Abstract
The Escherichia coli transcription factor sigma 32 binds to core RNA polymerase to form the holoenzyme responsible for transcription initiation at heat shock promoters, utilized upon exposure of the cell to higher temperatures. We have developed two ways to assay sigma 32-dependent RNA synthesis in E. coli. The plasmid-borne reporter gene for both is lacZ (β-galactosidase), driven by the groE promoter. In one application, the cells are exposed to a temperature of 42°C in order to induce accumulation of endogenous sigma 32. The other involves isopropylthiogalactopyranoside (IPTG)-induced synthesis of sigma 32 at 30°C from a gene contained on a second plasmid. The latter employs DnaK− cells, which additionally contained a second mutation, inactivating the endogenous sigma 32 gene (Bukau and Walker, EMBO J. 9:4027-4036, 1990). These assays were used to delineate the sequences CTTGA (−37 to −33) and GNCCCCATNT (−18 to −9) as important for sigma 32 promoter activity. At each of the specified base pairs, substitutions were found which reduced promoter activity by greater than 75%. Activity was also dependent upon the number of base pairs separating the two regions.
RNA synthesis in prokaryotes is carried out by a multisubunit RNA polymerase commonly referred to as the core enzyme (E) (5). For promoter recognition, a sigma (initiation) factor is required. It interacts with the core polymerase to yield the holoenzyme (Eσ), which is able to form an initiation-competent complex at promoter sequences in a multistep process involving conformational changes in both the RNA polymerase and the promoter DNA (4, 5). In Escherichia coli, seven different species of the σ subunit have been identified, each directing transcription of a specific set of genes. The predominant sigma factor in E. coli is σ70, which imparts on RNA polymerase the ability to recognize promoters of housekeeping genes. The heat shock sigma factor of E. coli (σ32) is responsible for transcription of genes encoding proteins that promote survival of the cell at elevated temperatures (9, 24, 27, 35). σ32 is a single polypeptide chain of 284 amino acids in length (19, 35); it is a member of the σ70 family of sigma factors, which share considerable homology in four distinct regions (21).
The activity of σ32 is increased at elevated growth temperatures and under conditions of nutrient starvation (7, 27, 34) as a result of multiple modes of regulation at the transcriptional, translational, and posttranslational levels (9, 24, 27, 34). The last involves the DnaK chaperone. When a component of the holoenzyme, σ32 is afforded protection against degradation by membrane-bound FtsH protease (30) and diffusible proteases (15). During cell growth at low temperature, DnaK binds to σ32 (8, 20), interfering with the binding of σ32 to RNA polymerase core enzyme. Upon exposure to increased temperature, σ32 dissociates from DnaK due both to recruitment of DnaK (34) by the newly generated unfolded protein in the cell and to the intrinsic weakening of the interaction between σ32 and DnaK at the higher temperature (2). The released σ32 then associates with core RNA polymerase, enabling the ensuing holo-RNA polymerase to initiate transcription of the approximately 30 E. coli heat shock proteins, including GroEL and GroES (9).
From analysis of the sequences of 18 heat shock promoters, the consensus sequences CTTGAAA in the −35 region and CCCCATNT in the −10 region were derived (9). To date, no studies on the correlation of the consensus sequences and Eσ32 promoter function have been published. Thus, it was of interest to identify the sequences required for heat shock promoter function. We here describe a refinement of the consensus sequence as well as the development of two different assays for studying Eσ32 promoter activity in vivo which have enabled us to determine the functionally important sequences of an Eσ32 promoter.
MATERIALS AND METHODS
Chemicals and enzymes.
Oligodeoxyribonucleotide primers were synthesized by Invitrogen. Materials for plasmid purification and extraction of DNA from agarose gels were purchased from Qiagen. T4 DNA ligase, T4 polynucleotide kinase, and restriction enzymes were purchased from New England Biolabs or Roche. Medium reagents were purchased from Gibco-BRL. Other chemicals were bought from Sigma.
Plasmids and strains.
