Abstract
Defining the gene products that play an essential role in an organism's functional repertoire is vital to understanding the system level organization of living cells. We used a genetic footprinting technique for a genome-wide assessment of genes required for robust aerobic growth of Escherichia coli in rich media. We identified 620 genes as essential and 3,126 genes as dispensable for growth under these conditions. Functional context analysis of these data allows individual functional assignments to be refined. Evolutionary context analysis demonstrates a significant tendency of essential E. coli genes to be preserved throughout the bacterial kingdom. Projection of these data over metabolic subsystems reveals topologic modules with essential and evolutionarily preserved enzymes with reduced capacity for error tolerance.
Sequencing and comparative analysis of multiple diverse genomes is revolutionizing contemporary biology by providing a framework for interpreting and predicting the physiologic properties of an organism. A variety of emerging postgenomic techniques such as genome-wide expression profiling and monitoring of macromolecular complex formation can reveal the detailed molecular compositions of cells. New computational approaches to exploring the inherent organization of cellular networks, the mode and dynamics of interactions among cellular constituents, are in early stages of development (14, 22, 23). These techniques allow us to begin unraveling a major paradigm of cellular biology: how biological properties arise from the large number of components making up an individual cell.
Defining which gene products play an essential role and under what conditions is vital to understanding the complexity of living organisms. Although methods to rapidly and systematically determine genome-wide gene essentiality are less advanced than other functional genomic techniques, a number of essentiality surveys involving different species have been reported. Many experimental approaches have been used to produce such data, including individual knockouts in Saccharomyces cerevisiae (10, 38), Caenorhabditis elegans (21), and recently B. subtilis (22a), RNA interference in C. elegans (20), and whole-genome transposon mutagenesis studies with several microorganisms. In the latter group, complete or extensive lists of essential and dispensable genes are available for Mycoplasma pneumoniae and Mycoplasma genitalium (15), Mycobacterium tuberculosis (31), Haemophilus influenzae (1), and S. cerevisiae (30). However, as of yet relatively little effort has been committed to a system level interpretation of these data in terms of cellular function or evolutionary relationships with other organisms (19).
Escherichia coli has historically been the focus of intense biochemical, genetic, and physiologic scrutiny, but genomic essentiality data for this organism have remained incomplete. Systematic efforts to compile genome-wide collections of E. coli deletion mutants are under way. Two groups have reported Tn10 transposon-based genetic footprinting projects with E. coli, but essentiality data were revealed only for a limited set of genes (3, 13). Currently, the Profiling of E. coli Chromosome database (available at http://www.shigen.nig.ac.jp/ecoli/pec) is the most complete list of essential and dispensable genes in E. coli. This list is not based on direct experimental evidence but is derived from systematic review of the experimental literature. Although this compilation is of great value, the wide variety of strains, conditions, and types of mutations used in individual studies significantly complicates interpretation.
Here we report a genome-wide, comprehensive experimental assessment of the E. coli MG1655 genes necessary for robust aerobic growth in a rich, tryptone-based medium. Of the 4,291 protein-encoding genes in E. coli, we assessed the essentiality of 3,746 genes (∼87% of the total). Individual assessments were projected onto a whole-cell functional reconstruction model including both metabolic and nonmetabolic systems. Distribution of conditionally essential and dispensable E. coli genes within functional systems was analyzed with respect to the occurrence of putative orthologs across a broad range of diverse bacterial genomes. This analysis demonstrates a significant tendency of experimentally identified essential E. coli genes to be evolutionarily preserved throughout the bacterial kingdom, especially a subset of genes representing key cellular processes such as DNA replication and protein synthesis. Finally, we analyzed the conditional essentiality of metabolic enzymes from the perspective of cellular system level organization, demonstrating enrichment with those enzymes that catalyze reactions within evolutionarily conserved topologic modules in the complex metabolic web of E. coli.
MATERIALS AND METHODS
Transposon mutagenesis.
E. coli strain MG1655 (F− λ− ilvG rfb-50 rph-1) (16) was used throughout this work. Genetic footprinting with the use of the plasmid pMOD<MCS> containing the artificial transposon EZ::TN<KAN-2> (Epicentre Technologies, Madison, Wis.) and identification of chromosomal insertion sites were previously described (9) and are detailed in the supplementary data (supplementary data for this paper are available at http://www.integratedgenomics.com/online_material/gerdes and on the University of Notre Dame and Northwestern University websites [http://www.umsl.edu/∼balazsi/JBact2003/ and http://www.oltvailab.northwestern.edu/Pubs/JBact2003/]). Cells were grown in an enriched Luria-Bertani (LB) medium composed of 10 g of tryptone/liter, 5 g of yeast extract/liter, 50 mM NaCl, 9.5 mM NH4Cl, 0.528 mM MgCl2, 0.276 mM K2SO4, 0.01 mM FeSO4, 5 × 10−4 mM CaCl2, and 1.32 mM K2HPO4. The growth medium also included the following micronutrients: 3 × 10−6 mM (NH4)6(MoO7)24, 4 × 10−4 mM H3BO3, 3 × 10−5 mM CoCl2, 10−5 mM CuSO4, 8 × 10−5 mM MnCl2, and 10−5 mM ZnSO4. The following vitamins were added (concentrations are in milligrams per liter): biotin, 0.12; riboflavin, 0.8; pantothenic acid, 10.8; niacinamide, 12.0; pyridoxine, 2.8; thiamine, 4.0; lipoic acid, 2.0; folic acid, 0.08; and p-aminobenzoic acid, 1.37. Kanamycin was added to 10 μg/ml.
As with any high-throughput technique, genetic footprinting is subject to a certain degree of experimental and analytical error. A variety of validation techniques indicate the overall error rate of our assignments to be well within 10% (9). The actual experimental detection and insert mapping error rate is much lower (within 1 to 2%). The major source of ambiguity is associated with data interpretation (see below). In the supplementary data, we include the insert distribution within each open reading frame (ORF) (raw data, including insert distribution within intergenic regions, are available upon request).
Statistical analyses of transposon insertion frequency.
Essential and ambiguous ORFs introduce a bias into the density of transposon insertions due to the fact that they “lose” the insertions incorporated within them during selective outgrowth. There were also unmapped genomic regions where transposon insertions could not be detected. To reconstruct insert distribution prior to selective outgrowth, and to account for the contribution of unmapped regions, we removed from the E. coli chromosomal map every ORF with a function asserted to be essential, ambiguous, or not determined, as well as the regions not covered by the mapping process, and joined together the rest of the chromosome. We analyzed the original and corrected insertion location data assuming that the insertions appear as a result of a Poisson process with an overall rate r of 3.218/kb. Based on this hypothesis, the probability to find M insertions within a DNA region of length L is given by
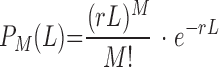 |
The P values corresponding to this hypothesis for the corrected data were calculated to estimate the statistical significance of the deviations from a Poisson process, for a threshold of P of 10−5 (see Fig. 1).
