Abstract
Improvements in purification of membrane-associated methane monooxygenase (pMMO) have resulted in preparations of pMMO with activities more representative of physiological rates: i.e., >130 nmol · min−1 · mg of protein−1. Altered culture and assay conditions, optimization of the detergent/protein ratio, and simplification of the purification procedure were responsible for the higher-activity preparations. Changes in the culture conditions focused on the rate of copper addition. To document the physiological events that occur during copper addition, cultures were initiated in medium with cells expressing soluble methane monooxygenase (sMMO) and then monitored for morphological changes, copper acquisition, fatty acid concentration, and pMMO and sMMO expression as the amended copper concentration was increased from 0 (approximately 0.3 μM) to 95 μM. The results demonstrate that copper not only regulates the metabolic switch between the two methane monooxygenases but also regulates the level of expression of the pMMO and the development of internal membranes. With respect to stabilization of cell-free pMMO activity, the highest cell-free pMMO activity was observed when copper addition exceeded maximal pMMO expression. Optimization of detergent/protein ratios and simplification of the purification procedure also contributed to the higher activity levels in purified pMMO preparations. Finally, the addition of the type 2 NADH:quinone oxidoreductase complex (NADH dehydrogenase [NDH]) from M. capsulatus Bath, along with NADH and duroquinol, to enzyme assays increased the activity of purified preparations. The NDH and NADH were added to maintain a high duroquinol/duroquinone ratio.
Methanotrophs are a group of gram-negative bacteria that utilize methane or methanol as the sole source of carbon and energy (1, 20). The initial oxidation of methane to methanol is catalyzed by methane monooxygenase (MMO). In some methanotrophs, two different MMOs can be expressed, depending on the copper concentration during growth (11, 37, 39): a soluble cytoplasmic MMO (sMMO) and a membrane-associated, or particulate, MMO (pMMO). In cells cultured under low copper/biomass ratios (≤0.9 nmol of Cu/mg of cell protein), the sMMO is expressed (20, 28). Cells cultured under higher copper/biomass ratios express pMMO, and there is no detectable sMMO expression (35, 43). While sMMO is a well-characterized enzyme that consists of a hydroxylase component composed of three polypeptides and a hydroxo-bridged binuclear iron cluster—an NADH-dependent reductase component composed of one polypeptide containing both FAD and [Fe2S2] cofactors and a regulatory polypeptide (18, 26, 27, 31, 47)—information on the molecular properties of pMMO is limited due to the instability of pMMO in cell-free fractions.
Purification of the pMMO has been reported from Methylococcus capsulatus Bath (2, 25, 33, 52) and M. trichosporium OB3b (30, 44). The reporting laboratories agree that pMMO is a copper-containing enzyme composed of three polypeptides with molecular masses of approximately 45,000 (α subunit), 26,000 (β subunit), and 23,000 Da (γ subunit) with an (αβγ)2 molecular structure (15). However, researchers in the field disagree on the number and type of metal centers associated with the pMMO as well as the nature of the physiological electron donor. One model proposes pMMO as an enzyme made up of 10 to 15 Cu atoms and 2 Fe atoms. In this model, two type II copper atoms and two EPR-silent iron atoms are associated with the αβγ complex (52). The remaining 8 to 13 copper atoms are bound to a small, 1,218-Da copper binding peptide/compound (cbc) that copurifies with the pMMO (14, 52). The role of copper-containing cbc (Cu-cbc) is not known; it may be involved in electron flow to the active site or serve a secondary role, such as copper acquisition, maintenance of a particular redox state, protection against oxygen radicals, or serving as a copper chaperone. The second theory proposes a 15- to 21-Cu-atom enzyme in which the coppers are coordinated into 5 to 7 spin-coupled trinuclear copper atom clusters, in which 2 to 3 clusters are catalytic and 3 to 4 clusters are involved in electron transfer from NADH to the catalytic centers (32-34). The third model proposes the pMMO as a 2-Cu-atom and 1- to 2-Fe-atom enzyme (2, 25, 46). The first and third models also propose that the pMMO is linked to the electron transfer chain at the quinone level (2, 11, 13, 25, 52), while the second theory proposes the enzyme utilizes NADH as the physiological reductant (33).
One of the main limitations to all of the models presented above is the use of low-activity preparations in the characterization of this novel enzyme. The reported purified preparations show activities of ≤17 nmol · min−1 · mg of protein−1, representing 1 to 5% of the physiological rates. Recent attempts by Basu et al. (2) have resulted in partially purified preparations with activities in the range of 50 nmol · min−1 · mg of protein−1, but they were unable to purify an active form of the enzyme. This paper describes an improved purification procedure resulting in purified preparations with activities of >130 nmol · min−1 · mg of protein−1. This report examines the growth conditions resulting in the stabilization of cell-free pMMO activity and the effect of detergent concentration on the metal composition of the pMMO, as well as addressing the issue of the physiological reductant of the pMMO.
MATERIALS AND METHODS
Organism and cultivation.
M. capsulatus Bath cells cultured for enzyme isolations were grown in nitrate mineral salts medium (NMS) with 5 μM CuSO4 and a vitamin mixture (24) at 42°C in shake flasks under an atmosphere of 30% methane and 70% air (vol/vol) to an optical density at 600 nm (OD600) of 1.5 to 2.0. One liter of flask culture was used to inoculate 2 liters of medium in a 14-liter BioFlo fermentor (New Brunswick, Edison, N.J.). Cells were cultured in the fermentor at 42°C and sparged at flow rates between 180 and 200 ml · min−1 for methane and between 800 and 1,200 ml · min−1 for air. The pH of the chemostat was maintained at 7.0 using potassium phosphate monobasic and sodium phosphate dibasic. When the culture reached an OD600 of 1.8 to 2.0, the concentrations of copper (added as a 500 μM CuSO4 stock solution) and iron (added as a 200 μM NaFe EDTA stock solution) in the culture medium were increased continuously at rates of 1.6 ± 0.2 μM Cu per h and 0.64 ± 0.08 μM Fe per h while maintaining an OD600 of 1.8 to 2.0. The feed rate of the medium was set to double the culture volume every 10 to 12 h. Upon reaching a working volume of 10 liters, the system was operated as a chemostat. Media and the copper addition rate were adjusted to maintain a constant cell density between 1.8 and 2.0. Cells were harvested from continuous cultures by centrifugation at 14,000 × g for 15 min at 4°C and resuspended in 10 mM 3-[N-morpholino]propanesulfonic acid (MOPS) (pH 7.3) buffer followed by subsequent centrifugation at 14,000 × g for 15 min. Washed cells were resuspended in 30 mM MOPS (pH 7.3)-1 mM benzamidine buffer.
M. capsulatus Bath cells were cultured to monitor the effect of copper addition during growth as described above with the following modifications. All glassware was acid washed in 0.1 N HNO3, and copper was omitted from the medium in flask cultures as well as in the initial fermentation medium. Culture samples were taken before the addition of copper (0 μM copper) and when the amended copper concentration in the chemostat reached approximately 1, 5, 6, 15, 20, 25, 35, 45, 60, 70, 75, 80, and 95 μM and were subsequently harvested and washed as described above.
Enzyme activity.
MMO activity was determined by the epoxidation of propylene as previously described (12). The reductants used were NADH (7 mM) and/or duroquinol (approximately 30 mM) for cell extracts and formate (2.5 mM) for whole-cell samples. The reduction of duroquinone to duroquinol was performed as described by Shiemke et al. (40). Both the duroquinol and NADH were lyophilized to remove residual ethanol. Following lyophilization, both reductants were checked for the presence of ethanol on a SRI 8610C gas chromatography system (SRI Instruments, Las Vegas, Nev.) equipped with a flame ionization detector and an 8-by-0.085-in. HaySep D column. Additional lyophilization steps were added if ethanol was detected. In reaction mixtures containing NADH dehydrogenase (NDH), approximately 12 μg of NDH, which would reduce 1 μmol of duroquinone · min−1 (10), was added. For optimal propylene oxidation rates, 0.4 ± 0.2 mol of Cu per mol of αβγ pMMO subunit was added to reaction mixtures containing purified pMMO or purified NDH-pMMO complex. The exact concentration of copper added was determined empirically for each sample. All reactions were initiated by the addition of 2 ml of propylene and 2 ml of air. Reaction mixtures were incubated at 42°C on a rotary shaker at 250 rpm. In addition to propylene oxidation activity in the soluble fraction, sMMO activity was monitored by the formation of naphthol from naphthalene as described by Brusseau et al. (6).
