ genes to VGJφ morphogenesis. Actually, Peru-15 was efficiently infected by VGJφ and normally produced phage particles (>1010 particles/ml), indicating that VGJφ is an autonomous phage.
Nucleotide sequence and genomic organization.
The nucleotide sequence of VGJφ consisted of 7,542 nucleotides and had a G+C content of 43.4%, which is slightly different from the G+C content of the host, V. cholerae (47.7% for chromosome I and 46.9% for chromosome II). The sequence deposited in the GenBank database corresponded to the plus DNA strand of the phage, the same strand that is packaged in the phage capsid. This was confirmed because only oligonucleotides hybridizing with this DNA strand exhibited sequence reactions when phage genomic ssDNA was used as the template.
A computer-aided homology search was performed for the ORFs presumed to encode functional viral genes in VGJφ. The ORFs were designated according to the numbers of amino acids specified by them (Fig. 1A). Of the 13 ORFs identified in VGJφ, 10 were homologous to ORFs encoding previously reported peptides of other filamentous phages. These ORFs had a modular genomic organization (22) similar to that of previously characterized filamentous phages, such as phages of the Ff group of E. coli and phage CTXφ of V. cholerae (40, 44, 56), in which genes with similar functions are grouped (Fig. 1A).
Within the putative replication module (Fig. 1A) we identified ORF359 and ORF112. The peptide encoded by ORF359 is homologous to the RstA protein of CTXφ (40% identity over 264 residues), which is necessary for phage replication (57). ORF359 is also similar in terms of size and position to genes of previously reported filamentous phages, which mapped at the same relative position as the gII gene in Ff phages (Fig. 1A). This gene encodes the pII protein, which is necessary for rolling-circle replication of phage genomes (44). Like RstA, ORF359 of VGJφ could be a homologue of pII of Ff phages. The deduced amino acid sequence encoded by ORF112 was homologous to peptides that mapped at the same position as the ssDNA binding protein of Ff phages, and its size was similar to the size of this binding protein (Fig. 1A).
Within the putative structural module of VGJφ we identified ORF81, ORF44, ORF29, ORF493, and ORF80, whose sizes and positions are similar to those of genes encoding the capsid structural proteins of Ff phages and other previously described filamentous phages. These ORFs are predicted to encode proteins with hydrophobic transmembrane domains whose N termini are oriented to the outside of the cytoplasmic membrane, which is reminiscent of coat proteins of Ff phages (39). These results support the hypothesis that ORF81, ORF44, ORF29, ORF493, and ORF80 could form the module of structural genes (Fig. 1A), which encode the capsid proteins of VGJφ. ORF493 is located at the same relative position as gIII of the Ff phages and gIIICTX of CTXφ, and its size is comparable to the sizes of these genes, which encode pIII, a minor capsid protein that recognizes and interacts with the receptors and coreceptors of these phages (1, 21, 38) (Fig. 1A). Therefore, ORF493 could be a homologue of gIII in VGJφ.
Within a third putative module of VGJφ, the assembly module, we found ORF384, whose product is homologous to the pI protein of φLF phage of X. campestris (31% identity over 164 residues) (6) and to the Zot protein of CTXφ (25% identity over 247 residues) (56). Based on its size and position, ORF384 of VGJφ is also homologous to the gI gene, which encodes pI in Ff phages and is needed for assembly and secretion of the viral particles (39, 51, 52). Thus, it is probable that the product of ORF384 plays the same role in VGJφ. Since ORF384 showed homology with the Zot protein of CTXφ, which is necessary for morphogenesis and also functions as a toxin (18, 56), we performed a more detailed analysis of this peptide. The N-terminal region of the peptide encoded by ORF384 (from residue 7 to residue 233) exhibited homology with the N-terminal region of Zot (from residue 8 to residue 242). The enterotoxic function of Zot has been assigned to its C-terminal region (15). Since in the rest of the sequence the peptide encoded by ORF384 is not homologous to Zot and the C-terminal region of Zot in turn is not homologous in the C-terminal regions of other known or putative pI proteins of other phages (34), the enterotoxic activity seems to be an accidental and specific function of Zot. Thus, a Zot-like enterotoxic activity of the ORF384 product, as has been suggested for the homologous protein in fs1 phage (16), is unlikely.
In a fourth putative module, the regulator module, we identified ORF136 and ORF154. The peptides encoded by these ORFs exhibited homology with several putative transcriptional regulators of phages. Also, the peptide encoded by ORF154 showed homology with putative repressors encoded chromosomally, like a putative repressor of Rhodococcus equi, and to a LacI family transcriptional regulator of Thermotoga maritima. ORF136 and ORF154 are transcribed in the direction opposite the direction of transcription of most of the ORFs of VGJφ (Fig. 1A). In CTXφ, rstR is also transcribed in the direction opposite the direction of transcription of the rest of the phage genes; this gene is known to encode a repressor that regulates transcription of the initiator replication protein, RstA, and also seems to regulate the expression of all phage genes (12, 33). Probably, the ORF136 and ORF154 products play a similar role in regulating the expression of VGJφ genes.
The following three additional ORFs could not be clearly included in any of the four putative modules described above: ORF67, ORF58, and ORF104. ORF67 is conserved only in fs1 phage, and the ORF58-encoded peptide is not related to any of the previously reported products of filamentous phages but has a region that is similar to a region of the E1 protein of human papillomavirus type 60, an ATP-dependent DNA helicase required for initiation of viral DNA replication (41). Finally, the product of ORF104 did not show homology with any known peptide. This ORF is located in a distinctive region of the VGJφ genome which contains 775 nucleotides (from base 4908 to base 5683) not homologous to any entry in the databases (DR1 in Fig. 1B).
The nucleotide sequence revealed that VGJφ is a close relative of the filamentous phages VSK and fs1 of V. cholerae, exhibiting 82.8 and 77.8% of DNA homology, respectively, while VSK and fs1 exhibit 87.4% DNA homology (Fig. 1B). The differences in the DNA sequences of these phages are mainly concentrated in the distinctive DR1 region and/or DR2 region of the VGJφ genome (Fig. 1B). The DR1 region is absent in the VSK and fs1 phages, and the DR2 region is absent only in the fs1 phage. Additionally, there is an important accumulation of nucleotide changes in ORF384 (Fig. 1B). These results indicated that VGJφ, VSK, and fs1 probably evolved from a common ancestor.
