Gram-negative bacteria have, in general, much higher intrinsic levels of resistance to various antibiotics, antiseptics, dyes, and detergents than do gram-positive bacteria. This is, in part, due to the effectiveness of the outer membrane as a barrier. The porin channels exclude large compounds (for example, vancomycin) and drastically slow down the influx of most antibiotics, which are usually much larger than common nutrients. Most antibiotics and chemotherapeutic agents that act on targets in the cytosol must cross the inner, cytoplasmic membrane, usually by spontaneous diffusion, and this necessitates their being at least moderately lipophilic. These compounds can in principle diffuse across the lipid bi-layer domain of the outer membrane. Transmembrane diffusion rates across this domain, however, are about two orders of magnitude slower than through the conventional phospholipid bi-layers (29), because the outer leaflet, composed exclusively of lipopolysaccharides (11), acts as an effective barrier.
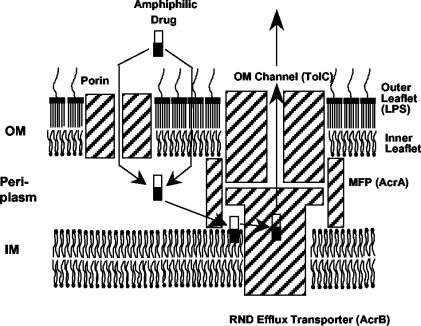
The outer membrane barrier alone, however, only slows down the influx of most of the noxious agents, and the gram-negative bacteria need the additional contribution of multidrug efflux pumps in order to achieve their characteristic levels of intrinsic resistance (24). The pumps belonging to the resistance-nodulation-division (RND) family are especially effective in generating resistance, as they form a tripartite complex together with the periplasmic proteins belonging to the membrane-fusion-protein (MFP) family and the outer membrane channels (Fig. 1), so that drugs are pumped out directly into the external medium. The RND pumps often have a very wide substrate specificity (23). An extreme case is the AcrB pump of Escherichia coli, which by forming a complex with an MFP, AcrA, and an outer membrane channel TolC pumps out tetracycline, chloramphenicol, β-lactams, novobiocin, fusidic acid, nalidixic acid, and fluoroquinolones among antibiotics and chemotherapeutic agents, SDS, Triton X-100, and bile salts among detergents, various cationic dyes and disinfectants, and even solvents (23, 40, 42). Examination of the structures of these substrates and the finding that carbenicillin and ceftriaxone, which cannot penetrate into the cytoplasm, were good substrates for an AcrB homolog, MexB (15), suggested in 1994 that a major pathway for the capture of substrates consists of the partial partitioning of the substrates into the outer leaflet of the plasma membrane, followed by the lateral entry of substrates from the lipid bi-layer into AcrB and its homologs (15). This model, shown in Fig. 1, has since been supported by several additional pieces of evidence. For example, Zgurskaya and Nikaido (46) showed that purified and reconstituted AcrB catalyzed the export of fluorescence-labeled phospholipids from within the bi-layer. Nikaido et al. (25) showed that among β-lactams only those with lipophilic side chains were efficiently pumped out by AcrB. On the other hand, the efflux of aminoglycosides, which are completely hydrophilic molecules, by RND transporters MexY (1) and AcrD (32) could not easily be explained by this model.
FIG. 1.
Lateral capture model of RND family pump function. This model is slightly modified from that of the original publication (15) and shows specific components of the AcrAB-TolC complex of E. coli. Abbreviations: OM, outer membrane; IM, inner membrane. The substrates, which are usually amphiphilic (the lipophilic portions of the substrates are drawn as black rectangles), are thought to partition spontaneously into the outer leaflet of IM and then are hypothesized to be captured laterally by the AcrB pump. The outer leaflet of OM is composed of lipopolysaccharides (LPS), which produce a strong diffusion barrier, presumably because of their less fluid interior.
STRUCTURES OF THE COMPONENTS OF THE TRIPARTITE EFFLUX COMPLEX
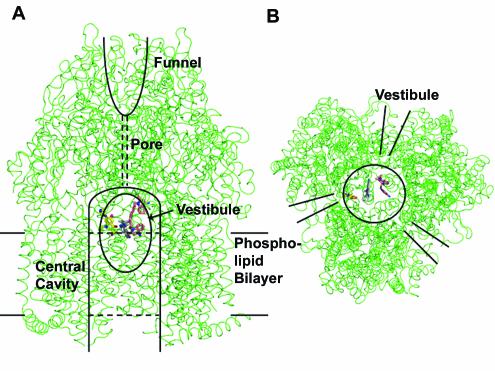
Among the components of the AcrAB-TolC complex, the crystallographic structure of TolC was elucidated by Koronakis et al. in 2000 (13). This trimeric protein not only spans the thickness of the outer membrane but also forms a 100-Å-long tunnel that covers at least half the depth of the periplasmic space. The periplasmic linker protein, AcrA, has been shown to be an elongated protein of 100 to 200 Å in length (45), and its elongated shape was confirmed by electron crystallography (2). However, its high resolution structure is not yet available. Last year, Murakami et al. (20) reported the crystallographic structure of AcrB. This was the first report of the crystallographic structure of a pump driven by the proton-motive force. The elucidation of the structure of this gigantic trimeric protein, each subunit of which contains more than 1,000 amino acid residues, was a remarkable achievement. Pumps of the RND superfamily, including AcrB, contain 12 transmembrane helices (per monomer) and characteristically have two large periplasmic loops (each with more than 300 amino acid residues) between transmembrane helices 1 and 2 and between 7 and 8 (39). The crystal structure immediately gave us several clues to the function of the efflux complex (Fig. 2). First, the periplasmic domain is huge, as was predicted earlier from the transmembrane topology data just mentioned. Second, the top of the trimeric periplasmic domain forms a funnel, and the edge of the funnel has dimensions that would fit with the tip of the periplasmic helical barrel of TolC, suggesting that the top of the AcrB periplasmic domain would directly contact TolC. Third, the funnel is connected to the very large central cavity between the transmembrane domains of the three protomers, a cavity with a diameter of 35 Å. In contrast, the connection, or pore, is very narrow, or essentially closed. Fourth, just outside the external surface of the membrane bi-layer there is a conspicuous opening between protomers, and this opening, called a vestibule, leads to the central cavity. Murakami et al. (20) pointed out that these vestibules are likely to be utilized for the capture of substrates from the outer leaflet of the lipid bi-layer, a finding consistent with the earlier model mentioned above. The central cavity was so large that it was assumed to be filled with lipid bi-layers. However, it was not possible to locate the substrates within the AcrB structure.