Plasmid pLC412 carrying N-terminally hexahistidine-tagged σ32 was obtained from Cathy Chang and Carol Gross. pSAKT32 is a derivative of pSAK15-70/32 (18), a pACYC-derived vector with a p15A origin of replication. It was modified by insertion of an ampicillin resistance cassette into the chloramphenicol resistance gene. It carries the σ32 gene (with the hexahistidine tag removed) under isopropylthiogalactopyranoside (IPTG)-inducible wild-type Plac control (see Fig. 1), as well as the lac repressor gene under control of the stronger (iq) mutant promoter. pSAKT32 was used as an extrachromosomal, intracellular source of σ32.
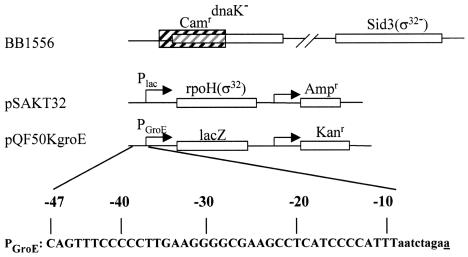
FIG. 1.
Components of the in vivo assay systems investigated. pQF50KgroE, shown here with the wild-type groE promoter, is the reporter plasmid used in both the heat and IPTG induction assays. The numbering shown here for the groE promoter was used throughout this paper to refer to particular conserved positions at other Eσ32 promoters as well. The sequence of the groE promoter is shown in capital letters. Vector sequences in the region downstream of the −10 position are shown in lowercase letters, with the putative transcription start site underlined. For the IPTG induction assay, DnaK− BB1556 cells that also have a mutated σ32 gene were used (see text), making the endogenous σ32 prone to degradation. In addition, they carried the pSAKT32 plasmid, with a σ32 gene under control of the lac promoter.
pQF50K is a derivative of the β-galactosidase promoter probe vector pQF50, for which the β-lactamase gene was inactivated by insertion of a kanamycin resistance cassette. It was further modified to yield pQF50KgroE (see Fig. 1) by using the BglII and XbaI sites for cloning synthetic DNA spanning both strands from positions −47 to −9 of wild-type or mutant groE promoters. We use the groE numbering throughout to refer to particular conserved sequences. All constructs were verified by DNA sequencing.
Strain BB1556 (MC4100 ΔdnaK52::Cmr sidB3) (1) was obtained from B. Bukau and M. P. Mayer (see Fig. 1). Its dnaK gene was inactivated by insertion of DNA containing a chloramphenicol resistance cassette (25). BB1556 cells contain an additional, spontaneous suppressor mutation in the σ32 gene, which leads to a greatly increased growth rate (1). It is a frameshift mutation in the σ32 coding region extending the N-terminal region of the protein by 38 amino acids and likely rendering it more sensitive to proteolysis. The resultant, much reduced intracellular σ32 levels alleviate many of the deleterious effects of the dnaK inactivation. All antibiotics were added to 50 μg/ml.
groE promoter activity driven by endogenous σ32.
DH5α cells carrying pQF50KgroE with wild-type or variant groE promoter sequences were grown in Luria-Bertani (LB) medium at 37°C. A 0.5-ml aliquot from an overnight culture was diluted 10-fold with fresh LB or M9 medium, and the cells were grown at 37°C to an optical density at 600 nm of 0.7. The temperature was then shifted to 42°C to effect greater levels of endogenous σ32. Cell growth was stopped 1 h later, and β-galactosidase assays were carried out with the Miller protocol (26). Every mutant was subjected to two to four independent determinations.
groE promoter activity driven by plasmid-encoded σ32.
pQF50KgroE and pSAKT32 were both transformed into the ΔdnaK sidB3 mutant strain BB1556 (1). Cells harboring both plasmids were selected with three different antibiotics: chloramphenicol, kanamycin, and ampicillin. This served to ensure the growth of only those cells carrying the transposon that inactivates the dnaK gene by insertion and to select the pQF50KgroE (kanamycin) and pSAKT-σ32 (ampicillin) plasmids. Cells were grown at 30°C with extensive aeration. A 0.5-ml aliquot of an overnight culture was diluted 10-fold in fresh LB medium, and when the culture reached optical density at 600 nm of 0.7, the synthesis of σ32 was induced by addition of IPTG to 1 mM. Cell growth was stopped 2.5 h later, and β-galactosidase assays were carried out. Each mutant was tested two to four times.
