Abstract
Many eubacteria are resistant to the toxic oxidizing agent potassium tellurite, and tellurite resistance involves diverse biochemical mechanisms. Expression of the iscS gene from Geobacillus stearothermophilus V, which is naturally resistant to tellurite, confers tellurite resistance in Escherichia coli K-12, which is naturally sensitive to tellurite. The G. stearothermophilus iscS gene encodes a cysteine desulfurase. A site-directed mutation in iscS that prevents binding of its pyridoxal phosphate cofactor abolishes both enzyme activity and its ability to confer tellurite resistance in E. coli. Expression of the G. stearothermophilus iscS gene confers tellurite resistance in tellurite-hypersensitive E. coli iscS and sodA sodB mutants (deficient in superoxide dismutase) and complements the auxotrophic requirement of an E. coli iscS mutant for thiamine but not for nicotinic acid. These and other results support the hypothesis that the reduction of tellurite generates superoxide anions and that the primary targets of superoxide damage in E. coli are enzymes with iron-sulfur clusters.
The cytoplasm is a reducing environment, and many oxidizing agents can cause cellular damage by covalently modifying intracellular targets. Among these, the tellurite oxyanion (TeO32−) is toxic to most microbes. Tellurite can cross the gram-negative membrane using systems involved in phosphate uptake (28) and is a substrate for nitrate reductase, which can reduce the anion to tellurium, which is insoluble and nontoxic (3).
To understand the basis of tellurite toxicity at the molecular level, we are exploring the mechanisms by which microbes are resistant to this anion. Several bacteria are naturally resistant to potassium tellurite, and both the genetic and biochemical bases of this resistance appear to be diverse. Tellurite resistance determinants are found both in bacterial chromosomes and in plasmids (22, 27).
The gram-positive bacterium Geobacillus stearothermophilus V, formerly Bacillus stearothermophilus V (16), is naturally resistant to high levels of tellurite (30, 31). Our work has focused on the identification and characterization of G. stearothermophilus genes that confer tellurite resistance when expressed in Escherichia coli. We have constructed gene libraries from G. stearothermophilus in high-copy-number plasmids, transformed sensitive E. coli hosts with these libraries, and selected for tellurite-resistant clones. Using this strategy, Vásquez et al. have found that the cysK gene of G. stearothermophilus confers a tellurite resistance phenotype in E. coli (30, 31). CysK catalyzes the synthesis of cysteine from O-acetyl serine and sulfide as substrates, the terminal, rate-limiting step in cysteine biosynthesis. The cysK genes from other microorganisms have also been shown to confer tellurite resistance in E. coli (1, 17).
In this paper, we show that the expression of G. stearothermophilus cysteine desulfurase (IscS), a second enzyme involved in cysteine metabolism, also confers tellurite resistance in E. coli. Cysteine desulfurases sack the sulfur atom from cysteine to construct and repair [Fe-S] clusters in protein substrates that, in turn, catalyze essential redox reactions in critical metabolic pathways. We have sequenced the G. stearothermophilus iscS gene, expressed its product in E. coli, and purified the IscS enzyme to homogeneity. We show that tellurite resistance depends on the activity of the IscS enzyme, supporting the hypothesis that essential proteins with iron-sulfur [Fe-S] clusters are among the main targets of the oxidative damage caused by tellurite in E. coli.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
G. stearothermophilus V was from our collection (30). The E. coli strains used in the study were derivatives of K-12 and included JM109 {endA1 recA1 gyrA96 thi hsdR17 (rK− mK+) relA1 supE44 Δ(lac-proAB) [F′ traD36 proA+B+ lacIqZM15]} and E. coli JM109(DE3) {endA1 recA1 gyrA96 thi hsdR17 (rK− mK+) relA1 supE44 Δ(lac-proAB) [F′ traD36 proA+B+ lacIqZM15 (DE3)]} from Promega, PK6311 (iscS::Kanr zff-208::Tn10) and its otherwise isogenic parent RZ4500 from Patricia Kiley, University of Wisconsin, and QC774 (F− ΔlacU169 rpsL sodA::lacZ49 sodB::kanΔ2) from Akiko Nishimura, National Institute of Genetics, Japan. Cells were grown at 37°C in Luria broth (LB) or M9 (minimal) medium (19) supplemented with 0.2% glucose, thiamine (2 μg/ml), and/or nicotinic acid (12.5 μg/ml). When appropriate, tetracycline (10 μg/ml), kanamycin (40 μg/ml), and/or ampicillin (100 μg/ml) was added to the medium. MICs of potassium tellurite and hydrogen peroxide were determined in liquid medium after 24 h of growth as described previously (4).
Plasmid constructions and genetic manipulations.
To clone the fragment of the G. stearothermophilus genome with the iscS gene, G. stearothermophilus DNA was cleaved with HindIII and ligated to plasmid vector pSP72 (Promega). Ligation mixes were electroporated into E. coli JM109, and tellurite-resistant clones were selected (30, 31). The sequence of the 3,461-bp G. stearothermophilus insert in p2VH (a plasmid that confers resistance to tellurite) was determined. DNA fragments used to subclone ORF687, ORF1200, and ORF963 of G. stearothermophilus were amplified using PCR with primer pair 5′-GGAATTCCATATGTATAAGGATGATCAGGAAATAGA and 5′-ACCCAAGCTTCTAGAGCGTGATCAGGTCTTTGT, primer pair 5′-GGAATTCCATATGAATCTTGAACAAATAAGAAAAGATACC and 5′-ACCCAAGCTTTCATTGCTGTCCCTCTTTCCTTAT, and primer pair 5′-GGAATTCCATATGAATAAATTTTTGCTTGAATCTGC and 5′-ACCCAAGCTTTCATATCAGAATCTTCCCATCCT, respectively (boldface characters indicate NdeI [CATATG] and HindIII [AAGCTT] restriction sites). PCR products were cleaved with NdeI and HindIII and ligated to the same sites of pET21b (Novagen) to generate plasmids pJT687, pJT1200, and pJT963, respectively. pJT1200-Pr, which has the G. stearothermophilus iscS gene with its own promoter, was made by amplifying iscS with primers ACCCAAGCTTAAAACCGGGAATGGCATTCAC and ACCCAAGCTTCATTGCTGTCCCTCTTTCCTTAT, cleaving the product with HindIII, and ligating the product to plasmid pBluescript.
