Abstract
Background
Two recombinant, live attenuated human parainfluenza virus type 1 (rHPIV1) mutant viruses have been developed, using a reverse genetics system, for evaluation as potential intranasal vaccine candidates. These rHPIV1 vaccine candidates have two non-temperature sensitive (non-ts) attenuating (att) mutations primarily in the P/C gene, namely CR84GHNT553A (two point mutations used together as a set) and CΔ170 (a short deletion mutation), and two ts att mutations in the L gene, namely LY942A (a point mutation), and LΔ1710–11 (a short deletion), the last of which has not been previously described. The latter three mutations were specifically designed for increased genetic and phenotypic stability. These mutations were evaluated on the HPIV1 backbone, both individually and in combination, for attenuation, immunogenicity, and protective efficacy in African green monkeys (AGMs).
Results
The rHPIV1 mutant bearing the novel LΔ1710–11 mutation was highly ts and attenuated in AGMs and was immunogenic and efficacious against HPIV1 wt challenge. The rHPIV1-CR84G/Δ170HNT553ALY942A and rHPIV1-CR84G/Δ170HNT553ALΔ1710–11 vaccine candidates were highly ts, with shut-off temperatures of 38°C and 35°C, respectively, and were highly attenuated in AGMs. Immunization with rHPIV1-CR84G/Δ170HNT553ALY942A protected against HPIV1 wt challenge in both the upper and lower respiratory tracts. In contrast, rHPIV1-CR84G/Δ170HNT553ALΔ1710–11 was not protective in AGMs due to over-attenuation, but it is expected to replicate more efficiently and be more immunogenic in the natural human host.
Conclusion
The rHPIV1-CR84G/Δ170HNT553ALY942A and rHPIV1-CR84G/Δ170HNT553ALΔ1710–11 vaccine candidates are clearly highly attenuated in AGMs and clinical trials are planned to address safety and immunogenicity in humans.
Background
Human parainfluenza virus type 1 (HPIV1) is responsible for approximately 6% of pediatric hospitalizations due to respiratory tract disease with significant illness occurring predominantly in infants and young children [1]. Clinical manifestations range from mild disease, including rhinitis, pharyngitis, and otitis media, to more severe disease, including croup, bronchiolitis, and pneumonia [1-6]. Collectively, human parainfluenza virus serotypes 1, 2 and 3 (HPIV1, 2 and 3) are the second leading causative agents of pediatric hospitalizations due to respiratory disease following respiratory syncytial virus (RSV) [7,1]. However, a licensed vaccine is currently not available for the prevention of illness caused by any HPIV.
HPIV1 is an enveloped, non-segmented, single-stranded, negative-sense RNA virus belonging to the family Paramyxoviridae, genus Respirovirus, of which HPIV3 is also a member. The HPIV1 genome is 15,600 nucleotides in length and contains six genes in the order 3'-N-P/C-M-F-HN-L-5', which encode three nucleocapsid-associated proteins including the nucleocapsid protein (N), the phosphoprotein (P), and the large polymerase protein (L) and three envelope-associated proteins including the internal matrix protein (M) and the fusion (F) and hemagglutinin-neuraminidase (HN) transmembrane surface glycoproteins [8]. F and HN are the two viral neutralization antigens and are major viral protective antigens. The P/C gene of HPIV1 contains a second open reading frame (ORF) that encodes up to four accessory C proteins, C', C, Y1 and Y2, that initiate at four separate translational start codons in the C ORF and are carboxy co-terminal [1]. However, it is unclear whether the Y2 protein is actually expressed during HPIV1 infection [9]. The HPIV1 C proteins have recently been shown to act as antagonists of the innate immune response during virus infection by inhibiting type 1 interferon (IFN) production and signaling of IFN through its receptor [10].
Our laboratory is developing a live attenuated virus vaccine for HPIV1 for intranasal administration to infants and young children. The intranasal route of administration is needle-free and has the advantage of direct stimulation of local immunity as well as induction of a substantial systemic immune response [11]. Furthermore, compared to an inactivated vaccine, a live virus vaccine stimulates a broader spectrum of innate and adaptive immune responses [11]. The recent licensure of the trivalent live attenuated influenza virus vaccine (Flumist™) indicates that it is possible to achieve an acceptable balance between attenuation and immunogenicity with a live attenuated respiratory virus vaccine [12].
Reverse genetics provides a method for introducing attenuating mutations in desired combinations into wild type (wt) HPIV1 [13-16]. Temperature sensitive (ts) attenuating (att) and non-ts att mutations have been developed that, in combination, can enhance both the phenotypic and genetic stability of a HPIV1 vaccine candidate. The licensed cold-adapted influenza A viruses contain similar non-ts and ts att mutations [17,18]. In the case of HPIV1, non-ts att mutations have been introduced into the P/C gene that inactivate the anti-IFN activities of the C accessory proteins [10]. One of these mutations (CΔ170) is a deletion mutation that affects codon 170 of the HPIV1 C protein; deletion mutations are desirable because they are essentially free of same-site reversion and thus provide for enhanced genetic and phenotypic stability. The CΔ170 mutation inhibited both the production of Type 1 IFN and the signaling of IFN through its receptor and specified an att phenotype in hamsters and African green monkeys (AGMs) [10,16]. A second non-ts att mutation involves a pair of amino acid substitutions, CR84G and HNT553A, that attenuates HPIV1 for AGMs when they are present together but not individually. This attenuating pair of mutations was not further genetically stabilized, i.e., it is possible to revert to a wt phenotype with a single nucleotide substitution at either mutation. A substitution at amino acid position 942 of L, LY942A, generated a ts att mutation that was engineered for increased genetic and phenotypic stability by the strategy of identifying a codon whose amino acid assignment yielded a ts att phenotype and which would require three nucleotide substitutions for reversion [13]. A virus bearing this stabilized mutation was attenuated in both AGMs and hamsters [15].
The present study consists of two parts. First, we developed an additional ts att mutation involving a small deletion in the HPIV1 L protein. This mutation was originally identified as a ts att point mutation in the bovine PIV3 (BPIV3) L protein (LS1711I) [19]. The corresponding site in the HPIV1 L protein was identified as position 1710 by sequence alignment, and this codon and its downstream neighbor (codon 1711) were deleted to yield the LΔ1710–11 mutation. This gave us two genetically stabilized ts att mutations in L, the LΔ1710–11 and the LY942A mutations. In the second part of the study, the two non-ts att mutations in C, namely the CR84G/HNT553A set and the CΔ170 mutation, were combined with each other and with either the LΔ1710–11 mutation or the LY942A mutation to develop two live intranasal HPIV1 vaccine candidates. Each of these vaccine candidates contained at least one genetically stabilized ts and non-ts att mutation. These viruses were evaluated for their in vitro attenuation phenotype and for replication, efficacy and immunogenicity in AGMs.
Results
Construction and recovery of mutant rHPIV1 viruses
Point and deletion mutations in the P/C, HN and L genes that attenuate HPIV1 for replication in the respiratory tract of hamsters or AGMs are indicated in Table 1[13-16]. The CR84G mutation is a single nucleotide substitution mutation that affects both the P and C proteins and that results in amino acid substitutions of R84 to G in C, and E87 to G in P (Table 1) [15]. The CR84G mutation is attenuating in the upper respiratory tract (URT) of AGMs, but only in the presence of the HNT553A point mutation indicated in Table 1[15]. The CR84G and HNT553A mutations are each based on single nucleotide substitutions (Table 1), and thus the att phenotype would be lost by reversion at either position. The CΔ170 deletion mutation in HPIV1 involves a six-nucleotide deletion, a length that was chosen to comply with the "rule of six" [20]. This deletion results in a loss of two amino acids and substitution of a third at codon positions 168–170 in C (RDF to S), and a deletion of amino acids GF in P at codon positions 172–173 (Table 1) [16]. The changes in the C protein also would be present in the nested C', Y1, and Y2 proteins (not shown) [16]. The Y942A mutation in L has three nucleotide changes in codon 942 and specifies a genetically and phenotypically stabilized ts att phenotype [13].
Table 1.
