Abstract
Frequent spontaneous loss of the magnetic phenotype was observed in stationary-phase cultures of the magnetotactic bacterium Magnetospirillum gryphiswaldense MSR-1. A nonmagnetic mutant, designated strain MSR-1B, was isolated and characterized. The mutant lacked any structures resembling magnetosome crystals as well as internal membrane vesicles. The growth of strain MSR-1B was impaired under all growth conditions tested, and the uptake and accumulation of iron were drastically reduced under iron-replete conditions. A large chromosomal deletion of approximately 80 kb was identified in strain MSR-1B, which comprised both the entire mamAB and mamDC clusters as well as further putative operons encoding a number of magnetosome-associated proteins. A bacterial artificial chromosome clone partially covering the deleted region was isolated from the genomic library of wild-type M. gryphiswaldense. Sequence analysis of this fragment revealed that all previously identified mam genes were closely linked with genes encoding other magnetosome-associated proteins within less than 35 kb. In addition, this region was remarkably rich in insertion elements and harbored a considerable number of unknown gene families which appeared to be specific for magnetotactic bacteria. Overall, these findings suggest the existence of a putative large magnetosome island in M. gryphiswaldense and other magnetotactic bacteria.
Magnetotactic bacteria are capable of forming magnetosomes, which are specific intracellular structures that enable the cells to orient along magnetic field lines (3, 4, 41). The superior crystalline and magnetic properties of magnetosomes make them potentially useful as a highly ordered biomaterial in a number of applications, e.g., in the immobilization of bioactive compounds, magnetic drug targeting, or as a contrast agent for magnetic resonance imaging (24, 41). Recently, the characteristics of bacterial magnetosomes have even been considered for use as biosignatures to identify presumptive Martian magnetofossils (49). Moreover, understanding bacterial magnetosome formation is expected to provide insights into more complex biomineralization systems in higher organisms (19). The biomineralization of magnetosome particles is achieved by a complex mechanism with control over the uptake, accumulation, and precipitation of iron, which, however, is poorly understood at the molecular and biochemical level.
The magnetotactic α-proteobacterium Magnetospirillum gryphiswaldense microaerobically produces up to 60 cubo-octahedral magnetosomes, which are approximately 45 nm in size and consist of membrane-bounded crystals of the iron mineral magnetite (Fe3O4) (34, 42). In contrast to most other magnetotactic bacteria, methods for mass culture and genetic manipulation of M. gryphiswaldense are available (17, 38, 44), which has facilitated its analysis in a number of studies (37, 39, 40, 43).
In Magnetospirillum species, the deposition of the mineral particle occurs within a specific compartment, which is provided by the magnetosome membrane (5, 35, 36). Recently, a number of magnetosome membrane-specific polypeptides were identified in isolated magnetosomes from M. gryphiswaldense (15). Cloning and sequencing of the genes encoding several of the most abundant magnetosome membrane-associated proteins (mamA, mamB, mamC, and mamD) revealed that these are arranged in two operon-like gene clusters. One major gene cluster containing several magnetosome genes, including mamA and mamB, was found to be highly conserved between M. gryphiswaldense and other magnetotactic bacteria according to the comparative sequence analysis of preliminary genome assemblies, which became available for M. magnetotacticum strain MS-1 and a magnetic coccus, strain MC-1 (http://www.jgi.doe.gov/tempweb/JGI_microbial/html/index.html).
The mamAB cluster comprises 15 and 9 colinear open reading frames in M. magnetotacticum and strain MC-1, respectively, and is characterized by a set of genes which are shared by all three magnetotactic strains. The corresponding gene products are homologous to several protein families, for example, TPR proteins (28), CDF transporters (30) and PDZ proteins resembling HtrA-like serine proteases (29) with speculated functions in iron transport into the magnetosome vesicles (CDF) and protein protein interactions (TPR and PDZ proteins). A second identified chromosomal locus comprised the mamD and mamC genes, both of which encode abundant magnetosome-associated proteins with unknown functions. These findings suggested that the genetic determination of magnetosome formation is complex and involves several different genomic loci. However, the overall organization of the clusters identified in the genome has not been determined because of the lack of contiguous sequence information. In addition, the essential function of mam genes in magnetosome formation has not been proven because of the unavailability of mutants.
In this study, we estimated the spontaneous mutability of the magnetic phenotype and isolated and characterized a spontaneous nonmagnetic mutant of M. gryphiswaldense which harbors a large chromosomal deletion of approximately 80 kb comprising all identified mam genes. In addition, the sequence of a 35-kb genomic fragment from this region was analyzed, including the complete mamAB, mamDC, and mms6 gene clusters and several additional hitherto unknown genes belonging to these clusters. We present data that indicate the existence of a large supercluster in M. gryphiswaldense that harbors all of the known mam genes and that may represent a putative magnetosome island.
MATERIALS AND METHODS
Bacterial strains.
Magnetospirillum gryphiswaldense strain MSR-1 (DSM 6361) (34, 42) was used in this study. A spontaneous nonmagnetic mutant, designated strain MSR-1B, was isolated by plating on ACA medium (44) from a stock culture of M. gryphiswaldense MSR-1 which had been subjected to a number of serial subcultures in slush agar medium (0.4% agar) (34) and stored at 4°C between transfers.
Flask cultivation.
For small culture volumes (10 to 500 ml), M. gryphiswaldense strains MSR-1 and MSR-1B were cultured in flask standard medium (FSM) as described previously elsewhere (17). The medium contained 0.3% (wt/vol) sodium pyruvate as a carbon source. Iron was added as ferric citrate before autoclaving as specified. Alternatively, the medium was supplemented with α,α′-dipyridyl to chelate residual iron in a physiologically unavailable form (40). Flask cultures of M. gryphiswaldense strains were carried out at 170 rpm (aerobic conditions) and 100 rpm (microaerobic conditions) in loosely stoppered 1-liter bottles containing 500 ml of FSM in an incubator shaker (New Brunswick).
Oxystat cultivation.
Growth experiments at various O2 tensions were performed in a modified dual-vessel laboratory fermentor system (Biostat A Twin; B. Braun Biotech. International, Melsungen, Germany) equipped for the automatic control of pH, temperature, and dissolved oxygen concentration (Oxystat) as described previously (17). Soy bean peptone was omitted from large-scale medium in order to create iron-deficient conditions because of the significant intrinsic iron content of peptone. Iron was added to FSM as ferric citrate as specified in the experiments. The medium (4 liters) was inoculated with 400 ml of a microaerobic flask culture. The initial cell number in the experiments was approximately 108/ml.
Iron transport assay.
The incorporation of 55Fe was measured essentially as described previously (40). For uptake measurements, cells in spent growth medium at an optical density at 565 nm of 0.3 to 0.35 were used. After 5 min of incubation of the cells at 30°C, transport was started by adding 55FeCl3 to a final concentration of between 0.5 and 50 μM. An activity of approximately 50 kBq was used per experiment. At intervals, samples of 0.2 ml were withdrawn, added to 5 ml of 0.1 M LiCl-5 mM EDTA, filtered on a 0.45-μm-pore-size cellulose nitrate filter (Sartorius), and washed once with the same buffer. The filters were dried at 50°C, and the radioactivity was determined in a liquid scintillation counter. Inhibition studies were performed in the same way with a final concentration of 100 μM carbonylcyanide-m-chlorophenylhydrazone (CCCP) and 1 mM 2,4-dinitrophenol (DNP).
Analytical methods.
Cell growth and magnetism were measured turbidimetrically at 565 nm. The average magnetic orientation of cell suspensions (magnetism) was assayed by an optical method as described previously (17, 43). Iron measurements were made with an atomic absorption spectrometer (3110; Perkin-Elmer, Überlingen, Germany) as described elsewhere (15, 17).
