Abstract
The OmpD porin is the most abundant outer membrane protein in Salmonella enterica serovar Typhimurium and represents about 1% of total cell protein. Unlike the case with the less abundant OmpC and OmpF porins, the stoichiometry of OmpD in the outer membrane does not change in response to changes in osmolarity. The abundance of OmpD increases in response to anaerobiosis and decreases in response to low pH, conditions encountered by serovar Typhimurium during the infection of its murine host. By constructing an operon fusion of the lacZY genes with the ompD promoter, we show that the abundance of OmpD in the outer membrane is regulated primarily at the level of transcription and is subject to catabolite repression. In response to anaerobiosis, the abundance of OmpD in the outer membrane also appears to be controlled posttranscriptionally by a function dependent on Fnr.
The Salmonella enterica serovars Typhi and Typhimurium cause lethal systemic infections in humans and mice, respectively. Serovar Typhi causes typhoid fever in humans, which kills more than 600,000 people annually (18). Because these serovars are close genetic relatives, the study of the pathogenic mechanisms by which serovar Typhimurium causes a lethal infection in mice serve as a model for understanding the pathogenic mechanisms of serovar Typhi in its exclusive human host.
Although these serovars are closely related, the determination of the complete genome sequences of strains of serovar Typhi (19) and serovar Typhimurium (13) show that about 20% of their genes are different and unique to each species. These differences are clustered mainly into islets and islands of genes that represent insertions or substitutions in one genome sequence with respect to the other (2).
Recently we have shown that serovar Typhi has a deletion of an islet of genes present in serovar Typhimurium and in all other serovars of S. enterica that we have tested. This region, which maps at minute 33.7 on the serovar Typhimurium chromosome, includes two adjacent genes, ompD and yddG, which are not present on the serovar Typhi genome (21, 22). The ompD gene encodes the most abundant protein in the outer membrane, a porin similar in primary amino acid sequence to the major porins OmpC, OmpF, and PhoE. Together, OmpD, OmpC, and OmpF account for about 1 × 105 to 2 × 105 porin molecules per cell (17). Under favorable growth conditions, the OmpD porin represents about half of the porins and has a stoichiometry comparable to that of the ribosome. The yddG gene encodes an inner membrane protein that is a member of the drug/metabolite transporter superfamily of proteins. We have shown that both ompD and yddG are required for resistance to methyl viologen. These results suggest that YddG may function together with OmpD in the efflux of methyl viologen from the cytoplasm to the extracellular space and that the porins may play critical roles in the efflux of toxic substrates, as well as in the influx of substrates across the outer membrane (21).
Like Escherichia coli K-12, both serovar Typhimurium and serovar Typhi also have OmpC and OmpF as abundant proteins in their outer membranes. The E. coli and serovar Typhimurium OmpF and OmpC porins are regulated reciprocally by the osmolarity of the growth medium. With high osmolarity, OmpC is more abundant in the bacterial outer membrane, whereas OmpF is less abundant; the converse is true at low osmolarity (5, 9). Regulation of the relative abundance of OmpC and OmpF depends on the ompB locus, which includes the genes envZ and ompR, encoding a sensor kinase and its cognate response regulator, respectively. The phosphorylated form of OmpR binds sites within the ompF and ompC promoters to regulate the transcription of these genes (6, 15).
To understand how the expression of the OmpD porin is regulated in wild-type serovar Typhimurium strain LT2, we began by examining environmental factors known to be involved in the regulation of expression of the major porins of E. coli K-12, including osmolarity, oxygen availability, and low pH (1, 5, 16). To extend our analysis, we also made mutants of LT2 defective in potential regulators of ompD expression. We constructed isogenic derivatives of LT2 carrying combinations of mutations in the fnr (formerly oxrA), ompD, envZ, cya, and crp genes by generalized transduction with P22 HT105/1 int201 (23), using donor strains MST2944 (ompD159::Tn10 galE496), MST2400 (envZ1005::Mud-P), MST2970 (fnr2::Tn10 leu485), MST2596 (cya961::Tn10), and MST2882 (crp773::Tn10). (A significant part of this work was presented as the undergraduate thesis of C.A.S. as part of the requirements for the Master of Science degree in biochemistry from the University of Chile.)