Isolation of membranes and soluble fractions.
All manipulations were performed at 4°C under anaerobic conditions (95% argon and 5% hydrogen [vol/vol]). DNase I (1 μg/ml) was added to the washed cell suspension and then deoxygenated by 3 to 5 cycles of vacuum followed by purging with oxygen-free argon. Prepared cells were lysed with three passes on a constant-flow French pressure cell at 18,000 lb/in2. The cell lysate was centrifuged at 14,000 × g for 15 min to remove unlysed cells and cell debris. The supernatant was taken and centrifuged at 150,000 × g for 1 h to sediment membranes. The membranes were resuspended using a Dounce homogenizer in a mixture containing 30 mM MOPS (pH 7.3), 1 M KCl, and 1 mM benzamidine buffer and centrifuged for 1 h at 150,000 × g. The washed membrane pellet was resuspended in a minimal volume of 30 mM MOPS (pH 7.3)-1 mM benzamidine buffer.
Solubilization of pMMO.
A 10% (wt/vol) solution of dodecyl β-d-maltoside was added to the washed membrane fraction to final concentrations of 0.25, 0.5, 0.75 1.0, 1.25, 1.5, 1.75, 2.0, 3.0, and 4.0 g of dodecyl β-d-maltoside per g of protein, and this mixture was stirred for 1 h and then centrifuged at 150,000 × g for 1 h at 4°C. Propylene oxidation activity was determined before centrifugation and in the particulate and soluble fractions following centrifugation.
Purification of pMMO, NDH, and NDH-pMMO complex.
The detergent-solubilized fraction following centrifugation of the suspension of 1.2 g of dodecyl β-d-maltoside per g of membrane protein was loaded on a 5.0- by 7-cm DEAE-Sepharose Fast Flow (FF) column equilibrated with buffer containing 30 mM MOPS (pH 7.3), 1 mM benzamidine, and 0.1% dodecyl-β-d-maltoside. The column was washed with buffer containing 30 mM MOPS (pH 7.3), 100 mM KCl, 1 mM benzamidine, and 0.1% dodecyl-β-d-maltoside. The NDH-pMMO complex remained bound to the column and eluted with buffer containing 30 mM MOPS (pH 7.3), 250 mM KCl, 1 mM benzamidine, and 0.1% dodecyl-β-d-maltoside. For separation of NDH and pMMO, samples from the DEAE-Sepharose FF column were diluted with an equal volume of buffer containing 30 mM MOPS (pH 7.3), 1 mM benzamidine, and 0.1% dodecyl-β-d-maltoside and loaded onto a 2.6- by 20-cm DEAE-Sepharose FF column equilibrated with buffer containing 30 mM MOPS (pH 7.3), 125 mM KCl, 1 mM benzamidine, and 0.1% dodecyl-β-d-maltoside. The loaded column was washed with 1 column volume of buffer containing 30 mM MOPS (pH 7.3), 125 mM KCl, 1 mM benzamidine, and 0.1% dodecyl-β-d-maltoside and eluted with buffer containing 30 mM MOPS (pH 7.3), 250 mM KCl, 1 mM benzamidine, and 0.1% dodecyl-β-d-maltoside. The fraction containing pMMO was then concentrated under nitrogen gas (99.99%) by ultrafiltration with a YM-50 filter (Millipore Corp., Bedford, Mass.). NDH was also isolated as previously described by Cook and Shiemke (10).
Dodecyl β-d maltoside treatment of pMMO.
The dodecyl β-d-maltoside concentration in purified pMMO samples was increased to 0.1, 0.2, 0.3, 0.4, or 0.5 mg/mg of protein. The samples were then incubated under anaerobic conditions for 30 min and applied to a 2.6- by 55-cm Superdex 30 column equilibrated with buffer containing 30 mM MOPS (pH 7.3), 1 mM benzamidine, and 0.1% dodecyl-β-d-maltoside. The pMMO samples were collected from the void volume, and Cu-cbc was collected from the included volume.
Electrophoresis and immunoblot analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed on precast 10 or 12% Bis-Tris gels with MOPS or on precast 10 or 12% 2-(morpholino) ethanesulfonic acid (MES)-SDS running buffer as specified by the manufacturer (Invitrogen, Carlsbad, Calif.), or SDS-PAGE was performed by the method of Laemmli (22). Gels were stained for total protein with Coomassie brilliant blue R or blotted for immunoassays. Densitometry was performed with a Bioimaging Technologies Biovideo MP1000 gel documentation system.
Proteins were blotted onto polyvinylidene difluoride Plus transfer membranes (Micron Separations, Inc., Westboro, Mass.) using an Xcell II Blot Module (Invirogen) according to manufacturer's specifications unless otherwise noted. For transfer of the α subunit of pMMO, the concentration of methanol in the transfer buffer was reduced to 5% and the concentration of SDS was increased to 0.02%. The transfer time for the α subunit of pMMO was 90 min at 30 V, while the transfer time for the β and γ subunits was 60 min at 30 V. Following treatment with serum raised against purified protein, antibodies were detected with the Opti-4CN substrate kit according to the manufacturer (Bio-Rad Laboratories, Hercules, Calif.).
Preparation of antibodies against the pMMO.
Antiserum against pMMO was raised in one New Zealand White rabbit by Animal Pharm Services, Inc., (Healdsburg, Calif.). Immunoglobin G was purified from the serum by using a protein A Sepharose CL-4B column as specified by manufacturer (Amersham Biosciences Corp., Piscataway, N.J.).
Protein, cell enumeration, and metal determinations.
Samples were assayed for protein by the method of Lowry et al. (29) using bovine serum albumin as a standard. Cells were enumerated on a Beckman Coulter Epics XL Flow Cytometer (Beckman Coulter, Inc., Allendale, N.J.).
Sample preparations for metal analysis were determined as previously described (52) and analyzed for copper or iron either by inductively coupled plasma atomic emission-mass spectroscopy (ICP-MS) with a model 4500 ICP-MS (Hewlett-Packard, St. Paul, Minn.) or with a Perkin-Elmer 1100B atomic absorption spectrophometer (Sheldon, Conn.).
The percentage of copper bound to cbc in the spent medium was determined by the ratio of copper that bound to a Dianion HP-20 column (Supelco, Bellefonte, Pa.) versus the copper in the void volume from the Dianion HP-20 column. The spent medium was loaded onto a 1.5- by 7-, 2- by 11-, or a 7- by 20-cm Diaion HP-20 column, depending on the sample size. The column was washed with 2 column volumes of H2O, and the organic phase was eluted from the Dianion HP-20 columns with 50% methanol-50% acetonitrile (vol/vol). The organic phase consisted of approximately 80% cbc, 12% protoheme IX, and 8% trace proteins and pyrroloquinoline quinone. The eluant containing the cbc fraction was rotary evaporated at 50°C, freeze-dried, and resuspended in 2 mM ammonium phosphate buffer (pH 7.0). For the percentage of copper bound to cbc, the concentration of copper was determined in the spent medium, in the void volume from the Dianion HP-20 column (soluble copper), and in the fraction that bound to the Dianion HP-20 column (copper bound to cbc).
Superoxide dismutase assay.
Superoxide anion radicals were generated using phenazine methosulfate and NADH as previously described (38). Generation of superoxide anion was monitored by observing the reduction of nitroblue tetrazolium to blue formazan at 560 nm (4, 7, 38).
Fatty acid analysis.