Electron microscopy.
Negative staining was performed as described previously (51). For ultrathin sectioning, cells were washed and suspended in 50 mM potassium phosphate buffer (pH 6.8), fixed in the presence of a mixture of 0.2% (vol/vol) glutaraldehyde, and embedded in Spurr's low-viscosity resin as described previously (53). Micrographs were taken with a Philips EM301 electron microscope at an acceleration voltage of 80 kV. Magnifications were calibrated with a cross-lined grating replica (Balzers).
DNA techniques.
Total DNA from M. gryphiswaldense strains was isolated as described previously (23). Other DNA manipulations were carried out essentially by standard methods (33). Primers used for PCR (Table 1) were purchased from MWG Biotech (Berlin, Germany).
TABLE 1.
Primers usd in this study
| Primer | Sequence (5′→3′) | Size (bp) of amplified DNA | Amplified gene/positionsa |
|---|---|---|---|
| CW1_2F | ACCTCGGTTGGGATTCTC | 23256-23273 | |
| CW1_1R | GTACATCGCCGTTCTCG | 420 | mamN/23660-23676 |
| S16 | TGTGGTCAAGGTGCCTGTG | 30219-30237 | |
| CW10_3R | CCTTATCCGAGCCTGTTTCG | 507 | mamU/30707-30726 |
| DS24NF | ATGTCTAGCAAGCCGTCG | 26498-26515 | |
| CW4_1R | CCAATGAACTCGATGAACG | 640 | mamA/27120-27138 |
| CW7_4F | TTCAAAGGCATCTTGGGGC | 28346-28364 | |
| CW7_3R | CTCCGTGGATACCGAACTGT | 548 | mamB/28894-28913 |
| SSC_f+6/+24 | CTTTCAACTTGCGCCGTA | 5095-5113 | |
| SSC_r+355/+337 | ATGTCTTCGTCGGACGCT | 351 | mamC/5428-5446 |
| SSD_f−15/+3 | GGAAAGGCCAATACCATG | 3655-3673 | |
| SSD_r+950/+932 | TCAGGCTTATTCCTCGCC | 966 | mamD/3976-3994 |
| SSF_f−5/+13 | AAGCAATGGCCGAGACTA | 3655-3673 | |
| SSF_r+315/+335 | TCAGATCAGGGCGACTACAT | 339 | mamF/3976-3994 |
| SSX_f−15/+3 | CTTGCCGGAGATCAGATG | 3377-3395 | |
| SSX_r+291/+274 | TTGCTTTGCCCTCGCTTA | 282 | mamG/3643-3658 |
| GFCD_f | TTAGGTTCAATCCGGGGC | 5724-5742 | |
| GFCD_r | TCGGGACAATGCGACATC | 435 | 6141-6159 |
| GFCD3′_f | AATTGGGCGTGTCGATCA | 7010-7028 | |
| GFCD3′_r | GCCTTGTCGATGACGAAG | 336 | 7328-7346 |
| GFCD5′_f | TTGTGGACACAGCGAAGC | 2978-2996 | |
| GFCD5′_r | ATGCACGATTCCCTCTCT | 396 | 3356-3374 |
| mms16_r | GGCACGAAGCTTACTTCT | ||
| mms16_f | ACGAAGTGGCCGTGGTGT | 244 | mms16 |
| bfr1_f | CAGTATTTCCTGCACGCC | ||
| bfr1_r | ACAGTGACCCACAATTGG | 495 | bfr1 |
| bfr2_f | TGCCCGGTTACTTAAGGA | ||
| bfr2_r | TCTCCTTACGCGATCTCG | 510 | bfr2 |
| SS14_f | ATGCACTGGCTCGAGGTT | 31074-31091 | |
| SS15_r | TCGCGCCAAAAGTATCAG | 365 | 31422-31439 |
| SS16_f | AGTTCATCGATCCGG | 33060-33077 | |
| SS17_r | ACGCGCTGTTCGAGATCG | 432 | 33474-33492 |
Positions refer to the sequence numbering shown in Fig. 6.
Pulsed-field gel electrophoresis.
For agarose plug preparation, mid-log-phase cells of strains MSR-1 and MSR-1B were harvested, washed, and resuspended in 1× PBS. Low-melting-point agarose (Invitrogen) was added to a final concentration of 1%. Cells in agarose plugs were lysed overnight at 55°C in 0.5 M EDTA-1% N-lauroylsarcosine (Sigma)-1 mg of proteinase K per ml (Merck) and then washed six times with Tris-EDTA (pH 8.0). For restriction digests, single plugs were equilibrated with the appropriate restriction enzyme buffer for 10 min and then digested overnight with 20 units of enzyme (MBI Fermentas). Electrophoresis was performed with the Chef-DRIII System (Bio-Rad). Pulsed-field certified agarose (Bio-Rad) gels (1% in 0.5× Tris-borate-EDTA) were run at 14°C, 6 V/cm, and an angle of 120°. Pulse times varied according to the size of the fragments to be resolved. Digitized gels were analyzed by the ImageMaster1D software (version 3.0; Amersham-Pharmacia).
Generation and screening of a bacterial artificial chromosome library.
For isolation and size fractionation of genomic DNA, preparative pulsed-field gel electrophoresis was performed essentially as described above. DNA greater than 600 kb was excised and dialyzed against 1× Tris-EDTA. Following HindIII digestion, the DNA was size selected by a further preparative pulsed-field gel electrophoresis run, and DNA fragments of 50 to 100 kb and 100 to 150 kb were excised from the gel. Agarose plugs were dialyzed against 1× Tris-EDTA, digested with Gelase (Epicentre), and concentrated and dialyzed on VSWP filters (Millipore) against 30% PEG8000 and 0.5× Tris-EDTA.The DNA was ligated to the HindIII-digested pIndigoBAC-5 cloning vector (Epicentre) and transformed into Escherichia coli DH10B (Invitrogen) with a Biometra cell porator and voltage booster system (350 V, 330 μF, 4 kΩ). Transformed cells were plated onto Luria-Bertani agar plates containing chloramphenicol, 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal), and isopropylthiogalactopyranoside (IPTG) in standard concentrations. White colonies were initially screened by colony hybridization with a digoxigenin-labeled mamA probe. Positive clones were subsequently screened by PCR with all available primers for the amplification of various mam genes (Table 1). Out of four clones that proved positive for all mam genes tested, one clone harboring a 68-kb insert was subjected to sequence analysis.
Generation and analysis of genome sequence data from M. gryphiswaldense.
Sequence data were obtained from the selected bacterial artificial chromosome and whole genome by a shotgun approach. Bacterial artificial chromosome DNA was isolated by alkaline lysis and purified on CsCl by standard procedures (33). For subcloning, DNA was sonicated, fragment ends were polished with T4 and Klenow polymerase (New England Biologicals), size selected, ligated in pUC19, transformed into E. coli DH10B (Invitrogen), and selected on ampicillin (33). For the whole genome, shotgun DNA was prepared (genomic kit; Qiagen) from strain MSR-1 and processed in the same way. In both cases, plasmid libraries with 1.5-kb and 3.5-kb inserts were obtained. The inserts of the libraries were amplified by PCR (31) as templates for sequencing. End sequences were performed with Big Dye chemistry (ABI), M13 primers, and ABI 3700 capillary sequencers (ABI), resulting in more than 10-fold coverage for the bacterial artificial chromosome insert and 4.2-fold coverage for the genome, with a total contig length of 4.1 Mb.