DNA fragments of G. stearothermophilus from which 90, 150, and 210 bp of the 3′ end of the iscS gene were deleted were amplified by PCR and ligated to the NdeI and HindIII sites of pET21b to construct plasmids pJT1200(Δ90), pJT1200(Δ150), and pJT1200(Δ210). A mutant version of the iscS gene predicted to encode a product in which lysine-213 is replaced by alanine (K213A) was made by amplifying chromosomal DNA as the template with primer pair GGAATTCCATATGAATCTTGAACAAATAAGAAAAGATACC and CTCAGGACCTCTAAGCCATGCTCTCCCGCACGCTGCAAG and primer pair CTTGCAGCGTGCGGGAGAGCATGGCTTAGAGGTCCTGAG and ACCCAAGCTTTCATTGCTGTCCCTCTTTCCTTAT (underlined characters represent the Ala codon and the complementary sequence in the other mutagenic oligonu-cleotide, respectively). Products were annealed and reamplified with the first and last of these primers, and the product was cleaved with NdeI and HindIII and ligated to plasmid pET21b to make plasmid pJT1200K213A. Standard molecular biology procedures were performed as described previously (19).
Overexpression and purification of cysteine desulfurase.
E. coli JM109(DE3) cells carrying pJT1200 were grown in LB with ampicillin at 37°C to an optical density at 600 nanometers of 0.6. Expression of iscS was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for an additional 5 h. Cells were harvested by centrifugation at 4,500 × g for 10 min and stored at −20°C or used immediately with identical results. Induced cells showed a characteristic intense yellow color due to the presence of the pyridoxal phosphate (PLP) cofactor in the overproduced cysteine desulfurase.
Cell pellets were thawed on ice and resuspended in 5 ml of buffer A (20 mM Tris-HCl [pH 8.0], 5% glycerol). Cells were sonicated in the presence of 10 μg of phenylmethylsulfonyl fluoride/ml and centrifuged at 13,000 × g for 15 min, and supernatants were diluted with an equal volume of buffer A and incubated at 70°C for 20 min with gentle swirling. After chilling on ice, extracts were centrifuged at 13,000 × g for 15 min and supernatants were applied to a series of AffiGel Blue (Bio-Rad; 2 ml), CM-Sepharose (Whatman; 4 ml), and DEAE-Sepharose (Whatman; 3 ml) columns. IscS flowed through the first two columns and adsorbed to the DEAE-Sepharose column, which was washed with buffer B (10 mM Tris-HCl [pH 8.0], 5% glycerol) prior to elution using a linear gradient of 0 to 0.15 M NaCl in buffer B. Protein in eluted fractions (1.0 ml) was monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and UV spectrophotometry; fractions with IscS were pooled and applied to a BioHTP (Bio-Rad; 1 ml) column for concentration, washed, eluted with 0.25 M potassium phosphate buffer, and dialyzed against buffer A. Using bovine serum albumin as a standard, protein concentrations were determined with a Bradford protein assay kit (Bio-Rad). The purity of IscS was estimated to be >98% as judged by SDS-PAGE.
The apparent molecular mass of the IscS monomer was determined by SDS-PAGE using alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa), ovalbumin (45 kDa), carbonic anhydrase (29 kDa), lysozyme (14 kDa), and cytochrome c (12.4 kDa) (Sigma) as prestained protein standards. The apparent molecular mass of the native enzyme in buffer A containing 250 mM NaCl was estimated by size exclusion chromatography using a Sephadex G 50-100 column (1 by 45 cm) and the same standards.
Antibody preparation and Western immunoblotting.
Antiserum against IscS was prepared by immunizing a female Rockefeller mouse intraperitoneally. Polyacrylamide gel slices containing IscS were fragmented, incubated with 0.5 ml of 0.9% NaCl for 24 h at 4°C, and centrifuged at 10,000 × g for 5 min at 4°C. A total of 100 μl of the supernatant (with about 100 μg of IscS) was mixed with an equal volume of Freund's incomplete adjuvant (Gibco-BRL) for each of three injections given to the mouse at 10-day intervals. At 1 week after the last injection, serum was obtained from blood by low-speed centrifugation.
Western blotting experiments were performed as described previously (14). Membranes were blocked with a suspension of 1% powdered milk in IS buffer (15 mM sodium phosphate, 1% NaCl, 0.002% KCl, pH 7.4) for 45 min, incubated with mouse anti-IscS antiserum (diluted 1:2,500 in IS buffer) for 45 min, washed three times with 0.02% Tween 20 in IS buffer for 5 min, incubated with alkaline phosphatase-conjugated goat anti-mouse immunoglobulin G serum (diluted 1:10,000) for 45 min, and washed three times for 5 min. Blots were developed in 20 ml of 0.1 M Tris-HCl (pH 9.5)-0.1 M NaCl-5 mM MgCl2-0.1 mg of Nitro Blue Tetrazolium/ml-0.05 mg of BCIP (5-bromo-4-chloro-3-indolylphosphate)/ml for 10 min at 37°C, washed with water, and dried. The anti-IscS antiserum cross-reacts with only one protein band in extracts from E. coli JM109(DE3)/pJT1200; this band is not present in extracts from E. coli JM109(DE3).
