Abstract
The proteomes of cultured Pseudomonas aeruginosa isolates from chronically infected cystic fibrosis (CF) lungs were compared by using genetically divergent clones and isogenic morphotypes of one strain. Cellular extracts gave very similar protein patterns in two-dimensional gels, suggesting that the conserved species-specific core genome encodes proteins that are expressed under standard culture conditions in vitro. In contrast, the protein profiles of extracts of culture supernatants were dependent on the growth phase, and there were significant differences between clones. The profiles also varied within clonally related morphotypes from one CF patient, including a hyperpiliated small-colony variant. Mass spectrometry revealed that this variant overexpressed proteins secreted by the type I secretion system (including proteins involved in iron acquisition) and by the type III secretion system. Furthermore, the proteins in the supernatant extracts from the small-colony variant which were recognized by sera from different CF patients varied greatly. We concluded that the secretome expression is a sensitive measure of P. aeruginosa strain variation.
The gram-negative bacterium Pseudomonas aeruginosa can be found in a wide variety of terrestrial and aquatic habitats (17, 19). Moreover, this bacterium is an opportunistic pathogen of plants, animals, and humans. Its ecological success is based on a remarkable degree of genomic flexibility (16) and phenotypic adaptation. Adaptation of Pseudomonas fluorescens to changing environmental conditions has been shown to be facilitated by the emergence of variant subpopulations due to selection in a heterogeneous habitat (15). This morphological diversity leads to establishment of niche specialists with gains of function. Mutations of single genes have also been found to lead to the evolution of P. aerugniosa phenotypic variants (6). The presence of such variants in the chronically infected cystic fibrosis (CF) lung habitat is a common finding and reflects the outstanding ability of P. aeruginosa to adapt. The adaptational process might even be supported by the emergence of so-called “hypermutator” strains in the CF lung, which facilitates morphological diversity (14).
In this study we analyzed the proteomes of various clinical P. aeruginosa strains and of isogenic P. aeruginosa morphotypes. Proteome analysis of clonal P. aeruginosa morphotypes isolated from an in vivo habitat is a new approach for gaining insight into the global adaptation of P. aeruginosa to hostile environments and is complementary to studies of adaptation of single P. aeruginosa strains to various culture conditions. Recently, in our laboratory a hyperpiliated small-colony variant (SCV) morphotype was identified, which exhibited increased twitching motility, autoaggregative growth behavior, increased fitness under stationary growth conditions, and an enhanced ability to form biofilms compared to the ability of a fast-growing phenotype of clonal origin that was isolated from the respiratory tract of the same CF patient (8, 9). Other groups have described similar biofilm-forming small variants of P. aeruginosa that emerged under favorable environmental conditions, possibly due to phase variation (2, 3). We compared this stable hyperpiliated SCV to isogenic P. aeruginosa morphotypes as a model to demonstrate how phenotypic conversion is reflected in the protein profile. Moreover, we analyzed the profile of immunoreactive proteins. P. aeruginosa grown in vitro was probed with CF sera, which elucidated the humoral response of a chronically infected human host to P. aeruginosa proteins expressed in vivo.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Strain PAO1 (= DSM 1707) and five minimally passaged clinical P. aeruginosa strains (20265, 61, 211, 17997, and 33) were used in this study. The clinical strains were isolated from sputum samples or deep throat swabs of CF patients (ages, 18 to 33 years) who attended the CF clinic at Hannover Medical School, Hannover, Germany. For comparative proteome analysis three different P. aeruginosa morphotypes were used: the SCV 20265 morphotype, which formed small (diameter, ∼1 mm), convex, opaque, circular colonies after 2 days of incubation at 37°C; a P. aeruginosa isolate that formed irregularly shaped colonies that were ∼3 mm in diameter (referred to as the wild type) and was recovered within 3 months as a sequential isolate from the same CF patient; and a fast-growing revertant isolated from the SCV 20265 population after serial passage in brain heart infusion medium, whose colonies had a flat and irregular surface and were ∼10 mm in diameter (9). The three morphotypes produced indistinguishable SpeI restriction profiles in pulsed-field gel electrophoresis (9), indicating clonal identity. For preparation of protein extracts, P. aeruginosa strains were grown in modified Vogel-Bonner minimal medium (3.3 mM MgSO4, 10 mM citric acid, 28 mM NaNH4HPO4, 37 mM K2HPO4, 214 mM potassium d-gluconate; pH 7.2). Portions (400 ml) of this medium were inoculated with 800 μl of overnight cultures of the P. aeruginosa strains and cultured at 37°C with shaking. In this study mid-exponential-phase cultures, as well as late-exponential-phase cultures and stationary-phase cultures, were used. Before preparation of the protein samples aliquots of the bacterial cultures were subcultured on agar plates to confirm that the colony morphology was unchanged.
Preparation of protein samples.
Two-dimensional (2-D) gel electrophoresis was performed with whole-cell and culture supernatant extracts. To prepare extracts of cellular proteins, bacterial cells were washed twice in phosphate-buffered saline and resuspended in solubilization solution, which contained 7 M urea, 2 M thiourea, 4% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 30 mM dithiothreitol (DTT), 0.5% (wt/vol) Pharmalyte (pH 3 to 10), 1 mM Pefabloc SC, and 2 μM leupeptin (18, 19). After lysis of the cells by sonication with a Branson Sonifier 250, each suspension was centrifuged (2,500 × g, 15 min, 15°C) to remove the cell debris and unbroken cells. The supernatant was subjected to a second centrifugation (50,000 × g, 40 min) to remove the insoluble components, and the protein concentration in the resulting supernatant was determined by a Bradford assay (Bio-Rad Protein assay).