All raw sequences were processed by Phred (11), controlled for vector or E. coli contamination, and assembled by Phrap (46). Analyzed regions were manually edited in GAP4 (http://www.sanger.ac.uk/Software/sequencing/docs/phrap2gap/). The quality of these sequence data was finished to justify the Bermuda rules (http://www.ornl.gov/hgmis/research/Bermuda.html#1). Finishing of analyzed sequences was done by resequencing clones and primer walking. Open reading frame (ORF) finding and annotation of M. gryphiswaldense genome sequences were performed with GenDB (25).
Nucleotide sequence accession numbers.
The nucleotide sequence of the 35-kb M. gryphiswaldense genomic region containing the complete mamAB and mamDC gene clusters has been deposited in the GenBank, EMBL, and DDJB libraries with accession number BX571797. The sequences for bfr1, bfr2, and mms16 M. gryphiswaldense genes were deposited under BX571782 and BX571783, respectively.
RESULTS
Estimation of spontaneous mutation frequency of the magnetic phenotype and isolation of the nonmagnetic mutant strain M. gryphiswaldense MSR-1B.
We repeatedly observed that long-term cultures of M. gryphiswaldense were completely taken over by nonmagnetic mutants after only a few transfers. Plating of cells on ACA medium revealed that these cultures consisted homogeneously of nonmagnetic cells which formed white colonies, versus the dark brown colonies of magnetic cells. In order to estimate the frequency with which spontaneous loss of cellular magnetism occurred, cells from a magnetic colony from the wild type were serially subcultured for 12 passages in 50 ml of FSM medium under continuous agitation and incubated at 28°C before plating. In repeated experiments, we failed to detect any nonmagnetic colonies among approximately 5 × 105 clones if the cells were kept under conditions of continuous growth. In contrast, nonmagnetic colonies were repeatedly isolated from other cultures which were grown to saturation and subsequently aged by keeping them for several days at 4°C without agitation to mimic storage conditions. Although the proportion of nonmagnetic clones varied between independent experiments, up to 0.5% of the total colonies from those stationary-phase cultures were nonmagnetic. One clone from a nonmagnetic long-term culture was selected for further characterization after verification of its identity as M. gryphiswaldense by sequencing of the 16S rRNA gene (not shown). The mutant strain was designated MSR-1B.
Phenotypic characterization of strain MSR-1B.
The morphology of MSR-1B cells appeared to be very similar to that of the wild-type cells by phase-contrast microscopy. We were unable to detect any particles resembling native or aberrant magnetosome crystals in electron micrographs of MSR-1B cells. In addition, intracellular membrane structures which might represent empty, preformed magnetosome membrane vesicles were absent from ultrathin sections of embedded cells. Loss of magnetism was permanent, and no reversions to the wild-type phenotype were observed. Due to the lack of magnetosomes, cell pellets of the mutant were rusty red in color, versus the blackish appearance of magnetic wild-type cells.
In addition to a lack of a magnetic reaction, MSR-1B was substantially impaired in motility. Less than 0.1% of the population simultaneously displayed brief swimming runs, as observed by microscopy. Consistently, the cells failed to form aerotactic bands in semisolid agar and in swarm plates (not shown). The reduced motility was accompanied by the lack of flagella (Fig. 1). Although the loss of motility in the population apparently was not total, we failed to detect any flagellum-like structures in numerous cells inspected by electron microscopy.
FIG. 1.
(A) Electron micrograph of negatively stained cells of M. gryphiswaldense MSR-1B. The nonmagnetic mutant MSR-1B exhibits the characteristic morphology of magnetic spirilla but lacks electron-dense magnetite crystals and flagella. Bar, 1 μm. (B) Ultrathin section of MSR-1B. Bar, 400 nm. (C) Ultrathin section of a wild-type M. gryphiswaldense MSR-1 cell, showing the characteristic magnetosome chain (M). Bar, 400 nm. PHA, polyhydroxyalkanoate; PPH, polyphosphate.
Growth and iron uptake characteristics of strain MSR-1B.
The ability to form magnetosomes could not be restored by any of the growth conditions tested in an oxystat-controlled fermentor (e.g., increased iron concentration and growth at various oxygen concentrations; data not shown). Under microaerobic (0.25 mbar of O2), iron-depleted (<1 μM Fe) conditions, MSR-1B grew slightly slower (average doubling time, 9.5 h) than the wild type (average doubling time, 8.9 h; Fig. 2A, B). However, iron-replete medium (80 μM Fe) inhibited the growth of MSR-1B (average doubling time, 11.1 h), whereas the growth of the wild type was significantly enhanced (average doubling time, 6.1 h) under these conditions (Fig. 2C, D). While the intracellular iron content of iron-deprived cells was similar both in the wild type (0.07% of dry weight) and in the mutant (0.06% of dry weight), wild-type cells accumulated significantly higher amounts of iron (1.3% of dry weight) than MSR-1B (0.4% of dry weight) when grown in iron-replete medium.
FIG. 2.
Growth (▪) and magnetism (•) of M. gryphiswaldense strains MSR-1 (wild type) and MSR-1B (nonmagnetic mutant) in the oxystat at microaerobic conditions (0.25 mbar of O2). (A) MSR-1B, <1 μM Fe; (B) MSR-1, <1 μM Fe; (C) MSR-1B, 80 μM Fe; (D) MSR-1, 80 μM Fe.
To analyze whether the nonmagnetic mutant strain MSR-1B was affected in its iron uptake kinetics, the incorporation of 55Fe was measured at various iron concentrations (Fig. 3). Compared to the wild type, strain MSR-1B displayed significantly reduced iron uptake at all concentrations tested. A Vmax of 0.46 nmol of Fe min−1 (mg [dry weight])−1 and a Km of 21.6 μM Fe were calculated from these experiments for the wild-type MSR-1, compared to a Vmax of 0.18 nmol of Fe min−1 (mg [dry weight])−1 and a Km of 11.6 μM Fe determined in the mutant strain MSR-1B. Generally, the Km and Vmax values determined for iron uptake in the wild type were slightly different from those in a previous study (40), which might be due to the different growth conditions used in this study. In both strains, the addition of 100 μM CCCP or 1 mM DNP resulted in an approximately 45 to 70% and 25 to 40% inhibition of iron uptake, respectively (data not shown). These results indicate that the mutant strain is still capable of energy-dependent iron uptake, but at a drastically reduced rate.
FIG. 3.
Iron uptake kinetics of strains MSR-1 (A) and MSR-1B (B). Cells were incubated with various concentrations of 55FeCl3: ○ 0.5 μM; • 1 μM; ▿ 2 μM; ▾ 5 μM; □ 10 μM; ▪ 20 μM; and ⋄ 50 μM Fe.
Large deletion in strain MSR-1B comprises all previously identified mam genes.
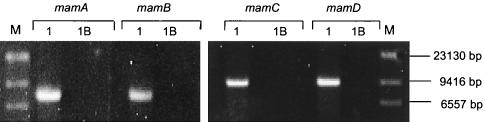
To test whether genes of the previously identified mam clusters were affected by the mutation, their presence was analyzed in strain MSR-1B. Southern blots with probes derived from mamA and mamB revealed the absence of these genes in genomic digests of the mutant. In addition, mamC and mamD probes also failed to recognize a hybridizing band in MSR-1B, indicating that both genes were deleted in the mutant strain (Fig. 4).
FIG. 4.
Southern blot analysis of EcoRI-digested MSR-1 and MSR-1B genomic DNAs subsequently blotted and hybridized with digoxigenin-labeled mamA, mamB, mamC, and mamD gene-specific probes. Lanes: 1, strain MSR-1; 1B, strain MSR-1B; M, digoxigenin-labeled DNA size standards.
