Abstract
The enzyme 2-phosphoglycolate phosphatase from Escherichia coli, encoded by the gph gene, was purified and characterized. The enzyme was highly specific for 2-phosphoglycolate and showed good catalytic efficiency (kcat/Km), which enabled the conversion of this substrate even at low intracellular concentrations. A comparison of the structural and functional features of this enzyme with those of 2-phosphoglycolate phosphatases of different origins showed a high similarity of the sequences, implying the use of the same catalytic mechanism. Western blot analysis revealed constitutive expression of the gph gene, regardless of the carbon source used, growth stage, or oxidative stress conditions. We showed that this housekeeping enzyme is involved in the dissimilation of the intracellular 2-phosphoglycolate formed in the DNA repair of 3′-phosphoglycolate ends. DNA strand breaks of this kind are caused by agents such as the radiomimetic compound bleomycin. The differential response between a 2-phosphoglycolate phosphatase-deficient mutant and its parental strain after treatment with bleomycin allowed us to connect the intracellular formation of 2-phosphoglycolate with the production of glycolate, which is subsequently incorporated into general metabolism. We thus provide evidence for a salvage function of 2-phosphoglycolate phosphatase in the metabolism of a two-carbon compound generated by the cellular DNA repair machinery.
In Escherichia coli, the gene gph, encoding 2-phosphoglycolate phosphatase (PGPase) (EC 3.1.3.18), is integrated in the dam-containing operon (20), which is located at the 3.51-Mb position of the genome (3) and contains the following genes (clockwise): aroK, encoding shikimic acid kinase (18); aroB, encoding 3-hydroquinate synthase (24); damX, encoding a protein of unknown function; dam, encoding DNA adenine methyltransferase (15, 20); rpe, encoding ribulose-5-phosphate 3-epimerase (21); gph, encoding PGPase (22); and trpS, encoding tryptophanyl-tRNA synthetase (12). Despite the presence of several internal promoters, there is evidence that these seven apparently unrelated genes are cotranscribed into the same mRNA and thus constitute an operon, which spans 7 kb (20). An analysis of extracts of cells defective in the gph gene and cells overexpressing the enzyme showed that the 27-kDa protein encoded by this gene is endowed with PGPase activity (22). These authors reported that a PGPase deficiency had no effect on growth rate, replication kinetics, or cell size.
The PGPases from several species, including tobacco (7), spinach (14), maize (13), and Chlamydomonas reinhardtii (23), as well as from red blood cells have been purified and characterized structurally and kinetically (27, 39). They share several features, a fact which has led some authors to propose a common mechanism of 2-phosphoglycolate dephosphorylation, in which an intermediate phosphoenzyme is generated and subsequently hydrolyzed (33). Bacterial PGPases are closely related, as suggested by the similarity of their amino acid sequences. Noteworthy is the conservation of a motif close to the N terminus, (F or L)DLDGTL, which has been suggested to be necessary for the activities of several phosphatases acting on carbon compounds or P-serine proteins (9).
PGPase participates in CO2 assimilation in chemoautotrophic organisms. In these organisms, it dephosphorylates the 2-phosphoglycolate formed as a by-product of Calvin cycle ribulose-1,5-bisphosphate carboxylase/oxygenase activity and converts it to glycolate, which can then be incorporated into general metabolism (34). PGPase also modulates the affinity of hemoglobin for oxygen by modification of the bisphosphoglycerate shunt in red blood cells (27). However, its specific physiological role in E. coli and other chemoheterotrophic organisms has not been elucidated so far (10, 22).
Here we purified and characterized the PGPase of E. coli and assigned it the physiological function of metabolizing the 2-phosphoglycolate produced in the repair of a major class of DNA lesions induced by oxidative stress.
MATERIALS AND METHODS
Bacterial strains, phages, and plasmids.
All of the strains used were E. coli K-12 derivatives. MC4100 (araD Δlac rpsL flbB deoC ptsF rbsR) (6) was used as the parental strain. XL1Blue (recA1 lac endA1 gyrA96 thi hsdR17 supE44 relA1 [F′ proAB lacIQ lacZΔM15 Tn10]) (Stratagene) was used as the host strain for recombinant plasmids in pBluescript SK (Stratagene). Strain BL21(DE3) (Amersham Pharmacia Biotech) was used to express glutathione S-transferase (GST)-PGPase. Strain ANL9 (20) was used to transduce the knockout mutation gph::Kanr into the genetic background of strain MC4100, yielding strain JA210. The transcriptional fusion of the glc operon, φ(glcD-lacZ) (26), was introduced by transduction into various genetic backgrounds. All transductions were performed with phage P1 as described by Miller (25).
pGEX-3X was used as a vector for GST fusion proteins (Amersham Pharmacia Biotech). Plasmid pTP100, bearing the gph gene, was constructed by cloning into pBluescript SK the 1,440-bp PCR fragment obtained with primers gph-BS1 (5′-GATGGATCCCGTCGATCGCATTGTGC-3′) and gph-BS2 (5′-TGGAATTCGGATCGATACCACAAGCC-3′), harboring, respectively, BamHI and EcoRI restriction sites (underlined). All constructs obtained by PCR were sequenced to confirm that no unwanted mutations had been introduced.
Cell growth and preparation of cell extracts.