Whole-cell samples were normalized to two different protein concentrations (1.8 and 3.6 mg of protein per ml) for each cell suspension. The cell suspensions were saponified, methylated, and extracted by the methods described by MIDI Corp. (Newark, Del.). In addition to the standards obtained from MIDI Corp. (Newark, Del.), quantification standards were obtained from Supelco 37-component fatty acid methyl ester (FAME) mix (Bellefonte, Pa.) and prepared in hexane. FAME samples were separated by using an Agilent 6890 gas chromatograph and identified with the Microbial Identification System (version 4.0) from MIDI Corp.
Amino acid analysis and sequence.
Amino acid sequence analysis was performed by Edman degradation with an Applied Biosystems 477A protein sequencer coupled to a 120A analyzer. Sequence analysis was performed on samples electroblotted to polyvinylidene difluoride membranes as described above.
Spectroscopy.
Optical absorption spectroscopy and X-band EPR spectra were obtained as previously described (48, 52).
Total RNA extraction from pure cultures.
Care was taken in handling RNA to prevent RNA degradation by RNase as described earlier (19). Briefly, RNase activity was removed through oven baking and the use of either RNase-free compounds or diethyl pyrocarbonate (DEPC; Sigma)-treated water, or RNase was treated with DEPC. RNA extractions were performed with the Qiagen Total RNeasy kit (Qiagen, Inc.,Valencia, Calif.), and cells were lysed by bead beating. After being treated with RNase-free DNase I (Promega), total RNA was extracted with the Qiagen Total RNeasy kit.
Homologous internal RNA standards were designed with similar base sequences that could be amplified with the same primers used for target mRNA and for competitive reverse transcription-PCR (RT-PCR) by using the QIAGEN OneStep RT-PCR kit. The RT-PCR was carried out in 50 μl consisting of 5 μl of standard RNA, 5 μl of total RNA, 10 μl of 5× RT-PCR buffer, 10 μl of 5× Q solution, 400 μM (each) deoxynucleoside triphosphates (dNTPs), 0.6 μM each primer, and 2 μl of Qiagen OneStep RT-PCR enzyme. The sequences of steps to reverse transcribe and amplify both targets and standards were as follows. Following an RT incubation at 50°C for 30 min and heating to 95°C for 15 min, 30 PCR cycles with the following amplification profile were conducted: 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min. All samples were finally extended at 72°C for 10 min. PCR products were analyzed quantitatively by capillary electrophoresis on a Beckman P/ACE MDQ instrument (Beckman Instruments, Palo Alto, Calif.).
RESULTS
Effect of copper and iron in the culture medium on pMMO activity.
A key step in the initial stabilization and isolation of pMMO was the cultivation of cells under high (i.e., toxic)-copper conditions (52). Since the initial isolation, pMMO isolation procedures have used a variety of methods to maintain a high copper/cell density ratio during growth (2, 25, 33, 44). To optimize the stabilizing effect of medium copper on cell-free pMMO activity, a series of continuous and discontinuous copper addition schemes were attempted. The optimal copper addition rate was determined to be 1.6 ± 0.2 μM/h in chemostats where the generation time was approximately 10 h and the OD600 of the culture was 1.9 ± 0.1. Under these culture conditions, whole-cell propylene oxidation rates increased with increasing copper concentration up to approximately 24 μM amended copper (Fig. 1). At concentrations between 24 and 80 μM amended copper, whole-cell propylene oxidation rates per milligram of protein showed a slight decrease, followed by an accelerated decrease at copper concentrations above 80 μM. In contrast to whole-cell propylene oxidation rates, propylene oxidation activity in the washed membrane fraction was barely detectable below 24 μM amended copper, but increased linearly between 20 and 80 μM amended copper. At higher copper concentrations, pMMO activity in the membrane fraction also decreased, although the decrease in the rate was small compared to the decreased rates observed in whole cells.
FIG. 1.
Effect of copper concentration in the culture medium on the rates of propylene oxidation in whole-cell (▵), washed membrane (○), and soluble (□) fractions. The rate of copper addition was 1.7 μM · h−1, while the cell density was maintained at an OD600 of between 1.8 and 2.0.
As previously observed (41), the iron concentration had to be amended to obtain high-activity pMMO preparations (>10 nmol · min−1 · mg of protein). In our studies, we have found that maximal cell-free pMMO activity is observed at iron/copper ratios above 1 iron atom to 2.5 copper atoms. Only low-activity (i.e., ≤20 nmol · min−1 · mg of protein) pMMO preparations were obtained in the absence of supplemental iron.
Effect of copper in the culture medium on pMMO expression and membrane development.
To document the physiological events that occur during growth under these culture conditions, the initial copper concentration in the medium was changed from 5 μM to 0 μM, thus allowing the cells to express sMMO. As the copper concentration was increased in these chemostats, samples were taken periodically and analyzed as follows: (i) by morphology; (ii) by copper concentration in the spent medium, whole-cell, soluble, and membrane fractions; (iii) by propylene oxidation activity in the whole-cell, soluble, and membrane fractions; (iv) by SDS-PAGE of the whole-cell, soluble, and membrane fractions; (v) by immunoblotting with antibodies specific for pMMO and methanol dehydrogenase (MDH), (vi) by transcript levels of the genes encoding polypeptides for both sMMO and pMMO using RT-PCR and capillary electrophoresis; and (vii) by fatty acid concentration via MIDI-FAME analysis. The fatty acid concentration was used as an estimate of phospholipid concentration.
Figure 2 shows copper acquisition in cells cultured under increasing-copper conditions. Under the culture conditions described above, the copper concentration in the spent medium was constant (0.8 ± 0.3 μM) until the copper addition reached 59 μM and then increased at higher concentrations. Of the copper recovered in the spent medium, <4% was associated with the organic fraction (i.e., the cbc fraction in the spent medium). Between 1 and 20 μM amended copper, the copper associated with the whole-cell and membrane fractions increased proportionately with copper addition (Table 1). Between 20 and 60 μM copper addition, the copper concentration in the spent medium remained 0.8 ± 0.3 μM, but there was a less-than-expected increase in copper concentration per milligram of cell or membrane protein (Fig. 2A). Above 60 μM copper, copper accumulated in the spent medium, and the copper concentration per milligram of protein in the whole-cell and membrane fractions decreased. The results were surprising considering the average copper mass balances of the cell fractionation studies were close to 70% (68% ± 12%) and the copper mass balance of the whole-cell (i.e., whole-cell and spent medium fractions) component of the studies was close to 80% (78% ± 16%). The discrepancy between the copper lost from the spent medium and cell-associated copper was resolved when the cell-associated copper was calculated on a per-cell-number basis and not via protein concentration (Fig. 2B). When plotted against cell number, the results show the cell-associated copper increased until the amended copper reached 80 μM. Taken as a whole, the results from Fig. 2 and Table 1 indicate that in cultures with cell densities of 1.9 × 109 ± 0.3 × 109 cells per ml, the cells took up essentially all of the amended copper until the amended copper concentrations exceeded 60 μM. The results also indicate that the affinity of copper acquisition systems in M. capsulatus Bath is ≥0.5 μM.
FIG. 2.
(A) Copper concentrations associated with the whole-cell fraction (□) and membrane fraction (○) of cells cultured in chemostats with increasing copper concentrations. The right-hand axis shows the copper concentration in the spent medium following cell separation (▵). (B) Percent increase in copper per cell (○) and copper per milligram of whole-cell protein (•). Chemostat conditions and cell samplings were as described in the legend to Fig. 1.
TABLE 1.