Summary of the mutations introduced into the rHPIV1 genomea.
| Gene | Mutationb | ORF | nt changes wt → mutant c | Type of mutation | Codon position | Amino acid change | # nt changes for reversion to wt |
| P/C | R84G | C | AGA → GGA | point | 84 | R → G | 1 |
| P | GAG → GGG | point | 87 | E → G | 1 | ||
| Δ170d | C | AGG GAT TTC → AGC | deletion | 168–170 | RDF → S (D deletion; 3 nt deletions in the flanking R-F codons results in a S substitution) | 6 (insertions)d | |
| P | GGA TTT→ deletion | deletion | 172–173 | GF deletion | 6 (insertions) | ||
| HN | T553A | HN | ACC → GCC | point | 553 | T → A | 1 |
| L | Y942Ae | L | TAT→ GCG | point | 942 | Y → A | 3e |
| Δ1710–11d | L | GCT GAG→ deletion | deletion | 1710–11 | AE deletion | 6 (insertions)d |
a HPIV1 strain Washington/1964, GenBank accession no. NC_003461.
b The nomenclature used to describe each mutation indicates the wt amino acid, the codon position and the new amino acid, or the position of the deletion (Δ), with respect to the C, HN or L protein.
c The nucleotides (nt) affected by substitution or deletion are shown underlined and in bold type.
d Designed for increased genetic stability by use of a deletion. Deletions involved six nt to conform to the rule of six [20].
e Designed for increased genetic stability by the use of a codon that differs by three nucleotides from codons yielding a wild type assignment.
In the present study, the LΔ1710–11 deletion mutation in HPIV1 was created at a site that corresponds by sequence alignment to a ts att point mutation originally identified in BPIV3 [19]. Importation of this BPIV3 point mutation has previously been shown to attenuate HPIV2 [21]. Here, the LΔ1710–11 mutation contains a six-nucleotide deletion that results in a deletion of amino acids AE at codon positions 1710–11 of the L gene of HPIV1 (Table 1).
The mutations in Table 1 were introduced into the HPIV1 antigenomic cDNA individually or in combinations to yield the panel of rHPIV1 viruses listed in Table 2. These viruses were recovered following transfection of cDNAs into HEp-2, BHK-T7 or Vero cells and biologically cloned in LLC-MK2 cells, and each was sequenced in its entirety to confirm the presence of the engineered mutation(s) and the absence of adventitious mutations. Unexpectedly, we were unable to isolate rHPIV1 containing the LΔ1710–11 mutation by itself and without adventitious mutations despite four attempts to do this using multiple replicates each time. However, we were able to recover virus bearing LΔ1710–11 in the presence of CR84G without adventitious mutations. Thus, our analysis of the phenotype of the LΔ1710–11 mutation was performed in the presence of the CR84G mutation, which is neither ts nor att [15].
Table 2.
Level of temperature sensitivity of replication of rHPIV1 mutants in vitro.
| Mean reduction (log10) in virus titer ± S.E. at the indicated temperature compared to 32°C c | |||||||||
| Virus a | Virus titer ± S.E. at 32°C b | 35°C | 36°C | 37°C | 38°C | 39°C | 40°C | Shut-off (°C) d | |
| 1 | HPIV1 wt | 7.7 ± 0.1 | 0.1 ± 0.1 | 0.1 ± 0.1 | 0.2 ± 0.1 | 0.7 ± 0.1 | 1.3 ± 0.1 | 3.0 ± 0.3 | - |
| 2e | rHPIV1-CR84G | 9.2 ± 0.4 | 0.4 ± 0.2 | 0.4 ± 0.6 | 0.8 ± 0.5 | 0.3 ± 0.4 | 1.8 ± 0.6 | 4.5 ± 0.9 | - |
| 3e | rHPIV1-CR84GHNT553A | 7.8 ± 0.1 | -0.3 ± 0.2 | -0.3 ± 0.2 | -0.2 ± 0.2 | 0.1 ± 0.2 | 0.7 ± 0.2 | 2.5 ± 0.6 | - |
| 4e | rHPIV1-CΔ170 | 7.9 ± 0.3 | 0.2 ± 0.2 | 0.7 ± 0.8 | 0.5 ± 0.2 | 1.0 ± 0.3 | 2.6 ± 0.7 | 4.5 ± 1.0 | - |
| 5 | rHPIV1-LY942A | 8.0 ± 0.1 | 0.2 ± 0.3 | 1.2 ± 0.3 | 2.6 ± 1.1c,d | 6.4 ± 0.4 | ≥6.8 f | ≥6.8 | 37°C |
| 6e | rHPIV1-CR84GHNT553ALY942A | 7.4 ± 0.2 | 0.4 ± 0.4 | 0.5 ± 0.4 | 2.3 ± 0.4 | 4.0 ± 0.6 | 6.0 ± 0.4 | ≥6.4 | 37°C |
| 7 | rHPIV1-CR84GLΔ1710–11 | 7.5 ± 0.7 | 0.8 ± 0.7 | 3.0 ± 0.6 | 4.8 ± 0.2 | ≥6.3 | ≥6.3 | ≥6.3 | 36°C |
| 8 | rHPIV1-CR84G/Δ170HNT553ALY942A | 6.3 ± 0.1 | 0.3 ± 0.2 | 0.9 ± 0.6 | 2.0 ± 0.3 | 4.9 ± 0.2 | ≥5.1 | ≥5.1 | 38°C |
| 9 | rHPIV1-CR84G/Δ170HNT553ALΔ1710–11 | 6.4 ± 0.3 | 2.6 ± 0.6 | 4.0 ± 0.4 | ≥5.2 | ≥5.2 | ≥5.2 | ≥5.2 | 35°C |
a Data are the mean of three to sixteen experiments.
b Viruses were titrated on LLC-MK2 cells at either permissive (32°C) or potentially restrictive (35°C – 40°C) temperatures for 7 days and virus titers are expressed as the mean ± standard error (S.E.). The limit of detection was 1.2 log10 TCID50/ml.
c Values in bold indicate restricted replication, where the mean log10 reduction in virus titer at the indicated temperature vs 32°C was 2.0 log10 or greater than the difference in titer of HPIV1 wt at the same temperature vs 32°C. A virus is designated ts if restricted replication at 35°C–40°C is observed.
d Underlined values indicate viral shut-off temperature, the lowest temperature at which restricted replication is observed.
e These data have been previously published [13] [15] [16] and are included here for the purposes of comparison.
f The symbol "≥" indicates that virus titers were at the limit of detection and therefore the reduction in virus titer versus 32°C is greater than or equal to the indicated value. There is no S.E. value for viruses at the limit of detection.
Characterization of rHPIV1s containing single att mutations
We first sought to characterize the rHPIV1 mutants bearing the four single att mutations (the CR84GHNT553A set, CΔ170, LY942A, and LΔ1710–11) to define the contributions of the individual mutations to the phenotypes of the rHPIV1 mutants (Groups 3, 4, 5, 7 in Tables 2 and 3). We previously generated and evaluated the rHPIV1-CR84GHNT553A and rHPIV1-CΔ170 viruses (each containing a single non-ts att mutation) in vitro and in vivo [13,15,16]. These previously evaluated single-mutation viruses were included here for the purpose of comparison with viruses containing the other individual mutations as well as combinations of mutations. An rHPIV1 mutant, rHPIV1-LY942A, bearing the Y942A mutation in L was generated for the present study. We had previously generated and characterized a virus, rHPIV1-CR84GHNT553ALY942A, containing the LY942A mutation in combination with the CR84GHNT553A pair of mutations [13]. The newly generated rHPIV1-LY942A virus would permit evaluation of its specific contribution to the level of temperature sensitivity in vitro and attenuation in vivo. The rHPIV1 mutant bearing the individual att mutation LΔ1710–11 (rHPIV1-CR84GLΔ1710–11) also contained the CR84G mutation, although this latter mutation is phenotypically silent on its own, as already noted.
Table 3.
Level of replication of HPIV1 vaccine candidates in the upper and lower respiratory tract of African green monkeys.
| Mean peak virus titer (log10 TCID50/ml)c | Mean sum of the daily virus titers (log10 TCID50/ml)d | atte | |||||||
| Virusa | Shut-off temperature b | No. of animals | NP swabf | TL g | NP swabf | TL g | URT | LRT | |
| 1 | HPIV1 wt | - | 14 | 4.2 ± 0.2 | 3.9 ± 0.3 | 26.4 ± 1.5 | 12.2 ± 1.6 | - | - |
| 2h | rHPIV1-CR84G | - | 4 | 3.6 ± 0.4 | 4.0 ± 0.5 | 21.0 ± 1.7 | 11.7 ± 2.5 | No | No |
| 3h | rHPIV1-CR84GHNT553A | - | 12 | 2.1 ± 0.2i | 4.8 ± 0.3 | 10.5 ± 0.9 | 14.3 ± 1.1 | Yes | No |
| 4h | rHPIV1-CΔ170 | - | 6 | 3.4 ± 0.5 | 2.3 ± 0.5 | 14.8 ± 1.9 | 5.1 ± 0.8 | Yes | Yes |
| 5 | rHPIV1-LY942A | 37°C | 4 | 2.3 ± 0.1 | 2.3 ± 0.2 | 16.9 ± 0.7 | 8.4 ± 1.2 | Yes | Yes |
| 6h | rHPIV1-CR84GHNT553ALY942A | 37°C | 8 | 2.4 ± 0.2 | 2.1 ± 0.3 | 12.9 ± 1.0 | 5.1 ± 0.6 | Yes | Yes |
| 7 | rHPIV1-CR84GLΔ1710–11 | 36°C | 4 | 1.5 ± 0.4 | 0.9 ± 0.2 | 8.6 ± 1.8 | 3.2 ± 0.6 | Yes | Yes |
| 8 | rHPIV1-CR84G/Δ170HNT553ALY942A | 38°C | 4 | 1.2 ± 0.3 | 0.6 ± 0.1 | 5.9 ± 0.5 | 2.6 ± 0.1 | Yes | Yes |
| 9 | rHPIV1-CR84G/Δ170HNT553ALΔ1710–11 | 35°C | 4 | 0.9 ± 0.3 | ≤0.5 ± 0.0 | 6.3 ± 0.5 | ≤2.5 ± 0.0 | Yes | Yes |
a Monkeys were inoculated i.n. and i.t. with 106 TCID50 of the indicated virus in a 1 ml inoculum at each site. Data are representative of one to five experiments.