FIG. 2.
3-D Structure of the trimeric AcrB and the binding of three substrates, ethidium, dequalinium, and ciprofloxacin. (A) A side view. Two of the three protomers are in front, and the remaining protomer is in the back. Part of the central cavity is visible through the vestibule (shown as an ellipse) that exists between the two protomers in front. Other features of the transporter, such as the central cavity, funnel, and pore, are described in the text. In the real complex, three drug molecules of the same structure were found in the AcrB trimer. In order to save space, a composite figure was generated from the structures bound with ethidium (gray), dequalinium (pink), and ciprofloxacin (yellow) (Protein Data Bank [PDB] files 1OY9, 1OYD, and 1OYF). We do not mean to imply that the binding cavity binds three different drug molecules at once (although this could occur). (B) A bottom view. The bound drug molecules can be seen within the central cavity. The positions of the vestibules are indicated. This figure was drawn with the program PyMOL (W. L. Delano, The PyMOL Molecular Graphics System; Delano Scientific, San Carlos, Calif.; www.pymol.org).
BINDING OF SUBSTRATES TO THE CENTRAL CAVITY
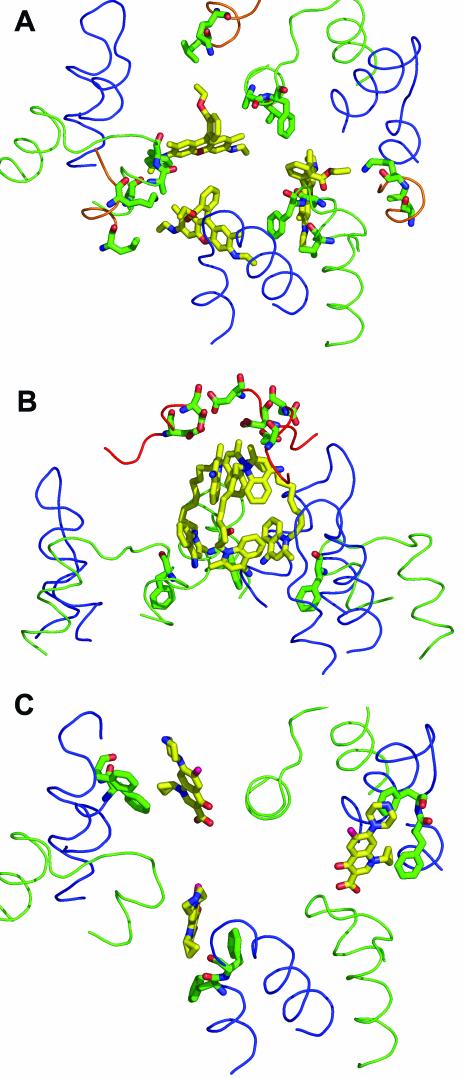
We have crystallized the native AcrB (without the addition of any tags) in the presence of various ligands and solved their structures by X-ray crystallography (44). All the ligands were used at a concentration of 50 μM, which is not excessive when we consider that half-inhibition of the pumping activity of reconstituted AcrB requires about 100 μM of conventional antibiotics (46). As far as we are aware, these are the first known liganded structures of any active transporter, if we exclude those of the outer membrane channels, such as FecA (7), FhuA (17), and LamB (34). All four ligands examined—rhodamine 6G, ethidium, dequalinium, and ciprofloxacin—bound to the periphery of the central cavity. In every case, the trimeric AcrB bound three drug molecules. One surprising feature of the binding was that both rhodamine 6G and ethidium, in spite of their cationic charge, did not bind to the ceiling of the cavity, where six negatively charged amino acid residues (Asp99 and Asp101 from each subunit) are located, but became bound at a location far away (by about 20 Å) from the ceiling. With dequalinium, which contains two cationic quinolinium moieties linked together with a decane chain, the top quinolinium bound to the ceiling but the bottom one was far away from any negatively charged residue (Fig. 3). These points will be discussed below.
FIG. 3.
Details of the substrate-binding interactions in AcrB. The ligand is shown as a yellow stick model, and the interacting amino acid side chains (within 6 Å of the ligand molecule) are shown as green stick models. The loop between transmembrane helices 3 and 4 (residues 380 to 401) is shown as a green line, and the loop between transmembrane helices 5 and 6 (residues 452 to 475) is shown as a blue line. (A) Binding of rhodamine 6G (from PDB file 1OY8). The ligand interacts mainly with Phe386, Val382, and Ala385. The additional participation of Leu25 and Lys29 in the loop between the transmembrane helix 1 and the periplasmic domain (residues 24 to 31) (shown as an orange line) is also seen. (B) Binding of dequalinium (PDB file 1OYD). The quinolinium moiety at the top interacts with Asp99 and Asp101 from the periplasmic domain. There are no amino acid residues within the 6-Å radius of the lower quinolinium moiety. Phe386, which is 6.3 Å away, is shown. (C) Binding of ciprofloxacin (PDB file 1OYF). The ligand interacts mainly with Phe458 and Phe459, as shown. For simplicity, Leu25, Lys29, and Ala385, which are also nearby (44), are not shown here. This figure was drawn with the program PyMOL.
DETAILS OF BINDING INTERACTIONS
When amino acid residues, any atoms of which are within a 6-Å distance from any atoms of the ligand, were identified, we found mostly amino acids with lipophilic side chains (Fig. 3). Beside rhodamine 6G, Phe386, Val382, and Leu25 were found in this range. In addition, Lys29 pointed its ɛ-amino group toward the ester oxygen atom of the ligand (44) (Fig. 3A). Ethidium bound nearby, and in this case Phe386 was essentially the only residue whose side chain seemed to interact with the ligand; although some atoms of the neighboring residues, Ala385 and Gly387, were within the 6-Å distance, the Cα and Cb atoms were facing away from the ligand (Fig. 4B). With dequalinium, the top quinolinium moiety was close to Asp99 and Asp101, indicating electrostatic interactions, but the bottom quinolinium moiety was close to only Phe386 (Fig. 3B). Finally, for ciprofloxacin, Phe458 and Phe459 appeared to sandwich the cyclopropyl moiety of the drug (Fig. 3C) and Lys29 and Leu25 were fairly close to the drug, but no acidic residues were found nearby.