For preparation of extracellular protein extracts the supernatants obtained after centrifugation of the bacterial cultures at 6,000 × g for 15 min at 4°C were passed through a 0.2-μm-pore-size filter. Deoxycholic acid (sodium salt) was added to a final concentration of 0.2 mg/ml. After 30 min of incubation on ice, the proteins were precipitated by addition of 6% (wt/vol) trichloroacetic acid and incubated at 4°C for 2 h. After centrifugation at 18,000 × g for 30 min the precipitated proteins were resuspended in distilled water, and 8 volumes of acetone (−20°C) was added. After incubation at −20°C for 2 h, the mixture was centrifuged at 3,500 × g for 20 min at 4°C, and the pellet was allowed to dry for 5 min before it was dissolved in an appropriate amount of solubilization buffer. After centrifugation at 50,000 × g for 40 min at 15°C to remove the insoluble components, the protein concentration of the remaining supernatant was determined. Protein extracts were either used immediately for 2-D gel electrophoresis or stored at −70°C.
2-D gel electrophoresis.
Isoelectric focusing (first dimension) was performed with the IPGphor system and Immobiline DryStrip gel strips (Amersham Biosciences). Equal quantities (as determined by a Bradford assay) of solubilized proteins from the different P. aeruginosa strains were diluted to obtain a final volume of 360 μl with solubilization solution and applied to the Immobiline gel strips (18 cm) by in-gel rehydration. Linear immobilized pH gradients (pH 4 to 7) and nonlinear pH gradients (pH 3 to 10) were used. Thirty to 50 μg of protein was applied for analytical gels (silver staining), and 200 to 500 μg of protein was loaded for micropreperative electrophoresis (Coomassie staining). After rehydration under silicone oil for 10 h, the proteins were focused for a total of 120 kV · h at 20°C. The proteins were reduced by equilibration of the strips in equilibration solution (6 M urea, 30% glycerol, 2% [wt/vol] sodium dodecyl sulfate [SDS], and 1% [wt/vol] DTT in 0.05 M Tris-HCl buffer, pH 8.8) for 15 min and then carbamidomethylated in the same solution containing 260 mM iodoacetamide but no DTT for 15 min. The second dimension (SDS-polyacrylamide gel electrophoresis) was performed by using the ISO-DALT electrophoresis system (Amersham Biosciences). The strips were transferred to 12 to 15% acrylamide gradient gels. A low-molecular-weight calibration standard (Amersham Biosciences) was loaded on each gel, and electrophoresis was performed overnight at 125 V and 10°C. The gels were either stained or prepared for electroblotting. Staining of the analytical gels was performed by using the silver staining protocol of Blum and Beier (1), while micropreparative gels were stained with colloidal Coomassie brilliant blue G-250 as described by Neuhoff et al. (11).
Western blot analysis.
For protein blotting, the protein-containing areas of the 2-D gels were excised and blotted onto polyvinylidene difluoride membranes (Roti-PVDF; Roth, Karlsruhe, Germany) under semidry conditions for 1.5 h at 0.9 mA/cm2 and room temperature with Tris-glycine transfer buffer (25 mM Tris, 190 mM glycine, 0.1% [wt/vol] SDS, 20% [vol/vol] methanol; pH 8.5). After protein transfer, the membranes were blocked in blocking solution (5% [wt/vol] nonfat dry milk powder, 1% [wt/vol] bovine serum albumin, 0.2% [vol/vol] Tween 20, and 0.02% gelatin in Tris-buffered saline [TBS]) at room temperature for 2 h. For antigen detection two pooled sera, each from six CF patients chronically infected with P. aeruginosa, and one control pooled serum from six CF patients from whom P. aeruginosa was not recovered were used after they had been immunoprecipitated to reduce background signals to nonprotein antigens (especially exopolysaccharides) as follows. A bacterial pellet from a stationary 5-ml P. aeruginosa culture (strain SCV 20265) was suspended in 750 μl of H2O, digested with proteinase K (Sigma) at 55°C overnight, and heated at 95°C for 2 h to inactivate the enzyme. Then 250 μl of this sample was incubated with 750 μl of each of the pooled sera overnight at 5°C in a rotary shaker. Precipitated antibodies were removed by ultracentrifugation at 50,000 × g for 45 min. The supernatant (diluted 1:250 to 1:750 in blocking solution) was used for detection of protein antigens on the polyvinylidene difluoride membranes with gentle agitation at room temperature for 2 h. After three washing steps (with TBS-Tween, TBS-Tween containing 0.5 M NaCl, and TBS-Tween containing 0.1% Triton X-100; 15 min each), a peroxidase-coupled secondary antibody (AffiniPure goat anti-human immunoglobulin A [IgA] plus IgG plus IgM[H+L]); Jackson ImmunoResearch), diluted 1:8,000 in blocking solution, was applied for 2 h. After three additional washes as described above, the membranes were rinsed with TBS and incubated in a chemoluminescent peroxidase substrate solution (Lumi-Light POD substrate; Roche Molecular Biochemicals, Mannheim, Germany) for 3 min. For detection of the chemoluminescence we used an LAS-1000 charge-coupled device camera (Fujifilm) in a dark box with exposure times between 10 s and 5 min.