Osmolarity does not affect OmpD regulation.
To measure the relative abundance of OmpD in the serovar Typhimurium outer membrane, we prepared outer membrane fractions from cells grown at 37°C in Luria-Bertani (LB) medium as described by Schnaitman (24) and modified by Lobos and Mora (11), resolved the proteins in these fractions by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis in 12.5% slab gels, and revealed these proteins by staining with Coomassie blue. Figure 1 shows that when wild-type strain LT2 is grown aerobically in LB medium at 37°C, it produces four abundant porins, OmpC, OmpF, OmpD, and OmpA, among which OmpD is the most abundant. For serovar Typhimurium, as for E. coli K-12, the abundance of OmpC relative to that of OmpF in the outer membrane decreases in response to decreasing osmolarity. The production of both OmpC and OmpF by serovar Typhimurium is dependent on envZ. A mutant derivative of LT2, SC2, with an envZ1005::Mud-P insertion makes neither OmpC nor OmpF, as reported previously (10). In contrast, the relative abundance of OmpD in the outer membrane does not change in response to changes in osmolarity, nor is it dependent on envZ function. This is also the case when cells are grown in LB medium at very high osmolarity (10% sucrose or 0.3 M NaCl; data not shown).
FIG. 1.
The abundance of OmpD in the outer membrane of serovar Typhimurium is not affected by changes in osmolarity or by a mutation in the envZ gene. At the top, the gel shows that otherwise isogenic envZ+ and envZ1005::Mud-P strains have similar proportions of OmpD in their outer membranes under conditions of moderate (+ NaCl) and low (− NaCl) osmolarity. Outer membrane proteins were prepared from cells grown in rich (LB) medium with and without NaCl. Only the portions of 12.5% polyacrylamide gels with bands corresponding to the major porins are shown. At the bottom, the levels of β-galactosidase activity made by otherwise isogenic ompD1::Mud-J (SC7) (open bars) and ompD1::Mud-J envZ1005::Mud-P (SC9) (shaded bars) strains are compared. Cultures were grown to an optical density at 600 nm of 0.2 and permeabilized with chloroform and 0.1% SDS, and β-galactosidase activity was measured by the method of Miller (14). Activities are expressed in Miller units, 103(A420 − [1.75 × A550])/(ml × min × A600−1), and represent the averages of at least three independent determinations.
The abundance of OmpD in the outer membrane increases in response to anaerobiosis.
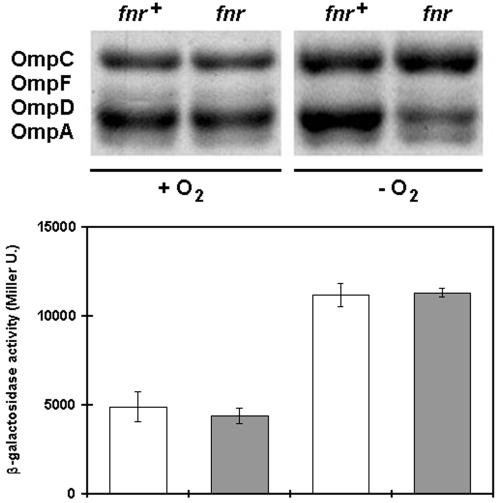
During the infection of its murine host, serovar Typhimurium must survive environments low in oxygen and pH. Because the outer membrane provides the first line of defense against unfavorable extracellular environments, the ability to change the composition of the outer membrane rapidly in response to environmental changes may be critical for virulence. Previously, we have found that the expression of a variety of serovar Typhimurium outer membrane proteins is induced by anaerobiosis (unpublished results). As shown in Fig. 2, in response to anaerobiosis, the relative abundance of OmpD in the outer membrane of wild-type cells roughly doubles with respect to other outer membrane proteins. Because OmpD is the major porin present in about 105 copies/cell under aerobic conditions, this twofold increase in OmpD corresponds to a dramatic change in the protein composition of the outer membrane. This result suggests that transcription of the ompD gene may be induced when oxygen is limiting.