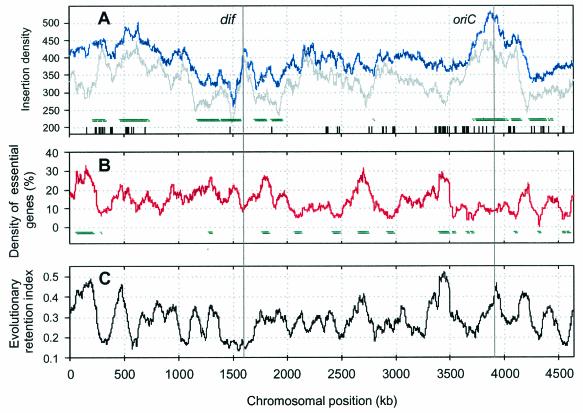
FIG. 1.
Distribution of transposon insertion densities, densities of essential genes, and ERIs along the E. coli chromosome. (A) Gray lines show the transposon insertion densities calculated as the number of transposition events per 100-kb sliding window over the entire E. coli MG1655 chromosome. Values indicated by the blue lines were computed in a similar manner, except that all chromosomal regions corresponding to essential and ambiguous genes were excluded from the calculations in order to reconstruct insert distribution prior to selective outgrowth (see also Materials and Methods). Gaps in the data (chromosomal regions where transposition events could not be detected due to technical reasons) are indicated by short vertical lines along the x axis. These regions were excluded from all analyses. Nucleotide positions of the E. coli genome sequence correspond to those in reference 4. The regions where the distributions of transposition events significantly deviate (P < 0.01) from a Poisson process are marked by horizontal green lines. oriC shows the origin of chromosomal replication, and dif denotes the dif locus within the replication termination area. (B) Distribution of essential genes along the E. coli chromosome, defined as a percentage of essential genes in the total number of genes within a 100-kb-long chromosomal region (calculated per sliding window as described above). The regions where the numbers of essential genes significantly deviate (P < 0.01) from values that could arise by chance are marked by horizontal green lines. (C) ERIs along the E. coli chromosome, defined as the average ERI for all genes within each 100-kb region. The ERI for a gene is defined as the fraction of organisms in a diverse set of 33 bacterial species which contain an ortholog of the gene in their genomes.
If the insertion locations are approximated by a Poisson process, the statistical reliability of essentiality calls depends on two factors: the overall insertion density r in the region where the ORF is located and the length L of the ORF. More specifically, the probability that an ORF is missed by chance is given as follows: P0(L) = e−rL, where r is the corrected density of insertions in the 10-kb region centered on the ORF on the chromosome. For example, to assure that the probability P0 that no transposon insertion is detected in the given gene by chance alone is <0.5, we need the following: rL > log(2) = 0.639. In our case, 604 of the 620 genes asserted to be essential satisfy this condition with rL of >0.639, indicating that ∼97% of all essential genes have a reliability of essentiality calls expressed by a P0 of <0.5. The number of essential genes with P0 smaller than a fixed value is given in Table 1. A detailed list for each gene is presented in the supplementary data (see Table S1).
TABLE 1.
Number of essential genes with P0 smaller than a fixed value
| P0 | No. of essential genes | % of essential genes |
|---|---|---|
| <0.01 | 159 | 26 |
| <0.05 | 281 | 45 |
| <0.1 | 367 | 59 |
| <0.2 | 476 | 77 |
| <0.3 | 550 | 89 |
| <0.4 | 587 | 95 |
| <0.5 | 604 | 97 |
| <0.6 | 610 | 98 |
Identification of putative orthologs of E. coli genes in diverse set of microbial genomes.
Putative orthologs of E. coli genes were identified by using the ERGO database (http://ergo.integratedgenomics.com/ERGO/) (26). Protein families in ERGO correspond to homologous ORFs with identical assigned functions (24). With each update of the database, grouping of proteins into families is refigured through a multistep process including (i) formation of a family core from proteins corresponding to several ORFs that are bidirectional best FASTA hits for one another in their respective genomes, (ii) family extension by adding proteins with identical assigned functions and by performing FASTA searches (27) and adding matches with expectation values of less than a preset threshold, as described earlier (12), and (iii) refinement of a family grouping based on multiple ClustalW alignments (36) of all included sequences. To identify putative orthologs of E. coli proteins, all protein families in ERGO were automatically queried for the simultaneous presence of a protein(s) corresponding to an E. coli ORF(s) and proteins corresponding to ORFs from the genomes of 32 diverse bacterial species (Agrobacterium tumefaciens, Anabaena sp., Aquifex aeolicus, Bacillus subtilis, Borrelia burgdorferi, Brucella melitensis, Buchnera sp., Campylobacter jejuni, Caulobacter crescentus, Chlamydia trachomatis, Clostridium acetobutylicum, Corynebacterium glutamicum, Deinococcus radiodurans, Fusobacterium nucleatum, Haemophilus influenzae, Helicobacter pylori, Listeria monocytogenes, Mesorhizobium loti, Mycobacterium tuberculosis, Mycoplasma pneumoniae, Neisseria gonorrhoeae, Pseudomonas aeruginosa, Ralstonia solanacearum, Rickettsia prowazekii, Sinorhizobium meliloti, Staphylococcus aureus, Streptococcus pneumoniae, Synechocystis sp., Thermotoga maritima, Treponema pallidum, Vibrio cholerae, and Xylella fastidiosa). Results of this search were further supplemented by addition of ORFs from each of these genomes that are bidirectional best FASTA hits with corresponding E. coli genes.
Densities of essential genes and evolutionary retention indexes (ERIs) along the chromosome.
The densities of essential genes along the E. coli chromosome (see Fig. 1B) were calculated within overlapping 100-kb regions displaced 1 kb from one another. For each 100-kb region, the essentiality was defined as the ratio of the number of essential genes to the total number of genes found in the region (NE/NT). The significance of essentiality for each 100-kb region was determined based on the hypergeometric distribution. Given that 620 of 4,291 E. coli genes were found to be essential, the probability of having NE essential genes out of a total number of NT genes within a 100-kb region is given by
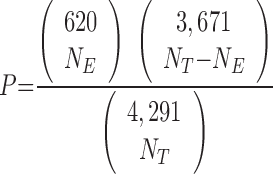 |
where 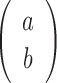 denotes the number of ways to choose b out of a elements.
denotes the number of ways to choose b out of a elements.
We determined the ERI for each of the 4,291 E. coli ORFs by calculating the fraction of genomes in the group that have an ortholog of the given ORF, with the number of representative organisms (NO) equal to 33. Thus, if the number of organisms that contain an ortholog of the E. coli ORF is NC, the ERI is given by the following formula: ERI = NC/NO. The ERIs along the E. coli chromosome were calculated within overlapping 100-kb chromosomal regions, displaced 1 kb from one another (see Fig. 1C). The ERI of each 100-kb region was determined by calculating the average of the ERIs for all ORFs located completely inside the region.