Bivariate correlations of copper concentration in the culture media to methane oxidation activity and concentrations of copper, pMMO peptide and transcript, and fatty acids per cell
| Measurement and fraction | Copper concn (μM) with copper additiona:
|
||
|---|---|---|---|
| 0-80 μM | 0-59 μM | 60-80 μM | |
| Methane oxidation | |||
| Whole cell | 0.91** | 0.91** | 0.416 |
| Soluble | −0.342 | −0.382 | ND |
| Membrane | 0.959** | 0.921** | 0.957** |
| Copper | |||
| Spent medium | 0.754* | 0.644 | 0.898** |
| Whole cell | 0.766** | 0.848** | −0.937 |
| Soluble | 0.902** | 0.787* | 0.983* |
| Membrane | 0.846** | 0.867** | −0.258 |
| Peptide | |||
| sMMO | −0.78 | −0.83 | ND |
| pMMO | 0.93* | 0.980** | 0.673 |
| Transcript | |||
| sMMO | −0.568 | −0.568 | ND |
| pMMO | 0.977* | 0.977* | ND |
| Fatty acids | |||
| 14:00 | 0.871** | 0.911** | −0.811 |
| 16:1 cis9 | 0.626 | 0.898** | −0.917 |
| 16:1 cis11 | 0.911** | 0.838** | −0.473 |
| 16:00 | 0.695* | 0.872** | −0.993 |
| 17:00 | 0.742* | 0.437 | −0.132 |
| 16:0 3OH | 0.616 | 0.906** | −0.955 |
| 15:00 | 0.549** | 0.670 | −0.948 |
| Total | 0.710* | 0.870** | −0.973 |
*, P ≤ 0.05; **, P ≤ 0.01. ND, not determined.
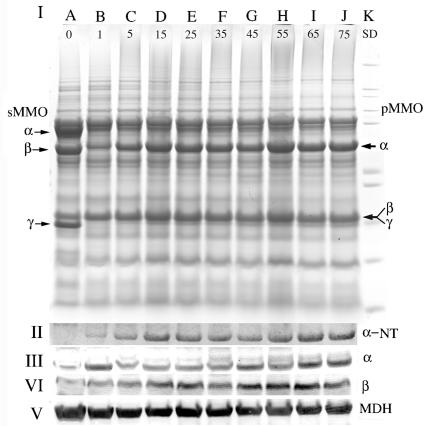
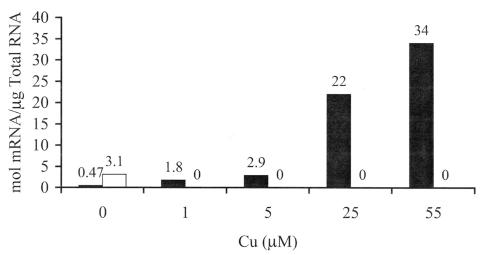
The results from Fig. 2 suggest the presence of highly expressed copper-regulated proteins in M. capsulatus Bath. pMMO was the obvious candidate for this copper-regulated protein, and the concentrations of both pMMO polypeptides (Fig. 3) and transcript (Fig. 4) were monitored. The peptide and transcript concentrations per cell both demonstrated that the expression levels of pMMO continued to increase even after the switchover from expression of sMMO to pMMO. Between 1 and 59 μM added copper, the concentration of pMMO polypeptides increased proportionally with copper (Fig. 3 and 4 and Table 1). At copper concentrations above 59 μM, a decrease in the concentration of pMMO polypeptides per cell was observed. Comparison of cells expressing pMMO cultured with 1 and 59 μM copper showed a 15-fold increase in the three pMMO polypeptides, an 18-fold increase in pMMO transcript concentration, and a 2.6-fold increase in the concentration of fatty acids per cell.
FIG. 3.
(I) SDS-PAGE of whole-cell samples from M. capsulatus Bath. Cell samples were taken before the addition of copper (0.3 μM copper) (lane A) and when the copper concentration in the chemostat reached 1 (lane B), 5 (lane C), 15 (lane D), 25 (lane E), 35 (lane F), 45 (lane G), 55 (lane H), 65 (lane I), and 75 (lane I) μM. Molecular mass standards (Invitrogen Mark 12 standards; 200, 116.3, 97.4, 66.3, 55.4, 36.3, 21.5, 14.4, 6, 3.5, and 2.5 kDa) are shown in lane K. Chemostat conditions and cell samplings were as described in the legend to Fig. 1. The cell sample in each lane was standardized to 1.3 × 108 cells per lane. (II) α-NT, Coomassie-stained gel illustrating the protein remaining in the 43-kDa region of an SDS-polyacrylamide gel following blotting for 1 h. The Coomassie-stained gel following transfer is included because of the poor transfer of the α subunit of the pMMO. (III and IV) Immunoblot analysis of M. capsulatus Bath cell fractions with antibodies against the α (III) and β (IV) subunits of the pMMO and with antibodies against MDH. (V) MDH was used as a non-copper-regulated protein control. Arrows to the left indicate sMMO hydroxylase polypeptides, and those to the right indicate pMMO polypeptides.
FIG. 4.
Transcript concentration of pmoA (▪) and mmoX (□) in M. capsulatus Bath cells cultured in medium not supplemented with copper; expressing sMMO; and following the addition of 1, 5, 25, or 55 μM CuSO4. Chemostat conditions and cell samplings were as described in the legend to Fig. 1.
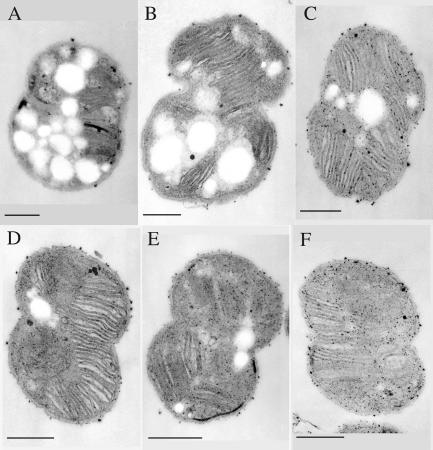
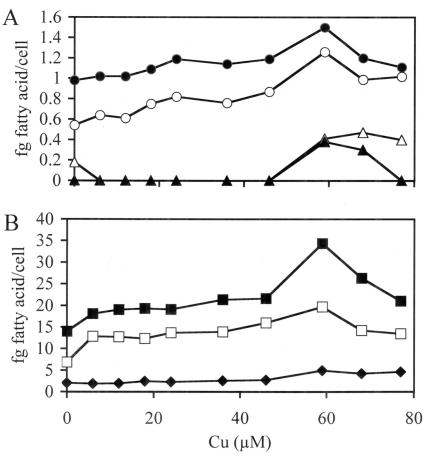
Previous results from this and S. Chan's laboratory have shown that when measured per milligram of membrane protein, the concentration of pMMO was essentially constant with increasing copper concentrations (32, 36, 52). To resolve the difference between the results presented above and those from previous studies, ultrastructural examination (Fig. 5) and fatty acid concentrations per cell (Fig. 6) were determined. Fatty acid concentrations per cell were used as an indicator of membrane density. Both studies demonstrated that a direct relationship exists between membrane content per cell and pMMO peptide concentration (P = 0.928, r = 0.01), pMMO transcript concentration (P = 1.00), and the concentration of copper in the culture medium (Table 1). The results also demonstrate that maximal membrane development and pMMO concentration per cell occurs at approximately 60 μM Cu (Fig. 5 and 6, Table 1). Between 1 and 59 μM amended copper, the percent increase in pMMO to percent increased membrane content per cell was approximately 1 to 0.17. Previous studies have shown a relationship between copper and membrane development in methanotrophs (3, 5, 7, 27-29). However, these studies compared only a few copper concentrations, with 10 μM amended copper as the maximum concentration examined.
FIG. 5.
Thin-section transmission electron micrograph of M. capsulatus Bath cultured as described in the legend to Fig. 1. Chemostat conditions were as described in the legend to Fig. 1. Cell samples were taken when the added copper concentration in the culture medium reached 5 (A), 20 (B), 40 (C), 60 (D), 80 (E), and 89 (F) μM. Marker bar, 200 nm.
FIG. 6.
Effect of copper concentration during growth of M. capsulatus Bath with low (A) and high (B) concentrations of fatty acids. The following fatty acids were monitored: 16:0 3-OH FAME (•), 14:0 FAME (○), 17:0 FAME (▵), 15:0 FAME (▴), 16:0 (▪), 16:1 cis 9 FAME (□), and 16:1 cis 11 (⧫). The chemostat conditions and cell samplings were as described in the legend to Fig. 1.