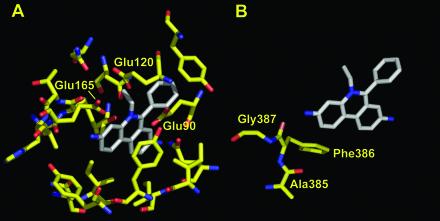
FIG. 4.
Details of the binding of ethidium to the QacR regulatory protein (A) and AcrB (B). Residues within a 6-Å distance from the bound ethidium are shown, based on PDB file 1JTY for QacR and PDB file 1OY9 for AcrB. This figure was drawn with the program PyMOL.
COMPARISON WITH THE BINDING SITES OF REGULATORY PROTEINS
Earlier, the binding sites of the regulatory proteins for multidrug efflux systems were investigated because these proteins also bind diverse ligands and because, being soluble proteins, they are easier to crystallize. The first study on the ligand-binding domain of the regulator BmrR of Bacillus subtilis showed that the binding of tetraphenylphosphonium occurred mainly via hydrophobic interactions, together with a crucial electrostatic interaction from a glutamate residue (47). This study also showed the flexibility of the binding pocket. Indeed, the ligand binding could not occur unless a helix-to-coil transition pushed a segment of the protein away to widen the binding site. Although the protein could not be crystallized with another ligand, it was possible to see how rhodamine 6G could also be accommodated at this site. The second study, by using the QacR repressor of Staphylococcus aureus (36), elucidated the X-ray crystallographic structure of the protein bound to six different ligands, rhodamine 6G, ethidium, dequalinium, crystal violet, malachite green, and berberine. This study definitely showed that different ligands bound to different parts of a rather large binding pocket. Again, the binding site was expanded by the expulsion of tyrosine side chains, and the binding was mediated mostly by hydrophobic interactions, with key electrostatic contributions from three glutamic acid residues (Fig. 4A). The principles involved in these binding interactions have been reviewed (22, 35). Neyfakh (22) argues that the tight binding of water-soluble substrates to the binding sites of enzymes is necessitated by the energetic burden of removing these substrates from the extensively hydrogen-bonded environment in aqueous solutions. For lipophilic ligands, which are the inducers and substrates of multidrug efflux pumps, there is little of this burden, and therefore they can bind to the proteins through mostly loose, hydrophobic interactions. One example cited by Neyfakh (22) is remarkable; the same aromatic ligand binds to the binding pocket of porcine-odorant-binding protein in more than one orientation (41), showing the looseness and flexibility of the binding interaction. Additional, similar examples exist. The binding of lipophilic substrates to the naphthalene 1,2-dioxygenase involves a large (6 × 8 × 10-Å) binding cavity that can accommodate a few hundred different substrates, including both planar (e.g., naphthalene) and nonplanar (e.g., biphenyl, whose phenyl rings are on perpendicular planes) substrates (3). A large, loose-binding pocket presumably also explains the observation that an enzyme catalyzing a similar reaction, toluene 2-monooxygenase, acts not only on aromatic compounds but also on small, chlorinated aliphatic compounds such as trichloroethylene (21).
Our results are consistent with these principles discovered for soluble proteins. Thus, the binding cavity is large, and the interaction between the ligands and the protein appears to be mostly hydrophobic (Fig. 3) (44). However, the details are strikingly different. (i) The ligand-binding sites of the regulators, such as QacR, have been called large in comparison with the tight-fitting substrate-binding sites of enzymes. However, they are small and narrow in comparison with the vast central cavity of the AcrB trimer, which has a diameter of 35 Å and whose upper portion alone has a volume of 5,000 Å3. With the regulators, expansion of the pocket was necessary in order to create enough space to accommodate the ligands (36). No such expansion is necessary with AcrB. (ii) In the regulators, the ligand interacts with many residues of the protein, although the interaction seems loose in comparison with the usual substrate-binding interactions that often involve dozens of hydrogen bonds. Thus, QacR, for example, contains more than a dozen amino acid residues within 6 Å of the bound ethidium molecule (Fig. 4A). In contrast, with AcrB, the only residues of which any atom comes within 6 Å of any atom of the bound ethidium are Ala385, Phe386, and Gly387 (Fig. 4B). The other sides of the ethidium molecule are completely open. Nevertheless, Phe386 appears to play a crucial role in the export (and presumably the binding) of ethidium, rhodamine 6G, and dequalinium, because conversion of this residue to alanine by site-directed mutagenesis nearly totally abolishes resistance to these compounds (J. R. Aires and H. Nikaido, unpublished data). (iii) Most importantly, with QacR there are three acidic residues, Glu90, Glu120, and Glu165, which undoubtedly interact electrostatically with the cationic dye ethidium (Fig. 4A). In striking contrast, there is no acidic residue within 6 Å (Fig. 4B), or even within 10 Å, of ethidium bound to AcrB trimer. Farther away, the closest carboxylate moiety is that of Asp99, which is about 12 Å away. The situation is similar with rhodamine 6G and ciprofloxacin (44). Although one of the two cationic, quinolinium moieties of dequalinium is close to the ceiling of the cavity where Asp99 and Asp101 are located (the distances of the carboxylate oxygens from dequalinium are 3.5 and 5.3 Å, respectively), the other quinolinium moiety is very far away from any acidic residue (Fig. 3B). How can we explain these unexpected observations?