RESULTS
Starting with plasmid pQF50KgroE, a collection of 48 variants of the groE promoter were constructed and used to define the sequences important to the function of this σ32-dependent promoter. Promoter activity was determined upon effecting an increase in the intracellular levels of σ32, either in DH5α subjected to a temperature increase to 42°C or in strain BB1556 additionally containing plasmid pSAKT32 (see Fig. 1) by addition of IPTG.
groE promoter activity driven by endogenous σ32 at 42°C.
In preliminary experiments, the activity of the wild-type groE promoter was determined as a function of the duration of exposure to 42°C, initiated when the optical density at 600 nm of the culture growing at 37°C reached 0.7. It was found that the reporter gene activity (i.e., β-galactosidase) was still increasing after 2 h, reflecting the stability of β-galactosidase, as well as the elevated levels of σ32.
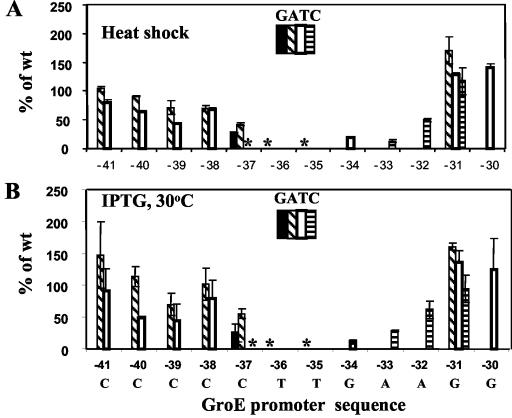
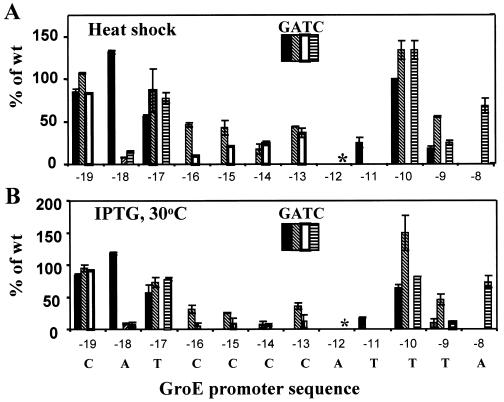
We chose 1 h as the incubation time for our assays. The total activity for the wild-type groE promoter prior to heat shock was 50 to 60 Miller units, and the activity after 1 h of heat shock was 85 to 100 Miller units. This observation is consistent with the fact that there is a significant amount of σ32 in the cell even at 37°C (33). The data obtained for variants with mutations in the −35 region of the promoter are shown in Fig. 2A, and those for variants with mutations in the −10 region are shown in Fig. 3A. At several positions, at least one substitution caused a drastic reduction in promoter activity, likely indicative of an important DNA-RNA polymerase contact. In addition, at other positions, substitutions reproducibly increased the level of promoter activity, indicating that the particular base pair in the groE promoter is suboptimal.
FIG. 2.
Sequence dependence of groE promoter activity in the −35 region. Activities are shown as a percentage of the measured β-galactosidase activity for the wild-type groE promoter. The positions at which sequence variants were introduced are shown for each panel, with the groE promoter sequence shown at the bottom. Sequence changes are coded as indicated; the bar graphs at each position represent, from left to right, substitutions of G, A, T, and C. The absence of bars indicates that the particular substitution was not tested. The asterisks indicate the C-37T, T-36G, and T-35G substitutions, for which the activity was 1% or less of wild-type activity in both assays. (A) Endogenous σ32, heat shock induction at 42°C. (B) Plasmid-encoded σ32, IPTG induction at 30°C.
FIG. 3.
Sequence dependence of groE promoter activity in the −10 region. Activities are shown as a percentage of measured β-galactosidase activity for the wild-type groE promoter. The positions at which sequence variants were introduced are shown for each panel, with the groE promoter sequence shown at the bottom. Sequence changes are coded as indicated; the bar graphs at each position represent, from left to right, substitutions of G, A, T, and C. The absence of bars indicates that the particular substitution was not tested. The asterisks indicate the A-12C substitution, for which the activity was less than 1% wild-type activity in both assays. (A) Endogenous σ32, heat shock induction at 42°C. (B) Plasmid-encoded σ32, IPTG induction at 30°C.
groE promoter activity driven by plasmid-encoded σ32 at 30°C.