Enzyme assays for cysteine desulfurase activity.
The activity of G. stearothermophilus IscS was determined by measuring the rate of production of thiocyanate from reaction mixtures (100 μl) containing 50 mM Tris-HCl (pH 8.0), 10 mM cysteine, 1 mM MgCl2, 10 mM KCN, and enzyme, as described previously (32). After incubation for 2 h at 50°C, 250 μl of 15% formaldehyde and 750 μl of ferric nitrate [6.67% Fe(NO3)3 · 9H2O (wt/vol), 8.67% HNO3 (vol/vol) in H2O] were added, the mixtures were centrifuged at 13,000 × g for 3 min, and the concentrations of ferric thiocyanate were determined by absorbance at 460 nm. One unit of enzyme activity is defined as the amount of enzyme that catalyzes the formation of 1 nmol of thiocyanate/min; specific activities are given as units per milligram of total protein.
Nucleotide sequence accession number.
The sequence determined for the 3,461-bp G. stearothermophilus insert in p2VH has been assigned GenBank accession no. AF533655.
RESULTS
Tellurite leads to oxidative damage in E. coli.
Several different mechanisms have been proposed to account for the toxicity of tellurite. Tellurium may replace sulfur and/or selenium in critical metabolites or enzymes, abating their essential functions (10). Alternatively, tellurite is a strong oxidizing agent that may cause general oxidative damage (25) or tellurite may cause specific damage to critical thiol groups or [Fe-S] clusters present in essential enzymes (29).
We favor a combination of the last two mechanisms named, in which tellurite toxicity results from the generation of O2− radicals formed upon tellurite reduction (27) which in turn causes specific damage to one or more critical proteins with [Fe-S] clusters. Two lines of evidence support this hypothesis. As shown in Table 1, a mutant of E. coli deficient in superoxide dismutase activity was hypersensitive to tellurite. Because superoxide is the substrate of superoxide dismutase, this result demonstrates genetically that tellurite toxicity results from the generation of superoxide radicals. Consistent with this genetic result, when E. coli cells were grown anaerobically they were less sensitive to tellurite and this toxic compound had a MIC 10-fold higher than that seen when cells were grown aerobically (data not shown).
TABLE 1.
MICs of potassium tellurite for E. coli strains used in this work
| E. coli strain | Plasmid | MIC (μg/ml) |
|---|---|---|
| JM109 | None | 1.25 |
| pSP72 | 1.25 | |
| pBluescript | 1.25 | |
| p2VH | 12.5 | |
| pJT1200-Pr | 12.5 | |
| JM109(DE3) | None | 1.25 |
| pET21b | 1.25 | |
| pJT687 | 1.25 | |
| pJT963 | 1.25 | |
| pJT1200 | 12.5-25.0 | |
| pJT1200-Δ90 | 1.25 | |
| pJT1200-Δ150 | 1.25 | |
| pJT1200-Δ210 | 1.25 | |
| pJT1200/K213A | 1.25 | |
| PK6311 (iscS) | None | 0.25 |
| pBluescript | 0.25 | |
| pJT1200-Pr | 12.5 | |
| RZ4500 | None | 1.25 |
| QC774 (sodA sodB) | None | 0.125-0.25 |
| pJT1200-Pr | 12.5 |
What are the principal targets of superoxide damage in E. coli? It has been shown that O2− radicals can damage a subset of proteins with [Fe-S] clusters, including dihydroxy acid dehydratase (8, 12), aconitase (11), and others. In hyperbaric oxygen-treated cells, both O2 and O2− (as well as other oxidants) inactivate dihydroxy acid dehydratase (2,3-dihydroxy acid hydrolyase EC 4.2.1.9) through the destruction of its sensitive [Fe-S] cluster, which can be then reactivated in vitro (7). [Fe-S] clusters are important components of additional proteins that participate in critical intracellular processes, including nucleotide biosynthesis, amino acid biosynthesis, DNA repair, transcriptional regulation, and energy metabolism (9).
When E. coli is grown in minimal medium, the principal target of superoxide damage is dihydroxy-acid dehydratase (8, 12). This result suggests that when E. coli is grown in rich medium, other, essential enzymes with [Fe-S] clusters are the principal targets of superoxide damage.
The G. stearothermophilus V iscS gene confers tellurite resistance in E. coli.
G. stearothermophilus is naturally resistant to potassium tellurite. To identify potential genes involved in this resistance, we constructed libraries of G. stearothermophilus genomic DNA in several different plasmid vectors. One of these libraries was made with plasmid vector pSP72 and electroporated into E. coli strain JM109. Subclones with a tellurite resistance phenotype were selected as colonies able to grow on plates with 10 μM potassium tellurite. As shown in Table 1, the host E. coli K-12 strain JM109, as well as JM109 with plasmid pSP72, is sensitive to the toxic anion tellurite, which has a MIC of 1.25 μg/ml for this strain. In contrast, strain JM109 carrying plasmid p2VH, a derivative of pSP72 containing an insert of 3.5 kb, is resistant to tellurite (MIC of 12.5 μg/ml).
The sequence of the insert in this plasmid contains three large open reading frames (ORFs) of 687, 1,200, and 963 bp. BLASTp analysis (2) showed that the predicted products of these ORFs were most similar to a Bacillus anthracis A2012 metallopeptidase (53% identity/69% similarity) (18), a putative cysteine desulfurase of Methanosarcina mazei Goe1 (35% identity/57% similarity) (6), and a Methanopyrus kandleri AV19 aminodipeptidase (29% identity/42% similarity) (24).