FIG. 2.
The abundance of OmpD in the outer membrane of serovar Typhimurium doubles during anaerobic growth, dependent on fnr function. At the top are shown the outer membrane proteins prepared from otherwise isogenic fnr+ and fnr2::Tn10 cells grown in rich medium with (+ O2) and without (− O2) oxygen. At the bottom, the levels of transcription of the ompD1::Mud-J operon fusion in the same genetic backgrounds (fnr+ [open bars] and fnr2::Tn10 [shaded bars]) are represented as β-galactosidase activities, as in the legend to Fig. 1. Note that although the level of transcription of the operon fusion doubles in both wild-type and fnr mutant genetic backgrounds (bottom), the abundance of OmpD in the outer membrane doubles only in the wild-type genetic background (top right) under anaerobic conditions.
In the absence of oxygen, serovar Typhimurium responds by utilizing alternative terminal electron acceptors or by activating fermentation pathways. Many of these responses to anaerobiosis are dependent on the global transcriptional regulator encoded by fnr (8, 25). Figure 2 shows that, in contrast with its wild-type parent, strain SC3 (fnr2::Tn10) does not have a higher percentage of OmpD in its outer membrane under anaerobic growth conditions. This result suggests that the transcription of the ompD gene is dependent on fnr function.
The relative abundance of OmpD that we observe in the outer membrane is the end product of a series of events dependent on the rates of ompD transcription and OmpD translation and the efficiency of insertion of OmpD in the outer membrane (as well as the stability of intermediates in this process). Because gene expression is regulated primarily at the level of transcription initiation, we tested whether the rate of transcription of the ompD gene is dependent on fnr. To do this, we constructed an operon fusion in which the ompD promoter is fused to the lacZY structural genes, by isolating a mutant with an ompD::Mud-J insertion.
The increase in abundance of OmpD, but not in the rate of ompD transcription, in response to anaerobiosis is dependent on Fnr.
To make a serovar Typhimurium ompD::Mud-J mutant, we used the method of transitory cis complementation to generate a pool of mutants of serovar Typhimurium with Mud-J insertions (7). The donor serovar Typhimurium strain TT10288 (hisD9953::Mud-J hisD9941::Mud-1) was grown in LB medium with ampicillin (30 μg/ml) and kanamycin sulfate (40 μg/ml) to exponential density and infected with phage P22 HT105/1 int201. Ten pools of each of 1,600 Kanr transductants of recipient strain LT2 colonies were made on LB plates (1.5% agar), and each pool was infected with P22 to make a series of secondary transducing lysates. These secondary donor lysates were used to transduce the Tetr recipient strain SC1 (ompD159::Tn10), and Tets Kanr recombinants that form blue colonies on plates with fusaric acid and chlortetracycline to counterselect resistance to tetracycline (12) and 40-μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside were selected. Overnight cultures were prepared from four Lac+ Kanr Tets colonies and the profiles of their outer membrane proteins were analyzed by SDS-polyacrylamide gel electrophoresis, and two were found to be missing OmpD in their outer membrane. The ompD1::Mud-J mutation in one of these strains was backcrossed into a wild-type LT2 recipient to yield strain SC7. To confirm that the Mud-J insertion in SC7 is in ompD, the junctions of the Mud-J insertion with the chromosome were amplified using the PCR with primer pairs MUR (5′-TTCGCATTTATCGTGAAACGCTTCC) and OMPD1 (GACAAAGACAAAACCCGTT) as well as MUL (TTCGTACTTCAAGTGAAT) and OMPD2 (CGTCCAGCAGGTTGATTTT). The sizes of the products of these amplifications, as well as the results of Southern analysis, show that the Mud-J insertion is in the center of ompD.