A second method for increasing the intracellular concentration of σ32 was by IPTG induction of expression of the pSAKT32-borne σ32 gene. In preliminary experiments, DH5α cells were used, and no effect on β-galactosidase activity was observed upon IPTG addition. In view of the fact that DnaK is known to act as an anti-sigma factor (8, 20), we attempted the same experiment in the DnaK− BB1556 strain (1). The result was markedly different, with a clear promoter sequence-dependent increase in reporter gene expression. We monitored β-galactosidase activity as a function of incubation time at 30°C in the presence of IPTG. Based on the results of this experiment (data not shown), we chose to determine promoter function after a standard 2.5-h incubation of the cells with IPTG. Under these conditions, the activity of the wild-type groE promoter was found to be 200 to 250 Miller units. In control experiments without IPTG addition, 40 to 50 units of reporter gene activity was still observed, probably due to the presence of some endogenous sigma 32 activity at 30°C in the BB1556 cells. The results of experiments with the groE promoter variants are shown in Fig. 2B and 3B.
Comparison of in vivo assays for σ32 activity.
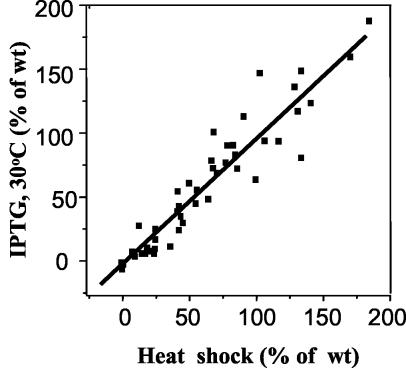
As can be seen in Fig. 2 and 3, the results obtained with the plasmid-expressed σ32 at 30°C closely paralleled those seen with the heat shock assay at 42°C. This is seen more clearly in Fig. 4, where the normalized results obtained for each promoter variant with both assays are plotted against each other. The points define a straight line, with a correlation coefficient of 0.93, demonstrating that the data obtained by the two methods reflect the same process.
FIG. 4.
Comparison of relative promoter activities (expressed as a percentage of wild-type [wt] promoter activity) measured with endogenous σ32 at 42°C (x axis) and with plasmid-encoded σ32 upon IPTG induction at 30°C (y axis). All data points included in Fig. 2 and 3 are shown. The data for the wild-type groE promoter were not included in the plot, as normalization results in activities of 100% for the wild-type promoter on both axes, which would artificially inflate correlation between the two sets of data. The linear least-squares line through the data has a correlation coefficient of 0.93.
Distance between the two recognized elements is important.
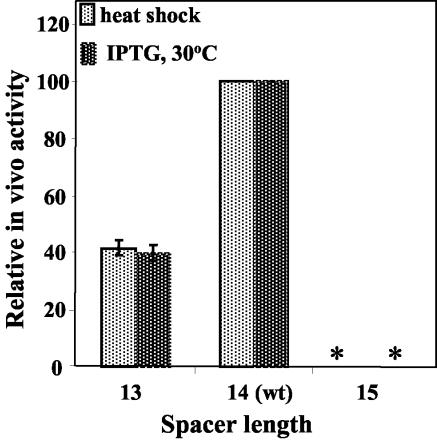
For promoters recognized by Eσ70, a clearly defined optimal distance is found between the upstream (−35) and downstream (−10) recognition elements. Deviation from this optimal distance by insertion or addition of only one base pair may affect promoter activity by as much as a factor of 10 (11, 29, 32). To test whether a similar pattern would also be displayed by Eσ32 promoters, we either inserted or deleted an A at position −19 in the wild-type sequence to yield promoters with AAA or A in the nontemplate strand, whereas the wild-type promoter has AA at this location. The data in Fig. 5 show that promoter activity decreased in both cases, being particularly sensitive to insertion of a base pair. The behavior of promoters recognized by Eσ70 and Eσ32 was similar in that both displayed a clear optimal distance, but the pronounced asymmetric pattern was atypical for Eσ70 promoters. In the few cases where asymmetry was observed with the latter, the promoter with the shorter spacer was the one with the lowest activity (22, 32). The precipitous decline in promoter activity for the longer spacer length observed in Fig. 5 could be the reason that of 20 known Eσ32 promoters (9, 16), only one (htgAp1, also called htpY) has a spacer length longer than that of the groE promoter, while for seven promoters it is similar and for 11 it is shorter.