To determine which of these three ORFs conferred tellurite resistance, DNA fragments corresponding to each ORF were amplified using the PCR and ligated to plasmid vector pET21b. Derivatives of E. coli JM109(DE3) with the recombinant plasmids, pJT687, pJT1200 and pJT963, were tested for tellurite resistance. Plasmid pET21b placed the expression of a cloned gene under the transcriptional control of the coliphage T7 early promoter. In the host strain E. coli JM109(DE3), the addition of IPTG induced the expression of T7 RNA polymerase, which in turn directed the high-level expression of a subcloned gene. In the absence of IPTG, there was a basal level of transcription of genes cloned in the proper orientation in this plasmid from fortuitous promoters overlapping the early T7 promoter and a low level of expression of their products. As shown in Table 1, tellurite had a low MIC of 1.25 μg/ml for the E. coli K-12 host strain JM109(DE3) as well as for JM109(DE3) carrying pET21b (and similar to the tellurite MIC for JM109). Among the three plasmids with subcloned inserts, only plasmid pJT1200 (with the iscS gene) conferred tellurite resistance (Table 1).
To support our gene assignments, derivatives of JM109(DE3) carrying each plasmid derivative of pET21b were grown in liquid culture to the exponential phase and the production of proteins encoded by each inserted ORF was induced by the addition of IPTG. Samples were taken before and after induction; intracellular proteins were resolved by SDS-PAGE, and their patterns were revealed by staining with Coomassie blue. After induction, each protein profile contained a single additional band corresponding in apparent molecular mass (35, 45, and 25 kDa) to the molecular mass of the predicted product of each ORF (34.5, 44.8, and 25.2 kDa, respectively) (data not shown).
As we show below by the results of a combination of biochemical and genetic experiments, the iscS gene, ORF1200, encodes a cysteine desulfurase. Unlike the case for other microbes, the G. stearothermophilus iscS gene does not appear to be within an operon containing other genes involved in de novo [Fe-S] cluster formation. In the genomes of most microbes, homologues of iscS are adjacent to homologues of the icsU gene; however, this is not the case for several gram-positive bacterial genomes, including that of G. stearothermophilus.
The G. stearothermophilus IscS protein is a cysteine desulfurase.
The G. stearothermophilus iscS gene, subcloned in plasmid pJT1200, codes for a protein that has 35% identity and 57% similarity with the putative cysteine desulfurase of M. mazei and lower identities with other cysteine desulfurases. We purified G. stearothermophilus IscS to homogeneity and demonstrated that it has cysteine desulfurase activity.
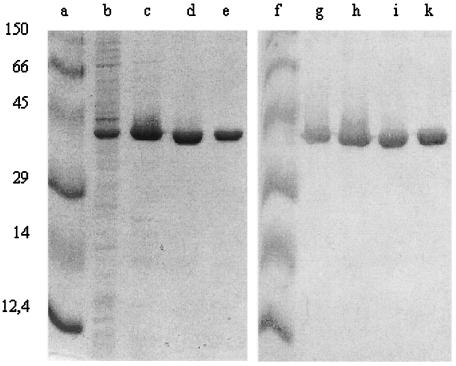
To purify G. stearothermophilus IscS, we grew E. coli strain JM109(DE3) with plasmid pJT1200 to exponential density and induced the expression of the iscS gene from the T7 promoter by the addition of IPTG. As shown in Fig. 1, after induction, a band of the apparent molecular mass of the predicted product of iscS represented about 10 to 20% of the total protein in soluble cell extracts made from induced cells. Because G. stearothermophilus is a thermophile, the next step in this purification involved the incubation of soluble cell extracts at 70°C for 20 min. This step eliminated almost 75% of the starting total protein in these extracts without an appreciable loss of IscS protein or cysteine desulfurase activity. Subsequent column chromatography steps resulted in enzyme preparations that were >98% pure (Fig. 1).
FIG. 1.
Purification of G. stearothermophilus V IscS protein. (Left panel) Electrophoretogram of proteins in the crude extract (lane b), after heat treatment (lane c), after DEAE-Sepharose treatment (lane d), and after BioHTP concentration (lane e). Prestained protein standards are indicated to the left (lane a); their molecular masses are given in kilodaltons. (Right panel) Western blot of the same gel stained with mouse anti-IscS as the primary antibody.
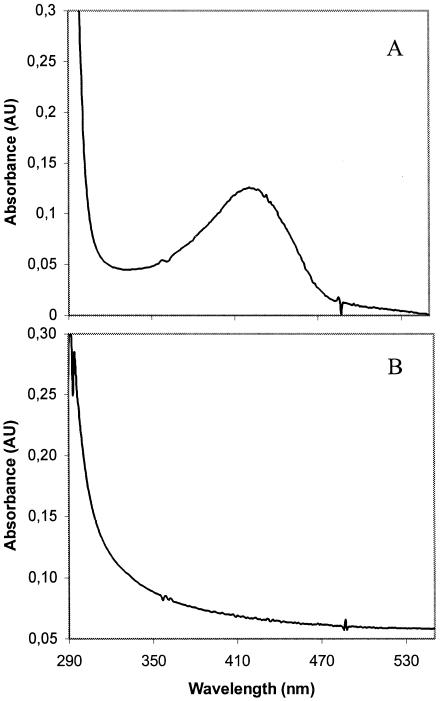
The amino terminus of the purified IscS protein (MNLEQIRKDTPLHKKYSYIN) was determined by Edman degradation and matched precisely the predicted primary sequence of the product of iscS. The native form of the enzyme is a homodimer with an apparent molecular mass of 93 to 97 kDa, as judged by size exclusion chromatography using a Sephadex G50-100 column (data not shown). The monomer has an apparent molecular mass of 45 kDa, as analyzed by SDS-PAGE (Fig. 1, left panel). This cysteine desulfurase belongs to the α family of PLP-dependent enzymes and exhibited the UV-visible spectrum characteristic of other cysteine desulfurases, with an absorbance maximum for PLP centered at 420 ± 2 nm (Fig. 2A).