Cells were grown aerobically in Luria broth (LB) or minimal medium and harvested as described elsewhere (4). For growth in minimal medium, carbon sources were added at 60 mM unless otherwise specified. Casein acid hydrolysate (Caa) was used at 0.5% (wt/vol) or at 1% (wt/vol) for the growth of transformed cells. The following antibiotic concentrations were used, unless otherwise noted: ampicillin, 100 μg/ml; tetracycline, 12.5 μg/ml; and kanamycin, 50 μg/ml. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and isopropyl-β-d-thiogalactopyranoside (IPTG) were used at 30 and 10 μg/ml, respectively. Kanamycin and tryptophan (42 μg/ml) were added to cultures of cells bearing φ(glcD-lacZ).
Extracts were prepared as described elsewhere (4) with 20 mM Tris-HCl buffer (pH 7.5) containing 10 mM MgCl2 and 1 mM dithiothreitol.
Cell treatments and survival estimation.
To select the dose of bleomycin-Fe(II) or H2O2 for use in the oxidative stress assays, MC4100 φ(glcD-lacZ) and JA210 φ(glcD-lacZ) cells were grown aerobically in minimal medium with 0.5% Caa in overnight cultures at 37°C. Cells were diluted into the same fresh medium the next day, grown to mid-exponential phase (optical density at 600 nm, 0.5) at 37°C, and then treated with various drug concentrations for 60 min. An aliquot of cells was used to obtain genomic DNA, while the rest of the withdrawn culture was collected and washed twice with minimal medium. Survival was estimated by diluting cells into the same medium, followed by plating on LB-kanamycin plates and overnight incubation at 37°C to determine CFU.
In a typical experiment, bleomycin was used at 5 μg/ml in the presence of 50 μM FeSO4, and H2O2 was used at 2 mM. Bleomycin sulfate (Almirall Prodesfarma, Barcelona, Spain) was freshly prepared with 50 mM phosphate buffer (pH 7.5). FeSO4 solutions were prepared immediately before use.
DNA manipulation.
Bacterial genomic DNA was obtained with a Wizard SV Plus genomic DNA kit, and plasmid DNA was routinely prepared with a Wizard SV Plus DNA purification system (Promega Corporation). DNA manipulations were performed as described by Sambrook and Russell (29). DNA sequencing was carried out with a dye terminator kit and an automated ABI 377 DNA sequencer. DNA fragments were amplified by PCR with E. coli chromosomal DNA as a template. PCR was performed with Pfu DNA polymerase under standard conditions.
Expression and purification of the gph-encoded enzyme.
PGPase was purified by using a GST gene fusion system with recognition sites for factor Xa cleavage. The gph gene was amplified by PCR with primers GphN-GST (5′-GGCGGATCCTTATGAATAAGTTTGAAGATATTCGCGGC-3′) and GphC-GST (5′-GCGGAATTCGGGCTTAGTCATTTTTCGATTC-3′), bearing, respectively, BamHI and EcoRI restriction sites (underlined). The PCR fragment was digested and cloned into the BamHI and EcoRI restriction sites of plasmid pGEX-3X, yielding plasmid pGEX-Gph. Primer GphN-GST was designed to fuse the reported ATG start codon of gph (20) in frame with GST.
PGPase was overexpressed in strain BL21(DE3) carrying recombinant plasmid pGEX-Gph after IPTG (0.5 mM) induction in LB-ampicillin medium for 3 to 4 h at 37°C. The GST fusion protein was purified by affinity chromatography with glutathione-Sepharose 4B resin (Amersham Pharmacia Biotech). Centrifugation was carried out at 4°C, and column chromatography was performed at room temperature. To purify PGPase, the cell pellet from cultures of strain BL21(DE3) bearing pGEX-Gph was suspended in phosphate-buffered saline (PBS; 140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 [pH 7.3]) and sonicated on ice. The cell extract, previously diluted 10-fold, was applied to a column containing glutathione-Sepharose 4B resin. After the column was washed with PBS, factor Xa cleavage buffer was passed through it to equilibrate the matrix before cleavage of the bound GST fusion protein. Digestion was performed by application of factor Xa solution (50 U) to the column and subsequent overnight incubation at room temperature. The cleaved PGPase protein was eluted with PBS and dialyzed against 20 mM Tris-HCl (pH 7.5) containing 10 mM MgCl2 and 1 mM dithiothreitol. The dialyzed sample was applied to a DEAE-Sepharose column equilibrated with the latter buffer. The column was washed with the same buffer, and proteins were eluted with a linear gradient of 0 to 0.5 M NaCl at a flow rate of 1 ml/min. The active fractions were combined, concentrated, and stored at −20°C in the presence of 20% (wt/vol) glycerol.
The GST-PGPase fusion protein was purified by following the same procedure, except that the factor Xa cleavage step was omitted and the GST fusion protein was eluted with 10 mM glutathione in 50 mM Tris-HCl (pH 8.0).
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) was performed as described by Laemmli (16).
Preparation of PGPase antisera and Western blot analysis.