Data analysis within the context of system level metabolic organization.
Using the information about the E. coli enzymes for all metabolic reactions available in the ERGO database, together with the essentiality data for the corresponding genes, we analyzed the correlation of enzyme essentialities within the known hierarchical structure of the E. coli metabolic organization. We have previously established a global topologic representation of the E. coli metabolic network, in which each branch on the hierarchical tree corresponds to a group of metabolites that are at its endpoints. Thus, each junction represents the module made up of the substrates that were clustered together up to that stage (28). For each branch, we can define an essentiality ratio based on the metabolic reactions present among the group of metabolites it represents.
To treat each reaction equally, we considered all links present between any two metabolites in the group, and for each of these links we took into account all the reactions that created the link. Specifically, for all pairs in the group, we included those metabolic reactions that transformed one of the substrates into another, according to a reaction list in which generic donor and acceptor moieties, such as H2O and ATP, are not considered (see reference 28 for details) and to which an unambiguous insertion phenotype has been assigned (NRall). Next, we counted those reactions whose corresponding catalytic enzymes proved to be essential (NRlethal). Note that since the hierarchical tree is constructed according to a two-step network complexity reduction procedure (28), there can be arcs between pairs of substrates that the tally does not include. To account for these, we examined each metabolic reaction with a known catalytic enzyme insertion phenotype on these internal arcs and incorporated them into the analysis. The essentiality of the branch (or module) is given by the fraction NRlethal/NRall and represents the fraction of essential enzymes of all biochemical reactions within a given metabolic module (branch). For additional details, see the supplementary data.
RESULTS
Genome-scale genetic footprinting in E. coli.
Genetic footprinting was first introduced for analysis of gene essentiality in S. cerevisiae (33). A modification of this technique using a Tn5-based in vitro transposome system (11) in E. coli was previously described, and gene essentiality within three cofactor biosynthetic pathways has been analyzed (9). Here we have extended this pilot analysis to the whole-genome level by using the same standardized growth conditions. The general experimental scheme is illustrated in the supplementary data. Briefly, following transposon mutagenesis, a population of ∼2 × 105 independent mutants was grown aerobically for 23 doublings in enriched LB medium supplemented with kanamycin. Genomic DNA was isolated from the whole population and used to map individual transposon inserts with a nested PCR approach.
Distribution of the 1.8 × 104 distinct insert locations detected along the E. coli chromosome is illustrated in Fig. 1A. The densities of transposon insertion events are randomly distributed, with two notable exceptions: an overall maximum around the origin of replication (oriC) and a minimum around the terminus (dif). This may reflect increased target copy number at the origin of replication in the actively dividing bacterial population used in this experiment. The overall insertion density is 3.218/kb, without appreciable variation between coding (3.221/kb) and noncoding (3.193/kb) regions.
Assessment of conditional gene essentiality based on genetic footprinting data.
Unambiguous essentiality assessments were made for 3,746 (or 87% of the total) E. coli protein-encoding genes or ORFs (Table 2). Of these, 620 (14%) were asserted to be essential, and 3,126 (73%) were asserted to be nonessential (dispensable) based on the occurrence of transposon inserts within each ORF and the overall insertion density in the local environment, as described in the supplementary data. The complete essentiality list is reported in the supplementary data (see Table S1). No assertions could be made for 327 genes for technical reasons, such as limited efficiency of PCRs in certain regions of the E. coli chromosome or nonspecific primer annealing in areas of DNA repeats. For 218 genes, we considered the evidence to be insufficient for a specific conclusion about essentiality. These genes were systematically called ambiguous, according to the criteria listed in the supplementary data. For example, ORFs shorter than 240 bp (<80 aa) and with no inserts were consistently classified as ambiguous rather than essential. In certain cases, relatively long ORFs (>900 bp) containing only a single transposon were designated ambiguous rather than nonessential.
TABLE 2.
Distribution of essential and nonessential genes and average ERIs in selected functional categoriesa
| Functional category | Description | Total no. of ORFs | No. E | No. N | No. ? | No. ND | % E | Mean ERI |
|---|---|---|---|---|---|---|---|---|
| AAM | Amino acid metabolism | 138 | 21 | 108 | 1 | 8 | 15 | 0.5 |
| CHM | Carbohydrate metabolism | 219 | 21 | 178 | 3 | 17 | 10 | 0.4 |
| NCM | Nucleotide and cofactor metabolism | 181 | 53 | 119 | 2 | 7 | 29 | 0.5 |
| LPC | Lipid, lipopolysaccharide, lipoprotein, peptidoglycan, and cell wall biosynthesis | 126 | 34 | 73 | 5 | 14 | 27 | 0.5 |
| NAM | Nucleic acid metabolism | 156 | 43 | 96 | 5 | 12 | 28 | 0.6 |
| PMS | Protein metabolism and secretion | 167 | 80 | 57 | 18 | 12 | 48 | 0.7 |
| MSM | Miscellaneous metabolism | 94 | 14 | 72 | 2 | 6 | 15 | 0.4 |
| BEN | Bioenergetics | 100 | 15 | 75 | 4 | 6 | 15 | 0.3 |
| SMC | Signaling, motility, and chemotaxis | 153 | 12 | 125 | 3 | 13 | 8 | 0.3 |
| RCD | Expression regulation and cell cycle and division | 177 | 30 | 118 | 13 | 16 | 17 | 0.2 |
| MTR | Membrane transport | 276 | 22 | 244 | 3 | 7 | 8 | 0.3 |
| PHT | Phage- and transposase-related processes | 62 | 12 | 35 | 2 | 13 | 19 | 0.3 |
| CAT | All categorized | 1,849 | 357 | 1,300 | 61 | 131 | 19 | 0.4 |
| UNC | Uncategorized | 2,442 | 263 | 1,826 | 157 | 196 | 11 | 0.2 |
| Total | Categorized and uncategorized | 4,291 | 620 | 3,126 | 218 | 327 | 14 | 0.3 |
Abbreviations are as follows: E, essential; N, nonessential; ND, not determined; ?, ambiguous.
Our results are generally consistent with previously published data on individual genes and with data from currently available collections of systematic gene deletions in E. coli. For example, of the 1,379 individual gene deletion mutants listed at the University of Wisconsin E. coli Genome Project website (http://www.genome.wisc.edu/functional/tnmutagenesis.htm), only 12% produced apparently conflicting designations of genes as essential (for a detailed list of the discrepancies, see Table S2 in the supplementary data). Although we have not attempted to reconcile each individual case, several reasons for discrepancies can be envisioned. Most importantly, the term essential, which intuitively suggests an absolute requirement for cell viability, also applies to any gene that imparts a substantial fitness advantage. Thus, mutants lacking gene products necessary for maintaining vigorous growth fall into the same category as those with “true lethal” mutations. Therefore, certain genes may be classified as essential by genetic footprinting, yet corresponding viable deletion mutants may be obtained. In addition, differences in medium compositions, aeration levels, temperatures, and cell densities may account for many inconsistencies. Surprisingly, polar effects, in which transposon insertion into dispensable genes disrupts transcription of essential genes, are relatively rare in genetic footprinting. This may be due to the presence of weakly active promoter-like sequences within the transposon used in these experiments (9, 11). Most examples of polar effects are associated with genes that may require high levels of expression to sustain rapid growth rates.