In addition to the polypeptides associated with the two MMOs, polypeptides with molecular masses of 36,000, 20,000, and 14,000 Da also appear to be regulated by copper (Fig. 2). N-terminal sequencing of all three polypeptides shows that the 20,000- and 14,000-Da polypeptides are breakdown products of the α subunit of the pMMO. The 36,000-Da polypeptide was a mixture of a breakdown product of the α subunit of the pMMO and type 2 NADH:quinone oxidoreductase (NDH) (6).
Detergent/protein ratio.
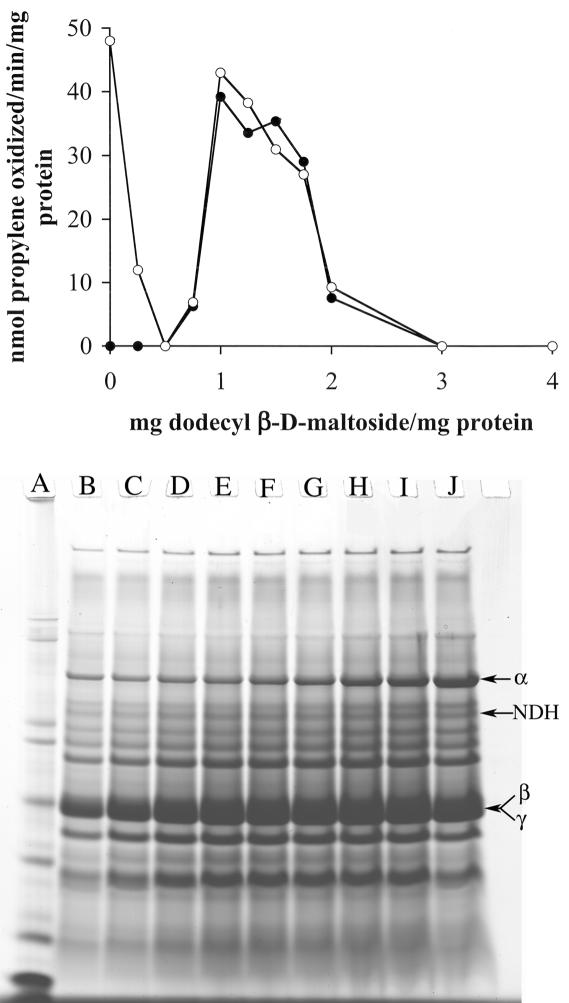
Previous studies on the solubilization of the pMMO using dodecyl β-d-maltoside focused on the activity in the detergent-solubilized fraction, with an optimal detergent/protein ratio of 1.6 mg of dodecyl β-d-maltoside per mg of protein (41, 52). With the higher-activity membrane preparations described above, the optimal detergent/protein ratio was reexamined. In contrast to previous studies, the propylene oxidation activity was measured before centrifugation and in the particulate and the detergent-solublized fractions following centrifugation. The results are shown in Fig. 7. Two basic conclusions were obtained from the results of the detergent series. First, the optimal detergent/membrane protein ratio for solublization of propylene oxidation activity was 1 to 1.25 mg of dodecyl β-d-maltoside per mg of protein. Above 0.3 mg of dodecyl β-d-maltoside per mg of membrane protein, solubilization of the αβγ polypeptides of the pMMO was proportional and increased with increasing detergent concentrations, with maximal solubilization at 4.6 mg of dodecyl β-d-maltoside per mg of membrane protein (Fig. 7). However, no activity was observed at detergent/protein ratios above 2.0. Second, both low and high detergent concentrations resulted in the irreversible inactivation of the pMMO. This inactivation was observed within 15 min of detergent addition.
FIG. 7.
(Top) Effect of dodecyl β-d-maltoside on membrane-associated propylene oxidation activity in M. capsulatus Bath. Shown are the rates of propylene oxidation following incubation in dodecyl β-d-maltoside for 1 h under anaerobic conditions before (○) and the soluble fraction after (•) centrifugation at 150,000 × g for 90 min. (Bottom) SDS-PAGE of solubilized protein (2-μl volume) samples following 150,000 × g centrifugation of washed membrane samples incubated for 1 h with 0.3 (B), 0.5 (C), 0.7 (D), 1.0 (E), 1.4 (F), 1.8 (G), 2.0 (H), 3.0 (I), or 4.0 (J) mg of dodecyl β-d-maltoside per mg of membrane protein.
To examine the effect of dodecyl β-d-maltoside concentration on pMMO, pMMO solublized at different detergent concentrations was purified (Fig. 8). As expected from the results presented in Fig. 7, pMMO isolated from both low-detergent (i.e., 0.5 and 0.75 mg of dodecyl β-d-maltoside per mg of membrane protein) and high-detergent (i.e., 2.0 and 3.0 mg of dodecyl β-d-maltoside per mg of membrane protein) extractions were inactive. The protein profiles of pMMO purified following each detergent extraction were identical; however, the metal composition of the samples varied. The metal content of inactive pMMO isolated from both low- and high-detergent extractions was approximately 2 copper and 2 iron atoms per αβγ subunit. In contrast, pMMO isolated following extractions of 1.0, 1.2, 1.4, and 1.6 mg of dodecyl β-d-maltoside per mg of membrane protein was active and contained 10 to 15 copper atoms and 2 iron atoms per αβγ subunit.
FIG. 8.
Effect of the dodecyl β-d-maltoside concentration used in the initial solubilization of pMMO on the propylene oxidation activity (□) and copper (○) and iron (▵) composition per αβγ subunit of purified pMMO.
Effect of detergent concentration on purified pMMO.
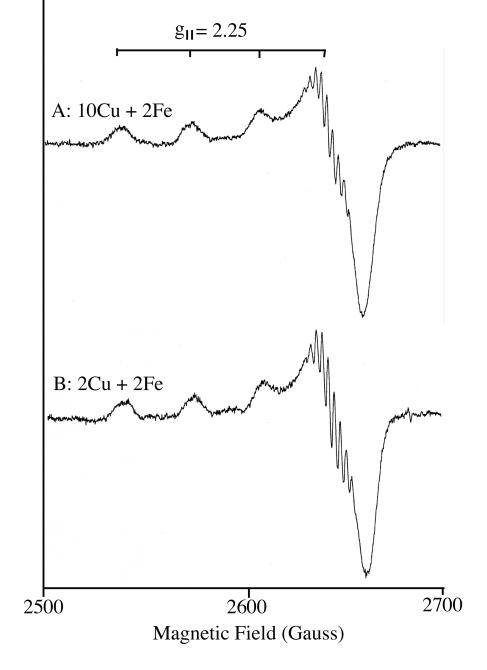
To examine the relationship between detergent and metal content of pMMO further, the effect of increasing detergent concentration on purified pMMO samples was tested. The addition of 0.1 and 0.2 mg of dodecyl β-d-maltoside per mg of protein had no effect on the purified sample. pMMO retained activity and the copper and iron content remained the same (Fig. 8). However, at concentrations ≥0.3 mg of dodecyl β-d-maltoside per milligram of pMMO, complete enzyme inactivation was observed and the copper atom concentration per pMMO αβγ subunit was approximately 2. The EPR spectra of the purified pMMO sample containing 10 copper atoms before (Fig. 9, trace A) and after (Fig. 9, trace B) the 0.3-mg dodecyl β-d-maltoside treatment, in which the copper concentration was reduced to 2 copper atoms, were essentially identical. The spectra from both samples showed the type 2 Cu(II) signals associated with the αβγ subunits of the pMMO were present and of similar intensities (2, 23, 25, 48-50, 52). As in previous studies using Triton X-100, the copper dissociated from the αβγ polypeptides, was associated with the cbc (52). Over 70% of the copper atoms bound by cbc are Cu(I) and thus EPR silent (14, 52), which explains the similarities in the spectra before and after detergent treatments.
FIG. 9.
X-band EPR spectra at 77 K of the cupric site in high-detergent-concentration-treated purified active pMMO (79 mU/mg of protein) containing 10 Cu and 2 iron atoms per αβγ complex (A) and following detergent treatment and containing 2 copper and 2 iron atoms per αβγ complex (B). The following experimental conditions were used: modulation frequency, 100 kHz; modulation amplitude, 5 G; time constant, 100 ms; microwave frequency, 9.191 GHz; and microwave power, 5.0 mW.