Serovar Typhimurium does not have the lac operon. This fact enables us to use lacZ as a reporter for transcription in both wild-type and mutant serovar Typhimurium strains by introducing Mud-J insertions. For example, the otherwise isogenic envZ+ and envZ1005::Mud-P strains whose outer membrane proteins are shown in Fig. 1 show no detectable β-galactosidase activity. When the ompD1::Mud-J insertion is introduced into these genetic backgrounds, the recombinant ompD1::Mud-J and ompD1::Mud-J envZ1005::Mud-P strains are found to produce similar levels of β-galactosidase activity under conditions of both low and high osmolarity. This result supports the idea that the transcription of ompD does not change in response to changes in osmolarity.
As shown in Fig. 2, we compared the levels of β-galactosidase activity made by otherwise isogenic ompD1::Mud-J fnr+ (SC7) and ompD1::Mud-J fnr2::Tn10 (SC8) strains under aerobic and anaerobic growth conditions. Under anaerobic growth conditions, the β-galactosidase activity shown by the mutant ompD1::Mud-J strain is slightly more than twice that shown by the same strain under aerobic conditions, a result that reflects what we observe when we assay the abundance of OmpD protein in the outer membrane of the wild-type parent of this strain under anaerobic versus aerobic growth conditions. In contrast, when we compare the levels of β-galactosidase activity shown by the ompD1::Mud-J fnr2::Tn10 strain under aerobic and anaerobic growth conditions, we find a different result. The fnr mutation has no effect on the expression of β-galactosidase activity from the operon fusion under anaerobic conditions compared to expression under aerobic conditions. These results show that fnr function does not play a direct role in the regulation of ompD transcription. Rather, the increase in the abundance of OmpD in the outer membrane in response to anaerobiosis must be due to an Fnr-dependent posttranscriptional regulatory mechanism. One possible step at which this fnr-dependent posttranscriptional control may occur is at the level of control of mRNA stability. The turnover of the transcripts of many genes is slowed under anaerobic conditions (4), and the extended half-life of mRNA species observed under anaerobic conditions may depend on an fnr-regulated function. Alternatively, the rate of degradation of OmpD may depend on an fnr-regulated function.
OmpD expression decreases in response to low pH.
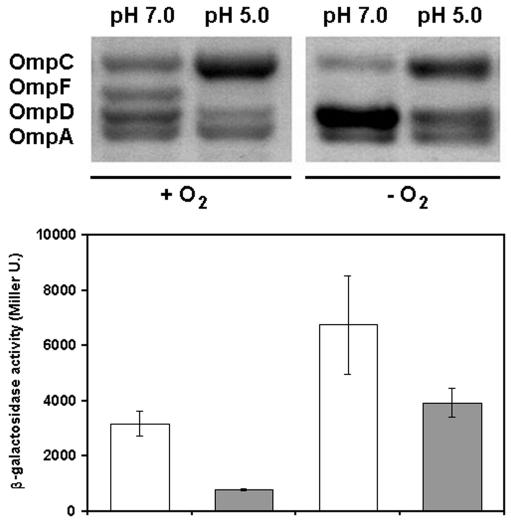
As shown in Fig. 3, when serovar Typhimurium is grown in LB medium at pH 5.0 versus pH 7.0, there is an increase in the abundance of OmpC relative to that of OmpF at this lower pH, consistent with previously published results (3). Also, we find that OmpD is present in the outer membrane in significantly lower abundance at the lower pH. This down-regulation of the abundance of OmpD in response to low pH is most likely due to decreased transcription of the ompD gene. At the lower pH, the strain with the ompD1::Mud-J reporter fusion shows about one-half the β-galactosidase activity as at the higher pH. Thus, the relative change in abundance of OmpD in response to pH is regulated at the level of transcription initiation. When cells are grown anaerobically at pH 5.0, however, the abundance of OmpD remains about the same as in cells grown aerobically at pH 7.0 (Fig. 3). This result shows that the mechanism of fnr-dependent posttranscriptional control of OmpD in response to anaerobiosis apparently acts independently of the mechanism of transcriptional control that results in a decrease in its abundance in response to low pH.