Antisera against E. coli PGPase were raised in New Zealand White rabbits as described elsewhere (1) by using purified GST-PGPase as an antigen. For Western blot analysis, protein samples were electrophoresed in an SDS-10% polyacrylamide gel and transferred to a HyBond-P polyvinylidene difluoride membrane by using a Bio-Rad MiniTransblot apparatus. The membrane was blocked in 100 mM Tris-HCl (pH 7.4)-100 mM MgCl2-0.5% (vol/vol) Tween 20-1% Triton X-100-1% (wt/vol) bovine serum albumin-5%(vol/vol) fetal calf serum (blocking solution) for 1 h at 4°C. It was then incubated with antisera against PGPase (1:1,000 dilution in blocking solution) for 16 h at 4°C. The PGPase-antibody complex was visualized by using an ECL Plus Western blotting system (Amersham Pharmacia Biotech) in accordance with the manufacturer's instructions. The secondary antibody was goat anti-rabbit immunoglobulin G-peroxidase conjugate diluted 1:2,000 in blocking solution.
Enzyme assays.
PGPase activity was assayed by determining the amount of inorganic phosphate released upon hydrolysis of 2-phosphoglycolate at 25°C (32). One unit was defined as the amount of enzyme that catalyzed the formation of 1 μmol of inorganic phosphate per min. The kinetic parameters were determined for purified PGPase in the presence of 10 substrate concentrations ranging from 10 μM to 1 mM. Each assay was performed in triplicate, and the values for Km and Vmax were obtained by linear regression analysis of the data plotted as described by Lineweaver and Burk (17).
Specific β-galactosidase activity was assayed at 28°C by using cells permeabilized with chloroform and SDS and by using o-nitrophenyl-β-d-galactopyranoside as a substrate; the activity was expressed in Miller units (25).
The protein concentration was determined as decribed by Lowry et al. (19) with bovine serum albumin as a standard.
2-Phosphoglycolate determination.
The concentration of 2-phosphoglycolate in cell extracts was determined enzymatically by using PGPase purified as indicated above. Samples of cell extracts for 2-phosphoglycolate determination were obtained as described above and filtered through Amicon YM-10 membranes to eliminate proteins. The assay mixture (0.3 ml) contained 0.75 U of purified PGPase, 3 mM MgCl2, and 0.2 ml of the filtered extracts in 40 mM morpholinepropanesulfonic acid (MOPS) buffer (pH 6.9). The enzyme was omitted from the blank control mixtures. The inorganic phosphate released after 10 min of incubation was determined as indicated above (32) and used to calculate the amount of 2-phosphoglycolate.
Statistical analysis.
The results of β-galactosidase assays were expressed as means and standard deviations for at least six independent experiments performed in triplicate. Data were analyzed by using a Kruskal-Wallis one-way analysis of variance followed by the Student-Newman-Keuls test.
RESULTS
Cloning and characterization of the gph gene.
We amplified the gph gene and cloned it into pBluescript SK, generating plasmid pTP100, as described above. The cloned gene was overexpressed in MC4100 cells transformed with this plasmid, and PGPase activity in the corresponding extracts was tested. The specific activity was 1,970 mU/mg of protein, more than 80 times the basal values obtained in strain MC4100 (24.3 mU/mg). Therefore, the gph gene cloned in pTP100 encoded a protein with PGPase activity. This conclusion was further supported by an experiment in which plasmid pTP100 was introduced into mutant strain JA210, deficient in PGPase. In this experiment, PGPase activity was also close to 2,000 mU/mg of protein.
A Blast search analysis of the amino acid sequence of the E. coli gph-encoded protein showed the following degrees of similarity with various PGPases: Shigella flexneri, 98%; Salmonella enterica serovar Typhimurium, 95%; Klebsiella aerogenes, 89%; Haemophilus influenzae, 60%; Ralstonia eutropha, 50%; and Agrobacterium tumefaciens, 49%. Regarding the amino acid sequence, two domains, one at the N terminus and the other at the C terminus, had the largest number of conserved residues. The N terminus contained the conserved sequence (F,L)DLDGTL, present in bacterial and yeast phosphatases (9) and suggested to be necessary for their phosphatase activities (Fig. 1). Furthermore, highly conserved clusters were also observed when the gph DNA sequences of several members of the Enterobacteriaceae were compared, reinforcing the conservation of this gene along the evolution of this phylogenetic group.
FIG. 1.
Alignment of amino acid sequences of PGPases from several organisms. The amino acid sequences of E. coli (p32662), K. aerogenes (Q9EYY5), H. influenzae (p44755), R. eutropha (p40852), and A. tumefaciens (AAL42616) are shown. Asterisks indicate residues that were conserved in at least four of the aligned sequences. Shaded residues correspond to the putative catalytic domain for phosphatase activity described by Galinier et al. (9). Alignments of the amino acid sequences were generated by using the PILEUP program of the Genetics Computer Group software package (University of Wisconsin).
Purification of PGPase and preparation of specific antibodies.