Superoxide dismutase-like activity of Cu-cbc.
Studies on the physiological role of cbc indicate it is a component of a high-affinity copper uptake system in methanotrophs that express both forms of MMO (14, 17, 21, 45, 52). However, as in previous preparations from this laboratory, the Cu-cbc copurified with pMMO. Previous studies had speculated that Cu-cbc may provide protection from oxygen radicals (52). The structural characterization of Cu-cbc (21) also indicates that this molecule may function as an oxygen radical scavenger: i.e., the copper atom is in an open coordination sphere with a tetrahedral geometry on the surface of the molecule (3, 8, 15, 16, 38). To test this activity, the O2 · − scavenging properties of Cu-cbc were examined according to its ability to inhibit the reduction of nitroblue tetrazolium (7, 8) (Fig. 10). Copper-free cbc has little to no superoxide dismutase-like activity, but Cu-cbc has superoxide dismutase-like activity similar to or greater than that of superoxide dismutase or copper complexes designed or examined for this activity (3, 4, 8, 15, 16, 38).
FIG. 10.
Inhibition of nitroblue tetrazolium reduction in the presence of Cu-cbc.
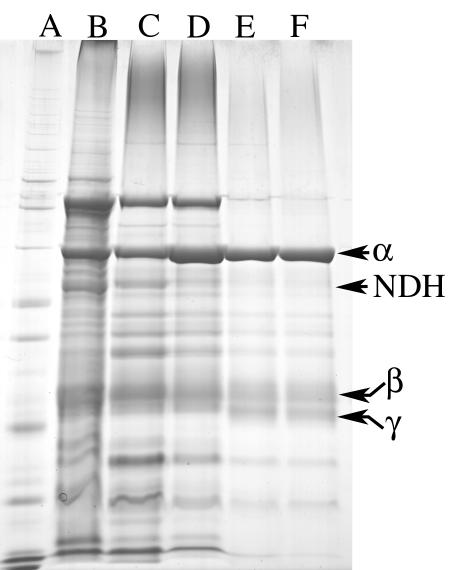
Purification of pMMO and NDH-pMMO complex.
The purification of pMMO and the NDH-pMMO complex from M. capsulatus Bath was performed as described in Materials and Methods, and the results are summarized in Table 2 and Fig. 11. The major loss of activity was observed during cell lysis, which can be minimized if the procedure is carried out under anaerobic conditions. Recent studies have suggested pMMO activity is not oxygen sensitive (25). However, the pMMO activities in the washed membrane fraction in studies reporting the absence of oxygen sensitivity in the washed membrane fraction were less than 15% of the activities reported in this study. In membrane fractions with propylene oxidation activity below 40 nmol of propylene oxidized · min−1 · mg of protein−1, the activity was stable following 3 h of exposure to air. However, in washed membrane fractions with propylene oxidation activity above 40 nmol of propylene oxidized · min−1 · mg of protein−1, loss of activity was often observed following exposure to air for 3 h. Following detergent solubilization, all pMMO samples tested were oxygen sensitive. A modest increase in activity was observed at each purification step after detergent solubilization, with final propylene oxidation activities of 147 ± 43 and 134 ± 36 nmol of propylene oxidized · min−1 · mg of protein−1 for the NDH-pMMO complex and purified pMMO, respectively.
TABLE 2.
Purification of pMMO from M. capsulatus Bath using duroquinol, NADH, NADH plus duroquinol or duroquinol alone, NADH, and NDH as the reductantsa
| Fraction | Copper concn (nmol/mg of protein) | Iron concn (nmol/mg of protein) | Propylene oxidation rate (nmol · min−1 · mg of protein−1)
|
||||
|---|---|---|---|---|---|---|---|
| Formate | Duroquinol | NADH | Duroquinol + NADH | Duroquinol + NADH + NDH | |||
| Whole cell | 121 ± 21 | 21 ± 8 | 235 ± 68 | NDb | ND | ND | ND |
| Washed membrane | 237 ± 42 | 54 ± 13 | ND | 63 ± 23 | 81 ± 37 | 78 ± 45 | ND |
| Dodecyl β-d-maltoside extract | 193 ± 23 | 46 ± 16 | ND | 51 ± 27 | 25 ± 14 | 74 ± 21 | ND |
| First DEAE-Sepharose FF | 117 ± 14 | 16 ± 3 | ND | 73 ± 34 | 36 ± 13 | 101 ± 33 | 147 ± 43 |
| Second DEAE-Sepharose FF | 101 ± 16 | 18 ± 5 | ND | 95 ± 31 | 0 | 97 ± 32 | 134 ± 36 |
Values represent results from four different preparations.
ND, not determined.
FIG. 11.
SDS-PAGE (slab gel) of fractions during the purification of NDH-pMMO complex and of pMMO from M. capsulatus Bath. Lanes: A, whole-cell fraction; B, washed membrane fraction; C, detergent-solubilized fraction (1.2 g of dodecyl β-d-maltoside per g of membrane protein); D, first DEAE-Sepharose FF eluate; E, second DEAE-Sepharose FF eluate.
Physiological reductant.
The initial isolation of the pMMO utilized duroquinol as a reductant (52). Propylene oxidation activity in the precolumn steps was higher if NADH was used as a reductant; however, NADH-driven propylene oxidation activity was lost in the final purification steps. Thus, duroquinol was used as the reductant in cell-free assays, despite the fact that this reductant became insoluble before saturation (52). In an attempt to increase the concentration of reduced duroquinone in reaction mixtures, NADH and NDH from M. capsulatus Bath were added along with duroquinol to pMMO activity assays. The additions of NADH and NDH to pMMO assays resulted in a 20 to 35% increase in enzyme activity (Table 2).
The purification procedure for pMMO was also modified to copurify pMMO with NDH. In the current purification procedure, the concentration of NDH peptide added to pMMO polypeptides is low, and the NDH activity in the NDH-pMMO complex represents <80% of the pMMO activity. Thus, this fraction still requires the addition of NDH for optimal activity.
DISCUSSION
In addition to the stabilization of cell-free pMMO activity, the results presented in this paper demonstrate that copper not only regulates a metabolic switch between the two methane MMOs, it also regulates the level of expression of the pMMO as well as the concentration of internal membranes. Previous studies have shown this relationship; however, only a few copper concentrations were examined, and the maximal levels of expression were never determined (5, 9, 11, 31, 37, 39, 42). The results presented here show that optimal expression of pMMO is observed in medium containing approximately 60 μM copper, when the cell density is 1.9 × 109 ± 0.3 × 109 per ml. However, for optimal cell-free pMMO activity, the concentration of copper in the culture medium was increased to 80 μM. Between 60 and 80 μM amended copper, the concentration of pMMO per cell is essentially constant, but the copper concentration per cell continues to increase. This increase in copper concentration per cell probably results from the increased saturation of the copper sites in the pMMO fraction. As observed in other laboratories, copper often stimulates the propylene oxidation activity in cell-free and purified pMMO samples (2, 25, 33, 44). An increase in the type 2 Cu(II) signal was also observed following the addition of copper to purified pMMO preparations (W. E. Antholine, D.-W. Choi, E. S. Boyd, and A. A. DiSpirito, unpublished results). Again the results suggest that the copper sites in cellular as well as purified preparations of pMMO are rarely saturated.
In addition to growth conditions, the results reported here illustrate the importance of detergent/protein ratios during pMMO solublization and purification. The inactivation of pMMO at detergent/membrane protein ratios of ≤0.75 and ≥2.0 apparently resulted from the differential solubilization of the pMMO and Cu-cbc. At detergent/membrane protein ratios of ≤0.75, the αβγ polypeptides solublized are Cu-cbc free. Detergent/membrane protein ratios of ≥2.0 result in the dissociation of the αβγ polypeptides and Cu-cbc. The results suggest the minimal metal content for active purified pMMO as described here is 8 to 10 Cu atoms and 2 iron atoms, with 2 Cu and 2 Fe atoms associated with the αβγ polypeptides and 6 to 8 atoms associated with cbc. The stabilizing effect of Cu-cbc on cell-free pMMO activity is probably the result of its superoxide dismutase-like activity. It is possible that this may be a secondary function of the Cu-cbc in vivo, but it may prove important in protecting this oxygen-utilizing enzyme following solublization from the membrane fraction.