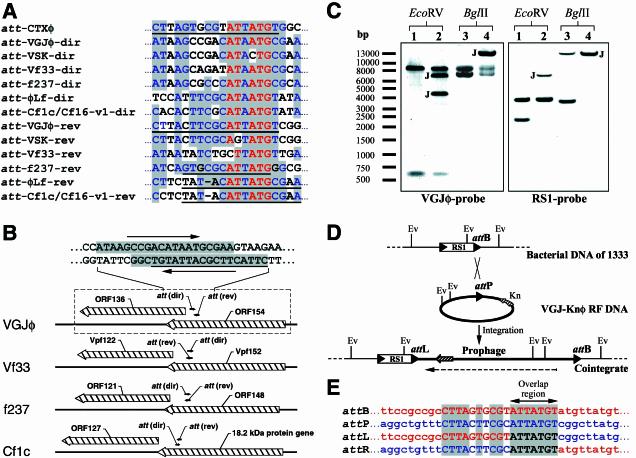
The DNA sequence of VGJφ also revealed the presence of two sites homologous to att sequences known to function in integrative filamentous phages, such as CTXφ of V. cholerae, f237 of V. parahaemolyticus, and the Cf1c, Cf16-v1, and φLf phages of X. campestris (10, 24, 25, 36, 37). The att-like sites in VGJφ were found to overlap in opposite directions (Fig. 2A and B) and to map to similar relative positions with respect to the att sites of all these phages (Fig. 2B). The att site in the viral plus strand was designated att-VGJφ-dir, and the opposite site was designated att-VGJφ-rev; both sites mapped inside ORF154 (Fig. 2B). By analogy with VGJφ, we also found att-like sites in the integrative phages Vf33 of V. parahaemolyticus (5) and VSK of V. cholerae (31); these sites are arranged in similar structures (Fig. 2A, B) that are absent in the fs1 and CTXφ phages.
FIG. 2.
att site-containing region of VGJφ and site-specific integration of VGJ-Knφ into chromosome I of strain 1333. (A) Sequence alignment of the att regions of the CTXφ (accession number AF220606), VGJφ (AY242528), and VSK (NC_003327) phages of V. cholerae, the Vf33 (AB017573) and f237 (NC_002362) phages of V. parahaemolyticus, and the Cf1c (NC_001396), Cf16-v1 (M23621), and φLf (X70328) phages of X. campestris. Levels of identity greater than 90% are indicated by red letters with a gray background, and levels of identity greater than 45% are indicated by blue letters with a gray background. The underlined sequences indicate known functional att sites of the phages. (B) Genomic region containing the att-like sites shown in panel A. In all cases att sites map inside VGJφ ORF154 homologues, near VGJφ ORF136 homologues, which partially overlap ORF154 homologues. The region enclosed in a box (dashed lines) is the region enclosed in a box in Fig. 1A. (C) Southern blot showing the cointegrate structure in 1333/VGJK2, a representative clone. The left panel was probed with the VGJφ-specific probe (lanes 1 and 3, purified RF DNA of VGJ-Knφ; lanes 2 and 4, total DNA of the cointegrate clone). The right panel was hybridized with the RS1-specific probe (lanes 1 and 3, total DNA of strain 1333; lanes 2 and 4, total DNA of the cointegrate clone). DNA samples were treated with the enzymes indicated at the top. Bands containing the novel junctions are indicated by J. Note that the 3.5-kb BglII fragment of 1333 (right panel, lane 3) was about 9.1 kb larger in 1333/VGJK2 (double band in lane 4 of the right panel), indicating that only one copy of VGJ-Knφ was integrated. (D) Schematic representation of the recombination between the VGJ-Knφ RF (thick line) and the bacterial chromosome of 1333 (thin line). The RS1 element is represented by an open box flanked by the nearly identical end repeats (solid arrowheads). The downstream end repeat serves as an attB site for integration of VGJ-Knφ. attP (att-VGJφ-rev) and the novel junction attL and attR sites are also represented by solid arrowheads. Ev, EcoRV sites involved in the analysis of the Southern blot (panel A). The Knr gene (Kn) is represented by a cross-hatched arrow. The dashed arrow indicates the direction of the plus strand of VGJφ. (E) DNA region with attP, attB, attL, and attR sites as determined by sequencing. The core homologous sequences are indicated by uppercase letters, and identical nucleotides are indicated by a gray background. DNA sequences of bacterial origin and DNA sequences of phage origin are indicated by red and blue letters, respectively; black letters indicate sequences of unknown origin at the moment of recombination. The arrow indicates the putative overlap region, where the cutting and rejoining of the recombination partners took place.
Titration of VGJφ and VGJ-Knφ.
VGJφ is able to form opaque plaques in soft LB agar overlays seeded with a sensitive strain, which allows phage titration. SG25-1, from which VGJφ was initially isolated, produced maximum titers of 109 PFU/ml in the medium; however, 569B infected with VGJφ could produce a titer of about 3 × 1011 PFU/ml in the medium after 6 h of infection (average of three independent experiments). VGJ-Knφ was easier to titrate by counting the number of Knr CFU of the infected strain, which in theory was equal to or lower than the number of Knr transducing phage particles. 569B infected with VGJ-Knφ was able to produce about 1.8 × 1011 Knr transducing particles/ml in the medium after 6 h of infection (average of three independent experiments). Therefore, insertion of the Knr cassette did not seriously impair the replication and morphogenesis of the phage, showing that the XbaI site is permissive for cloning of foreign DNA fragments.
Integration of VGJφ.