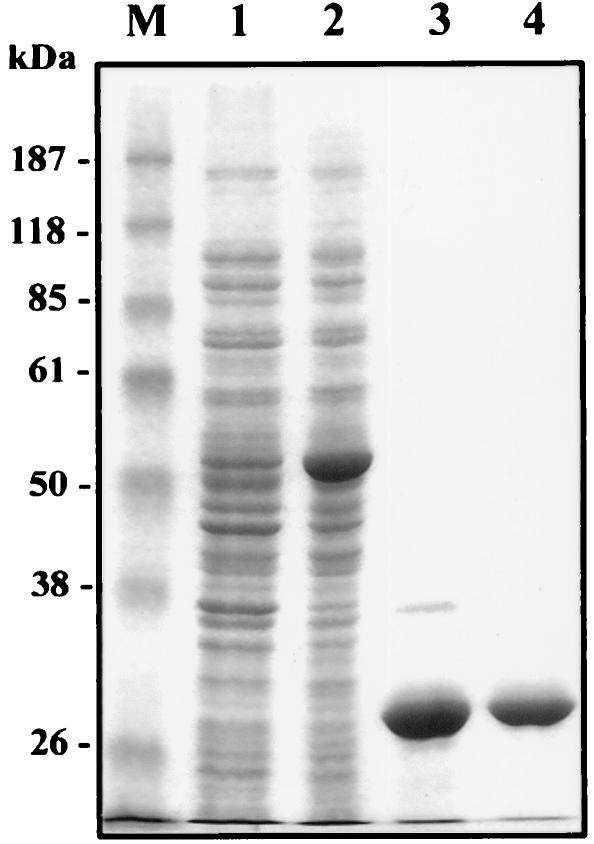
PGPase was purified by the procedure described in Materials and Methods, which yielded a homogeneous PGPase preparation, as determined by SDS-PAGE and Coomassie brilliant blue staining (Fig. 2).
FIG. 2.
SDS-PAGE of purified E. coli PGPase. Lane 1, crude extract of noninduced BL21(DE3) cells bearing pGEX-Gph; lane 2, crude extract of BL21(DE3) cells bearing pGEX-Gph and induced with IPTG; lane 3, fraction eluted from a glutathione-Sepharose 4B column after digestion with factor Xa; lane 4, fraction eluted from DEAE-Sepharose chromatography; lane M, prestained molecular mass markers (Gibco BRL). Proteins in the gel were visualized by Coomassie brilliant blue staining.
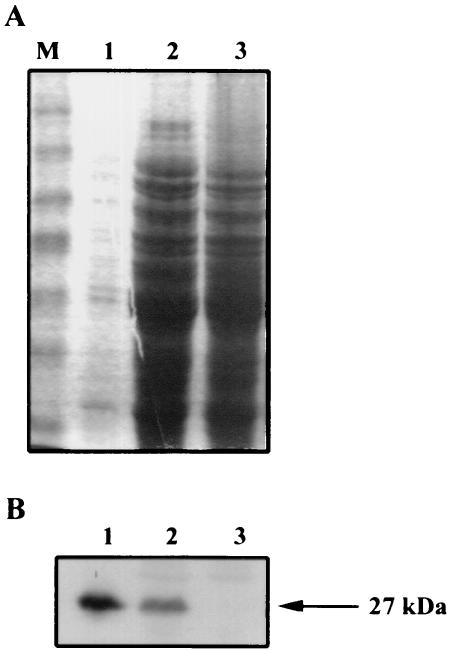
Antibodies against purified PGPase were obtained for characterization of the enzyme. As the native purified enzyme did not generate any immune response, antibodies against PGPase were obtained by using the GST-PGPase fusion, in which GST acts as a carrier. The specificity of the antibody preparation was assessed by an analysis of gph expression in crude extracts of strains MC4100, JA210, and JA210 transformed with pTP100 (overexpressing native PGPase), all grown on Caa (Fig. 3A). An immunodetected band of PGPase protein appeared in the lanes corresponding to MC4100 and JA210 transformed with pTP100, whereas no band was detected in the lane corresponding to the gph disruption mutant (Fig. 3B). No other reaction was seen over the rest of the immunoblot. These results showed that our antibody preparation specifically recognized PGPase and that the intensity of the immunodetected band was proportional to the amount of PGPase in the blot.
FIG. 3.
Analysis of the specificity of anti-PGPase serum. (A) SDS-PAGE of crude extracts obtained from various strains. Lane 1, 0.5 μg of JA210 harboring pTP100; lane 2, 50 μg of MC4100; lane 3, 50 μg of JA210; lane M, prestained molecular mass markers (Gibco BRL). Proteins in the gel were visualized by Coomassie brilliant blue staining. (B) Western blot analysis of samples from panel A.
Characterization of PGPase.
The molecular mass of the native enzyme was about 27 kDa, as deduced from the elution of the purified enzyme applied to an Ultrogel ACA44 column calibrated at several molecular masses (data not shown). The coincidence of this value with the mass deduced from the PGPase amino acid sequence (27,389 Da) and also with the mass determined by SDS-PAGE (Fig. 2) confirms the identity proposed for the gph gene product and indicates a monomeric structure for this enzyme.
The purified enzyme was assayed at various pH conditions ranging from 6.0 to 8.0 by using MOPS or Tris-HCl buffers to bracket a wider pH range. The enzyme activity was maximal at pH 6.9. The catalytic activities of PGPases from several organisms require chloride and magnesium (7, 23, 27). To test whether the same was true for our preparation, we studied enzymatic activity in the presence and absence of Cl− or Mg2+. The enzyme was purified as described above, but Tris-HCl buffers were replaced by 100 mM HEPES-NaOH (pH 7.0) and the activity was measured by using the same buffer. Maximal activity was achieved upon the addition of MgCl2. In the absence of both cofactors, activity was diminished by 40-fold. The addition of Mg2+ alone (MgSO4) had an almost negligible effect, whereas the addition of Cl− alone (KCl) increased the activity by 11-fold, indicating a cooperative effect between the two cofactors (Table 1).