Discrepancies resulting from inserts detected in the genes otherwise considered to be essential also occur. In some cases, single inserts occur close to protein termini or in interdomain boundary regions in multidomain proteins. For proteins consisting of two or more independently functioning domains, inserts may be tolerated within the 3′ portion of the gene if the C-terminal domain of the protein it encodes is associated with a dispensable function. This can occur even when a function associated with the N-terminal domain (from the 5′ region of the gene) is genuinely essential (as with ftsX [9]). Small, localized chromosomal duplications may account for inserts in genes otherwise recognized as essential (2). In this scenario, one copy of a duplicated gene provides the essential function while the other copy containing the transposon is stabilized by selection for kanamycin resistance. Large genes with only a small number of inserts may fall into this category since the total number of specific duplications within the population prior to transformation is probably very small (25).
Functional context analyses of essentiality data.
The interpretation of genomic essentiality data can be approached in a number of alternate ways, such as by using chromosomal (positional), functional (system level), or phylogenetic (evolutionary) context analysis. In addition to refining initial essentiality assignments and reconciling apparent discrepancies with existing knowledge, such analyses can improve and expand existing understanding of the systemic behavior of the cell at various levels. Without attempting a comprehensive analysis, we have limited the scope of our efforts to (i) prototyping and illustrating such analysis by using selected examples from various functional systems, (ii) evaluating the internal consistency of our data, and (iii) developing preliminary observations at the system level, as presented below.
Initially, we analyzed the data in a functional context, which involved dividing the overall physiology of the organism into smaller, internally coherent subsystems such as amino acid biosynthesis, nucleotide metabolism, and other broad functional categories (Table 2). This approach mirrors the standard didactic subdivision of microbial biochemistry and physiology. It also provides an organizational framework with which to analyze total genomic data and allows specific metabolic questions to be addressed.
For consistency, our functional analysis is based exclusively on SWISS-PROT functional annotations (8). Each of the 1,849 gene products with specific SWISS-PROT annotations and defined biochemical functions supported by solid experimental evidence was placed into one of the 12 functional categories (Table 2 and supplementary data [see Table S1]). Among the remaining 2,242 uncategorized protein-encoding genes, many have been tentatively annotated in SWISS-PROT and other databases, but most of these annotations either fall short of giving a specific testable function or have not been confirmed by direct experiments. As expected, the ratios of essential genes within various functional categories are rather uneven (Table 2). Categories that include gene products involved with key aspects of cellular metabolism (such as nucleic acid and protein metabolism) contain a substantially higher percentage of essential genes (28 and 48%, respectively) than the average for the entire genome (14%). The percentages of essential genes in categories such as signaling, motility, and chemotaxis (8%) and membrane transport (8%) are substantially below the whole-genome benchmark. The average essentiality for the subset of 2,242 uncategorized genes (11%) is substantially lower than the average for the subset of categorized genes (19%). Several representative metabolic and nonmetabolic systems (7 of 12 functional categories) were selected for use as examples of functional context analysis and for evaluation of the internal consistency of the data. Here we describe one such analysis, with additional detailed interpretations presented in the supplementary data.
Amino acid metabolism: lysine biosynthesis.
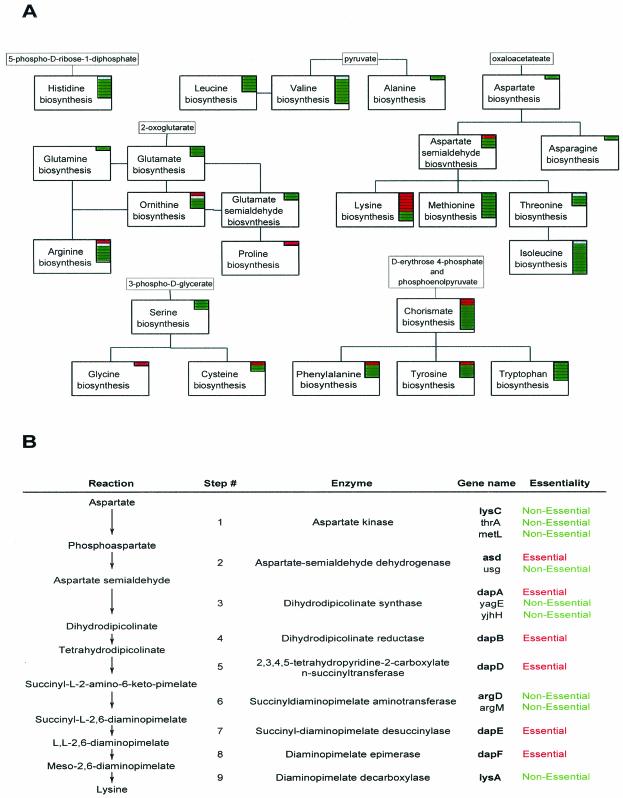
Most of the genes responsible for biosynthesis of various amino acids were expected to be nonessential since the medium contains most of the amino acids required for growth. With a few notable exceptions, this expectation was confirmed by our results. Of the 91 genes with specific SWISS-PROT annotations indicating involvement in amino acid biosynthesis, only 16 appear to be essential (Fig. 2A). Six of these genes are involved in lysine biosynthesis. E. coli produces lysine from aspartate via the nine-step pathway (Fig. 2B). Although lysine is available in the growth medium, its immediate precursor, diaminopimelate (DAP), which is required for cell wall biosynthesis, is not. The lysA gene encoding the enzyme that converts DAP to lysine at the last step of this pathway is dispensable. Analysis of DAP-lysine biosynthesis provides an example of refining pathway reconstruction and individual functional assignments based on genome-scale essentiality data. Genes (asd, dapA, dapB, dapD, dapE, and dapF) encoding most of the enzymes leading to DAP production are essential. The first gene in this pathway (lysC), encoding aspartokinase III, is dispensable due to the functional redundancy of the additional aspartokinase isozymes (encoded by metL and thrA). In contrast, the asd and dapA genes involved with the second and the third steps of DAP-lysine biosynthesis are essential in spite of the existence of apparent paralogs. Proteins encoded by the yjhH and yagE functionally uncharacterized genes are often annotated as potential dihydrodipicolinate synthases based on their high sequence similarities with the dapA gene product (BLAST E scores of 4e−33 and 2e−28, respectively). However, genetic footprinting data suggest that under our experimental conditions neither is capable of complementing loss of the essential dapA function. The opposite situation is observed with succinyl-DAP aminotransferase (encoded by argD), which is firmly defined as dispensable in our data. This apparent inconsistency can be resolved by assuming functional complementation by the argM gene product. The argM gene is known to encode succinyl-ornithine transaminase, which is primarily involved in arginine biosynthesis. However, this enzyme is closely related to succinyl-DAP aminotransferase by sequence, and the aminotransferases are known to possess rather broad substrate specificities, especially for structurally similar substrates (such as succinyl-DAP and succinyl-ornithine). Overexpression of the argM gene has been demonstrated to suppress an argD mutation in E. coli (32).