PROPOSAL FOR A COMPOSITE BINDING SITE
One possible explanation for the long distance between the negatively charged ceiling of the cavity and the positively charged ligands is that, because three ligand molecules bind to the AcrB trimer simultaneously, the ligand molecules are already in contact with each other at the positions shown in Fig. 2 and their closer approach to the ceiling of the cavity is prevented because the cavity becomes narrower at the top. However, this idea can be ruled out when we examine the binding of ciprofloxacin, where each drug molecule is far away from other bound drug molecules (Fig. 3C). The carboxylic oxygens, the carboxylic carbon, C-3 and C-4 of the quinolone ring, and the carbonyl oxygen at position 4 are all in a single perfect plane. This is against the idea that the carboxylic acid is deprotonated, because if so, the repulsion between the carbonyl oxygen at C-4 and these resonating, negatively charged oxygens would result in rotation around the C-3-carboxyl C bond. The planarity of the structure is convincing evidence that the carboxylic acid moiety of fluoroquinolones is an exceptionally weak acid, thanks to the stabilization of the protonated form via the formation of a six-membered, hydrogen-bonded ring system (Fig. 5) (reviewed in reference 26). In contrast, the 4-amino group of the piperazine ring is a fairly strong base and is expected to be largely protonated. Thus, ciprofloxacin should carry a positive charge, yet again it binds very far away from the ceiling of the cavity (Fig. 2). As pointed out above, there is no drug-to-drug contact and there should be no steric hindrance for the molecules to reach the negatively charged ceiling.
FIG. 5.
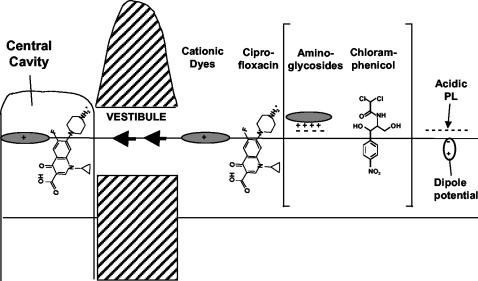
Hypothetical path of substrate capture by AcrB and related pumps. The crystal structure of AcrB trimer bound to cationic dyes (44) suggests strongly that such dyes, bound to the acidic surface of the bi-layer, travel through the vestibule and bind to the wall of the central cavity without much change in its location or orientation in relation to the bi-layer surface. A similar mechanism is likely to apply for ciprofloxacin, which may partially partition into the outer leaflet of the bi-layer before diffusing through the vestibule and binding to the cavity. For the other substrates shown in the brackets, the crystal structure of the liganded AcrB is not yet available. However, it seems likely that the polycationic aminoglycosides become adsorbed to the polyanionic surface of the bi-layer and are captured by some RND pumps. Similarly, amphiphilic molecules with dipolar lipophilic domains, such as chloramphenicol, may partition partially into the outer leaflet of the bi-layer and be captured by lateral diffusion through vestibules. PL, phospholipids.
These considerations suggest that there are likely to be other sources of negative charges that interact with these cationic ligands. When Fig. 2 is examined, it is striking that the ligands bind at the level where the head groups of the outer leaflet of phospholipid bi-layers are expected to be present. Murakami et al. (20) already suggested that the cavity is so wide that it must be filled with the lipid bi-layer. Indeed, at least the outer leaflet of this hypothetical bi-layer within the cavity seems contiguous with that of the bulk bi-layer outside, because the openings of the vestibules extend downward (Fig. 6A). It is thus most likely that the ligand binding observed in AcrB involves not only the AcrB protein but also the phospholipids within the central cavity. We propose that the ligand-binding site in AcrB is in this sense composite, containing both protein and lipid components. One of the most important implications of this concept is that it will help explain the extraordinarily wide substrate range of AcrB (and similar) pumps. They can thus bind cationic ligands easily by utilizing the head groups of acidic phospholipids. The presence of these anionic phospholipids will not hinder the binding of acidic ligands (such as penicillins) because of the lateral mobility of the lipids within the bi-layer. The system is thus extremely flexible, largely due to the participation of mobile and flexible lipids, and can thus accommodate a very wide range of substrates. It must be stated, however, that the presence of phospholipids within the cavity, although expected, has not yet been demonstrated experimentally. Furthermore, even if lipids are proven to be present in the cavity, it is an open question whether they remain associated with the AcrB protein during its purification in detergents.
FIG. 6.
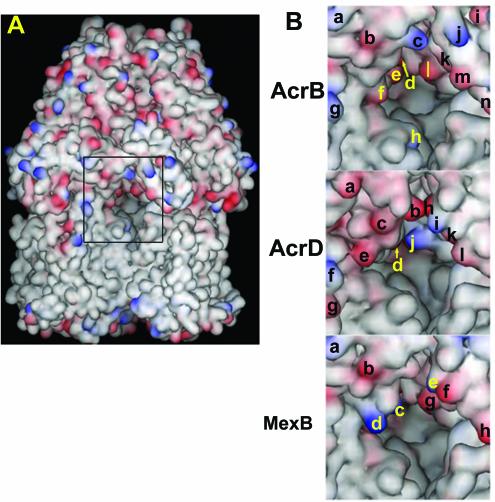
Vestibules of AcrB, AcrD, and MexB. (A) The trimeric AcrB pump is viewed from the outside, so that the two protomers are seen in front and the third protomer is mostly hidden in the back. The vestibule, at the interface of the two protomers, is indicated by the rectangle in the center. Acidic residues are colored red, and the basic residues are in blue. (B) The central region of the view shown in panel A, containing the area surrounding the vestibule, is shown in a larger magnification. Charged amino acid residues on the surface are identified by letters. For AcrB, Lys322 (a), Glu314 (b), Lys312 (c), Asp101 (d), Asp99 (e), Asp301 (f), Lys334 (g), Lys29 (h), Glu705 (i), Lys850 (j), Asp99 (k) (from the protomer on the right), Glu95 (l), Glu842 (m), and Glu839 (n) are indicated. For AcrD, Glu322 (a), Glu312 (b), Asp311 (c), Asp99 (d), Glu304 (e), Lys334 (f), Glu338 (g), Glu856 (h), Lys847 (i), Lys95 (j), Glu843 (k), and Asp840 (l) are shown. For MexB, Lys322 (a), Glu314 (b), Lys170 (c), Lys304 (d), Lys848 (e), Glu845 (f), Glu95 (g), and Asp838 (h) are indicated. The hypothetical structures of AcrD and MexB were generated by the SWISS-MODEL program (http://www.expasy.ch) by using the crystal structure of AcrB as the template. The drawing was made with the program DS Viewer Pro 5.0 (Accelrys, San Diego, Calif.).