b Shut-off temperature is defined in footnote d, Table 2.
c Virus titrations were performed on LLC-MK2 cells at 32°C and expressed as the mean ± S.E of the individual peak virus titers for the animals in each group irrespective of day. The limit of detection was 0.5 log10 TCID50/ml.
d Mean sum of the daily virus titers: the sum of the titers for all of the days of sampling was determined for each animal individually, and the mean was calculated for each group. On days when virus was not detected, a value of was 0.5 log10 TCID50/ml was assigned for the purpose of calculation. The mean sum of the lower limit of detection was 5.0 log10 TCID50/ml for NP swabs and 2.5 log10 TCID50/ml for TL samples.
e Virus is designated att in the URT or LRT based on a significant reduction in either mean peak titer or mean sum of daily titers compared to the HPIV1 wt group (see footnote h).
f Nasopharyngeal (NP) swab samples were collected on days 1–10 post-infection.
g Tracheal lavage (TL) samples were collected on days 2, 4, 6, 8, and 10 post-infection.
h These data have been previously published [13] [15] [16] and are included here for the purposes of comparison.
i Underlined values indicate a statistically significant reduction compared to corresponding HPIV1 wt titer, P < 0.05 (Student-Newman-Keuls multiple comparison test).
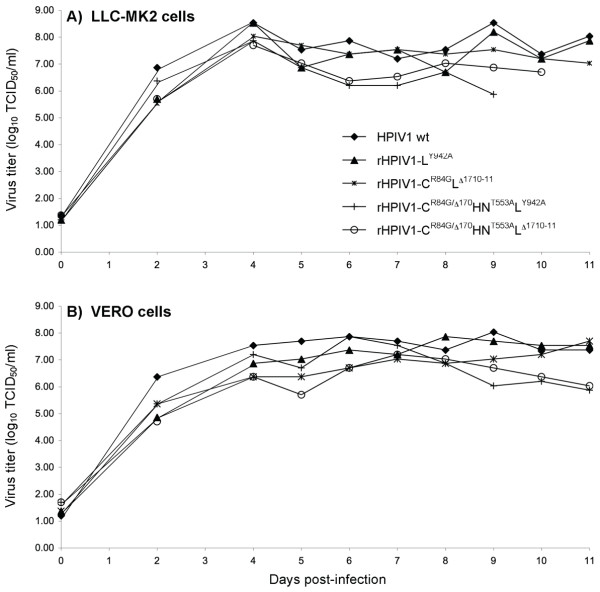
The level of temperature sensitivity of replication of the four viruses with single att mutations was first studied (Table 2, groups 3, 4, 5, and 7) and compared to that of rHPIV1 wt and rHPIV1-CR84G. Viruses containing only P/C gene mutations with or without the HN mutation were non-ts, whereas each of the L gene mutations specified a ts phenotype in vitro. The single LY942A mutation specified a shut-off temperature of 37°C, a level of temperature sensitivity that was equivalent to that previously observed for rHPIV1-CR84GHNT553ALY942A (Table 2, compare Groups 5 and 6). These data indicate that the LY942A mutation is responsible for the observed ts phenotype of rHPIV1-CR84GHNT553ALY942A (Table 2). The LΔ1710–11 mutation specified an even stronger ts phenotype than the LY942A mutation (Table 2). The LΔ1710–11 mutation clearly contributes significantly to the ts property of rHPIV1-CR84GLΔ1710–11 since rHPIV1-CR84G was confirmed to be non-ts (Table 2, compare Groups 2 and 7). Therefore, both LY942A and LΔ1710–11 are ts mutations in HPIV1. In a multiple cycle growth curve, the two newly generated rHPIV1 mutants with single att mutations, rHPIV1-LY942A and rHPIV1-CR84GLΔ1710–11, reached a titer equivalent to that of rHPIV1 wt in both LLC-MK2 and Vero cells (Figure 1). Thus, these individual mutations do not significantly restrict replication in vitro at the permissive temperature of 32°C and therefore could be useful mutations in vaccine candidates.
Figure 1.
Comparison of the replication of HPIV1 wt and rHPIV1 mutant viruses containing the indicated mutations in the P/C, HN and L genes in a multiple cycle growth curve. Monolayer cultures of LLC-MK2 cells and Vero cells were infected at a multiplicity of infection of 0.01 TCID50/cell and incubated at 32°C. The medium was removed on days 0 (residual inoculum), 2 and 4–11 post-infection, frozen for later determination of virus titers, and replaced by fresh medium containing trypsin. The virus titers shown are the means of 3 replicate cultures.
The level of replication of rHPIV1-LY942A and rHPIV1-CR84GLΔ1710–11 in AGMs was next evaluated and compared to that of rHPIV1 wt and the other two single att mutants (Table 3, Groups 1, 3, 4, 5, 7). A rHPIV1 mutant was considered attenuated if it exhibited a significant (P < 0.05) reduction in replication in either the mean peak virus titer or the mean sum of the daily virus titers (a measure of the total amount of virus shed over the duration of the infection) in either the nasopharyngeal (NP) swab (representative of the upper respiratory tract, URT) or tracheal lavage (TL) samples (representative of the lower respiratory tract, LRT) compared to the HPIV1 wt group. We have previously demonstrated that rHPIV1-CR84G replicates to levels equivalent to HPIV1 wt in AGMs, whereas rHPIV1-CR84GHNT553A and rHPIV1-CR84GHNT553ALY942A were attenuated in AGMs [15,16]. Here, both rHPIV1-LY942A and rHPIV1-CR84GLΔ1710–11 were significantly attenuated in the URT and LRT of AGMs in comparison to HPIV1 wt. The levels of attenuation of rHPIV1-LY942A and rHPIV1-CR84GHNT553ALY942A were comparable, indicating that the LY942A mutation is an attenuating mutation by itself and that the attenuation specified by the LY942A mutation is not additive to that specified by the CR84GHNT553A att mutation. The rHPIV1-CR84GLΔ1710–11 mutant also was significantly attenuated in AGMs, reducing virus titer in comparison to HPIV1 wt by 2.7 and 3.0 log10 50%-tissue-culture-infectious-doses (TCID50)/ml in the URT and LRT, respectively (Table 3). Since rHPIV1-CR84G was confirmed not to be attenuated in AGMs (Table 3, Group 2) [16], this suggests that the LΔ1710–11 mutation contributes significantly to the observed attenuation phenotype.
The immunogenicity and protective efficacy resulting from immunization with rHPIV1s containing single att mutations were evaluated in AGMs by measuring post-immunization HPIV1 hemagglutination inhibiting (HAI) serum antibody titers and by challenging immunized and control animals with HPIV1 wt 28 days following immunization and determining challenge virus titers in the URT and LRT (Table 4). AGMs immunized with rHPIV1s containing single att mutations (Groups 3, 4, 5, and 7) developed post-immunization HAI serum antibodies and manifested resistance to replication of the challenge virus. The rHPIV1-CR84GLΔ1710–11 mutant, which showed a strong level of attenuation following immunization of AGMs, was protective only at a low level in the URT.
Table 4.