FIG. 2.
Essentiality of genes controlling amino acid biosynthesis in E. coli. (A) Functional overview of amino acid biosynthesis. Each block represents one or more pathways leading to production of a particular amino acid or its key intermediates (shown in smaller boxes). Within each block, stacked bars represent the gene products involved in the pathway (according to SWISS-PROT release of June 2002). Bars are colored according to gene essentiality (green, nonessential; red, essential; gray, undefined). (B) Detailed representation of the lysine biosynthetic pathway. Genes predicted in the ERGO database to be paralogs in this pathway are shown, in addition to genes whose roles in the biosynthesis of lysine have been experimentally verified (in bold).
Phylogenetic analysis of essentiality data within functional groups.
To assess the data set from an evolutionary perspective, we examined the distribution of conditionally essential and dispensable E. coli genes with respect to the occurrence of putative orthologs across a broad range of diverse bacterial genomes. Putative orthologs within a reference set of 32 complete bacterial genomes chosen to represent maximum phylogenetic diversity were identified based on protein families, supplemented by bidirectional best hits (see Materials and Methods). For this analysis we introduce a simple parameter: an ERI computed for each E. coli gene as the fraction of genomes from the reference set containing a putative ortholog of the gene. ERI values varying from 0 (for genes unique to E. coli) to 1.0 (for omnipresent genes) are provided in the supplementary data (see Table S1). In a recent study, the Profiling of E. coli Chromosome data (http://www.shigen.nig.ac.jp/ecoli/pec) were used to demonstrate a remarkable tendency of essential gene sequences to be more evolutionarily conserved than those of nonessential genes (19). In our analysis, we used ERI values to focus on occurrence of essential and nonessential genes (preservation of orthologs) rather than on conservation of their respective sequences.
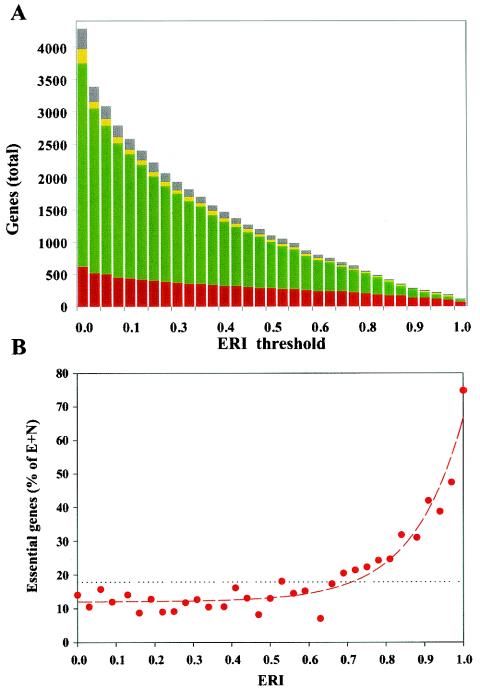
Figure 3A depicts the overall number of E. coli genes in decreasing order over the range of ERI values. An initial sharp decrease in the number of preserved genes (∼40%) occurs over a rather small phylogenetic distance of less than four genomes in our reference set (ERI ≤ 0.1). Further decay is at much lower rates, and orthologs of ∼10% of E. coli genes are preserved in at least 25 diverse genomes (ERI ≥ 0.8). This reflects a nonrandom ortholog preservation pattern, characterized by a highly conserved core group of genes. This core is highly enriched by genes identified as essential in our study. The tendency of essential genes to be evolutionarily preserved is also reflected in Fig. 1, demonstrating a significantly positive correlation (0.5240) between essentiality (Fig. 1B) and ERIs (Fig. 1C) along the E. coli chromosome. Similarly, plotting the fraction of essential genes at different ERI values demonstrates that the relationship between the two parameters has the following form: y = yo + aebx, implying that the essentiality of genes with a given ERI is due partly to a very strong tendency of essential genes to be retained by evolution (the exponential behavior dominant above an ERI of 0.6) and partly to an essential gene fraction of ∼10% that is present among genes within any ERI value group (Fig. 3B).
FIG. 3.
Distribution of E. coli genes as a function of ERIs. (A) Total number of genes with an ERI above the threshold plotted versus the ERI threshold. Color coding within bars represents fractions of essential (red), nonessential (green), ambiguous (yellow), and missing (gray) genes for each incremental increase of ERI threshold (with 33 diverse genomes in the reference set). (B) Fractions of essential genes at different ERI values. The data were fitted with the following function: y = yo+aebx, where yo is 12.0 ± 0.9, a is 0.023 ± 0.019, and b is 7.8 ± 0.8 (dashed red line). The dotted line represents the fractions of essential genes for the whole genome. (The fractions plotted are defined as the number of essential genes versus the number of essential (E) and nonessential (N) genes. Unknown or ambiguous genes are not taken into account.)
Comparison of average essentiality and ERI values between different functional categories reveals significant correlation (Table 2). Functional categories including highly specialized proteins such as transporters, regulators, and signaling molecules are characterized by average ERI values close to the average for the whole genome (∼0.3). Average essentiality within these groups also does not exceed an overall whole-genome level (∼14%). The least essential group of all uncategorized proteins with historically elusive functions has the lowest average ERI, ∼0.2. Therefore, many of these proteins are likely to be specific to the environmental and phylogenetic niches of E. coli. On the other hand, the bulk of cellular intermediary metabolism (categories AAM, CHM, NCM, LPC, and MSM [Table 2]) is associated with ERI values of 0.4 to 0.5. Essentiality within these metabolic categories varies depending on the levels of functional redundancy of their constituents in rich medium. Not surprisingly, the highest ERI values (up to 0.7) as well as the highest ratio of essential genes (up to 48%) occurs in functional categories that include replication, transcription, and translation, i.e., cellular processes that are conserved and unconditionally essential in most organisms.
Figure 4 illustrates the changes in distribution of essential genes between functional categories depending on their tendencies to be evolutionarily preserved. An initial bias in distribution of all categorized essential genes towards those involved with synthesis and processing of informational macromolecules increases dramatically at higher ERI values. The fraction of all essential genes contributed jointly by the functional categories PMS and NAM (Table 2) (∼30%) increases almost twofold (up to ∼60%) for a subset of essential genes with ERIs of >0.8, ultimately exceeding 90% as the ERI approaches 1.0.