Total DNA of SG25-1 analyzed by Southern blotting with the VGJφ-specific probe produced additional bands not expected from the RF of VGJφ (data not shown), suggesting that this strain contained both replicating and integrated phage DNA. This finding and the presence of two potential att sites in the VGJφ genome prompted us to examine the possibility of phage integration into the host chromosome. We used the Knr-tagged version of the phage, VGJ-Knφ, and the indicator strain 1333 to study the integration. Strain 1333 is an attenuated strain constructed in our lab for vaccine purposes, and it lacks the entire CTXφ prophage but has a copy of the satellite phage RS1 at the same chromosomal site (Fig. 2D). This strain was infected with VGJ-Knφ and then orogastrically inoculated into BALB/c suckling mice; after 7 days, Knr colonizing vibrios were recovered. Twelve independent Knr clones were analyzed by Southern blotting, which showed that a single copy of VGJ-Knφ DNA was integrated into the bacterial chromosome and was also present as a RF. The same banding pattern was obtained for the 12 clones, suggesting that the integration took place by site-specific recombination. The results of the Southern blot analysis of one representative Knr clone (1333/VGJK2) are shown in Fig. 2C. The analysis also revealed that the recombination event took place downstream of the EcoRV site of the RS1 element, probably by the flanking 3′ end repeat (attRS1) (Fig. 2C and D). This result was confirmed by PCR and sequencing of the novel junctions between the phage and the bacterial genomic DNA (Fig. 2E).
The same procedure was done with strain N16961, which contained one copy of the CTXφ prophage, followed by an RS1 element (19). Analysis by Southern blotting of 12 Knr clones produced results similar to those obtained with 1333 (data not shown). The structure of one Knr representative clone, N16/VGJK5, was also determined by sequencing the novel junctions. As occurred in 1333, VGJ-Knφ integrated at the flanking attRS1 site downstream of the RS1 element (data not shown). Interestingly, the sequences of the novel junctions revealed that att-VGJφ-rev (Fig. 2A), which was oriented in the same direction as the minus strand of the phage RF, was the site used for the recombination event. Therefore, VGJφ integrated in the orientation opposite that of RS1 in 1333 (Fig. 2D) and the CTXφ prophage in N16961. The att-VGJφ-rev site differed at only three bases from the unique attP site of the CTXφ phage (Fig. 2A), which was identical to the downstream end repeat of the RS1 (attRS1) (Fig. 2E).
We analyzed whether strains with integrated phage DNA still secreted phage particles. This could not be assessed in the strains described above, which contained VGJφ that was integrated as well as replicating. We obtained the integrated state alone (i.e., without the RF) in strain KHT46, in which VGJφ transmission was blocked by a lack of the phage receptor (see below). Thus, KHT46 electroporated with DNA of the VGJ-Knφ RF and passed by the mouse gastrointestinal tract contained solitary VGJ-Knφ prophages integrated at the attRS1 site (data not shown). These lysogens did not produce phage particles, in contrast to KHT46 containing replicating VGJ-Knφ, which normally produced virions (1010 particles/ml). Studies are in progress to assess if lysogens containing tandem VGJφ prophages are able to secrete the phage, as occurs with CTXφ.
Partial protein profile of VGJφ phage and VGJφ-infected V. cholerae.
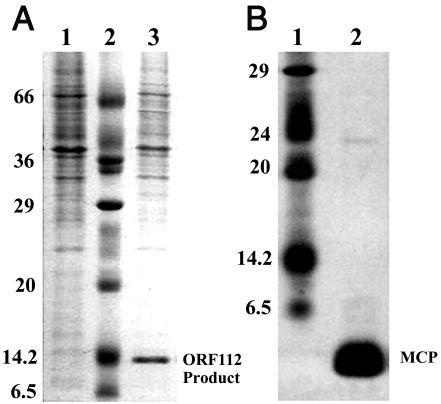
SDS-PAGE analysis of whole-cell lysates of 569B infected with VGJφ resulted in a strong band at an apparent molecular mass of about 14 kDa not observed when uninfected 569B cells were examined (Fig. 3A). A mass spectrometry analysis of this band revealed that it corresponded to the peptide encoded by ORF112 (Fig. 1A). The analysis also revealed that the mature peptide lacked the initial methionine and had a molecular mass of 12.72 kDa. This protein was fully conserved in the fs1 phage and was homologous to the products of ORFs Vpf117 and Vpf119 of the Vf33 and f237 phages of V. parahaemolyticus, respectively (5, 47). All these homologous ORFs are located at the same relative position within their genomes. The same position is occupied by the gV gene, which encodes the ssDNA binding protein (pV) in several filamentous phages (40, 44) (Fig. 1A). Thus, we anticipate that ORF112 could encode the ssDNA binding protein, pV, of VGJφ.
FIG. 3.
Partial protein profile of VGJφ as determined by SDS-PAGE. (A) Protein encoded by ORF112. Lane 1, total cell proteins of uninfected 569B; lane 2, molecular weight marker; lane 3, total cell proteins of 569B infected with VGJφ, in which the product of ORF112 could be observed. The quantity of cells applied in lane 1 was twice the quantity of cells applied in lane 3. (B) Major capsid protein (MCP) of VGJφ. Lane 1, molecular weight marker; lane 2, purified phage particles. The numbers on the left of each panel represent molecular masses in kilodaltons of markers.
The protein profile of purified particles of VGJφ showed that there was a major protein band at an apparent molecular mass of about 2.5 kDa as determined by conventional SDS-PAGE (Fig. 3B) and at about 3.5 kDa as determined by Tris-Tricine SDS-PAGE (data not shown), which provides better estimates of the molecular masses of small proteins. This protein is predicted to correspond to the product encoded by ORF44 of the structural module, which exhibited homology with major coat proteins of previously described filamentous phages. A sequence comparison with some known major coat proteins revealed that the ORF44-encoded peptide conserves the central hydrophobic domain, which has been implicated in membrane anchoring, surrounded by charged N and C termini (data not shown) (39). Like the major coat protein of the Pf3 and C2 phages (35, 39), no leader peptide seems to be present in the ORF44 product.
Identification of VGJφ receptor.