Sample preparation for mass spectrometry.
Selected protein spots were excised from the Coomassie brilliant blue-stained 2-D gels, washed with 50 mM NH4HCO3, destained and dehydrated with 50% acetonitrile twice, and dried in a SpeedVac concentrator. The dried gel pieces were rehydrated in a 2-μg/ml solution of sequencing-grade modified porcine trypsin (Promega) and crushed; 50 mM NH4HCO3 was added to a microcentrifuge tube to prevent drying of the samples during the subsequent cleavage reaction (overnight, 37°C). The resulting peptides were collected by two extractions, one with 50 mM NH4HCO3 and one with 50% acetonitrile-5% formic acid. The extracts were pooled in a microcentrifuge tube, and the peptide extracts were dried in a SpeedVac concentrator. The pellets were resuspended in 10 μl of 0.5% HCOOH-5% methanol and purified on RP18 material (ZipTipC18; Millipore Corp., Bedford, Mass.) as described by the manufacturer.
MALDI-TOF MS analysis and protein identification.
The molecular masses of ZipTip18-purified peptides were determined by positive-ion matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) by using a Bruker Reflex instrument equipped for delayed extraction and with an N2 laser (337 nm). For each sample 1 μl of matrix solution (10 mg of α-cyano-4-hydroxycinnamic acid in 1 ml of 60% methanol-0.1% formic acid) was placed on the scout ion source and crystallized as a thin layer. Then 1 to 2 μl of sample was pipetted directly on top of the thin matrix layer, and cocrystallization was carried out at room temperature. Spectra were recorded in reflection mode with an acceleration voltage of 20 kV and a reflection voltage of 21.5 kV. Monoisotopic masses from spectra were selected either manually or by the Knexus software (Proteometrics, New York, N.Y.). The proteins were identified by peptide mass fingerprinting by performing an automated search in the NCBInr database either with the Knexus software or with the software tool MS-Fit (http://prospector.ucsf.edu).
RESULTS AND DISCUSSION
Interstrain variability in proteome profiles and dynamics of protein expression.
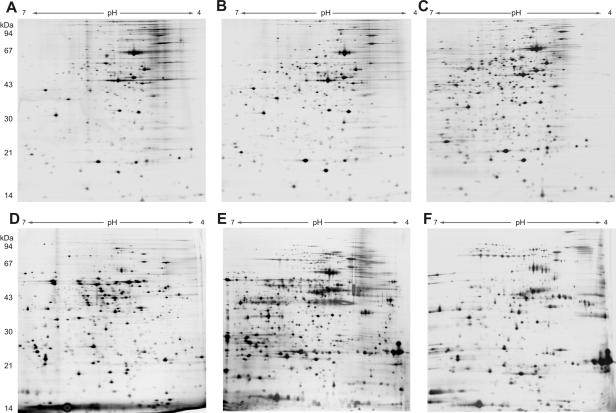
In this study we used a combination of 2-D gel electrophoresis and the ability to identify a protein on the basis of a predicted gene product in the genome database by mass spectrometry as a powerful method to analyze the global protein expression patterns of P. aeruginosa. To address the question of how genome characteristics of different P. aeruginosa strains are reflected in their protein expression profiles, the proteome profiles of five P. aeruginosa strains and the PAO1 strain were analyzed by 2-D gel electrophoresis. Not surprisingly, the protein patterns of the cellular extracts of the six strains resembled each another, and many protein spots identified in the extract of one strain could be found in the extracts of other strains. Figures 1A to C show the results for the cellular fractions of three representative P. aeruginosa strains that had very similar patterns. This strain-independent homogeneous expression of cellular proteins implies that there is a core genome shared by all P. aeruginosa strains. In contrast, analysis of the extracellular subproteome (secretome) revealed marked interstrain differences in the expression profiles of the six strains, which made assignment of spots identified in one strain to another strain nearly impossible. Figures 1D to F show the results for the extracellular protein fractions of the three selected strains. A high level of variability in secretomes, in contrast to the conserved membrane protein expression profile, has been observed previously in a comparison of an invasive P. aeruginosa strain and a cytotoxic P. aeruginosa strain (13).
FIG. 1.
Comparison of cellular extracts (A to C) and extracellular protein extracts (D to F) of P. aeruginosa PAO1 (A and D), SCV 20265 (B and E), and strain 17997 (C and F) by 2-D gel electrophoresis. The protein extracts were prepared after growth to the late exponential phase. Thirty micrograms of protein was separated by 2-D gel electrophoresis and silver stained (pH gradient for isoelectric focusing, pH 4 to 7).