This observation differs from those of two recent reports by Basu et al. (2) and Lieberman et al. (25), which presented partially purified and purified preparation of pMMO containing 2 Cu atoms and 1 to 2 Fe atoms per αβγ subunit. The difference in metal composition of the pMMO in these studies and the results reported here appear to be due to the isolation of pMMO with associated Cu-cbc, as reported in this study, and pMMO isolated in the absence of Cu-cbc. If the samples isolated by Basu et al. (2) and Lieberman et al. (25) are Cu-cbc free, the superoxide-dismutase-like activity of the Cu-cbc may be one of the reasons for higher specific activity of the preparations reported in this study.
The nature of the physiological reductant to the pMMO has been a source of controversy. Most of the available data from this and other laboratories indicate that the pMMO is coupled to the electron transport chain at the cytochrome bc1 complex, probably via the quinone (2, 11, 13, 25, 44, 51, 52), although NADH has also been proposed as the electron donor (33). The reports of NADH-driven pMMO activity are probably due to contaminating NDH in pMMO preparations. The NDH-pMMO complex did show activity with NADH as the sole reductant. However, the activity was only 30% of the activity following the addition of duroquinol. Thus, duroquinol-free activity may have resulted from contaminating ubiquinone in pMMO preparations. Trace contamination of the cytochrome bc1 complex has been shown to be a common trace contaminant in pMMO preparations (23, 41). The ability to separate the NDH and methane (as measured by propylene) oxidation activity indicates the findings of previous reports using NADH as a reductant in pMMO assays were probably due to the presence of NDH in these preparations (33).
In summary, the higher-activity pMMO preparations reported here were the result of several modifications to previous purification attempts. Of the parameters examined, the growth conditions appear to be the most critical factor in stabilizing cell-free pMMO activity. For high cell-free activities, the cells should be maintained in log phase throughout the copper and iron addition series. Copper addition is still critical, which not only results in increased pMMO expression but also increases the concentration of Cu-cbc in the membrane fraction. With respect to stabilizing cell-free pMMO activity, Cu-cbc probably serves a secondary function as an oxygen radical scavenger. Finally, the detergent/enzyme ratio is also critical to activity.
Acknowledgments
This work was supported by the Department of Energy grant 02-96ER20237 to A.A.D.S. and National Science Foundation grant 9708557 to J.D.S.
REFERENCES
- 1.Anthony, C. 1982. The biochemistry of methylotrophs. Academic Press, London, United Kingdom.
- 2.Basu, P., B. Katterle, K. A. Anderson, and H. Dalton. 2002. The membrane-associated form of methane monooxygenase from Methylococcus capsulatus (Bath) is a copper/iron protein. Biochem. J. 369:417-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bielski, B. H., and H. W. Richter. 1977. A study of superoxide radical chemistry by stopped-flow radiolysis and radiation induced oxygen consumption. J. Am. Chem. Soc. 99:3019-3023. [Google Scholar]
- 4.Bonomo, R. P., E. Conte, R. Marchelli, A. M. Santoro, and G. Tabbi. 1994. O2− scavenger properties of copper(II) m complexes with diamino-diamide-type ligands. J. Inorg. Biochem. 53:127-138. [Google Scholar]
- 5.Brantner, C. A., L. A. Buchholz, C. L. McSwain, L. L. Newcomb, C. C. Remsen, and M. L. P. Collins. 1997. Intracytoplasmic membrane formation in Methylomicrobium album BG8 in the growth medium. Can. J. Microbiol. 43:672-676. [Google Scholar]
- 6.Brusseau, G. A., H.-C. Tsien, R. S. Hanson, and L. P. Wackett. 1990. Optimization of trichloroethylene oxidation by methanotrophs and the use of a colorimetric assay to detect soluble methane monooxygenase activity. Biodegradation 1:19-29. [DOI] [PubMed] [Google Scholar]
- 7.Bury, A., and A. E. Underhill. 1987. Metal complexes of anti-inflammatory drugs. IV. Tenoxicam complexes of manganese(II), iron(III), cobalt(II), nickel(II), and copper(II). Inorg. Chim. Acta 138:85-89. [Google Scholar]
- 8.Casanova, J., G. Alzuet, J. Borras, J. Timoneda, S. Garcia-Granda, and I. Candano-Gonzalez. 1994. Coordination behavior of sulfathiazole. Crystal structure of dichloro-disulfathiazole ethanol Cu(II) complex. Superoxide dismutase activity. J. Inorg. Biochem. 56:65-76. [Google Scholar]
- 9.Collins, M. L. P., L. A. Buchholz, and C. C. Remsen. 1991. Effect of copper on Methylomonas albus BG8. Appl. Environ. Microbiol. 57:1261-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cook, S. A., and A. K. Shiemke. 2002. Evidence that a type-2 NADH:quinone oxidoreductase mediates electron transfer to particulate methane monooxygenase in Methylococcus capsulatus. Arch. Biochem. Biophys. 398:32-40. [DOI] [PubMed] [Google Scholar]
- 11.Dalton, H., S. D. Prior, D. J. Leak, and S. H. Stanley. 1984. Regulation and control of methane monooxygenase, p. 75-82. In R. L. Crawford and R. S. Hanson (ed.), Microbial growth on C1 compounds. American Society for Microbiology, Washington, D.C.
- 12.DiSpirito, A. A., J. Gulledge, A. K. Shiemke, J. C. Murrell, and M. E. Lidstrom. 1992. Trichloroethylene oxidation by the membrane-associated methane monooxygenase in type I, type II, and type X methanotrophs. Biodegradation 2:151-164. [Google Scholar]
- 13.DiSpirito, A. A., R. C. Kunz, D. W. Choi, and J. A. Zahn. Electron flow during methane oxidation in methanotrophs. In D. Zannoni (ed.), Respiration in Archaea and Bacteria, vol. 2, in press. Kluwer Scientific, Dordrecht, The Netherlands.
- 14.DiSpirito, A. A., J. A. Zahn, D. W. Graham, H. J. Kim, C. K. Larive, T. S. Derrick, C. D. Cox, and A. Taylor. 1998. Copper-binding compounds from Methylosinus trichosporium OB3b. J. Bacteriol. 180:3606-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El-Naggar, M. M. 1997. Protective action of some Cu(II) complexes against photohemolysis induced by m. chloroperbenzoic acid. J. Inorg. Biochem. 64:263-266. [DOI] [PubMed] [Google Scholar]
- 16.Fee, J. A. 1981. The copper/zinc superoxide dismutase, p. 259-298. In H. S. Sigel and A. Sigel (ed.), Metal ions in biological systems, vol. 13. Marcel Dekker, Inc., New York, N.Y. [Google Scholar]
- 17.Fitch, M. W., D. W. Graham, R. G. Arnold, S. K. Agarwal, P. Phelps, G. E. Speitel, Jr., and G. Georgiou. 1993. Phenotypic characterization of copper-resistant mutants of Methylosinus trichosporium OB3b. Appl. Environ. Microbiol. 59:2771-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green, J., and H. Dalton. 1985. The biosynthesis and assembly of protein A of soluble methane monooxygenase from Methylococcus capsulatus Bath. J. Biol. Chem. 260:15795-15802. [PubMed] [Google Scholar]
- 19.Han, J.-I., and J. D. Semrau. Quantification of the expression of pmoA in methanotrophs using RT-PCR. Proc. Am. Chem. Soc., in press.
- 20.Hanson, R. S., and T. E. Hanson. 1996. Methanotrophic bacteria. Microbiol. Rev. 60:439-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, H. J. 2003. Isolation, structural elucidation, and characterization of a novel copper-binding compound from Methylosinus trichosporium OB3b: its 1.1 A crystal structure. Ph.D. thesis. University of Kansas, Lawrence.