FIG. 5.
Dependence of relative groE promoter activity on the length of the spacer DNA separating the −10 and −35 regions. Changes in spacer length were made as indicated in the text. The numbers on the x axis refer to the length in base pairs between the two functionally defined regions, CTTGA and GNCCCCATNT; by this criterion, the wild-type groE promoter has a spacer length of 14 bp. Activities (expressed as a percentage of wild-type promoter activity) were measured following heat shock induction of endogenous σ32 at 42°C (left bar) or IPTG induction of the plasmid-borne σ32 gene at 30°C (right bar). The asterisks for the promoter with the 15-bp spacer are used to indicate that the measured activity was <1% of that of the wild-type promoter.
DISCUSSION
In vivo systems.
Although the protein now known as σ32 was discovered as a sigma factor 20 years ago (10, 19), relatively little is known about its structure-function relationships. Perhaps this is due to difficulties in purifying and storing active σ32 (our unpublished observations). The work described here on the sequence-activity relationship for a σ32-dependent promoter also demonstrates that σ32 provided on a plasmid behaves similarly at low temperatures to the endogenous protein produced in the cell following heat shock. As heat shock conditions are compatible with σ32 function in the cell, the use of the plasmid-encoded gene for studying σ32 function in vivo has been validated. This experimental system will facilitate the probing of structure-function relationships in vivo, obviating the need for σ32 purification.
The IPTG-inducible σ32 assay requires the presence of two compatible plasmids inside the same E. coli cell. As used here, the reporter plasmid (pQF50KgroE) is present at a much greater copy number than pSAKT32, containing the σ32 gene. This arrangement was chosen to avoid nonphysiological consequences of high intracellular σ32 levels, a goal that was at least partially attained in view of the similarity of the results obtained with the two assays. However, it would be useful to also test the feasibility of maintaining the σ32 gene on higher-copy-number plasmids, as higher levels of expression would afford advantages, such as greater ease of in vivo footprinting at the groE promoter with dimethyl sulfate or KMnO4.
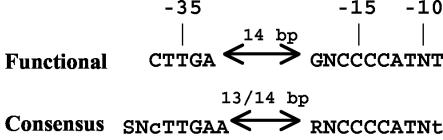
Comparison of the consensus and functional DNA sequences.
The functional Eσ32 promoter sequence determined in these studies can be compared to a consensus sequence displayed in Fig. 6, determined based on 20 heat shock promoters (18 from reference 9) and two others from the overlapping EcoCyc database (16). In this refinement of a prior sequence (9), the dnaKp3 promoter from the EcoCyc database was not included due to lack of homology to the other 20 promoters. Only minor differences are evident between the functional and the consensus sequences (see Fig. 6). At position −18 in the −10 region, even the degeneracy in the consensus sequence is reflected in the results of the functional studies: −18 G led to marginally greater reporter gene activity than A, and both were much better than T or C. In the promoter database, at this position 8 G's and 7 A's are found. Similarly, at position −10, A is marginally better than the other three in the functional studies, while A is prevalent among the 20 promoters analyzed (9 out of 20).
FIG. 6.
Comparison of functional and consensus Eσ32 promoter sequences. The top line displays the sequence determined in this work (positions where at least one base pair change reduces promoter activity by at least 75%). The bottom line is a consensus sequence for 20 promoters in two largely overlapping databases (9, 16) (only the dnaKp3 promoter [16] was omitted due to lack of overall homology with the others). Capital letters represent positions conserved in at least 15 of the promoters, lowercase letters represent conservation in 12 to 14 of the 20 promoters (N indicates no base preference; S = C or G; R = A or G). The spacer lengths, measured as indicated by the double arrows, are as defined in the legend to Fig. 5.