Filamentous phages generally use type IV pili as receptors to infect their hosts. Previously described V. cholerae-specific filamentous phages use TCP or MSHA pili as receptors (29, 56). Therefore, two mutants of El Tor strain C6706 with mutations for these pili, KHT52 (ΔtcpA10) and KHT46 (ΔmshA), were used to determine if any of the pili was the receptor of VGJφ. While parental strain C6706 and its TCP mutant KHT52 were sensitive to infection with VGJφ, the MSHA mutant KHT46 was completely resistant to the phage, indicating that MSHA was the receptor of VGJφ (data not shown). Complementation of KHT46 with the wild-type mshA structural gene (from parental strain C6706) carried on plasmid pJM132 restored phage sensitivity (data not shown), confirming that MSHA is the receptor for VGJφ. Besides, vaccine strain 638 is VGJφ resistant. This strain is known to express the MshA subunit but not to hemagglutinate chicken erythrocytes, indicating that some defect is present in it that impairs correct assembling of the MSHA pilus on the bacterial surface (3, 17, 55). Furthermore, no Knr CFU were recovered after infection of 108 cells of KHT46 with 1010 phage particles of VGJ-Knφ (Table 1), while more than 108 Knr CFU were obtained after infection of C6706 or KHT52 with the same phage preparation (Table 1). These data suggest that infection with VGJφ is very effective and depends on the MSHA pilus.
Host range.
We used the Knr version of the phage, VGJ-Knφ, to assess the susceptibility of each strain. All V. cholerae O1 and O139 strains tested in this study were sensitive to VGJ-Knφ, but the level of susceptibility was highly variable (Table 1). Classical strains CA385 and NIH35A3, as well as O139 strains MO45 and MDO12C and SG25-1, were much less susceptible than the rest of the strains tested (Table 1). We analyzed the causes for the relative resistance of these strains. In the case of MO45 and MDO12C we noticed the presence of one episome, which showed the same restriction map for both of these strains but a map that was clearly distinct from the VGJφ RF map (data not shown). However, the episomes cross-hybridized with a VGJφ-specific probe, suggesting that a phage related to VGJφ was present in these strains (data not shown). Furthermore, ssDNA could be recovered from cell-free supernatants of these strains, which also cross-hybridized with the same probe, supporting the hypothesis that a related filamentous phage is present in MO45 and MDO12C. Thus, the relative resistance of these strains to the VGJφ phage could be related to the presence of this related phage, which we called VEJφ. Maybe this phage, which was already established in the strains, interfered with the establishment of VGJφ. The same phenomenon seemed to occur with SG25-1, which already contained VGJφ. In the case of the classical strains CA385 and NIH35A3 the resistance seemed to be related to a defect in MSHA pilus assemblage, because although these strains expressed normal levels of an MshA subunit, as judged by Western blotting, they did not hemagglutinate chicken erythrocytes (data not shown), suggesting that correct assemblage of MSHA was impaired in these strains.
DISCUSSION
In this study we characterized VGJφ, a novel V. cholerae-specific filamentous phage related to (but clearly distinct from) the previously described phages fs1 and VSK (23, 31). VGJφ may also be related to phage 493, because 493 DNA cross-hybridizes with VSK DNA (28); unfortunately, the nucleotide sequence of phage 493 has not been published, so we could not examine this possibility in detail. Although VGJφ infects the same host that the CTXφ and fs2 filamentous phages infect, it seems to be more closely related evolutionarily to phage Vf33 of V. parahaemolyticus, because peptides encoded by VGJφ exhibited more extensive identity over longer stretches with proteins of Vf33 than with proteins of CTXφ and fs2 (data not shown).
It is proposed that VGJφ has a modular genome organization like that of previously described filamentous phages belonging to the Ff group of E. coli phages and like that of CTXφ of V. cholerae. The following three usual modules of filamentous phages are distinguishable: the replication module, the structural module, and the assembly and secretion module (Fig. 1A). Also, an additional regulatory module is distinguished in VGJφ, in which putative transcriptional repressor genes are grouped. A regulatory module like this seems to be present in other lysogenic filamentous phages, such as CTXφ of V. cholerae, Vf33 and f237 of V. parahaemolyticus, and Cf1c, Cf16-v1, and φLf of X. campestris, but to be absent from nonlysogenic phages (Fig. 1A and 2B). It would be interesting to study whether the presence of a regulatory module is correlated with the lysogenic phenotype.
The proposed assembly and secretion module of VGJφ does not have a homologue of the gIV gene of the Ff phages. Since the gIV gene is critical for phage morphogenesis of filamentous phages (51, 52), VGJφ probably uses a cell substitute like another phage of V. cholerae, the filamentous phage CTXφ, which also lacks a gIV homologue and uses the host protein EpsD for phage morphogenesis. EpsD is a secretin of the type II secretion system of V. cholerae that has also been shown to be a bacterial homologue of protein IV (11).
VGJφ also has a distinctive region of 775 nucleotides that is not significantly homologous to any entry in the databases. Therefore, the origin of this DNA region is unknown, but a plausible hypothesis is that VGJφ moved horizontally among different bacterial species, integrating and excising from the chromosomes of different hosts, where it picked up a fragment that has not been sequenced yet. The role (if any) of ORF104, which is located inside this region, is unknown.
The product of ORF112 of the putative replication module is expressed at a high level in VGJφ-infected V. cholerae. This high level of expression is compatible with the ssDNA binding activity proposed for this protein. Since the titer of VGJφ is very high, large quantities of the ssDNA binding protein are needed to maintain the phage genome as ssDNA until packaging. Although positional homologues of ORF112 in other filamentous phages have ssDNA binding activities, this ORF maps to the same relative position as the rstB gene of CTXφ, and its size is similar to the size of this rstB gene, whose product (RstB) has been suggested to play a role in CTXφ integration (57), which is different from the function proposed for ORF112. Since the ORF112 product and RstB do not have a significant level of homology, one possible explanation is that they encode different functions. However, since positional homologues usually code for equivalent functions, ORF112 and RstB could have dual functions, but the following two findings argue against integrative functions of RstB: (i) RstB exhibits no significant similarity with any known recombinase (57), and (ii) CTXφ DNA integration is dependent on the host recombinases XerCD (24), so the role of RstB in CTXφ integration is not clear. Thus, studies are in progress in our lab to clearly determine if the products of ORF112 of VGJφ and rstB of CTXφ function as ssDNA binding proteins.