To determine the extent of the deletion, a number of additional genes from both the mamAB and mamDC clusters and sequences neighboring to them were analyzed by PCR. In addition to mamA, mamB, mamC, and mamD, all genes tested were detected in the wild type but deleted in strain MSR-1B (Table 2). A further set of tested genes were mms16, bfr1, and bfr2, which were previously suggested to be involved in magnetite synthesis in Magnetospirillum strain AMB-1 and M. magnetotacticum MS-1 (6, 27). Homologues with high similarity (90 to 98%) to mms16, bfr1, and bfr2 were found in the M. gryphiswaldense genome and could also be detected in the mutant strain MSR-1B. To further determine the extent of the deletion, the genome sizes of the mutant and the wild type were compared by pulsed-field gel electrophoresis (Table 3). Genomic DNA from MSR-1B and MSR-1 clearly yielded different restriction patterns in digests with various enzymes. For the wild type, genome sizes of 4.59 Mb and 4.63 Mb were calculated from SwaI and PmeI digests, respectively, while strain MSR-1B yielded sizes of 4.52 and 4.54 Mb, respectively, with the same enzymes. This implied that a fragment of approximately 80 (50 to 110) kb that comprises all genes from both the mamDC and mamAB loci but not the mms16 and bfr genes is deleted from the MSR-1B genome.
TABLE 2.
Presence of magnetosome genes in strains MSR-1 and MSR-1B
| Gene or position | Gene present
|
|
|---|---|---|
| MSR-1 | MSR-1B | |
| mamA | + | − |
| mamB | + | − |
| mamC | + | − |
| mamD | + | − |
| mamF | + | − |
| mamG | + | − |
| mamN | + | − |
| mamU | + | − |
| GFDC5′ | + | − |
| GFDC | + | − |
| GFDC3′ | + | − |
| bfr1 | + | + |
| bfr2 | + | + |
| mms16 | + | + |
TABLE 3.
Sizes of restriction fragments generated by SwaI and PmeI digestion of genomic DNA and estimated genome sizes of strains MSR-1 and MSR-1Ba
| Fragment | Size (kb)
|
|||
|---|---|---|---|---|
|
SwaI
|
PmeI
|
|||
| MSR-1 | MSR-1B | MSR-1 | MSR-1B | |
| A | 1,420 | 1,420 | 1,510 | |
| B | 1,020 | 1,030 | 1,400 | |
| C | 760 | 770 | 1,180 | 1,160 |
| D | 400 | 710 | 720 | |
| E | 380 | 410 | ||
| F | 360 | 280 | ||
| G | 350 | 280 | ||
| H | 310 | 270 | ||
| I | 300 | 260 | ||
| J | 300 | 220 | ||
| K | 290 | 210 | ||
| L | 150 | |||
| M | 140 | |||
| N | 140 | |||
| O | 130 | |||
| Total | 4,590 | 4,520 | 4,630 | 4,540 |
Fragment sizes unique to each strain are in bold.
As the extent of the large deletion suggested the loss of a considerable number of genes, the protein composition of MSR-1B cells grown under different conditions was analyzed for the absence of protein bands by one-dimensional polyacrylamide gel electrophoresis (20). Under conditions of both iron excess and iron deficiency, the protein patterns of the mutant and the wild type were virtually identical. Likewise, no significant differences were detectable in microaerobically versus aerobically grown cells of both the mutant and the wild type (data not shown but available on request). These results indicated that no abundant proteins were among the products of the deleted genes.
Identification of a 50-kb genomic fragment harboring the mamAB and mamDC gene clusters in the M. gryphiswaldense wild-type strain.
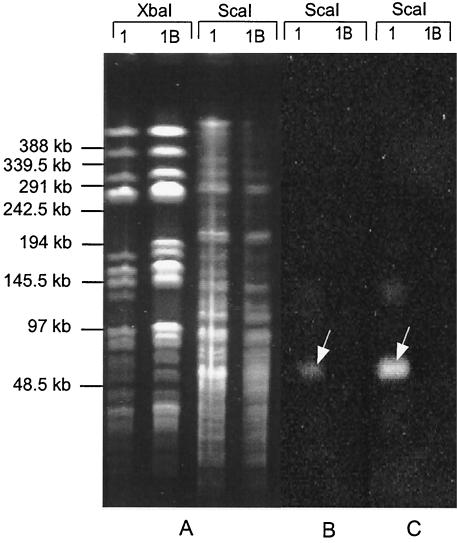
To localize the genomic region corresponding to the identified clusters, genomic digests of MSR-1 and MSR-1B were resolved by pulsed-field gel electrophoresis and subsequently hybridized with probes corresponding to mamB and mamD (Fig. 5). In ScaI digests of MSR-1, both probes recognized an identical fragment of approximately 50 kb that was missing in the MSR-1B mutant, as expected. Hence, it could be concluded that the mamAB and mamDC clusters are adjacent within less than 50 kb of the genome of MSR-1.
FIG. 5.
Southern blot analysis of ScaI-digested genomic DNA from M. gryphiswaldense MSR-1 and MSR-1B. Restriction fragments were resolved by pulsed-field gel electrophoresis (A) and subsequently blotted and hybridized with digoxigenin-labeled mamB (B) and mamD (C) gene-specific probes. Arrows indicate a single genomic 50-kb ScaI fragment cohybridizing to both mamB and mamD. Lanes: 1, strain MSR-1; 1B, strain MSR-1B.
Genomic organization of the mamAB and mamDC clusters in M. gryphiswaldense MSR-1.
After we had found evidence for a close genetic linkage of the mamAB and mamDC clusters, a genomic bacterial artificial chromosome library of M. gryphiswaldense MSR-1 was constructed and screened. A single clone harboring both the mamAB and mamDC clusters on a 68-kb fragment was identified and subjected to sequence analysis. The molecular organization of a 35-kb subsequence is presented in Fig. 6, and the characteristics of the annotated open reading frames are summarized in Table 4. A total of 37 genes were predicted. Although the extent of the deletion exceeds the size of the genomic region covered by the cloned fragment, all tested genes that proved missing in the mutant were among the identified genes. Genes that encode known magnetosome-bound proteins are located within three different operon-like clusters:
FIG. 6.
Molecular organization of the M. gryphiswaldense MSR-1 35-kb genomic region comprising all identified magnetosome genes. Names of ORFs encoding known magnetosome-associated proteins are shown in bold.
TABLE 4.