FIG. 4.
Distribution of essential genes among functional categories as a function of ERI thresholds. Functional categories are color coded and specified by three-letter designations as in Table 2. Within every threshold group, each bar represents the fraction (percent plotted on y axis) of all categorized essential genes corresponding to the number of essential genes in a given category (x axis) with ERI values above the set threshold (z axis).
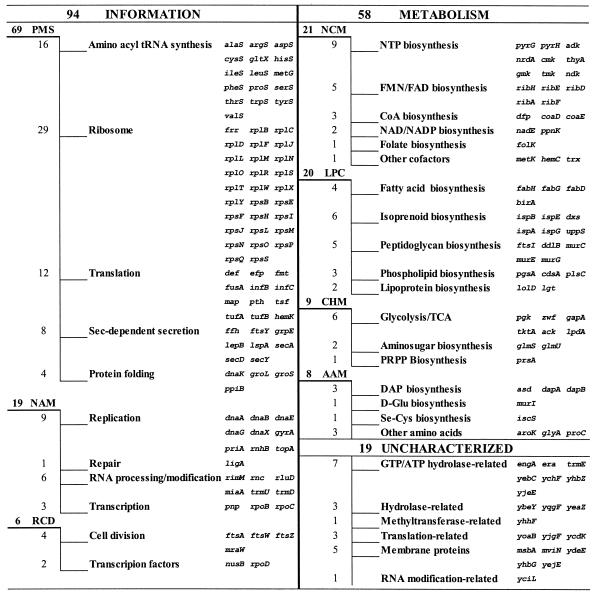
This analysis reveals two distinct classes of essential genes, which may be referred to as broadly preserved essential genes and species-specific essential genes. A subset of less than 180 genes (∼4% of the genome) with ERIs of >0.8 accounts for ∼25% of all of the essential genes revealed in this study, and it appears to provide an approximation of broadly preserved essential genes. Functional content analysis of this subset (Fig. 5) strongly supports the expectation that these genes represent universally and unconditionally essential constituents of cellular central machinery. This notion is in good agreement with available complete and partial gene essentiality datasets for Mycoplasma pneumoniae and Mycoplasma genitalium (15), Haemophilus influenzae (1), Staphylococcus aureus (7, 18), and Streptococcus pneumoniae (35). The overwhelming majority (70 to 87%) of assigned genes in these data, which correspond to E. coli genes listed in Fig. 5, appear to be essential (see Table S5 in the supplementary data for details). Of note, many of these broadly preserved essential genes, including those with yet undefined functions, may be considered potential broad-spectrum anti-infective drug targets (9, 29).
FIG. 5.
E. coli genes found to be essential and preserved in over 80% of diverse bacterial genomes (ERI > 0.8). These universal essential genes are grouped by functional categories (described in Table 2). NTP, nucleotide triphosphate; FMN, flavin mononucleotide; FAD, flavin adenine dinucleotide; CoA, coenzyme A; TCA, tricarboxylic acid cycle; PRPP, phosphoribosyl pyrophosphate.
In contrast, more than 75% of genes within the set of species-specific essential genes (which account for ∼30% of all essential E. coli genes with ERI values of <0.1) encode uncategorized proteins with poorly defined or completely unknown functions. Many of the genes with known functions within this class are related to transcription regulation, membrane transport, signaling, and other cellular processes whose essentiality is either strictly condition dependent or limited to a set of very specific needs of E. coli and closely related species.
Among the 263 essential genes marked in our analysis as uncategorized (see Table S1 in the supplementary data), 19 genes have specific functions assigned to them while 73 genes have putative assignments (according to SWISS-PROT and other public archives). Those include assignments indicating just an element of possible function, such as “probable GTP-binding protein” (ychF). For the remaining 171 genes, we were unable to find any reliable functional assignments. These genes may be qualified as essential unknowns (at least at the time when this analysis was performed). The list of these genes along with their respective ERI values is provided in the supplementary data (see Table S6). Only 10 (yciL, yjeE, ybeY, yebC, yjgF, ydeE, yoaB, yqgF, ycdK, and yhbC) of the essential unknowns (<6%) are broadly conserved in bacteria (ERIs of 0.8 to 1). In contrast, more than 60% of genes in this set are poorly conserved across our reference set of diverse genomes (108 genes with ERIs of 0 to 0.1). Less than half of them (42 genes) are conserved in most Enterobacteriaceae, while others are present only in E. coli and some closely related species.
System level analysis of essentiality data within topologic modules of E. coli metabolism.
It is widely recognized that the thousands of components of a living cell are dynamically interconnected, so that cellular functional properties are a result of the complex intracellular web of molecular interactions within the cell (14, 22, 23). This is perhaps most evident with intermediary metabolism, in which hundreds of metabolic substrates are densely integrated through biochemical reactions (17). Metabolic networks are organized into many small, highly connected topologic modules that combine in a hierarchical manner into larger, less cohesive units, with their numbers and degrees of clustering following a power law, as previously demonstrated for 43 reference organisms (28). Within E. coli, hierarchical modularity closely overlaps with known metabolic functions (28).
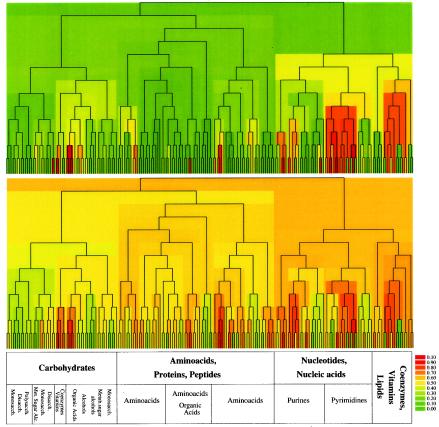
To comprehend the results of individual gene essentiality in the context of cellular system level functional organization, we projected the essentiality phenotype of metabolic enzymes onto a global topologic representation of the E. coli metabolic network (28). As shown in Fig. 6, the overall essentiality ratio of metabolic enzymes within the full metabolic network is relatively low, with essential enzymes limited to a subset of modules. Visual inspection of the figure indicates that while many metabolic modules are almost entirely nonessential, at the lowest hierarchical level several branches corresponding to small topologic modules appear to be essential, i.e., they are composed of biochemical reactions catalyzed by predominantly essential enzymes. Of these, the largest fractions are within the topologic modules related to nucleotide, coenzyme, and lipid metabolism. The pyrimidine metabolic module appears to contain the highest level of essential reactions.
FIG. 6.