VGJφ is able to integrate, via its own attP sequence, into the same chromosomal attB site where CTXφ integrates its DNA. Since the att structure of VGJφ is also present in the Cf1c, Cf16-v1, φLf, Vf33, f237, and VSK phages (Fig. 2A and B), the structure of this region may be important for recognition of the recombination partners at att sites by the recombinases that mediate the site-specific recombination process. As VGJφ does not encode any product homologous to known recombinases and given the similarity of the functional attP site of VGJφ to the attP site of CTXφ, it is highly probable that the integration process of VGJφ (and the rest of the phages that have a similar att structure) depends on XerCD recombinases of their hosts, as occurs with CTXφ (24). Support for this hypothesis comes from the fact that both CTXφ and VGJφ integrate at the same site of chromosome I of V. cholerae.
The action of XerCD in the integration process implies that there is cutting and rejoining of both DNA strands in a precise region of the recombination partners, with the resulting formation of two novel junctions. Alignment of the attP site of VGJφ and the novel junctions of the prophage allowed identification of the region where the cutting and rejoining take place. This was possible because of minor differences between the attP and att B sites; accordingly, the cutting and rejoining happen at the 7-mer DNA sequence (ATTATGT) shown in Fig. 2E. We do not know the exact position of the cutting and rejoining, but usually recombinases of the integrase family, to which XerCD belong, produce a staggered cut 6 to 8 bp apart in the overlap region of a functional core sequence (46). If XerCD recombinases mediate integration of VGJφ, the 7-bp region (ATTATGT) contains the overlap region where a similar cut occurs.
The attP site of VGJφ is inside ORF154, a putative transcriptional repressor of the regulatory module. Similarly, in several other filamentous phages the position of the attP site is conserved inside VGJφ ORF154 homologues (Fig. 2B), which in turn have a conserved genomic position. The presence of the att region inside a putative repressor gene may exert important regulatory effects, because in the lysogenic state this gene or its homologues would be interrupted. VGJφ was found to use att-VGJ-rev as the functional attP (Fig. 2A and E). This attP site is in the minus viral strand, as are functional attP sites of phages Cf1c, Cf16-v1, and φLf (10, 36, 37). However we cannot rule out the possibility that att-VGJ-dir is used for integration, since CTXφ uses an attP site oriented in the same direction as att-VGJ-dir. Also, the use of att-VGJ-dir may allow integration into a different chromosomal site of the same host or another host.
It is known that the VGJφ-related phages VSK and Vf33 are able to integrate into the genomes of their hosts, but the precise sequence with which they integrate has not been determined yet (5, 31). By analogy with VGJφ, we found potential att sites in the genomes of these phages (Fig. 2A), suggesting that they use these equivalent sites for integration. This conclusion awaits experimental confirmation; however, in the case of Vf33, we found that the att sites map in the same EcoRI fragment through which this phage has been reported to integrate into the genome of V. parahaemolyticus (5). Therefore, we anticipate that like VGJφ, Vf33 and VSK may use one of the two potential att sites for the integration process (probably att-Vf33-rev and att-VSK-rev, respectively) (Fig. 2A).
Like CTXφ, VGJφ integrates preferentially into the right junction (attB site) of previously resident CTXφ or RS1 prophages (Fig. 2D) (13) and not into the left junction, where there is a practically identical att site. This is compatible with our finding that VGJφ may use the XerCD recombinases for chromosomal integration, since only the attB site at the right junction is a substrate for this recombination system. The fact that the left junction is not a substrate for this system precludes integration of VJGφ into the left att site. The attB site at the right junction overlaps with the dif site of chromosome I of V. cholerae, which is also preserved after any new integration event with either of these phages. The dif site is a 28-bp sequence at which XerCD recombinases resolve chromosome dimers. Since strains containing an integrated copy of VGJφ have normal morphology and growth (data not shown), the integration of this phage does not seem to interfere with XerCD-dependent resolution of chromosomal dimers.
We did not detect integration of VGJφ in vitro (data not shown); however, we easily detected phage integration in vivo after passage of the infected bacterium through the intestines of suckling mice. These results suggest that the intestinal conditions might induce an increase in the recombination activity involved in phage integration.
We found that MSHA is the receptor for VGJφ, as it is for phage 493 (29). MSHA is a ubiquitous pilus found on the bacterial surface of many V. cholerae strains. MSHA seems to be expressed in a wide spectrum of culture conditions, including laboratory media and natural conditions. Since MSHA has also been implicated in biofilm formation and adherence to planktonic surfaces (58), our interest in VGJφ was substantially increased because this phage can mediate horizontal transfer of genes in the environment with great potential. Additionally, because of the putative role of MSHA in environmental survival, we consider it essential for naturally occurring V. cholerae. In this study we found that four of six classical strains are highly sensitive to VGJφ, suggesting that they assemble MSHA pili normally at the bacterial surface. This made us reconsider some findings concerning the lack of MSHA on the bacterial surface of classical strains of V. cholerae. One of the VGJφ derivatives constructed in this work, VGJ-Knφ, will be used as a more sensitive tool to measure the presence of MSHA pili in several strains of V. cholerae.
MSHA as a receptor must have a counterpart in the phage for direct interaction. In filamentous phages this role is played by the adsorption protein pIII, a structural minor capsid protein encoded by gene gIII (1, 21, 38). Our results suggest that ORF493 in VGJφ could be the homologue of the gIII gene of other known filamentous phages, like Ff, Ike, and I2-2 of E. coli and CTXφ of V. cholerae (1, 21, 38). Protein pIII is highly divergent in filamentous phages, which is consistent with the existence of different receptors for each of the phages. However, the sequence of the product encoded by ORF493 is homologous to the protein sequences encoded by ORF257 and ORF193 of fs1 phage, ORF208 and ORF302 of VSK, ORF498 of the fs2 phages of V. cholerae, and ORF491of the Vf33 phage of V. parahaemolyticus. Thus, like VGJφ, it is probable that the fs1, VSK, and fs2 phages of V. cholerae use MSHA as a receptor and that the Vf33 phage of V. parahaemolyticus uses an MSHA homologue to enter its host. This hypothesis is supported in part by previous results, which suggested that MSHA was the receptor for fs1 and fs2, although this was not fully demonstrated (16).