TABLE 1.
Dependence of PGPase activity on chloride and magnesium ions
| Presence (+) or absence (−) of: | PGPase activity
|
||
|---|---|---|---|
| Cl− | Mg2+ | mU/mg | % |
| − | − | 5.2 | 2.5 |
| + | − | 58.2 | 27.9 |
| − | + | 8.5 | 4.1 |
| + | + | 208.1 | 100.0 |
The enzyme was highly specific for 2-phosphoglycolate, consistent with data on PGPases from higher plants (7) and C. reinhardtii (23). No phosphatase activity was detected on the following phosphorylated compounds tested at concentrations of up to 5 mM: d-2 phosphoglycerate, d-3-phosphoglycerate, glycerol-3-phosphate, phosphoenolpyruvate, d-ribose-5-phosphate, d-fructose-6-phosphate, and d-fructose-1,6-bisphosphate. 2-Phosphoglycolate concentration kinetics determinations with purified preparations yielded a Km of 210 μM and a Vmax of 208 μmol min−1 mg−1 of protein from the Lineweaver-Burk plot. The kcat was 94 s−1, and the calculated catalytic efficiency (kcat/Km) for this substrate was 4.4 × 105 s−1 M−1.
gph gene expression.
To understand the physiological role of the gph gene product, the patterns of expression of the gph gene under various growth conditions were analyzed by Western blotting with anti-PGPase serum. We studied the effect on gph expression of several carbon sources, such as Caa, LB, glucose, glycerol, d-xylose, succinate, or d-ribose. In all cases, Western blot analysis of crude extracts obtained from exponential cultures of MC4100 cells showed PGPase immunodetected bands of similar intensities (data not shown). Consistently, PGPase activities measured in these crude extracts under all conditions were about 25 mU/mg. PGPase expression levels were also similar when the cells studied were in stationary phase or under oxidative stress induced by bleomycin treatment (see below). As expected, no band of specific immunodetected protein appeared when the extracts were obtained from mutant JA210 under any of the tested conditions (data not shown).
In vivo determination of the role of PGPase.
The physiological and metabolic roles of PGPase were determined by studying glycolate formation from 2-phosphoglycolate in cells challenged by oxidative stress mediated by the radiomimetic drug bleomycin, which breaks DNA, generating 3′-phosphoglycolate ends (35). The free 2-phosphoglycolate generated by the cellular DNA repair machinery needs to be dephosphorylated to glycolate before its introduction into general metabolism. The glc operon transcriptional fusion, known to be induced by glycolate in E. coli (26), was used to determine glycolate formation under these conditions.
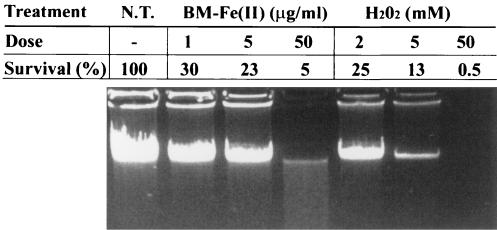
First, MC4100 cells bearing φ(glcD-lacZ) were treated for 1 h with various concentrations of bleomycin or hydrogen peroxide. The genomic DNA damage in treated cells was estimated by gel electrophoresis and correlated with the percentage of cell survival under each condition (Fig. 4). Similar results were obtained when this analysis was performed with cells of strain JA210 φ(glcD-lacZ) (data not shown), suggesting that the mutation in gph did not change the susceptibility of these cells to the oxidizing agents. In this way, 5 μg of bleomycin/ml and 2 mM H2O2 rendered the best survival/damage ratio and were selected for use in the following experiments.
FIG. 4.
Effect of oxidative stress on DNA integrity and cell survival for cells treated with bleomycin or H2O2. MC4100 cells (1 ml) grown on Caa and treated for 1 h with the indicated doses of bleomycin [BM-Fe(II)] or H2O2 were harvested, and genomic DNA was obtained. Five microliters of each DNA preparation was electrophoresed in a 1% agarose gel and visualized with ethidium bromide. The percent cell survival is shown for each treatment. N.T., no treatment.
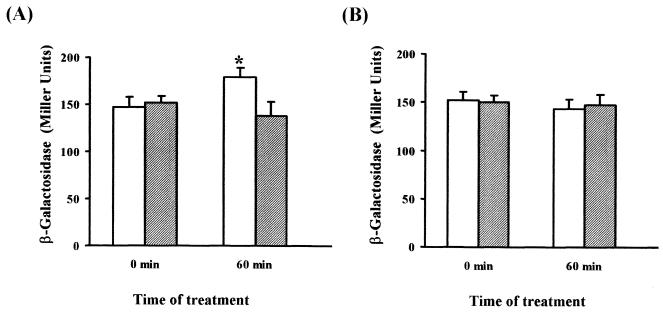
MC4100 φ(glcD-lacZ) and JA210 φ(glcD-lacZ) cells were grown on 0.5% Caa to mid-exponential phase. Both cultures were then treated with 5 μg of bleomycin/ml, and aliquots were withdrawn at 60 min for β-galactosidase determinations. β-Galactosidase activity increased in wild-type cells challenged with bleomycin (22%) (P < 0.05) (Fig. 5A). In the mutant strain, deficient in PGPase and hence unable to generate glycolate from 2-phosphoglycolate, β-galactosidase activity did not increase but slightly decreased with time (Fig. 5A). In a parallel experiment, oxidative stress was achieved by the addition of 2 mM H2O2, reported to produce different kinds of DNA breaks. Nevertheless, some authors have reported that this agent preferentially generates 3′-phosphate termini (2, 11), the repair of which renders free inorganic phosphate but not 2-phosphoglycolate. Consistently, analysis of β-galactosidase activities in wild-type and mutant strains showed no differences between the cultures of these strains after H2O2 treatment (Fig. 5B).