IMPLICATIONS OF THE MODEL
In addition to explaining the substrate specificity of AcrB, our proposed model is consistent with many other properties of AcrB and its homologs. (i) This model is consistent with the substrate specificity of the AcrB system. Thus, cationic dyes become concentrated in the head group area of the phospholipid bi-layer owing to the attraction by the head group of the anionic phospholipids and to their difficulty in entering the interior of the bi-layer due to the presence of an interior-positive dipole potential, which has a large value, about 200 to 300 mV (8). These molecules will reach the central cavity via lateral diffusion through the vestibule (as predicted by Murakami et al. [20]) and will become bound to the protein wall (Fig. 5). Fluoroquinolones, which behave as amphiphilic cations, as mentioned above, will partition partly into the outer leaflet of the plasma membrane by inserting their carboxyl ends into the membrane. Again, lateral diffusion will bring them to the central cavity, where binding will occur without altering the orientation of the molecule in relation to the membrane (Fig. 5). Perhaps the presence of membrane dipole potential explains why chloramphenicol is a good substrate for many multidrug efflux pumps. The p-nitrophenyl group in chloramphenicol has an exceptionally large dipole moment (4.9 Debye unit), and this feature will guide the insertion of this part of the molecule into the external edge of the bi-layer, with the hydrophilic portion sticking out into the aqueous phase. This will facilitate the lateral diffusion and binding of the drug in the central cavity (Fig. 5). One of us has shown earlier that among β-lactams only those compounds with lipophilic side chains are good substrates of AcrB (25). Such compounds are expected to partition partly into the bi-layer with their carboxylate groups exposed in the aqueous phase, and diffusion and binding may occur in a similar manner. We predict that anionic detergents, such as SDS or bile salts, will be captured in the same manner.
(ii) Our model also explains the behavior of some homologs of AcrB. Thus, AcrD, which was initially reported as an efflux pump of very hydrophilic compounds, aminoglycosides (32), was later shown to also pump out amphiphilic compounds such as SDS, bile salts, and novobiocin (27). This substrate range appeared puzzling. However, if we consider that polycationic aminoglycosides are likely to be adsorbed to the polyanionic surface of the lipid bi-layer, which in E. coli is about one-third acidic phospholipids, then both aminoglycosides and conventional, amphiphilic substrates may be thought to reach the central cavity in the same manner (Fig. 5). Similarly, cationic peptides, which are known to be extruded by at least one RND pump (37), may become associated with the outer surface of the plasma membrane and may be captured in the same manner. Even the divalent metal efflux pumps, such as CzcA (9), might utilize the same mechanism if such metal ions become bound to the acidic head groups of phospholipids on the outer surface of the cytoplasmic membrane.
Many RND pumps extrude solvents (12, 19, 31, 40). An especially interesting observation came from the study of Ramos et al. (31), who examined the inhibition of an RND-type solvent efflux pump, TtgB, by various compounds. Toluene and m-xylene, which have modest dipole moments, inhibited the efflux reaction, presumably because they competed as substrates, but benzene, which has no dipole moment, showed no inhibition. Thus, even with solvents, effective removal by RND pumps may be aided by the presence of dipole moments and their possible preferential partition into the boundary region of the bi-layer. However, AcrB was reported to pump out simple aliphatic solvents such as hexane and heptane (40). Unfortunately, these solvents were not compared with solvents with some dipole moments.
BINDING SITE RESIDUES IN AcrB HOMOLOGS AND SUBSTRATE SPECIFICITY
When we examine the multidrug efflux pumps that are homologs of AcrB, we find that the residues at the binding site are well conserved (Table 1). This is not surprising, because the binding region, as we have seen, is constructed in a way that allows the binding of ligands of vastly different structures. One metal-pumping homolog, CusA of E. coli (10), included for comparison, showed a wider divergence from the drug efflux transporters (Table 1), an observation that reinforces our conclusion that these residues are important in the binding (and subsequent transport) of the drug substrates.
TABLE 1.
Conservation of binding-site residues among RND multidrug efflux pumps
| Pump | Residue
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Phe386 | Phe388 | Phe458 | Phe459 | Leu25 | Lys29 | Asp99 | Asp101 | |
| AcrB | Phe | Phe | Phe | Phe | Leu | Lys | Asp | Asp |
| AcrF | Phe | Tyr | Phe | Phe | Leu | Gln | Asp | Asp |
| AcrD | Phe | Tyr | Phe | Phe | Leu | Ser | Asp | Asp |
| YhiV | Val | Phe | Phe | Met | Leu | Asn | Ser | Asp |
| MexB | Phe | Phe | Phe | Phe | Leu | Ser | Asp | Asp |
| MexD | Leu | Phe | Phe | Phe | Leu | Lys | Asp | Asp |
| MexF | Leu | Phe | Phe | Phe | Ile | Gln | Asp | Asp |
| MexY | Leu | Phe | Phe | Ile | Leu | Phe | Asn | Asp |
| CusAa | Gln | Leu | Thr | Leu | Trp | Asn | Asp | Tyr |
The CusA (YdbE) protein, an E. coli RND transporter that is involved in copper ion efflux (10), was included for comparison.
If the substrate specificity is not determined entirely by the binding-site residues, which other residues contribute to the specificity? One obvious possibility is that the substrates are selected as they pass through the vestibule on their way to the binding region in the central cavity. In fact, comparison of the entrances of the vestibules in AcrD (which transports aminoglycosides) and AcrB (which does not) (Fig. 6B) shows that this area in AcrD is lined with many more acidic residues that may attract the polycationic substrates. Comparison between Pseudomonas aeruginosa MexB and AcrB (Fig. 6B) shows that an acidic residue, Asp301 (Fig. 6B, top panel, f) in AcrB is replaced by a couple of basic residues, Lys304 and Lys170 (Fig. 6B, bottom panel, d and c) in MexB. The substrate specificities of AcrAB-TolC and MexAB-OprM are not easy to compare in intact cells, where efflux competes with spontaneous influx through the outer membrane, which occurs at different rates in E. coli and P. aeruginosa. However, when MexAB-OprM is expressed in AcrAB-deficient E. coli, such comparisons can be made. In this system, MexAB-OprM is less efficient in pumping out cationic dyes (e.g., ethidium) and cationic antibiotics (oleandomycin, erythromycin, and puromycin) than AcrAB-TolC and is more efficient in extruding weakly acidic quinolones such as cinoxacin and nalidixic acid, which lack the positively charged piperazine substituent of modern fluoroquinolones (38). These results are exactly as predicted from the more basic interior of the MexB vestibule. We are currently examining the role of charged residues at the vestibule entrance in substrate selection through site-directed mutagenesis.