FIG. 3.
The abundance of OmpD in the outer membrane of serovar Typhimurium decreases in response to lower pH. At the top are shown the profiles of outer membrane proteins prepared from cells grown in LB medium at pH 7.0 or at pH 5.0, aerobically (+ O2) or anaerobically (− O2). At the bottom, β-galactosidase activities made by the ompD1::Mud-J strain grown under the same conditions (pH 7.0 [open bars] and 5.0 [shaded bars]) are presented as in Fig. 1 and 2.
Transcription from the ompD promoter is activated by the cAMP/Crp complex.
Catabolite repression, or the glucose effect, is a global regulatory system that coordinates the balanced expression of genes involved in the catabolism of alternative carbon sources. Because the OmpD porin, like the OmpC and OmpF porins, may be involved in the influx of nutrients including saccharides, we also tested whether ompD expression is regulated by catabolite repression. When the mutant ompD1::Mud-J strain is grown in defined, minimal medium (E medium) (0.02 g of MgSO4 · 7H2O/liter, 2 g of citric acid · H2O/liter, 13.1 g of Na2HPO4 · 3H2O/liter, 3.3 g of NaNH4HPO4 · 4H2O/liter) supplemented with glucose (2 g/liter) or glycerol (2 g/liter), the levels of β-galactosidase activity made by the mutant under the two different growth conditions differ by about twofold (4,200 ± 700 versus 7,700 ± 1,500 Miller units, respectively), suggesting that the ompD promoter is sensitive to catabolite repression.
Catabolite repression in enteric gram-negative bacteria is dependent on the interaction of cyclic AMP (cAMP) with cAMP receptor protein (Crp) to form an active DNA-binding complex that activates transcription from some promoters and represses transcription at others. Assembly of this transcription factor requires the products of the cya and crp genes, encoding adenylyl cyclase and cAMP receptor protein, respectively (20). We constructed two double mutants of serovar Typhimurium carrying the ompD1::Mud-J mutation paired with either the cya961::Tn10 or crp773::Tn10 mutation. When we grow the ompD1::Mud-J cya961::Tn10 double mutant (SC12) aerobically in minimal glucose medium, we find that this mutant shows extremely low levels of β-galactosidase activity, less than 1/10 the level of activity made by the otherwise isogenic wild-type parent. This effect can be reversed by the addition of 1 mM cAMP to the medium. Similarly, the ompD::Mud-J crp773::Tn10 double mutant (SC13) also shows extremely low levels of β-galactosidase activity when grown in minimal glucose medium, a phenotype that cannot be masked by the addition of cAMP (Table 1). Thus, the transcription of the ompD gene is dependent on positive activation by the cAMP/Crp complex. Direct measurements of the abundance of OmpD in outer membrane preparations of the same cells are consistent with this interpretation (data not shown). The sequence of the region upstream of the serovar Typhimurium ompD gene (bp 1655173 to 1656261 in GenBank NC_003197) includes a near-consensus cAMP/Crp binding site located 160 to 139 bp upstream of the ATG start codon of ompD; this may be the site required for the positive regulation of the ompD gene by Crp.
TABLE 1.
Transcription of the ompD gene is regulated by catabolite repressiona
| Genotype | β-Galactosidase activity
|
|
|---|---|---|
| + glucose | + glucose, cAMP | |
| ompD1::Mud-J | 4,200 ± 700 | ND |
| ompD1::Mud-J cya961::Tn10 | 170 ± 50 | 3,800 ± 400 |
| ompD1::Mud-J crp773::Tn10 | 240 ± 50 | 270 ± 90 |
Serovar Typhimurium strains carrying the ompD1::Mud-J fusion in wild-type and mutant cya or crp genetic backgrounds were grown to exponential density at 37°C in minimal medium supplemented with 0.2% glucose or 0.2% glucose plus 1 mM cAMP, and β-galactosidase activities were determined as described in the text. Values (β-galactosidase activities in Miller units) represent the averages and standard deviations of at least three determinations. ND, not determined.