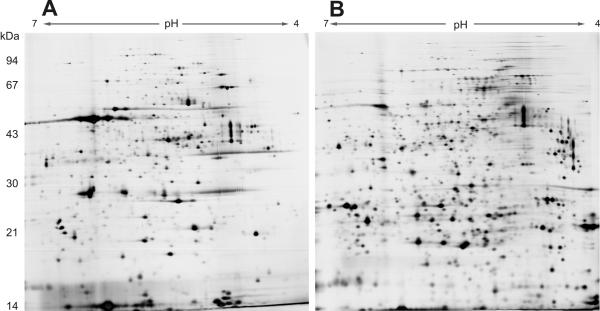
Monitoring the proteomes of the six P. aeruginosa strains during growth revealed relatively constant protein profiles for cellular extracts, and up to 80% of the spots could be detected over a broad incubation interval from early exponential growth (optical density at 600 nm, 0.5) to the stationary phase of growth after 2.5 days (optical density at 600 nm, 1.2 to 2.7) (data not shown). However, dramatic changes in the extracellular subproteome were identified by 2-D gel electrophoresis. Prolonged incubation led to a more complex protein pattern in the extracellular fraction, and the total number of proteins increased significantly, even though equal amounts of protein were loaded onto the gels (Fig. 2). It is known that many proteins are not secreted into the bacterial culture supernatant before the late exponential growth phase. However, contaminating cytosolic proteins were also increasingly detected, and together with proteolysis that might have been enhanced due to augmented protease secretion these proteins could have contributed to the higher diversity. Also, degradation fragments of proteins were observed after prolonged incubation times.
FIG. 2.
Dynamics of protein expression in the extracellular fraction of P. aeruginosa SCV 20265 over time. The extracellular fractions were prepared after growth to the mid-exponential phase (A) and the late exponential phase (B). Thirty-five micrograms of protein was separated by 2-D gel electrophoresis and silver stained (pH gradient for isoelectric focusing, pH 4 to 7).
Differential protein expression of isogenic P. aeruginosa morphotypes.
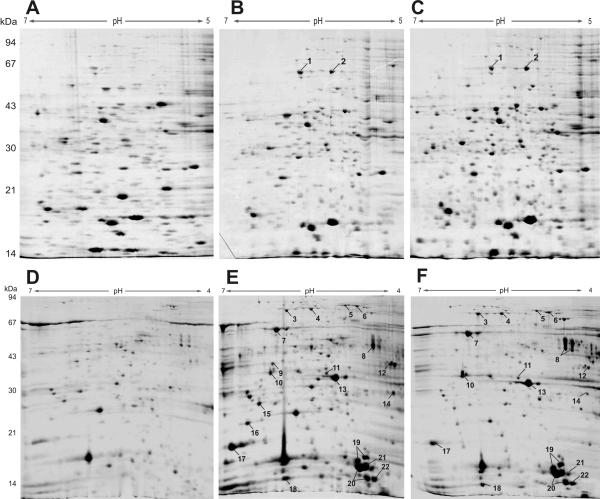
The observed interstrain variability of the secretome may reflect the differential, strain-specific regulation of gene expression and/or the utilization of genes that are not encoded by the core genome but are encoded by the highly dynamic accessory genome. To differentiate between the two mechanisms, we analyzed the proteomes of isogenic P. aeruginosa morphotypes recovered from the respiratory tract material of CF patients. For other morphotypes, such as mucoid variants, the CF airways provide a niche for a subgroup of hyperpiliated and autoaggregative P. aeruginosa SCVs with increased ability to form biofilms (9). To analyze intrastrain differential protein expression, we compared the hyperpiliated SCV 20265 strain with a fast-growing wild type (recovered as a sequential isolate of clonal origin from the respiratory tract material of the same CF patient) and a fast-growing revertant isolated in vitro from the SCV population. Similar to the observations made during interstrain comparisons, the expression profiles of cellular extracts of the three morphotypes revealed only slight differences (Fig. 3A to C), in contrast to the marked differences in the expression profiles of the extracellular fractions. As the P. aeruginosa morphotypes were isogenic, this finding implies that the variability of the secretome is due to differential regulation of protein expression, possibly as a consequence of small adaptational mutations. Remarkably, the extracellular proteins of the SCV 20265 strain in the mid-exponential growth phase showed great diversity and high abundance compared to the revertant, and the differences were even more pronounced when the data were compared to the data for the wild type. The protein profile of the revertant at later stages of growth resembled the early secretome profile of the SCV 20265 strain (data not shown). Figures 3 to F show the profiles of the extracellular protein fractions. Although equal amounts of protein were applied to the 2-D gels, as determined by a Bradford assay, the overall Coomassie brilliant blue-stained protein content of the wild type seemed to be less than the Coomassie brilliant blue-stained protein contents of the revertant and the SCV. As the difference in protein staining intensity could also be observed for normalized protein samples of the morphotypes separated by one-dimensional SDS-polyacrylamide gel electrophoresis (data not shown), it seems unlikely that there was variation in portions of the 2-D gels not shown. The wild type might produce substances that interfere with the Bradford assay, mimicking an organism with a higher protein content. However, to normalize the amount of protein loaded onto the gels, about twice as much culture supernatant of the wild type as of the SCV and revertants had to be extracted, implying that the total amount of protein in the culture supernatant of the wild type was distinctly less than the total amounts of protein in the culture supernatants of the SCV and revertant.
FIG. 3.
Comparison of cellular extracts (A to C) and extracellular protein extracts (D to F) of the clinical P. aeruginosa 20265 wild-type (A and D), SCV (B and E), and revertant (C and F) morphotypes. The extracts were prepared after growth to the late exponential phase. Four hundred micrograms of protein was separated by 2-D gel electrophoresis and stained with colloidal Coomassie brilliant blue G-250 (pH gradient for isoelectric focusing, pH 4 to 7). Differentially expressed proteins were selected for MALDI-TOF MS analysis. The numbers correspond to those in Tables 1 and 2.