ROLES OF OTHER RESIDUES KNOWN TO AFFECT SUBSTRATE SPECIFICITY
Several recent studies showed that the periplasmic domain plays an important role in determining the specificity of the pump. Elkins and Nikaido (6) showed, by exchanging domains between AcrB and AcrD, that the characteristically wide substrate specificity of AcrB that forms a contrast to a more limited substrate range of AcrD is determined largely by the periplasmic domains of these pumps. This is not surprising in view of the hypothesis presented above that passage through the vestibule has a large effect on substrate selection, as most hydrophilic residues lining the wall of the vestibule come from the periplasmic domain. Tikhonova et al. (38) constructed chimeras of AcrB and its P. aeruginosa homolog, MexB, which produces a somewhat different spectrum of resistance, as mentioned above. When the hybrid containing 849 N-terminal residues of AcrB was compared with that containing 612 N-terminal residues, the former was more like AcrB in showing somewhat higher resistance to cationic agents. (However, the situation was more complicated, because the former showed a more MexB-like, higher resistance to the weakly acidic quinolones cinoxacin and nalidixic acid.) The role of the region between residues 612 and 849 of AcrB in the selection of substrates is consistent with the fact that residues 830 to 849 form the right wall of the vestibule entrance. As seen in Fig. 6B, this wall is significantly different between AcrB and MexB. However, the left wall of the vestibule of chimeric proteins still retains the more basic residues characteristic of MexB (see above), and this may explain the complex pattern of the alteration of substrate specificity found in this study. The study with the highest resolution was carried out by Mao et al. (18). They used the knowledge that the MexCD-OprJ system of P. aeruginosa cannot pump out most β-lactams and selected for point mutants of mexD that allowed the efflux of carbenicillin. Among the mutants obtained, Q34K is on the left side of the vestibule entrance, and a positive charge there is likely to enhance the entry of acidic β-lactams. The other mutations, however, are not in the immediate vicinity of the vestibule. Interestingly, some of the mutations obtained, E89K and N673K, occur in residues lining the deep external depression in the periplasmic domain, which Murakami et al. (20) postulated might accommodate the AcrA molecule. Since we now know that AcrA is essential for the pumping function of AcrB (Aires and Nikaido, unpublished), this finding suggests an intriguing possibility that the range of substrates might be altered through the interaction between the pump and the MFP.
MECHANISMS FOR OTHER EFFLUX PUMPS
It seems likely that other RND pumps of bacteria, especially those catalyzing the efflux of amphiphilic and lipophilic ligands, would use a similar mechanism of substrate capture. The RND transporter superfamily is now known to include animal and human proteins, including the Niemann-Pick type C disease protein (39). The Niemann-Pick C1 protein was suspected to be involved in the intracellular movement of cholesterol (28) and was indeed shown to transport such lipophilic ligands as fatty acids upon its expression in E. coli (5). Since cholesterol is unlikely to exist in the aqueous phase, the C1 protein most likely captures this compound from within the lipid bi-layer. It is not known if there is a preference between the outer and the inner leaflets.
A similar mechanism of ligand binding involving a very large cavity, and possibly phospholipids as well, may be operating in efflux pumps of lipophilic substrates outside the RND superfamily. Low-resolution images of human MDR pump or P-glycoprotein, an ABC family multidrug pump catalyzing the efflux of amphiphilic and lipophilic agents, show a very large cavity (33). It is interesting that among the two bacterial ABC transporters whose X-ray structures have been elucidated, MsbA, which transports lipid A, has a large opening of about 25-Å diameter in the transmembrane region (4), whereas no such opening is found for a vitamin B12 transporter, BtuCD (16).
There are kinetic data that suggest simultaneous binding of more than one ligand molecule and thus may explain the large size of the binding pocket. For example, chloramphenicol and tetraphenylphosphonium appear to bind to an E. coli major facilitator family pump, Cmr (also called MdfA) (14).
QUESTIONS FOR FUTURE RESEARCH
Even with elucidation of the liganded structure of the pump, we still have many unanswered questions. The binding of the substrates to the central cavity obviously corresponds to the first step of the transport process. This is clear from the observation that ligand binding caused only a very small change in the conformation of AcrB; there appeared to be an outward tilting of the periplasmic domain, but the extent of this tilting was small, about 1° (44). This may not be so unexpected, because the ligands do not appear to bind tightly to AcrB (46). However, ligands of outer membrane gated channels, which bind very tightly, also produce only small conformational changes (7, 17), and this characteristic may be a general feature of most transporters. Furthermore, the central pore, which is the most likely path of travel for the substrates, is nearly completely closed in both the unliganded and liganded structures of AcrB. In order for the pore to open up for the passage of ligand(s), an extensive conformational change, which is probably caused by proton flux, must occur. The precise nature of this and the subsequent steps is unclear at the moment. Another outstanding question concerns the mechanism of ligand capture from the cytosol. Reconstitution experiments with CzcA (9) and AcrD (Aires and Nikaido, unpublished) suggest that such a capture mechanism does occur with these RND transporters. How the ligands can reach the top portion of the central cavity under these conditions is not known at present.