The serovar Typhimurium ompD gene is regulated similarly in serovar Typhi.
Prior to the determination of the complete and draft genome sequences of many serovars of S. enterica, we found that the ompD gene is present in all serovars of S. enterica with the exception of serovar Typhi (22), results consistent with all currently available genome sequence data. As a complementary approach to understand the function of ompD, we constructed a serovar Typhi hybrid carrying the serovar Typhimurium ompD gene. This serovar Typhi hybrid expresses the OmpD porin in its outer membrane and has a gain-of-function phenotype, increased resistance to methyl viologen (21, 22).
To determine how the ompD gene is regulated in the serovar Typhi genetic background, we constructed eight derivatives of serovar Typhi with the genotypes ompD+, ompD159::Tn10, ompD+ fnr2::Tn10, ompD+ envZ1005::Mud-P, ompD1::Mud-J, ompD1::Mud-J fnr2::Tn10, ompD1::Mud-J envZ1005::Mud-P, ompD1::Mud-J cya961::Tn10, and ompD1::Mud-J crp773::Tn10, by transferring alleles from serovar Typhimurium donor strains to serovar Typhi recipients using P22-mediated generalized transduction. We repeated each of the experiments described above for serovar Typhimurium with these serovar Typhi strains and find that ompD is regulated in serovar Typhi in a manner identical to its regulation in serovar Typhimurium (data not shown).
In summary, we have found that the relative abundance of the OmpD porin in the outer membrane of serovar Typhimurium is regulated by a multiplicity of environmental factors, including anaerobiosis, lower pH, and catabolite repression. Unlike the porins OmpC and OmpF, the abundance of OmpD does not change under different conditions of osmolarity and does not depend on the envZ-ompR two-component regulatory system.
What is the physiological function of OmpD? We have shown that the ompD gene is necessary for resistance to the toxic compound methyl viologen (21). More recently, we have shown that at least two other porins, OmpA and OmpW, also are critical for resistance to toxic compounds (unpublished results). The regulation of the ompD gene may provide us with some clues to the function of OmpD. Because the transcription of ompD is dependent on catabolite repression and increases in response to anaerobiosis, we suspect that OmpD may be involved in the efflux of toxic compounds made during the catabolism of alternative, poorer carbon sources.
Acknowledgments
We are indebted to Stanley Maloy for generous gifts of bacterial strains.
This work was funded by FONDECYT (Chile) grant 1020485 (to G.C.M.) and NIH Fogarty Center Senior International Fellowship TW05645 (to P.Y.). C.A.S. was supported by fellowships from CONICYT and DIPUC.
REFERENCES
- 1.Delihas, N., and S. Forst. 2001. MicF: an antisense RNA gene involved in response of Escherichia coli to global stress factors. J. Mol. Biol. 313:1-12. [DOI] [PubMed] [Google Scholar]
- 2.Edwards, R. A., G. J. Olsen, and S. R. Maloy. 2002. Comparative genomics of closely related salmonellae. Trends Microbiol. 10:94-99. [DOI] [PubMed] [Google Scholar]
- 3.Foster, J., Y. K. Park, L. Bang, K. Kamer, H. Betts, H. Hall, and E. Shaw. 1994. Regulatory circuits involved with pH-regulated gene expression in Salmonella typhimurium. Microbiology 140:341-352. [DOI] [PubMed] [Google Scholar]
- 4.Georgellis, D., T. Barlow, S. Arvidson, and A. von Gabain. 1993. Retarded RNA turnover in Escherichia coli: a means of maintaining gene expression during anaerobiosis. Mol. Microbiol. 9:375-381. [DOI] [PubMed] [Google Scholar]
- 5.Hall, M. N., and T. J. Silhavy. 1981. The ompB locus and the regulation of the major outer membrane porin proteins of Escherichia coli K12. J. Mol. Biol. 146:1-15. [DOI] [PubMed] [Google Scholar]