A recent study of Nouwens et al. (12) highlighted the influence of the las and rhl quorum-sensing systems on the extracellular protein profile determined by 2-D gel electrophoresis. All quorum-sensing mutants showed a significant reduction in the amount of secreted protein, suggesting that a lack of the las or rhl system severely disrupts protein secretion and/or expression of previously abundant extracellular constituents. The diversity and great abundance of extracellular proteins in the SCV might reflect increased expression of quorum sensing-regulated genes. Fifty-two proteins were found to be differentially regulated in the hyperpiliated SCV 20265 strain or the clonal revertant compared to the regulation in the wild type. Most of these proteins were shown to be overexpressed in the SCV 20265 strain (31 proteins, 5 of which did not have homologues with clear functions), whereas 14 of the proteins identified (2 of which were classified as hypothetical) exhibited reduced expression in the SCV 20265 strain compared to the expression in the wild type. Table 1 summarizes a selection of proteins (some of which are shown in Fig. 3) that are differentially expressed in the SCV 20265 strain or the revertant compared to the expression in to the wild type. Three proteins known to be secreted via the type I secretion apparatus in P. aeruginosa, HasAp, AprA, and AprX (4), were strongly expressed in the mid-exponential growth phase of SCV 20265. The latter two proteins and PA0752 (which also produced more intense protein spots in the secretome analysis of the SCV) have been shown to be quorum sensing regulated by proteome and transcriptome analyses, indicating that there is a possible causal relationship between protein expression in the SCV and quorum sensing (12, 18, 21). Moreover, proteins of the type III secretion system were more abundant in the secretome of the SCV morphotype. With the exception of ExoU (not all P. aeruginosa strains express ExoU), the secreted effector proteins (ExoS and ExoT), as well as structural components of the secretion apparatus (PcrV, PopB, PopD, and PopN), were shown to be overexpressed in the SCV morphotype. The type III secretion system injects effector proteins directly into the cytosol of eukaryotic host cells (10) and has been shown to contribute to the pathogenicity of P. aeruginosa (5, 7). We found increased expression of the type III secretion system in the SCV 20265 strain; however, normally the CF lung habitat selects for P. aeruginosa strains with attenuated virulence (20). Future studies will have to show whether the type III expression in the hyperpiliated SCV is indeed associated with increased virulence (e.g., in a mouse model of infection).
TABLE 1.
Selected proteins found to be differentially expressed in P. aeruginosa SCV 20265 or revertants compared to the clonal wild type by 2-D electrophoresis
| PA no. | Gene | Protein and/or function | Classification | cla | Position(s) in Fig. 3 | Upregulated compared to wild type |
|---|---|---|---|---|---|---|
| PA0044 | exoT | Exoenzyme T, ADP-ribosyltransferase | Secreted factors | 1 | 21b | SCV, early to late log phase; Revertant, early to late log phase |
| PA0315 | Hypothetical protein | Unclassified | 4 | 17 | SCV, early log phase | |
| PA0572 | Hypothetical protein | Unclassified | 4 | SCV, early log phase | ||
| PA0781 | Hypothetical protein, 37% similar to PhuR, outer membrane hemin receptor (P. aeruginosa) | Unclassified outer Membrane protein | 4 | 6 | SCV, early to late log phase; revertant, early to late log phase | |
| PA1086 | flgK | Flagellar hook-associated protein 1 FlgK | Motility and attachment; cell wall LPS, capsulec | 2 | Revertant, early to late log phase | |
| PA1092 | fliC | Flagellin type A, flagellar filament protein | Motility and attachment | 1 | 8 | Revertant, early to late log phase |
| PA1094 | fliD | Flagellar capping protein FliD, hook-associated protein | Motility and attachment: cell wall LPS capsule | 1 | Revertant, late log phase | |
| PA1245 | aprX | Hypothetical protein, secreted by type 1 pathway | Secreted factors | 4 | SCV, early log phase; revertant, early log phase | |
| PA1249 | aprA | Type I secreted alkaline metalloproteinase | Secreted factors | 1 | 12, 14b | SCV, early log phase; revertant, early log phase |
| PA1698 | popN | Type III secretion outer membrane protein PopN | Membrane proteins; protein secretion-export apparatus | 1 | 11 | SCV, early to late log phase; revertant, early to late log phase |
| PA1706 | pcrV | Type III secretion protein PcrV | Protein secretion-export apparatus | 1 | 13 | SCV, early to late log phase; revertant, early to late log phase |
| PA1708 | popB | Type III secretion translocator protein PopB | Protein secretion-export apparatus | 1 | 22b | SCV, early to late log phase; revertant, early to late log phase |
| PA1709 | popD | Type III secretion translocator outer membrane protein PopD | Protein secretion-export apparatus | 10, 18b | SCV, early to late log phase; revertant, early to late log phase | |
| PA2395 | Hypothetical protein | Unclassified | 4 | SCV, late log phase | ||
| PA2398 | fpvA | Outer membrane ferripyoverdine receptor | Transport of small molecules | 1 | 5 | SCV, early to late log phase; revertant, early to late log phase |
| PA2452 | Hypothetical protein | Unclassified | 4 | SCV, early log phase | ||
| PA3407 | hasAp | Heme acquisition protein HasAp, secreted by type I pathway | Transport of small molecules, secreted factors | 1 | SCV, early log phase | |
| PA3785 | Conserved hypothetical protein | Unclassified | 4 | 16 | SCV, early to late log phase | |
| PA3790 | oprC | Putative copper transport outer membrane porin OprC | Transport of small molecules | 1 | 1, 3 | SCV, early to late log phase |
| PA3841 | exoS | Type III secreted exoenzyme S, ADP-ribosyltransferase | Secreted factors | 1 | 7, 19b | SCV, early to late log phase |
| 20b | Revertant, early to late log phase | |||||
| PA4063 | Hypothetical protein | Unclassified | 4 | 15 | SCV, early log phase | |
| PA4221 | fptA | Fe(III)-pyochelin outer membrane receptor precursor | Transport of small molecules | 1 | 2, 4 | SCV, early to late log phase; revertant, early to late log phase |
| PA5217 | Probable binding protein component of ABC iron transporter | Transport of small molecules | 3 | 9 | SCV, early to late log phase |
d, confidence level of the genome annotation from the Pseudomonas genome database (http://www.pseudomonas.com).