The evolutionary retention and essentiality ratio of enzymes in the topologic modules of E. coli metabolism. The hierarchical tree derived from the topologic overlap matrix of E. coli metabolism that quantifies the relation between the various modules is shown, as previously described (28). The branches of the tree are color coded according to the fraction of essential enzymes (top panel) and the average ERI score of enzymes (bottom panel) catalyzing the biochemical reactions within a given topologic module. Red indicates a 100% essentiality/conservation ratio within a module. Note that essentiality is not uniformly distributed across all modules (branches), but we observe a few small modules with very high fractions of essential enzymes, while the majority of modules contain no or only a few essential enzymes. A similar segregation of modules with high evolutionary conservation is observed in the second panel, with their locations often correlating with those of the high essentiality modules. The predominant biochemical classes of substrates used to group the metabolites are shown. Polysacch., polysaccharide; disacch., disaccharide; monosacch., monosaccharide; met. sugar alc., metabolic sugar alcohols.
A significant correlation between essentiality and ERI values is apparent within metabolic modules, and many of the highly essential modules also contain metabolic enzymes with the highest ERI values (Fig. 6). Generally, essentiality and evolutionary retention of metabolic enzymes correlate, although exceptions are also evident as illustrated in detail for the pyrimidine module (supplementary data [see Fig. S3]). Pyrimidine metabolism, however, represents a special case in E. coli MG1655, since the rph-1 mutation in this strain depresses expression of the downstream pyrE gene (16). This strain is prototrophic for pyrimidines but grows significantly better in uracil-supplemented media. Although our studies were performed with rich media containing significant amounts of exogenous pyrimidines, the low level of pyrE transcription may have affected the ability of cells to efficiently adjust the relative levels of the pyrimidine nucleotides. This may explain the relatively high level of gene essentiality within the pyrimidine-related topologic module. These observations, however, may also reflect a hypothesized generic feature of metabolic networks: their limited ability to fully compensate for perturbations by reorganization of metabolic fluxes within evolutionarily conserved topologic modules.
DISCUSSION
A genetic footprinting technique was used to assess gene essentiality in E. coli K-12 across the entire genome under uniform growth conditions (logarithmic aerobic growth of strain MG1655 in enriched LB medium). This approach generated an internally coherent data set, which was examined at increasingly abstract levels to refine models of cellular organization. At the finest level, individual gene essentiality reveals basic physiologic information about cellular metabolism under specific growth conditions. At a more abstract level, the data can be used for focused comparative genomic analysis to define the core bacterial genetic repertoire, while at the highest level of abstraction, the data can be used to detect organizational principles of cellular networks.
Functional context analysis based on projection of the gene essentiality data across a whole-genome functional reconstruction (metabolic and nonmetabolic pathways and networks) provides a powerful way to refine and interpret the results of genetic footprinting. This type of analysis, previously described only for a limited set of metabolic pathways (9) and extended here to the whole-genome level, reveals a remarkable consistency between experimental observations and our present understanding of biochemical pathways and individual gene functions. Based on the overall consistency, one can resolve ambiguities, reconcile conflicting essentiality data, and even make tentative assignments for individual uncharacterized genes if they occur within well-known functional contexts (pathways).
Additionally, functional context analysis improves and extends our understanding of the systemic behavior of the cell at all levels: from individual genes and gene products to large functional systems and networks. Global projection of experimentally determined gene essentiality over a functional reconstruction model bridges the gap between two fundamentally different but related concepts: essential functions and essential genes. For example, essentiality data can distinguish functional (mutually complementing) and nonfunctional (noncomplementing) paralogs of genes with essential functional roles.
Analysis of essentiality data in a physiological context as a function of various factors and conditions, such as medium composition, aeration, growth phase, and temperature, etc., provides an opportunity to connect large functional modules with particular types of physiological states. Performing such analyses for a variety of conditions will provide critical support to systemic modeling efforts, such as flux-balance (6) and elementary mode analyses (34), and to our understanding of topologic modules (28). In this respect, the unexpected number of essential enzymes within the pyrimidine metabolic module in a pyrE-challenged E. coli strain reveals a significantly reduced ability of this module to tolerate additional gene inactivation, even in rich media. This suggests that the capacity for reorganization of metabolic fluxes within evolutionarily conserved, and presumably universally important, metabolic modules may be reduced, as a consequence either of their less evolved connectivity (37) or the performance of their functions at near optimality with corresponding innate fragility to uncommon error (5). The validity of these hypotheses will need to be tested by future experiments.
Acknowledgments
We thank W. Reznikoff for the gift of Tn5 transposase, L. Galtseva for design and implementation of the online supplementary data, and D. Frick for permission to reproduce the illustration in Fig. S2.
This work was supported by Integrated Genomics, Inc., and by grants from the National Institutes of Health and the Department of Energy to A.-L.B. and Z.N.O.
Footnotes
The author-arranged URLs for supplemental material provided in the article are no longer active. The supplemental material may now be found at http://tubic.tju.edu.cn/deg/ecoli or https://www.genome.wisc.edu/Gerdes2003/. This page was posted online on October 11, 2017 to give readers access to the new URLs.