FIG. 2.
UV absorption spectra of purified IscS enzymes from G. stearothermophilus V. Protein solutions (1.5 mg/ml) of wild-type IscS (A) and mutant IscS/K213A (B) were prepared in 10 mM Tris-HCl (pH 8.0)-5% glycerol and scanned using an HP Agilent 8453 spectrophotometer. Wild-type IscS has an absorption maximum at 421 nm characteristic of many PLP-dependent enzymes.
To demonstrate that IscS has cysteine desulfurase activity, we used an assay in which the production of sulfur from the desulfuration of l-cysteine in a first step was measured as the formation of thiocyanate from sulfur, formaldehyde, and nitrate in a second step (32), with the modification that the initial step in this assay was made at 50°C. At this temperature, extracts prepared from the host E. coli strain had no detectable cysteine desulfurase activity. As shown in Table 2, the IscS protein shows no significant loss of activity after both heat treatment and chromatography steps, which together result in about an eightfold purification of the enzyme.
TABLE 2.
Purification of G. stearothermophilus V IscS protein
| Fraction | Protein (mg) | Activity (U) | Specific activity (U/mg) | Yield (%) | Purifi- cation (fold) |
|---|---|---|---|---|---|
| Crude extract | 275 | 38.2 | 0.14 | 100 | 1.0 |
| Treated at 70°C | 76 | 45.8 | 0.60 | 120 | 4.3 |
| Column chromatographya | 28 | 35.0 | 1.25 | 92 | 8.9 |
See Materials and Methods for details. Enzyme units are expressed as nanomoles of KSCN/minute at 50°C.
Mutations in the G. stearothermophilus iscS gene abolish cysteine desulfurase activity.
To confirm that the iscS gene is responsible for tellurite resistance in E. coli and that tellurite resistance is a consequence of cysteine desulfurase activity, we constructed a mutant derivative of the iscS gene without enzyme activity. We found that plasmids from which 90, 150, and 210 bp of the 3′ end of the iscS gene were deleted did not confer resistance to tellurite in E. coli (Table 1) or produce IscS activity in crude extracts (data not shown). Microscopic analysis of these extracts showed that the induced mutant proteins formed inclusion bodies. These results suggest that the carboxyl terminus of IscS is essential for its proper folding, dimerization, or function.
Then we elected to make a directed change of residue Lys-213, a residue that likely binds the PLP cofactor, because it is conserved in the predicted primary sequences of the cysteine desulfurases from both gram-positive and -negative bacteria as well as Saccharomyces cerevisiae (32). The codon for this residue was replaced with a codon for alanine to make the mutant iscS-K213A gene, which was then cloned into plasmid expression vector pET21b and introduced into host E. coli JM109(DE3).
Table 1 shows that a strain carrying this plasmid did not confer increased tellurite resistance in E. coli. This strain was grown to exponential density, production of the mutant K213A protein was induced with IPTG, and the mutant K213A protein was purified to homogeneity. To purify this protein we used the procedure used for the wild-type enzyme with the exception that the step involving the initial heat treatment of the crude lysate was omitted, because the mutant protein could not be recovered if this step was included. Unlike extracts containing the wild-type IscS protein, extracts with the mutant K213A protein did not have an intense yellow color (consistent with the idea that residue Lys213 is critical for PLP adduction). The UV-visible spectrum of the purified mutant protein was missing the absorbance peak characteristic of PLP-containing enzymes (Fig. 2B), and enzyme assays showed that the purified mutant protein had less than 10% of the specific activity of the wild-type protein (data not shown).
The G. stearothermophilus V iscS gene partially complements an E. coli iscS defect.
Cysteine desulfurases are required for the assembly of [Fe-S] clusters in other enzymes, including dehydratases, aconitase (11), and fumarase (13). In E. coli K-12, the predominant cysteine desulfurase activity is the product of the E. coli iscS gene. Under aerobic growth conditions, mutations in E. coli iscS confer auxotrophy for thiamine and nicotinic acid in minimal medium and slow its rate of growth in rich medium (20, 23). To confirm by a genetic test that the G. stearothermophilus IscS protein is a cysteine desulfurase, we asked whether a plasmid with the G. stearothermophilus iscS gene could complement an E. coli iscS defect.
As shown in Table 1, PK6311, a strain of E. coli with a substitution of a kanamycin resistance cassette for the iscS gene (21), was hypersensitive to tellurite. In contrast, a recombinant derivative of this strain with plasmid pJT1200-Pr (carrying both the G. stearothermophilus iscS gene and its upstream promoter) was resistant to tellurite (Table 1). In addition, the recombinant strain retained the auxotrophy for nicotinic acid but not for thiamine (Table 3). These results show that the G. stearothermophilus iscS gene can complement only one of the two auxotrophies of the E. coli iscS mutant. Although the overproduction of another cysteine desulfurase in E. coli has been shown to be toxic (26), it is unlikely that the constitutive expression of the G. stearothermophilus iscS gene from plasmid pJT1200-Pr is toxic under these conditions. An iscS+ E. coli strain with this plasmid grew in rich medium at the same rate as otherwise isogenic strains that included the plasmid vector used to clone the G. stearothermophilus iscS gene or that had no plasmid (unpublished results).