At position −32 in the −35 region, the consensus sequence shows a clear preference for A (16 of 20). We only substituted C here, for only a slight reduction in activity. In the absence of a full set of substitutions, our data do not allow conclusions to be made concerning the functional importance of the −32 A. At position −37, a T was shown to be deleterious, and indeed T is excluded at this position (1 of 20). Early consensus sequences (3, 34) showed some conservation of C residues at positions −38 to −41, where in the groE promoter C's are indeed found. Among the 20 promoters, significant conservation in this region is only found at position −39 (9 C's and 7 G's). We observed only relatively small effects of A or T substitutions at each of the four base pairs corresponding to the −38 to −41 positions. We did not systematically study the effects of G substitutions, but from the activities of a collection of randomly substituted promoters, we conclude that they would not have major deleterious effects either. Promoters with GACT, AGGC, and CCCG in this region had activities of 31%, 35%, and 30%, respectively, of the wild-type groE promoter (Wang, Kronholz, and deHaseth, unpublished data) in the heat shock assay. We conclude that major RNA polymerase-DNA contacts likely do not occur in this region.
At four base pairs of the functionally defined promoter regions, substitutions were found which essentially abolished promoter activity (<1% of wild-type promoter activity): A-12C, C-37T, T-36G, and T-35G. With the exception of C-37, these correspond to highly conserved bases in the consensus sequence. We speculate that −12A is equivalent to the −11A of Eσ70 promoters and that DNA melting would initiate at this position (12). A C at −37 immediately upstream of the −35 region was found to also be important for both UP element-dependent and -independent expression of the Eσ70 promoter, rrnBP1 (13). Interestingly, the hierarchy of activities was similar for both promoters: for groE, substitutions at C-37 and relative activities were T, 0%; G, 26%; and A, 49% (percentages of wild-type values are averages for the two types of in vivo assays) and for rrnBP1, they were T, 10%; G, 37%; and A, 57% (percentages of wild-type are averages for promoters with and without the UP element).
As we studied only one substitution each for T-36 and T-35, it was not possible to compare our results with the hierarchies of activities in the extensive data set for Eσ70 promoters (17, 23). The presence of highly conserved and functionally important TTGA motifs in the −35 regions of both Eσ70 and Eσ32 might well indicate that similar contacts with both sigma factors occur in this region, as proposed by Gross and coworkers (28).
In the 20 heat shock promoters analyzed to yield the consensus sequence, there was a great disparity in the extent to which each promoter resembled the consensus or functional promoter sequences shown in Fig. 6. On the one hand, there was the htgAp1 (−35 TTTGA, −10 TTCCCCGGTT, 16-bp spacer) with five deviations from the optimal functional sequences (italic); on the other, the htpG1 (−35 CTTGA, −10 GTCCCCATCT, 14-bp spacer) and groE (−35 CTTGA, −10 ATCCCCATTT, 14-bp spacer) promoters. The latter two also have perfect alignment with the consensus sequence in Fig. 6 and are expected to be much stronger promoters than htgAp1. Thus, as with Eσ70 promoters, the basal level is likely an important determinant of transcription activity for Eσ32 promoters as well.
Our results with heat shock promoters can be summarized as consensus is best in terms of the amount of RNA synthesized. This statement has, with very few exceptions (6, 14, 31), also been found to be applicable to Eσ70 promoters (23), even though no naturally occurring promoters exactly match the actual consensus sequence and (nearly) all have evolved under constraints other than maximizing the amounts of RNA made. Determination of a consensus sequence involves comparison of the sequences of naturally occurring promoters in a database, all of which have evolved to contain a subset of the particular bases that are functionally optimal for promoter activity. Apparently, on the average, in each promoter, a sufficient number of bases represent the functionally optimal sequence to allow the latter to reemerge as the consensus sequence above the background of nonconserved sequences. For Eσ70 promoters, the exception to the consensus-is-best rule involves mutations to consensus sequence, causing impaired promoter clearance (6, 14, 31) and thus a reduction in the synthesis of full-length RNA. Our work did not provide any indication for the existence of such effects within the strong Eσ32 promoter groE.
Acknowledgments
This work was supported by NIH grant GM 31808 to P.L.D.
We thank M. P. Mayer and B. Bukau for strain BB1556, C. Gross and C. Chang for plasmid pLC42, C. Gross and V. Rhodius for help with promoter databases, Melissa Kronholz for sequencing some promoter variants, and D. Samols as well as members of our laboratory for critically reading the manuscript.