CONCLUSIONS
The recent elucidation of the structure of the liganded AcrB transporter suggests that the ligands first bind close to the outer surface of the phospholipid bilayer by processes influenced by the lipophilicity and charge and probably by the dipole moment of the ligands. They may then diffuse laterally through the vestibules that exist between the AcrB protomers, reach the central cavity, and finally become bound to the wall of the cavity, presumably before becoming pumped out by the energy supplied by proton influx. In this preferred mode of operation of the AcrB pump, the ligands always stay outside the plasma membrane barrier, a mechanism that explains why AcrB can pump out substrates of diverse ionic characters (23, 24). The E. coli Lol system is another example of a transport system whose substrate, in this case lipoprotein, is always kept in the periplasm (43). The “membrane vacuum cleaner” mechanism also seems to operate in multidrug efflux pumps of gram-positive bacteria (30). However, in this case the substrate capture occurs from the inner leaflet of the membrane, presumably because in the absence of the outer membrane barrier the slow spontaneous flipping of the substrate from the outer to the inner leaflet acts as a barrier that would limit the number of substrate molecules reaching the pump and prevent overwhelming of the pump function.
Acknowledgments
E. W. Yu thanks Daniel E. Koshland, Jr., for constant support and encouragement. We thank Helen Zgurskaya for critical reading of the manuscript.
This work was supported by an NIH grant (AI-09644) to H.N. and by an NIH postdoctoral fellowship to E.W.Y.
REFERENCES
- 1.Aires, J. R., T. Köhler, H. Nikaido, and P. Plésiat. 1999. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 43:2624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avila-Sakar, A. J., S. Misaghi, E. M. Wilson-Kubalek, K. H. Downing, H. Zgurskaya, H. Nikaido, and E. Nogales. 2001. Lipid-layer crystallization and preliminary three-dimensional structural analysis of AcrA, the periplasmic component of a bacterial multidrug efflux pump. J. Struct. Biol. 136:81-88. [DOI] [PubMed] [Google Scholar]
- 3.Carredano, E., A. Karisson, B. Kauppi, D. Choudhury, R. E. Parales, J. V. Parales, K. Lee, D. T. Gibson, H. Eklund, and S. Ramaswamy. 2000. Substrate binding site of naphthalene 1, 2-dioxygenase: functional implications of indole binding. J. Mol. Biol. 321:621-632. [DOI] [PubMed] [Google Scholar]
- 4.Chang, G., and C. B. Roth. 2001. Structure of MsbA from E. coli: a homolog of the multidrug resistance ATP binding cassette (ABC) transporters. Science 293:1793-1800. [DOI] [PubMed] [Google Scholar]
- 5.Davies, J. P., F. W. Chen, and Y. A. Ioannou. 2000. Transmembrane molecular pump activity of Niemann-Pick C1 protein. Science 290:2295-2298. [DOI] [PubMed] [Google Scholar]
- 6.Elkins, C., and H. Nikaido. 2002. Substrate specificity of RND-type multidrug efflux pumps AcrB and AcrD of Escherichia coli is determined predominantly by two large periplasmic loops. J. Bacteriol. 184:6490-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferguson, A. D., R. Chakraborty, B. S. Smith, L. Esser, D. van der Helm, and J. Deisenhofer. 2002. Structural basis of gating by the outer membrane transporter FecA. Science 295:1715-1719. [DOI] [PubMed] [Google Scholar]
- 8.Flewelling, R. F., and W. L. Hubbell. 1986. The membrane dipole potential in a total membrane potential model. Applications to the hydrophobic ion interactions with membranes. Biophys. J. 49:541-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldberg, M., T. Pribyl, S. Juhnke, and D. H. Nies. 1999. Energetics and topology of CzcA, a cation/proton antiporter of the resistance-nodulation-cell division protein family. J. Biol. Chem. 274:36065-36070. [DOI] [PubMed] [Google Scholar]
- 10.Grass, G., and C. Rensing. 2001. Genes involved in copper homeostasis in Escherichia coli. J. Bacteriol. 183:2145-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamio, Y., and H. Nikaido. 1976. Outer membrane of Salmonella typhimurium: accessibility of phospholipid head groups to phospholipase c and cyanogen bromide activated dextran in the external medium. Biochemistry 15:2561-2570. [DOI] [PubMed] [Google Scholar]
- 12.Kieboom, J., J. J. Denis, J. A. M. de Bont, and G. J. Zylstra. 1998. Identification and molecular characterization of an efflux pump involved in Pseudomonas putida S12 solvent tolerance. J. Biol. Chem. 273:85-91. [DOI] [PubMed] [Google Scholar]
- 13.Koronakis, V., A. Sharff, E. Koronakis, B. Luisi, and C. Hughes. 2000. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405:914-919. [DOI] [PubMed] [Google Scholar]
- 14.Lewinson, O., and E. Bibi. 2001. Evidence for simultaneous binding of dissimilar substrates by the Escherichia coli multidrug transporter MdfA. Biochemistry 40:12612-12618. [DOI] [PubMed] [Google Scholar]
- 15.Li, X.-Z., D. Ma, D. M. Livermore, and H. Nikaido. 1994. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: active efflux as a contributing factor to β-lactam resistance. Antimicrob. Agents Chemother. 38:1742-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Locher, K. P., A. T. Lee, and D. C. Rees. 2002. The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science 296:1091-1098. [DOI] [PubMed] [Google Scholar]
- 17.Locher, K., B. Rees, R. Koebnik, A. Mitschler, L. Moulinier, J. P. Rosenbusch, and D. Moras. 1998. Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell 95:771-778. [DOI] [PubMed] [Google Scholar]
- 18.Mao, M., M. S. Warren, D. S. Black, T. Satou, T. Murata, T. Nishino, N. Gotoh, and O. Lomovskaya. 2002. On the mechanism of substrate specificity by resistance nodulation division (RND)-type multidrug resistance pumps: the large periplasmic loops of MexD from Pseudomonas aeruginosa are involved in substrate recognition. Mol. Microbiol. 46:889-901. [DOI] [PubMed] [Google Scholar]