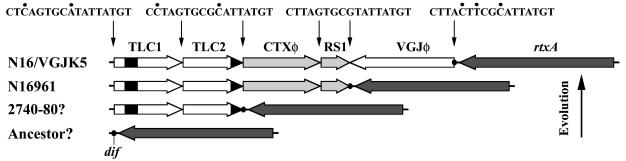
Integrating filamentous phages are more abundant than previously recognized. Many of them seem to exploit the XerCD recombination system to integrate into a site that overlaps the dif site of the bacterial chromosome (24). In V. cholerae the dif site seems to be a hot spot for multiple-phage integration. Several integration events may have contributed to the evolutionary history of the current circulating toxigenic strains of V. cholerae. An analysis of the complete genome sequence of the El Tor strain N16961 showed that it has two tandem imperfect repeats of the toxin-linked cryptic (TLC) element adjacent to the CTXφ prophage (49), which are flanked by att-like sites (Fig. 4). The TLC element is closely associated with the CTXφ prophage in toxigenic strains and has filamentous phage-related DNA sequences that can produce a cryptic plasmid (49). We speculate that TLC elements are the vestiges of an ancestral phage (TLCφ) that infected V. cholerae, integrated at the dif site, and subsequently became defective by loss of morphogenesis and structural modules, which impaired new transduction events of the TLC element by itself. Alternatively, the TLC element underwent deletion of its morphogenesis and structural modules in other hosts, from which it was transferred into V. cholerae in a foreign capsid. Once the TLC element was established, the bacterium was infected by CTXφ, which integrated into the TLC element attR site. Subsequently, a similar integration event with the RS1 satellite phage resulted in the current structure of N16961. V. cholerae strain 2740-80 (24, 48), which lacks CTXφ and RS1 prophages but has TLC element sequences, is reminiscent of the ancestral intermediate before acquisition of CTXφ by El Tor strains. Additionally, it is highly probable that VGJφ (or other related phages) is also lysogenized with this scenario in currently circulating toxigenic strains, forming a structure similar to that found in strain N16/VGJK5 obtained in this study (Fig. 4). Such lysogens might be more likely to be found in O139 strains, from which these phages have been isolated.
FIG. 4.
Hypothesis for the evolution of the dif region in toxigenic El Tor strains. Multiple phage integration events occurred during evolution. First, two events of integration of the hypothetical phage TLCφ into the dif site of an unknown ancestor (bottom map) gave rise to a structure similar to that of current strain 2740-80 (24, 48). Subsequently, integration of CTXφ and RS1 occurred, resulting in the structure of N16961. Finally, phages like VGJφ could also integrate downstream of RS1, resulting in a structure like that of strain N16/VGJK5 obtained in this study (top map). Thus, the dif site (dot) moved to the right from its original position. The region indicated by black inside TLC1 corresponds to an insertion sequence that could be inserted after TLC acquisition. Several copies of homologous insertion sequences are present in the V. cholerae chromosome. The black region in TLC2 corresponds to the eio region (49), which could be acquired with ancestral TLCφ. The rtxA gene is shown as a reference. The structure assigned to 2740-80 is hypothetical; we know that this strain must contain at least one TLC copy, so we represent it with the same TLC structure as N16961 to better illustrate the hypothesis. Surrounding att sequences for each prophage in N16/VGJK5 are shown. Differences from the att sequence downstream of RS1 are indicated by dots above the bases.
As VGJφ is a very prolific phage which usually produces titers greater than 1010 phage particles/ml/unit of optical density of infected culture, it seems to be more useful for applications than CTXφ, which produces relatively low titers in infected cultures and is harder to manipulate. Therefore, VGJφ is a potentially suitable vector for presenting heterologous antigens to the gut immune system if the phage is modified to display such antigens genetically fused to one of its capsid proteins. Live attenuated vaccine strains of V. cholerae could deliver such a phage in the intestinal mucosa.
One of the important conclusions of this paper is that there is a need for more detailed studies of phages which are able to integrate and transfer genetic information among bacterial populations. As CTXφ and VGJφ are both able to integrate their DNAs into the same site of the genome of V. cholerae, they can coexist adjacent to each other in the integrated state. Thus, it is possible that cholera toxin genes could be transduced by VGJφ by an anomalous excision event or another mechanism that has not been described yet. This possibility is being explored in our laboratory.
Acknowledgments
We are grateful to Richard A. Finkelstein for providing SG25-1 and many other V. cholerae strains used in this study, to Ronald K. Taylor for providing the KHT46 and KHT52 strains, to G. B. Nair for providing strains CRC262 and CRC266, to Luis Javier González for performing mass spectrometry identification of proteins, to Aminael Sánchez for providing technical assistance and a critical review of the manuscript, to María Cristina de la Rosa for providing electron microscope phage sample preparations, and to Tania Valdés, Sandra Rodríguez, and Odelsa Ancheta for providing electron micrographs.
REFERENCES
- 1.Armstrong, J., R. N. Perharm, and J. E. Walker. 1981. Domain structure of bacteriophage fd adsorption protein. FEBS Lett. 135:167-172. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1995. Short protocols in molecular biology. John Wiley & Sons Inc., New York, N.Y.