TABLE 3.
Complementation by the G. stearothermophilus V iscS gene of the auxotrophic requirement for thiamine of an E. coli PK6311 iscS mutant
| Strain/plasmid | Growth mediuma
|
|||
|---|---|---|---|---|
| M9 | M9 plus Thi | M9 plus NA | M9 plus Thi plus NA | |
| RZ4500 | + | + | + | + |
| RZ4500/pBluescript | + | + | + | + |
| RZ4500/pJT1200-Pr | + | + | + | + |
| PK6311 | − | − | − | + |
| PK6311/pBluescript | − | − | − | + |
| PK6311/pJT1200-Pr | − | − | + | + |
M9 minimal plates were supplemented with thiamine (Thi) and/or nicotinic acid (NA) as indicated and incubated at 37°C for 24 h; growth was scored as the ability to form single colonies. +, growth; −, no growth.
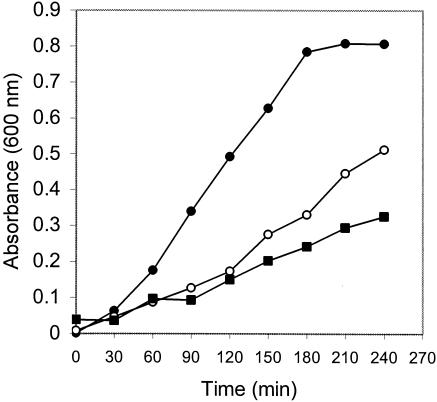
As shown in Fig. 3, expression of G. stearothermophilus iscS in E. coli increased the growth rate of an E. coli iscS mutant in rich medium but the complemented mutant strain did not grow as fast as an otherwise isogenic parental strain. Plasmids carrying the E. coli iscS gene also complemented incompletely this phenotype of the E. coli iscS mutant. For E. coli, the substitution of the kanamycin resistance cassette for the iscS gene may have pleiotropic effects on the expression of other downstream genes (including iscU, iscA, hscB, hscA, and fdx) involved in [Fe-S] cluster synthesis and repair in the same transcription unit (20).
FIG. 3.
Expression of the G. stearothermophilus V iscS gene partially complements the slow-growth phenotype of an E. coli iscS mutant. Growth curves were determined in LB medium at 37°C. •, E. coli RZ4500 (iscS+); ▪, E. coli PK6311 (iscS::Kan); ○, E. coli PK6311 (iscS::Kan) with plasmid pJT1200-Pr, which expresses the G. stearothermophilus V iscS gene.
Furthermore, as was the case for an E. coli iscS mutant, expression of G. stearothermophilus iscS in an E. coli sodA sodB double mutant conferred tellurite resistance (Table 1). This result argues that the presence of O2− radicals caused specific damage to one or more critical proteins with [Fe-S] clusters. In addition, we found that although E. coli strains with plasmids that expressed G. stearothermophilus iscS acquired increased resistance to tellurite, they did not acquire increased resistance to peroxide (data not shown). Taken together, these results support the hypothesis that tellurite generates O2− radicals which in turn cause specific damage to one or more critical proteins with [Fe-S] clusters.
DISCUSSION
Unlike E. coli K-12, the thermophilic, gram-positive bacterium G. stearothermophilus V can grow in the presence of high concentrations of potassium tellurite. In this paper, we have shown that the G. stearothermophilus iscS gene may be one of the genes responsible for the resistance phenotype. G. stearothermophilus IscS is a homodimer of about 92 kDa with a monomer molecular mass of 45 kDa (Fig. 1, left panel) and has temperature and pH optima of 50°C and 8.0 (data not shown). IscS (due to its bound PLP cofactor) exhibited a UV-visible spectrum (Fig. 2A) similar to those reported for other cysteine desulfurases (7, 32), and site-directed mutagenesis of iscS showed that residue Lys-213 (conserved among the primary sequences of other cysteine desulfurases) is necessary for PLP binding and enzyme activity (Fig. 2). The cysteine desulfurase activity of IscS is required for tellurite resistance in E. coli, because a recombinant E. coli strain that expressed a mutant K213A enzyme did not show increased tellurite resistance.
Why does the overexpression of G. stearothermophilus IscS confer tellurite resistance in E. coli? Three other metabolic enzymes have been shown to contribute to tellurite resistance in eubacteria: nitrate reductase (3), thiopurine methyltransferase (5), and cysteine synthase (1, 15, 31). Both nitrate reductase (3) and thiopurine methyltransferase (5) catalyze covalent modifications of tellurite which result in the direct detoxification of this strong oxidizing agent. The overexpression of cysteine synthase results in increased resistance to tellurite in E. coli (31), likely because cysteine synthase immediately precedes cysteine desulfurase in the pathway for de novo [Fe-S] cluster biosynthesis. Cysteine synthase catalyzes the rate-limiting step in the biosynthesis of cysteine, a substrate of cysteine desulfurase, which may be a rate-limiting step in the pathway for de novo [Fe-S] cluster biosynthesis. Alternatively, iron availability may be rate limiting for this step in [Fe-S] cluster formation. These results support the idea that the primary targets for damage by superoxide radicals are one or more essential proteins with [Fe-S] clusters.
The simplest version of this hypothesis is that the primary targets for oxidative damage caused by tellurite in E. coli are essential proteins containing [Fe-S] clusters. Expression of G. stearothermophilus iscS in an E. coli iscS mutant conferred increased resistance to tellurite, suggesting that G. stearothermophilus V IscS can replenish or repair these essential target proteins. An E. coli iscS mutant showed increased sensitivity to tellurite, arguing that E. coli IscS also replenishes or repairs these essential targets. However, mutations in the E. coli iscS gene are not lethal. An E. coli iscS mutant was able to grow in rich medium, arguing that if this hypothesis is correct, E. coli must make a cysteine desulfurase in addition to IscS that can insert [Fe-S] clusters into these essential target proteins.