Characteristics of gene products deduced from ORFs identified within the 35-kb genomic region of M. gryphiswaldense MSR-1
| ORFa | Size of gene product (amino acids) | Best BLAST hitb (accession no.) | E value | Homologues in magnetotactic bacteriac (accession no.) | E value | Proposed function of BLAST homolog | |
|---|---|---|---|---|---|---|---|
| ORF 1 | 230 | M. mag. (ZP_00055313.1) | 4e-05 | Unknown | |||
| MC-1 (ZP_00047740.1) | 0.003 | ||||||
| ORF 2 | 347 | r03081 Sinorhizobium meliloti (AL591792) | 1e-05 | M. mag. (ZP_00053421.1) | 8e-77 | Unknown transmembrane | |
| ORF 3 | 107 | — | M. mag. (ZP_00053420.1) | 6e-43 | Unknown | ||
| MC-1 (ZP_00042693.1) | 2e-07 | ||||||
| ORF 4 | 136 | — | mms6 AMB-1 (BAC65162.1) | 7e-11 | Iron binding | ||
| M. mag. (ZP_00053419.1) | 7e-11 | ||||||
| mamG | 84 | — | M. mag. (ZP_00050810.1) | 0.37 | Unknown | ||
| mamF | 111 | abf1 Kluyveromyces marxianus (Z19865) | 0.003 | M. mag. (ZP_00053416.1) | 9e-41 | Transcription regulation | |
| MC-1 (ZP_00042693.1) | 3e-09 | ||||||
| mamD | 314 | — | M. mag. (ZP_00053415.1) | 4e-88 | Unknown | ||
| mms7 AMB-1 (BAC65161.1) | 1e-87 | ||||||
| MC-1 (ZP_00043592.1) | 2e-11 | ||||||
| mamC | 125 | — | M. mag. (ZP_00053414.1) | 4e-16 | Unknown | ||
| mms13 AMB-1 (BAC65160.1) | 4e-16 | ||||||
| MC-1 (ZP_00042848.1) | 2e-04 | ||||||
| ORF 9 | 278 | idiA Synechococcus sp. (Z48754) | 9e-75 | — | Ferric iron transport | ||
| ORF 10 | 214 | TRm2011 Sinorhizobium meliloti (NP_435256.1) | 2e-30 | MC-1 (ZP_00049672.1) | 9e-41 | Transposase | |
| M. mag. (ZP_00052583.1) | 2e-37 | ||||||
| ORF 11 | 436 | istA Agrobacterium tumefaciens (AE009415) | 1e-90 | MC-1 (ZP_00042589.1) | 5e-25 | IS21 family transposase | |
| ORF 12 | 525 | PA1673 Pseudomonas aeruginosa (AE004595) | 8e-11 | M. mag. (ZP_00054420.1) | 2e-17 | Hemerythrin-like | |
| MC-1 (ZP_00043360.1) | 4e-07 | ||||||
| ORF 13 | 149 | PA1673 Pseudomonas aeruginosa (AE004595) | 3e-07 | M. mag. (ZP_00052998.1) | 1e-35 | Hemerythrin-like | |
| MC-1 (ZP_00044836.1) | 9e-11 | ||||||
| ORF 14 | 317 | Acetobacter xylinus (S35004) | 3e-46 | M. mag. (ZP_00052299.1) | 3e-17 | Transposase_11 IS1031C | |
| MC-1 (ZP_00048870.1) | 2e-09 | ||||||
| ORF 15 | 76 | MSR8681 Rhizobium loti (Q98AK2) | 1e-20 | M. mag. (ZP_00052258.1) | 2e-07 | Unknown | |
| ORF 16 | 415 | MLL1146 Rhizobium loti (Q98L77) | 3e-45 | M. mag. (ZP_00053281.1) | 0 | Unknown | |
| mamH | 431 | CT1475 Chlorobium tepidum (Q8KCE7) | 8e-35 | M. mag. (ZP_00053280.1) | e-170 | Sugar (and other) transporter | |
| MC-1 (ZP_00043594.1) | 5e-77 | ||||||
| mamI | 87 | MC-1 (ZP_00042845.1) | 0.002 | Unknown | |||
| mamE | 772 | MLL5022 Rhizobium loti (Q98CS8) | 9e-35 | mamE M. mag. (ZP_00054403.1) | 0 | Serine protease | |
| MC-1 (ZP_00042846.1) | 5e-55 | ||||||
| mamJ | 466 | SP1772 Streptococcus pneumoniae (E95206) | 0.001 | mamJ M. mag. (ZP_00054404.1) | 2e-32 | Cell wall surface anchor family | |
| mamK | 347 | mreB Methanopyrus kandleri (QSTYX3) | 2e-17 | mamK M. mag. (ZP_00054405.1) | e-159 | Actin-like | |
| MC-1 (ZP_00042854.1) | 6e-84 | ||||||
| mamL | 123 | — | mamL M. mag. (ZP_00054405.1) | 3e-07 | Unknown | ||
| MC-1 (ZP_00042854.1) | 0.048 | ||||||
| mamM | 318 | BH1238 Bacillus halodurans (F83804) | 1e-33 | mamM M. mag. (ZP_00054406.1) | e-171 | Cation efflux | |
| mamM MC-1 (ZP_00043611.1) | 4e-77 | ||||||
| mamN (ORF 1) | 437 | TM0934 Thermotoga maritima (AE001757) | 6e-30 | mamN M. mag. (ZP_00054407.1) | 0 | Transporter | |
| mamO (ORF 2) | 632 | CC1282 Caulobacter crescentus (C87408) | 6e-15 | mamO M. mag. (ZP_00054408.1) | 0 | Serine protease | |
| mamO MC-1 (ZP_00043610.1) | 7e-77 | ||||||
| mamP (ORF 3) | 270 | OrfE0 Rhodobacter capsulatus (CAA72164) | 0.042 | mamP M. mag. (ZP_0054409.1) | e-106 | Serine protease | |
| mamP MC-1 (ZP_00043609.1) | 1e-32 | ||||||
| mamA (ORF 4) | 217 | MM2348 Methanosarcina mazei (AE013478) | 3e-15 | mamA M. mag. (BAA11643.2) | e-111 | TPR protein | |
| mamA MC-1 (ZP_00043608.1) | 2e-35 | ||||||
| mamQ (ORF 5) | 272 | lemA Themotoga maritima (AE001759) | 3e-16 | mamQ M. mag. (ZP_00054411.1) | e-109 | Unknown | |
| mamQ MC-1 (ZP_0043606.1) | 1e-35 | ||||||
| mamR (ORF 6) | 72 | mamR M. mag. (ZP_00054412.1) | 4e-29 | ||||
| mamB (ORF 7) | 297 | ydfM Bacillus subtilis (C69781) | 1e-37 | mamB M. mag. (ZP_00054413.1) | e-156 | Cation efflux | |
| mamB MC-1 (ZP_00043605.1) | 7e-76 | ||||||
| mamS (ORF 8) | 180 | — | mamS M. magn. (ZP_00054414.1) | 1e-58 | Unknown | ||
| — | mamS MC-1 (ZP_00043604.1) | 3e-11 | |||||
| mamT (ORF 9) | 174 | mamT M. mag. (ZP_00054415.1) | 2e-81 | Unknown (heme binding) | |||
| mamT MC-1 (ZP_00043609.1) | 5e-04 | ||||||
| mamU (ORF 10) | 297 | MLL9677 Rhizobium loti (AP003017) | 6e-22 | mamU M. mag. (ZP_00054416.1) | e-115 | Unknown | |
| ORF 34 | 411 | SP1776 Streptococcus pneumoniae (E95206) | 7e-05 | — | Cell wall surface anchor family | ||
| ORF 35 | 254 | y4hP Rhizobium sp. (T10850) | 3e-76 | M. mag. (ZP_00052071.1) | 2e-78 | Transposase_25 IS66 | |
| MC-1 (ZP_000442447.1) | 8e-37 | ||||||
| ORF 36 | 104 | Agrobacterium tumefaciens (Q52599) | 4e-16 | MC-1 (ZP_00049269.1) | 2e-07 | Transposase_25 IS66 | |
| ORF 37 | 341 | y4hP Rhizobium sp. (T10850) | 1e-108 | M. mag. (ZP_00052071.1) | 2e-76 | Transposase_25 IS66 | |
| MC-1 (ZP_00047840.1) | 3e-41 |
Genes which encode known magnetosome-associated proteins are in bold. Numbers in parentheses refer to the previously described ORF in M. gryphiswaldense (16).
Only hits in organisms other than those shown in the homologues column are shown.
MC-1, Magnetococcus sp. strain MC-1; AMB-1, Magnetospirillum sp. strain AMB-1; M. mag., Magnetospirillum magnetotacticum strain MS-1.
mamAB cluster.