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London). 227:680-685. [DOI] [PubMed] [Google Scholar]
- 23.Lemos, S. S., M. L. P. Collins, S. S. Eaton, G. R. Eaton, and W. E. Antholine. 2000. Comparison of EPR-visible Cu2+ sites in pMMO from Methylococcus capsulatus (Bath) and Methylomicrobium album BG8. Biophys. J. 79:1085-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lidstrom, M. E. 1988. Isolation and characterization of marine methanotrophs. Antonie Leeuwenhoek J. Microbiol. Serol. 54:189-199. [DOI] [PubMed] [Google Scholar]
- 25.Lieberman, R. L., D. B. Shrestha, P. E. Doan, B. M. Hoffmann, T. L. Stemmler, and A. C. Rosenzweig. 2003. Purified particulate methane monooxygenase from Methylococcus capsulatus (Bath) is a dimer with both mononuclear copper and a copper-containing cluster. Proc. Natl. Acad. Sci. USA 100:3820-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lipscomb, J. D. 1994. Biochemistry of the soluble methane monooxygenase. Annu. Rev. Microbiol. 48:371-399. [DOI] [PubMed] [Google Scholar]
- 27.Liu, K. E., and S. J. Lippard. 1991. Redox properties of the hydroxylase component of methane monooxygenase from Methylococcus capsulatus (Bath). Effects of protein B, reductase, and substrate. J. Biol. Chem. 266:12836-12839. [PubMed] [Google Scholar]
- 28.Lontoh, S., and J. D. Semrau. 1998. Methane and trichloroethylene degradation by Methylosinus trichosporium expressing particulate methane monooxygenase. Appl. Environ. Microbiol. 64:1106-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurment with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 30.Miyaji, A., T. Kamachi, and I. Okura. 2002. Improvement of the purification method for retaining the activity of the particulate methane monooxygenase from Methylosinus trichosporium OB3b. Biotechnol. Lett. 24:1883-1887. [Google Scholar]
- 31.Murrell, J. C., I. R. McDonald, and B. Gilbert. 2000. Regulation of expression of methane monooxygenases by copper ions. Trends Microbiol. 8:221-225. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen, A.-N., A. K. Schiemke, S. J. Jacobs, B. J. Hales, M. E. Lidstrom, and S. I. Chan. 1994. The nature of the copper ions in the membranes containing the particulate methane monooxygenase from Methylococcus capsulatus (Bath). J. Biol. Chem. 269:14995-15005. [PubMed] [Google Scholar]
- 33.Nguyen, H.-H., S. J. Elliott, J. H.-K. Yip, and S. I. Chan. 1998. The particulate methane monooxygenase from Methylococcus capsulatus (Bath) is a novel copper-containing three-subunit enzyme. J. Biol. Chem. 273:7957-7966. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen, H.-H., K. H. Nakagawa, B. Hedman, S. J. Elliott, M. E. Lidstrom, K. D. Hodgson, and S. I. Chan. 1996. X-ray absorption and EPR studies on the copper ions associated with particulate methane monooxygenase from Methyococcus capsulatus Bath. Cu(I) ions and their implications. J. Am. Chem. Soc. 118:12766-12776. [Google Scholar]
- 35.Nielsen, A. K., K. Gerders, and J. C. Murrell. 1997. Copper-dependent reciprocal transcriptional regulation of methane oxidation genes in Methylococcus capsulatus Bath and Methylosinus trichosporium OB3b. Mol. Microbiol. 25:399-409. [DOI] [PubMed] [Google Scholar]
- 36.Peltola, P., P. Priha, and S. Laakso. 1993. Effect of copper on membrane lipids and on methane monooxygenase activity of Methylococcus capsulatus (Bath). Arch. Microbiol. 159:521-525. [Google Scholar]
- 37.Prior, S. D., and H. Dalton. 1985. Copper stress underlies the fundamental change in intracellular location of methane monooxygenase in methane oxidizing organisms: studies in batch and continuous culture. J. Gen. Microbiol. 131:155-163. [Google Scholar]
- 38.Ramadan, A. M., and M. M. El-Naggar. 1996. Synthesis, characterization and demonstration of superoxide dismutase-like activity of copper(II) chloride, bromide, nitrate, thiocyanate, sulphate, and perchlorate complexes with 2-methyl-amino pyridine. J. Inorg. Biochem. 63:143-153. [Google Scholar]
- 39.Scott, D., J. Brannan, and I. J. Higgins. 1981. The effect of growth conditions on intracytoplasmic membranes and methane monooxygenase activities in Methylosinus trichosporium OB3b. J. Gen. Microbiol. 125:63-72. [Google Scholar]
- 40.Shiemke, A. K., S. A. Cook, T. Mily, and P. Singleton. 1995. Detergent-solubilization of membrane-bound methane monooxygenase requires plastoquinol analogues as electron donors. Arch. Biochem. Biophys. 321:521-528. [DOI] [PubMed] [Google Scholar]
- 41.Smith, D. D. S., and H. Dalton. 1989. Solubilization of methane monooxygenase from Methylococcus capsulatus (Bath). Eur. J. Biochem. 182:667-671. [DOI] [PubMed] [Google Scholar]
- 42.Stanley, S. H., S. D. Prior, D. J. Leak, and H. Dalton. 1983. Copper stress underlies the fundamental change in intracellular location of methane monooxygenase in methane-oxidising organisms: studies in batch and continuous cultures. Biotechnol. Lett. 5:487-492. [Google Scholar]
- 43.Stolyar, S., A. M. Costello, T. L. Peeples, and M. E. Lidstrom. 1999. Role of multiple gene copies in particulate methane monooxygenase activity in the methane-oxidizing bacterium Methylococcus capsulatus Bath. Microbiology 132:1235-1244. [DOI] [PubMed] [Google Scholar]
- 44.Takeguchi, M., K. Miyakawa, and I. Okura. 1998. Purification and properties of particulate methane monooxygenase from Methylosinus trichosporium OB3b. J. Mol. Catal. 132:145-153. [DOI] [PubMed] [Google Scholar]
- 45.Telléz, C. M., K. P. Gaus, D. W. Graham, R. G. Arnold, and R. Z. Guzman. 1998. Isolation of copper biochelates from Methylosinus trichosporium OB3b and soluble methane monooxygenase mutants. Appl. Environ. Microbiol. 64:1115-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tukhvatullin, I. A., R. I. Gvozdev, and K. K. Andersson. 2000. The structure of the active center of β-peptide membrane-bound methane monooxygenase (pMMO) from Methylococcus capsulatus Bath. Biochem. Biophys. Mol. Biol. 374:177-182. [PubMed] [Google Scholar]
- 47.Wallar, B. J., and J. D. Lipscomb. 1996. Dioxygen activation by enzymes containing binuclear non-heme iron clusters. Chem. Rev. 96:2625-2657. [DOI] [PubMed] [Google Scholar]
- 48.Yuan, H., M. L. P. Collins, and W. A. Antholine. 1998. Concentration of Cu, EPR detectable Cu, and formation of cupric-ferrrocyanide in membranes with pMMO. J. Inorg. Biochem. 72:179-185. [DOI] [PubMed] [Google Scholar]
- 49.Yuan, H., M. L. P. Collins, and W. A. Antholine. 1997. Low frequency EPR of the copper in particulate methane monooxygenase from Methylomicrobium albus BG8. J. Am. Chem. Soc. 119:5073-5074. [Google Scholar]
- 50.Yuan, H., M. L. P. Collins, and W. A. Antholine. 1999. Type 2 Cu(2+) in pMMO from Methylomicrobium albus BG8. Biophys. J. 76:2223-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zahn, J. A., D. J. Bergmann, J. M. Boyd, R. C. Kunz, and A. A. DiSpirito. 2001. Membrane-associated quinoprotein formaldehyde dehydrogenase from Methylococcus capsulatus Bath. J. Bacteriol. 183:6832-6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zahn, J. A., and A. A. DiSpirito. 1996. Membrane-associated methane monooxygenase from Methylococcus capsulatus (Bath). J. Bacteriol. 178:1018-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]