Immunogenicity and protective efficacy of rHPIV1 vaccine candidates in AGMs.
| Mean peak challenge virus titer (log10TCID50/ml) c | Mean sum of the daily challenge virus titers (log10TCID50/ml)d | Post-challenge serum HAI titerb | ||||||
| Virus a | No. animals | Pre-challenge serum HAI titerb | NP swab | TL | NP swab | TL | ||
| 1 | HPIV1 wt | 12 | 6.7 ± 0.6 (12/12) | 0.8 ± 0.2f | 0.7 ± 0.1 | 2.3 ± 0.2 | 2.4 ± 0.2 | 6.6 ± 0.5 |
| 2e | rHPIV1-CR84G | 4 | 3.8 ± 0.9 (3/4) | ≤0.5 ± 0.0 | ≤0.5 ± 0.0 | ≤2.0 ± 0.0 | ≤2.0 ± 0.0 | 4.4 ± 1.2 |
| 3e | rHPIV1-CR84GHNT553A | 12 | 6.0 ± 0.6 (11/12) | 0.6 ± 0.1 | 0.6 ± 0.1 | 2.1 ± 0.1 | 2.1 ± 0.1 | 7.9 ± 0.4 |
| 4e | rHPIV1-CΔ170 | 6 | 5.5 ± 0.4 (6/6) | ≤0.5 ± 0.0 | ≤0.5 ± 0.0 | ≤2.0 ± 0.0 | ≤2.0 ± 0.0 | 6.5 ± 0.4 |
| 5 | rHPIV1-LY942A | 4 | 6.3 ± 1.2 (4/4) | 1.1 ± 0.2 | 1.2 ± 0.2 | 2.7 ± 0.3 | 2.8 ± 0.3 | 8.9 ± 1.1 |
| 6e | rHPIV1-CR84GHNT553ALY942A | 8 | 2.0 ± 0.0 (3/8) | 0.8 ± 0.2 | 0.8 ± 0.2 | 2.6 ± 0.3 | 2.4 ± 0.3 | 3.3 ± 0.7 |
| 7 | rHPIV1-CR84GLΔ1710–11 | 4 | 6.1 ± 1.8 (3/4) | 3.4 ± 0.6 | 3.0 ± 0.6 | 8.4 ± 2.0 | 8.3 ± 1.3 | 6.9 ± 1.5 |
| 8 | rHPIV1-CR84G/Δ170HNT553ALY942A | 4 | ≤1.0 ± 0.0 (0/4) | 2.2 ± 0.2 | 1.8 ± 0.5 | 5.1 ± 0.3 | 4.3 ± 1.3 | 5.5 ± 1.6 |
| 9 | rHPIV1-CR84G/Δ170HNT553ALΔ1710–11 | 4 | ≤1.0 ± 0.0 (0/4) | 4.5 ± 0.9 | 3.4 ± 0.4 | 11.8 ± 2.5 | 8.1 ± 1.3 | 7.5 ± 1.4 |
| 10 | Non-immune | 7 | ≤1.0 ± 0.0 (0/4) | 5.0 ± 0.6 | 3.9 ± 0.5 | 14.8 ± 1.2 | 11.0 ± 2.5 | 6.0 ± 1.3 |
a Monkeys were immunized i.n. and i.t. with 106 TCID50 of the indicated virus in a 1 ml inoculum at each site and were challenged on day 28 post-infection with HPIV1 wt.
b HAI titers to HPIV1 were determined by HAI assay of sera collected at day 28 (pre-challenge) and day 56 (post-challenge) in separate assays. Titers are expressed as mean reciprocal log2± S.E.; the limit of detection was 1.0 ± 0.0. The number of animals with a 4-fold or greater increase in pre-challenge antibody titers is shown in brackets for each group.
c Mean ± S.E of the individual peak virus titers for the animals in each group irrespective of day. Virus titrations were performed on LLC-MK2 cells at 32°C. The limit of detection was 0.5 log10 TCID50/ml. NP and TL samples were collected on days 2, 4, 6 and 8 post-challenge.
d Mean sum of the daily virus titers: the sum of the titers for all of the days of sampling was determined for each animal individually, and the mean was calculated for each group. On days when no virus was detected, a value of was 0.5 log10 TCID50/ml was assigned for the purpose of calculation. The mean sum of the lower limit of detection was 2.0 log10 TCID50/ml for NP swabs and TL samples.
e These data have been previously published [13] [15] [16] and are included here for the purposes of comparison.
f Underlined values indicate statistically significant reductions in mean peaks or sum of daily virus titers for HPIV1 wt titer compared to the corresponding non-immune group, P < 0.05 (Student-Newman-Keuls multiple comparison test).
Combination of three single att mutations into rHPIV1 to generate two live attenuated HPIV1 vaccine candidates
Having identified the in vitro and in vivo properties of the four single att mutations, we used this information to generate two live attenuated HPIV1 vaccine candidates containing both non-ts and ts attenuating mutations. These vaccine candidates were designed to incorporate a backbone containing one stabilized non-ts attenuating mutation, CΔ170, as well as the CR84GHNT553A att mutation. The addition of this second mutation (the CR84GHNT553A att mutation) would be expected to increase the overall stability of the virus by increasing the total number of attenuating mutations present in the vaccine candidate. To generate the two live attenuated HPIV1 vaccine candidates, either the stabilized ts att LY942A mutation or the LΔ1710–11 deletion mutation was added to the rHPIV1-CR84G/Δ170HNT553A backbone. We then evaluated the resulting combination mutants, rHPIV1-CR84G/Δ170HNT553ALY942A and rHPIV1-CR84G/Δ170HNT553ALΔ1710–11, as potential vaccine candidates.
These two viruses were first evaluated for their level of temperature sensitivity of replication in vitro (Table 2). The level of temperature sensitivity of rHPIV1-CR84G/Δ170HNT553ALY942A and rHPIV1-CR84G/Δ170HNT553ALΔ1710–11 (Groups 8 and 9 in Table 2) was equivalent to that of the corresponding L gene single-mutation viruses from which they were derived (namely rHPIV1-LY942A and rHPIV1-CR84GLΔ1710–1, Groups 5 and 7 in Table 2). This indicates that combining the non-ts and ts mutations in rHPIV1-CR84G/Δ170HNT553ALY942A and rHPIV1-CR84G/Δ170HNT553ALΔ1710–11 did not significantly alter their overall level of temperature sensitivity of replication in vitro. A multiple cycle growth curve at 32°C demonstrated that each virus achieved titers in Vero cells that will allow efficient manufacture. Specifically, the rHPIV1-CR84G/Δ170HNT553ALY942A and rHPIV1-CR84G/Δ170HNT553ALΔ1710–11 vaccine candidates reached peak titers of 7.9 and 7.2 log10 TCID50/ml, respectively, in Vero cells (Figure 1).
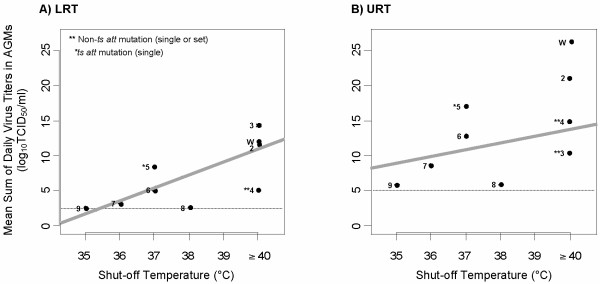
The level of replication of rHPIV1-CR84G/Δ170HNT553ALY942A and rHPIV1-CR84G/Δ170HNT553ALΔ1710–11 in AGMs were next evaluated and compared to that of rHPIV1 wt and the other two single att mutants (Table 3, Groups 1, 3, 4, 5, 7, 8, and 9). The rHPIV1-CR84G/Δ170HNT553ALY942A virus was strongly attenuated compared to rHPIV1 mutants bearing the corresponding single att mutations only in C/P, C/P/HN or L. The mean peak titer of rHPIV1-CR84G/Δ170HNT553ALY942A in the URT and LRT was reduced by 3.0 and 3.3 log10 TCID50/ml, respectively, in comparison to HPIV1 wt (Table 3). Similarly, the addition of the HNT553A and CΔ170 mutations to rHPIV1-CR84GLΔ1710–11 to generate the rHPIV1-CR84G/Δ170HNT553ALΔ1710–11 further attenuated the virus in AGMs, restricting virus replication in comparison to HPIV1 wt by 3.1 and 3.4 log10 TCID50/ml in the URT and LRT, respectively (Table 3). Therefore these two HPIV1 vaccine candidates demonstrate strong attenuation phenotypes in vivo. Considering the 9 viruses in Table 3 together, a relationship was found to exist between level of temperature sensitivity of replication in vitro and the attenuation manifested in vivo, i.e., the lower the shut off temperature, the higher the level of in vivo attenuation (Figure 2). Evaluation of these data using the Spearman rank test gives correlation coefficients of 0.47 and 0.67 for the URT and LRT, respectively, based on the mean daily sum of virus titers for individual AGMs. This indicates a moderate positive correlation with a stronger association between the level of temperature sensitivity and virus replication in the LRT. However, as might be expected, viruses bearing only the non-ts attenuating P/C gene mutations, including the CΔ170 and the CR84GHNT553A set of mutations, did not follow this pattern (Figure 2), and we would expect a higher correlation coefficient if these non-ts viruses were not included in the analysis.