The E. coli K-12 genome includes three genes, iscS, sufS, and nifS, predicted to encode cysteine desulfurases. These enzymes use as their substrates not only cysteine (and/or selenocysteine) but also the large number of proteins that accept [Fe-S] clusters. Therefore, members of the small family of cysteine desulfurases must have broad, overlapping protein substrate specificities. This conclusion is supported by the recent finding that pseudorevertants of mutants with an iscS defect overexpress the suf operon and that isc suf double mutants are likely synthetically lethal (26).
We have shown that although the cysteine desulfurase from G. stearothermophilus V can complement the thiamine auxotrophy, one of the metabolic defects due to a mutation in the E. coli iscS gene, it cannot complement the auxotrophy of this mutant for nicotinic acid and cannot fully restore the slow-growth phenotype of an E. coli iscS mutant grown in rich medium. On the other hand, it is surprising that G. stearothermophilus IscS, which shares little sequence similarity with E. coli IscS, was able to complement a subset of the defective functions in an E. coli iscS mutant. Thus, the cysteine desulfurases are likely to have different spectra of specificities for their protein substrates. We are now testing whether mutations in sufS confer increased sensitivity to tellurite, because this redundant desulfurase may also contribute to the kinetics of insertion or repair of [Fe-S] clusters in these essential target proteins.
Acknowledgments
We thank James Imlay for his critical reading of the manuscript and the Centro de Síntesis y Análisis de Biomoléculas, Universidad de Chile, Santiago, Chile, for performing the amino-terminal sequence analysis of G. stearothermophilus IscS.
J.C.T. was supported by a doctoral fellowship from the DAAD (Germany), and M.A.A. and D.E.F. were supported by doctoral fellowships from MECESUP (Chile). This work was supported by grants 1990917 and 1030234 from FONDECYT (Chile) to C.C.V. and by grant GM53392 from the National Institutes of Health to P.Y.
REFERENCES
- 1.Alonso, G., C. Gomes, C. González, and V. Rodríguez Lemoine. 2000. On the mechanism of resistance to channel-forming colicins (PacB) and tellurite, encoded by plasmid Mip233 (IncHI3). FEMS Microbiol. Lett. 192:257-261. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S., W. Gish, W. Miller, E. Myers, and D. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Avazéri, C., R. Turner, J. Pommier, J. Weiner, G. Giordano, and A. Verméglio. 1997. Tellurite reductase activity of nitrate reductase is responsible for the basal resistance of Escherichia coli to tellurite. Microbiology 143:1181-1189. [DOI] [PubMed] [Google Scholar]
- 4.Chiong, M., E. González, R. Barra, and C. Vásquez. 1988. Purification and biochemical characterization of tellurite-reducing activities from Thermus thermophilus HB8. J. Bacteriol. 170:3269-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cournoyer, B., S. Watanabe, and A. Vivian. 1998. A tellurite-resistance genetic determinant from phytopathogenic pseudomonads encodes a thiopurine methyltransferase: evidence of a widely conserved family of methyltransferases. Biochim. Biophys. Acta 1397:161-168. [DOI] [PubMed] [Google Scholar]
- 6.Deppenmeier, U., A. Johann, T. Hartsch, R. Merkl, R. A. Schmitz, R. Martinez-Arias, A. Henne, A. Wiezer, S. Baumer, C. Jacobi, H. Bruggemann, T. Lienard, A. Christmann, M. Bomeke, S. Steckel, A. Bhattacharyya, A. Lykidis, R. Overbeek, H. P. Klenk, R. P. Gunsalus, H. J. Fritz, and G. Gottschalk. 2002. The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J. Mol. Microbiol. Biotechnol. 4:453-461. [PubMed] [Google Scholar]
- 7.Flint, D. H., J. F. Tuminello, and T. Miller. 1996. Studies on the synthesis of the Fe-S cluster of dihydroxy-acid dehydratase in Escherichia coli crude extract. Isolation of O-acetylserine sulfhydrylases A and B and beta-cystathionase based on their ability to mobilize sulfur from cysteine and to participate in Fe-S cluster synthesis. J. Biol. Chem. 271:16053-16067. [DOI] [PubMed] [Google Scholar]
- 8.Flint, D. H., E. S. Randall, J. F. Tuminello, B. D. Lusiak, and O. R. Brown. 1993. The inactivation of dihydroxy-acid dehydratase in Escherichia coli treated with hyperbaric oxygen occurs because of the destruction of its Fe-S cluster, but the enzyme remains in the cell in a form that can be reactivated. J. Biol. Chem. 34:25547-25552. [PubMed] [Google Scholar]
- 9.Frazzon, J., and D. R. Dean. 2001. Feedback regulation of iron-sulfur cluster biosynthesis. Proc. Natl. Acad. Sci. USA 98:14751-14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garberg, P., L. Engman, V. Tolmachev, H. Lundqvist, R. Gerdes, and I. Cotgreave. 1999. Binding of tellurium to hepatocellular selenoproteins during incubation with inorganic tellurite: consequences for the activity of selenium-dependent glutathione peroxidase. Int. J. Biochem. Cell Biol. 31:291-301. [DOI] [PubMed] [Google Scholar]
- 11.Gardner, P. R., and I. Fridovich. 1991. Superoxide sensitivity of the Escherichia coli aconitase. J. Biol. Chem. 266:19328-19333. [PubMed] [Google Scholar]
- 12.Kuo, C. F., T. Mashino, and I. Fridovich. 1987. α,β-Dihydroxyisovalerate dehydratase. A superoxide-sensitive enzyme. J. Biol. Chem. 262:4724-4727. [PubMed] [Google Scholar]
- 13.Liochev, S. I., and I. Fridovich. 1993. Modulation of the fumarases of Escherichia coli in response to oxidative stress. Arch. Biochem. Biophys. 301:379-384. [DOI] [PubMed] [Google Scholar]
- 14.Matsudaira, P. T. (ed.). 1989. A practical guide to protein and peptide purification for microsequencing. Academic Press, Inc., San Diego, Calif.