Degradation fragment of protein identified.
LPS, lipopolysaccharide.
As iron is required for bacterial growth, bacteria have evolved a number of sophisticated mechanisms to acquire iron under the iron-limiting conditions within the host. In addition to the overexpression of HasAp and AprA in SCV 20265 mentioned above (both of these proteins not only are involved in iron acquisition but also are regulated in an iron-dependent manner), we also identified increased expression of membrane proteins in the mid-exponential growth phase which are involved in iron uptake, including receptors of the siderophores pyoverdin FpvA and pyochelin FptA. Moreover, sequence comparisons of PA0781 and PA4625 revealed homologies to tonB-dependent outer membrane receptors that may contribute to iron acquisition in SCV 20265. PA5217 exhibits sequence homology to a binding component of an ABC iron transporter, and PA4625 shows weak homology to the huxA-encoded protein from Haemophilus influenzae, which is required for cellular uptake of complexed iron. As iron sources are scarce within a eukaryotic host (22), the ability to efficiently acquire iron might provide a selective advantage to the SCV phenotype for persistence in CF lungs. The presence of membrane proteins in the extracellular environment is noteworthy, and the possibility that their occurrence in the extracellular fraction might be due to lysis or exocytosis of proteins in membrane vesicles has been discussed previously (13). To support the finding that the increased expression of even membrane proteins in the secretome was due to overexpression in the SCV morphotype, we performed a quantitative real-time PCR for two selected genes and determined their relative levels of expression as normalized to the level of expression the housekeeping PA1580 (citrate synthase) gene (gltA). We were able to show that relative up-regulation of the genes coding for the membrane protein FptA and the surface protein of the type III secretion apparatus (PopD) occurs at the transcriptional level (data not shown).
P. aeruginosa antigens detected by human sera.
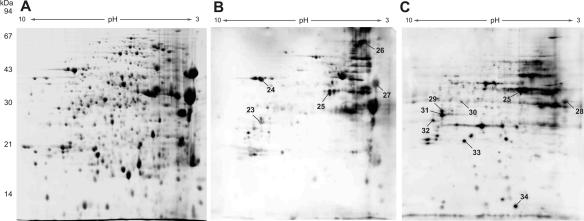
Our aim to identify P. aeruginosa strain-specific protein expression in vivo during chronic infection of the human host prompted us to perform an immunoblot analysis of 2-D gels using sera from CF patients. While the pooled sera from CF patients from whom P. aeruginosa was not recovered did not react with the P. aeruginosa antigens (data not shown), the sera from the chronically infected CF patients reacted with several protein spots (Fig. 4), although some specific anti-P. aeruginosa antibodies might have been removed by immunoprecipitation. However, although each specimen contained sera from six CF patients and the silver-stained protein patterns after 2-D gel electrophoresis were very similar in the experimental settings, analysis of the immunoblot patterns of the two CF serum pools revealed large differences (Fig. 4). This finding indicates that the immunoblot patterns of CF sera varied to such a large extent that even a pool of six sera was not able to normalize the variations. This might have been because not all patient sera reacted to all immunogenic spots. On the other hand, in addition to similar intensities for reactive proteins, we also found some clear spots in the immunoblot that were hardly detected or were not detected by silver staining, possibly indicating extreme antigenicity of these spots. However, we found that especially the secretomes of various clinical strains exhibited marked interstrain differences (Fig. 1). Therefore, the specific colonizing clones of single CF patients might not express identical immunogenic proteins within their secretomes in vivo. Consequently, the immune response to certain immunogenic proteins would be missing in the sera of CF patients colonized with a clone that does not express these proteins. On the other hand, the immune reactions of CF sera to proteins within the specific secretomes of the corresponding patient-colonizing clones might have been particularly strong when only small amounts of the protein were present in the 2-D gel of the P. aeruginosa strain that we selected for immunoblotting. Thus, the immunoblot pattern obviously depends on the colonizing P. aeruginosa clone exhibiting a highly variable, strain-specific secretome.
FIG. 4.
Antigens of the extracellular fraction of P. aeruginosa strain 20265 SCV grown to the late stationary phase detected by immunostaining. Extracellular extracts that also contained cytosolic proteins were separated by 2-D gel electrophoresis (A) (pH gradient for isoelectric focusing, pH 3 to 10: nonlinear; silver stained). Two pooled sera (B and C), each from six CF patients chronically infected with P. aeruginosa, were used for immunostaining.