Figure 2.
Representation of the association between the in vitro shut-off temperature and the attenuation phenotype in AGMs for HPIV1 wt (W) and rHPIV1 mutant viruses. For each virus (number designations correspond to the virus group numbers assigned in tables 2-4), the shut-off temperature (°C), as determined by an in vitro temperature sensitivity assay (Table 2), was plotted against the mean sum of daily virus titers (log10 TCID50/ml; Table 3) in the URT (A) and LRT (B) of AGMs. rHPIV1 wt and non-ts rHPIV1 mutants were assigned a shut-off temperature of 40°C for the purposes of this schematic. The limit of detection for the mean sum of daily virus titers is shown by a dashed line and viruses containing a single or set of non-ts attenuating mutation (**) or a single ts attenuating mutation (*) are highlighted, as shown. A linear trend line fit using the individual daily data is shown (solid line). The Spearman rank-correlation coefficient was determined to be 0.47 for the URT and 0.67 for the LRT, indicating a moderate positive correlation between shut-off temperature and mean daily sum of virus titer in the URT and a stronger association for the LRT.
The levels of immunogenicity and protective efficacy against HPIV1 wt challenge following immunization with rHPIV1-CR84G/Δ170HNT553ALY942A and rHPIV1-CR84G/Δ170HNT553ALΔ1710–11 were also determined (Groups 8 and 9 in Table 4). The two vaccine candidates failed to induce detectable HAI antibodies. However, immunization with the rHPIV1-CR84G/Δ170HNT553ALY942A was protective against HPIV1 wt challenge in both the URT and LRT (Table 4). In contrast, immunization with rHPIV1-CR84G/Δ170HNT553ALΔ1710–11 did not offer significant protection against HPIV1 wt challenge in the AGMs (Table 4), i.e., it appeared overattenuated in this animal model. A relationship was found between the level of replication of the immunizing virus and its ability to induce resistance to replication of the challenge virus (Tables 3 and 4), and this is graphically displayed in Figure 3.
Figure 3.
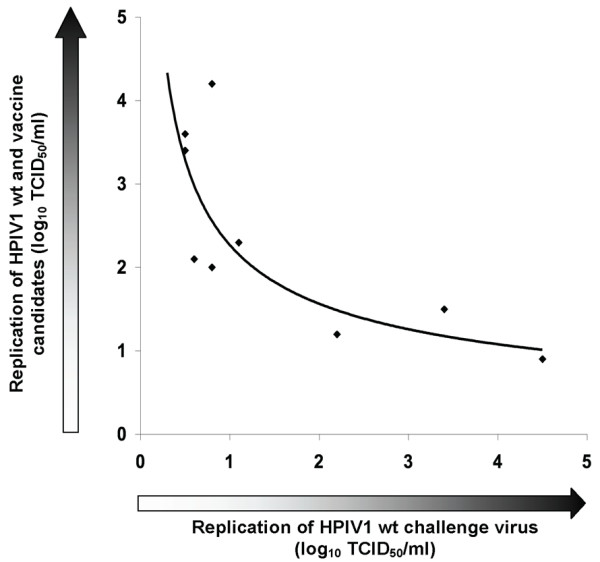
Representation of the relationship between the level of replication of HPIV1 wt and rHPIV1 mutants in AGMs and the subsequent level of replication of HPIV1 wt challenge virus in the immunized animals. The mean peak virus titer (log10 TCID50/ml) in the URT following immunization (y-axis) was plotted for viruses 1–9 (Table 3) against the mean peak challenge virus titers (log10 TCID50/ml; x-axis) in the same groups (Table 4). A curve of best fit has been inserted (solid line) to demonstrate the association between these two data sets.
Discussion
The advent of a reverse genetics system for the generation of infectious paramyxoviruses from full-length cDNA plasmids has greatly facilitated the development of live attenuated HPIV1 vaccine candidates [13-16]. The reverse genetics system for HPIV1 has allowed site-directed manipulation of the viral genome via cDNA intermediates, permitting the introduction of attenuating mutations in desired combinations into vaccine candidates. It has also been possible to genetically modify some of the attenuating mutations to optimize genetic and phenotypic stability of viruses bearing the mutations, both by the use of gene deletions and by using codons chosen for a low probability of reversion. This process enables us to optimize the safety profile of the live attenuated HPIV1 vaccine candidates before these viruses are tested in humans.
We are focusing our efforts on the development of live attenuated rHPIV1 vaccines since they have a number of advantages over inactivated or subunit vaccines, including the ability to: (i) induce the full spectrum of protective immune responses including serum and local antibodies as well as CD4+ and CD8+ T cells [11]; (ii) infect and replicate in the presence of maternal antibody permitting immunization of young infants [22,23]; (iii) cause an acute, self-limited infection that is readily eliminated from the respiratory tract; and (iv) replicate to high titers in cell substrates acceptable for products for human use, including qualified Vero cells, making manufacture of these vaccines commercially feasible. In the present study, two new rHPIV1 viruses containing single att mutations in L, LΔ1710–11 and LY942A, were generated and characterized, and these ts att mutations were used in combination with previously described non-ts att mutations in the P/C gene and HN gene to generate two new live attenuated HPIV1 vaccine candidates.
A major result of the present study was the creation of the LΔ1710–11 mutation that was found to specify a strong ts att phenotype. The LΔ1710–11 mutation was originally identified as an attenuating point mutation, LT1711I, in BPIV3 [19]. It was evaluated as a deletion mutation in the present study since a deletion mutation offers a higher level of genetic stability than a point mutation, a property that is desirable for mutations in a vaccine candidate. Indeed, since this deletion occurs in an ORF (in which the triplet nature of the codons must be maintained) and in a virus that conforms to the rule of six (in which the hexamer organization must be maintained), same-site reversion would require the precise restoration of six nucleotides. We unfortunately were not able isolate a rHPIV1 mutant with only the LΔ1710–11 mutation since each rHPIV1-LΔ1710–11 mutant that was isolated also possessed one or more adventitious mutations. The LΔ1710–11 mutation could only be recovered free of adventitious mutations when it was in combination with the CR84G mutation, and thus had to be studied in that context. We acknowledge that it is possible that the phenotypes that we observed for the rHPIV1-CR84GLΔ1710–11 are the result of an interaction between the CR84G and LΔ1710–11 mutations. However, we believe that this possibility is unlikely since the CR84G mutation does not contribute to the ts or att phenotype of HPIV1 as an independent mutation. Furthermore, the high level of temperature sensitivity and attenuation of rHPIV1-CR84GLΔ1710–11 versus that of rHPIV1-CR84G suggests a major independent role of the LΔ1710–11 mutation in these two phenotypes. rHPIV1-CR84GLΔ1710–11 manifested a shut-off temperature of 37°C in vitro and was restricted in replication in the URT and LRT of AGMs by 2.5 log10 or 3.0 log10, respectively. Therefore, we suggest that the LΔ1710–11 deletion mutation specifies a ts att phenotype for HPIV1, and, as such, it is a suitable mutation to include in a HPIV1 vaccine candidate.
The LY942A mutation was identified previously as an attenuating mutation for introduction into potential HPIV1 vaccine candidates and was stabilized by codon optimization studies [13]. These studies demonstrated that only three amino acids were shown to specify a wild type phenotype at this codon position (the wild type tyrosine, cysteine and phenylalanine) all of which would require three nucleotide changes to convert the GCG alanine to a codon specifying the wild type phenotype codon in the vaccine virus [13]. In addition, the LY942A mutation was shown to be highly stable under selective pressure during passage at permissive and restrictive temperatures [13]. Previous studies have evaluated the LY942A mutation only in the presence of the CR84GHNT553A set of mutations that attenuates HPIV1 for AGMs [13,15]. To determine the specific contribution of the LY942A mutation to the ts and att phenotypes associated with the rHPIV1-CR84GHNT553ALY942A virus, a rHPIV1 containing only the LY942A mutation was generated and was found to be as attenuated as rHPIV1-CR84GHNT553ALY942A for AGMs. This indicated that the LY942A mutation independently attenuated HPIV1 for AGMs and can be used in the absence of the CR84GHNT553A mutation to attenuate HPIV1 for AGMs. The attenuation specified by the CR84GHNT553A mutation was not additive with that of LY942A. This actually is a desirable property, since it permits the inclusion of a greater number of mutations while avoiding over-attenuation, and these additional mutations would become unmasked in the case of the loss of one or more other mutations and would thus maintain the att phenotype. Thus, LY942A is a stable mutation that specifies a ts att phenotype for HPIV1 and is suitable for introducing into a HPIV1 vaccine candidate as an independent attenuating mutation.