The mamAB cluster extends over 16.5 kb and comprises 17 consecutive, colinear ORFs that were designated mamH to mamU. The organization and sequence of the genes are similar to those in the homologous regions previously found in M. magnetotacticum MS-1 (15). However, one ORF located at the right border of the mamAB cluster of M. magnetotacticum encoding one of three CDF transporters in this strain was absent in M. gryphiswaldense. Two additional ORFs (mamH and mamI) were identified at the left boundary of the cluster in M. gryphiswaldense MSR-1.
mms6 cluster.
A putative 2.7-kb operon comprising four ORFs is located 15 kb upstream from the left border of the mamAB cluster. ORF4 is apparently homologous (78% similarity) to mms6, which has recently been identified as encoding a magnetosome-bound protein in Magnetospirillum sp. strain AMB-1 (2).
mamGFDC.
The putative mamGFDC operon is located 9.2 kb upstream of the mamAB cluster and extends over 2.1 kb. Two additional colinear ORFs preceding mamDC were identified and designated mamF and mamG. mamG appears to encode an equivalent of the Mms5 protein of Magnetospirillum sp. strain AMB-1 (2), for which the gene has not been identified so far. In addition, MamG shares partial homology with MamD of MSR-1 as well as Mms6 and Mms7 of Magnetospirillum sp. strain AMB-1, which appears to be restricted to repetitive stretches of hydrophilic amino acid residues with a speculated function in iron binding (2).
Remarkably, the regions flanking and between the clusters encoding magnetosome membrane proteins contain a considerable number of ORFs which have close homologues in the genomes of M. magnetotacticum MS-1 and strain MC-1 but yield no database hits to nonmagnetic organisms and hence can be considered specific to magnetotactic bacteria. One noticeable example of a gene with functional assignment outside of magnetotactic bacteria is idiA (ORF9), which is located between the mamDC and mamAB clusters. The IdiA (iron deficiency induced) protein is an iron-binding, thylakoid-associated protein involved in iron metabolism in Synechocystis spp. and other cyanobacteria (50). A further remarkable feature of the region sequenced is the presence of six ORFs with homology to mobile DNA elements such as insertion sequence elements and integrases. Preliminary sequence analysis of the region adjacent to the 35-kb fragment revealed further numerous representatives of these gene families (data not shown). In total, these genes represent 14% of the total sequence shown and include members of at least two different major transposase families (transposase 11 and transposase 25).
Identification of the right boundary of the deletion.
To pinpoint the excision site, a set of primer pairs were designed from the sequence. Primers SS16_f and SS17 yielded a PCR product in both strains MSR-1 and MSR-1B, while primers SS14_f and SS15_r failed to amplify a fragment from the mutant. We therefore concluded that the right boundary of the deletion is located between 31.4 kb and 33 kb of the bacterial artificial chromosome sequence (Fig. 6). Similar experiments revealed that the left boundary is not covered by the 68-kb bacterial artificial chromosome clone (data not shown).
DISCUSSION
The spontaneous loss of the ability to form magnetosomes has been observed occasionally in several different strains of magnetotactic bacteria in our laboratory and by others (8; B. L. Dubbels, A. A. Dispirito, J. D. Morton, J. D. Semrau, and D. A. Bazylinski, Abstr. 101st Annu. Meet. Am. Soc. Microbiol., 2001, p. 463-464). However, neither the frequency at which the loss of magnetism occurred nor the genotype of those mutations had been determined. In this study, we failed to reproduce the isolation of spontaneous nonmagnetic mutants from exponentially growing serial subcultures. However, after incubation and aging of cells under stationary-phase conditions, the frequency of nonmagnetic mutants in the cultures increased from virtually undetectable levels (<10−5) up to 0.5% of the population. Consistently, long-term stock cultures of several Magnetospirillum strains were occasionally found to be entirely taken over by nonmagnetic mutants after only a few serial transfers.
As the deletion mutant MSR-1B was impaired in growth in our experiments, the wild type would be expected to outcompete mutant cells in a mixed population. However, the conditions associated with the storage of cultures apparently not only favor the induction of mutations, but also select for growth of mutants. Although other mutants have not yet been characterized in as much detail as MSR-1B, preliminary analysis revealed their heterogeneous nature (unpublished data). An increase in genetic variability associated with the conditions of aging and stationary phase has been reported repeatedly for different bacteria. For instance, the spontaneous loss of virulence has been described for long-term stab and aging liquid cultures of Xanthomonas oryzae, a phenomenon that was referred to as stationary-phase variation (32).
The starvation conditions encountered during stationary-phase incubation were speculated to permit a transient increase in the mutation rate due to a variety of factors (13). In several cases, increased genetic variation could be clearly linked to the accumulation of insertion sequence element transpositions in stationary-phase cultures (26, 32). Insertion sequence elements have been associated with chromosome rearrangements and are often involved in assembling arrays of genes with so-called accessory functions (21, 22). The abundant occurrence of insertion sequence elements at a high density is a striking feature of the mam region. It will be interesting to see if the large number of mobile genetic elements accounts for the particularly high genetic plasticity and instability of this region. In fact, in MC-1 and M. magnetotacticum, a similar but not identical arrangement of the mam clusters can be inferred from the available data (http://www.jgi.doe.gov/tempweb/JGI_microbial/html/index.html), e.g., several sequences appear to be shuffled or exchanged between different sites, which implies that these regions might have undergone several rearrangements.
In strain MSR-1B, the large deletion of approximately 80 kb comprises all known mam genes. Sequence analysis revealed that the mamAB and mamDC loci in M. gryphiswaldense are separated by less than 10 kb and are in close proximity to a further putative operon encoding magnetosome membrane-associated proteins. Unlike these genes, mms16, encoding a magnetosome membrane-associated protein in Magnetospirillum strain AMB-1 (27), and the bacterioferritin genes bfr1 and bfr2, which were speculated to be involved in magnetite biomineralization (6), were unaffected by the deletion in MSR-1B. Data from Bertani and coworkers suggested a putative clustering of genes involved in magnetosome biomineralization in the genome of M. magnetotacticum MS-1 (7). However, our findings do not suggest that these genes are intimately linked to the mamAB and mamDC clusters in M. gryphiswaldense.
In addition to the inability to form magnetosomes, the mutant strain MSR-1B shows impaired growth depending on the extracellular iron concentration and displays reduced motility. The ultrastructural analysis (Fig. 1) revealed not only the total absence of any electron-dense structures resembling native or aberrant magnetosome crystals, but also of intracellular membrane structures presumably identical to the empty vesicular membrane structures found by Gorby et al. (14) in iron-deprived cells.
Whereas the uptake and growth kinetics of MSR-1B indicated that the mutant is still capable of energy-dependent iron uptake and growth in iron-replete conditions, the uptake and accumulation of iron were substantially reduced. Thus, it is likely that different uptake systems are involved to supply iron for growth and magnetosome formation. The deletion of mamB and mamM in the uptake-impaired mutant MSR-1B would be consistent with the presumptive function of these CDF transporters in magnetosome-directed iron transport (15). Interestingly, growth of MSR-1B appeared to be sensitive to elevated concentrations of iron. This might indicate a contribution of magnetite formation to iron homeostasis and detoxification of potentially harmful high intracellular levels, as, for instance, in the iron storage proteins ferritin and bacterioferritin (1). Apart from the functions discussed above, we failed to detect any further phenotypic traits associated with the deletion. This indicates that the lost genes are not essential for growth under laboratory conditions but involve multiple functions essential to magnetosome biomineralization.