REFERENCES
- 1.Bukau, B., and G. C. Walker. 1990. Mutations altering heat shock specific subunit of RNA polymerase suppress major cellular defects of E. coli mutants lacking dnaK chaperone. EMBO J. 9:4027-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chattopadhyay, R., and S. Roy. 2002. dnaK− sigma 32 interaction is temperature-dependent. Implication for the mechanism of heat shock response. J. Biol. Chem. 277:33641-33647. [DOI] [PubMed] [Google Scholar]
- 3.Cowing, D. W., J. C. A. Bardwell, E. A. Craig, C. Woolford, R. W. Hendrix, and C. A. Gross. 1985. Consensus sequence for Escherichia coli heat shock gene promoters. Proc. Natl. Acad. Sci. USA 82:2679-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cowing, D. W., J. Mecsas, J. M. T. Record, and C. A. Gross. 1989. Intermediates in the formation of the open complex by RNA polymerase holoenzyme containing the sigma factor sigma 32 at the groE promoter. J. Mol. Biol. 210:521-530. [DOI] [PubMed]
- 5.deHaseth, P. L., M. Zupancic, and M. T. Record, Jr. 1998. RNA polymerase-promoter interaction: the comings and goings of RNA polymerase. J. Bacteriol. 180:3019-3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellinger, T., D. Behnke, H. Bujard, and J. D. Gralla. 1994. Stalling of Escherichia coli RNA polymerase in the +6 to +12 region in vivo is associated with tight binding to consensus promoter elements. J. Mol. Biol. 239:455-465. [DOI] [PubMed] [Google Scholar]
- 7.Erickson, J. W., V. Vaughn, W. A. Walter, F. C. Neidhardt, and C. A. Gross. 1987. Regulation of the promoters and transcripts of rpoH, the Escherichia coli heat shock regulatory gene. Genes Dev. 1:419-432. [DOI] [PubMed] [Google Scholar]
- 8.Gamer, J., H. Bujard, and B. Bukau. 1992. Physical interaction between heat shock proteins dnaK, DnaJ, and GrpE and the bacterial heat shock transcription factor σ32. Cell 69:833-842. [DOI] [PubMed] [Google Scholar]
- 9.Gross, C. A. 1996. Function and regulation of the heat shock proteins, p. 1382-1399. In F. C. Neidhardt, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 10.Grossman, A. D., J. W. Erickson, and C. A. Gross. 1984. The htpR gene product of E. coli is a sigma factor for heat-shock promoters. Cell 38:383-390. [DOI] [PubMed] [Google Scholar]
- 11.Hawley, D. K., and W. R. McClure. 1983. Compilation and analysis of Escherichia coli promoter sequences. Nucleic Acids Res. 11:2237-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helmann, J. D., and P. L. deHaseth. 1999. Protein-nucleic acid interactions during open complex formation investigated by systematic alteration of the protein and DNA binding partners. Biochemistry 37:5959-5967. [DOI] [PubMed] [Google Scholar]
- 13.Josaitis, C. A., T. Gaal, W. Ross, and R. L. Gourse. 1990. Sequences upstream of the −35 hexamer of rrnB P1 affect promoter strength and upstream activation. Biochim. Biophys. Acta 1050:307-311. [DOI] [PubMed] [Google Scholar]
- 14.Kammerer, W., U. Deuschle, R. Gentz, and H. Bujard. 1986. Functional dissection of Escherichia coli promoters: information in the transcribed region is involved in the late steps of the overall process. EMBO J. 5:2995-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanemori, M., K. Nishihara, H. Yanagi, and T. Yura. 1997. Synergistic roles of HsIVU and other ATP-dependent proteases in controlling in vivo turnover of σ32 and abnormal proteins in Escherichia coli. J. Bacteriol. 179:7219-7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karp, P. D., M. Riley, M. Saier, I. T. Paulsen, J. Collado-Vides, S. M. Paley, A. Pellegrini-Toole, C. Bonavides, and S. Gama-Castro. 2002. The EcoCyc database. Nucleic Acids Res. 30:56-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi, M., K. Nagata, and A. Ishihama. 1990. Promoter selectivity of the Escherichia coli RNA polymerase: effect of base substitutions in the promoter −35 region on promoter strength. Nucleic Acids Res. 18:7367-7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar, A., B. Grimes, M. Logan, S. Wedgwood, H. Williamson, and R. S. Hayward. 1995. A hybrid sigma subunit directs RNA polymerase to a hybrid promoter in Escherichia coli. J. Mol. Biol. 246:563-571. [DOI] [PubMed] [Google Scholar]