The LY942A and LΔ1710–11 ts att mutations were used in conjunction with two of the non-ts att mutations, the CR84GHNT553A and CΔ170 mutations [16], to develop two live attenuated vaccine candidates for HPIV1, namely, rHPIV1-CR84G/Δ170HNT553ALY942A and rHPIV1-CR84G/Δ170HNT553ALΔ1710–11. These vaccine candidates thus each contain three independent attenuating mutations (two non-ts att and one ts att mutation), two of which have been genetically stabilized. The combination of mutations present in these two vaccine candidates should enhance the genetic and phenotypic stability of the viruses, although this will require formal demonstration in a clinical trial using clinical grade virus preparations.
Evaluation of the two vaccine candidates revealed that they are reasonable candidates for further study in clinical trials. Both candidates replicated well in Vero cells, a characteristic that is important for manufacturing purposes. Both viruses also demonstrated a strong ts phenotype in vitro (shut-off temperature of ≤38°C) that was similar to that of their ts parent virus, but the two viruses differ in their level of temperature sensitivity in vitro. Since the level of temperature sensitivity of respiratory viruses [24], including HPIV1 as demonstrated here, correlates with level of attenuation, it was anticipated that this difference in the ts phenotype would be reflected in a difference in the level of attenuation and immunogenicity in vivo, and this indeed was seen. The HPIV1 vaccine candidates were both strongly attenuated in the URT and LRT of AGMs, with rHPIV1-CR84G/Δ170HNT553ALY942A replicating to slightly higher levels than the more ts rHPIV1-CR84G/Δ170HNT553ALΔ1710–11. Both vaccines were weakly immunogenic and failed to induce a detectable level of serum HAI antibodies in AGMs. A low level of protective efficacy was observed in AGMs immunized with rHPIV1-CR84G/Δ170HNT553ALY942A, but the rHPIV1-CR84G/Δ170HNT553ALΔ1710–11 was not protective. This low level of immunogenicity and efficacy was not unexpected since each vaccine was highly restricted in replication and since there is a strong correlation between the level of replication of vaccine virus and its immunogenicity and ability to restrict replication of HPIV1 challenge virus. These results can be interpreted to indicate that the two vaccine candidates are over-attenuated, but we think that this conclusion would be premature. It is likely that these viruses will be more immunogenic, and therefore more efficacious, in humans compared to AGMs since they should replicate more efficiently in humans. The reasons for this are two-fold. First, HPIV1 is a human virus, and it should replicate more efficiently in its natural host in which it causes disease than in AGMs in which it causes only an asymptomatic infection. The actual level of replication of HPIV1 in seronegative humans is unknown, but it replicates efficiently even in adults with pre-existing immunity [25,26]. Second, these vaccine candidates are highly ts and should replicate more efficiently in humans, which have a lower body core temperature (36.7°C), than in AGMs (approximately 39°C). Therefore, although these vaccine candidates appear to be over-attenuated in AGMs, it is expected that the viruses should replicate somewhat more efficiently in humans and would be more immunogenic than in AGMs. It also is fortunate that the two vaccine candidates appear to differ somewhat in their level of attenuation, since this provides two chances to achieve an optimal balance between safety and efficacy.
Conclusion
The rHPIV1-CR84G/Δ170HNT553ALY942A and rHPIV1-CR84G/Δ170HNT553ALΔ1710–11 vaccine candidates are highly attenuated in AGMs. We plan to initiate studies in humans with the less attenuated vaccine candidate, rHPIV1-CR84G/Δ170HNT553ALY942A. If this virus proves to be insufficiently attenuated in the target population of young seronegative infants (following an initial step-wise progression of safety testing in adults, seropositive children, and seronegative children), we would proceed to evaluate the more attenuated rHPIV1-CR84G/Δ170HNT553ALΔ1710–11 vaccine candidate. If rHPIV1-CR84G/Δ170HNT553ALY942A is over-attenuated, then the LY942A mutation would be deleted and the rHPIV1-CR84G/Δ170HNT553A would be tested in humans. In this way, we will identify a HPIV1 vaccine candidate that exhibits a satisfactory balance between attenuation and immunogenicity for the target population of seronegative infants and young children.
Methods
Cells and viruses
LLC-MK2 cells (ATCCCCL7.1) and HEp-2 cells (ATCCCCL23) were maintained in Opti-MEM I (Gibco-Invitrogen, Inc. Grand Island, NY) supplemented with 5% FBS and gentamicin sulfate (50 μg/ml). Vero cells (ATCC CCL-81) were maintained in Opti-PRO SFM (Gibco-Invitrogen, Inc.) in the absence of FBS and supplemented with gentamicin sulfate (50 μg/ml) and L-glutamine (4 mM). BHK-T7 cells, which constitutively express T7 RNA polymerase [27], were kindly provided by Dr. Ulla Buchholz, NIAID, and were maintained in GMEM (Gibco-Invitrogen, Inc.) supplemented with 10% FBS, geneticin (1 mg/ml), MEM amino acids, and L-glutamine (2 mM). Biologically-derived wt HPIV1 Washington/20993/1964, the parent for the recombinant virus system, was isolated previously from a clinical sample in primary African green monkey kidney (AGMK) cells and passaged 2 additional times in primary AGMK cells [25] and once in LLC-MK2 cells [15]. This preparation has a wild type phenotype in AGMs, and will be referred to here as HPIV1 wt. It was previously described as HPIV1LLC1 [15]. HPIV1 wt and rHPIV1 mutants were grown in LLC-MK2 cells in the presence of 1.2% Tryple select, a recombinant trypsin (Gibco-Invitrogen, Inc.), as described previously [8].
Construction of mutant HPIV1 cDNA
P/C, HN and L gene mutations (Table 1) were introduced into the appropriate rHPIV1 subgenomic clones [14] using the Advantage-HF PCR Kit (Clontech Laboratories, Palo Alto, CA) with a modified PCR mutagenesis protocol described elsewhere [28]. The entire PCR amplified subgenomic clone was sequenced using a Perkin-Elmer ABI 3100 sequencer with the Big Dye sequencing kit (Perkin-Elmer Applied Biosystems, Warrington, UK) to confirm that the subclone contained the introduced mutation and to confirm the absence of adventitious mutations introduced during PCR amplification. Full-length antigenomic cDNA clones (FLCs) of HPIV1 containing the desired mutations were assembled using standard molecular cloning techniques [8], and the region containing the introduced mutation in each FLC was sequenced as described above to confirm the presence of the introduced mutation and absence of adventitious changes. Each virus was designed to conform to the rule of six, which is a requirement by HPIV1 and numerous other paramyxoviruses that the nucleotide length of their genome be an even multiple of six for efficient replication [20].
Recovery of rHPIV1 mutant viruses
Three different recovery methods were used to generate rHPIV1 mutants that differed in the source of the T7 polymerase needed to synthesize RNA from the transfected virus-specific plasmids and, in one case, a different transfection method was used. First, using previously described procedures [8], rHPIV1 virus was recovered from HEp-2 cells that were transfected with plasmids encoding the antigenome and N, P, and L support proteins and infected with an MVA-T7 vaccinia virus recombinant as a source of T7 polymerase. Second, Vero cells were grown to 80% confluency and transfection experiments were performed using the AMAXA Cell Line Nucleofector Kit V, according to manufacturer's directions (AMAXA, Koeln, Germany), as previously described [29]. Briefly, the cells were transfected with 5 μg each of the FLC and the pCL-Neo-BCI-T7 plasmid (expressing T7 polymerase under the control of a eukaryotic promoter) [30], 0.2 μg each of the N and P, and 0.1 μg of the L support plasmids. The transfection mixture was removed after 24 h at 37°C, and cells were washed and overlaid with Opti-PRO with L-glutamine (4 mM) supplemented with 1.2% Tryple select. The cells and supernatant were transferred to LLC-MK2 cells in T25 cm2 flasks (Corning, NY) 7 days following transfection. Third, BHK-T7 cells constitutively expressing T7 polymerase [27] were grown to 90 to 95% confluence in six-well plates. The cells were transfected with 5 μg of the FLC, 0.8 μg each of the N and P, and 0.1 μg of the L support plasmids in a volume of 100 μl of Opti-MEM per well. Transfection was carried out with Lipofectamine 2000 (Invitrogen, Inc., Carlsbad, CA), according to the manufacturer's directions. The transfection mixture was removed after a 24 h incubation period at 37°C, and the cells were washed and maintained in GMEM. On day 2 following transfection, the media was supplemented with 1.2% trypsin, and the recovered virus was harvested on days 2–4. All viruses were amplified by passage on LLC-MK2 cells, and each was cloned by two successive rounds of terminal dilution using LLC-MK2 monolayers in 96-well plates (Costar, Corning Inc., Acton, MA). To confirm that the recovered rHPIV1 mutants contained the appropriate mutations and lacked adventitious mutations, viral RNA (vRNA) was isolated from infected cell supernatants using the Qiaquick vRNA kit (Qiagen Inc., Valencia, CA), reverse transcribed using the SuperScript First-Strand Synthesis System (Invitrogen, Inc., Carlsbad, CA) and amplified using the Advantage cDNA PCR Kit (Clontech Laboratories). Each viral genome was sequenced in its entirety.