FIG. 5.
Analysis of expression of the glcD-lacZ transcriptional fusion under oxidative stress. Bleomycin at 5 μg/ml (A) or H2O2 at 2 mM (B) was added to aerobic cultures of strain MC4100 (white bars) or JA210 (grey bars) bearing φ(glcD-lacZ). β-Galactosidase activity was measured before and 60 min after the addition of the treatments. Values are means and standard deviations for at least six independent experiments performed in triplicate. A statistically significant difference (P < 0.05) with respect to the nontreated culture is indicated by an asterisk.
In the gph mutant, the 2-phosphoglycolate formed by treatment with bleomycin should accumulate inside the cell. To verify this hypothesis, the level of 2-phosphoglycolate in cell extracts was enzymatically determined with purified PGPase, which is highly specific for this substrate. To this end, strains MC4100 and JA210 were grown and treated with bleomycin for 1 h as indicated above for the assay of glc operon induction. Extracts of treated and untreated cells were used to determine 2-phosphoglycolate contents. Upon the addition of PGPase, only extracts of mutant cells treated with the drug revealed the release of phosphate, which allowed us to estimate a value of 4 to 5 nmol of 2-phosphoglycolate produced from 1010 treated cells. Control experiments performed with untreated cells showed that the basal repair of DNA damage produced during aerobic growth yields amounts of 2-phosphoglycolate that are below the level of detection for this enzymatic method. These results are in accordance with the proposed in vivo function of PGPase.
To test the effect on cell growth of bleomycin-induced 2-phosphoglycolate accumulation, cells were collected after 1 h of treatment and 100-fold diluted into fresh Caa to monitor culture growth. A significant retardation in the resumption of growth after treatment was observed in mutant cells compared to wild-type cells, strengthening the idea that PGPase is involved in a reaction relevant for DNA repair.
DISCUSSION
Lyngstadaas et al. (22) showed that the gph gene of E. coli encodes a protein with PGPase activity. Nevertheless, the characterization of this enzyme has not been reported, and its physiological role remains unknown. This work constitutes the first report on the purification and characterization of a PGPase of prokaryotic origin. From a functional point of view, the pH profile, dependence of catalytic activity on Cl− and Mg2+, substrate specificity, and kinetics revealed extensive similarity among the PGPases of different origins. This finding is consistent with the high percentage of similarity (more than 50%) found when the sequence of the E. coli PGPase or any other bacterial PGPase is compared to those of the other members of the family and with the presence of certain conserved motifs and amino acid residues. It is remarkable, however, that the E. coli enzyme has a monomeric structure, in contrast with the reported homodimeric structure of enzymes of eukaryotic origins (23).
The low Km for the substrate 2-phosphoglycolate and the good catalytic efficiency (kcat/Km) of the enzyme allow efficient conversion of the substrate even when it is present at relatively low intracellular concentrations. Under these conditions, the metabolic flow from 2-phosphoglycolate to malate depends only on the amount of 2-phosphoglycolate generated. This finding is further supported by the induction of the enzymes glycolate oxidase and malate synthase, both encoded by the glc operon (26), which does not permit the accumulation of the intermediates glycolate and glyoxylate. The efficiency in the dissimilation of 2-phosphoglycolate is also aided by the constitutive expression of the gph gene, which is not altered by carbon sources, the growth stage of the culture, or oxidative stress conditions. Constitutive expression is consistent with a housekeeping function for the gph gene.
The presence of the gph gene in all phyla indicates a paramount physiological function that may go beyond its recognized role in the Calvin cycle of autotrophic organisms or in the control of the bisphosphoglycerate shunt in red blood cells. Moreover, a pathogenic organism such as H. influenzae, whose genome has evolved to a minimum size, conserves a gene encoding PGPase. The E. coli enzyme has a high substrate specificity, a common feature of PGPases of other origins; this factor may place certain constraints on the possibility of physiological roles implying transformations other than the dephosphorylation of 2-phosphoglycolate. Thus, PGPase function may be connected to physiological processes leading to the formation of this phosphorylated compound within cells. One such situation is encountered when organisms live in aerobic conditions, which favor the formation of oxygen free radicals leading to DNA damage, such as strand breaks bearing 3′-phosphoglycolate termini (8). In vitro, the repair mechanism for these 3′-blocking ends releases free 2-phosphoglycolic acid. A large number of enzymes that repair these termini have been reported for bacteria (8, 31), yeasts (30), and higher eukaryotes (38).