- 6.Head, C. G., A. Tardy, and L. J. Kenney. 1998. Relative binding affinities of OmpR and OmpR-phosphate at the ompF and ompC regulatory sites. J. Mol. Biol. 281:857-870. [DOI] [PubMed] [Google Scholar]
- 7.Hughes, K. T., and J. R. Roth. 1988. Transitory cis complementation: a method for providing transposition functions to defective transposons. Genetics 119:9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jamieson, D. J., and C. F. Higgins. 1986. Two genetically distinct pathways for transcriptional regulation of anaerobic gene expression in Salmonella typhimurium. J. Bacteriol. 168:389-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jovanovich, S. B., M. Martinell, M. T. Record, Jr., and R. R. Burgess. 1988. Rapid response to osmotic upshift by osmoregulated genes in Escherichia coli and Salmonella typhimurium. J. Bacteriol. 170:534-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liljestrom, P., M. Luokkamaki, and E. T. Palva. 1987. Isolation and characterization of a substitution mutation in the ompR gene of Salmonella typhimurium LT2. J. Bacteriol. 169:438-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lobos, S. R., and G. C. Mora. 1991. Alterations in the electrophoretic mobility of OmpC due to variations in the ammonium persulfate concentration in sodium dodecylsulfate-polyacrylamide gel electrophoresis. Electrophoresis 12:448-450. [DOI] [PubMed] [Google Scholar]
- 12.Maloy, S. R., and W. D. Nunn. 1981. Selection for loss of tetracycline resistance by Escherichia coli. J. Bacteriol. 145:1110-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 14.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Mizuno, T., and S. Mizushima. 1990. Signal transduction and gene regulation through the phosphorylation of two regulatory components: the molecular basis for the osmotic regulation of the porin genes. Mol. Microbiol. 4:1077-1082. [DOI] [PubMed] [Google Scholar]
- 16.Mizuno, T., M. Y. Chou, and M. Inouye. 1984. A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript micRNA. Proc. Natl. Acad. Sci. USA 81:1966-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikaido, H., and M. Vaara. 1985. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 49:1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pang, T., M. Levine, B. Ivanoff, J. Wain, and B. Finlay. 1998. Typhoid fever—important issues still remain. Trends Microbiol. 6:131-133. [DOI] [PubMed] [Google Scholar]
- 19.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 20.Saier, M. H., Jr. 1998. Multiple mechanisms controlling carbon metabolism in bacteria. Biotechnol. Bioeng. 58:170-174. [DOI] [PubMed] [Google Scholar]
- 21.Santiviago, C. A., J. A. Fuentes, S. M. Bueno, A. N. Trombert, A. A. Hidalgo, L. T. Socias, P. Youderian, and G. C. Mora. 2002. The Salmonella enterica sv. Typhimurium smvA, yddG and ompD porin genes are required for the efficient efflux of methyl viologen. Mol. Microbiol. 46:687-698. [DOI] [PubMed] [Google Scholar]
- 22.Santiviago, C. A., C. S. Toro, S. A. Bucarey, and G. C. Mora. 2001. A chromosomal region surrounding the ompD porin gene marks a genetic difference between Salmonella typhi and the majority of Salmonella serovars. Microbiology 147:1897-1907. [DOI] [PubMed] [Google Scholar]
- 23.Schmieger, H. 1972. Phage P22 mutants with increased or decreased transducing abilities. Mol. Gen. Genet. 119:75-88. [DOI] [PubMed] [Google Scholar]
- 24.Schnaitman, C. A. 1971. Solubilization of the cytoplasmic membrane of Escherichia coli by Triton X-100. J. Bacteriol. 108:545-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strauch, K. L., J. B. Lenk, B. L. Gamble, and C. G. Miller. 1985. Oxygen regulation in Salmonella typhimurium. J. Bacteriol. 161:673-680. [DOI] [PMC free article] [PubMed] [Google Scholar]