Evaluation of recombinant HPIV1 vaccine candidates in a multiple cycle growth curve
The recombinant HPIV1 mutants were compared to HPIV1 wt on LLC-MK2 and Vero cells at 32°C in a multiple cycle growth curve. Confluent monolayer cultures in 6-well plates were infected in triplicate at a multiplicity of infection (MOI) of 0.01 50%-tissue-culture-infectious-doses (TCID50) per cell in media containing trypsin. The residual inoculum was withdrawn 2 h post infection as the day 0 sample and was replaced by medium with trypsin. On days 2, and 4–11 post-infection, the total medium supernatant was removed for virus quantitation and was replaced with fresh medium with trypsin. Supernatants containing virus were frozen at -70°C, and all samples were tested together for virus titer with endpoints identified by hemadsorption.
Characterization of the temperature sensitivity of the rHPIV1 vaccine candidates
The ts phenotype for each mutant rHPIV1 virus was determined by comparing its level of replication to that of HPIV1 wt at 32°C and at 1°C increments from 35°C to 40°C, as described previously [31]. Briefly, each virus was serially diluted 10-fold in 96-well LLC-MK2 monolayer cultures in L-15 media (Gibco-Invitrogen, Inc.) containing trypsin with four replicate wells per plate. Replicate plates were incubated at the temperatures indicated above for seven days, and virus infected wells were detected by hemadsorption with guinea pig erythrocytes. The virus titer at each temperature was determined in three to sixteen separate experiments and is expressed as the mean log10 TCID50/ml. The mean titer at an elevated temperature was compared to the mean titer at 32°C, and the reduction in mean titer was determined. The shut-off temperature of an rHPIV1 mutant is defined as the lowest temperature at which the reduction in virus titer compared to its titer at 32°C was 100-fold greater than the reduction in HPIV1 wt titer between the same two temperatures. A mutant is defined as having a ts phenotype if its shut-off temperature is ≤40°C.
Evaluation of replication of viruses in AGMs and efficacy against challenge
AGMs in groups of two to four animals at a time were inoculated intranasally (i.n.) and intratracheally (i.t.) with 106 TCID50 of either HPIV1 wt or mutant rHPIV1 in a 1 ml inoculum at each site. NP swab samples were collected daily from days 1 to 10 post-inoculation, and TL fluid samples were collected on days 2, 4, 6, 8 and 10 post-inoculation. The specimens were flash frozen and stored at -80°C and were subsequently assayed in parallel. Virus present in the samples was titered in dilutions on LLC-MK2 cell monolayers in 96-well plates and an undiluted 100 μl aliquot was also tested in 24-well plates. These were incubated at 32°C for 7 days. Virus was detected by hemadsorption, and the mean log10 TCID50/ml was calculated for each sample day. The limit of detection was 0.5 log10TCID50/ml. The mean peak titer for each group was calculated using the peak titer for each animal, irrespective of the day of sampling. The mean sum of the virus titers for each group was calculated from the sum, calculated for each animal individually, of the virus titers on each day of sampling, up to day 10. The sum of the lower limit of detectability was 5.0 log10 TCID50/ml for NP swabs and 2.5 log10 TCID50/ml for TL samples.
On day 28 post-inoculation, the AGMs were challenged i.n. and i.t. with 106 TCID50 of HPIV1 wt in 1 ml at each site. NP swab and TL samples were collected for virus quantitation on days 2, 4, 6 and 8 post-challenge.
All animal studies were performed under protocol LID22, as approved by the National Institute of Allergy and Infectious Disease (NIAID) Animal Care and Use Committee (ACUC).
Evaluation of immune responses in AGMs
Serum was collected from each monkey on days 0 and 28 post-immunization and on day 28 post-challenge (day 56 post-immunization). HPIV1 HAI antibody titers were determined at 21°C, as described previously [32], using 0.5% v/v guinea pig erythrocytes and HPIV1 wt as the antigen. The HAI antibody titer was defined as the end-point serum dilution that inhibited hemagglutination and is expressed as the mean reciprocal log2 ± standard error (SE).
Statistical Analysis
The Prism 4 (GraphPad Software Inc., San Diego, CA) one-way ANOVA test, (Student-Newman-Keuls multiple comparison test) was used to assess statistically significant differences between data groups (P < 0.05). The R software programme [33] was used to perform a Spearman rank test to determine correlation between data sets.
Competing interests
Patent applications for the vaccine candidates described here have been filed by NIH. In addition, the vaccine candidates are being developed under a Cooperative Research and Development Agreement (CRADA) between NIAID and MedImmune. NIAID investigators work under CRADAs as part of the normal responsibilities of their NIAID, NIH employment. Through the execution of licensing agreements, the NIAID makes the vaccine candidates available to parties interested in their further development and commercialization.
Authors' contributions
EB recovered viruses, performed in vitro and in vivo studies and drafted the manuscript. AC recovered virus and performed in vitro and in vivo studies. SRS recovered viruses and assisted with in vivo studies. PLC contributed to the study design and drafting of the manuscript. MHS and BRM supervised the study, participated in its design and planning and contributed to drafting of the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We thank Ernest Williams and Fatemeh Davoodi for performing the HAI assays and Emerito Amaro-Carambot for assistance with sequencing. We are grateful to Pamela Shaw and Dean Follman for assistance with statistical analysis. We also thank Brad Finneyfrock and Marisa St. Claire at Bioqual Inc. for carrying out the primate studies.
This project was funded as a part of the NIAID Intramural Program.
Contributor Information
Emmalene J Bartlett, Email: ebartlett@niaid.nih.gov.
Adam Castaño, Email: adam.castano@gmail.com.
Sonja R Surman, Email: SBarbagallo@niaid.nih.gov.
Peter L Collins, Email: PCOLLINS@niaid.nih.gov.
Mario H Skiadopoulos, Email: mskiadopoulos@niaid.nih.gov.
Brian R Murphy, Email: bmurphy@niaid.nih.gov.