It might be anticipated that a deletion equivalent to about 2.0% of the genome would result in a noticeable number of bands missing from the one-dimensional proteome of strain MSR-1B. The absence of a single periplasmic protein band could be linked to the nonmagnetic phenotype by the analysis of the one-dimensional protein profile of a spontaneous mutant derived from the magnetotactic bacterial strain MV-1 (B. L. Dubbels, A. A. Dispirito, J. D. Morton, J. D. Semrau, and D. A. Bazylinski, Abstr. 101st Annu. Meet. Am. Soc. Microbiol., 2001, p. 463-464). In our experiments, proteins affected by deletion apparently did escape detection by one-dimensional PAGE analysis and are not highly abundant cellular proteins. This was expected, at least for the known magnetosome membrane-associated proteins, as it has been estimated that all magnetosome membrane proteins constitutes less than 0.1% of the total cellular protein (15).
Recently, partial genome data for various magnetotactic bacteria have become available, and homologous genes encoding magnetosome proteins were identified in strains of Magnetospirillum and strain MC-1 (15). In this study, sequence information for the complete mamAB and mamDC regions of M. gryphiswaldense, which contain all genes encoding known magnetosome membrane proteins, is presented. The alignment of magnetosome genes is not contiguous but interrupted by genes of apparently unrelated functions, suggesting a mosaic-like structure. Interestingly, a considerable number of ORFs are located in this region, which appear to be specific to magnetotactic bacteria. Their universal but exclusive occurrence in magnetotactic bacteria as well as their colocation with the mam gene clusters suggest specific involvement in magnetosome formation. mamK of the mamAB cluster displays extensive similarity to mreB, which was previously characterized as a gene encoding an actin-like cytoskeletal protein (18, 52). Multiple homologues of mreB are present in the genomes of M. magnetotacticum MS-1, strain MC-1, and M. gryphiswaldense. As in M. gryphiswaldense, at least one homologue was found colocated with either the mamAB or mamDC cluster in the other two magnetotactic bacteria (unpublished data). It is therefore tempting to speculate that MreB may be associated with the formation of a cytoskeletal superstructure potentially involved in the organization and segregation of magnetosome chains (36).
Several of the characteristics mentioned above are strongly reminiscent of those described for genome islands in other bacteria (12). Genomic islands usually comprise large regions (10 to 500 kb in size) that are present, for instance, as pathogenicity islands in the genomes of pathogenic strains but absent from the genomes of nonpathogenic members of the same or related species. They often encode determinants required for pathogenicity but are nonessential for growth outside the host and are capable of horizontal gene transfer (9, 16). Recently, it has become apparent that genetic structures similar to pathogenicity islands are also parts of the genome of many nonpathogenic bacteria, where they carry gene functions required for certain metabolic activities, such as symbiosis (47, 48). The flanking regions are usually characterized by the presence of mobile genetic elements such as insertion sequences (21), which play a role in the mobilization and rearrangement of the island. In addition, many genomic islands have the tendency to delete spontaneously (16).
In conclusion, several of the common features of genomic islands are apparently shared by the deleted region in MSR-1B harboring the mam clusters. Thus, it can be inferred that most of the gene functions specifically required for magnetite synthesis are organized within a large genomic supercluster, which might be tentatively termed a magnetosome island, and putatively have been distributed by horizontal gene transfer. A growing number of bacterial isolates from different environments can be clearly identified as Magnetospirillum species based on 16S rRNA sequence analysis and morphological and physiological characteristics but lack the ability to form magnetosomes (10, 45). It will be interesting to see if these nonmagnetic magnetospirilla are distinguished from their magnetic relatives by the absence of the magnetosome island.
Acknowledgments
This study was supported by the BMBF and MPG.
We thank Thierry Lombardot (bioinformatics group, Max-Planck-Institut Bremen) for kind introduction to GenDB and Mario Sontag and Janina Thiel (sequencing group, Max-Planck-Institut Berlin) for help in finishing the DNA sequence and technical assistance. Continuing support by Friedrich Widdel (Dept. of Microbiology, Max-Planck-Institut Bremen) is gratefully acknowledged.
REFERENCES
- 1.Andrews, S. C. 1998. Iron storage in bacteria. Adv. Microb. Physiol. 40:281-351. [DOI] [PubMed] [Google Scholar]
- 2.Arakaki, A., J. Webb, and T. Matsunaga. 2003. A novel protein tightly bound to bacterial magnetic particles in Magnetospirillum magneticum strain AMB-1. J. Biol. Chem. 278:8745-8750. [DOI] [PubMed] [Google Scholar]
- 3.Balkwill, D., D. Maratea, and R. Blakemore. 1980. Ultrastructure of a magnetotactic spirillum. J. Bacteriol. 141:1399-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bazylinski, D. 1995. Structure and function of the bacterial magnetosome. ASM News. 61:337-343. [Google Scholar]
- 5.Bazylinski, D. A., A. Garratt-Reed, and R. B. Frankel. 1994. Electron-microscopic studies of magnetosomes in magnetotactic bacteria. Microsc. Res. Technol. 27:389-401. [DOI] [PubMed] [Google Scholar]
- 6.Bertani, L. E., J. S. Huangl, B. A. Weir, and J. L. Kirschvink. 1997.Evidence for two types of subunits in the bacterioferritin of Magnetospirillum magnetotacticum. Gene 201:31-36, [DOI] [PubMed] [Google Scholar]
- 7.Bertani, L. E., J. Weko, K. V. Phillips, R. F. Gray, and J. L. Kirschvink. 2001. Physical and genetic characterization of the genome of Magnetospirillum magnetotacticum, strain MS-1. Gene 264:257-263. [DOI] [PubMed] [Google Scholar]
- 8.Blakemore, R., D. Maratea, and R. S. Wolfe. 1979. Isolation and pure culture of a freshwater magnetic spirillum in chemically defined medium. J. Bacteriol. 140:720-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carniel, E. 2001. The Yersinia high-pathogenicity island: an iron-uptake island. Microbes Infect. 3:561-569. [DOI] [PubMed] [Google Scholar]
- 10.Coates, J. D., U. Michaelidou, R. A. Bruce, S. M. O'Connor, J. N. Crespi, and L. A. Achenbach. 1999. Ubiquity and diversity of dissimilatory (per)chlorate-reducing bacteria. Appl. Environ. Microbiol. 65:5234-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces with phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 12.Finan, T. 2002. Evolving insights: symbiosis islands and horizontal gene transfer. J. Bacteriol. 184:2855-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finkel, S. E., and R. Kolter. 1999. Evolution of microbial diversity during prolonged starvation. Proc. Natl. Acad. Sci. USA. 96:4023-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorby, Y. A., T. J. Beveridge, and R. P. Blakemore. 1988. Characterization of the bacterial magnetosome membrane. J. Bacteriol. 170:834-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grünberg, K., C. Wawer, B. M. Tebo, and D. Schüler. 2001. A large gene cluster encoding several magnetosome proteins is conserved in different species of magnetotactic bacteria. Appl. Environ. Microbiol. 67:4573-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 17.Heyen, U., and D. Schüler. 2003. Growth and magnetosome formation by microaerophilic Magnetospirillum strains in an oxygen-controlled fermentor. Appl. Microbiol. Biotechnol. 61:536-544. [DOI] [PubMed] [Google Scholar]
- 18.Jones, L. J., R. Carballido-Lopez, and J. Errington. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913-922. [DOI] [PubMed] [Google Scholar]
- 19.Kirschvink, J. L., and J. W. Hagadorn. 2000. A grand unified theory of biomineralization, p. 139-149. In E. Baeuerlein (ed.), Biomineralization. Wiley-VCH, Weinheim, Germany.