- 3.Benitez, J. A., L. Garcia, A. Silva, H. Garcia, R. Fando, B. Cedre, A. Perez, J. Campos, B. L. Rodriguez, J. L. Perez, T. Valmaseda, O. Perez, A. Perez, M. Ramirez, T. Ledon, M. D. Jidy, M. Lastre, L. Bravo, and G. Sierra. 1999. Preliminary assessment of the safety and immunogenicity of a new CTXφ-negative, hemagglutinin/protease-defective El Tor strain as a cholera vaccine candidate. Infect. Immun. 67:539-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campos, J., R. Fando, A. Silva, B. L. Rodríguez, and J. A. Benitez. 1998. Replicating function of the RS1 element associated with Vibrio cholerae CTXφ. FEMS Microbiol. Lett. 164:141-147. [DOI] [PubMed] [Google Scholar]
- 5.Chang, B., H. Taniguchi, H. Miyamoto, and S. Yoshida. 1998. Filamentous bacteriophages of Vibrio parahaemolyticus as a possible clue to genetic transmission. J. Bacteriol. 180:5094-50101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, K. H., F. S. Wen, T. T. Tseng, N. T. Lin, M. T. Yang, and Y. H. Tseng. 1998. Sequence analysis and expression of the filamentous phage φLf gene I encoding a 48-kDa protein associated with host cell membrane. Biochem. Biophys. Res. Commun. 245:313-318. [DOI] [PubMed] [Google Scholar]
- 7.Chopin, M. C., A. Rouault, S. D. Ehrlich, and M. Gautier. 2002. Filamentous phage active on the gram-positive bacterium Propionibacterium freudenreichii. J. Bacteriol. 184:2030-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Click, E. M., and R. E. Webster. 1997. Filamentous phage infection: required interactions with the TolA protein. J. Bacteriol. 179:6464-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Click, E. M., and R. E. Webster. 1998. The TolQRA proteins are required for membrane insertion of the major capsid protein of the filamentous phage f1 during infection. J. Bacteriol. 180:1723-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai, H., T. Y. Chow, H. J. Liao, Z. Y. Chen, and K. S. Chiang. 1988. Nucleotide sequences involved in the neolysogenic insertion of filamentous phage Cf16-v1 into the Xanthomonas campestris pv. citri chromosome. Virology 167:613-620. [PubMed] [Google Scholar]
- 11.Davis, B. M., E. H. Lawson, M. Sandkvist, A. Ali, S. Sozhamannan, and M. K. Waldor. 2000. Convergence of the secretory pathways for cholera toxin and the filamentous phage, CTXφ. Science 288:333-335. [DOI] [PubMed] [Google Scholar]
- 12.Davis, B. M., H. H. Kimsey, A. V. Kane, and M. K. Waldor. 2002. A satellite phage-encoded antirepressor induces repressor aggregation and cholera toxin gene transfer. EMBO J. 21:4240-4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis, B. M., H. H. Kimsey, W. Chang, and M. K. Waldor. 1999. The Vibrio cholerae O139 Calcutta bacteriophage CTXφ is infectious and encodes a novel repressor. J. Bacteriol. 181:6779-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis, B. M., K. E. Moyer, E. F. Boyd, and M. K Waldor. 2000. CTX prophages in classical biotype Vibrio cholerae: functional phage genes but dysfunctional phage genomes. J. Bacteriol. 182:6992-6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Pierro, M., R. Lu, S. Uzzau, W. Wang, K. Margaretten, C. Pazzani, F. Maimone, and A. Fasano. 2001. Zonula occludens toxin structure-function analysis. Identification of the fragment biologically active on tight junctions and of the zonulin receptor binding domain. J. Biol. Chem. 276:19160-19165. [DOI] [PubMed] [Google Scholar]
- 16.Ehara, M., S. Shimodori, F. Kojima, Y. Ichinose, T. Hirayama, M. J. Albert, K. Supawat, Y. Honma, M. Iwanaga, and K. Amako. 1997. Characterization of filamentous phages of Vibrio cholerae O139 and O1. FEMS Microbiol. Lett. 154:293-301. [DOI] [PubMed] [Google Scholar]
- 17.Falero, G., B. L. Rodríguez, T. Valmaseda, M. E. Pérez, J. L. Pérez, R. Fando, A. Robert, J. Campos, A. Silva, G. Sierra, and J. Benítez. 1998. Production and characterization of a monoclonal antibody against mannose-sensitive hemagglutinin of Vibrio cholerae. Hybridoma 17:63-67. [DOI] [PubMed] [Google Scholar]
- 18.Fasano, A., B. Baudry, D. W. Pumplin, S. S. Wasserman, B. D. Tall, J. M. Ketley, and J. B. Kaper. 1991. Vibrio cholerae produces a second enterotoxin, which affects intestinal tight junctions. Proc. Natl. Acad. Sci. USA 88:5242-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heilpern, A. J., and M. K. Waldor. 2000. CTXφ infection of Vibrio cholerae requires the tolQRA gene products. J. Bacteriol. 182:1739-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heilpern, A. J., and M. K. Waldor. 2003. pIIICTX, a predicted CTXφ minor coat protein, can expand the host range of coliphage fd to include Vibrio cholerae. J. Bacteriol. 185:1037-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill, D. F., N. J. Short, N. R. Perharm, and G. B. Petersen. 1991. DNA sequence of the filamentous bacteriophage Pf1. J. Mol. Biol. 218:349-364. [DOI] [PubMed] [Google Scholar]
- 23.Honma, Y., M. Ikema, C. Toma, M. Ehara, and M. Iwanaga. 1997. Molecular analysis of a filamentous phage (fs1) of Vibrio cholerae O139. Biochim. Biophys. Acta 1362:109-115. [DOI] [PubMed] [Google Scholar]
- 24.Huber, K. E., and M. K. Waldor. 2002. Filamentous phage integration requires the host recombinases XerC and XerD. Nature 417:656-659. [DOI] [PubMed] [Google Scholar]
- 25.Iida, T., K. Makino, H. Nasu, K. Yokoyama, K. Togomori, A. Hattori, T. Okuno, H. Shinagawa, and T. Honda. 2002. Filamentous bacteriophages of vibrios are integrated into the dif-like site of the host chromosome. J. Bacteriol. 184:4933-4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ikema, M., and Y. Honma. 1998. A novel filamentous phage, fs2, of Vibrio cholerae O139. Microbiology 144:1901-1906. [DOI] [PubMed] [Google Scholar]
- 27.Jacobson, A. 1972. Role of F pili in the penetration of bacteriophage f1. J. Virol. 10:835-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jouravleva, E. A., G. A. McDonald, C. F. Garon, M. B. Finkelstein, and R. A. Finkelstein. 1998. Characterization and possible function of a new filamentous bacteriophage from Vibrio cholerae. Microbiology 144:315-324. [DOI] [PubMed] [Google Scholar]