In this study, using E. coli as a model in in vivo experiments, we have shown that the 2-phosphoglycolate formed intracellularly through the DNA repair machinery after exposure to oxidative stress mediated by bleomycin is converted to glycolate by the action of PGPase. Through this pathway, the formed 2-phosphoglycolate is dephosphorylated, and its carbons are incorporated into general metabolism. Our results prove that a knockout mutation in gph impairs this salvage pathway and, consequently, that 2-phosphoglycolate accumulates in PGPase-deficient cells, suggesting that no other cellular reactions are involved in the metabolism of this compound.
As the presence of oxygen always causes DNA damage, the gph gene has been conserved in all aerobic organisms. In this sense, the strict anaerobe Methanococcus jannaschii, which has evolved in the complete absence of oxygen and has an unusual ribulose-1,5-bisphosphate carboxylase/oxygenase with a carboxylase activity extremely sensitive to oxygen (37), does not seem to contain any gene encoding a PGPase (5). In conclusion, the proposed role of PGPase in the metabolism of the 2-phosphoglycolate generated in the DNA repair process could be extended to any organism, from prokaryotes to higher eukaryotes, that live in the presence of oxygen.
The functional reasons to join in an operon structure the seven genes found in the dam-containing operon remain elusive. Up to now, some of the gene products have been related to DNA function. For instance, the aroK gene product could have a second activity possibly related to cell division (36); methylation mediated by dam is known to be involved in several processes, such as DNA replication, gene expression and mismatch repair, among others; and the epimerase encoded by the rpe gene has been reported to be involved not only in the nonoxidative branch of the pentose phosphate pathway but also in the control of chromosome replication (28). In this context, it is tempting to speculate that the physiological role proposed here for the gph gene product matches rather well the role in DNA-related processes suggested for most of the genes in the dam-containing operon, giving further support to the function proposed for the gph gene product.
Acknowledgments
We thank Erik Boye for kindly providing strain ANL9 and Joaquim Ros for critical reading of the manuscript.
M.F.N is a recipient of a predoctoral fellowship from the Generalitat de Catalunya. This work was supported by grant BMC 2001-3003 from the Ministerio de Ciencia y Tecnologia and grant SGR00128 from the Generalitat de Catalunya.
REFERENCES
- 1.Baldoma, L., and J. Aguilar. 1987. Involvement of lactaldehyde dehydrogenase in several metabolic pathways of Escherichia coli K12. J. Biol. Chem. 262:13991-13996. [PubMed]
- 2.Betti, M., S. Petrucco, A. Bolchi, G. Dieci, and S. Ottonello. 2001. A plant 3′-phosphoesterase involved in the repair of DNA strand breaks generated by oxidative damage. J. Biol. Chem. 276:18038-18045. [DOI] [PubMed] [Google Scholar]
- 3.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 4.Boronat, A., and J. Aguilar. 1979. Rhamnose-induced propanediol oxidoreductase in Escherichia coli: purification, properties, and comparison with the fucose-induced enzyme. J. Bacteriol. 140:320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bult, C. J., O. White, G. J. Olsen, L. Zhou, R. D. Fleischmann, G. G. Sutton, J. A. Blake, L. M. FitzGerald, R. A. Clayton, J. D. Gocayne, A. R. Kerlavage, B. A. Dougherty, J. F. Tomb, M. D. Adams, C. I. Reich, R. Overbeek, E. F. Kirkness, K. G. Weinstock, J. M. Merrick, A. Glodek, J. L. Scott, N. S. Geoghagen, and J. C. Venter. 1996. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273:1058-1073. [DOI] [PubMed] [Google Scholar]
- 6.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 7.Christeller, J. T., and N. E. Tolbert. 1978. Phosphoglycolate phosphatase: purification and properties. J. Biol. Chem. 253:1780-1785. [PubMed] [Google Scholar]
- 8.Demple, B., and L. Harrison. 1994. Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem. 63:915-948. [DOI] [PubMed] [Google Scholar]
- 9.Galinier, A., M. Kravanja, R. Engelmann, W. Hengstenberg, M. C. Kilhoffer, J. Deutscher, and J. Haiech. 1998. New protein kinase and protein phosphatase families mediate signal transduction in bacterial catabolite repression. Proc. Natl. Acad. Sci. USA 95:1823-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibson, J. L., and F. R. Tabita. 1997. Analysis of the cbbXYZ operon in Rhodobacter sphaeroides. J. Bacteriol. 179:663-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu, L., S. M. Huang, and M. Sander. 1994. Single amino acid changes alter the repair specificity of Drosophila Rrp1. Isolation of mutants deficient in repair of oxidative DNA damage. J. Biol. Chem. 269:32685-32692. [PubMed] [Google Scholar]
- 12.Hall, C. V., and C. Yanofsky. 1981. Cloning and characterization of the gene for Escherichia coli tryptophanyl-transfer ribonucleic acid synthetase. J. Bacteriol. 148:941-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardy, P., and P. Baldy. 1986. Corn phosphoglycolate phosphatase: purification and properties. Planta 168:245-252. [DOI] [PubMed] [Google Scholar]