- 15.Moore, M., and S. Kaplan. 1992. Identification of intrinsic high-level resistance to rare-earth oxides and oxyanions in members of the class Proteobacteria: characterization of tellurite, selenite, and rhodium sesquioxide reduction in Rhodobacter sphaeroides. J. Bacteriol. 174:1505-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nazina, T. N., T. P. Tourova, A. B. Poltaraus, E. V. Novikova, A. A. Grigoryan, A. E. Ivanova, A. M. Lysenko, V. V. Petrunyaka, G. A. Osipov, S. S. Belyaev, and M. V. Ivanov. 2001. Taxonomic study of aerobic thermophilic bacilli: descriptions of Geobacillus subterraneus gen. nov., sp. nov. and Geobacillus uzenensis sp. nov. from petroleum reservoirs and transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus thermoleovorans, Bacillus kaustophilus, Bacillus thermoglucosidasius and Bacillus thermodenitrificans to Geobacillus as the new combinations G. stearothermophilus, G. thermocatenulatus, G. thermoleovorans, G. kaustophilus, G. thermoglucosidasius, and G. thermodenitrificans. Int. J. Syst. Evol. Microbiol. 51:433-446. [DOI] [PubMed] [Google Scholar]
- 17.O'Gara, J., M. Gomelsky, and S. Kaplan. 1997. Identification and molecular genetic analysis of multiple loci contributing to high-level tellurite resistance in Rhodobacter sphaeroides 2.4.1. Appl. Environ. Microbiol. 63:4713-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Read, T. D., S. L. Salzberg, M. Pop, M. Shumway, L. Umayam, L. Jiang, E. Holtzapple, J. D. Busch, K. L. Smith, J. M. Schupp, D. Solomon, P. Keim, and C. M. Fraser. 2002. Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science 296:2028-2033. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Schwartz, C. J., O. Djaman, J. A. Imlay, and P. J. Kiley. 2000. The cysteine desulfurase, IscS, has a major role in in vivo Fe-S cluster formation in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:9009-9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwartz, C. J., J. L. Giel, T. Patschkowski, C. Luther, F. J. Ruzicka, H. Beinert, and P. J. Kiley. 2001. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc. Natl. Acad. Sci. USA 98:14895-14900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silver, S., and L. Phung. 1996. Bacterial heavy metal resistance: new surprises. Annu. Rev. Microbiol. 50:753-789. [DOI] [PubMed] [Google Scholar]
- 23.Skovran, E., and D. M. Downs. 2000. Metabolic defects caused by mutations in the iscS gene cluster in Salmonella enterica serovar Typhimurium: implications for thiamine synthesis. J. Bacteriol. 182:3896-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slesarev, A. I., K. V. Mezhevaya, K. S. Makarova, N. N. Polushin, O. V. Shcherbinina, V. V. Shakhova, G. I. Belova, L. Aravind, D. A. Natale, I. B. Rogozin, R. L. Tatusov, Y. I. Wolf, K. O. Stetter, A. G. Malykh, E. V. Koonin, and S. A. Kozyavkin. 2002. The complete genome of hyperthermophile Methanopyrus kandleri AV19 and monophyly of archaeal methanogens. Proc. Natl. Acad. Sci. USA 99:4644-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Summers, A., and G. Jacoby. 1977. Plasmid-determined resistance to tellurium compounds. J. Bacteriol. 129:276-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi, Y., and U. Tokomuto. 2002. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J. Biol. Chem. 277:28380-28383. [DOI] [PubMed] [Google Scholar]
- 27.Taylor, D. 1999. Bacterial tellurite resistance. Trends Microbiol. 7:111-115. [DOI] [PubMed] [Google Scholar]
- 28.Tomás, J. M., and W. W. Kay. 1986. Tellurite susceptibility and non-plasmid-mediated resistance in Escherichia coli. Antimicrob. Agents Chemother. 30:127-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turner, R., J. Weiner, and D. Taylor. 1999. Tellurite-mediated thiol oxidation in Escherichia coli. Microbiology 145:2549-2557. [DOI] [PubMed] [Google Scholar]
- 30.Vásquez, C., C. Saavedra, C. Loyola, H. Moscoso, and S. Pichuantes. 1999. Cloning of a tellurite resistance determinant from Bacillus stearothermophilus V in Escherichia coli. Biochem. Mol. Biol. Int. 47:171-175. [DOI] [PubMed] [Google Scholar]
- 31.Vásquez, C., C. Saavedra, C. Loyola, M. Araya, and S. Pichuantes. 2001. The product of the cysK gene of Bacillus stearothermophilus V mediates potassium tellurite resistance in Escherichia coli. Curr. Microbiol. 43:418-423. [DOI] [PubMed] [Google Scholar]
- 32.Zheng, L., R. H. White, V. L. Cash, R. F. Jack, and D. R. Dean. 1993. Cysteine desulfurase activity indicates a role for NifS in metallocluster biosynthesis. Proc. Natl. Acad. Sci. USA 90:2754-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]