The immunogenic proteins identified (Table 2) comprise secreted proteins (AprA, AprX, HasAp, and ExoS), surface proteins (PopB, PopD, FliC, OprF, YrbE, and SecD, as well as the putative porin PA0755), and, unexpectedly, some cytosolic proteins (GroEL, YhfP, and GshB). Moreover, five hypothetical proteins predicted from the genome sequence were found for the first time to be expressed in a habitat in vivo.
TABLE 2.
Immunogenic antigens identified in the proteome of P. aeruginosa SCV 20265
| PA no. | Gene | Protein and/or function | Classification | cla | Position(s) in Fig. 4 |
|---|---|---|---|---|---|
| PA0139 | ahpC | Alkyl hydroperoxide reductase subunit C | Adaptation, protection | 2 | |
| PA0244 | Hypothetical protein | Putative enzymes | 4 | ||
| PA0407 | gshB | Glutathione synthetase | Amino acid biosynthesis and metabolism; biosynthesis of cofactors, prosthetic groups, and carriers | 2 | |
| PA0552 | pgk | Phosphoglycerate kinase | Carbon compound catabolism; energy metabolism | 2 | |
| PA0660 | Hypothetical protein | Unclassified | 4 | 30 | |
| PA0755 | Probable porin | Membrane proteins; transport of small molecules | 3 | ||
| PA0953 | Probable thioredoxin | Putative enzymes | 3 | ||
| PA1092 | fliC | Flagellin type A, flagellar filament protein | Motility and attachment | 1 | |
| PA1245 | aprX | Hypothetical protein | Unclassified | 4 | 27 |
| PA1245 | aprA | Alkaline metalloproteinase | Secreted factors | 1 | 28b |
| PA1494 | Conserved hypothetical protein | Unclassified | 4 | 26 | |
| PA1708 | popB | Translocator protein PopB | Protein secretion-export apparatus | 1 | |
| PA1709 | popD | Translocator outer membrane protein PopD | Protein secretion-export apparatus | 1 | 23, 31, 34b |
| PA1777 | oprF | Major porin and structural outer membrane porin OprF | Membrane proteins: transport of small molecules | 1 | 25 |
| PA1833 | yhfP | Probable oxidoreductase | Putative enzymes | 3 | |
| PA1844 | Hypothetical protein | Unclassified | 4 | ||
| PA2491 | Probable oxidoreductase | Putative enzymes | 3 | ||
| PA3407 | hasAp | Heme acquisition protein HasAp | Transport of small molecules | 1 | |
| PA3821 | secD | Secretion protein SecD | Membrane proteins, protein secretion-export apparatus | 2 | |
| PA3841 | exoS | Exoenzyme S, ADP-ribosyltransferase | Secreted factors | 1 | 24 |
| PA3883 | Probable short-chain dehydrogenase | Putative enzymes | 3 | 29 | |
| PA4385 | groEL | GroEL protein, 60-kD chaperonin | Chaperones and heat shock proteins | 1 | |
| PA4401 | Probable glutathione S-transferase | Putative enzymes | 3 | 33 | |
| PA4455 | yrbE | Probable permease of ABC transporter | Membrane proteins; transport of small molecules | 3 | 32 |
| PA4917 | Hypothetical protein | Unclassified | 4 |
d, confidence level of the genome annotation from the Pseudomonas genome database (http://www.pseudomonas.com).
degradation fragment of protein identified.
Conclusion.
We anticipated that our comparative study of immunoprofiles and global proteomes in genetically divergent strains and isogenic morphotypes would provide insight into the relative effects of the conserved core genome, variable genetic elements, and phenotypic adaptation on the protein makeup of P. aeruginosa. Proteome analysis of several clinical P. aeruginosa strains revealed almost identical patterns for the cellular extracts, whereas interclonal diversity and intraclonal diversity were demonstrated for the secretomes of cultured P. aeruginosa. The diversity was even greater for the immunogenic protein patterns expressed in vivo. The finding that there were strain-independent conserved cellular extracts might have reflected the fact that the need for adaptation under in vitro culture conditions (although the strains have been cultured in minimal medium) is low, and strain-specific differences that reflect the versatility of niche specialists are not likely to be detected under these conditions in the cellular extracts. Moreover, the cellular proteome comprises mostly proteinaceous cell constituents that are expected to be species specific but not strain specific. However, we could demonstrate that under in vitro growth conditions secretome expression is strongly strain and morphotype specific. Since the secreted P. aeruginosa proteins come into direct contact with their environment, they could be especially important and thus be essential for bacterial adaptation. Moreover, the secretome includes important virulence factors essential for establishment of an infection within the human host. Thus, the need to adapt and survive even under harsh environmental conditions might necessitate differentiated regulation of secretome expression. One example of an adverse environment is the hostile in vivo habitat of the CF lung, where adaptation is essential for bacterial persistence. The extremely variable immunogenic protein pattern elucidated in this study by using sera from chronically infected CF patients implies that there is highly strain-specific regulation of the secretome under in vivo growth conditions. Future studies should determine the molecular mechanism underlying this differential regulation of secreted proteins that leads to evolution of P. aeruginosa variants with a gain of function. For the hyperpiliated SCVs analyzed in this study as model organisms for bacterial phenotypic adaptation, the enhanced secretion of extracellular proteins and adaptation to iron-limiting conditions might be selective advantages and might be essential for growth and persistence, possibly within a biofilm.