- 19.Mosqueda, G., and J. L. Ramos. 2000. A set of genes encoding a second toluene efflux system in Pseudomonas putida DOT-T1E is linked to the tod genes for toluene metabolism. J. Bacteriol. 182:937-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murakami, S., R. Nakashima, E. Yamashita, and A. Yamaguchi. 2002. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419:587-593. [DOI] [PubMed] [Google Scholar]
- 21.Newman, L. M., and L. P. Wackett. 1997. Trichloroethylene oxidation by purified toluene 2-monooxygenase: products, kinetics, and turnover-dependent inactivation. J. Bacteriol. 179:90-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neyfakh, A. A. 2002. Mystery of multidrug transporters: the answer can be simple. Mol. Microbiol. 44:1123-1130. [DOI] [PubMed] [Google Scholar]
- 23.Nikaido, H. 1998. Multiple antibiotic resistance and efflux. Curr. Opin. Microbiol. 1:516-523. [DOI] [PubMed] [Google Scholar]
- 24.Nikaido, H. 2001. Preventing drug access to targets: cell surface permeability barriers and active efflux in bacteria. Semin. Cell Dev. Biol. 12:215-223. [DOI] [PubMed] [Google Scholar]
- 25.Nikaido, H., M. Basina, V. Nguyen, and E. Y. Rosenberg. 1998. Multidrug efflux pump AcrAB of Salmonella typhimurium excretes only those β-lactam antibiotics containing lipophilic side chains. J. Bacteriol. 180:4686-4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nikaido, H., and D. G. Thanassi. 1993. Penetration of lipophilic agents with multiple protonation sites into bacterial cells: tetracyclines and fluoroquinolones as examples. Antimicrob. Agents Chemother. 37:1393-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishino, K., and A. Yamaguchi. 2001. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 183:5803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pentchev, P. G., R. O. Brady, E. J. Blanchette-Mackie, M. T. Vanier, E. D. Carstea, C. C. Parker, E. Goldin, and C. F. Roff. 1994. The Niemann-Pick C lesion and its relationship to the intracellular distribution and utilization of LDL cholesterol. Biochim. Biophys. Acta. 1225:235-243. [DOI] [PubMed] [Google Scholar]
- 29.Plésiat, P., and H. Nikaido. 1992. Outer membranes of gram-negative bacteria are permeable to steroid probes. Mol. Microbiol. 6:1323-1333. [DOI] [PubMed] [Google Scholar]
- 30.Poelarends, G. J., P. Mazurkiewicz, and W. N. Konings. 2002. Multidrug transporters and antibiotic resistance in Lactococcus lactis. Biochim. Biophys. Acta. 1555:1-7. [DOI] [PubMed] [Google Scholar]
- 31.Ramos, J. L., E. Duque, P. Godoy, and A. Segura. 1998. Efflux pumps involved in toluene tolerance in Pseudomonas putida DOT-T1E. J. Bacteriol. 180:3323-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenberg, E. Y., D. Ma, and H. Nikaido. 2000. AcrD of Escherichia coli is an aminoglycoside efflux pump. J. Bacteriol. 182:1754-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenberg, M. F., R. Callaghan, R. C. Ford, and C. F. Higgins. 1997. Structure of the multidrug resistance P-glycoprotein to 2.5 nm resolution determined by electron microscopy and image analysis. J. Biol. Chem. 272:10685-10694. [DOI] [PubMed] [Google Scholar]
- 34.Schirmer, T., T. A. Keller, Y.-F. Wang, and J. P. Rosenbusch. 1995. Structural basis for sugar translocation through maltoporin channels at 3.1 Å resolution. Science 267:512-514. [DOI] [PubMed] [Google Scholar]
- 35.Schumacher, M. A., and R. G. Brennan. 2002. Structural mechanisms of multidrug recognition and regulation by bacterial multidrug transcription factors. Mol. Microbiol. 45:885-893. [DOI] [PubMed] [Google Scholar]
- 36.Schumacher, M. A., M. C. Miller, S. Grkovic, M. H. Brown, R. A. Skurray, and R. G. Brennan. 2001. Structural mechanisms of QacR induction and multidrug recognition. Science 294:2158-2163. [DOI] [PubMed] [Google Scholar]
- 37.Shafer, W. M., X. D. Qu, A. J. Waring, and R. I. Lehrer. 1998. Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux family. Proc. Natl. Acad. Sci. USA 95:1829-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tikhonova, E. B., Q. Wang, and H. I. Zgurskaya. 2002. Chimeric analysis of the multicomponent multidrug efflux transporters from gram-negative bacteria. J. Bacteriol. 184:6499-6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tseng, T. T., K. S. Gratwick, J. Kollman, D. Park, D. H. Nies, A. Goffeau, and M. H. Saier, Jr. 1999. The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and developmental proteins. J. Mol. Microbiol. Biotechnol. 1:107-125. [PubMed] [Google Scholar]
- 40.Tsukagoshi, N., and R. Aono. 2000. Entry into and release of solvents by Escherichia coli in an organic-aqueous tow-liquid-phase system and substrate specificity of the AcrAB-TolC solvent-extruding pump. J. Bacteriol. 182:4803-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vincent, F., S. Spinelli, R. Ramoni, S. Grolli, P. Pelosi, C. Cambillau, and M. Tegoni. 2001. Complexes of porcine odorant binding protein with odorant molecules belonging to different chemical classes. J. Mol. Biol. 305:459-469. [DOI] [PubMed] [Google Scholar]
- 42.White, D. G., J. D. Goldman, B. Demple, and S. B. Levy. 1997. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J. Bacteriol. 179:6122-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yakushi, T., K. Masuda, S. Narita, S. Matsuyama, and H. Tokuda. 2000. A new ABC transporter mediating the detachment of lipid-modified proteins from membranes. Nat. Cell Biol. 2:212-218. [DOI] [PubMed] [Google Scholar]
- 44.Yu, E. W., G. McDermott, H. I. Zgruskaya, H. Nikaido, and D. E. Koshland, Jr. 2003. Structural basis of multiple drug binding capacity of the AcrB multidrug efflux pump. Science 300:976-980. [DOI] [PubMed] [Google Scholar]
- 45.Zgurskaya, H., and H. Nikaido. 1999. AcrA is a highly asymmetric protein capable of spanning the periplasm. J. Mol. Biol. 285:409-420. [DOI] [PubMed] [Google Scholar]
- 46.Zgurskaya, H., and H. Nikaido. 1999. Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc. Natl. Acad. Sci. USA 96:7190-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheleznova, E. E., P. N. Markham, A. A. Neyfakh, and R. G. Brennan. 1999. Structural basis of multidrug recognition by BmrR, a transcription activator of a multidrug transporter. Cell 96:353-362. [DOI] [PubMed] [Google Scholar]