- 29.Jouravleva, E. A., G. A. McDonald, J. W. Marsh, R. K. Taylor, M. Boesman-Finkelstein, and R. A. Finkelstein. 1998. The Vibrio cholerae mannose-sensitive hemagglutinin is the receptor for a filamentous bacteriophage from V. cholerae O139. Infect. Immun. 66:2535-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaper, J. B., J. G. Morris, Jr., and M. M. Levin. 1995. Cholera. Clin. Microbiol. Rev. 8:48-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kar, S., R. K. Ghosh, A. N. Ghosh, and A. Ghosh. 1996. Integration of the DNA of a novel filamentous bacteriophage VSK from Vibrio cholerae O139 into the host chromosomal DNA. FEMS Microbiol. Lett. 145:17-22. [DOI] [PubMed] [Google Scholar]
- 32.Kenner, J. R., T. S. Cosner, D. N. Taylor, A. F. Trofa, M. Barbera-Oro, T. Hyman, J. M. Adams, D. T. Bettie, K. P. Killeen, D. R. Spriggs, J. J. Mekalanos, and J. C. Sadoff. 1995. Peru-15, an improved live attenuated oral vaccine candidate for Vibrio cholerae O1. J. Infect. Dis. 172:1126-1129. [DOI] [PubMed] [Google Scholar]
- 33.Kimsey, H. H., and M. K. Waldor. 1998. CTXφ immunity: application in the development of cholera vaccines. Proc. Natl. Acad. Sci. USA 95:7035-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koonin, E. V. 1992. The second cholera toxin, Zot, and its plasmid-encoded and phage-encoded homologues constitute a group of putative ATPases with an altered purine NTP-binding motif. FEBS Lett. 312:3-6. [DOI] [PubMed] [Google Scholar]
- 35.Kostrikis, L. G., S. A. Reisberg, H. Y. Kim, S. Shin, and L. A. Day. 1995. C2, an unusual filamentous bacterial virus: protein sequence and conformation, DNA size and conformation, and nucleotide/subunit ratio. Biochemistry 34:4077-4087. [DOI] [PubMed] [Google Scholar]
- 36.Kuo, T. T., M. S. Tan, M. T. Su, and M. K. Yang. 1991. Complete nucleotide sequence of filamentous phage Cf1c from Xanthomonas campestris pv. citri. Nucleic Acids Res. 19:2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin, N. T., R. Y. Chang, S. J. Lee, and Y. H. Tseng. 2001. Plasmids carrying cloned fragments of RF DNA from the filamentous phage φLf can be integrated into the host chromosome via site-specific integration and homologous recombination. Mol. Genet. Genomics 266:425-435. [DOI] [PubMed] [Google Scholar]
- 38.Lubkowski, J., F. Hennecke, A. Puckthun, and A. Wlodawer. 1999. Filamentous phage infection: crystal structure of g3p in complex with its coreceptor, the C-terminal domain of TolA. Structure 7:711-722. [DOI] [PubMed] [Google Scholar]
- 39.Marvin, D. A. 1998. Filamentous phage structure, infection and assembly. Curr. Opin. Struct. Biol. 8:150-158. [DOI] [PubMed] [Google Scholar]
- 40.Marvin, D. A., and B. Hohn. 1969. Filamentous bacterial viruses. Bacteriol. Rev. 33:172-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsukura, T., T. Iwasaki, and M. Kawashima. 1992. Molecular cloning of a novel human papillomavirus (type 60) from a plantar cyst with characteristic pathological changes. Virology 190:561-564. [DOI] [PubMed] [Google Scholar]
- 42.McMaster, G. K., and G. G. Carmichael. 1977. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc. Natl. Acad. Sci. USA 74:4835-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mekalanos, J. J. 1983. Duplication and amplification of toxin genes in Vibrio cholerae. Cell 35:253-263. [DOI] [PubMed] [Google Scholar]
- 44.Model, P., and M. Russel. 1988. Filamentous bacteriophage, p. 375-456. In R. Calendar (ed.), The bacteriophages, vol. 2. Plenum Publishing Corporation, New York, N.Y.
- 45.Moyer, K. E., H. H. Kimsey, and M. K. Waldor. 2001. Evidence for a rolling-circle mechanism of phage DNA synthesis from both replicative and integrated forms of CTXφ. Mol. Microbiol. 41:311-323. [DOI] [PubMed] [Google Scholar]
- 46.Nash, H. A. 1996. Site-specific recombination: integration, excision, resolution, and inversion of defined DNA segments, p. 2363-2376. In C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D. C.
- 47.Nasu, H., T. Iida, T. Sugahara, Y. Yamaichi, K. S. Park, K. Yokoyama, K. Makino, H. Shinagawa, and T. Honda. 2000. A filamentous phage associated with recent pandemic Vibrio parahaemolyticus O3:K6 strains. J. Clin. Microbiol. 38:2156-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pearson, G. D., A. Woods, S. L. Chiang, and J. J. Mekalanos. 1993. CTX genetic element encodes a site-specific recombination system and an intestinal colonization factor. Proc. Natl. Acad. Sci. USA 90:3750-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rubin, E. J., W. Lin, J. J. Mekalanos, and M. K. Waldor. 1998. Replication and integration of a Vibrio cholerae cryptic plasmid linked to the CTX prophage. Mol. Microbiol. 28:1247-1254. [DOI] [PubMed] [Google Scholar]
- 50.Russel, M. 1991. Filamentous phage assembly. Mol. Microbiol. 5:1607-1613. [DOI] [PubMed] [Google Scholar]
- 51.Russel, M. 1995. Moving through the membrane with filamentous phages. Trends Microbiol. 3:223-228. [DOI] [PubMed] [Google Scholar]
- 52.Russel, M., N. A. Linderoth, and A. Sali. 1997. Filamentous phage assembly: variation on a protein export theme. Gene 192:23-32. [DOI] [PubMed] [Google Scholar]
- 53.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 54.Thelin, K. H., and R. K. Taylor. 1996. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect. Immun. 64:2853-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Valle, E., T. Ledon, B. Cedre, J. Campos, T. Valmaseda, B. Rodriguez, L. Garcia, K. Marrero, J. Benitez, S. Rodriguez, and R. Fando. 2000. Construction and characterization of a nonproliferative El Tor cholera vaccine candidate derived from strain 638. Infect. Immun. 68:6411-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous bacteriophage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]
- 57.Waldor, M. K., E. J. Rubin, G. D. Pearson, H. Kimsey, and J. J. Mekalanos. 1997. Regulation, replication, and integration functions of the Vibrio cholerae CTXφ are encoded by region RS2. Mol. Microbiol. 24:917-926. [DOI] [PubMed] [Google Scholar]
- 58.Watnick, P. I., K. J. Fullner, and R. Kolter. 1999. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J. Bacteriol. 181:3606-3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]