References
- Chanock RM, Murphy BR, Collins PL. Parainfluenza Viruses. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Strauss SE, editor. Fields Virology. 4. Vol. 1. Philadelphia: Lippincott Williams&Wilkins; 2001. pp. 1341–1379. [Google Scholar]
- Henderson FW, Collier AM, Sanyal MA, Watkins JM, Fairclough DL, Clyde WA, Jr, Denny FW. A longitudinal study of respiratory viruses and bacteria in the etiology of acute otitis media with effusion. N Engl J Med. 1982;306:1377–1383. doi: 10.1056/NEJM198206103062301. [DOI] [PubMed] [Google Scholar]
- Marx A, Torok TJ, Holman RC, Clarke MJ, Anderson LJ. Pediatric hospitalizations for croup (laryngotracheobronchitis): biennial increases associated with human parainfluenza virus 1 epidemics. J Infect Dis. 1997;176:1423–1427. doi: 10.1086/514137. [DOI] [PubMed] [Google Scholar]
- Reed G, Jewett PH, Thompson J, Tollefson S, Wright PF. Epidemiology and clinical impact of parainfluenza virus infections in otherwise healthy infants and young children <5 years old. J Infect Dis. 1997;175:807–813. doi: 10.1086/513975. [DOI] [PubMed] [Google Scholar]
- Heikkinen T, Thint M, Chonmaitree T. Prevalence of various respiratory viruses in the middle ear during acute otitis media. N Engl J Med. 1999;340:260–264. doi: 10.1056/NEJM199901283400402. [DOI] [PubMed] [Google Scholar]
- Counihan ME, Shay DK, Holman RC, Lowther SA, Anderson LJ. Human parainfluenza virus-associated hospitalizations among children less than five years of age in the United States. Pediatr Infect Dis J. 2001;20:646–653. doi: 10.1097/00006454-200107000-00003. [DOI] [PubMed] [Google Scholar]
- Murphy BR, Prince GA, Collins PL, Van Wyke Coelingh K, Olmsted RA, Spriggs MK, Parrott RH, Kim HW, Brandt CD, Chanock RM. Current approaches to the development of vaccines effective against parainfluenza and respiratory syncytial viruses. Virus Res. 1988;11:1–15. doi: 10.1016/0168-1702(88)90063-9. [DOI] [PubMed] [Google Scholar]
- Newman JT, Surman SR, Riggs JM, Hansen CT, Collins PL, Murphy BR, Skiadopoulos MH. Sequence analysis of the Washington/1964 strain of human parainfluenza virus type 1 (HPIV1) and recovery and characterization of wild-type recombinant HPIV1 produced by reverse genetics. Virus Genes. 2002;24:77–92. doi: 10.1023/A:1014042221888. [DOI] [PubMed] [Google Scholar]
- Power UF, Ryan KW, Portner A. The P genes of human parainfluenza virus type 1 clinical isolates are polycistronic and microheterogeneous. Virology. 1992;189:340–343. doi: 10.1016/0042-6822(92)90712-X. [DOI] [PubMed] [Google Scholar]
- Van Cleve W, Amaro-Carambot E, Surman SR, Bekisz J, Collins PL, Zoon KC, Murphy BR, Skiadopoulos MH, Bartlett EJ. Attenuating mutations in the P/C gene of human parainfluenza virus type 1 (HPIV1) vaccine candidates abrogate the inhibition of both induction and signaling of type I interferon (IFN) by wild-type HPIV1. Virology. 2006;352:61–73. doi: 10.1016/j.virol.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Murphy BR. Mucosal immunity to viruses. In: Ogra PL, Mestecky J, Lamm ME, Strober W, McGhee JR, Bienstock J, editor. Mucosal Immunology. Second. Academic Press, Inc; 1999. pp. 695–707. [Google Scholar]
- Mossad SB. Demystifying FluMist, a new intranasal, live influenza vaccine. Cleve Clin J Med. 2003;70:801–806. doi: 10.3949/ccjm.70.9.801. [DOI] [PubMed] [Google Scholar]
- McAuliffe JM, Surman SR, Newman JT, Riggs JM, Collins PL, Murphy BR, Skiadopoulos MH. Codon substitution mutations at two positions in the L polymerase protein of human parainfluenza virus type 1 yield viruses with a spectrum of attenuation in vivo and increased phenotypic stability in vitro. J Virol. 2004;78:2029–2036. doi: 10.1128/JVI.78.4.2029-2036.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JT, Riggs JM, Surman SR, McAuliffe JM, Mulaikal TA, Collins PL, Murphy BR, Skiadopoulos MH. Generation of recombinant human parainfluenza virus type 1 vaccine candidates by importation of temperature-sensitive and attenuating mutations from heterologous paramyxoviruses. J Virol. 2004;78:2017–2028. doi: 10.1128/JVI.78.4.2017-2028.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett EJ, Amaro-Carambot E, Surman SR, Newman JT, Collins PL, Murphy BR, Skiadopoulos MH. Human parainfluenza virus type I (HPIV1) vaccine candidates designed by reverse genetics are attenuated and efficacious in African green monkeys. Vaccine. 2005;23:4631–4646. doi: 10.1016/j.vaccine.2005.04.035. [DOI] [PubMed] [Google Scholar]
- Bartlett EJ, Amaro-Carambot E, Surman SR, Collins PL, Murphy BR, Skiadopoulos MH. Introducing point and deletion mutations into the P/C gene of human parainfluenza virus type 1 (HPIV1) by reverse genetics generates attenuated and efficacious vaccine candidates. Vaccine. 2006;24:2674–2684. doi: 10.1016/j.vaccine.2005.10.047. [DOI] [PubMed] [Google Scholar]
- Murphy BR, Park EJ, Gottlieb P, Subbarao K. An influenza A live attenuated reassortant virus possessing three temperature-sensitive mutations in the PB2 polymerase gene rapidly loses temperature sensitivity following replication in hamsters. Vaccine. 1997;15:1372–1378. doi: 10.1016/S0264-410X(97)00031-5. [DOI] [PubMed] [Google Scholar]
- Murphy BR, Coelingh K. Principles underlying the development and use of live attenuated cold-adapted influenza A and B virus vaccines. Viral Immunol. 2002;15:295–323. doi: 10.1089/08828240260066242. [DOI] [PubMed] [Google Scholar]
- Skiadopoulos MH, Schmidt AC, Riggs JM, Surman SR, Elkins WR, St Claire M, Collins PL, Murphy BR. Determinants of the host range restriction of replication of bovine parainfluenza virus type 3 in rhesus monkeys are polygenic. J Virol. 2003;77:1141–1148. doi: 10.1128/JVI.77.2.1141-1148.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolakofsky D, Pelet T, Garcin D, Hausmann S, Curran J, Roux L. Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited. J Virol. 1998;72:891–899. doi: 10.1128/jvi.72.2.891-899.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan SM, Surman SR, Amaro-Carambot E, Collins PL, Murphy BR, Skiadopoulos MH. Live-attenuated intranasal parainfluenza virus type 2 vaccine candidates developed by reverse genetics containing L polymerase protein mutations imported from heterologous paramyxoviruses. Vaccine. 2005;23:4765–4774. doi: 10.1016/j.vaccine.2005.04.043. [DOI] [PubMed] [Google Scholar]
- Wright PF, Karron RA, Belshe RB, Thompson J, Crowe JE, Jr, Boyce TG, Halburnt LL, Reed GW, Whitehead SS, Anderson EL, et al. Evaluation of a live, cold-passaged, temperature-sensitive, respiratory syncytial virus vaccine candidate in infancy. J Infect Dis. 2000;182:1331–1342. doi: 10.1086/315859. [DOI] [PubMed] [Google Scholar]
- Karron RA, Belshe RB, Wright PF, Thumar B, Burns B, Newman F, Cannon JC, Thompson J, Tsai T, Paschalis M, et al. A live human parainfluenza type 3 virus vaccine is attenuated and immunogenic in young infants. Pediatr Infect Dis J. 2003;22:394–405. doi: 10.1097/00006454-200305000-00002. [DOI] [PubMed] [Google Scholar]
- Richman DD, Murphy BR. The association of the temperature-sensitive phenotype with viral attenuation in animals and humans: implications for the development and use of live virus vaccines. Rev Infect Dis. 1979;1:413–433. doi: 10.1093/clinids/1.3.413. [DOI] [PubMed] [Google Scholar]
- Murphy BR, Richman DD, Chalhub EG, Uhlendorf CP, Baron S, Chanock RM. Failure of attenuated temperature-sensitive influenza A (H3N2) virus to induce heterologous interference in humans to parainfluenza type 1 virus. Infect Immun. 1975;12:62–68. doi: 10.1128/iai.12.1.62-68.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CB, Purcell RH, Bellanti JA, Chanock RM. Protective effect of antibody to parainfluenza type 1 virus. N Engl J Med. 1966;275:1145–1152. doi: 10.1056/NEJM196611242752101. [DOI] [PubMed] [Google Scholar]
- Buchholz UJ, Finke S, Conzelmann KK. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol. 1999;73:251–259. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller K, Duffy I, Duprex P, Rima B, Beschorner R, Fauser S, Meyermann R, Niewiesk S, ter Meulen V, Schneider-Schaulies J. Recombinant measles viruses expressing altered hemagglutinin (H) genes: functional separation of mutations determining H antibody escape from neurovirulence. J Virol. 2001;75:7612–7620. doi: 10.1128/JVI.75.16.7612-7620.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surman SR, Collins PL, Murphy BR, Skiadopoulos MH. An improved method for the recovery of recombinant paramyxovirus vaccine candidates suitable for use in human clinical trials. Journal of virological methods. 2007;141:30–33. doi: 10.1016/j.jviromet.2006.11.024. [DOI] [PubMed] [Google Scholar]
- Witko SE, Kotash CS, Nowak RM, Johnson JE, Boutilier LA, Melville KJ, Heron SG, Clarke DK, Abramovitz AS, Hendry RM, et al. An efficient helper-virus-free method for rescue of recombinant paramyxoviruses and rhadoviruses from a cell line suitable for vaccine development. Journal of virological methods. 2006;135:91–101. doi: 10.1016/j.jviromet.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Skiadopoulos MH, Tao T, Surman SR, Collins PL, Murphy BR. Generation of a parainfluenza virus type 1 vaccine candidate by replacing the HN and F glycoproteins of the live-attenuated PIV3 cp45 vaccine virus with their PIV1 counterparts. Vaccine. 1999;18:503–510. doi: 10.1016/S0264-410X(99)00227-3. [DOI] [PubMed] [Google Scholar]
- Clements ML, Belshe RB, King J, Newman F, Westblom TU, Tierney EL, London WT, Murphy BR. Evaluation of bovine, cold-adapted human, and wild-type human parainfluenza type 3 viruses in adult volunteers and in chimpanzees. J Clin Microbiol. 1991;29:1175–1182. doi: 10.1128/jcm.29.6.1175-1182.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The GNU Operating System http://www.gnu.org/