REFERENCES
- 1.Akerley, B. J., E. J. Rubin, V. L. Novick, K. Amaya, N. Judson, and J. J. Mekalanos. 2002. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae Proc. Natl. Acad. Sci. USA 99:966-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, R. P., and J. R. Roth. 1978. Tandem chromosomal duplications in Salmonella typhimurium: fusion of histidine genes to novel promoters. J. Mol. Biol. 119:147-166. [DOI] [PubMed] [Google Scholar]
- 3.Badarinarayana, V., P. W. Estep III, J. Shendure, J. Edwards, S. Tavazoie, F. Lam, and G. M. Church. 2001. Selection analyses of insertional mutants using subgenic-resolution arrays. Nat. Biotechnol. 19:1060-1065. [DOI] [PubMed] [Google Scholar]
- 4.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Csete, M. E., and J. C. Doyle. 2002. Reverse engineering of biological complexity. Science 295:1664-1669. [DOI] [PubMed] [Google Scholar]
- 6.Edwards, J. S., M. Covert, and B. Palsson. 2002. Metabolic modelling of microbes: the flux-balance approach. Environ. Microbiol. 4:133-140. [DOI] [PubMed] [Google Scholar]
- 7.Forsyth, R. A., R. J. Haselbeck, K. L. Ohlsen, R. T. Yamamoto, H. Xu, J. D. Trawick, D. Wall, L. Wang, V. Brown-Driver, J. M. Froelich, K. G. C., P. King, M. McCarthy, C. Malone, B. Misiner, D. Robbins, Z. Tan, Z. Y. Zhu Zy, G. Carr, D. A. Mosca, C. Zamudio, J. G. Foulkes, and J. W. Zyskind. 2002. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus Mol. Microbiol. 43:1387-1400. [DOI] [PubMed] [Google Scholar]
- 8.Gasteiger, E., E. Jung, and A. Bairoch. 2001. SWISS-PROT: connecting biomolecular knowledge via a protein database. Curr. Issues Mol. Biol. 3:47-55. [PubMed] [Google Scholar]
- 9.Gerdes, S. Y., M. D. Scholle, M. D'Souza, A. Bernal, M. V. Baev, M. Farrell, O. V. Kurnasov, M. D. Daugherty, F. Mseeh, B. M. Polanuyer, J. W. Campbell, S. Anantha, K. Y. Shatalin, S. A. Chowdhury, M. Y. Fonstein, and A. L. Osterman. 2002. From genetic footprinting to antimicrobial drug targets: examples in cofactor biosynthetic pathways. J. Bacteriol. 184:4555-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 11.Goryshin, I. Y., J. Jendrisak, L. M. Hoffman, R. Meis, and W. S. Reznikoff. 2000. Insertional transposon mutagenesis by electroporation of released Tn5 transposition complexes. Nat. Biotechnol. 18:97-100. [DOI] [PubMed] [Google Scholar]
- 12.Graham, D. E., R. Overbeek, G. J. Olsen, and C. R. Woese. 2000. An archaeal genomic signature. Proc. Natl. Acad. Sci. USA 97:3304-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hare, R. S., S. S. Walker, T. E. Dorman, J. R. Greene, L. M. Guzman, T. J. Kenney, M. C. Sulavik, K. Baradaran, C. Houseweart, H. Yu, Z. Foldes, A. Motzer, M. Walbridge, G. H. Shimer, Jr., and K. J. Shaw. 2001. Genetic footprinting in bacteria. J. Bacteriol. 183:1694-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasty, J., D. McMillen, F. Isaacs, and J. J. Collins. 2001. Computational studies of gene regulatory networks: in numero molecular biology. Nat. Rev. Genet. 2:268-279. [DOI] [PubMed] [Google Scholar]
- 15.Hutchison, C. A., S. N. Peterson, S. R. Gill, R. T. Cline, O. White, C. M. Fraser, H. O. Smith, and J. C. Venter. 1999. Global transposon mutagenesis and a minimal mycoplasma genome. Science 286:2165-2169. [DOI] [PubMed] [Google Scholar]
- 16.Jensen, K. F. 1993. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J. Bacteriol. 175:3401-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeong, H., B. Tombor, R. Albert, Z. N. Oltvai, and A.-L. Barabási. 2000. The large-scale organization of metabolic networks. Nature 407:651-654. [DOI] [PubMed] [Google Scholar]
- 18.Ji, Y., B. Zhang, S. F. Van Horn, P. Warren, G. Woodnutt, M. K. Burnham, and M. Rosenberg. 2001. Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science 293:2266-2269. [DOI] [PubMed] [Google Scholar]
- 19.Jordan, I. K., I. B. Rogozin, Y. I. Wolf, and E. V. Koonin. 2002. Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res. 12:962-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamath, R. S., A. G. Fraser, Y. Dong, G. Poulin, R. Durbin, M. Gotta, A. Kanapin, N. Le Bot, S. Moreno, M. Sohrmann, D. P. Welchman, P. Zipperlen, and J. Ahringer. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421:231-237. [DOI] [PubMed] [Google Scholar]
- 21.Kim, S. K., J. Lund, M. Kiraly, K. Duke, M. Jiang, J. M. Stuart, A. Eizinger, B. N. Wylie, and G. S. Davidson. 2001. A gene expression map for Caenorhabditis elegans Science 293:2087-2092. [DOI] [PubMed] [Google Scholar]
- 22.Kitano, H. 2002. Computational systems biology. Nature 420:206-210. [DOI] [PubMed] [Google Scholar]
- 22a.Kobayashi, K., et al. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 100:4678-4683. [DOI] [PMC free article] [PubMed]
- 23.Koonin, E. V., Y. I. Wolf, and G. P. Karev. 2002. The structure of the protein universe and genome evolution. Nature 420:218-223. [DOI] [PubMed] [Google Scholar]
- 24.Kyrpides, N., R. Overbeek, and C. Ouzounis. 1999. Universal protein families and the functional content of the last universal common ancestor. J. Mol. Evol. 49:413-423. [DOI] [PubMed] [Google Scholar]
- 25.Neidhardt, F. C., F. Curtiss, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. E. Umbarger. 1996. Escherichia coli and Salmonella typhimurium cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 26.Overbeek, R., N. Larsen, T. Walunas, M. D'Souza, G. Pusch, E. Selkov, Jr., K. Liolios, V. Joukov, D. Kaznadzey, I. Anderson, A. Bhattacharyya, H. Burd, W. Gardner, P. Hanke, V. Kapatral, N. Mikhailova, O. Vasieva, A. Osterman, V. Vonstein, M. Fonstein, N. Ivanova, and N. Kyrpides. 2003. The ERGO(TM) genome analysis and discovery system. Nucleic Acids Res. 31:164-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ravasz, E., A. L. Somera, D. A. Mongru, Z. N. Oltvai, and A.-L. Barabási. 2002. Hierarchical organization of modularity in metabolic networks. Science 297:1551-1555. [DOI] [PubMed] [Google Scholar]
- 29.Rosamond, J., and A. Allsop. 2000. Harnessing the power of the genome in the search for new antibiotics. Science 287:1973-1976. [DOI] [PubMed] [Google Scholar]
- 30.Ross-Macdonald, P., P. S. Coelho, T. Roemer, S. Agarwal, A. Kumar, R. Jansen, K. H. Cheung, A. Sheehan, D. Symoniatis, L. Umansky, M. Heidtman, F. K. Nelson, H. Iwasaki, K. Hager, M. Gerstein, P. Miller, G. S. Roeder, and M. Snyder. 1999. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature 402:413-418. [DOI] [PubMed] [Google Scholar]
- 31.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2001. Comprehensive identification of conditionally essential genes in mycobacteria. Proc. Natl. Acad. Sci. USA 98:12712-12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider, B. L., A. K. Kiupakis, and L. J. Reitzer. 1998. Arginine catabolism and the arginine succinyltransferase pathway in Escherichia coli J. Bacteriol. 180:4278-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, V., D. Botstein, and P. O. Brown. 1995. Genetic footprinting: a genomic strategy for determining a gene's function given its sequence. Proc. Natl. Acad. Sci. USA 92:6479-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stelling, J., S. Klamt, K. Bettenbrock, S. Schuster, and E. D. Gilles. 2002. Metabolic network structure determines key aspects of functionality and regulation. Nature 420:190-193. [DOI] [PubMed] [Google Scholar]
- 35.Thanassi, J. A., S. L. Hartman-Neumann, T. J. Dougherty, B. A. Dougherty, and M. J. Pucci. 2002. Identification of 113 conserved essential genes using a high-throughput gene disruption system in Streptococcus pneumoniae. Nucleic Acids Res. 30:3152-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagner, A. 2000. Robustness against mutations in genetic networks of yeast. Nat. Genet. 24:355-361. [DOI] [PubMed] [Google Scholar]
- 38.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, R. W. Davis, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]