Acknowledgments
We are very grateful to Dieter Bitter-Suermann for his continuous encouragement and support, and we thank Jaqueline Majewski, who is funded by the European Commission (contract QLG2-CT-1999-00932), for excellent technical assistance.
REFERENCES
- 1.Blum, H., and H. G. H. Beier. 2003. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8:93-99. [Google Scholar]
- 2.Deziel, E., Y. Comeau, and R. Villemur. 2001. Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J. Bacteriol. 183:1195-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drenkard, E., and F. M. Ausubel. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416:740-743. [DOI] [PubMed] [Google Scholar]
- 4.Duong, F., E. Bonnet, V. Geli, A. Lazdunski, M. Murgier, and A. Filloux. 2001. The AprX protein of Pseudomonas aeruginosa: a new substrate for the Apr type I secretion system. Gene 262:147-153. [DOI] [PubMed] [Google Scholar]
- 5.Fauvarque, M. O., E. Bergeret, J. Chabert, D. Dacheux, M. Satre, and I. Attree. 2002. Role and activation of type III secretion system genes in Pseudomonas aeruginosa-induced Drosophila killing. Microb. Pathog. 32:287-295. [DOI] [PubMed] [Google Scholar]
- 6.Govan, J. R., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hauser, A. R., E. Cobb, M. Bodi, D. Mariscal, J. Valles, J. N. Engel, and J. Rello. 2002. Type III protein secretion is associated with poor clinical outcomes in patients with ventilator-associated pneumonia caused by Pseudomonas aeruginosa. Crit. Care Med. 30:521-528. [DOI] [PubMed] [Google Scholar]
- 8.Haussler, S., B. Tummler, H. Weissbrodt, M. Rohde, and I. Steinmetz. 1999. Small-colony variants of Pseudomonas aeruginosa in cystic fibrosis. Clin. Infect. Dis. 29:621-625. [DOI] [PubMed] [Google Scholar]
- 9.Haussler, S., I. Ziegler, A. Lottel, F. Gotz, M. Rohde, D. Wehmhohner, S. Saravanamuthu, B. Tummler, and I. Steinmetz. 2003. Highly adherent small-colony variants of Pseudomonas aeruginosa in cystic fibrosis lung infection. J. Med. Microbiol. 52:295-301. [DOI] [PubMed] [Google Scholar]
- 10.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neuhoff, V., N. Arold, D. Taube, and W. Ehrhardt. 1988. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9:255-262. [DOI] [PubMed] [Google Scholar]
- 12.Nouwens, A. S., S. A. Beatson, C. B. Whitchurch, B. J. Walsh, H. P. Schweizer, J. S. Mattick, and S. J. Cordwell. 2003. Proteome analysis of extracellular proteins regulated by the las and rhl quorum sensing systems in Pseudomonas aeruginosa PAO1. Microbiology 149:1311-1322. [DOI] [PubMed] [Google Scholar]
- 13.Nouwens, A. S., M. D. Willcox, B. J. Walsh, and S. J. Cordwell. 2002. Proteomic comparison of membrane and extracellular proteins from invasive (PAO1) and cytotoxic (6206) strains of Pseudomonas aeruginosa. Proteomics 2:1325-1346. [DOI] [PubMed] [Google Scholar]
- 14.Oliver, A., R. Canton, P. Campo, F. Baquero, and J. Blazquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251-1254. [DOI] [PubMed] [Google Scholar]
- 15.Rainey, P. B., and M. Travisano. 1998. Adaptive radiation in a heterogeneous environment. Nature 394:69-72. [DOI] [PubMed] [Google Scholar]
- 16.Romling, U., K. D. Schmidt, and B. Tummler. 1997. Large genome rearrangements discovered by the detailed analysis of 21 Pseudomonas aeruginosa clone C isolates found in environment and disease habitats. J. Mol. Biol. 271:386-404. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt, K. D., B. Tummler, and U. Romling. 1996. Comparative genome mapping of Pseudomonas aeruginosa PAO with P. aeruginosa C, which belongs to a major clone in cystic fibrosis patients and aquatic habitats. J. Bacteriol. 178:85-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schuster, M., C. P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spiers, A. J., A. Buckling, and P. B. Rainey. 2000. The causes of Pseudomonas diversity. Microbiology 146:2345-2350. [DOI] [PubMed] [Google Scholar]
- 20.Tummler, B., J. Bosshammer, S. Breitenstein, I. Brockhausen, P. Gudowius, C. Herrmann, S. Herrmann, T. Heuer, P. Kubesch, F. Mekus, U. Romling, K. D. Schmidt, C. Spangenberg, and S. Walter. 1997. Infections with Pseudomonas aeruginosa in patients with cystic fibrosis. Behring Inst. Mitt. 98:249-255. [PubMed]
- 21.Wagner, V. E., D. Bushnell, L. Passador, A. I. Brooks, and B. H. Iglewski. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185:2080-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wandersman, C., and I. Stojiljkovic. 2000. Bacterial heme sources: the role of heme, hemoprotein receptors and hemophores. Curr. Opin. Microbiol. 3:215-220. [DOI] [PubMed] [Google Scholar]