- 20.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 21.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahillon, J., C. Leonard, and M. Chandler. 1999. insertion sequence elements as constituents of bacterial genomes. Res. Microbiol. 150:675-687. [DOI] [PubMed] [Google Scholar]
- 23.Marmur, J. 1961. A procedure for the isolation of deoxiribonucleic acid from microorganisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 24.Matsunaga, T., and H. Takeyama. 1998. Biomagnetic nanoparticle formation and application. Supramol. Sci. 5:391-394. [Google Scholar]
- 25.Meyer, F., A. Goesmann, A. C. McHardy, D. Bartels, T. Bekel, J. Clausen, J. Kalinowski, B. Linke, O. Rupp, R. Giegerich, and A. Pühler. 2003. GenDB-an open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 31:2187-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naas, T., M. Blot, W. M. Fitch, and W. Arber. 1994. Insertion sequence-related genetic variation in resting Escherichia coli K-12. Genetics 136:721-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okamura, Y., H. Takeyama, and T. Matsunaga. 2001. A magnetosome-specific GTPase from the magnetic bacterium Magnetospirillum magneticum AMB-1. J. Biol. Chem. 276:48183-48188. [DOI] [PubMed] [Google Scholar]
- 28.Okuda, Y., K. Denda, and Y. Fukumori. 1996. Cloning and sequencing of a gene encoding a new member of the tetratricopeptide protein family from magnetosomes of Magnetospirillum magnetotacticum. Gene 171:99-102. [DOI] [PubMed] [Google Scholar]
- 29.Pallen, M. J., and B. W. Wren. 1997. The HtrA family of serine proteases. Mol. Microbiol. 26:209-221. [DOI] [PubMed] [Google Scholar]
- 30.Paulsen, I. T., and M. H. Saier. 1997. A novel family of ubiquitous heavy metal ion transport proteins. J. Membr. Biol. 156:99-103. [DOI] [PubMed] [Google Scholar]
- 31.Radelof, U., S. Hennig, P. Seranski, M. Steinfath, J. Ramser, R. Reinhardt, A. Poustka, F. Francis, and H. Lehrach. 1998. Preselection of shotgun clones by oligonucleotide fingerprinting: an efficient and high throughput strategy to reduce redundancy in large-scale sequencing projects. Nucleic Acids Res. 26:5358-5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rajeshwari, R., and R. V. Sonti. 2000. Stationary-phase variation due to transposition of novel insertion elements in Xanthomonas oryzae pv. oryzae. J. Bacteriol. 182:4797-4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Schleifer, K., D. Schüler, S. Spring, M. Weizenegger, R. Amann, W. Ludwig, and M. Köhler. 1991. The genus Magnetospirillum gen. nov., description of Magnetospirillum gryphiswaldense sp. nov. and transfer of Aquaspirillum magnetotacticum to Magnetospirillum magnetotacticum comb. nov. Syst. Appl. Microbiol. 14:379-385. [Google Scholar]
- 35.Schüler, D. 2002. The biomineralization of magnetosomes in Magnetospirillum gryphiswaldense. Int. Microbiol. 5:209-214. [DOI] [PubMed] [Google Scholar]
- 36.Schüler, D. 2000. Characterization of the magnetosome membrane in Magnetospirillum gryphiswaldense, p. 109-118. In E. Baeuerlein (ed.), Biomineralization. Wiley-VCH, Weinheim, Germany.
- 37.Schüler, D., and D. Baeuerlein. 1997. Iron uptake and magnetite crystal formation of the magnetic bacterium Magnetospirillum gryphiswaldense, p. 159-185. In G. Winkelmann and C. J. Carrano (ed.), Transition metals in microbial metabolism. Harwood Academic Publishers, Amsterdam, The Netherlands.
- 38.Schüler, D., and E. Baeuerlein. 1998. Dynamics of iron uptake and Fe3O4 biomineralization during aerobic and microaerobic growth of Magnetospirillum gryphiswaldense. J. Bacteriol. 180(1):159-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schüler, D., and E. Baeuerlein. 1997. Iron transport and magnetite crystal formation of the magnetic bacterium Magnetospirillum gryphiswaldense. J. Phys. IV 7:647-650. [Google Scholar]
- 40.Schüler, D., and E. Baeuerlein. 1996. Iron-limited growth and kinetics of iron uptake in Magnetospirillum gryphiswaldense. Arch. Microbiol. 166:301-307. [DOI] [PubMed] [Google Scholar]
- 41.Schüler, D., and R. B. Frankel. 1999. Bacterial magnetosomes: Microbiology, biomineralization and biotechnological applications. Appl. Microbiol. Biotechnol. 52:464-473. [DOI] [PubMed] [Google Scholar]
- 42.Schüler, D., and M. Köhler. 1992. The isolation of a new magnetic spirillum. Zentralbl. Mikrobiol. 147:150-151. [Google Scholar]
- 43.Schüler, D., R. Uhl, and E. Baeuerlein. 1995. A simple light-scattering method to assay magnetism in Magnetospirillum gryphiswaldense. FEMS Microbiol. Lett. 132:139-145. [Google Scholar]
- 44.Schultheiss, D., and D. Schüler. 2003. Development of a genetic system for Magnetospirillum gryphiswaldense. Arch. Microbiol. 179:89-94. [DOI] [PubMed] [Google Scholar]
- 45.Shinoda, Y., Y. Sakai, M. Ue, A. Hiraishi, and N. Kato. 2000. Isolation and characterization of a new denitrifying spirillum capable of anaerobic degradation of phenol. Appl. Environ. Microbiol. 66:1286-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Staden, R., K. F. Beal, and J. K. Bonfield. 2000. The Staden package, 1998. Methods Mol. Biol. 132:115-130. [DOI] [PubMed] [Google Scholar]
- 47.Sullivan, J. T., and C. W. Ronson. 1998. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc. Natl. Acad. Sci. USA 95:5145-5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sullivan, J. T., J. R. Trzebiatowski, R. W. Cruickshank, J. Gouzy, S. D. Brown, R. M. Elliot, D. J. Fleetwood, N. G. McCallum, U. Rossbach, G. S. Stuart, J. E. Weaver, R. J. Webby, F. J. De Bruijn, and C. W. Ronson. 2002. Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J. Bacteriol. 184:3086-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomas-Keprta, K. L., S. J. Clemett, D. A. Bazylinski, J. L. Kirschvink, D. S. McKay, S. J. Wentworth, H. Vali, E. K. Gibson Jr., Jr., and C. S. Romanek. 2002. Magnetofossils from ancient Mars: a robust biosignature in the martian meteorite ALH84001. Appl. Environ. Microbiol. 68:3663-3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tölle, J., K. P. Michel, J. Kruip, U. Kahmann, A. Preisfeld, and E. K. Pistorius. 2002. Localization and function of the IdiA homologue Slr1295 in the cyanobacterium Synechocystis sp. strain PCC 6803. Microbiology 148:3293-3305. [DOI] [PubMed] [Google Scholar]
- 51.Valentine, R. C., B. M. Shapiro, and E. R. Stadtman. 1968. Regulation of glutamine synthetase. XII. Electron microscopy of the enzyme from Escherichia coli. Biochemistry 7:2143-2152. [DOI] [PubMed] [Google Scholar]
- 52.van den Ent, F., L. A. Amos, and J. Löwe. 2001. Prokaryotic origin of the actin cytoskeleton. Nature 413:39-44. [DOI] [PubMed] [Google Scholar]
- 53.Walther-Mauruschat, A., M. Aragno, F. Mayer, and H. G. Schlegel. 1977. Micromorphology of Gram-negative hydrogen bacteria. II. Cell envelope, membranes, and cytoplasmic inclusions. Arch. Microbiol. 114:101-110. [DOI] [PubMed] [Google Scholar]