- 14.Husic, H. D., and N. E. Tolbert. 1984. Anion and divalent cation activation of phosphoglycolate phosphatase from leaves. Arch. Biochem. Biophys. 229:64-72. [DOI] [PubMed] [Google Scholar]
- 15.Jonczyk, P., R. Hines, and D. W. Smith. 1989. The Escherichia coli dam gene is expressed as a distal gene of a new operon. Mol. Gen. Genet. 217:85-96. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 17.Lineweaver, H., and D. Burk. 1934. Determination of the enzyme dissociation constants. J. Am. Chem. Soc. 56:658-666. [Google Scholar]
- 18.Lobner-Olesen, A., and M. G. Marinus. 1992. Identification of the gene (aroK) encoding shikimic acid kinase I of Escherichia coli. J. Bacteriol. 174:525-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 20.Lyngstadaas, A., A. Lobner-Olesen, and E. Boye. 1995. Characterization of three genes in the dam-containing operon of Escherichia coli. Mol. Gen. Genet. 247:546-554. [DOI] [PubMed] [Google Scholar]
- 21.Lyngstadaas, A., G. A. Sprenger, and E. Boye. 1998. Impaired growth of an Escherichia coli rpe mutant lacking ribulose-5-phosphate epimerase activity. Biochim. Biophys. Acta 1381:319-330. [DOI] [PubMed] [Google Scholar]
- 22.Lyngstadaas, A., A. Lobner-Olesen, E. Grelland, and E. Boye. 1999. The gene for 2-phosphoglycolate phosphatase (gph) in Escherichia coli is located in the same operon as dam and at least five other diverse genes. Biochim. Biophys. Acta 1472:376-384. [DOI] [PubMed] [Google Scholar]
- 23.Mamedov, T. G., K. Suzuki, K. Miura, K. I. Kucho, and H. Fukuzawa. 2001. Characteristics and sequence of phosphoglycolate phosphatase from a eukaryotic green alga Chlamydomonas reinhardtii. J. Biol. Chem. 276:45573-45579. [DOI] [PubMed] [Google Scholar]
- 24.Millar, G., and J. R. Coggins. 1986. The complete amino acid sequence of 3-dehydroquinate synthase of Escherichia coli K12. FEBS Lett. 200:11-17. [DOI] [PubMed] [Google Scholar]
- 25.Miller, J. H. 1992. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Pellicer, M. T., C. Fernandez, J. Badia, J. Aguilar, E. C. C. Lin, and L. Baldoma. 1999. Cross-induction of glc and ace operons of Escherichia coli attributable to pathway intersection. Characterization of the glc promoter. J. Biol. Chem. 274:1745-1752. [DOI] [PubMed] [Google Scholar]
- 27.Rose, Z. B., D. S. Grove, and S. N. Seal. 1986. Mechanism of activation by anions of phosphoglycolate phosphatases from spinach and human red blood cells. J. Biol. Chem. 261:10996-11002. [PubMed] [Google Scholar]
- 28.Sakakibara, Y. 1997. Involvement of the ribulosephosphate epimerase gene in the dnaA and dnaR functions for initiation of chromosome replication in Escherichia coli. Mol. Microbiol. 24:793-801. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratoty manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Sander, M., and D. Ramotar. 1997. Partial purification of Pde1 from Saccharomyces cerevisiae: enzymatic redundancy for the repair of 3′-terminal DNA lesions and abasic sites in yeast. Biochemistry 36:6100-6106. [DOI] [PubMed] [Google Scholar]
- 31.Sandigursky, M., and W. A. Franklin. 1998. Exonuclease IX of Escherichia coli removes 3′-phosphoglycolate end groups from DNA. Radiat. Res. 150:609-611. [PubMed] [Google Scholar]
- 32.Schäferjohann, J., J. G. Yoo, and B. Bowien. 1993. The cbb operons of the facultative chemoautotroph Alcaligenes eutrophus encode phosphoglycolate phosphatase. J. Bacteriol. 175:7329-7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seal, S. N., and Z. B. Rose. 1987. Characterization of a phosphoenzyme intermediate in the reaction of phosphoglycolate phosphatase. J. Biol. Chem. 262:13496-13500. [PubMed] [Google Scholar]
- 34.Shively, J. M., G. van Keulen, and W. G. Meijer. 1998. Something from almost nothing: carbon dioxide fixation in chemoautotrophs. Annu. Rev. Microbiol. 52:191-230. [DOI] [PubMed] [Google Scholar]
- 35.Stubbe, J., and J. W. Kozarich. 1987. Mechanism of bleomycin-induced DNA degradation. Chem. Rev. 87:1107-1136. [Google Scholar]
- 36.Vinella, D., B. Gagny, D. Joseleau-Petit, R. D'Ari, and M. Cashel. 1996. Mecillinam resistance in Escherichia coli is conferred by loss of a second activity of the AroK protein. J. Bacteriol. 178:3818-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watson, G. M. F., J. P. Yu, and F. R. Tabita. 1999. Unusual ribulose 1,5-bisphosphate carboxylase/oxygenase of anoxic Archaea. J. Bacteriol. 181:1569-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu, Y. J., E. Y. Kim, and B. Demple. 1998. Excision of C-4′-oxidized deoxyribose lesions from double-stranded DNA by human apurinic/apyrimidinic endonuclease (Ape1 protein) and DNA polymerase beta. J. Biol. Chem. 273:28837-28844. [DOI] [PubMed] [Google Scholar]
- 39.Zecher, R., U. Schwulera, and H. U. Wolf. 1982. Purification, isolation and characterization of a phosphoglycolate phosphatase isoenzyme from human erythrocytes. Int. J. Biochem. 14:775-781. [DOI] [PubMed] [Google Scholar]