- 19.Landick, R., V. Vaughn, E. T. Lau, R. A. VanBogelen, J. W. Erickson, and F. C. Neidhardt. 1984. Nucleotide sequence of the heat shock regulatory gene of E. coli suggests its protein product may be a transcription factor. Cell 38:175-182. [DOI] [PubMed] [Google Scholar]
- 20.Liberek, K., T. P. Galitski, M. Zylicz, and C. Georgopoulos. 1992. The dnaK chaperone modulates the heat shock response of Escherichia coli by binding the σ32 transcription factor. Proc. Natl. Acad. Sci USA 99:11019-11023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lonetto, M., M. Gribskov, and C. A. Gross. 1992. The sigma 70 family: sequence conservation and evolutionary relationships. J. Bacteriol. 174:3843-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKane, M., and G. N. Gussin. 2000. Changes in the 17 bp spacer in the PR promoter of bacteriophage λ affect steps in open complex formation that precede DNA strand separation. J. Mol. Biol. 299:337-349. [DOI] [PubMed] [Google Scholar]
- 23.Moyle, H., C. Waldburger, and M. M. Susskind. 1991. Hierarchies of base pair preferences in the P22 ant promoter. J. Bacteriol. 173:1944-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narberhaus, F. 1999. Negative regulation of bacterial heat shock genes. Mol. Microbiol. 31:1-8. [DOI] [PubMed] [Google Scholar]
- 25.Paek, K.-H., and G. C. Walker. 1987. Escherichia coli dnaK null mutants are inviable at high temperature. J. Bacteriol. 169:283-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Platt, T., B. Muller-Hill, and J. H. Miller. 1972. Assays of the lac operon enzymes, p. 351-376. In J. H. Miller (ed.), Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Rosen, R., and E. Z. Ron. 2002. Proteome analysis in the study of the bacterial heat-shock response. Mass Spectrometry Rev. 21:244-265. [DOI] [PubMed] [Google Scholar]
- 28.Siegele, D. A., J. C. Hu, W. A. Walter, and C. A. Gross. 1989. Altered promoter recognition by mutant forms of the σ70 subunit of Escherichia coli RNA polymerase. J. Mol. Biol. 206:591-603. [DOI] [PubMed] [Google Scholar]
- 29.Stefano, J. E., and J. D. Gralla. 1982. Spacer mutations in the lacPs promoter. Proc. Natl. Acad. Sci. USA 79:1069-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomoyasu, T., J. Gamer, B. Bukau, M. Kanemori, H. Mori, A. Rutman, A. Oppenheim, T. Yura, K. Yamanaka, H. Niki, S. Hiraga, and T. Ogura. 1995. Escherichia coli FtsH is a membrane-bound, ATP-dependent protease which degrades the heat-shock transcription factor σ32. EMBO J. 14:2551-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vo, N. V., L. M. Hsu, C. M. Kane, and M. J. Chamberlin. 2003. In vitro studies of transcript initiation by Escherichia coli RNA polymerase. 3. Influences of individual DNA elements within the promoter recognition region on abortive intiation and promoter escape. Biochemistry 42:3798-3811. [DOI] [PubMed] [Google Scholar]
- 32.Warne, S. E., and P. L. deHaseth. 1993. Promoter recognition by Escherichia coli RNA polymerase. Effects of single base pair deletions and insertions in the spacer DNA separating the −10 and −35 regions are dependent on spacer DNA sequence. Biochemistry 32:6134-6140. [DOI] [PubMed] [Google Scholar]
- 33.Yamamori, T., and T. Yura. 1982. Genetic control of heat-shock protein synthesis and its bearing on growth and thermal resistance in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 79:860-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yura, T., H. Nagai, and H. Mori. 1993. Regulation of the heat-shock response in bacteria. Annu. Rev. Biochem. 47:321-350. [DOI] [PubMed] [Google Scholar]
- 35.Yura, T., T. Tobe, K. Ito, and T. Osawa. 1984. Heat shock regulatory gene (htpR) of Escherichia coli is required for growth at high temperature but is dispensable at low temperature. Proc. Natl. Acad. Sci. USA 81:6803-6807. [DOI] [PMC free article] [PubMed] [Google Scholar]